Abstract
Ty3/Gypsy long terminals repeat (LTR) retrotransposons are structurally and phylogenetically close to retroviruses. Two notable structural differences between these groups of genetic elements are 1) the presence in retroviruses of an additional envelope gene, env, which mediates infection, and 2) a specific dual ribonuclease H (RNH) domain encoded by the retroviral pol gene. However, similar to retroviruses, many Ty3/Gypsy LTR retrotransposons harbor additional env-like genes, promoting concepts of the infective mode of these retrotransposons. Here, we provide a further line of evidence of similarity between retroviruses and some Ty3/Gypsy LTR retrotransposons. We identify that, together with their additional genes, plant Ty3/Gypsy LTR retrotransposons of the Tat group have a second RNH, as do retroviruses. Most importantly, we show that the resulting dual RNHs of Tat LTR retrotransposons and retroviruses emerged independently, providing strong evidence for their convergent evolution. The convergent resemblance of Tat LTR retrotransposons and retroviruses may indicate similar selection pressures acting on these diverse groups of elements and reveal potential evolutionary constraints on their structure. We speculate that dual RNH is required to accelerate retrotransposon evolution through increased rates of strand transfer events and subsequent recombination events.
Keywords: convergent evolution, LTR retrotransposons, retroviruses, ribonuclease H
Introduction
Long terminal repeat (LTR) retrotransposons, transposable elements that mobilize via an RNA intermediate and contain LTRs, are structurally similar to retroviruses and usually contain two genes: gag and pol. Gag encodes a major structural protein that is processed by a transposon-encoded protease (PR) into capsid and nucleocapsid proteins during virus-like particle maturation (Kirchner and Sandmeyer 1993; Freed 1998). Pol encodes PR and other enzymes necessary for reverse transcription: reverse transcriptase (RT), ribonuclease H (RNH), and integrase (INT). Some LTR retrotransposons carry an additional gene that resembles the envelope (env) gene from vertebrate retroviruses (Xiong and Eickbush 1990; Malik et al. 2000; Kim et al. 2004). The two major types of LTR retrotransposons, Ty1/Copia and Ty3/Gypsy, are classified according to their origin and the order of functional domains within their pol gene (Kumar and Bennetzen 1999; Schulman 2013). Additional types of LTR retrotransposon-like elements, such as DIRS elements with tyrosine recombinase and caulimoviruses of plants, show a common origin but lack LTRs and have a distinct structure (Xiong and Eickbush 1990; Poulter and Goodwin 2005).
Evolutionary studies based on the analysis of different conserved domains have traced the origin of retroviruses to the ancient Ty3/Gypsy LTR retrotransposons (Xiong and Eickbush 1990; Llorens et al. 2008). Previous studies have also reported that the evolutionary history of the retroviral RNH domain appears to be complex and likely includes independent acquisitions and possible subfunctionalization (Malik and Eickbush 2001; Smyshlyaev et al. 2013). RNH activity is required for DNA strand transfer and processing of the polypurine tract primer that primes the synthesis of the second DNA strand during LTR retrotransposon and retroviral retrotransposition (Wilhelm et al. 2001; Lener et al. 2002; Basu et al. 2008). Both LTR retrotransposons and retroviruses carry a type I RNH domain that is closely related to the cellular RNHs involved in DNA replication in bacteria, archaea, and eukaryotes RNHs (Malik and Eickbush 2001; Ohtani et al. 2004; Cerritelli and Crouch 2009; Tadokoro and Kanaya 2009). Notably, retroviruses harbor an RNH domain that has a specific conserved region, similar to cellular type I RNHs. The LTR retrotransposon RNH domain, however, is divergent from other cellular-like RNHs and demonstrates a lack of conserved amino acid residues typical for retroviral and other type I RNHs. This RNH degeneration is believed to be advantageous because it allows preservation of the polypurine tract primer of LTR retrotransposons (Malik and Eickbush 2001).
It was previously proposed that the retroviral ancestor (ancient Ty3/Gypsy LTR retrotransposon) harbored an RNH domain typical of LTR retrotransposons and that the additional cellular-like RNH was acquired later in retroviral evolution. Subsequently, the original copy of RNH was subfunctionalized as the so-called tether domain, which likely modulates the activity of RNH in retroviruses (Malik and Eickbush 2001; Lapkouski et al. 2013). Such a complex dual RNH domain with modulated activity is believed to perform more fine-tuned RNA cleavage, leading to increased rates of DNA strand transfer during reverse transcription and more frequent recombination between two RNA molecules packed into a virus-like particle (Basu et al. 2008; Delviks-Frankenberry et al. 2008; Lapkouski et al. 2013).
In this study, we demonstrate that this dual RNH domain is also present in plant Ty3/Gypsy LTR retrotransposons. Our comparative and evolutionary analyses indicate that these Ty3/Gypsy LTR retrotransposons belong to the Tat clade. By tracing the putative evolutionary history of newly isolated Tat LTR retrotransposons, we identify events of cellular-like RNH domain acquisition and, similar to retroviruses, subsequent subfunctionalization of the original RNH. The resulting dual RNH from the Tat LTR retrotransposons strongly resembles retroviral “tether-RNH” fusion. Thus, the dual RNH domain from plant Tat LTR retrotransposons has the potential to act in a manner similar to that of the retroviral RNH domain, increasing the rates of DNA strand transfer and recombination between copies of LTR retrotransposons. The parallelism in the evolutionary history of retroviral tether-RNH and the dual RNH from plant Tat LTR retrotransposons described herein is striking. We propose that the complex features of the RNH domain in both cases were acquired independently as a result of convergent evolution toward, most likely, the “invention” of the intrinsic means for frequent recombination and, therefore, the potential for accelerated evolution.
Results
Identification and Classification of LTR Retrotransposons with a Cellular-Like RNH Domain
Our comprehensive analysis of plant non-LTR retrotransposons, which was performed earlier, allowed the identification of a new, unique RNH domain that is closely related to the “archaeal” type of cellular-like RNH (aRNH), as opposed to RNH domains from other non-LTR retrotransposons. We suggested that this RNH had been acquired by these retrotransposons from the host genome early in plant evolution (Smyshlyaev et al. 2013). Using these aRNH domain homologues as queries in a BLASTp search against the plant protein database in NCBI, we found that some of the hits were also derived from sequences related to Ty3/gypsy LTR retrotransposons, in addition to those generated by non-LTR retrotransposons. The LTR retrotransposons with a cellular-like RNH domain have never been described to date, and the closest phylogenetically related group carrying this type of RNH is vertebrate retroviruses (Xiong and Eickbush 1990; Malik and Eickbush 2001). We hypothesized that there are Ty3/gypsy LTR retrotransposons present in plant genomes that carry the aRNH domain. However, their identity, distribution, structure, and other characteristics remain unknown.
