SUMMARY
The microbiota stimulate inflammation, but the signaling pathways and the members of the microbiota involved remain poorly understood. We found that the microbiota induce interleukin-1β (IL-1β) release upon intestinal injury and this is mediated via the NLRP3 inflammasome. Enterobacteriaceae and in particular the pathobiont Proteus mirabilis, induced robust IL-1β release that was comparable to that induced by the pathogen Salmonella. Upon epithelial injury, production of IL-1β in the intestine was largely mediated by intestinal Ly6Chigh monocytes, required chemokine receptor CCR2 and was abolished by deletion of IL-1β in CCR2+ blood monocytes. Furthermore, colonization with P. mirabilis promoted intestinal inflammation upon intestinal injury via the production of hemolysin which required NLRP3 and IL-1 receptor signaling in vivo. Thus, upon intestinal injury, selective members of the microbiota stimulate newly recruited monocytes to induce NLRP3-dependent IL-1β release which promotes inflammation in the intestine.
INTRODUCTION
The intestine is inhabited by trillions of resident bacteria that can provide beneficial effects to the host (Kamada et al., 2013). For example, bacterial metabolites including vitamins and short chain fatty acids contribute to appropriate development of the host. Resident bacteria also confer resistance against pathogen infection and are critically involved in the development of lymphoid populations in the intestine (Kamada et al., 2013). Because commensal bacteria can stimulate the immune system, the host has evolved several mechanisms to prevent inappropriate activation of inflammatory responses in the intestine. For instance, the intestinal epithelium and resident macrophages are hyporesponsive to bacterial Toll-like receptor ligands such as lipopolysaccharides (Lotz et al., 2006; Smythies et al., 2005). In addition, several barriers including the mucus layer and antimicrobial peptides limit the contact between microbes and the host immune system and contribute to gut homeostasis (Kamada and Nunez, 2014). A role for the microbiota in eliciting intestinal inflammation is supported by findings that chemically-induced and spontaneous colitis are reduced or abolished in antibiotic-treated mice and germ-free mice (Garrett et al., 2007; Hudcovic et al., 2001; Kirkland et al., 2012; Kitajima et al., 2001; Vijay-Kumar et al., 2007). In mouse models, certain members of the microbiota have been linked to inflammatory responses and intestinal pathology. For example, Bacteroides species and members of the Enterobacteriaceae family including Klebsiella pneumoniae and Proteus mirabilis can promote colitis (Bloom et al., 2011; Garrett et al., 2010). However, the signaling pathways by which resident bacteria stimulate the host immune system to induce inflammation in vivo remain poorly understood.
A critical step in the activation of inflammatory responses is the recognition of microbes or endogenous molecules produced in the setting of infection or cellular injury by host pattern recognition receptors (Takeuchi and Akira, 2010). A major innate signaling pathway is the inflammasome, a multi-protein platform that activates caspase-1 leading to the proteolytic processing of pro-interleukin-1β (IL-1β) and pro-IL-18 into their mature active forms (Schroder and Tschopp, 2010). To date, four bona fide inflammasomes, have been described of which three, the NLRP1, NLRP3 and NLRC4 inflammasomes, contain a member of the intracellular Nod-like receptor (NLR) family (Franchi et al., 2012b). Among the NLR inflammasomes, NLRP3 is activated by multiple stimuli including bacterial pore-forming toxins, ATP, microbial RNA, and particulate matter (Hornung et al., 2008; Mariathasan et al., 2006). In contrast, NLRC4 activation is induced by several Gram-negative pathogens by the translocation of small amounts of flagellin or PrgJ-like rod proteins into the host cytosol (Franchi et al., 2006; Miao et al., 2010). In the intestine, NLRC4-dependent IL-1β production is triggered by Salmonella in resident phagocytes which contribute to host defense through the recruitment of neutrophils (Franchi et al., 2012a). The role of the NLRP3 inflammasome in intestinal inflammation is controversial. For example, some studies showed that mice lacking NLRP3 or caspase-1 were less susceptible to chemically-induced colitis (Bauer et al., 2012; Bauer et al., 2010), whereas other authors reported increased susceptibility and worsen pathology (Allen et al., 2010; Zaki et al., 2010). The role of IL-1β in colitis is also controversial. Some studies showed that blocking of IL-1β signaling ameliorate intestinal inflammation in different animal colitis models (Coccia et al., 2012; Saitoh et al., 2008), but one study showed that IL-1β-deficient mice are more susceptible to experimental colitis than wild-type (WT) mice (Bersudsky et al., 2014). The reasons for these contradictory results are unclear, but it may be caused, at least in part, by baseline differences in the gut microbiota (Bauer et al., 2012). While some intestinal commensals such as Escherichia coli can induce IL-1β via the NLRP3 inflammasome in bone marrow-derived macrophages (BMDM), the ability of resident bacteria to induce IL-1β in vivo and the innate immune cells involved in IL-β release in response to the intestinal microbiota remain unclear. In this study, we provide evidence that selective members of the microbiota and specifically P. mirabilis can induce robust IL-1β production via the NLRP3 inflammasome and promote IL-1β-dependent inflammation in the intestine. Production of IL-1β by the microbiota is mediated by CCR2+ monocytes that are recruited to the intestine in response to epithelial injury.
RESULTS
Intestinal resident bacteria induce IL-1β release and promote colitis via IL-1β
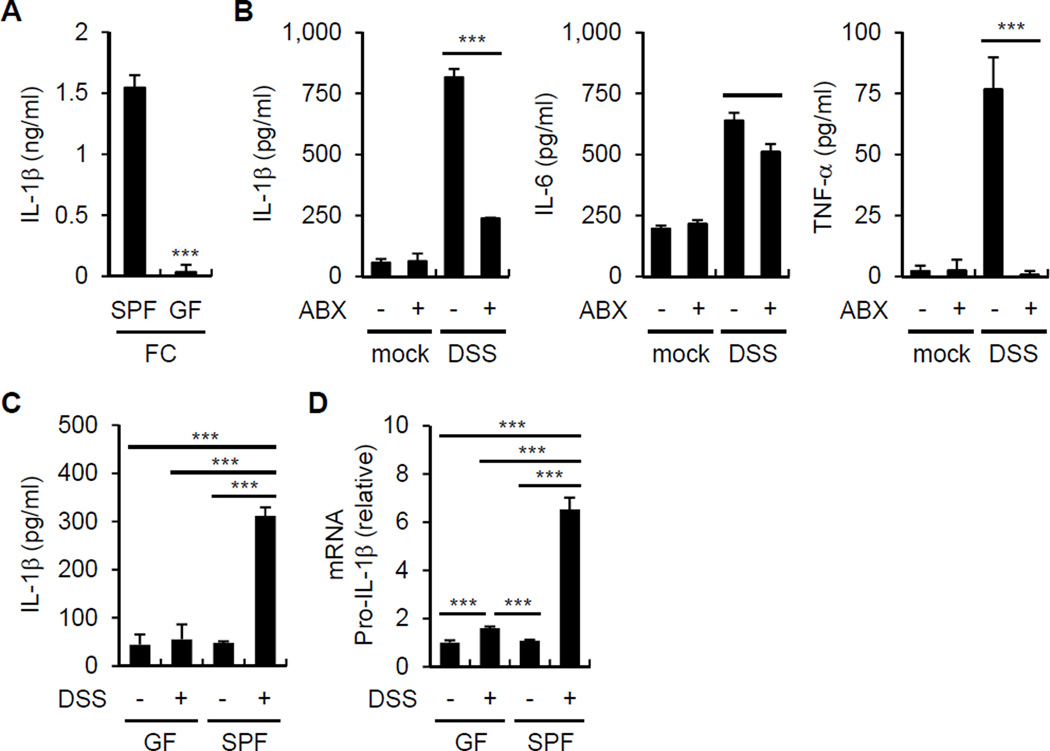
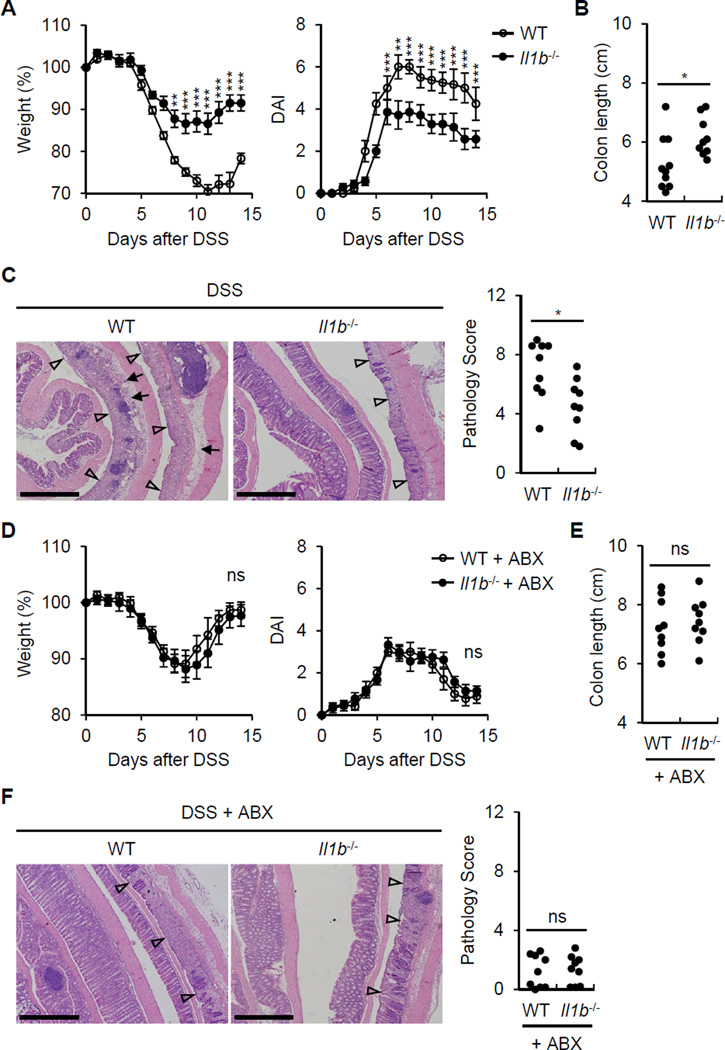
To determine whether the fecal microbiota induce IL-1β release, BMDM were stimulated with fecal contents from conventionally reared mice and germ-free (GF) mice and IL-1β was assessed in the culture supernatants. The feces of conventionally raised specific pathogen-free (SPF) mice, but not GF mice, induced robust amounts of IL-1β (Figure 1A). To determine whether the microbiota induces IL-1β in vivo, we treated mice with a cocktail of antibiotics or PBS and then orally with dextran sulfate sodium (DSS), a chemical that damages the epithelium resulting in exposure of lamina propia (LP) cells to luminal microbes. Treatment with antibiotics reduced the production of IL-1β, IL-6 and TNF-α by LP cells when compared to PBS-treated mice (Figure 1B). Consistently, LP cells from SPF mice produced more IL-1β and pro-IL-1β mRNA than from GF mice after DSS administration (Figure 1C and 1D). To determine whether microbiota-induced IL-1β is important for triggering intestinal inflammation in vivo, we assessed features associated with inflammation in WT and Il1b−/− mice after DSS administration. IL-1β deficiency was associated with reduced weight loss (Figure 2A) and lower disease activity index (DAI) (Figure 2A). In addition, WT mice had more colonic shortening, a marker of colitis (Figure 2B), and exhibited more inflammation in the colon upon DSS administration than Il1b−/− mice (Figure 2C). Depletion of the microbiota by treatment with antibiotics abolished the phenotype associated with IL-1β deficiency (Figure 2D-2F). These results indicate that the microbiota induces IL-1β upon epithelial damage and IL-1β promotes intestinal inflammation.
