Abstract
Toothed whales and two groups of bats independently acquired echolocation, the ability to locate and identify objects by reflected sound. Echolocation requires physiologically complex and coordinated vocal, auditory, and neural functions, but the molecular basis of the capacity for echolocation is not well understood. A recent study suggested that convergent amino acid substitutions widespread in the proteins of echolocators underlay the convergent origins of mammalian echolocation. Here, we show that genomic signatures of molecular convergence between echolocating lineages are generally no stronger than those between echolocating and comparable nonecholocating lineages. The same is true for the group of 29 hearing-related proteins claimed to be enriched with molecular convergence. Reexamining the previous selection test reveals several flaws and invalidates the asserted evidence for adaptive convergence. Together, these findings indicate that the reported genomic signatures of convergence largely reflect the background level of sequence convergence unrelated to the origins of echolocation.
Keywords: bat, dolphin, convergent evolution, neutral evolution
Echolocation originated independently in toothed whales and two groups of bats (fig. 1A). Understanding the molecular basis of this complex evolutionary innovation is of substantial interest. Comparing 22 mammalian genome sequences, Parker et al. (2013) reported hundreds of convergently evolving proteins among echolocators and suggested that genome-wide molecular convergence underlay the origins of echolocation and associated phenotypes. However, protein convergence could also occur by chance (Zhang and Kumar 1997; Castoe et al. 2009). Here, we show that the reported genomic signatures of convergence largely reflect such chance events that are unrelated to the origins of echolocation.
Fig. 1.
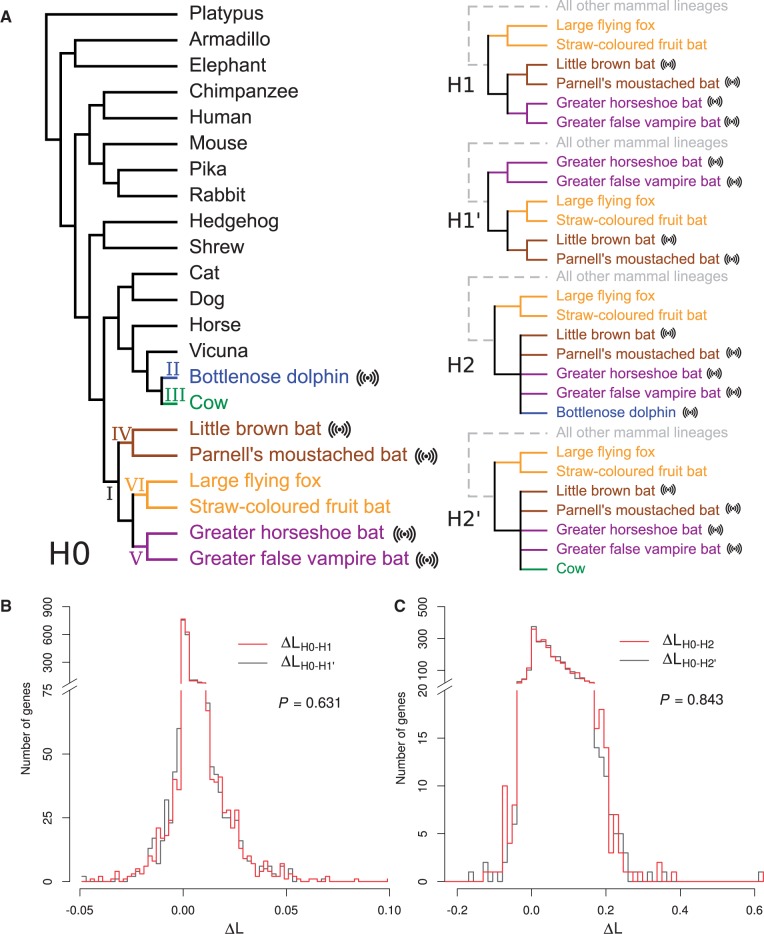
No genome-wide signatures of protein sequence convergence associated with echolocation. (A) Hypotheses and corresponding tree topologies. H0, species tree; H1, clustering of the two groups of echolocating bats; H1′, clustering of echolocating Yangochiroptera bats and nonecholocating Yinpterochiroptera bats; H2, clustering of echolocating bats and dolphin; H2′, clustering of echolocating bats and cow. In H1, H1′, H2, and H2′, the tree topology for “all other mammalian lineages” is the same as in the species tree. Echolocating species are indicated with an echo symbol. The six branches where convergent sites are counted in table 1 are marked by I–VI. (B) Frequency distributions of ΔLH0-H1 and ΔLH0-H1' among the 2,326 proteins are not significantly different. ΔL refers to the per site logarithm of the likelihood ratio between two hypotheses for a protein. (C) Frequency distributions of ΔLH0-H2 and ΔLH0-H2' among the 2,326 proteins are not significantly different. In (B) and (C), the P-values are from Kolmogorov–Smirnov tests.
