Abstract
The largest living carnivorous marsupial, the Tasmanian devil (Sarcophilus harrisii), is the sole survivor of a lineage originating about 12 Ma. We set out to investigate the spectrum of transposable elements found in the Tasmanian devil genome, the first high-coverage genome of an Australian marsupial. Marsupial genomes have been shown to have the highest amount of transposable elements among vertebrates. We analyzed the horizontally transmitted DNA transposons OC1 and hAT-1_MEu in the Tasmanian devil genome. OC1 is present in all carnivorous marsupials, while having a very limited distribution among the remaining Australian marsupial orders. In contrast, hAT-1_MEu is present in all Australian marsupial orders, and has so far only been identified in a few placental mammals. We screened 158 introns for phylogenetically informative retrotransposons in the order Dasyuromorphia, and found that the youngest SINE (Short INterspersed Element), WSINE1, is no longer active in the subfamily Dasyuridae. The lack of detectable WSINE1 activity in this group may be due to a retrotransposon inactivation event approximately 30 Ma. We found that the Tasmanian devil genome contains a relatively low number of continuous full-length LINE-1 (Long INterspersed Element 1, L1) retrotransposons compared with the opossum genome. Furthermore, all L1 elements in the Tasmanian devil appeared to be nonfunctional. Hidden Markov Model approaches suggested that other potential sources of functional reverse transcriptase are absent from the genome. We discuss the issues associated with assembling long, highly similar L1 copies from short read Illumina data and describe how assembly artifacts can potentially lead to erroneous conclusions.
Keywords: Sarcophilus, retrotransposon, DNA transposon
Introduction
The Tasmanian devil (Sarcophilus harrisii) became isolated on the Australian island of Tasmania approximately 400 years ago (Brown 2006). For the past 20 years, the Tasmanian devil population has been plagued by a contagious cancer—the devil facial tumor disease (DFTD) (Hawkins et al. 2006; Murchison 2008; Deakin and Belov 2012). DFTD spreads among Tasmanian devils through biting wounds and more than half of the population has disappeared since the disease was first discovered in 1996 (Deakin and Belov 2012; Murchison et al. 2012). To date, genomes from both healthy and DFTD-tissue have been sequenced to study the molecular basis of DFTD in an attempt to develop a cure and save the species from extinction (Miller et al. 2011; Murchison et al. 2012).
Transposable elements make up a significant fraction (up to 52%) of marsupial genomes (Mikkelsen et al. 2007; Renfree et al. 2011; Nilsson et al. 2012) and can be divided into class I and class II elements. Class II elements, or DNA transposons, are a type of transposable element that can relocate within the genome using a cut-and-paste mechanism. Class I elements, or retrotransposons, propagate using a copy-and-paste mechanism (Goodier and Kazazian 2008) through an RNA intermediate and have accumulated in significantly greater numbers in the genome than DNA transposons (Lander et al. 2001; Feschotte and Pritham 2007).
The essential autonomous driver of retrotransposition in marsupial and placental mammalian genomes is the Long INterspersed Element-1 (LINE-1, L1) (fig. 1), which encodes the protein machinery mediating retrotransposition of both L1 elements and Short INterspersed Elements (SINEs). LINE-2 (L2) and LINE-3 (L3) were active drivers of retrotransposition in the ancestor of placental and marsupial mammals, but appear to have stopped mobilizing more than 100 Ma in the therian mammmal lineage (Smit et al. 1995). In a few mammalian groups, a LINE-like element, the RetroTransposon-like Element (RTE), propagated to high genomic copy numbers (fig. 1) (Gogolevsky et al. 2008; Walsh et al. 2013). RTE elements are found in only a few placental mammalian groups (e.g., Ruminantia, Afrotheria), but are present in all marsupial orders (Gentles et al. 2007).
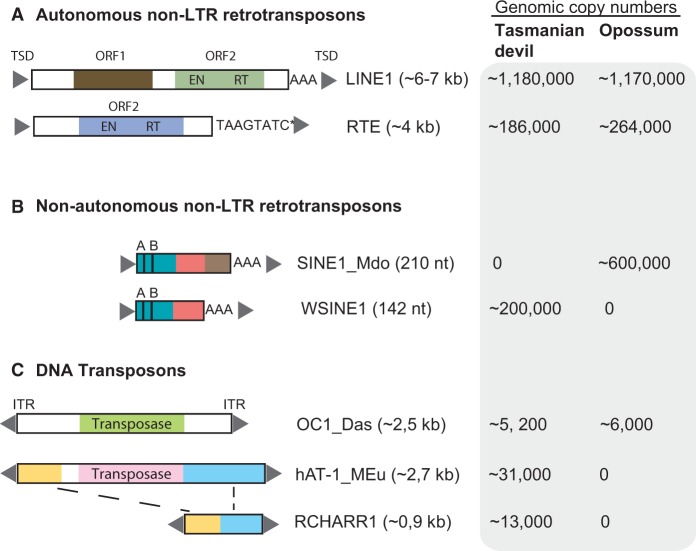
Fig. 1.
Structural organization of non-LTR retrotransposons (A, B) and DNA transposons (C) identified in the Tasmanian devil genome. ORFs are disrupted by frameshifts, nonsense mutations, and indels. We investigated L1-1_MD from opossum and L1-1_SH from Tasmanian devil. WSINE1 from Tasmanian devil and SINE1_Mdo from opossum are both CORE-SINEs and share a homologous head and body-region. Presented copy numbers of the different elements were published in Nilsson et al. (2012), Gentles et al. (2007), and Gilbert et al. (2013). For the nonautonomous non-LTR retrotransposons, the green rectangle indicates the tRNA-region, the pink rectangle is the CORE sequence, whereas the brown rectangle shows the opossum specific sequence. The 5′- and 3′-ends of RCHARR1 are 99% identical to the 5′ and 3′ of the hAT-1_MEu sequence (blue and yellow boxes). ORF1/2, open reading frames 1 and 2; EN, endonuclease; ITR, inverted terminal repeats; AB, A and B box of the RNA polymerase III promoter; AAA, poly(A)tail. *RTE elements do not include a poly(A)tail at their 3′-ends, but TAAGTATC tandem repeats.
Transposable elements are known to destabilize the human genome, cause mutations, affect gene expression, have the potential to cause disease in humans and can be transcriptionally activated in human tumors (Goodier and Kazazian 2008; Beck et al. 2011; Hancks and Kazazian 2012; Casacuberta and González 2013).
We aimed to study the activity of endogenous transposable elements in the Tasmanian devil genome. We used comparative genomics to understand the evolutionary activity of transposable elements in Tasmanian devil genomes using the South American opossum (Monodelphis domestica) genome as an outgroup. We applied comparative genomics within the order Dasyuromorphia to pinpoint retrotransposon inactivation events and the horizontal transmission of DNA transposons into the Tasmanian devil genome.
Results
Genomic Distribution and Properties of Autonomous Non-Long Terminal Repeat Retrotransposons in the Tasmanian Devil Genome
Our screening revealed that L1 and the LINE-like RTE elements comprise 16.9% (1,179,690 copies) and 1.82% (186,289 copies) of the Tasmanian devil genome (WTSI_Devil_ref v7.0), respectively. Of the 1,179,690 L1 copies in the Tasmanian devil genome, the most recently active element is L1-1_SH, with a consensus sequence length of 6,676 nucleotides (nt) (Jurka 2011a). The open reading frame 1 (ORF1) and ORF2 were identified in the L1-1_SH consensus sequence using orthologous ORFs from opossum and wallaby L1. The 5′-untranslated region (UTR) covers bases from positions 1–1158, ORF1 positions 1159–1914, and ORF2 positions 2425–6261. Thus, the 5′-UTR is 1,158 nt long, the 3′-UTR covers 415 nt including the poly(A)tail, and the spacer between ORF1 and ORF2 is 509 nt in length. We retrieved 384 L1 copies (>6,000 nt) from the Tasmanian devil genome (WTSI_Devil_ref v7.0), which were either full-length or 5′-truncated. The ORFs of all L1 copies were inactivated by frameshifts, nonsense mutations, and/or indels. Of the 384 L1 copies exceeding 6,000 nt, 303 copies contained nested integrations, consisting primarily of other L1 sequences. The 64 copies without additional transposable element insertions had long indels (>100 nt) and/or incomplete 5′- and 3′-UTRs. For our analysis, we chose to investigate the 17 L1-1_SH copies with intact 5′- and 3′-UTRs and no nested integrations of transposable element sequences. The 17 L1-1_SH copies range in size from 6,432 to 6,762 nt, and are flanked by 5–15 nt target site duplications (TSDs) (supplementary table S1, Supplementary Material online). We expanded the search for functional L1s by screening a second Tasmanian devil genome (a male donor “Cedric”: Devil_CABOG_asm.06) for intact ORF2 sequences, and found this genome to be similarly devoid of functional L1 copies.
We successfully retrieved 355 of the 384 L1 copies that range from 6,000–6,762 nt from the Cedric devil genome by comparison of orthologous flanking sequences in the reference genome. However, a number of copies (29) could not be retrieved with confidence due to the lower sequencing coverage of the Cedric genome. Of the selected 384 L1 copies, 193 copies were found with both flanks intact, 83 copies had flanking sequences located on two separate contigs, 81 copies had only one flank on a single contig, and 29 copies had flanks that could not be found anywhere in the Cedric genome.
