Abstract
This scientific commentary refers to ‘Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy’ by Miller-Delaney et al. (10.1093/brain/awu373).
This scientific commentary refers to ‘Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy’ by Miller-Delaney et al. (10.1093/brain/awu373).
Epigenetic modification of the genome is a powerful mechanism for regulation of RNA expression. For the large majority of non-neoplastic brain disorders, a lack of human brain tissue suitable for complementary epigenetic and RNA expression analyses poses a major challenge for translating data from experimental models to human disease. Temporal lobe epilepsy is a unique exception. Tissue from pharmacoresistant patients who undergo epilepsy surgery allow singular access for comprehensive molecular genetic studies. In this issue of Brain, Miller-Delaney and colleagues use this intriguing resource to report fundamental new insights into epigenetic regulation in human brain tissue (Miller-Delaney et al., 2014).
Epigenetic dynamics under disease conditions are generally thought to represent wide-scale alterations in the expression of multiple classes of RNA. In the context of epilepsy, however, most studies of epigenetic modifications have focused on distinct structural changes and alterations in the expression of specific mRNAs (Kobow et al., 2009; Ryley Parrish et al., 2013). Much less is known about genome-wide epigenetic alterations and corresponding patterns of RNA expression in the disease. Miller-Delaney and colleagues addressed the challenge of molecular genetic analyses in human brain tissue by comparing genome-wide DNA methylation patterns in the hippocampi of patients with pharmaco-refractory temporal lobe epilepsy with those in autopsy samples of brain tissue from individuals with non-neurological causes of death. The commonest lesion pattern in patients with temporal lobe epilepsy is hippocampal sclerosis, with segmental neurodegeneration and concomitant astrogliosis as pathological hallmarks (Fig. 1A–D). However, patterns of damage vary among individuals (Blümcke et al., 2013) and Miller-Delaney et al. (2014) therefore subdivided their temporal lobe epilepsy group accordingly. Only 146 protein-coding genes were found to show altered DNA methylation in temporal lobe epilepsy, a surprisingly low number when compared to the complexity and functional consequences of this disorder. The vast majority of affected promoters showed hypermethylation, an observation that is in good agreement with the epigenetic data from key animal models of epilepsy (Kobow et al., 2013). In a novel and intriguing finding, Miller-Delaney and colleagues also identified several methylation-sensitive microRNAs and long non-coding RNAs in the brain tissue.
Figure 1.
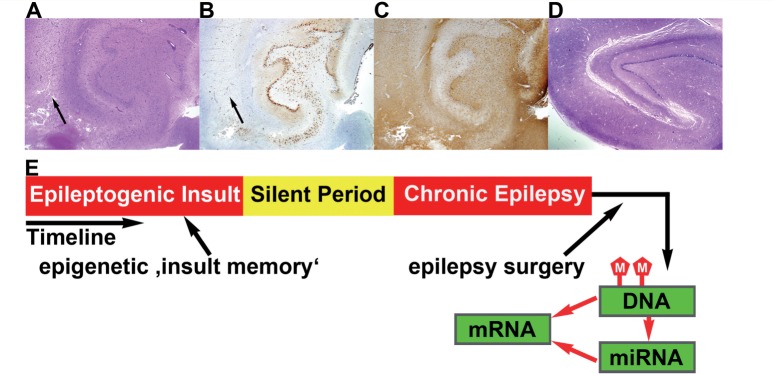
Epigenetic regulatory model in temporal lobe epilepsy. (A) Hippocampal biopsy specimen obtained during surgery in a patient with pharmacoresistant temporal lobe epilepsy (haematoxylin and eosin). The section shows hippocampal sclerosis, reflecting pathological alterations typical of those in the material used by Miller-Delaney et al. (2014). The main alterations comprise neuronal cell loss, particularly in the CA1 area (black arrows) visible in A and B (B: NeuN immunohistochemistry), and reactive astrogliosis (C: glial fibrillary acidic protein immunohistochemistry). (D) An autopsy section (haematoxylin and eosin) showing the hippocampal formation of a patient without neurological disorders, lacking segmental neurodegeneration and gliosis, and typical of an experimental control subject. (E) Differences in cellular composition between subjects with and without epilepsy are reflected in patterns of DNA methylation and mRNA and microRNA expression. The impact of other putative epileptogenic factors on hippocampal DNA methylation patterns constitutes a form of long-term ‘insult-induced memory’. (A–D) Magnification: ×1.25.
By correlating these differential patterns of genome methylation with the corresponding RNA expression patterns, Miller-Delaney and co-workers managed to fully exploit the potential of their experimental approach. Their analyses revealed a surprisingly weak overall correlation between promoter epigenetic dynamics and the expression of coding transcripts. However, for certain RNAs, promoter methylation patterns had a striking impact in human (epilepsy) brain tissue. This was true in particular for several microRNAs, as well as for certain non-coding RNAs. This difference in the degree of correspondence between epigenetic promoter programming and expression levels for coding mRNAs versus microRNAs might indicate that expression levels of the latter are more directly influenced by changes in promoter methylation—perhaps even under physiological conditions in the human brain. The abundance of coding mRNAs is regulated not only by promoter activity but also by factors such as the activity of microRNAs, which might complicate the link between epigenetics and mRNA levels. By contrast, the link between epigenetics and microRNA levels might be more direct. This differential impact of methylation on mRNA versus microRNA abundance could have important consequences for the development of novel therapeutic strategies, with respect to predicting the molecular effects of targeting different RNA classes by epigenetic means in CNS disorders.
