Abstract
The bacteriophage population is large, dynamic, ancient, and genetically diverse. Limited genomic information shows that phage genomes are mosaic, and the genetic architecture of phage populations remains ill-defined. To understand the population structure of phages infecting a single host strain, we isolated, sequenced, and compared 627 phages of Mycobacterium smegmatis. Their genetic diversity is considerable, and there are 28 distinct genomic types (clusters) with related nucleotide sequences. However, amino acid sequence comparisons show pervasive genomic mosaicism, and quantification of inter-cluster and intra-cluster relatedness reveals a continuum of genetic diversity, albeit with uneven representation of different phages. Furthermore, rarefaction analysis shows that the mycobacteriophage population is not closed, and there is a constant influx of genes from other sources. Phage isolation and analysis was performed by a large consortium of academic institutions, illustrating the substantial benefits of a disseminated, structured program involving large numbers of freshman undergraduates in scientific discovery.
DOI: http://dx.doi.org/10.7554/eLife.06416.001
Research organism: Virus
eLife digest
Viruses are unable to replicate independently. To generate copies of itself, a virus must instead invade a target cell and commandeer that cell's replication machinery. Different viruses are able to invade different types of cell, and a group of viruses known as bacteriophages (or phages for short) replicate within bacteria. The enormous number and diversity of phages in the world means that they play an important role in virtually every ecosystem.
Despite their importance, relatively little is known about how different phage populations are related to each other and how they evolved. Many phages contain their genetic information in the form of strands of DNA. Using genetic sequencing to find out where and how different genes are encoded in the DNA can reveal information about how different viruses are related to each other. These relationships are particularly complicated in phages, as they can exchange genes with other viruses and microbes.
Previous studies comparing the genomes—the complete DNA sequence—of reasonably small numbers of phages that infect the Mycobacterium group of bacteria have found that the phages can be sorted into ‘clusters’ based on similarities in their genes and where these are encoded in their DNA. However, the number of phages investigated so far has been too small to conclude how different clusters are related. Are the clusters separate, or do they form a ‘continuum’ with different genes and DNA sequences shared between different clusters?
Here, Pope, Bowman, Russell et al. compare the individual genomes of 627 bacteriophages that infect the bacterial species Mycobacterium smegmatis. This is by far the largest number of phage genomes analyzed from a single host species. The large number of genomes analyzed allowed a much clearer understanding of the complexity and diversity of these phages to be obtained. The isolation, sequencing and analysis of the hundreds of M. smegmatis bacteriophage genomes was performed by an integrated research and education program, called the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program. This enabled thousands of undergraduate students from different institutions to contribute to the phage discovery and sequencing project, and co-author the report. SEA-PHAGES therefore shows that it is possible to successfully incorporate genuine scientific research into an undergraduate course, and that doing so can benefit both the students and researchers involved.
The results show that while the genomes could be categorized into 28 clusters, the genomes are not completely unrelated. Instead, a spread of diversity is seen, as genes and groups of genes are shared between different clusters. Pope, Bowman, Russell et al. further reveal that the phage population is in a constant state of change, and continuously acquires genes from other microorganisms and viruses.
Introduction
Bacteriophages are the dark matter of the biological universe, forming a vast, ancient, dynamic, and genetically diverse population, replete with genes of unknown function (Pedulla et al., 2003). Phages are the most abundant organisms in the biosphere, and the ∼1031 tailed phage particles participate in ∼1023 infections per second on a global scale, with the entire population turning over every few days (Suttle, 2007). The population is not only vast and dynamic, but comparisons of virion structures suggest that it is also extremely old (Krupovic and Bamford, 2010). It is thus not surprising that bacteriophages are genetically highly diverse, although their comparative genomics has lagged behind that of other microbes, largely due to the lack of individual isolates for genomic analyses (Hatfull and Hendrix, 2011). To date, there are approximately 2000 completely sequenced bacteriophage genomes in the GenBank database, a small number relative to the more than 30,000 sequenced prokaryotic genomes (http://www.ncbi.nlm.nih.gov/genome/browse/), in spite of phage genomes being only 1–5% of the size of their host genomes.
Double-stranded DNA tailed phages are proposed to have evolved with common ancestry but with different phages having differential access to a large common gene pool (Hendrix et al., 1999). Phage genomes are typified by their mosaic architectures generated by gene loss and gain through horizontal genetic exchange; however, the parameters influencing access to the common gene pool are numerous and likely include host range, genome size, replication mode, and life style (temperate vs lytic). Migration to new hosts is probably common, but is affected by local host diversity and mutation rates, as well as resistance mechanisms such as receptor availability, restriction, CRISPRs, and abortive infection systems (Buckling and Brockhurst, 2012; Jacobs-Sera et al., 2012; Hoskisson et al., 2015). Constraints on gene acquisition may also be imposed by synteny—particularly among virion structural genes—and by size limits of DNA packaging (Juhala et al., 2000; Hatfull and Hendrix, 2011).
We have previously described comparative analyses of modest numbers of mycobacteriophages and shown that they can be sorted by nucleotide sequence and gene content comparisons into groups of closely related genomes referred to as ‘clusters’ (designated Cluster A, B, C, etc.); phages without any close relatives are referred to as ‘singletons’. Some of the clusters can be further divided into subclusters (e.g., Subcluster A1, A2, A3, etc.) according to nucleotide sequence relatedness (Pedulla et al., 2003; Hatfull et al., 2006, 2010; Pope et al., 2011b). The genomes are mosaic whereby individual phages are constructed as assemblages of modules, many of which are single genes (Pedulla et al., 2003). Each mycobacteriophage cluster has features particular to that cluster (e.g., regulatory systems, repeated sequences, tRNA genes, etc. [Pope et al., 2011a, 2011b, 2013, 2014a, 2014b]), but because of the pervasive mosaicism, the relationships among phages within clusters and between clusters are complex. Collections of phages have been isolated on other hosts such as Bacillus spp., Escherichia coli, Pseudomonas spp., Propionibacterium spp. and Staphylococcus spp. (Kwan et al., 2005, 2006; Kropinski et al., 2007; Marinelli et al., 2012; Hatfull et al., 2013; Grose and Casjens, 2014; Grose et al., 2014; Lee et al., 2014) and these can be similarly divided into clusters based on DNA similarity. Recent analysis of 337 phages infecting 31 bacterial species within the Enterobacteriaceae (Grose and Casjens, 2014) reveals 56 clusters of phage genomes. It is thus clear that there is substantial diversity within the phage population, even when comparing phages of a common host and which are expected to be in direct genetic contact with each other in their natural environment (Hatfull and Hendrix, 2011). Nonetheless, the numbers of genomes isolated on a particular host generally are too small to define the nature and the size of the populations at large with any substantial resolution.
Viral metagenomic studies provide valuable insights into phage diversity and population dynamics, but typically generate few complete genome sequences or any specific information relating viral genomes to specific bacterial hosts (Hambly and Suttle, 2005; Rodriguez-Brito et al., 2010; Mokili et al., 2012). A recent analysis of Synechococcus phages using metagenomic analysis coupled with viral tagging showed that there are multiple ‘populations’ of these phages (similar to the clusters described above), but suggested that these represent distinct groups of related phages rather than a continuous spectrum of diversity (Deng et al., 2014). This differs from prior predictions that the phage population as a whole likely spans a continuum of diversity—albeit with uneven representation of different groups of related phages—because of genomic mosaicism (Hendrix, 2003; Hatfull, 2010, 2012). However, as the Synechococcus phage data are derived from a single sample using a single host, it is unclear if this extends to phages of other hosts (Deng et al., 2014).
Here we describe the comparative analysis of a large number of completely sequenced mycobacteriophage genomes and demonstrate that they represent a spectrum of diversity and do not constitute discrete populations. Rarefaction analyses of their constituent genes are consistent with populations of gene families shared among mycobacteriophages being augmented by the introduction of new gene families from outside sources. The assembling of a large and highly informative collection of bacteriophages by a consortium of students and faculty at multiple institutions demonstrates that a course-based research experience (CRE) can be successfully implemented at large scale without compromising the authenticity or richness of a scientific investigation imbued with discovery and project ownership.
Results and discussion
A genome-by-genome approach to defining phage diversity
Exploring phage diversity using a genome-by-genome approach has notable advantages and some potential disadvantages. The main advantage is that complete genome sequences give information about genome length and composition, providing key insights into genome mosaicism and how genome segments are shared and exchanged. A difficulty is that there are not large extant phage collections available for most bacterial hosts, and isolation, purification, and characterization of phages can be slow and time-consuming. Because isolation typically requires plaque formation and growth in the laboratory, some naturally occurring phages may escape isolation using standard methods. Thus, although the diversity of phages isolated and propagated in the laboratory may not capture all types of phage, it represents a minimum, not a maximum, index of diversity.
Authentic research in a CRE
The 2012 report from the President's Council of Advisors on Science and Technology (PCAST) focused on the poor retention of undergraduate students in science, technology, engineering and mathematics (STEM) as an impediment to meeting US economic demands (PCAST, 2012). One of the PCAST recommendations is to replace traditional introductory laboratory courses with research-based experiences that would inspire freshman students and promote STEM retention. A powerful strategy is to engage students in scientific discovery through CREs. The successful implementation of this strategy depends on (i) identifying research questions that can engage students in contributing genuine advances in scientific knowledge without requiring prior expert knowledge, and (ii) designing the project so that large numbers of students can participate in a meaningful fashion.
We have previously described the Howard Hughes Medical Institute (HHMI) Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program, in which beginning undergraduate students isolate, purify, sequence, annotate, and compare bacteriophages, and have described its educational advantages (Jordan et al., 2014). By taking advantage of the massive diversity of the phage population so that each student can isolate a unique phage, the program encourages student ownership of their science. And because the collective discoveries by many students generate new scientific insights, the program creates a scientific community of students engaged in authentic research.
