Abstract
Two steroid acids, cephalosporin P1 and isocephalosporin P1, were isolated from Hapsidospora irregularis FERM BP-2511. These compounds are structurally related to fusidic acid. Their NMR data were completely assigned on the basis of the 2D NMR spectra. Incubation of these two compounds with Microbaterium oxydans CGMCC 1788 in Luria-Bertani broth yielded the same set of three new 3-dehydrogenated products, 3-keto-isocephalosporin P1, 3-keto-cephalosporin P1 and 6-deacetyl-3-keto-cephalosporin P1. The final pH of the bacterial culture was 9.0. Incubation of 3-keto-isocephalosporin P1 or 3-keto-cephalosporin P1 in Tris-HCl buffer (pH 9.0) revealed that these two compounds can convert to each other by shifting the acetyl group between C-6 and C-7. The acetyl group at C-6 or C-7 can also be removed by hydrolysis to yield the minor product 6-deacetyl-3-keto-cephalosporin P1. These fusidic acid derivatives were tested for the antibacterial activity against the Gram-positive pathogen Staphylococcus aureus. 3-Keto-cephalosporin P1 showed the highest activity among the five compounds, with a minimal inhibition concentration (MIC) of 4 μg/mL, which is more potent than the substrate cephalosporin P1. Both cephalosporin P1 and 3-keto-cephalosporin P1 were active against methicillin-resistant S. aureus, with the same MIC of 8 μg/mL.
Keywords: Antibacterial, Biotransformation, Dehydrogenation, Hapsidospora irregularis, Microbacterium oxydans, Fusidic acids
Infections are one of the top causes of death. Antibiotic drugs have been effective in treating bacterial infections and saving lives. However, extensive use of antibiotics may also induce the occurrence of resistant bacteria. For example, Staphylococcus aureus (SA) is recognized as one of the most common pathogenic bacteria that cause diseases in humans, such as pneumonia, surgical wound and bloodstream infections. Its variant methicillin-resistant Staphylococcus aureus (MRSA) is resistant to all beta-lactam antibiotics such as penicillin and methicillin. This strain is called “super bug” because its drug-resistance and serious threat to human lives. The spread of resistant bacteria has led to untreatable infection, which has become one of the world's most pressing public health threats.1 Thus, new antibacterial agents are needed.
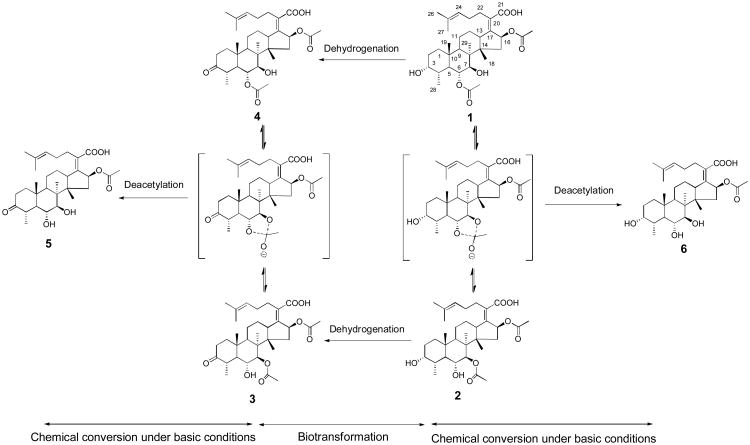
Most antibiotics were discovered from microorganisms such as soil-derived actinomycetes. Many of these antibiotics were isolated between 1940s and 1960s. After that, discovery of new antibacterial natural products slowed down. This creates a crisis of lacking effective anti-infectious drugs to combat infections, especially considering that antibiotic resistance is rising. Our group has been interested in discovering antibacterial natural products and creating new derivatives using biocatalytic tools for bioactivity screening. In the course of screening antibacterial microbial extracts, we found that Hapsidospora irregularis FERM BP-2511 is active against SA and MRSA, with the minimal inhibition concentrations (MICs) of 500 and 1000 μg/mL, respectively. H. irregularis belongs to the family of Pseudeurotiaceae.2 It is known to produce a cyclodepsipeptide, leualacin, which can inhibit the binding of 3H-nitrendipine to cardiac Ca2+ channel in a competitive manner.3, 4 No antibacterial compounds have ever been reported from this fungus. We thus grew H. irregularis FERM BP-2511 in K2 broth and isolated two metabolites 1 and 2 (Fig. 1) from the culture using open column chromatography and HPLC.5 Their structures were elucidated on the basis of the spectral data.