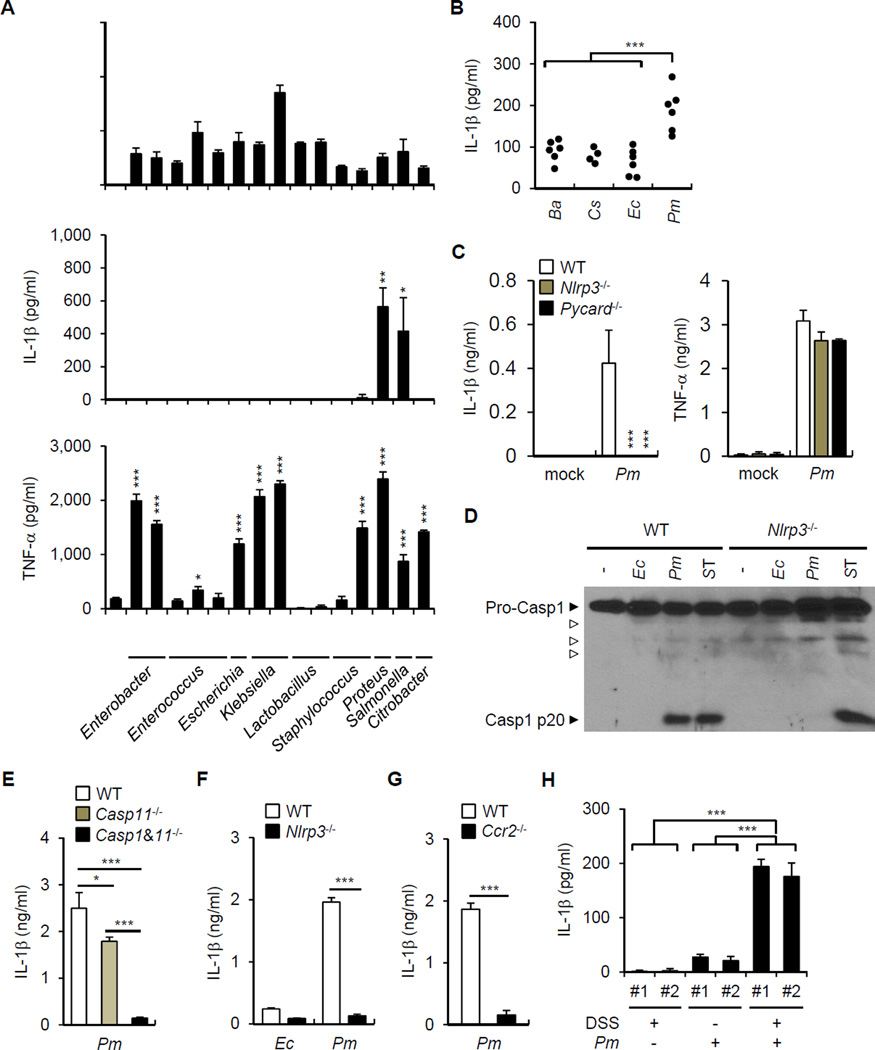
Figure 1. Commensal bacteria induce IL-1β.
(A) Fecal contents (FC) obtained from GF or SPF mice were used to stimulate BMDM for 18 hrs. IL-1β was measured from culture supernatant by ELISA.
(B) Total LP cells were isolated from PBS or antibiotic cocktail (ABX)-treated mice on day 7 after mock or 2.5% DSS treatment and cultured for 3 hrs. IL-1β, IL-6, and TNF-α from culture supernatant were measured by ELISA.
(C) and (D) GF and SPF mice were treated with 2% DSS from day 0 to 6. Total LP cells were isolated on day 7. (C) Total LP cells were cultured for 3 hrs and IL-1β was measured by ELISA.
(D) pro-IL-1β mRNA expression was analyzed by qRT-PCR and normalized relative to GAPDH expression.
(A)-(D) Data are representative of at least two individual experiments and values represent mean of triplicate samples ± SD. **p < 0.01, ***p < 0.001.
Figure 2. IL-1β induced by the commensal bacteria promotes colitis.
(A) WT and Il1b−/− mice were treated with 2% DSS from day 0 to day 6. Body weight and disease activity index (DAI) were measured daily for two weeks. Values represent means ± SEM.
(B) and (C) WT and Il1b−/− mice were treated with 2% DSS from day 0 to day 6 and euthanized at day 10. (B) Colon length was measured and (C) pathology score were assessed.
(D) WT and Il1b−/− mice were treated with 2% DSS from day 0 to day 6 and received PBS or ABX daily by oral gavage from day −1 to 7. Body weight and disease activity index (DAI) were measured daily for two weeks. Values represent means ± SEM.
(E) and (F) WT and Il1b−/− mice were treated with 2% DSS from day 0 to day 6 and and received PBS or ABX daily by oral gavage from day −1 to 7. Mice were euthanized at day 10. (B) Colon length was measured and (C) pathology score were assessed.
(C) and (F) Represenative images of the distal colon. Open arrowheads denote mucosal ulcer with total loss of epithelium. Solid arrows indicate presence of acute inflammatory cells and/or edema in the submucosa. Scale bar; 500 µm.
Every data are pooled from two separate experiments (n > 8, each group). *p < 0.05, **p < 0.01, ***p < 0.001.