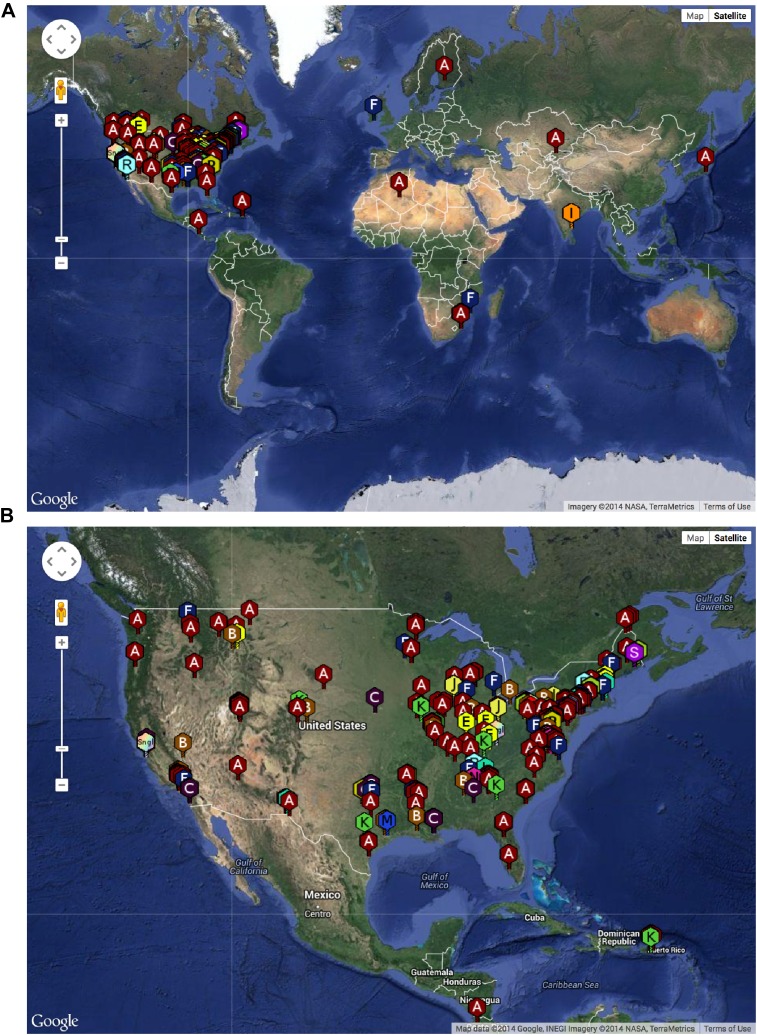
The SEA-PHAGES program has contributed to the growth of the collection of sequenced mycobacteriophages to nearly 700 individual isolates (http://phagesdb.org), of which 627 were selected for a detailed analysis (Supplementary file 1). This is by far the largest collection of sequenced phage genomes for any single host and thus promises to substantially advance our understanding of phage diversity. The phages were isolated using either direct plating or by enrichment using Mycobacterium smegmatis mc2155 as a host, and sequenced using next-generation approaches (see ‘Materials and methods’). More than 5000 students—primarily freshmen—at 74 institutions have been involved since inception of the SEA-PHAGES program in 2008, and the phages isolated represent a broad geographical distribution (Figure 1) and a variety of viral morphotypes (http://phagesdb.org). The new insights gained from comparative genomic analyses of these phages—as described below—demonstrate the effectiveness of viral discovery and genomics as a model for CRE development.
Figure 1. Geographical distribution of sequenced mycobacteriophages.
(A) Locations of sequenced mycobacteriophages across the globe. (B) Locations of sequenced mycobacteriophages across the United States. Colors and letter designations on the isolates refer to the cluster to which the genomes belong. Data from www.phagesdb.org.
Assembling mycobacteriophages into clusters and subclusters
Using previously reported parameters based primarily on nucleotide sequence similarity spanning >50% genome length (Hatfull et al., 2006), the 627 genomes were assembled into 20 clusters (A–T) and eight singletons (with no close relatives) (Figure 2, Supplementary file 1); 11 clusters were subdivided into 2 to 11 subclusters (Table 1). There is considerable variation in cluster size with substantial differences in the numbers of genomes in each cluster (2–232), but there is relatively little variation in either genome length or the numbers of genes per genome in any given cluster (Table 1). Cluster assignment is of practical utility and is generally robust, with clustered phages typically sharing genome architectures, as noted for the Enterobacteriacea (Grose and Casjens, 2014). For example, Cluster A phages are similar in size and transcriptional organization, and share an unusual immunity system (Brown et al., 1997; Pope et al., 2011b). Cluster M phages all contain large numbers of tRNA genes (Pope et al., 2014a), Cluster K (Pope et al., 2011a) and Cluster O (Cresawn et al., 2015) phages have different but characteristic repeated sequences, and Cluster J phages have an unusual capsid with a triangulation (T) number of 13 (Pope et al., 2013). Therefore, the organization of related mycobacteriophages into clusters provides a framework for identifying and interpreting gene trafficking within and among potentially distinct groups of genomes.
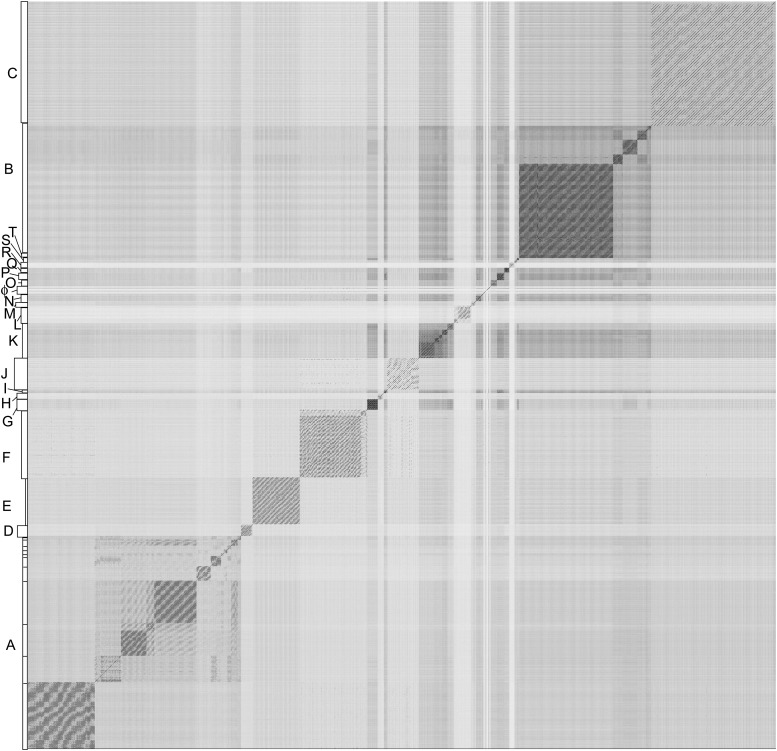
Figure 2. Nucleotide sequence comparison of 627 mycobacteriophages displayed as a dotplot.
Complete genome sequences of 627 mycobacteriophages were concatenated into a single file which was compared with itself using Gepard (Krumsiek et al., 2007) and displayed as a dotplot using default parameters (word length, 10). The order of the genomes is as listed in Supplementary file 1. Nucleotide similarity is a primary component in assembling phages into clusters, which typically requires evident DNA similarity spanning more than 50% of the genome lengths.
DOI: http://dx.doi.org/10.7554/eLife.06416.004
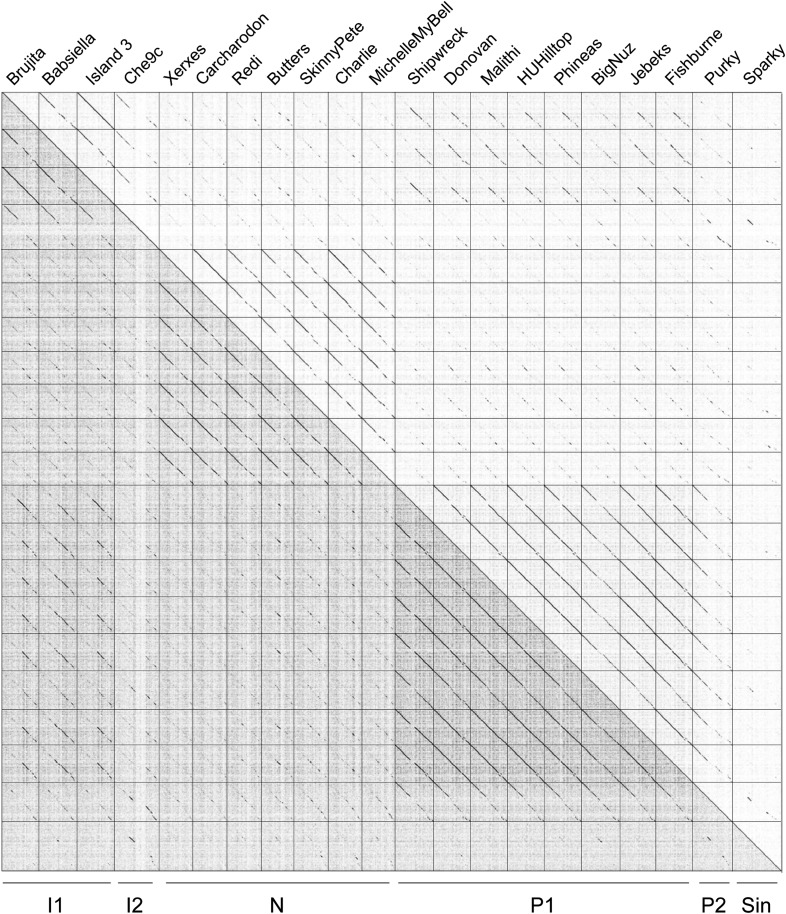
Figure 2—figure supplement 1. Dotplot of phages in Clusters I, N, P and the singleton Sparky.
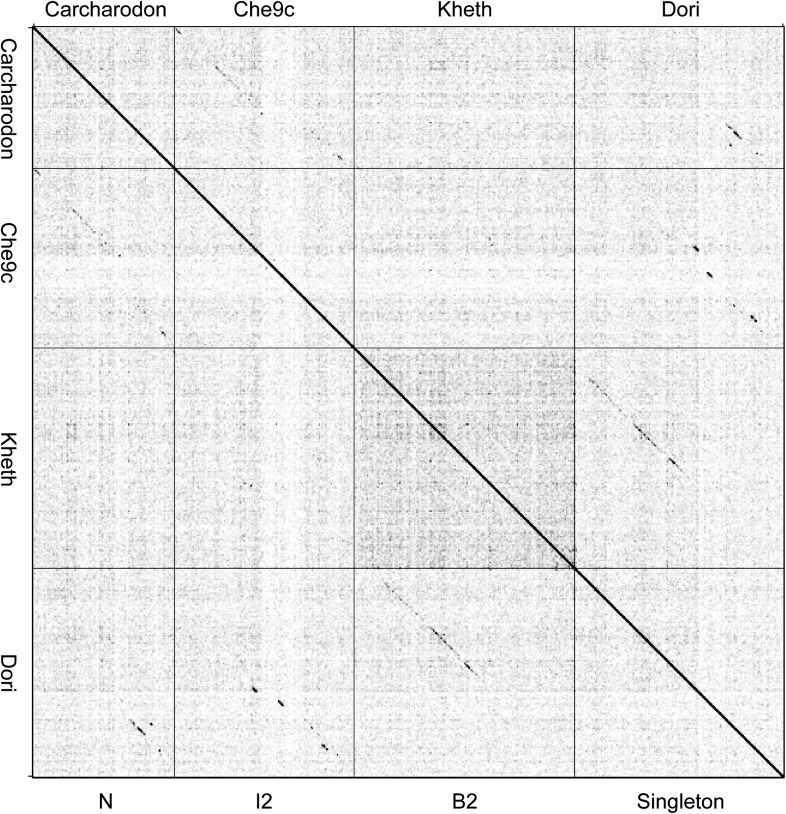
Figure 2—figure supplement 2. Dotplot of Carcharodon, Che9c, Kheth, and Dori.
Figure 2—figure supplement 3. Dotplot of Corndog, Brujita, SG4, Yoshi, and MooMoo.
Table 1.