Figure 1.
Synthesis of three new fusidic acid derivatives 3-5 from cephalosporin P1 (1) and isocephalosporin P1 (2).
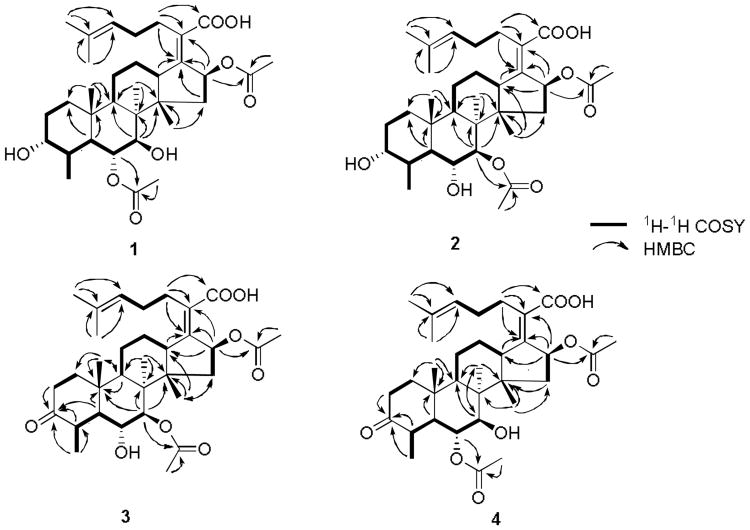
Both 1 and 2 have a molecular weight of 574 according to the [M-H]- ion peak at m/z 573.3 in their ESI-MS spectra. The 13C NMR spectra of these two compounds revealed that they have thirty three carbons including 3 carbonyls, 4 oxygenated methines, 4 olefinic carbons, and 22 sp3 carbons. Accordingly, the molecular formula of 1 and 2 was deduced to be C33H50O8, suggesting that they have nine degrees of unsaturation. Extensive analysis of the 2D NMR spectra including 1H-1H COSY, HSQC, and HMBC spectra (Fig. 2) allowed 1 and 2 to be identified as cephalosporin P1 (Fig. 1) and isocephalosporin P1 (Fig. 1), respectively. Their NMR data were assigned and are shown in Tables S1 and S2.
Figure 2.
Selected 1H-1H COSY and HMBC correlations for 1-4.
Cephalosporin P1 (1) and isocephalosporin P1 (2) are triterpenoid antibiotics that belong to the fusidic acid family of natural products. Fusidic acid is a bacteriostatic antibiotic from the fungus Fusidium coccineum and was developed by Leo Laboratories (Ballerup, Denmark) and released for clinical use in the 1960s. Its sodium salt has been used in many countries. Fusidic acid is primarily effective against Gram-positive bacteria. 1 was first previously isolated from a species of Cephalosporium,6 and 2 was produced by Cephalosporium acremonium.7 Early work demonstrated that 1 has an antibacterial spectrum that predominantly encompasses Gram-positive organisms, with particularly potent activity against SA.8, 9 This is the first time that 1 and 2 were isolated from H. irregularis. The NMR data of 2 were not completely assigned in the previous work. In this study, we conducted complete NMR analysis for these two molecules and assigned all the NMR signals for both 1 and 2.