A relatively large number of sequenced genomic fragments from diverse plants is available for the further in-depth mining of LTR retrotransposons (http://www.phytozome.net/, last accessed May 25, 2014). To perform a large-scale computational analysis of plant genomes, we created a pipeline for the automated identification of aRNH-containing LTR retrotransposons. First, the LTRharvest tool was used to extract copies of LTR retrotransposons from 57 plant genomes (a list of the analyzed genomes is presented in supplementary table S1, Supplementary Material online). Next, all identified sequences were filtered for the presence of both RT and aRNH domains and clustered based on their RT similarity for each individual genome. Finally, a full-length representative element was retrieved for each cluster (see Materials and Methods for details). The mining resulted in a set of 146 sequences of distinct LTR elements from 43 plant genomes (supplementary table S1, Supplementary Material online).
The preliminary phylogenetic analysis of the RT domain from newly identified representative copies of LTR elements containing aRNH and known LTR retrotransposons revealed that all novel sequences formed a monophyletic group that had been previously described as the Tat clade of Ty3/gypsy LTR retrotransposons (fig. 1). Tat LTR retrotransposons are recognized as potential plant retroviruses, as they are closely related to the Athila clade, which consists of plant LTR retrotransposons carrying an env-like gene (Wright and Voytas 1998; Llorens et al. 2011), and also harbor extra open reading frames (eORFs). However, the functions and origins of the eORFs of Tat LTR retrotransposons remain unknown. Both the Tat and Athila clades are widely distributed in plant genomes (Steinbauerová et al. 2011).
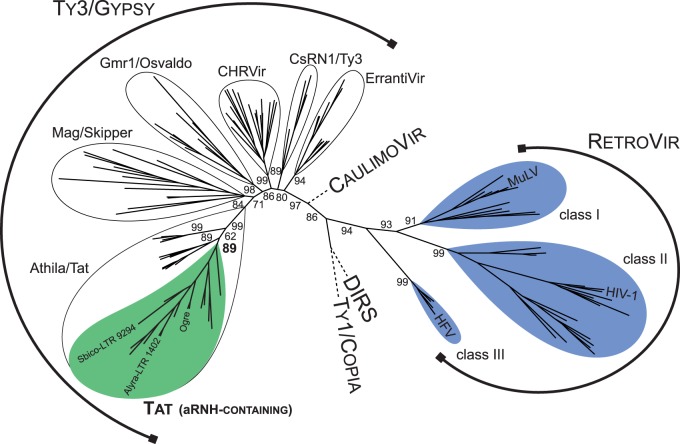
Fig. 1.
Maximum-likelihood tree based on the amino acid sequences of RT from diverse LTR retrotransposons, retroviruses (RetroVir), caulimoviruses (CaulimoVir), and DIRS elements. Statistical support was evaluated by the approximate likelihood-ratio test (aLRT) and is shown at the corresponding nodes of the tree. Retroviruses and aRNH-containing LTR retrotransposons are highlighted in blue and green, respectively. The complete phylogenetic tree with accession numbers and names of the elements is presented in supplementary figure S1, Supplementary Material online.
We distinguished six phylogenetic lineages among aRNH-containing Tat LTR retrotransposons based on their phylogenetic relationships (fig. 2A). The single LTR element from the spikemoss Selaginella moellendorffii (Viridiplantae; Lycopodiidae) formed lineage I on a phylogenetic tree reconstructed based on the multiple alignment of the amino acid sequences of the RT domain. Lineages II and III were represented by elements from gymnosperms. Finally, all aRNH-containing Tat LTR retrotransposons from angiosperms were clustered into three distinct lineages: IV–VI. Interestingly, after an additional analysis of moss genomes (Goffinet B, personal communication), we did find Tat-like LTR retrotransposons, but the aRNH domain was not present in their coding sequences, indicating that aRNH was likely acquired by plant Tat LTR retrotransposons after separation of the moss clade (fig. 2B).
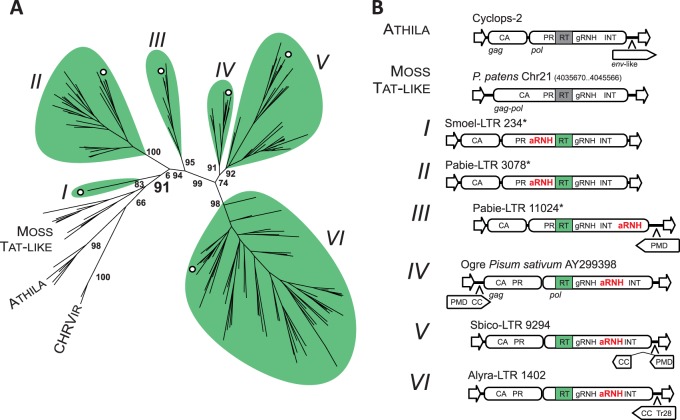
Fig. 2.
Diversity of Tat LTR retrotransposons. (A) Maximum-likelihood tree based on the amino acid sequences of RT from representatives of Tat LTR retrotransposons, Athila elements and chromoviruses. Statistical support was evaluated by the aLRT and is shown at the corresponding nodes of the tree. Six lineages of aRNH-containing Tat LTR retrotransposons are denoted by Latin numerals I–VI and are highlighted in green. Moss Tat-like elements, which are phylogenetically close to aRNH-containing Tat LTR retrotransposons but lack aRNH, are also denoted on the tree, as are Athila elements and chromoviruses (CHRVir). The branches corresponding to representative elements from figure 2B are denoted by white circles. The complete phylogenetic tree with accession numbers and names of the elements is presented in supplementary figure S2, Supplementary Material online. (B) Structural composition of the elements from identified lineages. Schemes of the structures of elements from corresponding lineages are shown. The structure of one of the representative copies was used, with the exception of those denoted by asterisks, for which multiple copies of the element were used to reconstruct the overall structure due to the lack of putatively intact copies. eORFs are denoted by pentagons with the orientation corresponding to the frame (sense/antisense). CA, capsid domain of Gag protein; gRNH, RNH of Ty3/gypsy LTR retrotransposons.
aRNH-Containing Tat LTR Retrotransposons Are Structurally Highly Diverse
A further in-depth comparative analysis showed that perturbation in the organization of the plant aRNH-containing Tat LTR retrotransposons could be easily traced from lineage to lineage. This includes the following: 1) protease (PR) domain allocation within Gag or Pol; 2) location/orientation of eORFs; and 3) the position of the aRNH domain in Pol (fig. 2B). As previously noted, LTR retrotransposon PR is usually encoded by pol. Our analysis revealed that PR is fused with Gag, and not Pol, in the Tat LTR retrotransposons of flowering plants (lineages IV–VI). The early lineages of Tat LTR retrotransposons, however, encode PR at a canonical location, the 5′-end of pol (fig. 2B), suggesting PR allocation to Gag as one of the evolutionary developments in plant Tat LTR retrotransposons.