Diversity and genetic isolation of mycobacteriophage genome clusters
| Cluster | # Subclusters | # Genomes | Average # genes* | Average length (bp) | Total phams† | Total genes | CLASP‡ | CAP§ | CCI# | CII¶ |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 11 | 232 | 90 ± 5.3 | 51,514 | 1085 | 20,880 | 38.3 | 12.4 | 0.08 | 80.2 |
| B | 5 | 109 | 100.4 ± 4.5 | 68,653 | 421 | 10,944 | 66.2 | 23.2 | 0.24 | 81.0 |
| C | 2 | 45 | 231 ± 5.9 | 155,504 | 486 | 10,395 | 89.3 | 29.4 | 0.48 | 84.6 |
| D | 2 | 10 | 89.3 ± 6.4 | 64,965 | 147 | 893 | 88.1 | 64.3 | 0.61 | 71.4 |
| E | 1 | 35 | 141.9 ± 3.4 | 75,526 | 236 | 4967 | 87.2 | 63.8 | 0.60 | 59.3 |
| F | 3 | 66 | 105.3 ± 5.3 | 57,416 | 658 | 6950 | 54.4 | 4.9 | 0.16 | 55.8 |
| G | 1 | 14 | 61.5 ± 1.2 | 41,845 | 72 | 861 | 96.0 | 91.1 | 0.85 | 55.6 |
| H | 2 | 5 | 98.4 ± 5.7 | 69,469 | 207 | 492 | 61.6 | 31.5 | 0.48 | 67.6 |
| I | 2 | 4 | 78 ± 3.7 | 49,954 | 147 | 312 | 58.9 | 35.0 | 0.53 | 23.8 |
| J | 1 | 16 | 239.8 ± 9.3 | 110,332 | 530 | 3776 | 70.8 | 40.1 | 0.45 | 58.5 |
| K | 5 | 32 | 95.7 ± 4.6 | 59,720 | 411 | 3069 | 51.8 | 20.0 | 0.23 | 73.5 |
| L | 3 | 13 | 127.9 ± 6.5 | 75,177 | 246 | 1663 | 78.2 | 50.8 | 0.52 | 72.4 |
| M | 2 | 3 | 141 ± 8.8 | 81,636 | 201 | 423 | 73.5 | 63.0 | 0.70 | 69.2 |
| N | 1 | 7 | 69.1 ± 2.2 | 42,888 | 152 | 484 | 64.1 | 45.6 | 0.45 | 40.8 |
| O | 1 | 5 | 124.2 ± 3.1 | 70,651 | 151 | 621 | 90.6 | 83.3 | 0.82 | 64.2 |
| P | 2 | 9 | 78.8 ± 2.1 | 47,668 | 159 | 709 | 76.1 | 42.3 | 0.50 | 34.0 |
| Q | 1 | 5 | 85.2 ± 3.7 | 53,755 | 90 | 426 | 96.6 | 90.4 | 0.95 | 73.3 |
| R | 1 | 4 | 101.5 ± 2.5 | 71,348 | 117 | 406 | 91.4 | 84.8 | 0.87 | 71.8 |
| S | 1 | 2 | 109 ± 2.0 | 65,172 | 117 | 218 | 91.7 | 91.7 | 0.93 | 70.9 |
| T | 1 | 3 | 66.7 ± 2.4 | 42,833 | 83 | 200 | 86.1 | 82.5 | 0.80 | 62.7 |
| Dori | 1 | 1 | 94 | 64,613 | 94 | 94 | N/A | N/A | N/A | 35.8 |
| DS6A | 1 | 1 | 97 | 60,588 | 96 | 97 | N/A | N/A | N/A | 58.3 |
| Gaia | 1 | 1 | 194 | 90,460 | 193 | 194 | N/A | N/A | N/A | 58.0 |
| MooMoo | 1 | 1 | 98 | 55,178 | 98 | 98 | N/A | N/A | N/A | 31.6 |
| Muddy | 1 | 1 | 71 | 48,228 | 70 | 71 | N/A | N/A | N/A | 71.4 |
| Patience | 1 | 1 | 109 | 70,506 | 109 | 109 | N/A | N/A | N/A | 57.8 |
| Sparky | 1 | 1 | 93 | 63,334 | 93 | 93 | N/A | N/A | N/A | 48.4 |
| Wildcat | 1 | 1 | 148 | 78,296 | 148 | 148 | N/A | N/A | N/A | 69.6 |
Average number of protein-coding genes per genome, with standard deviation.
Total phams is the sum of all phamilies (groups of homologous mycobacteriophage genes) in that cluster.
The Cluster Averaged Shared Phamilies (CLASP) index is the average of the percentages of phamilies shared pairwise between genomes within a cluster.
The Cluster-Associated Phamilies (CAP) index is the percentage of the average number of phamilies per genome within a cluster whose phamilies are present in every cluster member.
The Cluster Cohesion Index (CCI) is generated by dividing the average number of genes per genome by the total number of phamilies (phams) in that cluster.
The Cluster Isolation Index (CII) is the percentage of phams that are present only in that cluster, and not present in other mycobacteriophages.
N/A: Not applicable.
Gene content relationships among sequenced mycobacteriophages
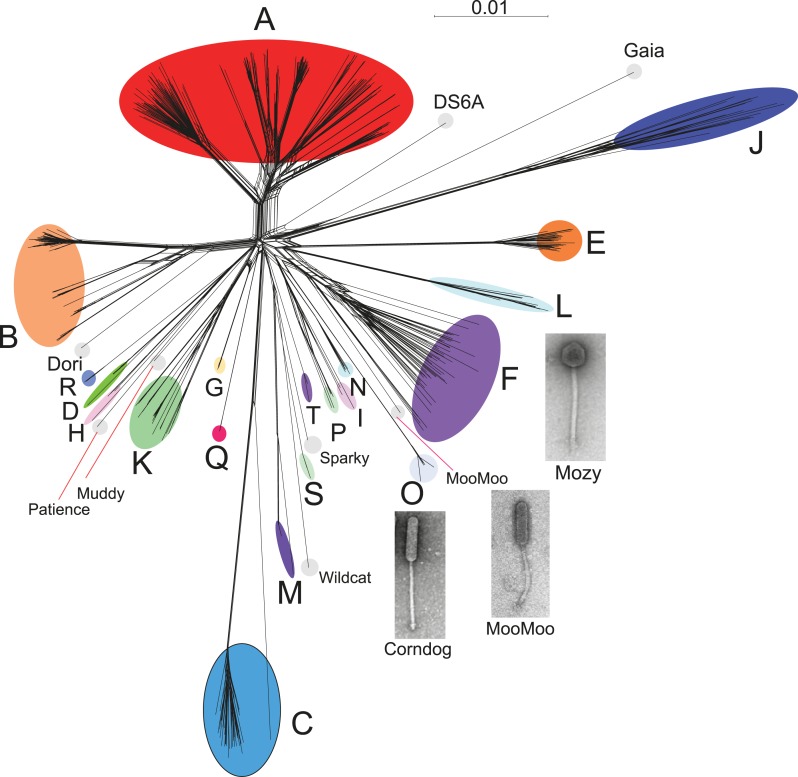
Genome mosaicism is more apparent from comparison of gene product amino acid sequences than nucleotide sequence comparisons because of the accumulation of genome rearrangements over a longer period of evolution, during which indications of DNA similarity are lost. To compare mycobacteriophage gene contents we grouped related genes into protein families (‘phamilies’ or ‘phams’) using Phamerator (Cresawn et al., 2011), which we modified to use kClust (Hauser et al., 2013) so as to easily accommodate the large numbers of comparisons. The 69,633 genes assembled into 5205 phams of which 1613 (31%) are orphams (single-gene phamilies [Hatfull et al., 2010]). Approximately 25% of phams can be assigned functions in viral structure and assembly, DNA metabolism, integration, lysis, and regulation, but the vast majority are of unknown function. Representation of gene content relationships among all 627 phages as a network phylogeny reveals relationships that are in accord with the cluster and subcluster designations derived from nucleotide sequence comparisons (Figure 3). The multiple branches between clusters/subclusters reflect the phylogenetic complexities that arise from genome mosaicism, where genes within a genome have distinct evolutionary histories.
Figure 3. Network phylogeny of 627 mycobacteriophages based on gene content.
Genomes of 627 mycobacteriophages were compared according to shared gene content using the Phamerator (Cresawn et al., 2011) database Mykobacteriophage_627, and displayed using SplitsTree (Huson and Bryant, 2006). Colored circles indicate grouping of phages labeled according to their cluster designations generated by nucleotide sequence comparison (Figure 2); singleton genomes with no close relatives are labeled but not circled. Micrographs show morphotypes of the singleton MooMoo, the Cluster F phage Mozy, and the Cluster O phage Corndog. With the exception of DS6A, all of the phages infect Mycobacterium smegmatis mc2155.
DOI: http://dx.doi.org/10.7554/eLife.06416.010
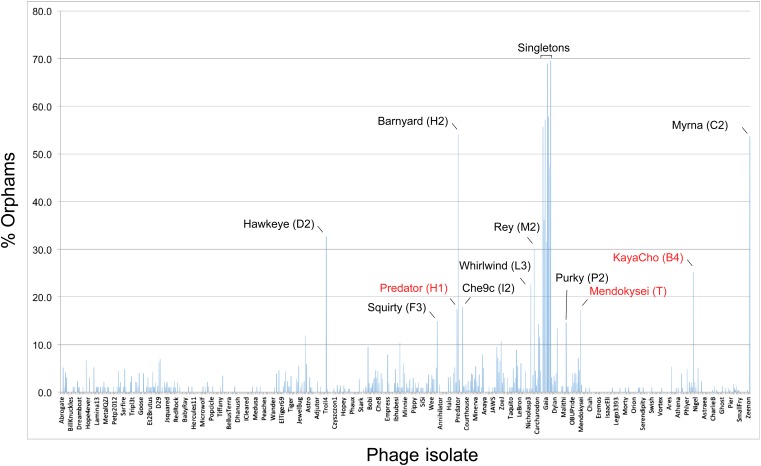
The distribution of orphams (genes without mycobacteriophage homologues) provides additional support for cluster/subcluster assignments; Figure 4). A relatively high proportion of orphams is a characteristic of both singleton genomes and single-genome subclusters (Figure 4). At least 30% of genes in all of the singleton genomes are orphams, and the single-genome subclusters have a minimum of 15% orphams; genomes in other clusters and subclusters typically have fewer than 10% orphams (Figure 4). The presence of numerous orphams ensures that the lack of cluster inclusion did not result from sequence errors or insufficient or inappropriate gene annotation. Notable exceptions are Predator (Subcluster H1) and Mendokysei (Cluster T), both of which are in very small clusters/subclusters, and KayaCho (Subcluster B4). KayaCho may warrant separation into a new subcluster (e.g., B6), but overall the orpham distribution is consistent with the cluster/subcluster designations.
Figure 4. Proportions of orphams in mycobacteriophage genomes.
The proportions of genes that are orphams (i.e., single-gene phamilies with no homologues within the mycobacteriophage dataset) are shown for each phage. The order of the phages is as shown in Supplementary file 1. All of the singleton genomes have >30% orphams, and most of the other genomes with relatively high proportions of orphams are the single-genome subclusters (Table 2) including Hawkeye (D2), Myrna (C2), Squirty (F3), Barnyard (H2), Che9c (I2), Whirlwind (L3), Rey (M2), and Purky (P2). Three phages shown in red type are not singletons or single-genome subclusters but have relatively high proportions of orphams. Predator and Mendokysei are members of the diverse and small clusters (five or fewer genomes) H and T, respectively; KayaCho is a member of Subcluster B4 but has a sufficiently high proportion of orphams to arguably warrant formation of a new subcluster, B6.