Commensal bacteria induce IL-1β release via the NLRP3 inflammasome
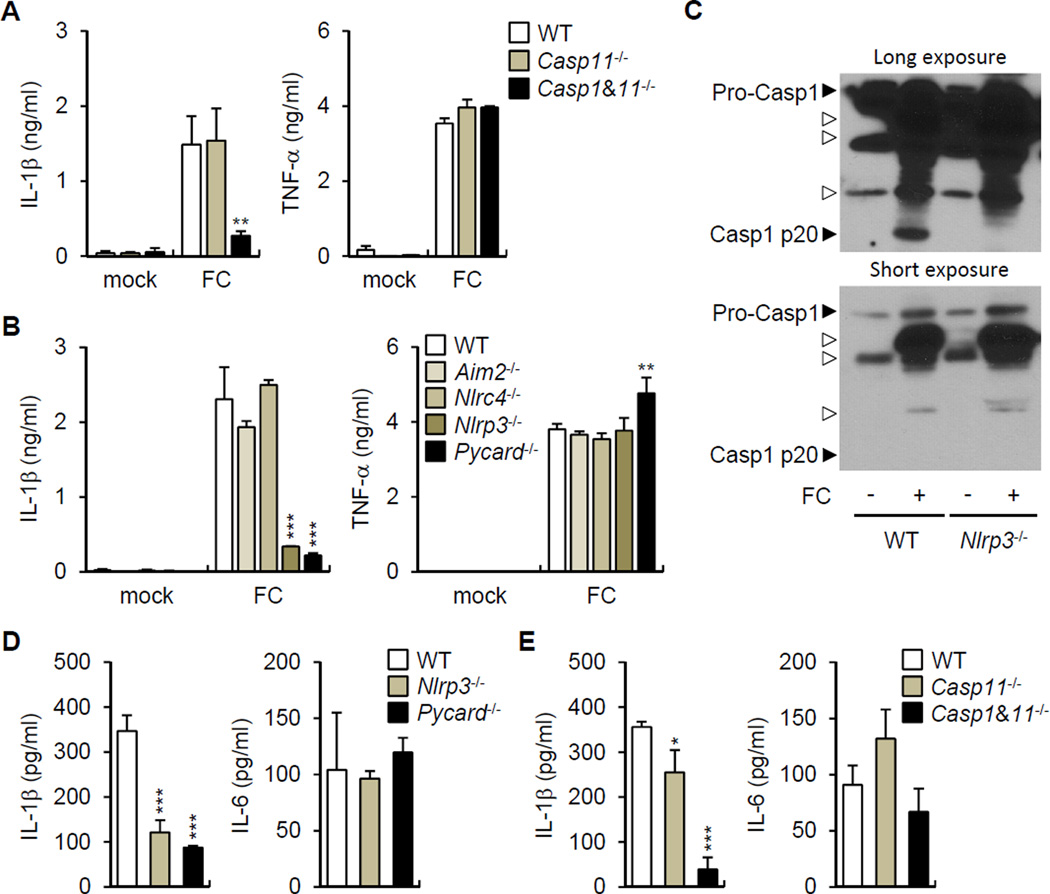
To begin to understand how the intestinal microbiota induces the release of IL-1β, we stimulated BMDM from WT and mice deficient in caspase-1, the essential adaptor ASC and different sensors that activate the inflammasome with fecal contents. The production of IL-1β, but not TNF-α, required caspase-1, NLRP3 or ASC (Figure 3A and 3B), but neither caspase-11 nor AIM2 or NLRC4 (Figure 3B). Consistently, caspase-1 activation as determined by the formation of the mature p20 subunit was induced by stimulation with fecal contents in WT, but not Nlrp3−/− macrophages (Figure 3C). To determine whether the NLRP3 inflammasome is important for the induction of IL-1β in vivo, we purified LP cells from DSS-treated WT mice and mutant mice deficient in inflammasome components and assessed the release of IL-1β by LP cells ex vivo. LP cells purified from the colonic LP of WT mice released spontaneously IL-1β and this response was impaired in Casp1−/−, Nlrp3−/− and Pycard−/− mice (Figure 3D and 3E). These results indicate that the microbiota induces IL-1β release via the NLRP3 inflammasome.
Figure 3. Commensal bacteria induce IL-1β through the NLRP3 inflammasome.
(A) and (B) Fecal contents (FC) obtained from SPF mice were used to stimulate WT, Casp11−/−, Casp1−/− Casp11−/− BMDM (A) and WT, Aim2−/−, Nlrc4−/−, Nlrp3−/−, Pycard−/− BMDM (B) for 18 hrs. IL-1β and TNF-α were measured from culture supernatant.
(C) WT and Nlrp3−/− BMDM were stimulated with FC obtained from SPF mice and caspase-1 was detected by immunoblotting. Open arrow heads indicate non-specific bands.
(D) and (E) WT, Casp11−/−, Casp1−/− Casp11−/−, Nlrp3−/−, and Pycard−/− mice were treated with 2.5% DSS from day 0 to 7. Total LP cells were cultured for 3 hrs and IL-1β and IL-6 were measured in culture supernatant.
Values represent mean of triplicate samples ± SD. Data are representative for three experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Recruited monocytes are the major source of IL-1β produced in response to the microbiota
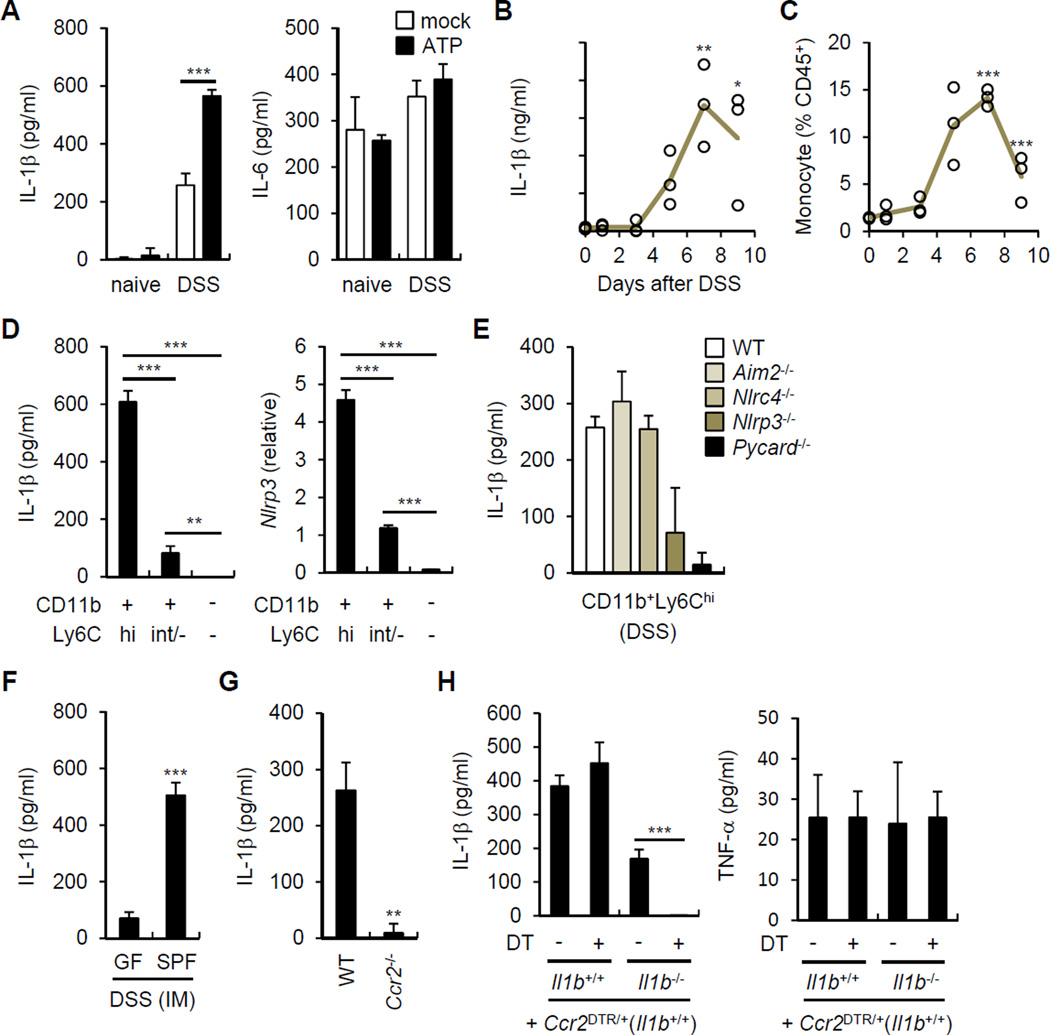
The normal intestine contains large numbers of resident mononuclear phagocytes including macrophages and dendritic cells. In addition, blood monocytes are rapidly recruited to the intestinal tissue in response to inflammatory stimuli (Zigmond and Jung, 2013). We determined next which population of mononuclear phagocytic cells produces IL-1β in response to the microbiota. We purified LP cells from naïve mice and mice harboring CD11b+Ly6Chigh inflammatory monocytes (IM) in response to DSS administration. In ex vivo experiments, intestinal phagocytic cells from DSS-treated mice, but not naïve mice, spontaneously produced IL-1β which was further enhanced by the addition of ATP, a signal that activates the NLRP3 inflammasome (Figure 4A). In contrast, production of IL-6 was comparable in naïve and DSS-treated mice and was not enhanced by ATP (Figure 4A). Notably, resident LP cells from naïve animals did not produce IL-1β even after addition of exogenous ATP (Figure 4A). Production of IL-1β by LP phagocytes was detected at day 3 and peaked around day 7 after DSS (Figure 4B). The kinetics of IL-1β production by LP cells correlated with the number of IM (CD11b+Ly6Chigh) recruited to the intestine (Figure 4B and 4C). To define the LP populations that produce IL-1β in response to the microbiota, LP cells from DSS-treated mice were sorted into CD11b+Ly6Chigh (IMs), CD11b+Ly6Clow (phagocytes) and CD11b−Ly6C− (non-phagocytes). CD11b+Ly6Chigh cells released more IL-1β and expressed higher amounts of Nlrp3 mRNA than phagocytes and non-phagocytic cells (Figure 4D). IMs isolated from the colon of DSS-treated Nlrp3−/− and Pycard−/−mice released less IL-1β than WT, Aim2−/− and Nlrc4−/− IMs (Figure 4E). The spontaneous release of IL-1β by CD11b+Ly6Chigh cells was largely triggered by the microbiota because it was greatly reduced in cells isolated from the intestine of DSS-treated GF mice (Figure 4F).