Parker et al. (2013) assembled and aligned the orthologous sequences of 2,326 proteins. For each protein, they estimated the log-likelihood differences per site (ΔL) between the known mammalian species tree (H0) and each of two alternative trees (H1 and H2). In H1, the two groups of echolocating bats are clustered, whereas in H2, the echolocating bats and the bottlenose dolphin, an echolocating whale, are grouped (fig. 1A). A negative ΔLH0-H1 (or ΔLH0-H2) indicates that the evolution of the protein favors H1 (or H2) over H0, which Parker et al. (2013) regarded as a signature of molecular convergence of echolocators.
However, because the null distribution of ΔLH0-H1 is unknown, it is necessary to set a negative control against which ΔLH0-H1 is compared. By exchanging in H1 the phylogenetic positions of nonecholocating (orange) and echolocating (purple) bats belonging to Yinpterochiroptera, we created H1′ (fig. 1A), which does not cluster the two groups of echolocating bats but otherwise exhibits the same amount of phylogenetic distortion from H0 as does H1. Significantly more negative ΔLH0-H1 values compared with ΔLH0-H1′ values across the 2,326 proteins would be consistent with Parker et al.’s (2013) claim of a genome-wide signature of protein convergence associated with bat echolocation. However, we found that the frequency distribution of ΔLH0-H1 is superimposed on that of ΔLH0-H1′ (P = 0.63, Kolmogorov–Smirnov test; fig. 1B). Similarly, we created H2′ by exchanging the phylogenetic positions of cow and dolphin in H2 (fig. 1A). Again, the frequency distribution of ΔLH0-H2 is superimposed on that of ΔLH0-H2' (P = 0.84; fig. 1C), suggesting just as much convergence between bats and cow as was observed between bats and dolphin. Note that, in the species tree, the branch length measured by the number of amino acid substitutions per site across all proteins analyzed is greater for the exterior branch leading to cow than that leading to dolphin (see figure 1A of Parker et al. 2013). We predicted and verified by computer simulation that this branch length difference results in a slightly more positive ΔLH0-H2' than ΔLH0-H2 on average, rendering our conclusion conservative. The branches leading to echolocating (purple) and nonecholocating (orange) bats of Yinpterochiroptera have similar lengths in the species tree (see figure 1A of Parker et al. 2013) and therefore the comparison between ΔLH0-H1' and ΔLH0-H1 is fair.
Earlier work identified seven hearing-related proteins that underwent convergent evolution in echolocators (Li et al. 2008, 2010; Liu et al. 2010, 2011, 2012; Davies et al. 2012; Shen et al. 2012). Parker et al. (2013) mentioned 22 additional proteins annotated as “hearing” or “deafness” in the data analyzed. We found that ΔLH0-H1 is smaller than ΔLH0-H1' for 59% of the 29 hearing proteins, not significantly more than the random expectation of 50% (P > 0.2, one-tail binomial test). When the seven known convergent proteins are excluded, ΔLH0-H1 is smaller than ΔLH0-H1' for only 45% of the 22 proteins (P > 0.7). Similarly, ΔLH0-H2 is smaller than ΔLH0-H2' for 59% of the 29 proteins (P > 0.2) and 50% of the 22 proteins (P > 0.5). Qualitatively identical results were obtained by paired t-tests. Hence, as a group, hearing proteins show no significant enrichment of phylogenetic signals for echolocator-specific convergence.