DOI: http://dx.doi.org/10.7554/eLife.06416.012
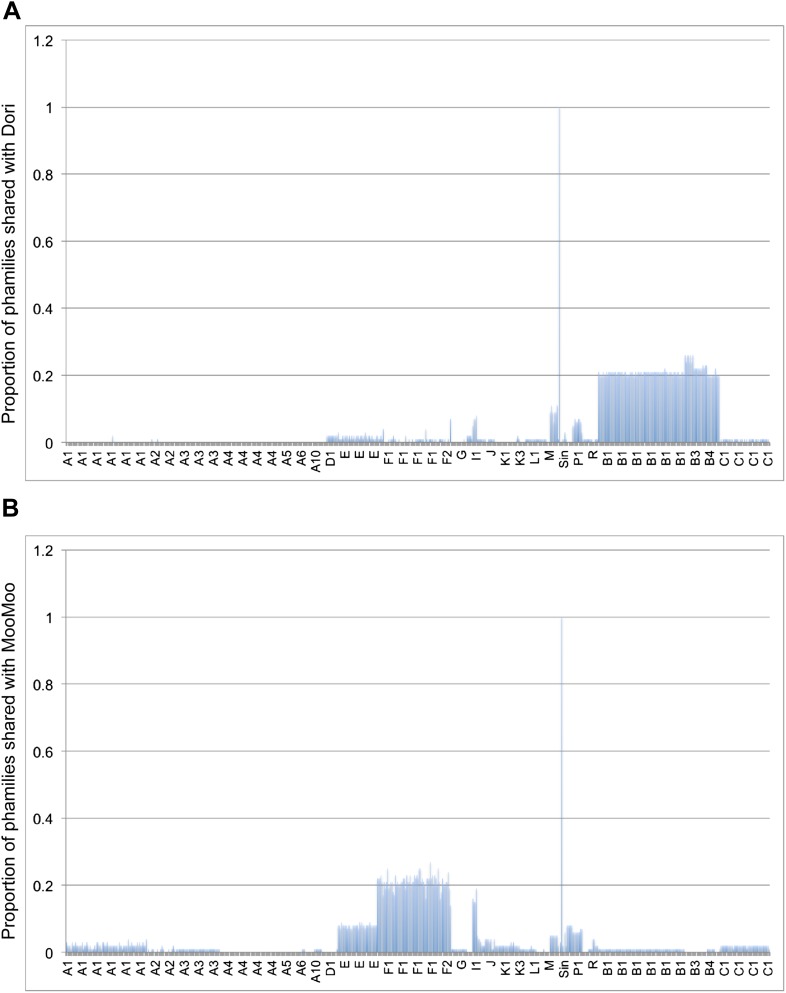
Figure 4—figure supplement 1. Shared gene content between Dori, MooMoo, and other mycobacteriophages.
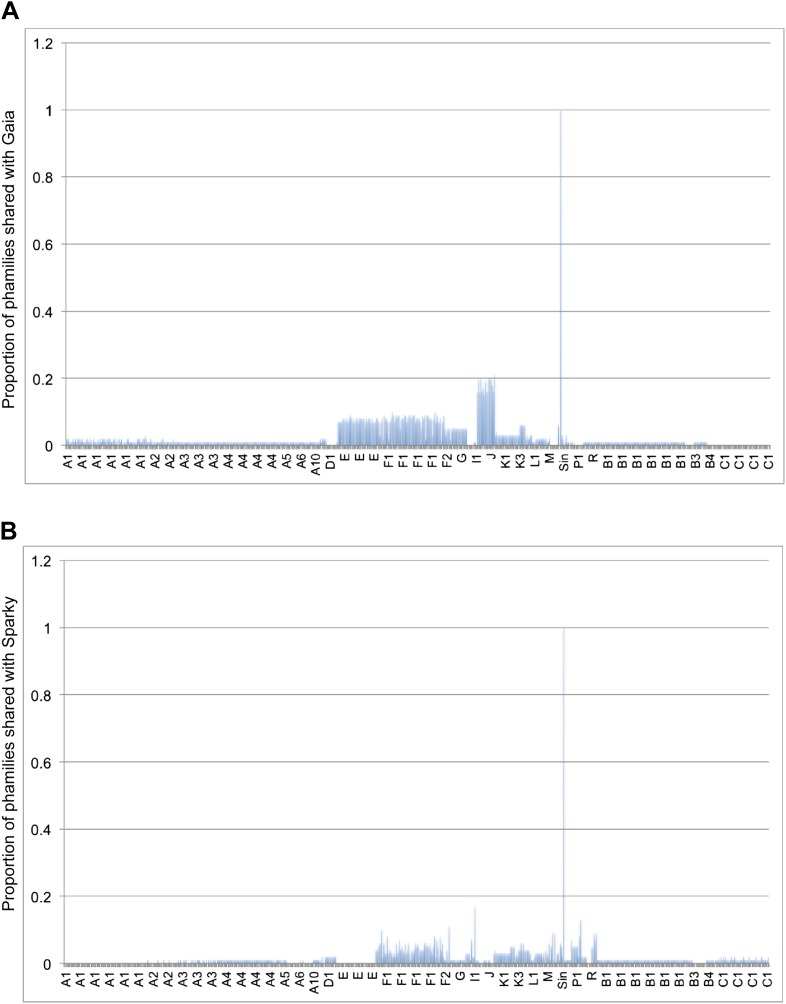
Figure 4—figure supplement 2. Shared gene content between Gaia, Sparky, and other mycobacteriophages.
The diversity of different clusters is highly varied
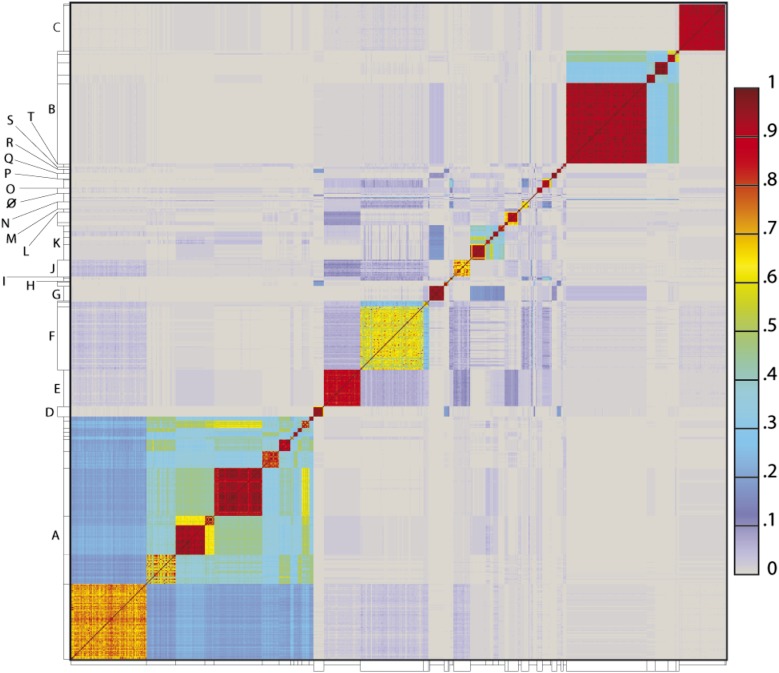
To determine the extent to which the various clusters/subclusters represent discrete groups, we generated a heat map showing pairwise shared gene content (Figure 5) and quantified the cluster/subcluster diversity (Table 1, Figure 6). The heat map strikingly illustrates that diversity is non-uniform, with genomes in some clusters (e.g., Subclusters B1, C1) being very closely related, whereas in others they display substantial differences (e.g., Subclusters A1, F1). The variation is also evident within the large Cluster A group, with some subclusters having low diversity (e.g., A4, A5, A6), some being highly diverse (e.g., A1, A2), and some plausibly further splitting into subgroups (A3) (Figure 5).
Figure 5. Heat map representation of shared gene content among 627 mycobacteriophages.
The percentages of pairwise shared genes was determined using a Phamerator (Cresawn et al., 2011) database (Mykobacteriophage_627) populated with 627 completely sequenced phage genomes. The 69,574 genes were assembled into 5205 phamilies (phams) of related sequences using kClust, and the average proportions of shared phams calculated. Genomes are ordered on both axes according to their cluster and subcluster designations (Supplementary file 1) determined by nucleotide sequence similarities (Figure 2). The values (proportions of pairwise shared phams averaged between each partner) are colored as indicated.
DOI: http://dx.doi.org/10.7554/eLife.06416.016
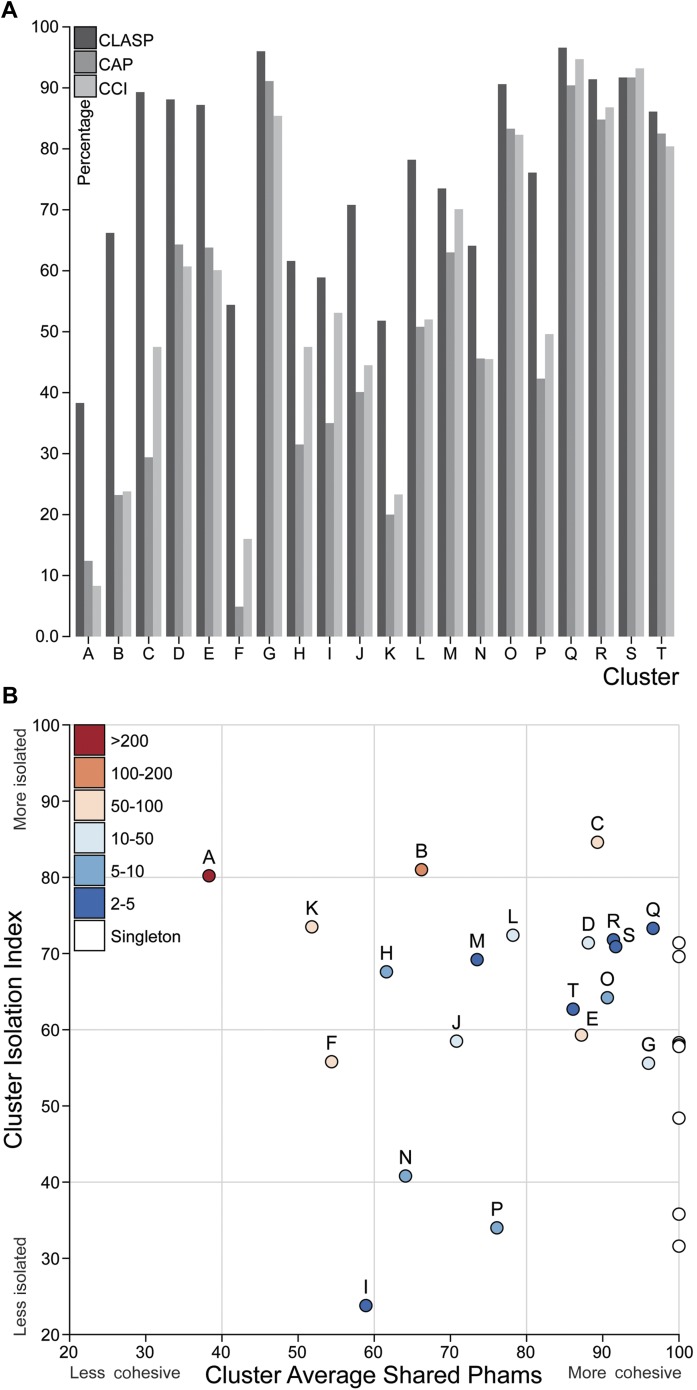
Figure 6. Cluster diversity and isolation.