Figure 4. Recruited inflammatory monocytes are the main source of IL-1β during DSS-induced colitis.
(A) Total LP cells isolated on day 0 (naive) and on day 7 (DSS) after DSS treatment were cultured for 3 hrs and left unstimulated or cultured in the presence of 5 mM ATP for the additional 30 min. IL-1β and IL-6 were measured from culture supernatant.
(B) and (C) Total LP cells were isolated at indicated times from WT mice given 2.5% DSS from day 0 to day 7. (B) IL-1β was measured from culture supernatant after 3 hrs of culture. (C) Percent of inflammatory monocytes (CD11b+Ly6Chigh) in CD45+ LP cells was determined by flow cytometry. Open circles represents means of values from individual mice.
(D) Total LP cells were isolated from DSS-treated mice on day 7 after 2.5% DSS treatment and CD45+ cells were sorted according to CD11b and Ly6C expression. Sorted cells were cultured for 3 hrs and IL-1β in culture supernatant and Nlrp3 mRNA expression was analyzed by ELISA and qRT-PCR, respectively.
(E) WT, Aim2−/−, Nlrc4−/−, Nlrp3−/−, and Pycard−/− mice were treated with 2.5% DSS for 7 days. Sorted inflammatory monocytes were cultured for 3 hrs and IL-1β was measured in culture supernatant.
(F) GF and SPF mice were treated with 2% DSS for 6 days. Sorted inflammatory monocytes (IM) were cultured for 3 hrs and IL-1β was measured in culture supernatant.
(G) WT and Ccr2−/− mice were treated with 2.5% DSS for 7 days. Total LP cells were cultured for 3 hrs and IL-1β was measured in culture supernatant.
(H) IL-1β release by total LP cells isolated from indicated chimera mice. Mice received PBS or diphtheria toxin (DT) on day 0, 3, and 6. LP cells were isolated on day 7 and cultured for 3 hrs to measure cytokine concentrations in culture supernatant.
Data are representative of at least two experiments. Values are means of triplicate samples ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S1.
To further verify the role of recruited IM in microbiota-induced IL-1β release, we used mice deficient in CCR2, which is an essential chemokine receptor for emigration of Ly6Chigh monocytes from the BM and recruitment to the intestine (Kim et al., 2011; Serbina and Pamer, 2006). Ccr2−/− mice exhibited impaired recruitment of IM to the intestine in response to DSS administration (Figure S1A). CCR2 deficiency abolished the release of IL-1β by LP cells isolated from DSS-treated mice (Figure 4G). Given that IMs can produce various different inflammatory mediators capable of activating surrounding cells and contribute to develop DSS-induced colitis (Waddell et al., 2011) (Figure S1B), we used CCR2-diphtheria toxin receptor (Ccr2DTR/+) depleter mice to determine the autonomous role of IMs in IL-1β production. We used Ccr2DTR/+ mice to generate mixed chimeras by transplanting mixtures of bone marrow (BM) from WT or Il1b−/− mice with BM from Ccr2DTR/+ mice at a 1:1 ratio into lethally-irradiated WT mice. Consistent with previous results (Hohl et al., 2009), we found efficient depletion (> 90%) of CCR2+ monocytes after a single i.p. administration of DT to Ccr2DTR/+mice (Figure S1C). LP cells from recipient mice reconstituted with WT (Il1b+/+) and Ccr2DTR/+ BM produced comparable amounts of IL-1β after DSS treatment regardless of DT treatment (Figure 4H). Consistent with mixed chimerism, LP cells from mice reconstituted with Il1b−/− and Ccr2DTR/+ BM released less IL-1β than mice reconstituted with WT BM (Figure 4H). Notably, the release of IL-1β, but not TNF-α, by LP cells from mice reconstituted with Il1b −/− and Ccr2DTR/+ bone marrow was abrogated after DT treatment (Figure 4H). Flow cytometric analysis revealed comparable numbers of CD11b+Ly6Chi IM in the LP of DT- and DSS-treated Il1b+/++Ccr2DTR/+ and Il1b−/−+Ccr2DTR/+ mice (Figure S1D), ruling out that the impairment of IL-1β release was caused by defective intestinal recruitment of IM in Il1b−/−+Ccr2DTR/+ chimeric mice. Collectively, these results indicate that IM recruited to the intestine are the major source of IL-1β produced in response to stimulation by the microbiota.
Selective resident bacteria elicit robust release of IL-1β
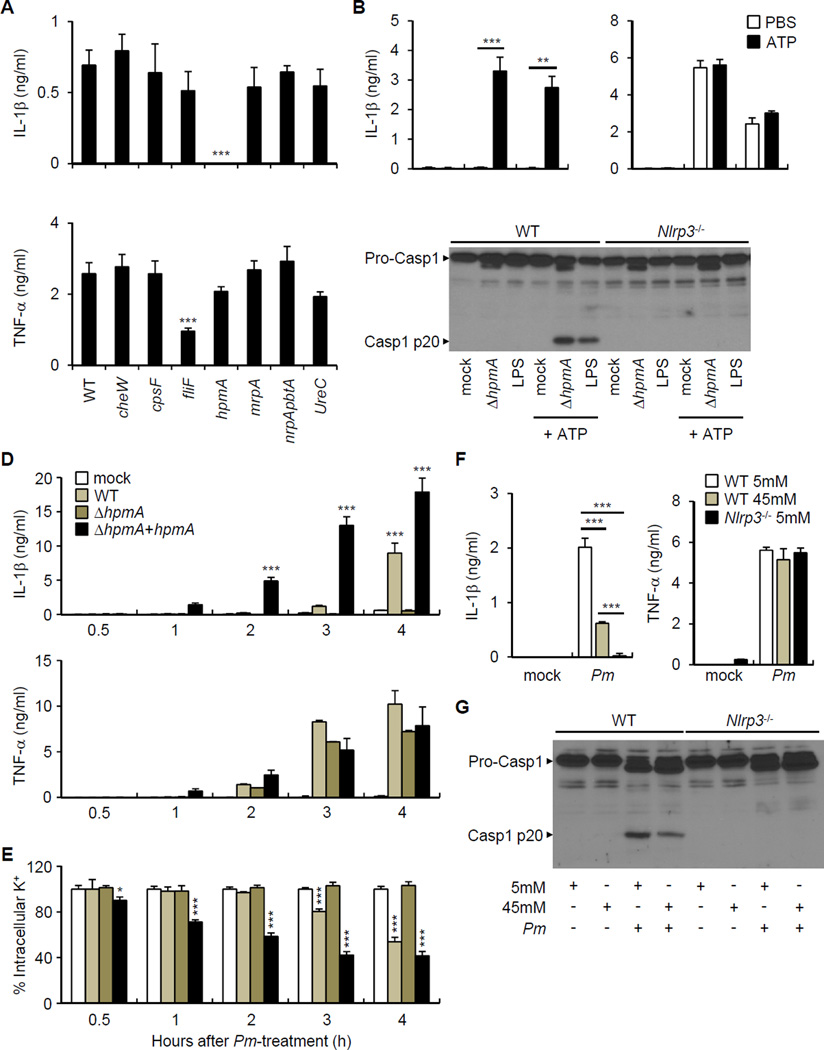
To identify commensal bacteria that induce IL-1β release, we screened a panel of bacterial species isolated from the mouse intestine for their ability to trigger IL-1β production in macrophages at a low bacteria-macrophage ratio. As fecal contents could induce IL-1β production under aerobic conditions, we first tested aerobic or facultative anaerobic commensal bacteria. All commensal bacteria tested could induce pro-IL-1β mRNA expression (Figure 5A, top panel). Notably, P. mirabilis, out of 13 tested, induced rapid and robust secretion of IL-1β (Figure 5A, middle panel). We also assessed the ability of Bacteroides and Clostridium species to induce IL-1β; however, these abundant anaerobic bacteria did not induce IL-1β release (data not shown). Remarkably, the kinetics and potency of P. mirabilis to induce IL-1β release was comparable to that of the pathogen Salmonella enterica Typhimurium (Figure 5A). After longer stimulation (18 hrs), the commensal E. coli and the pathogen Citrobacter rodentium induced IL-1β secretion, but at in lower amounts than P. mirabilis (Figure S2). The link between P. mirabilis and IL-1β was highly specific in that most commensal bacteria triggered TNF-α (Figure 5A, lower panel, and Figure S2). To test specificity of P. mirabilis to induce IL-1β production in vivo, GF mice were mono-associated with P. mirabilis, B. acidifaciens, C. sporogenes, or E. coli and their intestinal colonization was monitored in the feces before DSS treatment. Despite comparable bacterial colonization (Figure S3), LP cells isolated from mice mono-associated with P. mirabilis produced more IL-1β than LP cells from mice colonized with the other commensal bacteria (Figure 5B). Analyses of macrophages deficient in components of the inflammasome revealed that IL-1β release and caspase-1 activation, but not TNF-α, induced by P. mirabilis was abolished in Nlrp3−/− and Pycard−/− BMDM (Figure 5C and 5D). Similarly, P. mirabilis induced IL-1β release in LP cells isolated from DSS-treated WT mice after 3 hrs culture and this response was largely dependent on caspase-1 and NLRP3, but much less on caspase-11 (Figure 5E and 5F). Consistent with previous results, P. mirabilis induced more IL-1β release than E. coli in LP cells (Figure 5F). LP cells isolated from Ccr2−/− mice treated with DSS did not release IL-1β after P. mirabilis stimulation (Figure 5G). To determine whether P. mirabilis is sufficient to induce IL-1β production in vivo, we colonized GF mice with P. mirabilis and assessed the spontaneous production of IL-1β by LP cells purified from untreated and DSS-treated mice. Consistent with an important role for recruited monocytes, there was no or minimal IL-1β release by LP cells isolated from untreated mice colonized with P. mirabilis or GF mice treated with DSS (Figure 5H). In contrast, we found increased IL-1β production by LP cells from GF mice colonized with P. mirabilis and treated with DSS when compared to mice treated with the bacterium or DSS alone (Figure 5H).