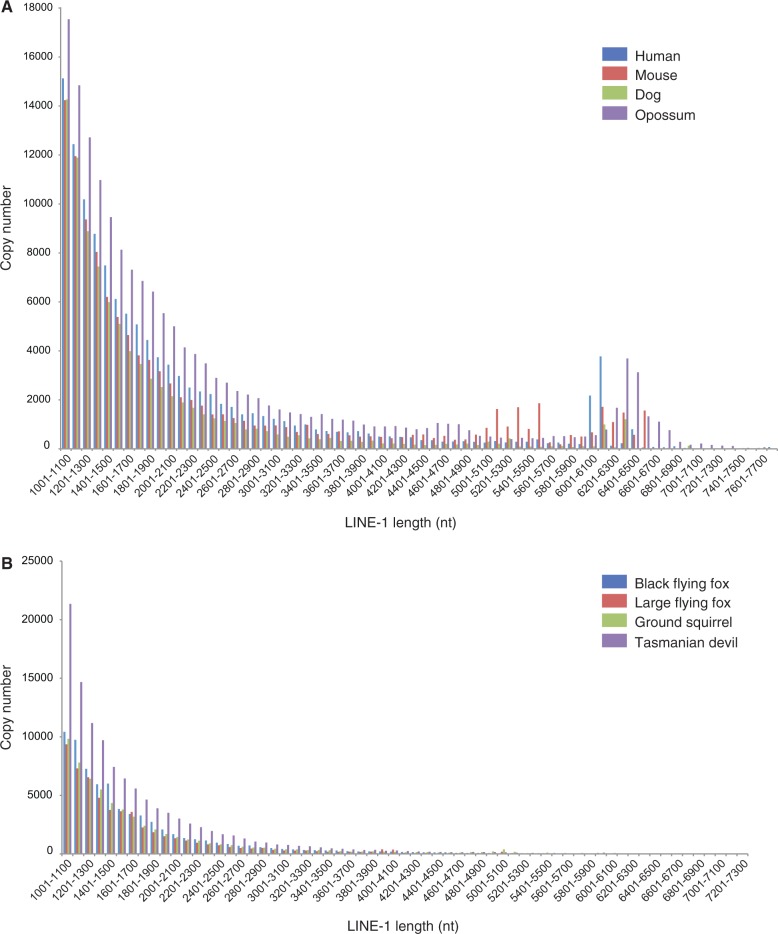
The majority of L1 and RTE copies in a genome are 5′–truncated; thus, all identified L1 and RTE copies longer than 1,000 nt (L1) or 500 nt (RTE), respectively, were plotted against their copy number. The graphs display the length distribution of L1 and RTE copies in the Tasmanian devil and opossum genomes in comparison to six different placental mammals (fig. 2 and supplementary fig. S1, Supplementary Material online). Three mammals with retrotranspositionally active L1s (human, mouse, and dog) were selected and compared to three mammalian genomes harboring only inactive L1s (ground squirrel, black flying fox, and large flying fox) (fig. 2). For mammals with retrotranspositionally active L1s, there are significant numbers of L1 copies longer than 6,000-nt. In contrast, mammals with retrotranspositionally inactive L1s lack the peak associated with greater than 6,000–nt-long copies (fig. 2).
Fig. 2.
(A) L1 copy lengths plotted against the respective L1 copy numbers identified in four mammals (human, mouse, dog, and opossum) with retrotranspositionally active L1 elements, (B) L1 copy lengths plotted against the respective L1 copy numbers identified in four mammals (ground squirrel [Platt and Ray 2012], Black flying fox [Pteropus alecto], Large flying fox [Pteropus vampyrus] [Cantrell et al. 2008; Zhang et al. 2013], and Tasmanian devil), in which functional L1 elements have not been identified. Peaks in the range of 6,000–6,500 nt indicating full-length L1 elements are only observed in the genomes of those mammals with functional, retrotransposition-competent L1 elements but are absent from the genomes of those mammals devoid of any functional L1 elements.
To measure the degree of substitution saturation among the nonfunctional L1-1_SHs in the Tasmanian devil genome, we aligned nonfunctional ORF2 cassettes from the previously identified 17 copies with the least amount of mutations. Twenty-seven L1-1_MD sequences with intact ORF1 and ORF2 were aligned from the opossum genome. Opossum L1-1_MD ORF2 sequences were compared with the sequence variation in the 17 L1-1_SH ORF2 copies (supplementary fig. S2, Supplementary Material online). The overall mean sequence distance among the 27 L1-1_MD copies was 0.013, indicating a divergence of 1.3%, which is consistent with a source element model involving active retrotransposition (Deininger et al. 1992). On the other hand, the mean sequence distance among the L1-1_SH copies was 0.171, indicating a divergence of 17.1%, which suggests that these L1 copies are retrotranspositionally inactive.
After screening all possible L1-1_SH 3′-ends in the Tasmanian devil genome and removing ambiguities, 165,380 copies with a length of 500 nt remained. None of these was found to be 100–98% identical, whereas 13 and 52 copies exhibited similarities of 97% and 96%, respectively. After decreasing the length of the 3′ end to 200 nt, 171,840 copies remained. Although there were no fragments with 100% sequence identity, 157 copies were similar at 99%. We found 270, 870, and 1,598 copies with sequence identities of 98%, 97%, and 96%, respectively. The same 3′-end analysis of L1 copies was done for panda (Ailuropoda melanoleuca), another mammal that has been sequenced using short-read approaches. L1-1_AMe was used as a query to identify the 500 nt of the 3′-end and we obtained 8,000 copies after removing sequences with ambiguities. Of the remaining 8,000 copies, 1,811 copies were identical (100%), whereas 3,920, 4,766 and 5,196 copies were 99%, 98% and 97% identical, respectively.
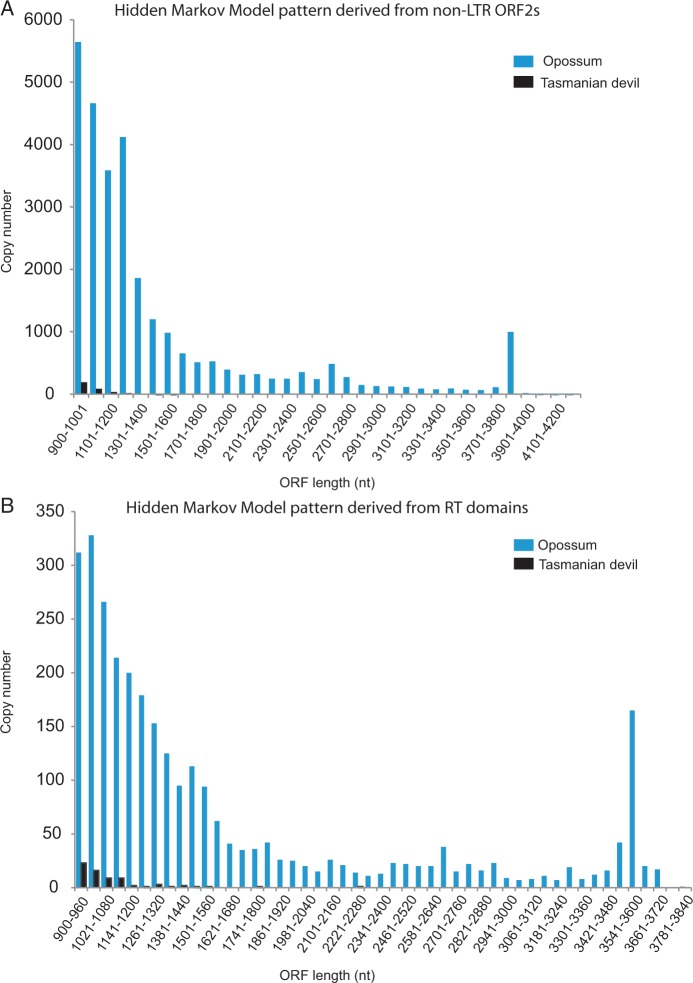
The reverse transcriptase (RT) is essential for both cis- and trans-mobilization of retrotransposons, because it reverse transcribes RNAs encoded by both autonomous and nonautonomous retrotransposons into cDNA. In order to test for the possibility that sources of RT other than L1-1_SH exist in the Tasmanian devil genome, two Hidden Markov Model (HMM) patterns were created based on retroviral RT domains and L1 ORF2 sequences. The patterns were searched against all possible ORFs encoding more than 300 amino acids (aa). The 300 aa value allows identification of ORFs that were recently inactivated and contain single stop-codons or frameshifts yielding translatable ORFs of greater than 900 nt (300 aa) as well as full-length RT-encoding ORFs. Each of the two HMM patterns identified different numbers of ORFs originating from RT in each species. HMM searches in the opossum genome yielded RT ORFs that were present in 3,012 copies (RT domain HMM search) and 28,665 copies (non-Long Terminal Repeat [LTR] ORF2 HMM search). Contrastingly, HMM searches in the Tasmanian devil genome identified RT ORFs that were present in 71 copies (RT domain HMM search) and 349 copies (non-LTR ORF2 HMM search) (table 1, fig. 3). The Tasmanian devil non-LTR ORF2 sequences contained a significant number (99 of 349 sequences) of ORF sequences containing stretches of N’s.
Table 1.
Percentages of Different Types of RT Derived ORFs in the Opossum and Tasmanian Devil Genomes Based on an HMM Approach.
| RT Domain HMM |
Non-LTR ORF2 HMM |
|||
|---|---|---|---|---|
| Opossum | Tas. Devil | Opossum | Tas. Devil | |
| Number of sequences >300 aa | 3,012 | 71 | 28,665 | 349 |
| SINEs | 0.0 | 0.1 | 0.1 | 0.2 |
| LINEs | 32.7 | 23.5 | 99.4 | 87.6 |
| L1 | 32.7 | 23.3 | 89.5 | 87.0 |
| L2 | 0 | 0 | 0.0 | 0.1 |
| L3 | 0 | 0.2 | 0.0 | 0.0 |
| RTE | 0.0 | 0 | 9.9 | 0.4 |
| LTRs | 66.0 | 54.2 | 0.1 | 0.1 |
| ERVL | 0 | 0 | 0 | 0 |
| ERV_MaLR | 0 | 0 | 0 | 0 |
| ERV_classI | 60.0 | 5.5 | 0.1 | 0.1 |
| ERV_classII | 6.0 | 48.7 | 0.0 | 0 |
| DNA transposons | 0 | 0 | 0 | 0 |
Fig. 3.