(A) The CLuster Averaged Shared Phamilies (CLASP; blue), Cluster Associated Phamilies (CAP; red) and Cluster Cohesion Index (CCI; green) values are plotted for each mycobacteriophage cluster. (B) The Cluster Isolation Index (CII) and CLASP values (both shown as percentages) are plotted for each phage cluster. Singletons (white circles) are not individually labeled but correspond to the values shown in Table 1.
DOI: http://dx.doi.org/10.7554/eLife.06416.018
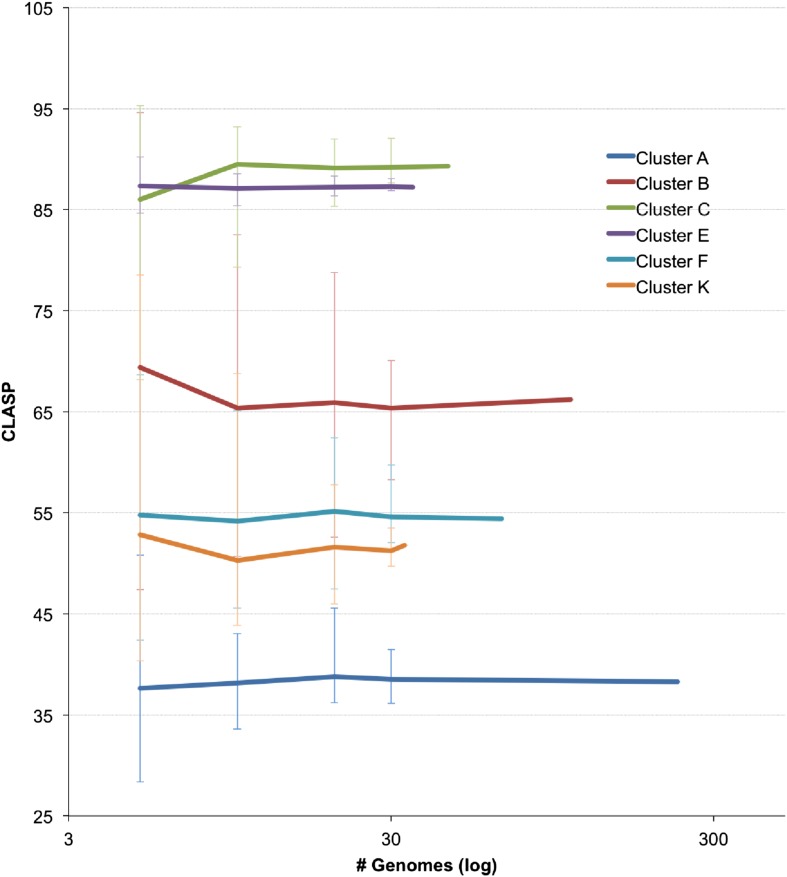
Figure 6—figure supplement 1. Resampling CLASP values for cluster diversity and size.
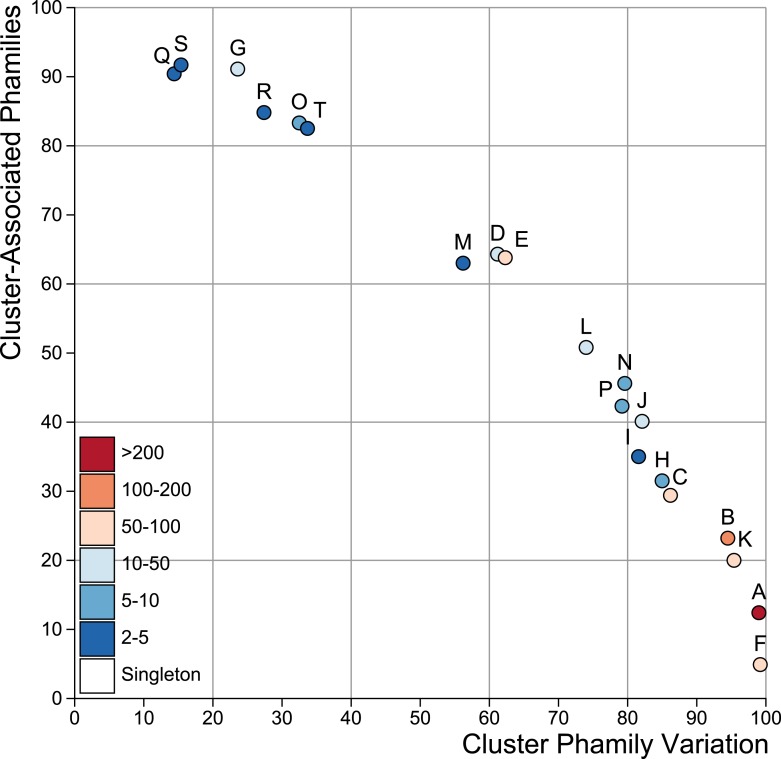
Figure 6—figure supplement 2. Cluster diversity shown by Cluster-Associated Phamilies (CAP) and Cluster Phamily Variation (CPV) indices.
We quantified the cluster diversity using three different measures, CLuster Average Shared Phamilies (CLASP), Cluster Associated Phamilies (CAP), and Cluster Cohesion Index (CCI) (Tables 1, 2, Figure 6A). Both CAP (the number of phams present in all genomes within a cluster divided by the average number of genes per genome) and CCI (the average number of genes per genome as a percentage of the total number of phams in that cluster) show substantial variation between clusters (Table 1, S2), and little evidence for commonly conserved ‘core genes’, as suggested for T4-related phages (Petrov et al., 2010). However, both of these parameters are somewhat influenced by cluster/subcluster size, which varies from cluster to cluster. In contrast, CLASP (the percentage of phamilies shared between two genomes, then averaged across all possible pairs within a cluster or subcluster) is relatively insensitive to cluster/subcluster size (as seen by a resampling analysis; Figure 6—figure supplement 1), but still shows substantial variation from one cluster to another (Table 1, Figure 6A).
Table 2.
Genometrics and Cluster Cohesion Indexes of mycobacteriophages
| Cluster | Subcluster | # Genomes | Average # genes | Average length (bp) | # Phams | CLASP* | CAP† | CCI‡ |
|---|---|---|---|---|---|---|---|---|
| A | 232 | 90.0 | 51,514 | 1085 | 38.3 | 12.4 | 8.0 | |
| A1 | 72 | 91.2 | 51,954 | 416 | 72.3 | 36.9 | 22.0 | |
| A2 | 28 | 93.4 | 52,805 | 312 | 64.7 | 30.1 | 30.0 | |
| A3 | 37 | 87.7 | 50,325 | 163 | 81.1 | 48.8 | 54.0 | |
| A4 | 46 | 87.4 | 51,376 | 125 | 92.7 | 70.6 | 70.0 | |
| A5 | 16 | 86.0 | 50,531 | 152 | 81.4 | 58.7 | 57.0 | |
| A6 | 11 | 97.8 | 51,677 | 128 | 90.2 | 75.1 | 76.0 | |
| A7 | 3 | 84.3 | 52,941 | 115 | 74.9 | 64.4 | 73.0 | |
| A8 | 4 | 97.8 | 51,597 | 107 | 93.5 | 86.8 | 91.0 | |
| A9 | 4 | 96.0 | 52,838 | 106 | 92.7 | 83.4 | 91.0 | |
| A10 | 7 | 80.0 | 49,174 | 112 | 81.6 | 60.9 | 71.0 | |
| A11 | 4 | 98.5 | 52,260 | 113 | 93.6 | 88.3 | 87.0 | |
| B | 108 | 100.4 | 68,653 | 421 | 66.2 | 23.2 | 24.0 | |
| B1 | 77 | 101.8 | 68,532 | 144 | 93.2 | 72.9 | 71.0 | |
| B2 | 8 | 89.9 | 67,267 | 101 | 94.9 | 84.6 | 89.0 | |
| B3 | 12 | 102.8 | 68,698 | 121 | 96.3 | 84.7 | 85.0 | |
| B4 | 8 | 96.1 | 70,619 | 166 | 79.9 | 45.8 | 58.0 | |
| B5 | 3 | 96.3 | 70,033 | 108 | 91.7 | 87.2 | 89.0 | |
| C | 45 | 231.0 | 155,504 | 486 | 89.3 | 29.4 | 48.0 | |
| C1 | 44 | 231.0 | 155,297 | 345 | 91.9 | 73.2 | 67.0 | |
| C2 | 1 | 229.0 | 164,602 | 227 | N/A | N/A | N/A | |
| D | 10 | 89.3 | 64,965 | 147 | 88.1 | 64.3 | 61.0 | |
| D1 | 9 | 87.3 | 64,697 | 100 | 94.9 | 88.8 | 87.0 | |
| D2 | 1 | 107.0 | 67,383 | 107 | N/A | N/A | N/A | |
| E | 35 | 141.9 | 75,526 | 235 | 87.2 | 63.8 | 60.0 | |
| F | 66 | 105.3 | 57,416 | 658 | 54.4 | 4.9 | 16.0 | |
| F1 | 60 | 104.8 | 57,486 | 573 | 59.6 | 20.6 | 18.0 | |
| F2 | 5 | 110.8 | 55,996 | 207 | 65.7 | 49.0 | 54.0 | |
| F3 | 1 | 107.0 | 60,285 | 105 | N/A | N/A | N/A | |
| G | 14 | 61.5 | 41,845 | 72 | 96.0 | 91.1 | 85.0 | |
| H | 5 | 98.4 | 69,469 | 207 | 61.6 | 31.5 | 48.0 | |
| H1 | 4 | 95.8 | 69,137 | 131 | 81.9 | 67.9 | 73.0 | |
| H2 | 1 | 109.0 | 70,797 | 110 | N/A | N/A | N/A | |
| I | 4 | 78.0 | 49,954 | 147 | 58.9 | 35.0 | 53.0 | |
| I1 | 3 | 76.0 | 47,588 | 101 | 77.5 | 66.7 | 75.0 | |
| I2 | 1 | 84.0 | 57,050 | 84 | N/A | N/A | N/A | |
| J | 16 | 239.8 | 110,332 | 530 | 70.8 | 40.1 | 45.0 | |
| K | 33 | 95.7 | 59,720 | 411 | 51.8 | 20.0 | 23.0 | |
| K1 | 15 | 94.3 | 59,877 | 166 | 85.5 | 47.9 | 57.0 | |
| K2 | 4 | 96.3 | 56,597 | 128 | 85.2 | 77.7 | 75.0 | |
| K3 | 3 | 98.2 | 61,322 | 111 | 92.2 | 89.5 | 88.0 | |
| K4 | 5 | 94.0 | 57,865 | 106 | 93.7 | 87.2 | 89.0 | |
| K5 | 6 | 98.2 | 62,154 | 144 | 82.1 | 68.2 | 68.0 | |
| L | 13 | 127.9 | 75,177 | 246 | 78.2 | 50.8 | 52.0 | |
| L1 | 3 | 123.7 | 74,050 | 135 | 92.6 | 88.8 | 92.0 | |
| L2 | 9 | 129.3 | 75,456 | 170 | 90.1 | 72.2 | 76.0 | |
| L3 | 1 | 128.0 | 76,050 | 126 | N/A | N/A | N/A | |
| M | 3 | 141.0 | 81,636 | 201 | 73.5 | 63.0 | 70.0 | |
| M1 | 2 | 135.0 | 80,593 | 138 | 96.6 | 96.6 | 98.0 | |
| M2 | 1 | 153.0 | 83,724 | 152 | N/A | N/A | N/A | |
| N | 7 | 69.1 | 42,888 | 152 | 64.1 | 45.6 | 45.0 | |
| O | 5 | 124.2 | 70,651 | 151 | 90.6 | 83.3 | 82.0 | |
| P | 9 | 78.8 | 47,668 | 159 | 76.1 | 42.3 | 50.0 | |
| P1 | 8 | 78.4 | 47,313 | 126 | 82.9 | 52.9 | 62.0 | |
| P2 | 1 | 82.0 | 50,513 | 82 | N/A | N/A | N/A | |
| Q | 5 | 85.2 | 53,755 | 90 | 96.6 | 90.4 | 95.0 | |
| R | 4 | 101.5 | 71,348 | 117 | 91.4 | 84.8 | 87.0 | |
| S | 2 | 109.0 | 65,172 | 117 | 91.7 | 91.7 | 93.0 | |
| T | 3 | 66.7 | 42,833 | 83 | 86.1 | 82.5 | 80.0 |
Cluster Averaged Shared Phamilies.