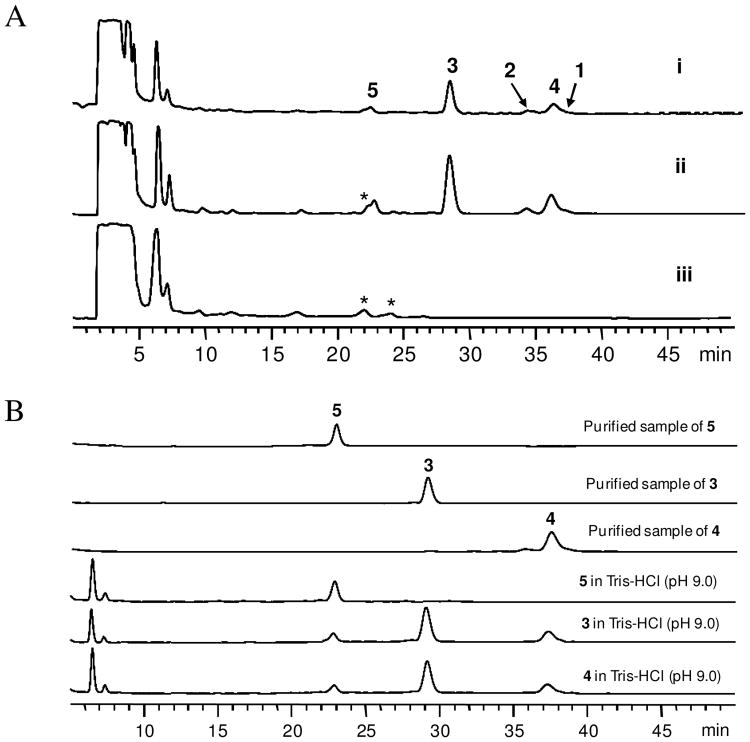
Biotransformation has shown its promise in creating chemical diversity in natural products.10, 11 Because 1 has been reported to inhibit Gram-positive bacteria,12 we chose a strain of Gram-negative bacterium Microbaterium oxydans CGMCC 1788 for the biotransformation experiment to prepare new analogs from 1 and 2. This strain was previously used for glycosylation of the plant natural product puerarin.13 HPLC analysis revealed that compared to the culture control (trace iii, Figure 3A), incubation of M. oxydans CGMCC 1788 with 1 in LB medium at 30°C for 4 days yielded three products 3-5 (trace i, Fig. 3A). The UV spectra of these products are similar to that of the substrate (Fig. S1), suggesting that they are derivatives of 1. Interestingly, incubation of the same strain with 2 gave rise to the same set of three products (trace ii, Fig. 3A). These compounds were then isolated from the extract of the biotransformation broth of 1 using open column chromatography and HPLC.14 The purified products were subjected to MS and NMR analyses.
Figure 3.
HPLC analysis of the biotransformation products of 1 and 2 by M. oxydans CGMCC 1788. (A) HPLC trace of the biotransformation products of 1 (i) and 2 (ii) by M. oxydans CGMCC 1788 at 210 nm. Trace iii is the blank culture control. The asterisked peaks are metabolites from the culture control. HPLC conditions: column: Agilent ZORBAX SB-C18, 5 μm, 4.6 mm × 250 mm, eluted with 44% acetonitrile-water with 0.05% formic acid for 15 minutes, 44%-50% acetonitrile-water with 0.05% formic acid over 5 minutes, then 50% acetonitrile-water with 0.05% formic acid for an additional 30 minutes. (B) HPLC analysis (210 nm) of the incubation of 3, 4 and 5 with Tris-HCl buffer (pH 9.0).
Compounds 3 and 4 showed the same molecular weight of 572 according to the [M-H]- ion peak at m/z 571.3 and [M+Na]+ peak at m/z 595.3 in the ESI-MS spectra. Both compounds showed a [M-H]- ion peak at m/z 571.3258 (C33H47O8, calc. 571.3276) in the HR-MS spectrum. Accordingly, the molecular formula was determined to be C33H48O8, suggesting that these two compounds are isomers and they are two protons less than the substrates 1 and 2.