Figure 5. Selective commensals induce IL-1β release through the NLRP3 inflammasome.
(A) BMDM were stimulated with indicated mouse commensal bacteria, Citrobacter rodentium (Cr), or Salmonella enterica serovar Typhimurium (ST) at a bacteria:macrophage ratio of 1. Ebc, Enterobacter cloacae; Ebh, E. hormaechei; Ecc, Enterococcus casseliflavus; Ecf, E. faecalis; Ecg, E. gallinarum; Ec, Escherichia coli; Ko, Klebsiella oxytoca; Kp, K. pneumoniae; Lm, Lactobacillus murinus; Lr, L. reuteri; Ss, Staphylococcus sciuri; Sx, S. xylosus; Pm, Proteus mirabilis. Levels of cytokines in culture supernatant and pro-IL-1β mRNA expression at 3 hrs were determined by ELISA and qRT-PCR, respectively. Values represent means of triplicate samples ± SD.
(B) GF mice were mono-associated with indicated mouse commensal bacteria. Ba, Bacteroides acidifaciens; Cs, Clostridium sporogenes; Ec, E. coli; Pm, P. mirabilis. Mice were treated with 1% DSS for 7 days and total LP cells were isolated and cultured for 3 hrs to assess IL-1β release in supernatant. Means of values from one or two mice are indicated. Results are pooled from three experiments with similar results.
(C) WT, Nlrp3−/−, and Pycard−/− BMDM were stimulated with mouse Pm and cytokines were measured in culture supernatant after 3 hrs. Values represent means of triplicate samples ± SD.
(D) WT and Nlrp3−/− BMDM were stimulated with Ec, Pm, and ST for 3 hrs and caspase-1 activation was detected by immunoblotting.
(E)-(G) WT or indicated mutant mice were treated with 2.5% DSS for 7 days and total LP cells were isolated and were stimulated with indicated bacteria at a bacteria: macrophage ratio of 1 for 3 hrs. Values represent means of triplicate samples ± SD.
(H) GF mice or gnotobiotic mice monocolonized with mouse P. mirabilis by oral gavage were treated with 2% DSS for 6 days. Total LP cells were isolated on day 7 and IL-1β was determined in culture supernatant after 3 hrs. Values represent means of triplicate samples ± SD. Data are representative of at least two experiments. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S2 and S3.
P. mirabilis HpmA hemolysin is required for the activation of the NLRP3 inflammasome
We sequenced the genome of the mouse P. mirabilis referred here as strain UM001 to identify candidate genes that may be involved in triggering NLRP3 activation. We obtained a 3,950,860 base-pair (bp)-long genomic sequence of the P. mirabilis strain UM001 which revealed 3,579 putative protein-coding and 84 noncoding genes within 28 contigs (Figure S4A). Comprehensive comparison analysis of the genomic sequences by RAST showed that the mouse P. mirabilis genome contains several virulence genes that are also present in the reference human P. mirabilis strain H14320 (Aziz et al., 2008; Pearson et al., 2008). These genes included cheW (a regulator of chemotaxis), fliF (flagellin), hpmA (hemolysin), mrpA (fimbria), genes encoding essential proteins of a T3SS such as spa47, and ureC (urease) (Figure S4B). Notably, the human P. mirabilis strain HI4320 also induced IL-1β release (Figure 6A). Analyses of several isogenic P. mirabilis mutants of HI4320 revealed that hpmA, but not cheW, cpsF, fliF, mrpA, or ureC, was required for the induction of IL-1β release, but not of TNF-α (Figure 6A). Expression of the hpmA gene was detected in both the mouse and human P. mirabilis strains (Figure S4C). Comparison of the HpmA amino acid sequence from the mouse and human strains revealed that they were 99% identical (Figure S4D). Secretion of IL-1β via the NLRP3 inflammasome requires signal 1 that induces pro-IL-1β and signal 2 that activates caspase-1 (Franchi et al., 2012b). To assess whether HpmA can provide signal 1 or signal 2, we stimulated BMDM with ΔhpmA P. mirabilis or LPS as an inducer of signal 1 in absence or presence of ATP which provides signal 2 for inflammasome activation. Both ΔhpmA P. mirabilis and LPS induced the release of IL-1β and caspase-1 activation, but only in the presence of ATP (Figure 6B and 6C). Together with results shown in Figure 6A, the data indicate that ΔhpmA P. mirabilis can only provide signal 1 and that HpmA is required for P. mirabilis to provide signal 2 for activation of the NLRP3 inflammasome. Complementation of the ΔhpmA mutant strain with a hpmA expression plasmid rescued the ability of the mutant strain to induce IL-1β release, but it did not affect its ability to release TNF-α (Figure 6D). K+ efflux is a common event that is required for NLRP3 activation induced by multiple stimuli (Munoz-Planillo et al., 2013). Consistently, WT P. mirabilis, but not the ΔhpmA mutant, induced K+ efflux which was enhanced in the ΔhpmA mutant reconstituted with hpmA plasmid (Figure 6E). The enhanced induction of IL-1β and K+ efflux by the complemented (ΔhpmA+hpmA) strain presumably results from the higher copy number of the plasmid-derived hpmA gene. Culture of macrophages in 45 mM K+ inhibited the ability of P. mirabilis to induce caspase-1 activation and the release of IL-1β, but not that of TNF-α (Figure 6F and 6G). Collectively, these results indicate that activation of the NLRP3 inflammasome by P. mirabilis is mediated by the HpmA hemolysin.
Figure 6. P. mirabilis HpmA mediates NLRP3 inflammasome activation.
(A) BMDM were pre-incubated with LPS (100 ng/ml) for 3 hrs and then stimulated with P. mirabilis HI4320 (WT) and indicated isogenic mutants for 3 hrs at bacteria:macrophage ratio of 1. Cytokines were measured in culture supernatant after 3 hrs culture.
(B) and (C) WT or Nlrp3−/− BMDM were pre-incubated with ΔhpmA Pm or LPS (100 ng/ml) for 3 hrs and left unstimulated or cultured in the presence of 5 mM ATP for additional 30 min. (B) IL-1β and TNF-α were measured from culture supernatant of WT BMDM. (C) Caspase-1 activation was detected by immunoblotting.
(D) and (E) LPS-treated BMDM were incubated with HpmA-deficient (ΔhpmA) P. mirabilis mutant, and the same strain complemented with hpmA plasmid (ΔhpmA+hpmA). Cytokines in culture supernatant (D) and intracellular K+ concentrations (E) were measured at different time points.
(F) and (G) WT and Nlrp3−/−BMDMs were stimulated with P. mirabilis (Pm) in culture medium containing the indicated concentration of extracellular K+. (D) Cytokine concentrations in culture supernatant were determined by ELISA. (E) Active caspase-1 in cell lysate and cell supernatant were detected by immunoblotting.
Values are means of triplicate samples ± SD. Data are representative of at least three experiments. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S4.