A second development was the capture of eORFs. Some Tat LTR retrotransposons were previously shown to carry eORFs at nonconventional locations (e.g., upstream of gag) or in an antisense orientation relative to the two main ORFs: gag and pol. The proteins encoded by these eORFs were reported to exhibit homology to plant mobile domain (PMD) and to transposase 28 (Tr28) domain (Kejnovsky et al. 2006; Steinbauerová et al. 2011). Our analysis revealed that elements from lineages I and II, as well as Tat-like LTR retrotransposons from mosses, did not appear to contain any eORFs, suggesting that the progenitor of plant Tat LTR retrotransposons did not carry these sequences and that the sequences were likely acquired later in plant evolution. The first eORF in the evolutionary history of Tat LTR retrotransposons appears in gymnosperm lineage III. This eORF is situated in an antisense orientation at the 3′-end of the element, and its protein product contains PMD (fig. 2B and supplementary fig. S4, Supplementary Material online). The eORF location and presence of PMD is retained in the elements from lineage V, and, in addition to PMD, the eORF encodes a putative coiled coil (CC) at the 3′-terminus. Elements from lineage IV also have “PMD-CC”-encoding eORFs, which are located at the 5′-end of the element in the sense orientation.
Our phylogenetic analysis suggests that PMD has a complex origin, is closely related to PMDs from mutator-like transposable elements (MULE) transposons and is also somewhat related to highly conserved PMDs associated with nontransposon domains, such as High Mobility Group box or Peptidase48 (supplementary figs. S4 and S5, Supplementary Material online). Although the function of PMDs is unknown, the PANTHER classification system places PMDs into the serine/threonine phosphatase family (PTHR11668) and the epididymal membrane protein E9-related family (PTHR16007), which may suggest its potential function (Mi et al. 2005). However, it is worth nothing that we were not able to identify significant sequence homology between PMD and representatives of the PTHR11668 and PTHR16007 protein families. Finally, instead of PMD, elements from lineage VI have a Tr28 domain fused with CC (fig. 2B and supplementary fig. S4, Supplementary Material online). Although the origin of the Tr28 domain remains elusive, some authors have suggested it to be a remnant of a previously inserted mobile element (Kejnovsky et al. 2006).
The most interesting feature of the newly identified Tat LTR retrotransposons, within the context of this research, was the presence of both the aRNH domain and RNH domain common to Ty3/gypsy LTR retrotransposons (gRNH) in the Pol protein (fig. 2B). The aRNH domain appears to be the most distinctive, in the sense that it was found in several different positions relative to other domains present in the Pol protein. Indeed, aRNH is located upstream relative to RT in lineages I and II but is always downstream relative to RT and gRNH in all other lineages including lineage III representatives, which harbor aRNH even further downstream, at the very C-terminus of their putative Pol proteins (fig. 2B and supplementary fig. S2, Supplementary Material online). In contrast, the gRNH domain was always found next to RT, suggesting the existence of functional and structural constraints. Additionally, the variations in the position of aRNH within the Pol protein might indicate a recent and complex evolutionary history of the aRNH domain within the Tat clade. The domains could be acquired independently by different lineages or be the result of the substantial shuffling within the sequence of one retrotransposon copy or between different copies or even between diverse retrotransposons present in the same genome. Duplications of the domains (e.g., original gRNH or preexisting aRNH) followed by subsequent diversification of the domain copies as well as degradation of one of the duplicates might potentially have led to the observed diversity in organization.
Acquisition of the aRNH Domain from the Host Genomes
To unveil the origin(s) and possible patterns of aRNH domain diversification, we performed comparative and phylogenetic analyses of cellular and retrotransposon-specific RNH domains from various sources (fig. 3 and supplementary fig. S3, Supplementary Material online). A multiple alignment of the amino acid sequences for RNH domains from non-LTR and LTR retrotransposons and cellular RNH proteins showed the presence of a conserved catalytic arginine in aRNH from Tat LTR retrotransposons, similar to the aRNHs genes from plant, bacterial, and archaeal genomes and the RNH domain from plant L1 non-LTR retrotransposons (fig. 3 and supplementary fig. S3, Supplementary Material online; Ohtani et al. 2004; You et al. 2007; Smyshlyaev et al. 2013). The standard gRNHs, similar to RNHs of other LTR retrotransposons, do not contain conserved amino acid residues at this position and therefore are assumed to possess lower catalytic activity (fig. 3 and supplementary fig. S3, Supplementary Material online, Malik and Eickbush 2001).
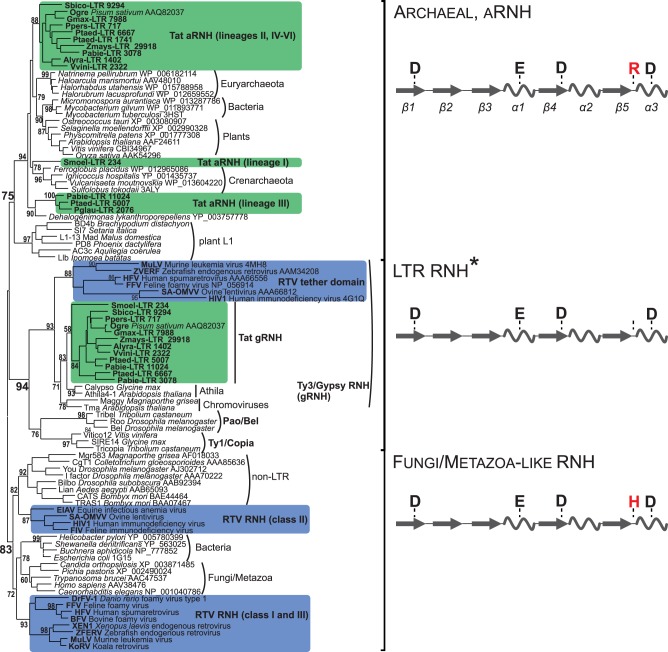
Fig. 3.