The 13C NMR spectra of 3 showed a low field carbon signal at δC 216.4, suggesting that a ketone group was generated during the biotransformation process. In the meantime, an oxygenated methine signal was missing compared to the substrates 1 and 2. This observation suggested that this oxygenated methine might have been converted to a ketone group through dehydrogenation. To find out which oxygenated methine was oxidized, 2D NMR including 1H-1H COSY, HSQC, and HMBC were recorded. 1H-1H COSY correlations revealed a spin system of CH3(28)-CH(4)-CH(5)-CH(6)-CH(7), which is different from the substrates 1 and 2, in which the spin system also includes CH2(1)-CH2(2)-CH(3) (Fig. 2). This suggested that the ketone group is at C-3. HMBC correlations of H-28, H-5, H-4, and H-2 to the ketone carbon signal at δC 216.4 confirmed the presence of the ketone group at C-3 (Fig. 2). Similar to 2, 3 has two acetyl groups at C-7 and C-16 according to the HMBC correlations of CH3 at δH 1.99 (3H, s) and H-7 at δH 4.91 (1H, d, 7.5) to the carbonyl signal of the acetyl group at δC 170.7, and CH3 at δH 1.88 (3H, s) and H-16 at δH 5.78 (1H, d, 8.6) to the carbonyl signal of the other acetyl group at δC 170.5 (Fig. 2). Therefore, 3 was characterized as the 3-keto derivative of 2 and is named as 3-keto-isocephalosporin P1 (Fig. 1). The proton and carbon signals were assigned and are listed in Table 1.
Table 1.
The 1H (300 MHz) and 13C (75 MHz) NMR data for 3-5 (3 and 5 in acetone-d6, 4 in CDCl3, δ in ppm, J in Hz).
| Position | δH | δC | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| 3 | 4 | 5 | 3 | 4 | 5 | |
| 1 | 1.55-1.62 (1H, m) | 1.68-1.74 (1H, m) | 1.42-1.54 (1H, m) | 32.5 | 33.1 | 31.9 |
| 2.08-2.14 (1H, m) | 1.88-1.92 (1H, m) | 2.00-2.09 (1H, m) | ||||
| 2 | 2.17-2.26 (1H, m) | 2.45-2.50 (2H, m) | 2.24-2.29 (1H, m) | 35.2 | 35.6 | 34.7 |
| 2.58-2.67 (1H, m) | 2.63-2.69 (1H, m) | |||||
| 3 | - | - | - | 216.4 | 215.0 | 217.3 |
| 4 | 2.35-2.41 (1H, m) | 2.40-2.45 (1H, m) | 2.31-2.39 (1H, m) | 46.2 | 45.1 | 46.3 |
| 5 | 2.01-2.08 (1H, m) | 2.01-2.07 (1H, m) | 1.88-1.97 (1H, m) | 48.7 | 47.0 | 48.4 |
| 6 | 3.72 (1H, dd, 7.9, 10.9) | 4.87 (1H, dd, 10.6, 5.9) | 3.60 (1H, dd, 11.0, 8.6) | 73.0 | 79.1 | 75.0 |
| 7 | 4.91 (1H, d, 7.5) | 3.64 (1H, d, 5.9) | 3.49 (1H, d, 8.6) | 85.1 | 82.1 | 86.7 |
| 8 | - | - | - | 45.9 | 45.1 | 45.8 |
| 9 | 2.01-2.08 (1H, m) | 2.07-2.15 (1H, m) | 2.03-2.17 (1H, m) | 43.4 | 41.8 | 43.2 |
| 10 | - | - | - | 37.5 | 37.0 | 38.0 |
| 11 | 1.68-1.74 (1H, m) | 1.59-1.74 (2H, m) | 1.68-1.80 (2H, m) | 23.4 | 23.2 | 23.2 |
| 1.47-1.53 (1H, m) | ||||||