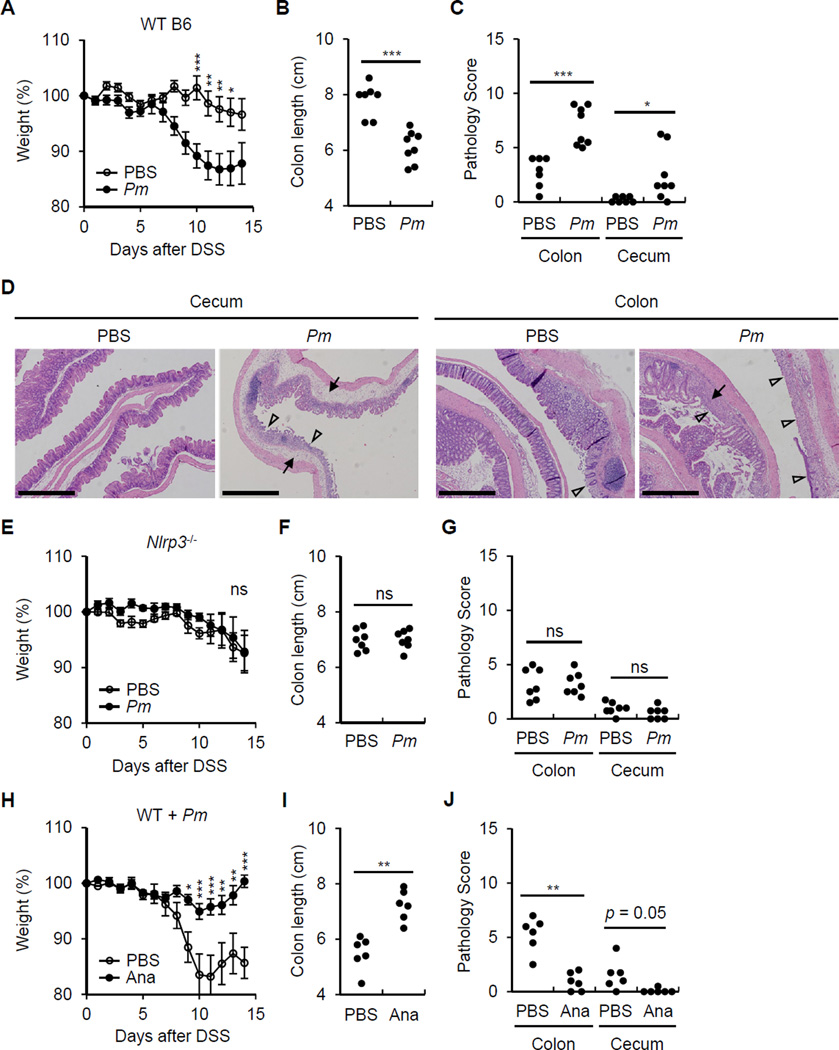
P. mirabilis colonization enhances DSS-induced colitis via NLRP3-mediated IL-1 signaling
To determine whether P. mirabilis can affect DSS-induced colitis, we generated a streptomycin-resistant P. mirabilis (StrR Pm) strain by culturing the mouse P. mirabilis bacterium with streptomycin. Like the parental P. mirabilis strain, StrR Pm induced IL-1β release and exhibited swarming capacity (Figure S5). When mice were treated with streptomycin and infected with StrR Pm, colonization of the StrR Pm strain was detected in the luminal contents of the ileum, cecum, and colon (Figure S6A) and the feces (Figure S6B). Notably, mice colonized with P. mirabilis did not display obvious signs of intestinal inflammation including loss of body weight, soft stools or intestinal inflammation by histological examination (Figure S6C, 6D and data not shown). However, mice colonized with P. mirabilis exhibited more loss of body weight and colonic shortening than control mice after DSS administration (Figure 7A and 7B). The colitis induced by DSS is typically restricted to the distal colon in normal mice (Brown et al., 2007). Remarkably, mice colonized with P. mirabilis exhibited marked inflammation in the cecum upon DSS administration (Figure 7C and 7D). Consistent with the results in Figure 4, enhancement of DSS-colitis induced by P. mirabilis colonization was not detected in Ccr2−/− mice (Figure S6E and S6F). Furthermore, the increase in body weight loss, colonic shortening and enhanced tissue pathology associated with P. mirabilis colonization were not observed in Nlrp3−/− mice (Figure 7E-7G) and were reversed by treatment with Anakinra, an antagonist of IL-1 receptor signaling (Figure 7H-7J). In order to rule out the possibility that enhanced colitis merely reflect higher numbers of bacteria in P. mirabilis colonized mice, we inoculated mice with the same number of WT or ΔhpmA P. mirabilis by gavage. Analysis of fecal samples of WT and ΔhpmA P. mirabilis showed comparable intestinal colonization (Figure S7). Importantly, colonization with AhpmA P. mirabilis resulted in less body weight loss and colonic shortening compared to colonization with WT P. mirabilis (Figure S7). Collectively, the results indicate P. mirabilis colonization enhances DSS-induced colitis via NLRP3-mediated IL-1 signaling in inflammatory monocytes.
Figure 7. P. mirabilis enhances DSS-induced inflammation via NLRP3 and IL-1R signaling.
Streptomycin-treated WT or Nlrp3−/− mice were gavaged with 1×109 cfu of streptomycin-resistant P. mirabilis (Pm) on day 0, 4, and 8. Colitis was induced by administration of 1% DSS from day 0 to day 10. Mice were euthanized for analyses on day 15.
(A)-(C) WT mice injected with PBS (n = 12) or inoculated with Pm (n = 15).
(D) Representative histology of cecum and distal colon in WT mice in the absence and presence of Pm colonization after 1% DSS administration on day 15 after DSS administration. Open arrowheads denote damaged or ulcerated mucosa with total loss of epithelium, solid arrowheads indicate presence of inflammatory cells in the submucosa and edema. Scale bar; 500 µm.
(E)-(G) Nlrp3−/− mice injected with PBS (n = 12) or inoculated with Pm (n = 11).
(H)-(J) Pm-colonized WT mice were injected daily with PBS (n = 9) or Anakinra (Ana, 50 mg/kg, n = 8) from day 4.
Body weight data were pooled from three (A) and (E) or two (H) separate experiments. Values represent means ± SEM. Colon length and pathology score were pooled from two experiments with similar results. *p < 0.05, **p < 0.01, ***p < 0.001, ns; not significant. See also Figure S5-S7.
DISCUSSION
Previous studies showed that the microbiota can promote intestinal inflammation and pathology in animal models (Hudcovic et al., 2001; Kirkland et al., 2012; Kitajima et al., 2001). A major inflammatory pathway induced in the intestine is that triggered by IL-1β. Clinical studies have suggested that IL-1β is important in disease because robust amounts of IL-1β are released by colonic LP cells from inflammatory bowel disease patients which correlates with lesional disease activity (Ligumsky et al., 1990). However, the role of the microbiota and the mechanism involved in triggering IL-1β production are unclear. Based on experiments in mice treated with antibiotics and GF mice, we show that the microbiota is critical for the induction of IL-1β release and this response is mediated by the NLRP3 inflammasome in vitro and in vivo. Remarkably, the great majority of the resident bacteria isolated from the mouse intestine induce little or no release of IL-1β, although they can elicit robust production of TNF-α. Although several intestinal bacteria including P. mirabilis and E. coli induce activation of the NLRP3 inflammasome in vitro, the ability of P. mirabilis to induce caspase-1 activation and IL-1β release was striking in that it was comparable to that of pathogenic Salmonella. P. mirabilis was found to induce robust IL-1β release; however, our results suggest that other bacteria including E. coli can induce IL-1β in vitro and possibly in vivo. Furthermore, commensals other than P. mirabilis, some of which may be uncultivable could be involved in regulating IL-1β release in vivo. Although a member of the intestinal microbiota in humans and mice (Armbruster and Mobley, 2012; Garrett et al., 2010), P. mirabilis is a major cause of urinary tract infection in humans (Armbruster and Mobley, 2012). In mouse model, P. mirabilis has been linked to the severity of colitis (Garrett et al., 2010). Thus, P. mirabilis acts as pathobiont in mice and humans. Consistently, sequence analysis of the mouse P. mirabilis genome revealed that the commensal bacterium harbors multiple virulence genes shared with human uropathogenic P. mirabilis strains (Pearson et al., 2008). One of the virulence genes, hpmA, was found to be critical for the activation of NLRP3 inflammasome. The hpmA gene encodes a secreted pore-forming cytolysin that is produced during bacterial growth and infection (Swihart and Welch, 1990). Although the function of HpmA in P. mirabilis is unknown, our study indicates that HpmA can induce K+ efflux and this activity is sensed by host macrophages to activate the NLRP3 inflammasome.
Several studies have assessed the role of the NLRP3 inflammasome in DSS-induced colitis with discordant results. For example, two studies reported increased susceptibility to DSS-induced colitis in Nlrp3−/− mice which was associated with decreased IL-1β and IL-18 production, loss of epithelial integrity, and increased colonic inflammation (Allen et al., 2010; Zaki et al., 2010). In contrast, another group found lower concentrations of several cytokines including IL-1β in colonic tissue and protection against colitis in Nlrp3−/− mice (Bauer et al., 2010). These contradictory results are paralleled by observations in Casp1−/− mice. For example, one study showed protection against DSS-induced colitis in Casp1−/− mice whereas another groups showed the opposite (Allen et al., 2010; Siegmund et al., 2001; Zaki et al., 2010). Although it is difficult to ascertain the reason for these contradictory results, recent studies showed that protection of Nlrp3−/− mice against colitis can be reversed by co-housing Nlrp3−/− mice with WT mice (Bauer et al., 2012). The latter results suggest that the differences in results regarding the role of the NLRP3 inflammasome might be explained, at least in part, by baseline differences in the intestinal microbiota among mouse colonies. Given the marked variation in the ability of different intestinal bacteria to activate the NLRP3 inflammasome, it is likely that mice with different microbiota composition will differ in the intestinal activation of the inflammasome induced upon epithelial injury.
