Abstract
Though widely assumed to bear a 5′-terminal triphosphate or monophosphate, recent evidence suggests that the 5′ end of bacterial RNA can sometimes bear a modification reminiscent of a eukaryotic cap. A new study has now identified E. coli RNAs that begin with a noncanonical cap resembling the redox cofactor nicotinamide adenine dinucleotide (NAD), as well as a cellular enzyme that can remove it. The biological function of such caps remains to be determined.
Keywords: NudC, RNA degradation, RNase E, RNA I, RppH, sRNA
A number of distinguishing characteristics have traditionally been thought to differentiate bacterial mRNA from eukaryotic mRNA, such as a 5′-terminal triphosphate versus an N7-methylguanosine triphosphate cap and the absence or presence of a 3′-terminal poly(A) tail. The latter distinction was overturned decades ago by the discovery of widespread if transient polyadenylation of bacterial transcripts [1]. However, 5′ caps had retained their status as an attribute unique to eukaryotic mRNA until a few years ago, when the laboratory of David Liu reported the mass spectrometric detection of a cap-like NAD moiety at the 5′ end of bulk RNA isolated from Escherichia coli and Streptomyces venezuelae [2] (Figure 1A). Fractionation by size showed that this modification was present primarily on RNAs shorter than 200 nucleotides, but specific transcripts bearing an NAD cap were not identified. In a recent issue of Nature, Andres Jäschke and his coworkers have now determined the identity of these RNAs and begun to investigate what the biological role of their caps may be [3].
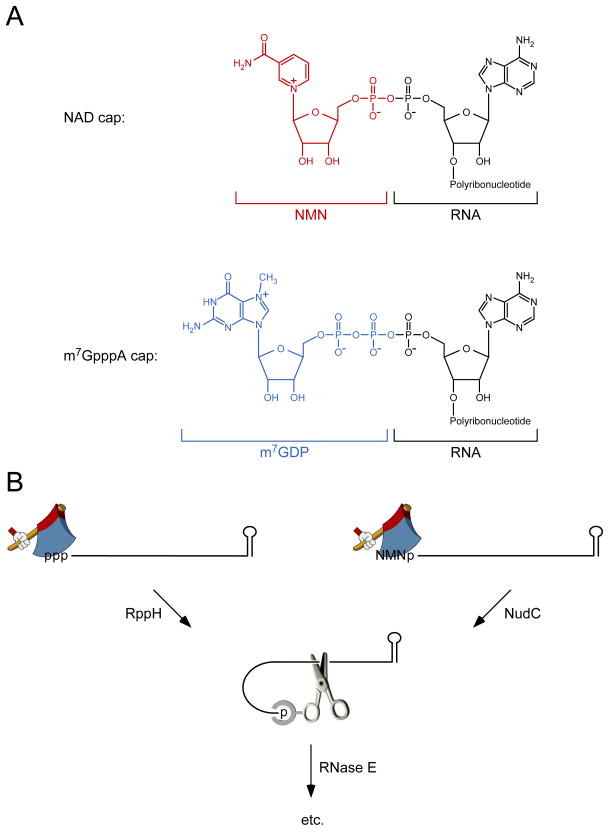
Figure 1. NAD caps and their removal by NudC.
(A) Structure of bacterial and eukaryotic RNA caps. A bacterial nicotinamide adenine dinucleotide (NAD) cap (top) comprises nicotinamide mononucleotide (NMN, red) covalently joined via its 5′ phosphate to the 5′ terminus of an RNA transcript that begins with adenylate, whereas a eukaryotic m7GpppA cap (bottom) comprises N7-methylguanosine diphosphate (m7GDP, blue) similarly joined to the 5′-terminal adenylate of RNA.
(B) 5′-end-dependent pathways for RNA degradation in E. coli. The 5′ terminus of triphosphorylated RNA (left) or NAD-capped RNA (right) is converted to a monophosphate by RppH or NudC, respectively. The resulting intermediate is then cut by the endonuclease RNase E, whose cleavage activity at internal sites is potentiated by the ability of this enzyme to bind the monophosphorylated 5′ end. ppp, triphosphate; p, monophosphate; NMN, nicotinamide mononucleotide; hatchet, RppH or NudC; scissors, RNase E; etc., additional degradative steps that are not shown.
To identify E. coli RNAs bearing an NAD cap, Jäschke’s team devised a way (NAD captureSeq) to selectively purify such RNAs from E. coli extracts. Central to their procedure was using an NAD(H)-specific enzyme (adenosine diphosphate ribosylcyclase from Aplysia californica) and click chemistry to selectively biotinylate NAD-modified RNAs so that they could be purified on streptavidin-coated beads, amplified by RT-PCR, and sequenced. The RNAs enriched in this manner included 15 noncoding RNAs, such as the plasmid replication inhibitor RNA I and the regulatory small RNA (sRNA) GcvB, as well as 5′-terminal fragments of 29 mRNAs. Mass spectrometry confirmed that the modification at the 5′ end of RNA I and GcvB was NAD. As might be expected for a 5′-terminal modification with an adenosine monophosphate component, the transcription units for each of the 44 NAD-capped RNAs begin with the nucleotide A. However, apart from this shared characteristic, these RNAs appear to have nothing else in common.