| 12 | 1.74-1.78 (1H, m) | 2.20-2.29 (1H, m) | 1.70-1.81 (1H, m) | 26.7 | 25.9 | 26.8 |
| 2.29-2.35 (1H, m) | 1.77-1.82 (1H, m) | 2.08-2.17 (1H, m) | ||||
| 13 | 2.66-2.71 (1H, m) | 2.54-2.58 (1H, m) | 2.63-2.69 (1H, m) | 50.7 | 50.4 | 50.7 |
| 14 | - | - | - | 48.6 | 48.4 | 48.9 |
| 15 | 2.14 (1H, dd, 6.8, 15.5) | 1.72-1.77 (1H, m) | 1.83-1.87 (1H, m) | 40.9 | 40.5 | 41.5 |
| 1.18 (1H, d, 15.1) | 2.16-2.24 (1H, m) | 2.17-2.25 (1H, m) | ||||
| 16 | 5.78 (1H, d, 8.6) | 5.85 (1H, d, 7.9) | 5.80 (1H, d, 8.6) | 74.6 | 74.4 | 74.6 |
| 17 | - | - | - | 147.4 | 150.0 | 148.3 |
| 18 | 1.17 (3H, s) | 1.20 (3H, s) | 1.23 (3H, s) | 20.9 | 21.5 | 21.9 |
| 19 | 0.98 (3H, s) | 1.15 (3H, s) | 0.90 (3H, s) | 23.6 | 23.7 | 23.4 |
| 20 | - | - | - | 132.0 | 130.3 | 130.2 |
| 21 | - | - | - | 171.3 | 173.7 | 174.3 |
| 22 | 2.41-2.56 (2H, m) | 2.34-2.48 (2H, m) | 2.34-2.53 (2H, m) | 29.5 | 28.9 | 29.3 |
| 23 | 2.05-2.19 (2H, m) | 2.05-2.16 (2H, m) | 2.03-2.14 (2H, m) | 29.1 | 28.5 | 28.5 |
| 24 | 5.15 (1H, t, 7.3) | 5.10 (1H, t, 7.2) | 5.15 (1H, t, 7.2) | 124.3 | 123.2 | 124.4 |
| 25 | - | - | - | 132.7 | 132.9 | 132.6 |
| 26 | 1.60 (3H, s) | 1.60 (3H, s) | 1.60 (3H, s) | 17.9 | 17.9 | 17.9 |
| 27 | 1.66 (3H, s) | 1.68 (3H, s) | 1.66 (3H, s) | 25.9 | 25.9 | 25.9 |
| 28 | 1.26 (3H, d, 6.9) | 1.11 (3H, d, 6.5) | 1.28 (3H, d, 7.6) | 18.8 | 16.9 | 19.5 |
| 29 | 1.36 (3H, s) | 1.15 (3H, s) | 1.29 (3H, s) | 20.3 | 19.9 | 20.5 |
| 6-OCOCH3 | - | 2.10 (3H, s) | - | - | 21.6 | - |
| 6-OCOCH3 | - | - | - | - | 171.7 | - |
| 7-OCOCH3 | 1.99 (3H, s) | - | - | 22.0 | - | - |
| 7-OCOCH3 | - | - | - | 170.7 | - | - |
| 16-OCOCH3 | 1.88 (3H, s) | 1.96 (3H, s) | 1.88 (3H, s) | 20.6 | 20.8 | 20.7 |
| 16-OCOCH3 | - | - | - | 170.5 | 170.8 | 170.5 |
Compared to 1 and 2, 4 also has an extra ketone carbon signal at δC 215.0 but lacks an oxygenated methine group. Similar to 3, the ketone group was found to be at C-3 according to the spin system of CH3(28)-CH(4)-CH(5)-CH(6)-CH(7) revealed by the 1H-1H COSY spectrum, which was confirmed by the HMBC correlations of H-28, H-4, and H-2 to the ketone signal (Fig. 2). There are two acetyl groups in 4. One acetyl group can be assigned to C-16 based on the HMBC correlations of a CH3 group at δH 1.96 (3H, s) and H-16 at δH 5.85 (1H, d, 7.9) to the carbonyl carbon of the acetyl group at δC 170.8. The other acetyl group was assigned to C-6 based on the HMBC correlations of a CH3 at δH 2.10 (3H, s) and H-6 at δH 4.87 (1H, dd, 10.6, 5.9) to the carbonyl signal of the acetyl group at δC 171.7. Accordingly, 4 was identified as 3-keto-cephalosporin P1. The proton and carbon signals were assigned and are listed in Table 1.