Maximum-likelihood tree based on the amino acid sequences of different types of type I RNHs. Statistical support was evaluated by the aLRT and is shown at the corresponding nodes of the tree. aRNH and gRNH from Tat LTR retrotransposons are highlighted in green. RNH and tether from retroviruses (RTV) are highlighted in blue. Schemes of the secondary structures of three subtypes of RNH with the corresponding active site residues are shown at the right of the tree. The α-helices are depicted as helices, and the β-sheets are shown as arrows. The conserved arginine (R) or histidine (H) residue of the active site, which is specific for different RNHs, is highlighted in red. *The positions corresponding to DEDD catalytic core residues are not conserved in the gRNHs of Tat LTR retrotransposons and tether domain of vertebrate retroviruses (see supplementary fig. S3, Supplementary Material online).
As expected based on the results of the comparative analysis, the aRNHs and gRNHs formed distinct groups on the phylogenetic tree (fig. 3). The aRNHs from plant Tat LTR retrotransposons formed a single monophyletic group together with the aRNH domains belonging to plant L1 non-LTR retrotransposons (Smyshlyaev et al. 2013) and the aRNH genes from the genomes of bacteria, archaea, and plants (Ohtani et al. 2004); gRNHs from the same retrotransposons fell into the monophyletic clade with the RNHs of other LTR retrotransposons (aRNH and LTR RNH subtypes, fig. 3). The most parsimonious explanation for the observed phylogenetic relationships is the acquisition of aRNH rather than duplication of the original gRNH domain followed by diversification. Moreover, because Tat LTR retrotransposons are found exclusively in plants and aRNH genes are also described in plant genomes (“plants” clade in fig. 3), the most likely sources of aRNH domains for Tat LTR retrotransposons are plant genomes. This is also supported by the recent finding that transposable elements have a tendency to take the necessary protein domains directly from their hosts (Abrusán et al. 2013).
Interestingly, our phylogenetic reconstruction did not support the monophyletic origin of aRNH domains from Tat LTR retrotransposons. Instead, three distinct clades were formed by the sequences analyzed, which might reflect the involvement of the multiple events of horizontal transfer or suggest that at least three independent acquisitions of the aRNH domain have occurred in the evolutionary history of Tat LTR retrotransposons (fig. 3). The largest clade of aRNH included sequences from Tat LTR retrotransposon lineages II and IV–VI. The aRNHs from linage III formed their own clade on the phylogenetic tree, together with the RNH sequence from the bacterium Dehalogenimonas lykanthroporepellens (Chloroflexi; Dehalococcoidetes), which was retrieved using a BLASTp search against the NCBI protein database and the aRNH of clade III representatives as a query. The aRNH domain from the S. moellendorffii Tat LTR retrotransposon (lineage I; see fig. 3) was also separated phylogenetically from the other aRNHs of Tat LTR retrotransposons and formed a single clade with the aRNHs from Crenarchaeota (fig. 3).
Dual “gRNH-aRNH” and Retroviral tether-RNH Show Remarkable Structural Similarity
As observed from the phylogenetic and structural analyses described above, aRNH-containing Tat LTR retrotransposons from flowering plants harbor the gRNH domain immediately followed by the aRNH domain (lineages IV-VI; fig. 2B). Such dual gRNH-aRNH organization is somewhat reminiscent of the retroviral-specific cellular-like RNH domain associated with the connection domain (tether), which also adopts an RNH-like fold (Malik 2005). To further explore the potentially significant structural similarity between dual RNHs of plant Tat LTR retrotransposons and tether-RNH from retroviruses, we performed a comparison of their secondary structures and revealed a remarkable resemblance between them (fig. 4). The RNH structure commonly consists of a 5-stranded β-sheet surrounded by a distribution of α-helices (Cerritelli and Crouch 2009). As expected, the presence of five β-strands, which can potentially form a β-sheet, and α-helices was predicted for aRNH and gRNH from the Tat LTR retrotransposons in our analysis. This dual RNH overlapped almost perfectly with the secondary structure of HIV-1 tether-RNH (PDB 4g1q).
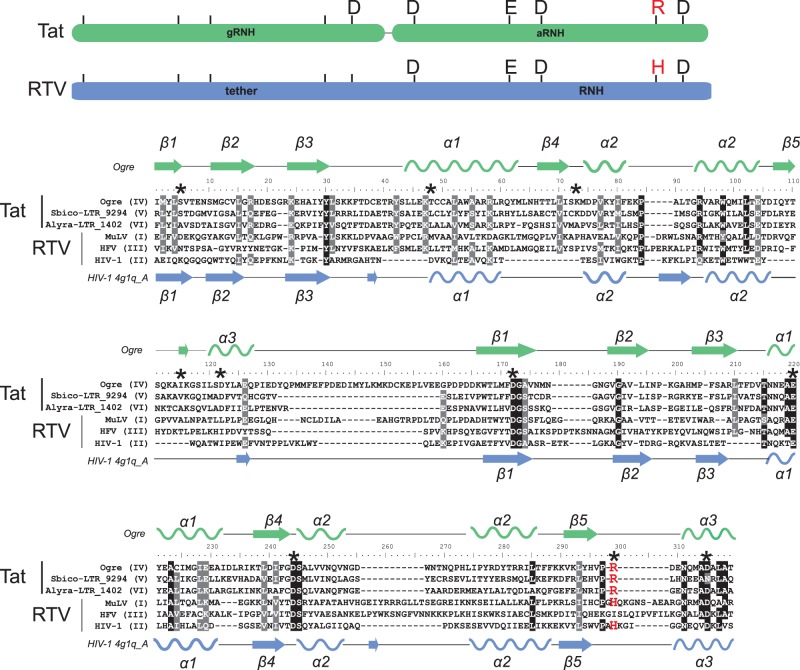
Fig. 4.
Alignment of dual RNHs of diverse Tat LTR retrotransposons and retroviruses (RTV). The names of the sequences used are presented at the left of the alignment and contain information on their lineage for Tat LTR retrotransposons (lineages IV-VI) or their class for retroviruses (classes I-III). Schemes of dual RNHs from Tat LTR retrotransposons (gRNH-aRNH) and retroviruses (tether-RNH) with the indicated active site residues are shown at the top of the figure and are highlighted in green and blue, respectively. The conserved R or H residue of the active site, which is specific for different RNH subtypes, is highlighted in red in the scheme. The secondary structure predicted for gRNH-aRNH of the Ogre element (NCBI accession number AAQ82037) is shown at the top of the alignment. The secondary structure of tether-RNH of HIV-1 retrieved from PDB (accession number 4g1q_A) is shown at the bottom of the alignment. The α-helices are depicted as helices, and the β-sheets are shown as arrows. Positions corresponding to residues of the active site are denoted by black asterisks in the alignment. R or H residues of the active site are highlighted in red.