Next, we inferred ancestral protein sequences for interior nodes in H0 and counted the number of sites with convergent amino acid substitutions along relevant sets of branches as a direct measure of protein convergence. In total, 2,270 proteins each with available sequences from at least dolphin, cow, and the six bat species in figure 1A were analyzed. For example, under the hypothesis of a single origin of bat echolocation, protein convergence associated with echolocation should occur between branches I and II but not between I and III (fig. 1A). However, we observed no more convergent sites in the former than the latter (table 1). It was previously shown that, under no adaptive convergence, the number of convergent sites in a branch set is expected to be proportional to the number of divergent sites in the same set (Castoe et al. 2009), where divergent sites are those at which divergent amino acid substitutions have occurred along the branches of interest. We thus asked if the number of convergent sites in branches I and II significantly exceeds that in branches I and III, given their numbers of divergent sites. The answer is clearly no, and in fact the opposite is true (table 1). Consistent observations were made in comparisons between branch sets (IV and II) and (IV and III) and between branch sets (V and II) and (V and III) (table 1). A similar result was found in an independent analysis of 6,400 genes from ten mammals (Thomas and Hahn 2015), although it is unknown why there are more convergent sites between bats and cow than between bats and dolphin, given their respective numbers of divergent sites. In two comparisons, however, we observed more convergent sites in echolocators than the controls. First, in the comparison between the three-branch sets (II, IV, and V) and (III, IV, and V) under the hypothesis of dual origins of bat echolocation, the branch set representing echolocators has more convergent sites than the control set, given their numbers of divergent sites, although the difference is not significant (table 1). Notably, of the 14 convergent sites among branches II, IV, and V, 12 fall in six of the seven known convergently evolving hearing proteins (prestin is not included in this analysis due to missing data), indicating that at most a few proteins were subject to convergent evolution in all three echolocating lineages. Second, the comparison between branch sets (IV and V) and (IV and VI) shows that the former set, representing echolocating bats, has significantly more convergent sites than the control set, given their numbers of divergent sites (table 1). But after the removal of the six known convergently evolving hearing proteins, the two branch sets are no longer significantly different (table 1). Together, these direct comparisons of the number of convergent sites between echolocating lineages and control lineages offer no evidence for genome-wide convergence in echolocating lineages beyond the background level.
Table 1.
Comparison in the Total Number of Sites that Have Experienced Convergent Substitutions from 2,270 Proteins.
| Comparison | Observed Number of Convergent Sites | Observed Number of Divergent Sites | P-Valuea |
|---|---|---|---|
| (I and II) vs. (I and III)b | 176 vs. 223 | 380 vs. 352 | 0.012 |
| (IV and II) vs. (IV and III) | 204 vs. 287 | 479 vs. 445 | 0.00022 |
| (V and II) vs. (V and III) | 152 vs. 183 | 325 vs. 270 | 0.0067 |
| (IV and V and II) vs. (IV and V and III) | 14 vs. 4 | 27 vs. 22 | 0.083 |
| (IV and V) vs. (IV and VI) | 93 vs. 207 | 75 vs. 273 | 0.0062* |
| (IV and V) vs. (IV and VI)c | 66 vs. 204 | 67 vs. 269 | 0.18 |
aG-test of the hypothesis that the number of convergent sites in a branch set is proportional to the number of divergent sites in the same branch set. An asterisk is given if the case branch set has significantly more convergent sites than expected.
bTwo sets of branches for which the numbers of amino acid sites that have experienced convergent substitutions are compared. Roman numbers refer to the branch labels in figure 1A.
cAfter the removal of six hearing genes previously reported to be subject to convergent evolution in echolocators.