ESI-MS spectra of 5 showed the ion peaks [M-H]- at m/z 529.3 and [M+Na]+ at m/z 553.3, indicating that its molecular weight is 530. The molecular formula was deduced to be C31H46O7 based on the [M-H]- peak at m/z 529.3165 (C31H45O7, calc. 529.3171) in the HR-MS spectrum. This is 42 mass units less than 3 and 4, suggesting that it lacks an acetyl group. This was supported by the 13C NMR spectrum, in which only thirty one carbons were observed, including 3 carbonyls, 4 olefinic carbons, 3 oxygenated methines, and 21 sp3 carbons. Compared to 3 and 4, this compound only had one acetyl carbonyl carbon signal at δC 170.5. In the 1H NMR spectrum, the chemical shifts of H-16 and H-24 were similar to those in 1-4, indicating that the acetyl group at C-16 is still present in 5. The chemical shifts of H-6 and H-7 are δH 3.60 (1H, dd, 11.0, 8.6) and δH 3.49 (1H, d, 8.6), respectively, suggesting that there is no acetyl group at C-6 and C-7. Instead, there are two free hydroxyl groups at these positions, which explains why 5 is more polar than 3 and 4 as it was eluted earlier than 3 and 4 on a reversed-phase HPLC column (Fig. 3A). Thus, 5 was characterized as 3-ketone-6-deacetylcephalosporin P1 (Fig. 1). The NMR signals were assigned based on the 1H-1H COSY and HMBC spectra and a comparison with those of 3 and 4, and the data are shown in Table 1.
Although M. oxydans CGMCC 1788 was known to glycosylate puerarin,13 incubation of 1 and 2 with M. oxydans CGMCC 1788 did not yield any glycosylated products, but three dehydrogenated derivatives. This is not surprising since 1 and 2 are structurally distinct from puerarin and lack the corresponding phenolic hydroxyl groups for the glycosylation. Generation of these three new products suggests that this bacterium has various modifying enzymes for different substrates.
It is interesting that both 1 and 2 can be converted to the same products 3-5 by M. oxydans CGMCC 1788. While enzymatic dehydrogenation of 3-OH of steroids by microbial strains has been reported before,15 the shift of the acetyl group between C-6 and C-7 is likely due to a chemical process. Further analysis of the biotransformation broth revealed that the pH value of the M. oxydans CGMCC 1788 culture in LB medium can reach above 9.0 after 48 hours. To test whether the movement of the acetyl group between C-6 and C-7 was caused by the high pH value, we incubated the dimethyl sulfoxide solution of 3-5 separately with Tris-HCl buffer (pH 9.0) at 30°C for 4 days, and checked the products by HPLC. As shown in Figure 3B, 3 and 4 are interchangeable in the basic solution. The acetyl group at C-6 or C-7 can also be hydrolyzed to generate 5 (3-ketone-6-deacetylcephalosporin P1) in the same solution. Incubation of 5 in the same solution did not yield any products (Fig. 3B), indicating that this compound is stable under the test conditions. Incubation of 1 and 2 with Tris-HCl buffer (pH 9.0) at 30°C also indicated that they are interconvertible at this pH and a minor hydrolyzed compound 6 with a molecular weight of 532 was detected (Fig. S2). This is consistent with the observation that 1 and 2 were found in each other's biotransformation broth (traces i and ii, Fig. 3A). Based on these results, a synthetic pathway of the three new products 3-5 are proposed in Figure 1. The co-presence of the three products in the biotransformation broth of 1 and 2 is a result of combined actions of the movement of the acetyl group at high pH value and enzymatic dehydrogenation by M. oxydans CGMCC 1788.