The predicted aRNH structure of Tat LTR retrotransposons strongly resembles the canonical cellular-like RNH fold with a DEDD catalytic core and, similar to all aRNHs, has an additional conserved arginine residue at the active site (figs. 3 and 4 and supplementary fig. S3, Supplementary Material online). Retroviral RNHs also have a canonical fold but have different origins. Instead of the arginine residue typical for aRNH, retroviral RNHs have a conserved histidine residue in that position. Retroviral RNHs form a single clade with non-LTR, bacterial and fungi/metazoa RNHs (fungi/metazoa-like subtype of RNH, fig. 3) and were previously proposed to emerge from two sources: The class I and III retroviruses more likely captured bacterial or fungi/metazoa RNH, whereas the class II retroviruses acquired RNH from non-LTR retrotransposons (Malik and Eickbush 2001; Malik 2005; Smyshlyaev et al. 2013).
In contrast to well conserved folding of retroviral RNH and aRNH of Tat LTR retrotransposons and the similarly to the tether from retroviruses, the folding of the gRNH domain of plant Tat LTR retrotransposons was found to be somewhat disorganized and divergent from the original RNH structure. Moreover, at the sequence level, the highly conserved DEDD motif, which is typical for all RNH folds and conserved in most LTR retrotransposon RNHs (supplementary fig. S3, Supplementary Material online), has degenerated in the gRNH of Tat LTR retrotransposons and the tether of retroviruses, with the exception of the last aspartic acid residue, which is conserved in Tat gRNHs (figs. 3 and 4 and supplementary fig. S3, Supplementary Material online).
Additionally, our phylogenetic reconstruction clustered gRNHs from diverse Ty3/gypsy LTR retrotransposons together with Tat elements, with the tether from retroviruses as the earliest branching clade (fig. 3). Such branching is in accordance with LTR retrotransposon RT phylogeny, supports the common origin of Ty3/gypsy LTR retrotransposons and retroviruses (Xiong and Eickbush 1990), and traces the ancestry of the tether to the gRNH of the precursor Ty3/gypsy LTR retrotransposon. Moreover, this branching also suggests that the gRNH from the precursor Ty3/Gypsy LTR retrotransposon contained an intact DEDD (fig. 5), which subsequently and independently from retroviruses degenerated in the gRNH of Tat LTR retrotransposons.
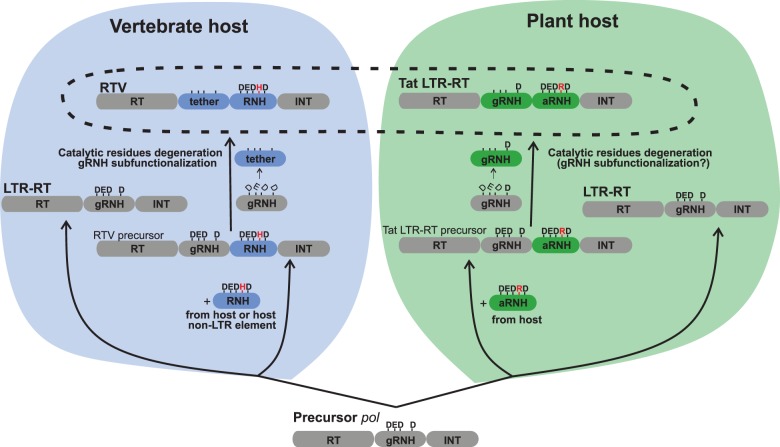
Fig. 5.
Convergent evolution of plant Tat LTR retrotransposons (Tat LTR-RTs) and retroviruses (RTV) through the acquisition of cellular-like RNHs. Putative structure of the precursor LTR retrotransposon (LTR-RT) pol of an ancient eukaryote is shown at the bottom. Each ellipse of the structure represents a functional domain and is correspondingly denoted. The residues of active site of RNH are shown by letters at the top. The left panel displays the origin of vertebrate RTV and is highlighted in blue. Some LTR-RTs in early fungi/metazoa sequestered a cellular-like RNH of the fungi/metazoa-like subtype with a characteristic histidine (H) residue in the active site either from a non-LTR retrotransposon or fungi/metazoa cellular RNH gene (RNH is highlighted in dark blue, and the H residue is highlighted in red; see figure 4 for more details) into their pol gene in a position following gRNH. The retained gRNH was converted into the tether domain, and all catalytic core residues of gRNH degenerated (the degeneration is depicted by unbalanced letters, and the tether domain functionally associated with RNH in HIV-1 is highlighted in dark blue). The resulting structure gave rise to RTV. The initial pol structure was, however, retained and gave rise to modern nonviral LTR retrotransposons in fungal and metazoan genomes. The right panel displays the evolution of LTR retrotransposons in plants and is highlighted in green. We propose that, similar to RTV, some of the ancient plant LTR retrotransposons also acquired a cellular-like RNH but of a distinct archaeal subtype (aRNH) with a characteristic arginine (R) residue in the active site (highlighted in dark green; the R residue is highlighted in red) into their pol gene at a position following gRNH. Similar to RTV, gRNH was retained, and most of its catalytic core residues (except for the C-terminal aspartate residue) degenerated (the degeneration is depicted by unbalanced letters). The resulting structure gave rise to the Tat LTR retrotransposons of flowering plants. The initial pol structure was retained and gave rise to other groups of LTR retrotransposons in plant genomes.
Thus, our data suggest that plant Tat LTR retrotransposons and retroviruses captured cellular-like RNHs independently: Tat LTR retrotransposons recurrently acquired aRNH from a plant host genome, whereas retroviruses obtained RNH either from a non-LTR retrotransposon or from a fungi/metazoa host genome. Furthermore, the original gRNH underwent subfunctionalization independently in Tat LTR retrotransposons and retroviruses (fig. 5).