Parker et al. (2013) assumed that a significant negative correlation between site-wise ΔL and ω (nonsynonymous/synonymous rate ratio) within a gene indicates adaptive convergence. Although adaptive convergence may lead to a negative correlation between ΔL and ω, there is no proof that neutral evolution cannot. Compared with low-ω sites, high-ω sites are more likely to experience convergence by chance (as well as divergence). Thus, one cannot exclude the possibility that a negative correlation results from neutral evolution, especially when ω is not significantly greater than 1. Furthermore, to estimate ω, Parker et al. (2013) used H1 or H2 instead of the species tree. Because the true evolutionary histories of all genes considered here are described by the species tree, the ω estimates based on the wrong trees are biologically meaningless. We thus reanalyzed the two genes (Rapgef1 and Cdkl5) presented in the insets of figure 2A of Parker et al. (2013) that were reported to show adaptive convergence for H1. Under the species tree and using the branch-site likelihood method (Zhang et al. 2005), we tested the action of positive selection in the foreground branches of IV and V (fig. 1A) whereas all other branches in the tree were treated as background branches (see Materials and Methods). But, in neither gene did we find signal for positive selection (table 2). We similarly analyzed the two genes (Bnipl and Nubp2) in the insets of figure 2B of Parker et al. (2013) that were claimed to show adaptive convergence for H2. We tested positive selection in branches I and II (under the model of a single origin of bat echolocation) or branches II, IV, and V (under the model of dual origins of bat echolocation). Again, there was no significant signal of positive selection (table 2). Regardless, for each of the four genes, we identified those sites that have a Bayes Empirical Bayes (BEB) probability of greater than 0.5 to be in class 2a or 2b, meaning that these sites likely have higher ω in the foreground branches than in the background branches. Only in two genes did we find more than one such site, but in neither gene was the correlation negative between ΔL and foreground ω for these sites (r = 0.30, P = 0.38 for Cdkl5 and r = 0.44, P = 0.33 for Bnipl with I and II being the foreground branches). Thus, even on the basis of Parker et al.’s (2013) own criterion, proper analysis reveals no adaptive convergence in these genes.
Table 2.
Branch-Site Likelihood Ratio Test of Positive Selection in Genes Claimed by Parker et al. (2013) to Have Undergone Adaptive Convergence.
| Gene | Foreground Branchesa | Log-Likelihood under Branch-Site Model A | Log-Likelihood under Null Model (ω2 = 1) | χ2 (df = 1) | P-Value |
|---|---|---|---|---|---|
| Rapgef1 | IV and V | −3462.452928 | −3462.452928 | 0.000 | 1.00 |
| Cdkl5 | IV and V | −6161.704025 | −6161.704025 | 0.000 | 1.00 |
| Bnipl | I and II | −3498.585575 | −3498.600281 | 0.029 | 0.59 |
| Bnipl | II, IV, and V | −3499.990739 | −3499.990739 | 0.000 | 1.00 |
| Nubp2 | I and II | −4527.964898 | −4527.964898 | 0.000 | 1.00 |
| Nubp2 | II, IV, and V | −4527.964898 | −4527.964898 | 0.000 | 1.00 |
aRoman numbers refer to the branch labels in figure 1A. All other branches are background branches.
In summary, our reanalyses of Parker et al.’s (2013) data showed that the genome-wide phylogenetic signal of molecular convergence is no stronger for echolocators than for comparable nonecholocators. Note that the phylogenetic test employed by Parker et al. (2013) is not a formal test of molecular convergence, because convergence does not necessarily result in a wrong phylogeny and a wrong phylogeny is not necessarily caused by convergence (Zhang and Kumar 1997; Castoe et al. 2009). Nonetheless, it is clear that Parker et al.’s (2013) conclusion is not supported even on the basis of this phylogenetic test. Furthermore, we found that echolocators experienced no more molecular convergence than nonecholocators in the absence of the seven hearing genes known to be subject to convergent evolution. Thus, the reported genomic signatures of protein convergence must largely reflect the background chance convergences that are unrelated to the independent origins of echolocation in mammals. This conclusion, however, does not preclude the possibility that the convergent substitutions previously identified from the case studies of a few hearing proteins are important for echolocation. But given the nonnegligible chance occurrence of molecular convergence, proof of adaptive convergence of a protein should include proper statistical tests and functional assays (Zhang 2006). In this regard, it is worth mentioning that both these requirements have been fulfilled for prestin, the motor protein of the outer hair cells of the inner ear of the mammalian cochlea (Li et al. 2010; Liu et al. 2014). Specifically, the Asn to Thr change at position 7 of prestin occurred three times, in branches II, IV, and V, respectively, and increased a key parameter of prestin function that is associated with high-frequency hearing in echolocating mammals. At the genomic scale, the rapid accrual of gene sequences has stimulated genome-wide detections of molecular convergence (Bazykin et al. 2007; Rokas and Carroll 2008; Parker et al. 2013), but efforts are also needed to establish the expected level of neutral molecular convergence, against which the observed levels can be compared such that adaptive molecular convergence may be inferred. In the absence of such neutral expectations, it is imperative to use appropriate negative controls in the study of potential adaptive molecular convergence.