Intestinal bacteria belonging to the Enterobacteriaceae family have been linked to colitis in mice and humans. For example, P. mirabilis acts in concert with other intestinal bacteria to induce colitis in TRUC mice, a spontaneous model of ulcerative colitis (Garrett et al., 2010). Although the mechanism that is responsible for TRUC colitis is not fully understood, recent studies have revealed an important role for group 3 innate lymphoid cells (ILC3) in the TRUC colitogenic phenotype (Powell et al., 2012). Because the activation of ILC3 is induced via IL-23 and IL-1β (Coccia et al., 2012; Takatori et al., 2009), it is possible that P. mirabilis contributes to TRUC colitis through the activation of ILC3 cells and induction of colitogenic cytokines such as IL-17A. Although there is no evidence as yet that P. mirabilis contributes to colitis in humans, adherent and invasive E. coli (AIEC) strains that accumulate in the intestine of patients with inflammatory bowel disease induce robust IL-1β release via the NLRP3 inflammasome (De la Fuente et al., 2014). Like P. mirabilis, AIEC is a pathobiont that expresses virulence factors and can enhance experimental colitis in mice (Drouet et al., 2012), although the host signaling pathways involved remain unclear. Because P. mirabilis colonization can promote colonic inflammation via NLRP3, we suggest that Enterobacteriaceae family members such as P. mirabilis and AIEC promote intestinal inflammation, at least in part, by inducing local IL-1β release.
Colonization of conventionally reared mice or GF mice with P. mirabilis alone did not cause overt colitis. The latter can be explained by the requirement of intestinal injury and recruitment of inflammatory monocytes for the activation of the NLRP3 inflammasome and release of IL-1β. It is possible that the HpmA is produced by P. mirabilis under steady-state conditions but its activity is antagonized by host factors such as intraluminal IgA. Unlike resident mononuclear phagocytes that are hyporesponsive to commensal bacteria, inflammatory monocytes recruited to the intestine express NLRP3 and pro-IL-1β in response to microbial stimulation and are responsive to NLRP3-activating signals such as ATP. Consistent with these studies, Ly6Chigh inflammatory monocytes recruited to the intestine in response to DSS express pattern recognition receptors such as TLR2 and NOD2 that sense bacteria and their deletion attenuates DSS-induced colitis (Zigmond et al., 2012). The lack of a functional NLRP3 inflammasome in resident phagocytes may ensure that inappropriate production of IL-1β is not induced under steady-state conditions or when there is minimal epithelial injury. In contrast, resident intestinal macrophages constitutively express NLRC4 and pro-IL-1β and release IL-1β upon infection with enteric pathogens such as Salmonella (Franchi et al., 2012a). These observations suggest that the NLRC4 inflammasome functions to detect pathogen invasion early whereas the NLRP3 inflammasome is activated at later stages in inflammatory monocytes and may serve to boost immune responses against invasive pathogens and/or to enhance tissue repair in the intestine.
EXPERIMENTAL PROCEDURES
Mice
Six to eight-week-old WT C57BL/6 (B6), Aim2−/−, Pycard−/−, Casp1−/−Casp11 −/−, Casp11−/−, Ccr2−/−, Il1b−/−, Nlrc4−/−, and Nlrp3−/− mice in B6 background were bred and kept under specific pathogen-free (SPF) condition in University of Michigan Cancer Center. CCR2 depleter (Ccr2DTR/+) mice were kindly provided by Dr. Eric Pamer, Memorial Sloan Kettering Cancer Center (Hohl et al., 2009). GF mice were bred and maintained at the GF Animal Core Facility of the University of Michigan. The animal studies were conducted under protocols approved by the University of Michigan Committee on Use and Care of Animals.
Bacteria
Commensal bacteria were isolated from the fecal content of B6 mice. For most commensals, the feces were suspended in PBS, plated on Brain Heart Infusion (BHI) plates at different dilutions under aerobic conditions and individual colonies isolated and frozen in glycerol. Lactobacillus spp. were plated on de Man, Rogosa and Sharpe (MRS) agar. For isolation of Bacteroides or Clostridia species, fecal samples were plated on Bacteroides Agar (Nissui, Japan), and BHI agar plates containing 5% sheep blood, respectively, under anaerobic conditions. The bacterial species were verified by 16S rRNA sequencing. Most of bacteria were inoculated in Luria-Bertani (LB) broth (MP Biomedicals) and cultured overnight at 37°C with shaking. Lactobacillus spp. were inoculated in MRS broth (Difco) and cultured for 2 days under anaerobic conditions. Bacteroides, and Clostridium were cultured in Chopped Meat Medium (Anaerobe systems) for 2 days under anaerobic conditions. To generate streptomycin-resistant mouse-isolated P. mirabilis, bacteria were cultured multiple cycles in broth containing increasing doses of streptomycin (Calbiochem). A single colony was picked from culture every 2 to 3 cycles using LB plates containing tetracycline (20 µg/ml) and streptomycin (5 to 200 µg/ml). P.mirabilis HI4320, a human urinary tract isolate has been described (Jones and Mobley, 1987). Isogenic P. mirabilis HI4320 mutants constructed using a TargeTron mutagenesis kit have been described (Alteri et al., 2013). To generate hpmA-complemented human P. mirabilis HI4320 strain (HI4320 ΔhpmA+pGENhpmA), the mutant HI4320 ΔhpmA strain was transformed with pGEN plasmid expressing hpmA and the bacteria selected with ampicillin (20 µg/ml).
Genomic Sequencing Analysis
Whole genome sequencing of P. mirabilis UM001 was performed on the Illumina HiSeq 2000 platform using paired-end reads with 200–300bp insert size and 100bp read length. Assembly of the 5.25 million reads obtained was carried out using Velvet Optimizer (Zerbino and Birney, 2008) to produce a final genome assembly 3,950,860bp in length comprised of 28 contigs with a contig N50 of 377,567bp. Annotation to identify coding sequences, assign predicted functions and to predict RNA structures was generated using Prokka (Seemann, 2014).
DSS-induced colitis and P. mirabilis colonization
Water containing 1 – 2.5% DSS (molecular weight, 36,000–50,000; MP Biomedicals) was administered for 6 – 10 days and the mice were switched to regular water. Body weight and stool consistency were checked daily. In some experiments, mice were given an antibiotic cocktail (500 µg of ampicillin, 250 µg of vancomycin, 250 µg of metronidazole, 250 µg of gentamycin, and 500 µg of neomycin) by gavage for 9 days starting one day prior to DSS treatment. For P. mirabilis colonization experiments, mice received 2 g/L streptomycin in the drinking water throughout the experiment starting 10 days before bacterial gavage. Mice were inoculated with 1 × 109 cfu of streptomycin-resistant P. mirabilis by gavage and all mouse groups received water containing 1% DSS for 10 days to induce colitis. To antagonize IL-1β, mice were intraperitoneally injected with Anakinra (50 mg/kg) daily from day 4 to day 14. Body weight measured every day and results are pool from 2 to 4 separate experiments which used at least 3 mice per group. Disease activity index (DAI) was monitored every day in DSS-treated mice as described (Kamada et al., 2005). Briefly, loss of body weight (0 = none, 1 = 1% to 5%, 2 = 5% to 10%, 3 = 10% to 20%, 4 = > 20%), stool consistency (0 = normal stool, 2 = loose stool, 4 = diarrhea), hemoccult (0 = normal. 2 = homoccult positive, 4 = gross blood). PBS-injected and P. mirabilis-infected mice were sacrificed on day 15 and histological score was evaluated blindly by one pathologist using a described scoring system (Chen et al., 2008). Briefly, severity of inflammation (0 = none, 1 = mild, 2 = moderate, 3 = severe), the level of involvement (0 = none, 1 = mucosa, 2 = mucosa and submucosa, 3 = transmural), and extent of epithelial/crypt damage (0 = none, 1 = basal 1/3, 2 = basal 2/3, 3 = crypt loss, 4 = crypt and surface epithelial destruction). Each variable was then multiplied by a factor reflecting the percentage of the colon involved (0–25%, 26–50%, 51–75%, 76–100%), and then summed to obtain the overall score.
Preparation and culture of macrophage and intestinal phagocytes
BMDM and LP cells were prepared as described (Franchi et al., 2012a). BMDM (2×105 cells/well) were suspended in IMDM supplemented with 2% FBS and seeded in 48-multiwell plates. In some experiments, BMDM were stimulated with Ultrapure E. coli LPS (100 ng/ml; Invivogen) for 3 hrs. Feces from SPF or GF mice were suspended in sterile PBS (100 mg/ml), homogenized, and spun at 1,000 rpm for 5 seconds. Fecal contents were passed through 40 µm cell strainer (BD Falcon) to remove aggregates and used to stimulate BMDM at a 1:200 dilution. BMDM were incubated with fecal contents for 3 hrs, the medium was replaced with fresh medium containing gentamicin (100 µg/ml) and the culture continued for an additional 15 hrs. BMDM were cultured with individual bacteria strains at a bacterial:macrophage ratio of 1 for 3 hrs. For long-term stimulation, the medium was replaced with fresh medium containing gentamicin (100 µg/ml) and the culture continued for an additional 15 hrs. Isolated LP cells were resuspended in RPMI medium containing 10% heat-inactivated fetal bovine serum (FBS), 2-β-mercaptoethanol (50 µM), L-glutamine (2 mM), sodium pyruvate (1 mM), MEM non-essential amino acids (Gibco). LP cells were pooled from 2 or 3 mice per group, and total LP cells (2× 105 cells/well) or sorted cells (1×105 cells/well) were seeded in 96-multiwell plates in triplicate. Cells were incubated for 3 hrs in 5% CO2 incubator and cell-free supernatant were subjected to further analysis. For ex vivo stimulation, cells were left unstimulated or incubated with 5 mM ATP for 30 min or with bacteria at bacteria:macrophage ratio of 1 for 3 hrs.