Length distribution of RT-encoding ORFs found in two HMM pattern searches in the opossum and Tasmanian devil genomes. All ORFs longer than 300 aa were screened in both marsupial genomes. (A) A non-LTR HMM pattern was derived from 42 phylogenetically diverse ORF2 sequences (from LINE1, LINE2, LINE3, RTE, and Penelope) and screened in opossum and Tasmanian devil genomes. (B) In total, 268 RT domain sequences from the Gypsy Database 2.0 were used to create an HMM pattern. The copy number was plotted against length for each HMM pattern and species. See table 1 for additional information regarding the resulting distribution of element classes from each HMM pattern.
The HMM pattern based on the non-LTR ORF2 sequence identified primarily RTE and L1 elements. This search showed that L1 ORF2 (L1-1_MD ORF2) is the most frequent RT-encoding ORF in the opossum genome, but found only fragmented L1 ORF2s in the Tasmanian devil. The HMM pattern based on RT-domain sequences identified mainly endogenous retrovirus (ERV) elements in both species, and found that the expansion of ERVs has been different between the opossum and Tasmanian devil lineages. The majority of ERV-derived ORFs in the opossum genome are class I ERVs (gamma retrovirus), whereas ERV class II (beta retrovirus) elements are the most frequent in Tasmanian devil (table 1).
No Detectable WSINE1 Activity in the Tasmanian Devil Genome
The most recently propagated SINE in the Tasmanian devil genome is WSINE1 (Wallaby SINE) (Nilsson et al. 2012). Based on the master gene model of retrotransposon propagation, suggesting a small number of active source elements (Deininger et al. 1992), the youngest copies in a genome will be virtually identical. Both Tasmanian devil and opossum genomes were screened for the presence of identical SINE copies (WSINE1 or SINE1_Mdo) to find evidence for active retrotransposition. We found several subfamilies with identical SINEs in opossum. The largest subfamily consisted of 175 identical SINE1_Mdo copies, and several subfamilies contained around 20 copies each. In contrast, all WSINE1 copies identified in the Tasmanian devil genome had accumulated significant numbers of mutations and indels (supplementary fig. S3, Supplementary Material online).
The approximately 200,000 pairwise alignments of WSINE1 insertions in two individual Tasmanian devil genomes (Cedric genome [Miller et al. 2011] and WTSI_Devil_ref v7.0) were screened for insertions or deletions, and 652 contained indels longer than 50 nt. All 652 pairwise alignments were manually screened and found to be assembly artifacts or partially sequenced WSINE1s.
Thus, our data suggest an absence of WSINE1 mobilization in the Tasmanian devil population, as all identified WSINE1 elements were present in both of the aligned genomes (i.e., no polymorphic loci were identified), and all copies have accumulated significant numbers of substitutions.
Dasyuromorphian Phylogeny and Evolutionary Activity of WSINE1 and L1-1_SH
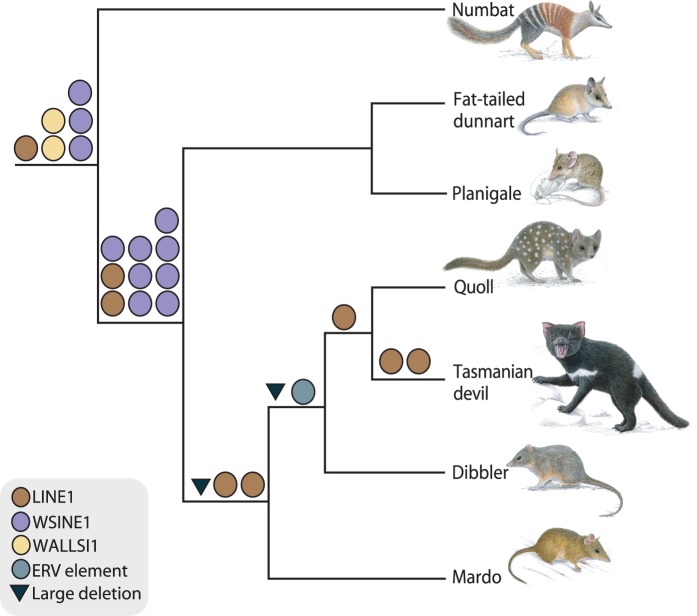
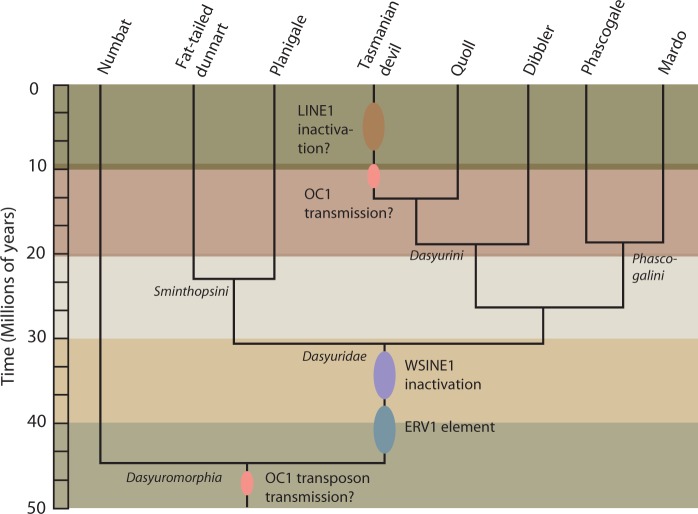
We analyzed the presence/absence patterns of retrotransposons to shed light on the phylogeny of Dasyuromorphia and obtained evidence for the silencing of WSINE1 and for the timing of this putative inactivation event. Polymerase chain reaction (PCR) analysis of 80 WSINE1-containing introns from different dasyuromorphian species showed that all elements clustered on the branch leading to Dasyuromorphia or Dasyuridae. As we were not able to resolve any branches younger than 30 My by analyzing SINE insertions, we screened for the autonomous retrotransposon L1-1_SH to analyze dasyuromorphian phylogeny. Several of the younger branches in the dasyuromorphian tree were supported by L1-1_SH retrotransposition events. All of the analyzed L1-1_SH insertions were 5′-truncated. Where possible, we selected a minimum of three informative markers from the 158 intronic loci containing informative retrotransposition events, in order to achieve statistically significant support for each branch (Waddell et al. 2001). The selected 18 introns were sequenced and analyzed in a larger taxon sampling representing all subfamilies of Dasyuromorphia as well as the necessary outgroup species (fig. 4, supplementary table S2 and data set S1, Supplementary Material online). Six insertions were recovered for the monophyly of the order Dasyuromorphia ([6 0 0] P = 0.0005), ten for the branch leading to Dasyuridae ([10 0 0] P = 0.0001), and two for the grouping of Dasyurini and Phascogalini ([2 0 0] P = 0.1111). Branches leading to Dasyurini and Dasyurus+Sarcophilus were supported by one insertion each ([1 0 0] P = 0.3333). Two large intronic deletions (>100 nt) occurred on the branches leading to Dasyurini+Phascogalini and Dasyurini and can be used as phylogenetic markers.
Fig. 4.
Phylogenetically informative retrotransposon insertions for the order Dasyuromorphia. Each circle represents a phylogenetically informative retrotransposon insertion. Colors refer to the element type. Two large deletions of more than 100 nt were found and are indicated as black triangles. Numbat, Myrmecobius fasciatus; Fat-tailed dunnart, Sminthopsis crassicaudata; Planigale, Planigale sp.; Quoll, Dasyurus geoffroii; Dibbler, Parantechinus apicalis; Mardo, Antechinus flavipes.
We did not recover any phylogenetically informative WSINE1 and WALLSI1 retrotransposition events for divergences younger than the split leading to Dasyuridae approximately 30 Ma. Our study uncovered 50 My of L1-1_SH retrotransposition activity from the origin of Dasyuromorphia to the Tasmanian devil branch (fig. 4).
Novel Tasmanian Devil ERV-I Element
A transposable element with similarities to an ERV in the opossum (LTR104d_Dm) was identified in the Tasmanian devil genome. The novel Tasmanian devil ERV1 element is present in 226 copies, flanked by 4-nt long TSDs, and can be divided into two subfamilies. Type 1 is composed of 139 copies divided into 1a and 1b (1a: 589 nt long and present in 75 copies; 1b: 596 nt long and present in 64 copies), and type 2 is 711 nt long and found in 87 copies (supplementary data set S2, Supplementary Material online). The novel ERV1 fragments are likely soloLTRs from an ERV1 expansion that occurred in the ancestor of Dasyuridae around 40–30 Ma.