Epigenetic regulation has some particular characteristics that make it exceptionally attractive as a putative pathomechanism of temporal lobe epilepsy. One of these is the potential for long-lasting, stable effects on gene expression that outlive an initial transient signal. This may be especially relevant for post-mitotic neurons, which are subject to various insults with short- to long-lasting effects on their activity and connectivity (Guan et al., 2002). In temporal lobe epilepsy, seizures do not generally start at birth in affected individuals but develop later in life (Pitkänen and Engel, 2014). Many patient histories reveal a transient insult in early childhood, which is followed by a silent interval, potentially lasting several years, before recurrent seizures emerge. What type of molecular modifications can be that long-lasting? Epigenetic remodelling could in fact constitute the brain’s ‘memory of transient epileptogenic insults’ (Fig. 1E). However, many short- and long-term influences on DNA methylation will be reflected in an epigenetic pattern. In addition to transient potentially epileptogenic insults, the pattern of DNA methylation could also signal an increased disease risk owing to, for example, genomic imprinting or the gestational environment, which has been suggested to have pathogenetic relevance in other CNS disorders (Jiao et al., 2013), or to differences in the cellular composition of the temporal lobes of patients with epilepsy versus control subjects (Fig. 1). More sophisticated experimental approaches that consider patients’ family epigenetics and that use bioinformatics to control for cellular admixtures might be useful in the future (Guintivano et al., 2013). Furthermore, epigenetic patterns may be rather dynamic (El-Osta et al., 2008), and can also reflect relatively short-term changes, which can complicate data interpretation.
The work has some limitations, which Miller-Delaney and colleagues have obviously considered. One inherent difficulty with approaches that analyse human brain tissue relates to the choice of controls. Miller-Delaney et al. used autopsy brain tissue, but while DNA-methylation analyses can be carried out with this material, this approach clearly limits the comparability of genomic methylation dynamics and RNA expression patterns. Furthermore, the control and temporal lobe epilepsy cohorts will be subject to the methodological and statistical limitations inherent in large-scale studies based on such small sized groups. Miller-Delaney and colleagues have taken great care to control for group effects by minimizing differences in key parameters such as gender and age, wherever possible. But this clearly has limitations. As a result of the rather young age of the patients with temporal lobe epilepsy, the control group contains individuals who died from cardiovascular disease at unusually young ages. This raises suspicions of inherited impairments that increase cardiovascular disease risk and burden—might these have a potential epigenetic impact? Furthermore, the effects of antiepileptic pharmacotherapy in the group with temporal lobe epilepsy cannot easily be controlled for, and may be substantial. Stressing the fact that the present work is a pilot study, Miller-Delaney and colleagues fully acknowledge these shortcomings and suggest follow-up multicentre studies, which we eagerly anticipate will provide more detailed insights into this fast developing field of research with intriguing clinical implications.
References
- Blümcke I, Thom M, Aronica E, Armstrong DD, Bartolomei F, Bernasconi A, et al. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: a task force report from the ILAE Commission on Diagnostic Methods. Epilepsia. 2013;54:1315–29. doi: 10.1111/epi.12220. [DOI] [PubMed] [Google Scholar]
- El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409–17. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–93. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Opal MD, Dulawa SC. Gestational environment programs adult depression-like behavior through methylation of the calcitonin gene-related peptide gene. Mol Psychiatry. 2013;18:1273–80. doi: 10.1038/mp.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobow K, Jeske I, Hildebrandt M, Hauke J, Hahnen E, Buslei R, et al. Increased reelin promoter methylation is associated with granule cell dispersion in human temporal lobe epilepsy. J Neuropathol Exp Neurol. 2009;68:356–64. doi: 10.1097/NEN.0b013e31819ba737. [DOI] [PubMed] [Google Scholar]
- Kobow K, Kaspi A, Harikrishnan KN, Kiese K, Ziemann M, Khurana I, et al. Deep sequencing reveals increased DNA methylation in chronic rat epilepsy. Acta Neuropathol. 2013;126:741–56. doi: 10.1007/s00401-013-1168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Delaney SF, Bryan K, Das S, McKiernan RC, Bray IM, Reynolds JP, et al. Differential DNA methylation profiles of coding and noncoding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain. 2015;138:601–16. doi: 10.1093/brain/awu373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitkänen A, Engel J., Jr Past and present definitions of epileptogenesis and its biomarkers. Neurotherapeutics. 2014;11:231–41. doi: 10.1007/s13311-014-0257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryley Parrish R, Albertson AJ, Buckingham SC, Hablitz JJ, Mascia KL, Davis Haselden W, et al. Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience. 2013;248:602–19. doi: 10.1016/j.neuroscience.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]