Compounds 1-5 were subjected to the antibacterial assay.16 All these samples cannot inhibit the Gram-negative bacterium Escherichia coli on LB agar plate. 1-4 were active against the Gram-positive bacterium SA (Table 2). While the MIC of 1 against SA was determined to be 8 μg/mL, its biotransformation product 4 showed stronger antibacterial activity, with a MIC of 4 μg/mL. 1 and 4 showed the same MIC of 8 μg/mL against MRSA. 2 and 3 displayed moderate activity against SA, but were less active against MRSA. 5 did not show any antibacterial activity against the two strains at the test concentrations.
Table 2.
In vitro antibacterial activity of 1-5 (MIC, μg/mL) against SA and MRSA.
| Strain | SA | MRSA |
|---|---|---|
| 1 | 8 | 8 |
| 2 | 64 | 125 |
| 3 | 64 | >125 |
| 4 | 4 | 8 |
| 5 | >125 | >125 |
| Kanamycin | 2 | >125 |
Fusidic acid is an antibiotic primarily effective against Gram-positive bacteria. 1 is structurally related to fusidic acid and can inhibit Gram-positive bacteria with low MICs. 2 also exhibits inhibitory activity against SA and MRSA, but its MICs were higher than those of 1. 4 is the dehydrogenated derivative of 1, which can inhibit SA more strongly than the substrate. In contrast, 2 and its dehydrogenated derivative 3 exhibited weaker or no activity against the two test strains. Without any acetyl group at C-6 or C-7, 5 did not show any antibacterial activity against MA and MRSA at the test concentrations. From these bioassay results, it can be concluded that the acetyl group plays a critical role in the antibacterial activity. The MIC data indicated that an acetyl group at C-6 is better than at C-7 for the antibacterial activity. If neither of these positions has an acetyl group, the compound will lose the antibacterial activity against the test strains.
In summary, two natural steroid acids 1 and 2 were isolated from H. irregularis. Three new fusidic acid derivatives 3-5 were prepared from these two compounds by microbial transformation, among which 4 has comparable or even higher antibacterial activity than the natural products 1 and 2.
Supplementary Material
Acknowledgments
This work was supported by Grant number AI065357 RM DP 008 from the National Institute of Allergy and Infectious Diseases. We thank Prof. Sheng Yuan of Nanjing Normal University, China for the gift of Microbaterium oxydans CGMCC 1788.