ERV1 was identified by the discovery of a shorter fragment located in the intron of CNOT1 (CCR4-NOT transcription complex, subunit 1) in the Tasmanian devil genome. This short fragment has characteristics of retrotransposition such as a poly(A)tail and TSDs. This is likely a result of an L1-mediated trans-mobilization event, where the L1 protein machinery recognized a short poly(A) sequence within the ERV RNA and retrotransposed the ERV1 fragment through interaction with this short stretch of A’s. The consequence of these trans-mobilization events was the five copies ranging in size from 188–335 nt (supplementary table S3, Supplementary Material online) due to incomplete retrotransposition which led to 5′-truncated ERV1 copies with TSDs and poly(A)tails. This element appears to have been inserted into the CNOT1 intron of the ancestral lineage leading to Dasyurinae approximately 18 Ma. Conserved PCR primers located inside the ERV1 element were used to amplify it in other dasyuromorphian species. Analysis of the PCR products indicated that the long ERV1 type is present only in Dasyuridae and not in Myrmecobiidae (numbat) (data not shown), suggesting an origin approximately 30 Ma.
DNA Transposon Activity in the Tasmanian Devil Genome
Only 1.13% of the Tasmanian devil genome is covered by DNA transposon sequences—a relatively low proportion compared with placental mammals. To investigate recent DNA transposon insertions in the Tasmanian devil genome, we extracted all copies longer than 1,500 nt, counted the total genomic copy number, and searched for ORFs in all six reading frames. We identified 198 copies of DNA transposons that exceeded 1,500 nt (supplementary table S4, Supplementary Material online) but only one retained an intact transposase-coding ORF. We selected two DNA transposons, OC1 (OposCharlie1) and the hAT-1_MEu, for further analysis as they had a high genomic copy number and retained full-length copies in the Tasmanian devil genome. The phylogenetic distribution of the hAT-1_MEu transposon had not been screened previously, whereas the OC1 transposon had been studied in a limited set of dasyuromorphian species (Gilbert et al. 2013).
Evidence for Recent and Ancient Mobilization of the hAT OC1 Transposon
The hAT OC1 type is present in approximately 5,200 copies in the Tasmanian devil genome (Gilbert et al. 2013), and 132 hAT OC1_Das sequences were longer than 1,500 nt. The average maximum likelihood (ML) sequence distance is 0.081 (gamma correction 1.65) among the 132 copies greater than 1,500 nt. We found one intact ORF (1,832 nt-610 aa) on a partial contig on chromosome 3 of OC1, suggesting a possible recent transmission of this DNA transposon into the Tasmanian devil genome. We PCR-amplified the ORF coding for OC1_Das transposase from cDNA generated from the fat-tailed dunnart kidney cell line (supplementary fig. S4, Supplementary Material online). Sequence analysis of 12 independent fat-tailed dunnart-specific cDNA products did not uncover any intact transposase-encoding ORF.
The evolutionary history of the OC1 transposon was evaluated by reconstructing a phylogenetic tree of the OC1 transposase-coding ORF. Forty-two copies of OC1 transposase-coding ORFs were extracted from the available Tasmanian devil genome sequence. In addition, 12 copies were PCR-amplified from cDNA from the fat-tailed dunnart, and consensus sequences were derived for both species. OC1 transposon sequences from Tasmanian devil, dunnart, six other mammalian species, and the green anole lizard (Anolis carolinensis) were used to calculate an ML phylogenetic tree. The data set was 1,802 nt long, and the best fitting evolutionary model was GTR + G(4) (Lanave et al. 1984). The ML tree uncovered a close relationship between the Tasmanian devil and the fat-tailed dunnart OC1 transposon, and showed that the OC1 sequences from opossum, placental mammals, and the green anole lizard are distantly related (supplementary fig. S5, Supplementary Material online). A phylogenetic screen for the presence of the OC1 transposon was performed using PCR. We attempted to amplify a specific 533-nt OC1 fragment from eight dasyuromorphians, three closely related Australian orders (Peramelemorphia, Notoryctemorphia, and Diprotodontia), and the South American order Didelphimorphia. The PCR amplification of OC1 was only successful in Dasyuromorphia and Tarsipes (Diprotodontia) (supplementary fig. S6A, Supplementary Material online). Our results suggest that the OC1 transposon was transmitted into the genome of the ancestor of Dasyuromorphia, as demonstrated by its presence in all investigated species from this order. Alternatively, OC1 may have been independently transmitted on multiple occasions. Based on our taxon sampling from Diprotodontia, Peramelemorphia, and Notoryctemorphia, it appears that OC1 has been transmitted to a lesser extent in the genomes of these orders compared with Dasyuromorphia. However, it has been shown that there can be vast differences in the amplification and fixation rate of DNA transposons between species and populations over evolutionary time scales that could influence our results (Feschotte and Pritham 2007).
Ancient hAT-1_MEu Transposons Are Present in All Australian Marsupials
We show that hAT-1_MEu DNA transposons (Jurka 2011b), originally identified in the tammar wallaby genome, are present in the Tasmanian devil genome. The hAT-1_MEu transposon has a consensus sequence of 2,787 nt and approximately 31,000 copies of hAT-1_MEu were identified in the Tasmanian devil genome. Of the 31,000 copies, only 34 copies were longer than 1,500 nt. The average ML distance among the 34 copies that are longer than 1,500 nt is 0.35 using an estimated gamma correction of 5.6. The high copy number as well as the high nucleotide distance among the greater than 1,500-nt-long copies suggests a more ancient transfer of hAT-1_MEu into the genome than OC1. The 3′-end of hAT-1_MEu (positions 1–492/2,320–2,784 nt) exhibits a high sequence similarity to the 5′-end (positions 1–494) and the 3′-end (positions 521–974) of the DNA transposon RodentCharlieR1 (RCHARR1) (Waterston et al. 2002; Pace 2008), suggesting that hAT-1_MEu is related to RCHARR1. RCHARR1 is most likely a nonautonomous Miniature Inverted-repeat Transposable Element (MITE) of the autonomous hAT-1_MEu. RCHARR1 exists in 13,600 copies in the Tasmanian devil genome, ranging in length from 81 to 458 nt. A phylogenetic PCR screening identified hAT-1_MEu in all investigated dasyuromorphians as well as the three other Australian orders Diprotdontia, Peramelemorphia, and Notoryctemorphia (supplementary fig. S6B, Supplementary Material online). The obtained amplification pattern suggests independent transmissions in the four orders, due to the lack of hAT-1_MEu amplification in Macrotis lagotis (Peramelemorphia) (supplementary fig. S6B, Supplementary Material online).
Discussion
Transposable elements can affect the genome in various ways, such as by gene silencing, adding new exons, contributing to alternative splicing, or by altering transcription depending on the region in which they insert (Beck et al. 2011). Several studies have described the impact of transposable elements on environmental adaptation and have suggested that they may play a role in species survival (Casacuberta and González 2013). Transposable elements make up approximately 52% of the marsupial genome (Mikkelsen et al. 2007; Renfree et al. 2011; Nilsson et al. 2012), suggesting that they may have significant influence on the genetics of this mammalian infraclass. Marsupial transposable elements have not been investigated to the same degree as placental mammals, in part due to limited sequencing efforts. Currently, only three marsupial genomes are available—the South American opossum, the Australian tammar wallaby and the Tasmanian devil (Mikkelsen et al. 2007; Renfree et al. 2011; Murchison et al. 2012).
WSINE1 Became Transpositionally Inactive 30 Ma
Currently available marsupial genome data has revealed that CORE-SINEs have been active in all marsupial lineages (Gentles et al. 2007; Munemasa et al. 2008; Nilsson et al. 2012). CORE-SINEs are tRNA-related SINEs characterized by a highly conserved 65-nt domain that have been inactive in placental mammals for more than 100 My (Jurka et al. 1995). WSINE1 is present in approximately 200,000 copies in the Tasmanian devil genome (Nilsson et al. 2012). WSINE1 is found in all Australian marsupial orders and has a highly conserved sequence while still proliferating in the different orders (Nilsson et al. 2010, 2012) and is thus not divided into specific subfamilies as for instance human Alu elements. Analyses of nested transposable element integrations have suggested that it is the youngest SINE in the Tasmanian devil genome (Nilsson et al. 2012). Thus, WSINE1 appears to fit the criteria for resolving relationships within Dasyuromorphia. Previously, retrotransposons have been successfully utilized as phylogenetic markers to resolve relationships among several mammalian groups (e.g., Shimamura et al. 1997; Kriegs et al. 2006; Nilsson et al. 2010).
We investigated 158 introns for the presence of phylogenetically informative retrotransposon insertions in the order Dasyuromorphia. Of these markers, 23 strongly supported the monophyly of the order Dasyuromorphia, the subfamily Dasyuridae, and the grouping of Dasyurini with Phascogalini. A single marker supported the grouping of quolls (Dasyurus) with Tasmanian devils (Krajewski et al. 2000; Meredith et al. 2008; Nilsson et al. 2012) (fig. 4).
Despite screening more than 80 introns with WSINE1 elements, we could not detect any activity of WSINE1 copies on any branch in the 30-My-old Dasyuridae lineage. An independent study that screened 160 introns containing WSINE1s did not recover any insertions that were younger than 30 My (Zeeman et al. 2013). This observation suggests that WSINE1 became inactive at about 30 Ma on the branch leading to Dasyuridae. Further evidence for an ancient inactivation event of WSINE1 comes from the source element model of SINE propagation (Deininger et al. 1992). After screening the opossum and Tasmanian devil genomes a contrasting pattern emerged between the two species. Our in silico screen of the opossum genome detected 175 identical SINE1_Mdo copies that most likely stem from recent integrations derived from the same source element. This is consistent with an independent study that showed that the opossum genome has transpositionally active L1 and SINE1_Mdo elements (Gu et al. 2007). Contrastingly, the Tasmanian devil genome does not contain identical WSINE1 copies, suggesting that these elements are no longer mobilized. This corroborates the finding from the screen of phylogenetically informative retrotransposons and a previous in silico estimate of an age of 23 My for the youngest WSINE1 sub-group in the Tasmanian devil genome (Nilsson et al. 2012). It has been observed in a group of species-rich (>360 species) South American rodents (Sigmodontinae) that SINES can stop mobilizing in a genome (Rinehart et al. 2005).