Materials and Methods
To calculate the mean site-wise ΔL for each protein, we followed the procedure described in Parker et al. (2013), using the 2,326 alignments provided by the authors. Soft polytomies in H2 and H2′ were resolved by RAxML (version 8.0.22; Stamatakis 2014) as previously described (Parker et al. 2013). Marginal ancestral sequences in H0 were inferred by the Bayesian method (Yang et al. 1995) implemented in PAML (version 4.7; Yang 2007), using the parameters that yielded the maximum likelihood of H0. Convergent substitutions at a site are those inferred substitutions that resulted in the same amino acid and occurred in all branches examined for convergence; they thus include both convergent and parallel substitutions defined in Zhang and Kumar (1997). Coding sequences were fitted to branch-site model A in PAML with site classes 2a and 2b having ω2 ≥ 1 in foreground branches and 0 < ω0 < 1 (class 2a) and ω1 = 1 (class 2b) in background branches. Model A was compared by a likelihood ratio test with the null model in which ω2 was fixed at 1. We estimated the ω value of a site by the mean of ω values of all site classes weighted by the BEB posterior probabilities with which the site belongs to these classes.
Acknowledgments
The authors thank the authors of Parker et al. (2013) for sharing the sequence alignments and Xiaoshu Chen, Matthew Hahn, Wei-Chin Ho, Stephen Rossiter, Jian-Rong Yang, and three anonymous reviewers for valuable comments on early drafts of this article. This work was supported in part by research grant R01GM103232 from the U.S. National Institutes of Health to J.Z.
References
- Bazykin GA, Kondrashov FA, Brudno M, Poliakov A, Dubchak I, Kondrashov AS. Extensive parallelism in protein evolution. Biol Direct. 2007;2:20. doi: 10.1186/1745-6150-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, Naylor G, Jiang ZJ, Parkinson CL, Pollock DD. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KT, Cotton JA, Kirwan JD, Teeling EC, Rossiter SJ. Parallel signatures of sequence evolution among hearing genes in echolocating mammals: an emerging model of genetic convergence. Heredity. 2012;108:480–489. doi: 10.1038/hdy.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang J, Rossiter SJ, Jones G, Cotton JA, Zhang S. The hearing gene Prestin reunites echolocating bats. Proc Natl Acad Sci U S A. 2008;105:13959–13964. doi: 10.1073/pnas.0802097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Z, Shi P, Zhang J. The hearing gene Prestin unites echolocating bats and whales. Curr Biol. 2010;20:R55–R56. doi: 10.1016/j.cub.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cotton JA, Shen B, Han X, Rossiter SJ, Zhang S. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 2010;20:R53–R54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- Liu Y, Han N, Franchini LF, Xu H, Pisciottano F, Elgoyhen AB, Rajan KE, Zhang S. The voltage-gated potassium channel subfamily KQT member 4 (KCNQ4) displays parallel evolution in echolocating bats. Mol Biol Evol. 2012;29:1441–1450. doi: 10.1093/molbev/msr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li S, Wang W, Xu D, Murphy RW, Shi P. Parallel evolution of KCNQ4 in echolocating bats. PLoS One. 2011;6:e26618. doi: 10.1371/journal.pone.0026618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Qi FY, Zhou X, Ren HQ, Shi P. Parallel sites implicate functional convergence of the hearing gene prestin among echolocating mammals. Mol Biol Evol. 2014;31:2415–2424. doi: 10.1093/molbev/msu194. [DOI] [PubMed] [Google Scholar]
- Parker J, Tsagkogeorga G, Cotton JA, Liu Y, Provero P, Stupka E, Rossiter SJ. Genome-wide signatures of convergent evolution in echolocating mammals. Nature. 2013;502:228–231. doi: 10.1038/nature12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Carroll SB. Frequent and widespread parallel evolution of protein sequences. Mol Biol Evol. 2008;25:1943–1953. doi: 10.1093/molbev/msn143. [DOI] [PubMed] [Google Scholar]
- Shen YY, Liang L, Li GS, Murphy RW, Zhang YP. Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 2012;8:e1002788. doi: 10.1371/journal.pgen.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GWC, Hahn MW. Determining the null model for adaptive convergence from genomic data: a case study using echolocating mammals. Mol Biol Evol. 32(5):1232–1236. doi: 10.1093/molbev/msv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet. 2006;38:819–823. doi: 10.1038/ng1812. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]