Having determined that a specific subset of E. coli RNAs bears an NAD cap, the authors then inquired about its function. One possibility they considered was an effect on RNA degradation. Previous research had demonstrated that 5′ termini can influence this important regulatory process in E. coli, where internal cleavage of transcripts by the endonuclease RNase E is often triggered by prior conversion of the 5′-terminal triphosphate to a monophosphate by the RNA pyrophosphohydrolase RppH [4,5] (Figure 1B). This modification creates a better substrate for RNase E, which, in addition to its active site, bears a discrete pocket for binding monophosphorylated RNA 5′ ends [6,7]. Jäschke’s group first examined the influence of an NAD cap on the action of these two enzymes in vitro, using RNA I as a model substrate. Like triphosphorylated RNA I, NAD-capped RNA I produced by in vitro transcription was found to be much less susceptible than monophosphorylated RNA I to cleavage by purified RNase E. However, unlike its triphosphorylated counterpart, NAD-capped RNA I was also resistant to purified RppH, raising the possibility that such a cap might protect transcripts from RppH-dependent degradation in vivo.
How, then, might NAD-capped RNAs be degraded in E. coli? Jäschke and his coworkers tested a hypothesis, first proposed by Alexander McLennan [8], that NudC, an E. coli enzyme previously shown to hydrolyze NAD so as to produce nicotinamide mononucleotide (NMN) and adenosine monophosphate [9], might also act on NAD-capped RNA, yielding NMN and monophosphorylated RNA as products. This hypothesis was confirmed by experiments with purified NudC, which was able to convert NAD-capped RNA I obtained by in vitro transcription into a product more vulnerable to rapid RNase E cleavage. They then asked whether the presence of NudC limits the abundance of NAD-capped RNA in E. coli. RNA was extracted from isogenic strains containing or lacking NudC, and the percentage of RNA I molecules bearing an NAD cap was examined by a procedure based on the ability of such a cap to resist treatment with alkaline phosphatase. The phosphatase-resistant RNA was decapped with purified NudC and detected by PABLO, a splinted ligation assay specific for monophosphorylated RNA [4]. In this manner, they determined that 13% of RNA I bears an NAD cap in wild-type cells. This amount doubles to 26% in ΔnudC cells, evidence that NudC acts to decap RNA I in vivo.
Together, these observations led Jäschke and his team to propose that NudC may trigger the degradation of NAD-capped RNA in E. coli by removing the cap so as to generate a monophosphorylated intermediate susceptible to rapid cleavage by RNase E (Figure 1B). If so, one might expect capped RNAs to decay more slowly in ΔnudC cells. However, the absence of NudC had no detectable effect on the lifetime of either RNA I or GcvB in E. coli. Given that RNA I is known to be degraded by a 5′-end-dependent mechanism involving RNase E [4,10], this finding suggests that the fraction of these RNAs that becomes capped is insufficient to influence the kinetics of degradation in vivo or that NAD caps can be removed not only by NudC but also by another, hypothetical enzyme with which NudC is functionally redundant.
What, then, are the biological implications of these findings? Although they suggest a potential new pathway for 5′-end-dependent decay, the physiological significance of NAD caps and their removal by NudC awaits empirical evidence of an effect on RNA lifetimes. Perhaps a larger percentage of E. coli transcripts become capped under other growth conditions, resulting in a greater effect on their rates of decay. It is also possible that sRNA decapping by NudC and the consequent generation of a monophosphorylated 5′ end that can recruit RNase E may be less important for accelerating the degradation of the sRNAs themselves than of the mRNAs with which they base pair. Alternatively, NAD caps may influence RNA function in unforeseen ways not related to degradation, perhaps by mediating binding to one of the many cellular enzymes that use NAD as a substrate.
Other interesting questions relate to how these caps are acquired in the first place and why they are present on only a small subset of RNAs. Previously, Liu’s group found that E. coli RNA polymerase is unable to incorporate NAD at the 5′ end of nascent transcripts by using it as a surrogate for ATP during transcription initiation [2]. This observation and the absence of caps on long E. coli RNAs suggest that NAD capping occurs post-transcriptionally, raising the possibility that these caps may be added to triphosphorylated RNA 5′ ends by one or more enzymes heretofore thought to participate only in NAD biosynthesis. Why they are found on relatively few transcripts and predominantly on RNAs shorter than 200 nucleotides are intriguing puzzles that await further investigation.
Acknowledgments
The authors gratefully acknowledge support by a research grant from the National Institutes of Health (R01GM035769 to J.G.B.) and by a Vilcek Fellowship (to D.J.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nakazato H, Venkatesan S, Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975;256:144–146. doi: 10.1038/256144a0. [DOI] [PubMed] [Google Scholar]
- 2.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahová H, Winz ML, Höfer K, Nübel G, Jäschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2014 doi: 10.1038/nature14020. [DOI] [PubMed] [Google Scholar]
- 4.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 6.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 7.Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 8.McLennan AG. Substrate ambiguity among the nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell Mol Life Sci. 2013;70:373–385. doi: 10.1007/s00018-012-1210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frick DN, Bessman MJ. Cloning, purification, and properties of a novel NADH pyrophosphatase. Evidence for a nucleotide pyrophosphatase catalytic domain in MutT-like enzymes. J Biol Chem. 1995;270:1529–1534. doi: 10.1074/jbc.270.4.1529. [DOI] [PubMed] [Google Scholar]
- 10.Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]