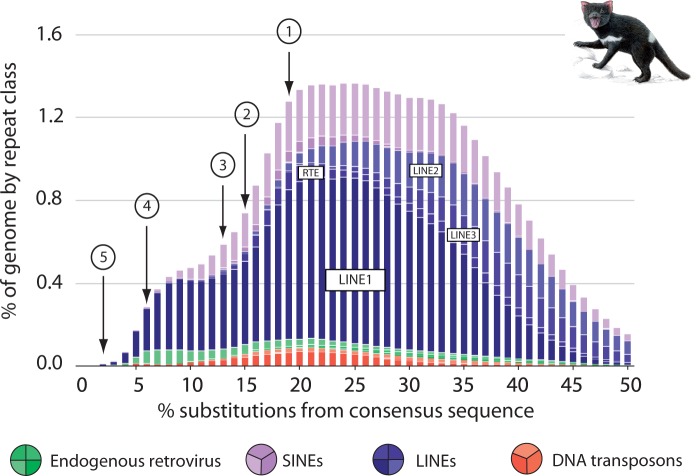
Interestingly, L1 copies in the dasyuromorphian lineage appear to have retained retrotransposition competence for several million years after the inactivation of WSINE1 (figs. 4 and 5). This is evident by the detection of L1 insertions for younger divergences, as well as two 5′-truncated L1s in the Tasmanian devil, that are absent from its closest relative, the quoll. Repeat landscape analysis of the Tasmanian devil genome shows how the activity of WSINE1 started to decline prior to L1 (fig. 6). Repeat landscapes are used to graphically represent the abundance of transposable elements plotted against divergence to their respective consensus sequences in a genome, and give an overview of the expansion and decline of transposable elements over time.
Fig. 5.
A time-calibrated tree of dasyuromorphian phylogeny (divergence times from Krajewski et al. 2000; Meredith et al. 2008; Nilsson et al. 2012). The WSINE1 inactivation event, DNA transposon transmissions, and the occurrence of the newly identified ERV1 have been mapped on the tree.
Fig. 6.
A repeat landscape of the Tasmanian devil genome showing the expansion and decline of transposable elements. The x axis shows the percentage of CpG adjusted Kimura-2-parameter substitutions to the consensus sequences in 1% bins. The y axis shows the relative amount of genome sequence covered by each transposable element group. The youngest elements have the least amount of substitutions whereas evolutionary older elements will have acquired more mutations. Trends showing a decline in various non-LTR retrotransposon groups have been indicated by arrows and numbers. (1) L3 inactivation, (2) L2 inactivation, (3) RTE inactivation, (4) SINE inactivation, (5) possible L1 inactivation. As in most mammals, the percentage of ERVs and DNA transposons relative to the entire genome is low. The colored circles refer to different types of elements found in the Tasmanian devil genome.
Degraded LINE1 and RTE Landscapes in the Tasmanian Devil
Evidence indicates that WSINE1 mobilization has stopped in the Tasmanian devil genome, and in all 69 species belonging to Dasyuridae. We retrieved 5′-truncated L1 insertions that are only present in the Tasmanian devil, suggesting that L1 was active during the last 12 My. However, the results from the in silico screening of the Tasmanian devil genome suggest that there are no functional L1 copies or any alternative sources of RT activity. In most mammals, L1 copies cover about 17–20% of the total genome (e.g., Lander et al. 2001; Mikkelsen et al. 2007). In the human genome, there are approximately 106 L1 copies of which approximately 7,000 are full-length (Lander et al. 2001; Khan et al. 2006). We identified approximately 13,000 full-length L1 copies in the opossum genome, all of which appeared to be inactive due to random mutations, and approximately 500 copies in which both ORFs were intact and possibly functional. In contrast, we observed significantly fewer numbers (∼380 copies) of full-length L1 or RTE retrotransposons in the Tasmanian devil genome, and no intact L1 ORFs could be identified as had been suggested previously (Jurka 2011a).
Although in silico analysis of full-length L1 elements in the Tasmanian devil genome showed that they are harboring numerous mutations that could be attributed to assembly artifacts, the analysis of 500 nt of the L1 3′-end sequences should provide a better resolution of the recent history of L1. Our screen revealed the absence of L1 3′-ends that are 100–98% identical. This is in sharp contrast to the panda genome where we find 1,811 L1 3′-ends with a length of 500 nt that are 100% identical. However, when we analyze the 3′-terminal 200 nt of these copies, we find 157 copies that are 99% identical in the Tasmanian devil genome. Our results suggest that there are short fragments (200 nt) of L1-1_SH 3′-UTRs that are highly similar (157 copies—99%) but these are found, compared with panda, in small numbers. We used an HMM approach to test for alternative sources of RT in the Tasmanian devil genome and were unable to detect any functional sources of RT.
Additionally, we analyzed three other mammalian genomes with transpositionally inactive L1s (ground squirrel and two megabat species) (Grahn et al. 2005; Cantrell et al. 2008; Platt and Ray 2012) (fig. 2) in a similar vein and observed a similar pattern of decline and erosion of L1 copies as in the case of the Tasmanian devil genome. However, we cannot exclude the possibility that artifacts during genome assembly might be the cause of the observed erosion of L1 in the Tasmanian devil (and in other mammalian genomes with apparent L1 inactivation). This suspicion is raised because those genomes with transpositionally active L1 copies, such as human, mouse, and dog (fig. 2), were generated using Sanger sequencing.
Do Next-Generation Sequencing Data Allow for Accurate Assembly of LINE Retrotransposon Sequences?
The advent of next-generation sequencing methods, relying on short reads (50–150 nt), has made it possible to obtain genome sequences faster than with traditional Sanger sequencing methods (Koboldt et al. 2013). However, with the exception of the human genome, most mammalian genomes remain poorly or incompletely sequenced due to limited sequencing efforts and/or assembly problems from repetitive sequences (Green 2007; Milinkovitch et al. 2010; Alkan et al. 2011; Birney 2011).
Short reads can pose problems for assembly with regard to so-called multireads, that is, reads, that are mapped to several locations within a genome (Treangen and Salzberg 2011). Multireads come from repetitive sequences, such as transposable elements, tandem repeats or segmental duplications. The multiread problem does not affect all transposable elements, rather only copies that are highly similar (>97%). As functional L1 elements in the mammalian genome are greater than 99% similar, they are especially susceptible to the multiread assembly problem. Furthermore, full-length mammalian L1s are between 6,000 and 8,000 nt long and are therefore too long to be covered by a single 454, Illumina, or Sanger sequence read. Additional sequencing problems come from homopolymeric poly(A) stretches which are located, for instance, at the 3′-ends of younger L1 insertions and range from 10–85 nt (Szak et al. 2002). This creates problems for both Sanger and Illumina sequencing, and can lead to the collapse of the entire L1 sequence during genome assembly. It is impossible to establish if an L1 sequence is following the poly(A) stretch when sequencing from the genomic flank into the potential L1 3′-UTR. Low coverage genomes present an additional challenge for the interpretation of transposable element data such as functional L1 copies. For example, zero to few full-length L1 copies with intact ORFs could be assembled in the initial release of the cat (Felis catus), tammar wallaby, and dog (Canis familiaris) genomes (Wang and Kirkness 2005; Pontius et al. 2007; Renfree et al. 2011). Nonetheless, despite these apparent difficulties in using short read data to assemble mammalian genomes, we were able to successfully retrieve 100 L1 copies, with two intact ORFs, from the panda genome—the first mammalian genome to be sequenced using only short reads (average read length: 52 nt) (Li et al. 2010). By doing so, we demonstrated the feasibility of assembling long, highly similar L1 insertions under certain conditions. However, current genome assemblers implement different algorithms and do not always use the same strategy to assemble sequences occurring on multiple locations in the genome (Henson et al. 2012). Both the sequencing strategy and assembly pipeline can influence transposable element identification, with recently inserted full-length L1 integrations being one of the most susceptible element.
We note that low copy numbers of functional L1s may actually be present in the Tasmanian devil genome, but may have eluded detection due to artifacts during the sequencing and/or assembly stages. Previous studies of mammalian L1 inactivation events have been based on trace Sanger sequences and experimental evidence (Grahn et al. 2005; Rinehart et al. 2005; Cantrell et al. 2008). L1 extinction was discovered for the first time in South American sigmodontine rodents (Casavant et al. 2000; Grahn et al. 2005). In order to screen genomic L1 copies for intact ORF2 sequences, which are crucial for retrotransposition, a part of the ORF2 region was PCR amplified and cloned in frame in the lacZ gene of a pBluescript vector (Cantrell et al. 2000). LacZ expression resulting in blue bacterial colonies is only possible if the inserted ORF2 sequence does not harbor any stop codons in the investigated reading frame. The presence of stop codons in ORF2 results in white bacterial colonies. The ratio of blue to white bacterial colonies on an agar plate would give an indication of the ratio of functional L1 copies in a genome (Cantrell et al. 2000). The experimental screening method was used to evaluate the evolution of L1 in sigmodontine rodents, and demonstrated that SINEs had become extinct prior to the L1 inactivation event 9 Ma (Grahn et al. 2005; Rinehart et al. 2005). The same screening method was used to identify another L1 inactivation event in megabats (family Pteropodidae), estimated to have occurred around 22 Ma (Cantrell et al. 2008). The L1 sequence present in megabats was synthesized, and retrotransposition assays confirmed that it was functional; however, the presence of a long spacer sequence (445–481 nt) between ORF1 and ORF2 was found to act as an inhibitor of retrotransposition (Yang et al. 2014). The synthesis of an L1 from the megabat genome shows that it is possible to revive inactivated retrotransposon fossils and test their functionality (Yang et al. 2014). An in silico screening of the 13-lined squirrel genome (Spermophilus tridecemlineatus) suggests a third documented L1 inactivation event (Platt and Ray 2012). Using a calculated neutral mutation rate, the L1 inactivation in the 13-lined squirrel genome was estimated to around 4–5 Ma, with a reduction in activity beginning as early as 19–26 Ma (Platt and Ray 2012).