Cluster Associated Phamilies.
Cluster Cohesion Index.
The discreteness of different clusters is highly varied
The heat map of genome comparisons (Figure 5) also illustrates the degrees to which clusters and subclusters share gene content, a reflection of cluster discreteness, or how isolated discrete clusters are from each other. For example, although the Cluster A phages are highly diverse, they also appear relatively isolated and share relatively few genes with other clusters (Figure 5). In contrast, phages in Cluster E share substantial numbers of genes with other clusters, including those in Clusters F, J, L, P, and several singletons. We have quantified these relationships with the Cluster Isolation Index (CII, the percentage of phams present within a cluster that are not present in other mycobacteriophage genomes), which demonstrates the considerable variation in isolation from phages of other clusters/subclusters (Table 1, Figure 6B). For example, at one extreme, 84.6% of Cluster C gene phamilies are found only in Cluster C and not elsewhere. At the other extreme, only 23.8% of Cluster I gene phamilies are constrained to that cluster, with the remainder having relatives present in genomes in other clusters. Other clusters form a spectrum of relationships between these extremes (Table 1, Figure 6B), and clusters such as I and P—which share recognizable DNA sequence similarity (Figure 2—figure supplement 1)—share >60% of their genes with other phages (low CII values; Table 1). Thus, although some clusters could be considered as discrete groups—as reported for the Synechococcus phages (Deng et al., 2014)—this is far from being a universal or characteristic feature of groups of related phages.
Cluster isolation analyses reveal additional complexities arising from highly mosaic genomes. For example, the singleton Dori is clearly related to Cluster B phages (Figure 3) with which it shares limited DNA similarity (Figure 2—figure supplement 2) with 20–26% of its genes (Figure 4—figure supplement 1), but also has nucleotide similarity and shares genes with Cluster N and I2 phages, among others (Figure 2—figure supplement 2, Figure 4—figure supplement 1), as reflected in its low CII (Table 1, Figure 6B). Likewise, the singleton MooMoo has segments of DNA similarity and shares ∼20% of its gene content (as determined by shared phams) with Cluster F phages (Figure 3, Figure 2—figure supplement 3, Figure 4—figure supplement 1), but also has similarity to Clusters N and I, as well as a low CII (Table 1, Figure 6B). It has low DNA similarity to Cluster O (Figure 2—figure supplement 3), but has several phams in common with the Cluster O phages, and has the same unusual prolate morphology (Figure 3). Complex relationships are also seen in the singletons Gaia and Sparky (Figure 4—figure supplement 2).
Taken together, the analyses of both cluster diversity and cluster isolation show that mycobacteriophage populations contain a continuum of diversity, with non-uniform abundance of different types of phages. The prevalence of isolated phages may not necessarily reflect the proportions of different types of phages in the environment, but the availability of a large collection of isolated phages enables capture and whole genome analysis of relatively rare phages that are critical to understanding the complexities of genome relationships. We recently reported genomic analysis of the singleton mycobacteriophage Patience, which has a substantially lower GC% than its host (50.3% vs 67.4%), has a different codon usage profile, but is undergoing codon selection for growth in a high GC% environment (Pope et al., 2014b). If there is a flux of phage genomes and genes entering the mycobacterial neighborhood, then we predict that the phages of a single host do not reflect a closed system with discrete populations, but one that is open with ever-expanding diversity.
The mycobacteriophage population is not a closed system
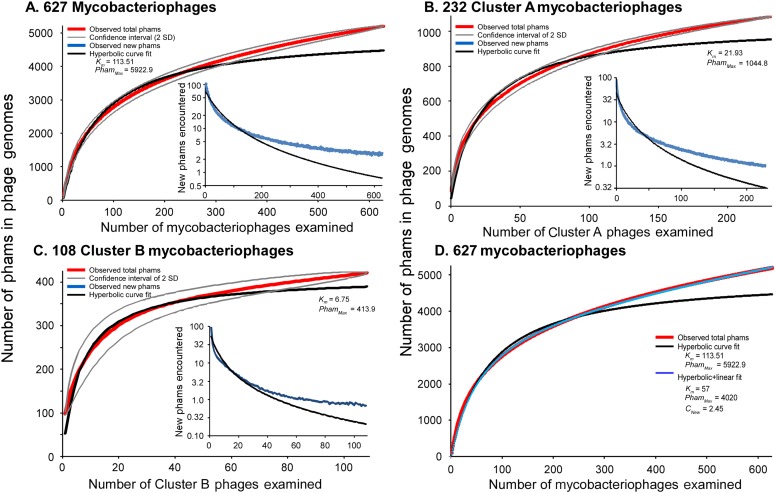
Both the huge diversity of phamilies in mycobacteriophages and the high frequency of orphams suggest that genes are constantly added to phage genomes from outside sources just as genes are added to the genomes of their bacterial hosts via horizontal gene transfer. Such gene influx—for example, from host-jumping phages such as Patience (Pope et al., 2014b)—would provide genetic novelty and enable phages to adapt to their ever-changing hosts. To examine gene flux into the mycobacteriophage population, we performed a rarefaction analysis by re-sampling the gene phamilies within the phage population (Figure 7). Remarkably, the rarefaction curves of the entire collection—including the 95% confidence limits—do not fit a hyperbola as would be expected if the mycobacteriophages were limited to an isolated set of genes, and about 2.5 new gene phamilies are predicted to be identified with each newly isolated phage (Figure 7A). Similar independent analyses on the phages of Cluster A or the phages of Cluster B show that this is also observed within these clusters (Figure 7B,C). Thus both individual clusters and the collection as a whole are not genetically fixed, but are in constant flux. While a hyperbola can model sampling of gene phamilies from a finite pool, it does not accommodate the influx of new phamilies. The addition of a linear term (see ‘Materials and methods’), representing the introduction of new phamilies from outside sources, results in a non-asymptotic curve which predicts the continual identification of new phams even after large numbers of genomes have been sampled (R > 0.999; Figure 7D). This linear term acts as a surrogate for the linear range of a second hyperbolic curve, one representing the resampling of a much larger set of gene phamilies available for introduction into mycobacteriophage genomes. Unfortunately, the current dataset remains insufficient to confidently extrapolate to give an estimate of the total number of viral protein families in the biosphere, which has been previously estimated to be anywhere between a half a million and 2 billion (Rohwer, 2003; Ignacio-Espinoza et al., 2013).
Figure 7. Rarefaction analysis of mycobacteriophage genomes.
(A) The numbers of phamilies are reported for between 1 and 627 phage genomes sampled at random without replacement; the mean of 10,000 iterations is shown in red; gray lines indicate a confidence interval of two standard deviations. The black line shows a hyperbolic curve fit to the data from phage counts 1 to 314. The inset shows the number of new phams encountered upon the inclusion of each phage, with the mean number for the 10,000 iterations shown in blue and the predicted value from the hyperbolic curve shown in black. (B) Rarefaction analysis of 232 Cluster A phages. The total numbers of phamilies are reported for between 1 and 232 phages sampled at random without replacement from Cluster A; the mean of 10,000 iterations is shown in red; gray lines indicate a confidence interval of two standard deviations. The black line shows a hyperbolic curve fit to the data from phage counts 1 to 117. The inset shows the number of new phams encountered upon the inclusion of each phage, with the mean number for 10,000 iterations shown in blue and the predicted value from the hyperbolic curve shown in black. (C) Rarefaction analysis of 108 Cluster B phages; the hyperbolic curve was fit to the data from phage counts 1 to 54. (D) Fits of the hyperbolic (Equation 1) and hyperbolic with linear (Equation 2) models for phamily identification within genome samples.
DOI: http://dx.doi.org/10.7554/eLife.06416.023
We note that because of the generally slow pace of the advancement of phage genomics, we have little insight into the phage populations of other hosts. We retrieved all double-stranded DNA tailed phage genomes in GenBank that we could identify (a total of 1781), corresponding to about 120 host bacterial genera, with a median number of phages per host genus of two. Using similar parameters for pham building as described above, the 181,717 predicted genes assemble into 47,479 phamilies. The relatively low representation of each phamily (3.8 genes/phamily) compared to the mycobacteriophages (13.4 genes/phamily) is a further reflection of the gross under-sampling of the phage population as a whole.
Implications for bacteriophage taxonomy
Bacteriophage taxonomic classification reflecting phylogeny presents substantial challenges because of genome mosaicism (Lawrence et al., 2002). Classification by viral morphology is well established, but may not accurately reflect the genetic relationships, as illustrated for the prolate-headed MooMoo (Figure 3). We also note that the mycobacteriophage myoviruses have a high CII and form a discrete group (Table 1) as do the Synechococcus myophages (Deng et al., 2014), perhaps reflecting a virulent lifestyle that constrains productive gene exchange; T4-related phages from diverse hosts share a core set of 15–20% of their genes, and whole genome comparisons reveal extensive mosaicism (Petrov et al., 2010). Host range mutability thus may differ in phages with different morphotypes, limiting access to the gene pool, and although grouping phages into clusters and subclusters provides analytical advantages because of the wide range in prevalence of different phages (Table 1), it is not suitable as a broadly applicable hierarchical taxonomic system. The comparative analysis of these mycobacteriophages thus supports reticulate taxonomies that more accurately reflect the phylogenetic complexities (Lawrence et al., 2002; Lima-Mendez et al., 2007).