Discussion
Acquisition of aRNH by Tat LTR Retrotransposons
In this study, we assess the diversity and distribution of aRNH-containing Tat LTR retrotransposons. The reconstruction of the phylogenies based on RT and RNH domain sequences suggests that aRNH originally appeared in plant Tat LTR retrotransposons after the divergence of mosses and more likely was sequestered from the host genome. The first Tat LTR retrotransposon carrying aRNH is found in the spikemoss S. moellendorffii, and the aRNH domain was likely reacquired or reshuffled later during the evolutionary history of Tat LTR retrotransposons. The monophyletic origin of aRNH from Tat LTR retrotransposons is not supported, as revealed by our reconstruction of the phylogenetic relationships among diverse RNH domains and cellular genes (fig. 3). The variable location of aRNH within Pol also indirectly suggests the nonmonophyletic origin of aRNH (fig. 2B). It is, therefore, possible that aRNH was indeed captured multiple times in the evolutionary history of Tat LTR retrotransposons. Alternatively, aRNH could be acquired once and subsequently shuffled within Pol. The patchy distribution and lack of monophyly could be explained by the multiple events of horizontal transfer. Indeed, the close phylogenetic relationships between plant, bacterial, and archaeal aRNHs clearly suggest the implication of multiple events of horizontal transfer (fig. 3). In support of this view, horizontal transfer is now recognized as a widespread and frequent phenomenon in the evolution of plant LTR retrotransposons (Novikova et al. 2008, 2010; El Baidouri et al. 2014), and the evidence of horizontal transfers between bacteria and plants have been recently reported (Richardson and Palmer 2007; Nikolaidis et al. 2014).
We suggest that Tat LTR retrotransposons initially carried aRNH upstream of RT, as observed in the retrotransposons from lineages I and II (fig. 2). Later in evolution, some of the retrotransposons captured the new aRNH or transferred the initial aRNH downstream of gRNH, giving rise to the lineage III Tat LTR retrotransposons with C-terminal aRNH and to the Tat LTR retrotransposons in flowering plants with dual gRNH-aRNH structure (fig. 2). Finally, some of the aRNHs were transferred to archaeal and bacterial genomes, shaping the observed RNH phylogeny (fig. 3).
Additional Structural Perturbations in Tat LTR Retrotransposons
The additional ORFs, eORFs, were found among the structural features of some Tat LTR retrotransposons. However, the origin and function of eORF-encoded proteins remains enigmatic. The eORF-encoded proteins from evolutionarily older lineages of Tat LTR retrotransposons contain a single PMD, and a CC accompanies PMD in recent lineages. The most recently emerged lineages of Tat LTR retrotransposons carry the Tr28 domain in their eORF-encoded proteins. Some hints on the role of PMD might be inferred from its allocation to the epididymal membrane protein superfamily, which includes diverse eukaryotic transmembrane proteins (PTHR16007). In retroviruses, the CC structure is an indispensable part of the env-encoded protein that catalyses membrane fusion (Weng and Weiss 1998). However, approximately 10% of all eukaryotic proteins are predicted to contain CCs and 5–30% to contain transmembrane helices (Liu and Rost 2001), a fact that hinders any speculation on the Env-like nature of the encoded proteins based solely on the presence of these two structures. Indeed, experimental studies are necessary to identify the precise role that eORF-encoded proteins play in the retrotransposon life cycle.
The Gag-PR fusion detected for some Tat LTR retrotransposons is not typical for retrotransposons. The gene encoding fused Gag-PR was previously described only in the Ogre LTR retrotransposon, which our analysis placed among the Tat elements of lineage IV (fig. 2A). The fusion gene was shown to be separated from pol by an intron that can be spliced to produce a Gag-Pol protein (Macas and Neumann 2007; Steinbauerová et al. 2008). In contrast, many class II retroviruses encode PR and Pol (RT, RNH, and INT) in different reading frames, except for lentiviruses, for which PR and Pol are always fused, as in most of LTR retrotransposons (Gifford et al. 2005). Moreover, similar to Tat LTR retrotransposons, some retroviruses, such as Rous sarcoma virus, encode PR in gag (Schwartz et al. 1983).
The differential regulation of gag and pol genes in retroviruses ensures the optimal production of the structural (Gag) and enzymatic (Pol) products. The Gag-PR fusion or at least PR separation from Pol might indicate the necessity of producing more retrovirus-encoded PR protein relative to other enzymatic products. For example, virus-encoded PR contributes not only to the processing of Gag but also that of Env in some retroviruses, such as murine leukemia virus and Mason-Pfizer monkey virus (Schultz and Rein 1985; Brody et al. 1992), but does not appear to participate in lentiviral Env processing, for which the cellular protease furin is sufficient (Murakami 2012; Haim et al. 2013). Because lentiviruses rely exclusively on furin for Env processing, they need less retrovirus-encoded PR, and the fusion of PR to Pol may reflect this requirement. Thus, the presence of the Gag-PR fusion in Tat LTR retrotransposons indirectly suggests that retrotransposon-encoded PR is involved in eORF-encoded protein processing.
aRNH as a Potential Device for the Accelerated Evolution of Tat LTR Retrotransposons
The dual gRNH-aRNH of Tat LTR retrotransposons in flowering plants bears a remarkable resemblance to retroviral tether-RNH (fig. 4). The functional advantages of such a dual RNH in retroviruses and plant Tat LTR retrotransposons in comparison to the single degenerate gRNH from other LTR retrotransposons is yet to be analyzed in depth. A structural study of HIV-1 revealed that the tether modulates the conformational changes that are necessary to orient the RNA strand for RNH cleavage (Lapkouski et al. 2013). This indicates that the tether may regulate RNH function, leading to specialized and more frequent cleavage events during primer generation and removal. In contrast, major conformational changes are required for substrate cleavage by Ty3 gRNH, as was shown previously based on the structure of the recombinant “RT-gRNH” protein from the yeast Ty3 LTR retrotransposon. Thus, RNA hydrolysis by gRNH alone would be infrequent due to the relative structural rigidity (Nowak et al. 2014).
RNA hydrolysis removes segments of the RNA template strand from the growing DNA strand, freeing a single-stranded region to anneal to the second site (strand transfer), which is required for both retroviral and retrotransposon strand transfers (Lener et al. 2002; Basu et al. 2008). The wide genetic diversity, a hallmark of HIV-1 infection, relies heavily on strand transfer-dependent retroviral recombination (Smyth et al. 2012). It is possible that dual RNH provides more effective recombination machinery in retroviruses in comparison to LTR retrotransposon gRNH. Although strand transfer efficiency and its role in generating LTR retrotransposons diversity is poorly understood, indirect evidence, such as extensive recombination between autonomous and nonautonomous Tat LTR retrotransposons detected in soybean (Du, Tian, Bowen, et al. 2010; Du, Tian, Hans, et al. 2010), supports the hypothesis that the dual RNH domain from Tat LTR retrotransposons is a potential device for accelerating evolution through a high rate of recombination. To unambiguously link the presence of a cellular-like RNH to the efficiency of recombination, it is necessary to perform a detailed comparison of the genetic variability resulting from recombination in LTR retrotransposons with and without a dual RNH.