We suggest that high throughput experimental approaches need to be developed to efficiently screen nonhuman mammalian genomes for functional L1s, in order to verify results from genome sequencing. One strategy is to involve pipelines such as VariationHunter (Hormozdiari et al. 2010), TIF (Nakagome et al. 2014) or RetroSeq (Keane et al. 2013) that are specifically developed to identify polymorphic transposable element insertions between individuals in a population to gather evidence about retrotransposition activity. However, information such as genomic location and the actual sequence of functional L1 copies cannot be elucidated using such in silico approaches. Rather, identification of functional L1 copies requires rigorous wet bench verification of the identified novel insertions as well as the reference genome sequence of the organism.
Evolutionary Activity of DNA Transposons in Australian Marsupials
In contrast to class I transposable elements that rarely seem to lose the ability to retrotranspose within a genome, class II elements are usually found as fragmented copies in mammalian genomes (Feschotte and Pritham 2007). To date, only one transpositionally active DNA transposon has been identified in mammals (Mitra et al. 2013). The limited numbers and fragmented nature of DNA transposons in most mammals may be a result of host-encoded silencing mechanisms that act on newly introduced elements. Very little is known about the evolutionary history and lateral transfer of DNA transposons in marsupials. We have investigated three different DNA transposons in the Tasmanian devil genome, of which only the OC1_Das transposon was likely mobilized recently (Gilbert et al. 2013). To date, the OC1 transposon was found in 12 distantly related mammals and the green anole lizard (Novick et al. 2010; Gilbert et al. 2013). After examining the phylogenetic distribution of OC1_Das in Dasyuromorphia, we concluded that it is present in all major subfamilies of this group, suggesting a single ancient insertion event prior to the diversification of all dasyuromorphian subfamilies. However, the number of transmission events into the genome remains unclear due to the fact that a complete OC1 ORF was detected in the Tasmanian devil, together with copies of 8% sequence divergence, suggesting a relatively recent insertion event. Phylogenetic screening of additional Australian marsupials suggests that OC1 is not present in the marsupial orders, except for the diprotodontian honey possum Tarsipes. The reason for the success of OC1 in dasyuromorphian genomes remains unclear.
The hAT-1_MEu DNA transposon was previously found in five placental mammalian species (mouse, rat, praire vole, tenrec, and shrew) and the anole lizard (Waterston et al. 2002; Pace 2008; data not shown). We showed that hAT-1_MEu (and its MITEs, e.g., hATA_ME) is present in 31,000 copies in the Tasmanian devil genome and has a broader phylogenetic distribution among marsupials than among placental mammals, because it is present in all four Australian orders. The four Australian marsupial orders share a common ancestor that lived approximately 60 Ma, and it is unclear whether the speciation events that lead to the extant orders took place in Antarctica or Australia (Woodburne and Case 1996). The absence of the hAT-1_MEu transposon from the Macrotis lagotis genome (Peramelemorphia) (supplementary fig. S6B, Supplementary Material online) suggests that several independent transmission events of this transposon may have taken place.
The limited distribution of the hAT-1_MEu element among placental mammalian orders compared with marsupials may suggest that hAT-1_MEu transmission was facilitated by a specific pathogen in a geographically localized region, over a short period of time.
Potential Causes of SINE and LINE Inactivation
Several lines of evidence suggest that WSINE1 stopped propagating 30 Ma in the ancestor of Dasyuridae; however, it remains unclear whether L1 is still functional given the quality of current genomic data of this species. Several hypotheses have been proposed to explain how and why SINEs and LINEs might go extinct in a genome. These include genetic drift, competition between retrotransposons, and evolution of host control mechanisms (Cantrell et al. 2008; Erickson et al. 2011; Platt and Ray 2012). Over millions of years, host genomes have evolved complex mechanisms of self-defense to control the activity of retrotransposons (Schumann et al. 2010; Levin and Moran 2011; Heras et al. 2014). It has been demonstrated in great apes that Alu elements (primate specific SINEs) accumulate at different rates among branches of the primate tree (e.g., orangutan genomes appear to have very limited SINE activity) (Hormozdiari et al. 2013). Contrastingly, the study also showed that L1 appears to accumulate at a similar rate across all primate branches (Hormozdiari et al. 2013). Thus, L1 activity appears to have remained constant in primates, whereas SINE activity has been more prone to bursts of activity followed by periods of quiescence for unknown reasons (Hormozdiari et al. 2013). The interplay between different transposable element groups and host genome factors is not well understood, as exemplified by the study of great apes.
Further genome sequences from Australian marsupials, and especially Dasyuromorphia, will add to the understanding of how transposable elements have evolved in this group compared with placental mammals.
Materials and Methods
Genomic Distribution of Transposable Elements
The reference genome assembly of the Tasmanian devil (WTSI_Devil_ref v7.0) (Murchison et al. 2012) and a Tasmanian devil genome from a male donor Cedric (Devil_CABOG_asm.06) (Miller et al. 2011) were screened for transposable elements using RepeatMasker (Repbase Update 20110920) (Smit et al. 1996) and CENSOR (Kohany et al. 2006). The Cedric genome assembly was sequenced with a Roche 454 GS FLX platform (short and long reads) and an Illumina platform (paired-end sequencing with 300 nt insert size) (Miller et al. 2011) and assembled with CABOG (Miller et al. 2008). “WTSI_Devil_ref v7.0” assembly was sequenced by applying Illumina using short insert and mate pair (3,000–10,000 nt insert sizes) libraries, and assembled with the Phusion2 genome assembly pipeline (Murchison et al. 2012). Additionally, the WTSI_Devil_ref v7.0 assembly is based on Illumina sequencing of individual chromosomes as well as transcriptomes (Murchison et al. 2012). The Cedric assembly consists of 148,891 scaffolds with an N50 size of 147,544 nt, whereas the WTSI_Devil_ref v7.0 assembly consists of 35,974 scaffolds with an N50 of 1,847,186 nt (Miller et al. 2011; Murchison et al. 2012). Genome assembly of the South American gray opossum (Monodelphis domestica), which is referred to in the manuscript as “opossum” (version monDom5) (Mikkelsen et al. 2007), was used for comparative analyses of L1 and SINE1_Mdo (Repbase ID SINE-1_MD) activities due to better sequence coverage than the Australian tammar wallaby (Macropus eugenii) (Renfree et al. 2011). Due to the use of different names for the same transposable element in different databases, both names have been indicated in the text where relevant.
Frequencies and distributions of L1 and RTE elements (supplementary fig. S1, Supplementary Material online) were investigated in the Tasmanian devil genome. The length distributions of L1 and RTE were calculated by plotting copy number against length using a custom PERL script. A repeat landscape was calculated for the WTSI_Devil_ref v7.0 Tasmanian devil genome due to the higher coverage of this individual relative to Cedric. The genome was repeatmasked with Cross_Match (Green 1996). Two different Perl-scripts (calcDivergencefromAlign.pl/createRepeatLandscape.pl) available with the RepeatMasker package were used to create the repeat landscape.
Six placental mammals were chosen to investigate the L1 length distribution in species with suspected L1 inactivation and known L1 retrotranspositional activity. Three species were chosen as it is confirmed that they have transpositionally active L1 elements: Human (Homo sapiens), mouse (Mus musculus), and dog (Canis familiaris), and an additional three species were selected as it has been suggested that they have transpositionally inactive L1 elements: Black flying fox (Pteropus alecto), Large flying fox (Pteropus vampyrus), and the ground squirrel (Spermophilus tridecemlineatus). As sequenced mammalian genomes with suggested L1 inactivation are rare, both available megabat species were included, despite being part of the same genus (Pteropus).
To find transposititonally active L1s in the Tasmanian devil genome, all L1s longer than 6,000 nt were extracted from the genome. By extracting and analyzing only those elements that were masked as L1, we avoided any potential bias by targeting specific L1 subfamilies. Marsupial L1 elements have not been subject to any detailed subfamily analysis as has been done with placental mammalian L1 elements earlier (Smit et al. 1995), and in particular Australian marsupial L1 subfamilies (i.e., tammar wallaby and Tasmanian devil) are poorly characterized. The longest L1 elements in the Tasmanian devil genome were all identified as L1-1_SH elements (Jurka 2011a), and these copies were analyzed for the presence of stop-codons, conservation, and TSDs. ORFs were identified by aligning homologous ORF1 from L1-1_ME (tammar wallaby) and ORF2 sequences from L1-1_MD (opossum) to the L1-1_SH consensus sequence (Jurka 2011a).