Immunoblotting
Cells were lysed in buffer containing 1% NP-40 supplemented with complete protease inhibitor cocktail (Roche, Mannheim, Germany) and 2 mM dithiothreitol. Lysates were resolved by SDS-PAGE and transferred to PVDF membranes by electro-blotting. The rabbit anti mouse caspase-1 antibody has been described (Franchi et al., 2012a). Protein bands were revealed with goat anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories) and enhanced chemiluminescent substrate (Thermo Scientific).
Intracellular K+ determination and cytokine detection
Intracellular K+ concentrations were measured as described previously (Munoz-Planillo et al., 2013). Mouse cytokines were measured in culture supernatants by enzyme-linked immuneabsorbent assay (ELISA) kits (R&D Systems, Minneapolis, MN).
Flow cytometry and cell sorting
LP cells were stained with the following antibodies: CD45 (30-F11), CD11b (M1/70), Ly6C (AL-21), Ly6G (1A8), all purchased from BD Pharmingen. Cells were stained and sorted using a FACSAria III instrument (BD Bioscience). Data were analyzed using FlowJo program (Tree Star).
Quantitative real-time PCR
RNA was extracted with E.Z.N.A. total RNA kit (Omega Biotek) according to the manufacturer’s instructions. RNA was reverse transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystem) and cDNA was then used for quantitative PCR analysis using SYBR Green Gene Expression Assay on an ABI 7900HT analyzer. The following primer sets were used for amplification: IL-1β-F; 5’-GATCCACACTCTCCAGCTGCA, IL-1β-R; 5’- CAACCAACAAGTGATATTCTCCATG, NLRP3-F; 5’-ATGGTATGCCAGGAGGACAG, NLRP3-R; 5’-ATGCTCCTTGACCAGTTGGA, GAPDH-F; 5’-TGCGACTTCAACAGCAACTC, GAPDH-R; 5’-GCCTCTCTTGCTCAGTGTCC. Samples were run in triplicate for each experimental condition and mean values were used to calculate statistics.
Generation of mixed chimeric mice
BM cells were obtained from Ccr2DTR/+ mice, mixed with equivalent number of WT or Il1b−/− BM cells and 1×107 donor cells were injected into 8-week-old recipient WT mice that were lethally irradiated with 1100 rads. Mice were used 8 weeks after reconstitution. To deplete CCR2+ cells, 10 µg/kg diphtheria toxin (DT) was intraperitoneally injected on day 0, 3, 6.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism program (GraphPad Software). Differences between two groups were evaluated using Student’s t test. For multiple comparison, one-way ANOVA or Kruscal-Wallis test were used. For body weight and DAI comparisons, two-way ANOVA with Bonferroni posttest was used. p < 0.05 were considered significant.
Supplementary Material
ACKNOWLEDGEMENT
We thank Grace Chen for critical review of the manuscript and Eric Pamer for proving mutant mice. We thank the University of Michigan Host Microbiome Initiative for support. This work is supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology 2013R1A6A3A03024348 (S.-U.S.), a Career Development award from the Crohn’s and Colitis Foundation of America (N.K.), the Michigan Gastrointestinal Peptide Research Center NIDDK 5P30DK034933 (N.K.), grants 098051 and WT086418MA from the Wellcome Trust (T.D.L) and DK091191 and DK095782 from the National Institutes of Health (G.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.-U.S., N.K. and G.N. conceived the study. S.-U.S. performed most of the experiments. R.M.-P., Y.-G.K., D.K., Y.K., M.H and S.D.H. provided critical reagents and helped with experiments, R.M.-P., H.L.T.M., H.P.B., T.D.L. and N.I. analyzed data. G.N. supervised all aspects of this study. S.-U.S. and G.N. wrote the manuscript with contributions from all authors.
REFERENCES
- Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster CE, Mobley HL. Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 2012;10:743–754. doi: 10.1038/nrmicro2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Lehr HA, Endres S, Schnurr M. Protective and aggravating effects of Nlrp3 inflammasome activation in IBD models: influence of genetic and environmental factors. Dig. Dis. 2012;1(30 Suppl.):82–90. doi: 10.1159/000341681. [DOI] [PubMed] [Google Scholar]
- Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59:1192–1199. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- Bersudsky M, Luski L, Fishman D, White RM, Ziv-Sokolovskaya N, Dotan S, Rider P, Kaplanov I, Aychek T, Dinarello CA, et al. Non-redundant properties of IL-1alpha and IL-1beta during acute colon inflammation in mice. Gut. 2014;63:598–609. doi: 10.1136/gutjnl-2012-303329. [DOI] [PubMed] [Google Scholar]
- Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr, Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, Stenson WF, Stappenbeck TS. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Invest. 2007;117:258–269. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia M, Harrison OJ, Schiering C, Asquith MJ, Becher B, Powrie F, Maloy KJ. IL-1beta mediates chronic intestinal inflammation by promoting the accumulation of IL-17A secreting innate lymphoid cells and CD4(+) Th17 cells. J. Exp. Med. 2012;209:1595–1609. doi: 10.1084/jem.20111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente M, Franchi L, Araya D, Diaz-Jimenez D, Olivares M, Alvarez-Lobos M, Golenbock D, Gonzalez MJ, Lopez-Kostner F, Quera R, et al. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int. J. Med. Microbiol. 2014;304:384–392. doi: 10.1016/j.ijmm.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet M, Vignal C, Singer E, Djouina M, Dubreuil L, Cortot A, Desreumaux P, Neut C. AIEC colonization and pathogenicity: influence of previous antibiotic treatment and preexisting inflammation. Inflamm. Bowel Dis. 2012;18:1923–1931. doi: 10.1002/ibd.22908. [DOI] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, Nunez G. NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol. 2012a;13:449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 2012b;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl TM, Rivera A, Lipuma L, Gallegos A, Shi C, Mack M, Pamer EG. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 2009;6:470–481. doi: 10.1016/j.chom.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudcovic T, Stepankova R, Cebra J, Tlaskalova-Hogenova H. The role of microflora in the development of intestinal inflammation: acute and chronic colitis induced by dextran sulfate in germ-free and conventionally reared immunocompetent and immunodeficient mice. Folia Microbiol. 2001;46:565–572. doi: 10.1007/BF02818004. [DOI] [PubMed] [Google Scholar]
- Jones BD, Mobley HL. Genetic and biochemical diversity of ureases of Proteus, Providencia, and Morganella species isolated from urinary tract infection. Infect. Immun. 1987;55:2198–2203. doi: 10.1128/iai.55.9.2198-2203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Inoue N, Hisamatsu T, Okamoto S, Matsuoka K, Sato T, Chinen H, Hong KS, Yamada T, Suzuki Y, et al. Nonpathogenic Escherichia coli strain Nissle1917 prevents murine acute and chronic colitis. Inflamm. Bowel Dis. 2005;11:455–463. doi: 10.1097/01.mib.0000158158.55955.de. [DOI] [PubMed] [Google Scholar]
- Kamada N, Nunez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146:1477–1488. doi: 10.1053/j.gastro.2014.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kim YG, Kamada N, Shaw MH, Warner N, Chen GY, Franchi L, Nunez G. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34:769–780. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland D, Benson A, Mirpuri J, Pifer R, Hou B, DeFranco AL, Yarovinsky F. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp. Anim. 2001;50:387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- Ligumsky M, Simon PL, Karmeli F, Rachmilewitz D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut. 1990;31:686–689. doi: 10.1136/gut.31.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J. Exp. Med. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 2008;190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell N, Walker AW, Stolarczyk E, Canavan JB, Gokmen MR, Marks E, Jackson I, Hashim A, Curtis MA, Jenner RG, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- Siegmund B, Lehr HA, Fantuzzi G, Dinarello CA. IL-1 beta -converting enzyme (caspase-1) in intestinal inflammation. Proc. Natl. Acad. Sci. USA. 2001;98:13249–13254. doi: 10.1073/pnas.231473998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swihart KG, Welch RA. Cytotoxic activity of the Proteus hemolysin HpmA. Infect. Immun. 1990;58:1861–1869. doi: 10.1128/iai.58.6.1861-1869.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O’Shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S, Williams IR, Gewirtz AT. Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell A, Ahrens R, Steinbrecher K, Donovan B, Rothenberg ME, Munitz A, Hogan SP. Colonic eosinophilic inflammation in experimental colitis is mediated by Ly6C(high) CCR2(+) inflammatory monocyte/macrophage-derived CCL11. J. Immunol. 2011;186:5993–6003. doi: 10.4049/jimmunol.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 2013;34:162–168. doi: 10.1016/j.it.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, Mack M, Shpigel N, Boneca IG, Murphy KM, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.