Putative Convergent Evolution of Tat LTR Retrotransposons and Retroviruses
The term “convergent evolution” describes the acquisition of the same biological trait in unrelated lineages. Recent results suggest convergent evolution as being a more widespread phenomenon than was previously thought, implying that genome evolution is not random and might be somewhat predictable (McCutcheon et al. 2009; Parker et al. 2013; Stern 2013). Convergent evolution was also proposed to contribute greatly to the evolution of viruses (Garamszegi et al. 2013). However, although a wide range of sequenced viral genomes is readily available, only a few examples of convergent evolution in viruses have been reported (Heldwein et al. 2006; Yutin and Koonin 2012; Eickbush et al. 2013). Furthermore, the remarkable plasticity of viruses and inability to identify precisely the origin of many viral genes are impediments to identifying the changes associated with convergent evolution (Koonin 2011). For instance, the structural similarity of viral capsids from diverse viruses has been previously noted (Bamford et al. 2005; Krupovic and Bamford 2008). However, the inability to reconstruct a conclusive phylogenetic tree of all viruses has made it impossible to imply convergent evolution as an explanation for this similarity (Holmes 2011).
By tracing structural changes in Tat LTR retrotransposons and retroviruses, we noted striking similarities in some key events of the evolutionary histories of these lineages, suggesting convergent evolution (fig. 5). The development of a dual RNH domain appears to be the first step in the process of the diversification of Tat LTR retrotransposons and retroviruses from their progenitors. Next, the acquisition of eORFs and the Gag-PR fusion occurred in some Tat LTR retrotransposon and retrovirus lineages; it is likely that the acquisition of dual RNHs triggered the subsequent restructuring. For example, dual RNHs might facilitate the capturing of additional genes through recombination during reverse transcription. As a consequence, eORFs in Tat LTR retrotransposons and env gene in retroviruses might have been easily obtained and fixed in populations. The necessity of effectively processing such newly acquired protein-coding sequences resulted in higher requirements for PR function and its subsequent fusion to Gag.
Thus, the persistence of dual RNH indicates the importance of this structure for both Tat LTR retrotransposons and retroviruses and showcases convergent evolution for virus-like systems through the independent acquisition of distantly related modules with similar function. However, it remains to be determined whether such convergence in structure correlates with convergence in function in this particular case of putative convergent evolution.
Materials and Methods
Computational Mining for aRNH-Containing LTR Retrotransposons
The plant genomic sequences used in this study were retrieved from databases, as listed in supplementary table S1, Supplementary Material online. To identify all LTR retrotransposons harboring aRNH, the following algorithm implemented in a single Python script was applied (the script is available from the authors by request). First, de novo identification of LTR retrotransposon-like sequences was performed using the LTRharvest software with the set of given constraints and parameters: -minlenltr 200, -maxlenltr 2000, -mindistltr 3000, -maxdistltr 22000, -similar 85.0, -overlaps no, -mintsd 3, -maxtsd 20 (Ellinghaus et al. 2008). Second, annotation of LTR retrotransposon-specific features was performed using the LTRdigest tool and a set of hidden Markov model (HMM) profiles available in Gypsy Database (Steinbiss et al. 2009; Llorens et al. 2011). In addition, an aRNH-specific HMM profile was generated based on the multiple alignment of the aRNH sequences identified previously (Smyshlyaev et al. 2013). Only sequences carrying both RT and aRNH domains were further analyzed. Finally, selected sequences were grouped into clusters based on the similarity of their RT domain using the Vmatch tool (http://www.vmatch.de/, last accessed May 31, 2014). The copy showing the highest score to an RT HMM profile from GyDB was selected as being representative of the cluster; if two or more copies in the cluster had the same score, the representative was selected based on the maximum similarity of the LTRs. To reduce the computational burden of the subsequent steps in the analysis, the LTR retrotransposons grouping out of clusters were not analyzed.
Characterization of the Structural Composition of aRNH-Containing LTR Retrotransposons
In the course of our initial analysis, the Gag, PR, RT, RNH and INT domains were predicted using the set of HMM profiles. However, information on domains encoded by eORFs was not found. Therefore, we used an additional round of structural domain prediction, and the representative LTR retrotransposons were scanned using HHpred, a server that performs homology detection and structure prediction (http://toolkit.tuebingen.mpg.de/hhpred/, last accessed August 10, 2014; Söding et al. 2005) and using COILS, which predicts CCs (http://toolkit.tuebingen.mpg.de/pcoils, last accessed August 10, 2014; Lupas et al. 1991). Secondary structure predictions were also performed using Ali2D (http://toolkit.tuebingen.mpg.de/ali2d, last accessed August 10, 2014).
Comparative and Phylogenetic Analysis
The RT amino acid sequences were aligned using MUSCLE software (Edgar 2004). The amino acid sequences of RNH and PMD are less conservative than RT, and a profile multiple alignment with the predicted local structures and 3D constraints (PROMALS3D) server was used to produce the alignment (Pei et al. 2008). The alignments were manually curated, and the phylogenetic tree was reconstructed using the maximum-likelihood algorithm implemented in the PhyML tool with the default parameters (Guindon et al. 2010). The approximate likelihood-ratio test of the branches was used for statistical support (Anisimova and Gascuel 2006).
Supplementary Material
Supplementary table S1 and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge Dr Bernard Goffinet and Dr Yang Liu for providing genomic sequences for moss species. This work was supported by Russian Foundation for Basic Research (grant number 14-04-01498) and by the State scientific project number VI.61.1.2.