All L1 ORFs are inactivated by frameshifts, nonsense mutations, and indels in the Tasmanian devil genome. Screening of both Tasmanian devil genome assemblies (WTSI_Devil_ref v7.0 and Devil_CABOG_asm.06) revealed the same L1 pattern. Similarly, L1-1_MDs with two intact ORFs from the opossum genome were extracted and analyzed. Conservation plots were based on ORF2 alignments from L1-1_SH and L1-1_MD using the EMBOSS software plotcon (Rice et al. 2000). Overall mean sequence distances of the 17 L1-1_SH copies and the 29 L1-1_MD copies were calculated using MEGA5 (Tamura et al. 2011). This estimates the mean distance among a group of sequences and not the pairwise distance to L1-1_SH. The pairwise distance for the individual 17 copies to the L1-1_SH consensus sequence was calculated with MEGA5 (Tamura et al. 2011). A phylogenetic tree of ORF2 from the 17 L1-1_SH copies and the L1-1_SH consensus sequence was made using ML in TreeFinder (Jobb et al. 2004). The best suited evolutionary model was calculated using ModelProposer to be GTR:G5 (Jobb et al. 2004) and the data set consisted of 4,234 nt. The opossum (L1-1_MD) and tammar wallaby (L1-1_ME) ORF2 sequences were used as outgroups for the L1-1_SH copies. The scaffold name is used as identifier for each L1 copy (supplementary table S1, Supplementary Material online) and the pairwise distances for each individual copy to the consensus sequence L1-1_SH is listed next to the name (supplementary fig. S7, Supplementary Material online).
The use of short read data for genome assembly can result in sequence artifacts, especially for genomes containing high amounts of repetitive sequences (Treangen and Salzberg 2011). In order to evaluate whether short read data can assemble repetitive sequences such as L1, we screened the panda genome (Ailuropoda melanoleuca) (Li et al. 2010) for potential functional L1 copies (i.e., those copies containing two functional ORFs). The panda genome was sequenced and assembled, using approximately 52-nt-long reads and mate-pair library sequencing (Li et al. 2010). Another approach to screen a genome for active L1s is to investigate the sequence conservation of the 3′-UTR between different L1 copies (Wang and Kirkness 2005). Due to the mechanism of L1 retrotransposition, most L1 insertions are 5′-truncated (fig. 2). Recent L1 retrotransposition events should be detectable by a high frequency of highly similar 3′-ends (Wang and Kirkness 2005). The assembly of L1 3′-ends is less problematic due to the short length and for Sanger sequenced genomes, the entire 3′-UTR and part of the genomic flank can be covered by one single read. We extracted all L1-1_SH 3′-ends (200 and 500 nt) from the Tasmanian devil genome and removed those copies that had ambiguity characters. The sequences were clustered using DNACLUST (Ghodsi et al. 2011) at different similarity cut-offs to determine the number of sequences with similarities at 100–96%. The percentage of similarity among these copies was calculated between 100% and 96%. The same analysis was done for the panda genome using the youngest L1 consensus sequence (L1-1_AMe).
HMM Search for RT Encoding Regions
Two different HMM patterns were constructed to search for RT sequences in each of the genomes. The first HMM pattern was based on 268 RT domain sequences from the Gypsy Database 2.0 (Llorens et al. 2011). A second, non-LTR retrotransposon-specific HMM pattern was created for ORF2 sequences from L1, L2, L3, RTE, and Penelope elements. To minimize computational time, all ORFs longer than 300 aa in the Tasmanian devil and opossum genomes were screened with the HMM profiles using HMMER3 (Eddy 2011). The resulting ORFs identified by the HMM patterns were extracted from the opossum and Tasmanian devil genomes and the sequences were masked using Repeatmasker (Smit et al. 1996) using the Mammalia library. The copy number of the RT-derived sequences was plotted against length.
Search for Recently Integrated SINEs in two Tasmanian Devil Genomes
Retrotransposition events can be detected by pairwise alignment between two closely related individuals. Recent retrotransposon insertions will be present in one genome and absent from the other. This method was used to identify recently integrated WSINE1 loci by aligning a masked Tasmanian devil genome (Murchison et al. 2012) to an unmasked Tasmanian devil genome (Miller et al. 2011). We then aligned flanking sequences (500 nt) to identify orthologous WSINE1 insertions between the two genomes. Pairwise alignments of insertions and genomic flanks were performed using the needle program from the EMBOSS package (Rice et al. 2000). All pairwise alignments were screened for gaps using a custom PERL script. In addition, a custom PERL script was used to calculate the number of identical full-length WSINE1s in the Tasmanian devil genome and the number of identical full-length SINE1_Mdos (supplementary fig. S3, Supplementary Material online) in the opossum genome.
Cell Culture, DNA, and RNA Isolation
The marsupial cell line SC-11 (Sminthopsis crassicaudata) from ECACC was grown to confluence in T-75 flasks in Dulbecco’s modified Eagle’s media supplemented with 10% FCS at 37°C with 5% CO2 and 80% humidity. RNA was isolated using the RNeasy Mini kit (Qiagen) and the samples were DNaseI treated prior to cDNA amplification. Genomic DNA was isolated from tissues of different marsupial species (supplementary table S5, Supplementary Material online) using a standard phenol–chloroform method (Sambrook and Russell 2001). Touchdown PCR was used to amplify intron, retrotransposon, or DNA transposon sequences with specific primers from genomic DNA using Taq polymerase (VWR) unless stated otherwise. The amplicons were sequenced using a BigDye terminator sequencing kit 3.1 (Applied Biosystems, Foster City, CA) and analyzed with an ABI 3730 DNA Analyzer (Applied Biosystems).
Phylogenetic Reconstruction of Dasyuromorphia Based on Retrotransposon Insertions
Exon-based primers were designed using the Tasmanian devil genome sequence spanning 158 introns, which were either containing retrotransposons (WSINE1, WALLSI1, L1-1_SH) or not including any retrotransposon sequences. The introns were experimentally investigated in eight dasyuromorphian species representing each of the major subfamilies (supplementary tables S5 and S6, Supplementary Material online). The WSINE1, WALLSI1, and L1 elements were chosen, based on transposition-in-transposition (TinT; Churakov et al. 2010) analysis, which identified them as the youngest elements in the Tasmanian devil genome (Nilsson et al. 2012). WALLSI2 is specific to the Australian order Diprotodontia and is not present in the Tasmanian devil genome (Nilsson et al. 2012). The introns were amplified using touch-down PCR and the resulting products were inspected on an agarose gel. Introns that yielded different sized PCR products, across species, were sequenced directly or cloned into the TA-vector pDRIVE (Qiagen). The resulting sequences were manually aligned using Se-Al (Rambaut 2002). We then assessed the value of potentially informative phylogenetic markers by inspecting the type and orientation of retrotransposon insertions, presence of TSDs, and the homology of the flanking introns and flanking exons. If the integrity of the flanks and TSDs could not be confirmed, the marker was discarded. In special cases where the element was obviously deleted in a species that was far from the point of insertion in the phylogeny (indicated by a missing part of the flank or both TSDs), it was denoted as a deletion event (D) (supplementary table S2, Supplementary Material online). The markers were plotted on a previously established (Meredith et al. 2008; Nilsson et al. 2012) phylogenetic tree of Dasyuromorphia (fig. 4). To provide statistically significant support for the investigated branches, at least three independent retrotransposon insertions were required (Waddell et al. 2001).
Evolutionary Investigation of DNA Transposons in Australian Marsupials
To investigate DNA transposon activity in the Tasmanian devil and opossum genomes, all DNA transposon sequences longer than 1,500 nt were extracted from the premasked genomes using a PERL script. To target recently active full-length DNA transposons, a cut-off value of 1,500 nt was chosen. The extracted DNA transposon sequences were grouped according to type and aligned using MAFFT (Multiple Alignment using Fast Fourier Transform). GETORF (EMBOSS; Rice et al. 2000) was used to identify ORFs for all six reading frames in the DNA transposon sequences. The nucleotide distance was estimated using ML applying gamma correction with MEGA5 (Tamura et al. 2011). We investigated the phylogenetic distribution of the hAT (hobo/Ac/Tam3) DNA transposon OposCharlie1_Das (Gilbert et al. 2013; RepBase ID hAT-2) among marsupials. We performed PCR amplification of a 533-nt-long fragment in eight dasyuromorphians and nine species from three other Australian orders, (Diprotodontia (five species), Peramelemorphia (three species), Notoryctemorphia (one species) as well as the South American opossum (Monodelphis domestica) and North American opossum (Didelphis virginiana).
cDNA was generated from total RNA isolated from the fat-tailed dunnart (Sminthopsis crassicaudata) cell line SC-11 using the Two-step Long Range cDNA kit (Qiagen). The ORF coding for OC1 transposase was amplified through RT-PCR with four different primers (supplementary table S7, Supplementary Material online) specific for the ORF using rTAQ DNA polymerase (TAKARA). The primer located in the 5′-terminal region of the transposase-coding ORF started with the ORFs ATG start codon, whereas the primer located in the 3′-region was located 10 nt upstream of the stop codon (supplementary table S7, Supplementary Material online). The 1,909-nt product was cloned into the TOPO TA vector, and 12 clones were sequenced from the fat-tailed dunnart. The resulting transposase sequences were inspected for ORFs using GETORF (Rice et al. 2000).
The Tasmanian devil and fat-tailed dunnart transposase sequences were aligned with other mammalian and reptile OC1 sequences and their relationships were analyzed using an ML approach in TREEFINDER (Jobb et al. 2004). An evolutionary model was selected using the ModelProposer function in Treefinder. We screened for the presence of the hAT-1_MEu transposon using a 611-nt-long fragment (supplementary table S7, Supplementary Material online) in the same 19 species as the OC1 DNA transposon.
Data Access
The sequences for each of the 18 introns containing phylogenetically informative retrotransposon insertions are deposited in GenBank under the accession numbers: LM651251–LM651362. All sequence alignments are available as supplementary data set S1, Supplementary Material online.