Footnotes
Supplementary material that may be helpful in the review process should be prepared and provided as a separate electronic file. That file can then be transformed into PDF format and submitted along with the manuscript and graphic files to the appropriate editorial office.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Lewis K. Nat Rev Drug Discov. 2013;12:371. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 2.Malloch D, Cain RF. Can J Bot. 1970;48:1815. [Google Scholar]
- 3.Hamano K, Kinoshita M, Furuya K, Miyamoto M, Takamatsu Y, Hemmi A, Tanzawa K. J Antibiot. 1992;45:899. doi: 10.7164/antibiotics.45.899. [DOI] [PubMed] [Google Scholar]
- 4.Hamano K, Kinoshita M, Tanzawa K, Yoda K, Ohki Y, Nakamura T, Kinoshita T. J Antibiot. 1992;45:906. doi: 10.7164/antibiotics.45.906. [DOI] [PubMed] [Google Scholar]
- 5.H. irregularis FERM BP-2511 was cultured in 3 L of K2 medium (2.4% potato dextrose broth powder, 1% casamino acid, 2% sucrose, 0.5% KH2PO4, and 0.125% MgSO4 in water) at 30°C with shaking at 280 rpm for 7 days. The fermentation broth was centrifuged at 3,500 rpm for 5 minutes to separate the supernatant and cells. The supernatant was loaded to a Diaion HP-20 column directly. The pelleted cells were extracted three times with 500 mL of methanol. After solvent evaporation, the residue was re-suspended in water and loaded to the same Diaion HP-20 column. The column was successively eluted with 0, 25%, 50%, 75%, and 100% aqueous methanol. The target compounds were eluted by 50% aqueous methanol. This fraction was further separated on an Agilent 1200 HPLC using an Agilent Eclipse XDB C-18 (5 μm, 4.6 mm × 150 mm), eluted with 70% methanol-water at 1 mL/min. The peaks at 19.9 and 27.8 minutes were collected to yield 1 (110.0 mg) and 2 (11.5 mg) in pure form.
- 6.Burton HS, Abraham EP. Biochem J. 1951;50:16874. doi: 10.1042/bj0500168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou TS, Eisenbraun EJ, Rapala RT. Tetrahedron. 1969;25:3341. [Google Scholar]
- 8.Ritchie AC, Smith N, Florey HW. Br J Pharmacol Chemother. 1951;6:430. doi: 10.1111/j.1476-5381.1951.tb00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Daehne W, Godtfredsen WO, Rasmussen PR. Adv Appl Microbiol. 1979;25:95. doi: 10.1016/s0065-2164(08)70148-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhan J, Guo H, Ning L, Zhang Y, Guo D. Planta Med. 2006;72:346. doi: 10.1055/s-2005-916231. [DOI] [PubMed] [Google Scholar]
- 11.Zhan J, Guo H, Dai J, Zhang Y, Guo D. Tetrahedron Lett. 2002;43:4519. [Google Scholar]
- 12.O'Neill AJ, Bostock JM, Morais Moita A, Chopra I. J Antimicrob Chemother. 2002;50:839. doi: 10.1093/jac/dkf248. [DOI] [PubMed] [Google Scholar]
- 13.Yu C, Xu H, Huang G, Chen T, Liu G, Chai N, Ji Y, Wang S, Dai Y, Yuan S. Appl Microbiol Biotechnol. 2010;86:863. doi: 10.1007/s00253-009-2341-9. [DOI] [PubMed] [Google Scholar]
- 14.The biotransformation broth of 1 was harvested by centrifugation at 3,500 rpm for 5 minutes. The supernatant was extracted three times with an equal volume of ethyl acetate. The solvent was evaporated in vacuo. The residue was re-dissolved in methanol and separated on an open MCI gel column, eluted with 0, 25%, 50%, 75%, and 100% aqueous methanol to afford five fractions. The target compounds were eluted with 75% aqueous methanol. This fraction was then further separated on an Agilent 1200 HPLC using an Agilent Eclipse XDB C-18 (5 μm, 4.6 mm × 150 mm), eluted with 50% acetonitrile-water containing 0.05% formic acid at 1 mL/min. The peaks at 6.9, 9.5 and 13.6 minutes were collected to yield 5 (2.4 mg), 3 (15.4 mg) and 4 (6.5 mg) in pure form.
- 15.Zhan J, Liu W, Guo H, Zhang Y, Guo D. Enzyme Microb Technol. 2003;33:29. [Google Scholar]
- 16.Escherichia coli XL-1 Blue, SA ATCC 25923 and MRSA ATCC 33591 were incubated in LB broth with the test samples at different concentrations (125, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125, and 0.064 μg/mL) at 37°C in 96-well plates, and the growth was analyzed after 20 hours.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.