References
- Abrusán G, Szilágyi A, Zhang Y, Papp B. Turning gold into “junk”: transposable elements utilize central proteins of cellular networks. Nucleic Acids Res. 2013;41:3190–3200. doi: 10.1093/nar/gkt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Brody BA, Rhee SS, Sommerfelt MA, Hunter E. A viral protease-mediated cleavage of the transmembrane glycoprotein of Mason-Pfizer monkey virus can be suppressed by mutations within the matrix protein. Proc Natl Acad Sci U S A. 1992;89:3443–3447. doi: 10.1073/pnas.89.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, Hughes SH, Coffin JM, Jere A, Pathak VK. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci U S A. 2008;105:10943–10948. doi: 10.1073/pnas.0804660105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Bowen NJ, Schmutz J, Shoemaker RC, Ma J. Bifurcation and enhancement of autonomous-nonautonomous retrotransposon partnership through LTR Swapping in soybean. Plant Cell. 2010;22:48–61. doi: 10.1105/tpc.109.068775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J. Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J. 2010;63:584–598. doi: 10.1111/j.1365-313X.2010.04263.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush DG, Burke WD, Eickbush TH. Evolution of the R2 retrotransposon ribozyme and its self-cleavage site. PLoS One. 2013;8:e66441. doi: 10.1371/journal.pone.0066441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M, Carpentier M-C, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014;24:831–838. doi: 10.1101/gr.164400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Garamszegi S, Franzosa EA, Xia Y. Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human-virus protein-protein interaction networks. PLoS Pathog. 2013;9:e1003778. doi: 10.1371/journal.ppat.1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R, Kabat P, Martin J, Lynch C, Tristem M. Evolution and distribution of class II-related endogenous retroviruses. J Virol. 2005;79:6478–6486. doi: 10.1128/JVI.79.10.6478-6486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haim H, Salas I, Sodroski J. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J Virol. 2013;87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Holmes EC. What does virus evolution tell us about virus origins? J Virol. 2011;85:5247–5251. doi: 10.1128/JVI.02203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovsky E, Kubat Z, Macas J, Hobza R, Mracek J, Vyskot B. Retand: a novel family of gypsy-like retrotransposons harboring an amplified tandem repeat. Mol Genet Genomics. 2006;276:254–263. doi: 10.1007/s00438-006-0140-x. [DOI] [PubMed] [Google Scholar]
- Kim FJ, Battini J-L, Manel N, Sitbon M. Emergence of vertebrate retroviruses and envelope capture. Virology. 2004;318:183–191. doi: 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E V. 2011 The logic of chance: The nature and origin of biological evolution. Upper Saddle River: FT Press. [Google Scholar]
- Krupovic M, Bamford DH. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Lapkouski M, Tian L, Miller JT, Le Grice SFJ, Yang W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat Struct Mol Biol. 2013;20:230–236. doi: 10.1038/nsmb.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lener D, Budihas SR, Le Grice SFJ. Mutating conserved residues in the ribonuclease H domain of Ty3 reverse transcriptase affects specialized cleavage events. J Biol Chem. 2002;277:26486–26495. doi: 10.1074/jbc.M200496200. [DOI] [PubMed] [Google Scholar]
- Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Fares MA, Moya A. Relationships of gag-pol diversity between Ty3/Gypsy and Retroviridae LTR retroelements and the three kings hypothesis. BMC Evol Biol. 2008;8:276. doi: 10.1186/1471-2148-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Futami R, Covelli L, Domínguez-Escribá L, Viu JM, Tamarit D, Aguilar-Rodríguez J, Vicente-Ripolles M, Fuster G, Bernet GP, et al. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 2011;39:D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P. Ogre elements—a distinct group of plant Ty3/gypsy-like retrotransposons. Gene. 2007;390:108–116. doi: 10.1016/j.gene.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Malik HS. Ribonuclease H evolution in retrotransposable elements. Cytogenet Genome Res. 2005;110:392–401. doi: 10.1159/000084971. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 2001;11:1187–1197. doi: 10.1101/gr.185101. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S, Eickbush TH. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T. Retroviral env glycoprotein trafficking and incorporation into virions. Mol Biol Int. 2012;2012:682850. doi: 10.1155/2012/682850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Doran N, Cosgrove DJ. Plant expansins in bacteria and fungi: evolution by horizontal gene transfer and independent domain fusion. Mol Biol Evol. 2014;31:376–386. doi: 10.1093/molbev/mst206. [DOI] [PubMed] [Google Scholar]
- Novikova O, Mayorov V, Smyshlyaev G, Fursov M, Adkison L, Pisarenko O, Blinov A. Novel clades of chromodomain-containing Gypsy LTR retrotransposons from mosses (Bryophyta) Plant J. 2008;56:562–574. doi: 10.1111/j.1365-313X.2008.03621.x. [DOI] [PubMed] [Google Scholar]
- Novikova O, Smyshlyaev G, Blinov A. Evolutionary genomics revealed interkingdom distribution of Tcn1-like chromodomain-containing Gypsy LTR retrotransposons among fungi and plants. BMC Genomics. 2010;11:231. doi: 10.1186/1471-2164-11-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak E, Miller JT, Bona MK, Studnicka J, Szczepanowski RH, Jurkowski J, Le Grice SFJ, Nowotny M. Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry. Nat Struct Mol Biol. 2014;21:389–396. doi: 10.1038/nsmb.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Yanagawa H, Tomita M, Itaya M. Identification of the first archaeal Type 1 RNase H gene from Halobacterium sp. NRC-1: archaeal RNase HI can cleave an RNA-DNA junction. Biochem J. 2004;381:795–802. doi: 10.1042/BJ20040153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin N V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter RTM, Goodwin TJD. DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet Genome Res. 2005;110:575–588. doi: 10.1159/000084991. [DOI] [PubMed] [Google Scholar]
- Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Schulman AH. Retrotransposon replication in plants. Curr Opin Virol. 2013;3:604–614. doi: 10.1016/j.coviro.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Schultz A, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Smyshlyaev G, Voigt F, Blinov A, Barabas O, Novikova O. Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proc Natl Acad Sci U S A. 2013;110:20140–20145. doi: 10.1073/pnas.1310958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth RP, Davenport MP, Mak J. The origin of genetic diversity in HIV-1. Virus Res. 2012;169:415–429. doi: 10.1016/j.virusres.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauerová V, Neumann P, Macas J. Experimental evidence for splicing of intron-containing transcripts of plant LTR retrotransposon Ogre. Mol Genet Genomics. 2008;280:427–436. doi: 10.1007/s00438-008-0376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbauerová V, Neumann P, Novák P, Macas J. A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica. 2011;139:1543–1555. doi: 10.1007/s10709-012-9654-9. [DOI] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37:7002–7013. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–1493. doi: 10.1111/j.1742-4658.2009.06907.x. [DOI] [PubMed] [Google Scholar]
- Weng Y, Weiss CD. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J Virol. 1998;72:9676–9682. doi: 10.1128/jvi.72.12.9676-9682.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Uzun O, Mules EH, Gabriel A, Wilhelm FX. Polypurine tract formation by Ty1 RNase H. J Biol Chem. 2001;276:47695–47701. doi: 10.1074/jbc.M106067200. [DOI] [PubMed] [Google Scholar]
- Wright DA, Voytas DF. Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics. 1998;149:703–715. doi: 10.1093/genetics/149.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D-J, Chon H, Koga Y, Takano K, Kanaya S. Crystal structure of type 1 ribonuclease H from hyperthermophilic archaeon Sulfolobus tokodaii: role of arginine 118 and C-terminal anchoring. Biochemistry. 2007;46:11494–11503. doi: 10.1021/bi700830f. [DOI] [PubMed] [Google Scholar]
- Yutin N, Koonin E V. Hidden evolutionary complexity of nucleo-cytoplasmic large DNA viruses of eukaryotes. Virol J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.