Supplementary Material
Supplementary data sets S1 and S2, tables S1–S7, and figures S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank P. Heinemann and N. Schreck for computational work, Z. Ivics for helpful discussions, and A.F.A. Smit with help to identify the ERV1 element. F. Lammers kindly calculated the repeat landscape of the Tasmanian devil genome. Jon Baldur Hlíberg (www.fauna.is) painted the animals in figure 4. The phylogenetic reconstruction of the order Dasyuromorphia using retrotransposons was part of a master thesis completed in September 2012 and presented as a poster at the Deutsche Zoologische Gesellshaft (DZG) conference at the Senckenberg Museum, Frankfurt in 2012. This work was supported by Deutsche Forschungsgemeinschaft (DFG) grant NI-1284/1-1 to M.N.
References
- Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E. Assemblies: the good, the bad, the ugly. Nat Methods. 2011;8:59–60. doi: 10.1038/nmeth0111-59. [DOI] [PubMed] [Google Scholar]
- Brown OJF. Tasmanian devil (Sarcophilus harrisii) extinction on the Australian mainland in the mid-Holocene: multicausality and ENSO intensification. Alcheringa. 2006;31:49–57. [Google Scholar]
- Cantrell MA, Grahn RA, Scott L, Wichman HA. Isolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques. 2000;29:1310–1316. doi: 10.2144/00296rr02. [DOI] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics. 2008;178:393–404. doi: 10.1534/genetics.107.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- Casavant NC, Scott L, Cantrell MA, Wiggins LE, Baker RJ, Wichman HA. The end of the LINE?: lack of recent L1 activity in a group of South American rodents. Genetics. 2000;154:1809–1817. doi: 10.1093/genetics/154.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Grundmann N, Kuritzin A, Brosius J, Makałowski W, Schmitz J. A novel web-based TinT application and the chronology of the Primate Alu retroposon activity. BMC Evol Biol. 2010;10:376. doi: 10.1186/1471-2148-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Belov K. A comparative genomics approach to understanding transmissible cancer in Tasmanian devils. Annu Rev Genomics Hum Genet. 2012;13:207–222. doi: 10.1146/annurev-genom-090711-163852. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA, Hutchison CA, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson IK, Cantrell MA, Scott L, Wichman HA. Retrofitting the genome: L1 extinction follows endogenous retroviral expansion in a group of muroid rodents. J Virol. 2011;85:12315–12323. doi: 10.1128/JVI.05180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Wakefield MJ, Kohany O, Gu W, Batzer MA, Pollock DD, Jurka J. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodsi M, Liu B, Pop M. DNACLUST: accurate and efficient clustering of phylogenetic marker genes. BMC Bioinformatics. 2011;12:271. doi: 10.1186/1471-2105-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Waters P, Feschotte C, Schaack S. Horizontal transfer of OC1 transposons in the Tasmanian devil. BMC Genomics. 2013;14:134. doi: 10.1186/1471-2164-14-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolevsky KP, Vassetzky NS, Kramerov DA. Bov-B-mobilized SINEs in vertebrate genomes. Gene. 2008;407:75–85. doi: 10.1016/j.gene.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Grahn RA, Rinehart TA, Cantrell MA, Wichman HA. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet Genome Res. 2005;110:407–415. doi: 10.1159/000084973. [DOI] [PubMed] [Google Scholar]
- Green P. University of Washington; 1996. Cross_match, version 0.990329. Available from: http://www.phrap.org. [Google Scholar]
- Green P. 2x genomes—does depth matter? Genome Res. 2007;17:1547–1549. doi: 10.1101/gr.7050807. [DOI] [PubMed] [Google Scholar]
- Gu W, Ray DA, Walker JA, Barnes EW, Gentles AJ, Samollow PB, Jurka J, Batzer MA, Pollock DD. SINEs, evolution and genome structure in the opossum. Gene. 2007;396:46–58. doi: 10.1016/j.gene.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH., Jr Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CE, Baars C, Hesterman H, Hocking GJ, Jones ME, Lazenby B, Mann D, Mooney N, Pemberton D, Pyecroft S, et al. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol Conserv. 2006;131:307–324. [Google Scholar]
- Henson J, Tischler G, Ning Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012;13:901–915. doi: 10.2217/pgs.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras SR, Macias S, Cáceres JF, Garcia-Perez JL. Control of mammalian retrotransposons by cellular RNA processing activities. Mob Genet Elements. 2014;4:e28439. doi: 10.4161/mge.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Hajirasouliha I, Dao P, Hach F, Yorukoglu D, Alkan C, Eichler EE, Sahinalp SC. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics. 2010;26:i350–i357. doi: 10.1093/bioinformatics/btq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Konkel MK, Prado-Martinez J, Chiatante G, Herraez IH, Walker JA, Nelson B, Alkan C, Sudmant PH, Huddleston J, et al. Rates and patterns of great ape retrotransposition. Proc Natl Acad Sci U S A. 2013;110:13457–13462. doi: 10.1073/pnas.1310914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobb G, von, Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jurka J. L1-1_SH. Repbase reports. 2011a;11:2168–2168. [Google Scholar]
- Jurka J. hAT-1_Meu. Repbase reports. 2011b;11:653–653. [Google Scholar]
- Jurka J, Zietkiewicz E, Labuda D. Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 1995;23:170–175. doi: 10.1093/nar/23.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Wong K, Adams DJ. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics. 2013;29:389–390. doi: 10.1093/bioinformatics/bts697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski C, Wroe S, Westerman M. Molecular evidence for the pattern and timing of cladogenesis in dasyurid marsupials. Zool J Linn Soc. 2000;130:375–404. [Google Scholar]
- Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. doi: 10.1007/BF02101990. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, Futami R, Covelli L, Dominguez-Escriba L, Viu JM, Tamarit D, Aguilar-Rodriguez J, Vicente-Ripolles M, Fuster G, Bernet GP, et al. The Gypsy Database (GyDB) of Mobile Genetic Elements: Release 2.0. Nucl Acids Res. 2011;39(Suppl. 1):D70–D74. doi: 10.1093/nar/gkq1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Westerman M, Case JA, Springer MS. A phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J Mamm Evol. 2008;15:1–36. [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Milinkovitch MC, Helaers R, Depiereux E, Tzika AC, Gabaldón T. 2X genomes—depth does matter. Genome Biol. 2010;11:R16. doi: 10.1186/gb-2010-11-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, et al. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil) Proc Natl Acad Sci U S A. 2011;108:12348–12353. doi: 10.1073/pnas.1102838108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A, Johnson J, Li K, Mobarry C, Sutton G. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Li X, Kapusta A, Mayhew D, Mitra RD, Feschotte C, Craig NL. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc Natl Acad Sci U S A. 2013;110:234–239. doi: 10.1073/pnas.1217548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa M, Nikaido M, Nishihara H, Donnellan S, Austin CC, Okada N. Newly discovered young CORE-SINEs in marsupial genomes. Gene. 2008;407:176–185. doi: 10.1016/j.gene.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Murchison EP. Clonally transmissible cancers in dogs and Tasmanian devils. Oncogene. 2008;27(Suppl. 2):S19–S30. doi: 10.1038/onc.2009.350. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagome M, Solovieva E, Takahashi A, Yasue H, Hirochika H, Miyao A. Transposon Insertion Finder (TIF): a novel program for detection of de novo transpositions of transposable elements. BMC Bioinformatics. 2014;15:71. doi: 10.1186/1471-2105-15-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MA, Churakov G, Sommer M, Tran NV, Zemann A, Brosius J, Schmitz J. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 2010;8:e1000436. doi: 10.1371/journal.pbio.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MA, Janke A, Murchison EP, Ning Z, Hallström BM. Expansion of CORE-SINEs in the genome of the Tasmanian devil. BMC Genomics. 2012;13:172. doi: 10.1186/1471-2164-13-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2010;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Pace JK, 2nd The evolutionary history and genomic impact of mammalian DNA transposons [PhD thesis] 2008 University of Texas at Arlington. [Google Scholar]
- Platt RN, 2nd, Ray DA. A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene. 2012;500:47–53. doi: 10.1016/j.gene.2012.03.051. [DOI] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Agencourt Sequencing Team, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Se-Al: sequence alignment editor. 2002. Available from: http://tree.bio.ed.ac.uk/software/seal.
- Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rinehart TA, Grahn RA, Wichman HA. SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet Genome Res. 2005;110:416–425. doi: 10.1159/000084974. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schumann GG, Gogvadze EV, Osanai-Futahashi M, Kuroki A, Münk C, Fujiwara H, Ivics Z, Buzdin AA. Unique functions of repetitive transcriptomes. Int Rev Cell Mol Biol. 2010;285:115–188. doi: 10.1016/B978-0-12-381047-2.00003-7. [DOI] [PubMed] [Google Scholar]
- Shimamura M, Yasue H, Ohshima K, Abe H, Kato H, Kishiro T, Goto M, Munechika I, Okada N. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- Smit AF, Tóth G, Riggs AD, Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J Mol Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010. Available from: http://www.repeatmasker.org.
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:research0052. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform Ser Workshop Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci U S A. 2013;110:1012–1016. doi: 10.1073/pnas.1205856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirkness EF. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Woodburne MO, Case JA. Dispersal, vicariance, and the Late Cretaceous to early Tertiary land mammal biogeography from South America to Australia. J Mammal Evol. 1996;3:121–161. [Google Scholar]
- Yang L, Brunsfeld J, Scott L, Wichman H. Reviving the dead: history and reactivation of an extinct l1. PLoS Genet. 2014;10:e1004395. doi: 10.1371/journal.pgen.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman A, Churkaov G, Donellan S, Grützner F, Fangqing Z, Brosius J, Schmitz J. Ancestry of the Australian termitivorous Numbat. Mol Biol Evol. 2013;30:1041–1045. doi: 10.1093/molbev/mst032. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.