Implications for student learning through research experiences
A research experience can be a powerful vehicle that enables a person to gain an understanding of the process of science (Hunter et al., 2007). When the research experience occurs early and at a large scale, as described here, the focus can shift from selecting a few ‘qualified’ students to exploring the potential interests of many students. Clearly, an essential ingredient is the nature of the research project, as definitions of research may vary from an inquiry-based exercise to authentic research with the potential to contribute publishable findings. To optimize the educational benefits, the research project must be intellectually and technically accessible to beginning students (i.e., few prerequisites) and scalable so that many students can simultaneously make progress in parallel, yet independently (Hatfull et al., 2006). Importantly, each student's findings should contribute to a scientific question with integration of all students' discoveries advancing a scientific question of significance, as judged by scientific peer review. This, we believe, defines an ‘authentic’ research experience. We note that in the SEA-PHAGES platform, substantial student effort is invested in arriving at high-quality genome annotations by close manual inspection followed by expert verification, a critical component of the detailed comparative analysis of phage gene content described here.
Concluding comments
Bacteriophage genomics has progressed relatively slowly compared to that of other microbes in spite of their relatively small genome sizes. Here we have demonstrated that programmatically integrating the research and education missions at large scale provides an effective solution to expanding our knowledge of viral diversity, with a multitude of insights gained as a consequence of the scale of phage discovery. The nature of different genomic types, the variations of the diversity both within clusters and shared genome content among clusters, and the expanse of the mycobacteriophage population can be viewed at an unprecedented level of resolution. Our conclusions align well with comparative analyses of phages of Enterobacteriacea (Grose and Casjens, 2014) and Bacillus spp. (Grose et al., 2014) and we predict that these are general parameters of bacteriophage diversity, at least when sampling broadly across the environment. Both the rarefaction analysis described here and preliminary analysis of phamilies of all sequenced DNA phages illustrate how little of the global phage population has been genomically sampled. With a near endless supply of diverse viruses readily accessible for isolation and analyses, integrated research/education programs will continue to play substantial roles in defining the nature of the virosphere.
Materials and methods
Phages and genomes
In addition to extant GenBank sequence information, mycobacteriophages were isolated, sequenced, and annotated in the Phage Hunters Integrating Research and Education (PHIRE) or SEA-PHAGES programs. Phage genomes were shotgun sequenced using either 454, Ion Torrent, or Illumina platforms to at least 20-fold coverage. Shotgun reads were assembled de novo with Newbler versions 2.1 to 2.9. Assemblies were checked for low coverage or discrepant areas, and targeted Sanger reads were used to resolve weak areas and identify genome ends. All genome sequences are publically available at phagesDB.org or in GenBank. Nucleotide comparisons used BLASTN or Gepard (Krumsiek et al., 2007).
Database construction
To create Phamerator database Mykobacteriophage_627, phamilies were constructed by first clustering the entire database of 69,574 genes using strict kClust parameters (70% clustering threshold and 0.25 alignment coverage of the longer sequence). This was followed by multiple sequence alignment of each preliminary cluster using Kalign (Lassmann and Sonnhammer, 2005). Consensus sequences were then extracted using HHmake and HHconsensus (Remmert et al., 2012). The resulting list of sequences was subjected to a second—and less strict—round of clustering via kClust (30% clustering threshold and 0.5 alignment coverage of the longer sequence) to obtain the final phamily assignments.
Network phylogeny constructions were made using the NeighborNet function with default parameters in SplitsTree (Huson, 1998; Huson and Bryant, 2006).
Cluster diversity and isolation indices
Four parameters were used to evaluate cluster diversity. The first is the CLASP index that calculates the percentage of phamilies shared between two genomes, then averages across all possible pairs within a cluster or subcluster. Because the pairwise similarities are averaged, CLASP is relatively insensitive to either the overall size of the cluster, or the heterogeneity of its diversity (such as in Cluster C in which of the 45 genomes in total, 44 are in Cluster C1, and only one is in Cluster C2). CLASP robustness with respect to cluster size was demonstrated through a resampling analysis. For each cluster with more than 30 members, a random subset (of 5, 10, 20, or 30 genomes) was selected and CLASP was calculated. For each sample size, 20 iterations were performed with replacement. As expected, there is substantial deviation among the iterations, especially at smaller sizes. However, there is little change in the average CLASP values with different sample sizes (Figure 4—figure supplement 1), showing that cluster size is not a primary driver of diversity. The resampling analyses also suggest that while a greater number of genomes helps refine the CLASP value, there is still predictive power when only 10 genomes are compared. On average, the maximum and minimum iteration values at a sample size of 10 genomes were within 8% of the whole-cluster CLASP value. This implies that, for example, increasing Cluster D from 10 to 50 or 100 genomes may raise or lower its current CLASP value of 88.1, but that value is likely to remain between ∼80 and ∼96.
The second measure used is the CAP, which is calculated as the number of phamilies present in all genomes within a cluster divided by the average number of phamilies per genome. These cluster-conserved genes could correspond to core genes that define a particular phage group such as cluster or subcluster. However, for those clusters with sufficient diversity to detect such core genes, these values are low. For example, among the 66 Cluster F genomes, only five phamilies are present in all genomes. None are virion structural genes, one is a glycosyltransferase whose role is unknown, one is a putative regulator, and the others are small proteins of unknown function. For the Cluster A genomes, 11 phamilies are conserved, seven of which are virion structural proteins, three are involved in DNA metabolism (DNA Pol, Helicase, Rec-Like protein), and one is of unknown function.
The third parameter is the Cluster Phamily Variation (CPV) index, which is the proportion of phams that are not present in all members of the cluster. CAP and CPV are inversely related but imperfectly as CPV varies with cluster size even among similarly diverse clusters; a plot of CAP values against CPV values is shown in Figure 6—figure supplement 2.
The CCI is calculated as the average number of genes per genome as a percentage of the total number of phams in that cluster. Thus if all genomes in a cluster are identical (and if phamilies occur only once in a genome), CCI would be 100; the CCI for two sets of five randomly chosen genomes is ∼2. CCI values correlate with cluster size, but similarly sized clusters as such G, J, and L, or E and K have substantially different CCI values (Table 1).
The CII is the percentage of phams present within a cluster that are not present in other mycobacteriophage genomes.
Rarefaction analysis
Rarefaction analysis was performed by randomly selecting subsets (without replacement) of between 1 and 627 (all), 232 (Cluster A) or 108 (Cluster B) mycobacteriophages and determining the numbers of phamilies represented. This was repeated 10,000 times to generate a mean number of phamilies observed given a number of phage genomes selected. The means of the accumulated numbers of phams and the numbers of new phages identified are plotted as the function of the number of genomes selected at random. The observed numbers were fit to a hyperbolic function for 50% of the sample (i.e., 1 to 314, 116 or 54 genomes for all, Cluster A or Cluster B phages, respectively); Hanes-Woolf regression was used to estimate PhamMax and Km of the hyperbola:
| (1) |
where NGenomes is the number of genomes sampled, NPhams is the number of total phams seen within those genomes, PhamMax is the total number of phams among all mycobacteriophage genomes, and Km is the number of genomes required to sample one half of PhamMax.
The lack of fit of the observed data to the hyperbola—with the observed data reflecting infinite size—suggests that the overall population is dynamic. The lack of hyperbolic fit of the data does not result from outliers such as phages with highly deviant GC%, because removing these does not improve the fit. The fit is also not substantially improved by analysis of the two largest clusters, Cluster A and Cluster B (Figure 7), suggesting that the dynamic nature of the gene pool is not an artifact of examining independent phage clusters with separate gene pools.
To model this behavior, we modified Equation 1 to include the introduction of novel phams via recombination with outside, non-mycobacteriophage genomes:
| (2) |
where CPhage is the number of outside phams seen in each phage. The value of CPhage was estimated from Figure 7B and new values for PhamMax and KPham were estimated by Hanes-Woolf regression following data normalization.
Acknowledgements
Students, faculty, and their contributions to authorship are listed in the Supplementary file 2. We thank Aileen Beard, Gerald Henkel-Johnson, and Larry McGahey at the College of St. Scholastica, the Core Facility for Imaging, Cellular and Molecular Biology at Queens College, Jennifer Kelly and Towanda Kirksey-Stanton at Jacksonville State Univeristy, Susan Crump at Merrimack College and Dr Gregory Hendricks at the University of Massachusetts Medical School Electron Microscopy Imaging Facility, Melissa Cox at North Carolina State University and Valerie Lapham at the NCSU Center for Electron Microscopy, Dr Karen M Snetselaar at Saint Joseph's University, and Rick Ellingworth at the University of Wisconsin-River Falls for excellent technical assistance. We also thank Drs Winston Anderson and Broderick Eribo for their roles as consultants at Howard University, and Dr R Edelmann and the Miami University Center for Advanced Microscopy and Imaging for their support and assistance with electron microscopy. We also thank John Morrell, Alicia Brighton, Joshua Fisher, Michael Shelfo, Brigham Wright, Jessica Engle, Brian Early, Kyle Smith, Kyler Haskell, Tambi Issac, Bryce Lunt, David Payne II, Lissenya Argueta, Bryan Merrill, Adam Gardner, Hailey Meadows, Adam Hansen, and Marshall Sheide for contributions to phage isolation.
This work was supported in part by the Howard Hughes Medical Institute SEA-PHAGES program, by the Howard Hughes Medical Institute through its Professorship grant to GFH, and by NIH grant GM51975 to GFH. Additional support was provided by the Department of Microbiology and Molecular Biology and the BYU College of Life Sciences; Cabrini College; NIH Grant No. P20 GM103408 to the College of Idaho; the National Science Foundation grant 0703449, the CUNY LSAMP program, the Office of the Provost, the Division of General Education, the Division of Mathematics and Natural Sciences, the Queens College UM/RE program and the Biology Department at Queens College; the Department of Biological Sciences at Lehigh University; the Center for Biotechnology and Biomedical Sciences of the Department of Biology in the School of Science and Engineering at Merrimack College; the NCSU Biotechnology Program and Department of Microbiology; a Davis Foundation grant, the Providence College Undergraduate Research Committee, RI-INBRE and NIGMS grant R15-GM094712 to Providence College; the Department of Biology, Saint Joseph's University; the Natural Science Department at the University of Houston-Downtown; the University of Maine Honors College, the University of Maine Department of Molecular and Biomedical Sciences, Maine-INBRE and NIH-INBRE Grant 8P20GM1003423-12; an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM0103423) to the University of Maine at Machias; the Howard Hughes Medical Institute, RISE and BRIC Programs, the Department of Biology, and the Offices of the Academic Dean and Chancellor at the University of Puerto Rico at Cayey; the University of Wisconsin River Falls Biology Department; the Gatton Academy of Science and Mathematics in Kentucky and the Western Kentucky University Bioinformatics and Information Science Center; Georgia College STEM Initiative and a Georgia College Faculty Research Grant; NSF Grant REVISION DUE-1205059 and the Department of Natural Sciences at Del Mar College; Miami University Department of Microbiology and the College of Arts and Science Dean's office and National Science Foundation ABI award 1146960; the HHMI Scicomp Project (52007572) to Xavier University of Louisiana; the Doris Duke Foundation; the Gonzaga University Biology Department, NSF-TUES grant DUE-1245778 and HHMI Undergraduate Science Education grant to Gonzaga University; the School of Biological Sciences and the School of Molecular Biosciences at Washington State University; and the Benjamin Harris Memorial Fund through the Pittsburgh Foundation.
Funding Statement
David Asai, Kevin Bradley, and Lucia Barker (formerly) are (or were) employees of Howard Hughes Medical Institute who also provided support for the SEA-PHAGES and PHIRE programs. DA, KB, and LB contributed to the design of the programs and the systems for data collection.
Contributor Information
Roberto Kolter, Harvard Medical School, United States.
Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science:
Patrick Abbazia, Kristia Abernathy, Andrew Abesamis, Syed Amaan Abidi, Mamon Abrahim, Colton Abrams, Alecia Achimovich, Brandon Ackerman, Jonuelle Acosta, Luis A Actis, Tamarah L Adair, Jaime Adame, Sandra D Adams, Jefferson Adams, Kenyeda B Adams, Rashidat F Adekunle, Christianah Ademuwagun, Eric J Adjei-Danquah, Nancy Adkins, Sheetal Agarwal, Riddhima Agarwal, Geovar Agbayani, Robert Agee, Sahil Aggarwal, Temitayo Agoro, Carmen Aguirre, Rachael Ahler, Salman Ahmad, Amiya Ahmed, Michelle Ahn, Stephen Aiken, Kara Aittama, Bisma Ahmed Ajaz, Alexandra Akins, Bukola Akintayo, Felix Akojie, Zein Al-Atrache, Ola-Oluwakiti Alabi, Olamide Alakija, Nitheesha Alapati, Christian Alba, Patrick Albertolle, Pedro Alejandro Ajsivinac, Cindy Alexander, Lisa M Alexander, Rush Alexander, Stephen Aley, Andrea Alfonso, Rebecca F Alford, Sarah Ali, Raul Alicea-Cabrera, Malak Alkanani, Dwa'a Alkhalaf, Brandon J Allen, Elizabeth A Allen, Elizabeth Allen, Venkata Alluri, Fernanda Alonzo, Erika Alvarado, Dymaries Alvarado-Vega, Amanda Alvelo-Aviles, Maria Alvisi, Kimberly Amick, Kimber M Amweg, Kirk R Anders, Alexander G Anderson, Alison Anderson, Kelly Anderson, Michael Anderson, Joseph Anderson, Kathryn M Anderson, Patrick Anderson, Sonya L Anderson, Joshua Andle, Nicole Anguiano, Nicolas Antis, Abigail Antoine, Tessa Anton, Ashley Anway, Callie Anyan, Juan Apiz-Saab, Javier Apodaca, Robert Appleyard, Saba Aqel, Marianne Arakelyan, Jobi Arceneaux, Jordan Archer, Kathleen Archer, Nelish S Ardeshna, Luke T Arduino, Manuel Ares, Jr, Lissenya B Argueta, Taylor Arhar, Jessica Arighi, Abigail JS Armstrong, Najealicka Armstrong, Chanarion Arnold, Eric Arnold, Kristin Arnold, Rachel Aron, Nikita Arora, Catlin Arrington, Lynda Asadourian, Kathyrn Asalone, Anthony Ascolillo, Jessica Ashcraft, Brandon D Ashley, Carmela Asinas, Christopher C Asuzu, Aliza Auces, Robin Audette, Matthew Aultman, Quenten Austin, Nicanor Austriaco, Michelle Averkiou, Taara Avery, Izma Aviles, Anamaris Aviles-Rivera, Lauren Awdziejczyk, Froogh Aziz, Rahat Aziz, Grace Babbs, Nikhil Babu, Stevie Bach, Megan Bachman, Tasha D Baer, Joanna Bagienska, Dadde Bah, Hana Baig, Andrew Bailey, Ryan Bailey, Paul Baker, Mitchell F Balish, Sarah Ball, Ellee Banaszak, Sophie Bandurski, Debarko Banerji, Laura Banken, Brittany Banks, William J Banning, Chen Bao, W Bradley Barbazuk, Nastassia R Barber, Joshua R Barber, Jessica Barber, Rafi Bari, Lucia Barker, Alexis Barna, Emily Barner, Ryan Barnes, Brooke H Barnhart, Stephanie N Barr, Alessandra L Barrera, Anne Barron, William D Barshop, Nicole Bartels, Kristi Bartholomay, Azhar Bashir, Kimberly Bastille, Steven Bateh, Tyler Bates, Neha Batra, Megan Batty, Rebecca Baudin, Victor Bauer, Cynthia Bauerle, Ian M Bayles, Aaron Beach, Gwendolyn Beacham, Lorenzo Bean, Shannon Beaty, Torri Beaudoin, Darius D Becker-Krail, Madison Beckman, Christina Beckwith, Blake Beehler, Bethany Beekly, Alissa Behl, Katherine Belfield, Campbell Belisle Haley, Abbey Bell, Bianca Bell, Devyn Bell, Mecca Bell, Trevene Bell, Adriano Bellotti, Ryan Benczik, Robert C Benjamin, Pilgrim Benjamin, Elizabeth Benner, Rebecca Benoit, Nicholas Bense, Brandon Bensel, Gabrielle Benson, Hannah Bergh, Rebecca E Berk, Charlotte Berkes, Janella Bermudez, Joshua Bernal, Daniel Bernal, Thomas J Bernardo, Anthony Bernicchi, Molly Berning, Jose Efrain Berrios-Lopez, Luis Berrios-Pagan, Johanna Berrios-Ruiz, Hannah Berry, Hannah Berry, Kara Beseler, Aaron A Best, Reba R Best, Nicolette Bestul, Victoria Betancourt, Andres Betancourt-Torres, Rudolf Beutner, Yachana Bhakta, Nazia Bhatti, Sonam B Bhimbra, Swapan Bhuiyan, Tiffany Bibeau, Mary Anthonette Binongcal, Monica Binsol, Miles Black, William Blaine, Cole Blair, Peter Blair, Aaron Blake, Samantha Blake, Bradley Blankenship, Michelle Blemker, Kathleen Blevins, Lawrence Blumer, Katherine Boas, Jon Lucas Boatwright, Brittany H Bodnar, Molly Bogolin, Alan Bohn, Anthony Bohner, Amy Bohner, Cayla Boisseranc, Dave Bollivar, Logan Bond, J Alfred Bonilla, James Bonner, Daniel Bonnette, Ashley Boone, Kyle Boone, Amanda Boozalis, Jaclyn Ann Boozalis, Elyse Borchik, Brooke Borgert, Kim M Borges, Denisse Borja, Julia Boroday, Dajana Borova, Mary Borque, Valerie Bostrom, Mara Bottomley, James Bowen, Ian N Boys, Kevin Bradley, Kosi Bradley, Jace Bradshaw, Judd Bragg, Kaitlyn Brahm, Veronica E Brandley, Andrew Brannan, Clinton Branton, Clayton Branton, Caitlyn B Brashears, Sara Bratsch, Edward L Braun, Mary A Braun, Gabriel Brautman, Donald P Breakwell, Mackenzie Bredereck, Lisa Brehove, Caroline Breitenberger, Jason Breithaupt, Joseph Bretzmann, Levi Brewer, Jerald S Bricker, Valerie C Briell, Alicia K Brighton, Kirsten Brink, Lauren Broadway, John W Brooker, Mia Broughton, Abigail Brown, Ariel Brown, Bryony Brown, Emma Brown, Gerald Brown, Heather Brown, Janaye Brown, Melissa Brown, Hilary A Brownstead, Claire Brownstone, Regina Bruce, Amy Bruckbauer, Laura Brumbaugh, Sarah Brusko, Anthony Brusnahan, Christian Brutofsky, Wesley Bryan, Hanna Bryant, Sarah Bryant, Ryann M Brzoska, Harman Bual, Blake Buchanan, Ciara Buechner, Daniel J Buhalo, Taylor Buhr, Duy Xuan Bui, Mary Bulfin, Sarah Bunker, Mary Burak, Sarah Burdette, Elizabeth Burger, Aaron Burghgraef, Kyle Burghgraef, Kevin Burke, Victoria Burkhead, Tate Burkholder, Andrew Burlingame, Sandra H Burnett, Angela Burr, Derek Burton, Tiffany Burton, Mathew Bushey, Kristina Busser, Nicholas Bussian, Maude Bute, Kristen A Butela, Mandy Butler, Dominique Bynum-Cooper, Meghan Byrne, Deanna Byrnes, Cristina Cabrera-Mino, Carla Caceres-Velazquez, Jacqueline Caelwarts, Grace Cain, Mario Caldararo, Darcie J Caldwell, Tomika S Caldwell, Czarina Calicdan, Christopher M Caliva, Caitlin J Callaghan, Brennan Calley, William Eamon Callison, Lee Calvert, Javier Camacho, Amanda Campbell, Warren Campbell, Ian W Campbell, Joshua L Campbell, Laura Campbell, Ross Campbell, Julie Anne Canter, Nico Carbone, Erick Cardona-Portalatin, Andrew Cardwell, Lydia Carlin, Emily Carlisi, Kristen Carlisle, Alexander Carlson, Courtney Carlstrom, Elizabeth Carpenter, James Carpino, Katherine Carr, Sophia Carroll, Connor Carry, Susan Carson, Jennifer Carter, Leah Carter, Lucas Carter, Morgan Carter, Steven M Caruso, Sarah Carzo, Danielle Cascione, Tarrin Casey, Marliz Casiano-Real, Tyler Caskin, Colleen Cassidy, James Cassoday, Paulina Castillo, Byron Castillo Silva, Anna Casto-Markosky, Erinleigh Caughron, Heather Caulkins, Mitchel D Cavallarin, William Cavedon, Elymae Cedeno Garcia, Francisco Cerda, Nicco Cerda, Logan Cerkovnik, Jasiel Cervantes, Juan Cervantes, James Cescon, Priya Chakrabarti, Sanjeev Chalissery, Molly Chamberlin, Codee Champney, Dong Woo Chang, Jee Yoon Chang, Michelle Chang, Ana Chaparro, Joshua Chappell, Reed Charlop, Anna Chase, Wrik Chatterjee, Kevin Chavez, Vivian Chavez, Alexandra Chen, Annie Chen, Xiaozhu Chen, Yue Ting Chen, Xi Chen, Jason Chia-Sheng Cheng, Alison A Chesky, Lashanda Cheston, Mahathee Meenakshi Chetlapalli, Darren Chew, Behroz Khushrav Chhor, Caitlyn M Chilinski, Pawan Chitta, Ariel W Cho, Leela D Chockalingam, Jeremy Chou, Tiffany Chow, Jordan Church, Richard Churchill, Bryce Churilla, Eric Ciardiello, Bryan Ciccarello, Varun Cidambi, Amy Cimo, Cassie Clark, Matthew Clark, Zachary Clark, Kari L Clase, Barbara Clement, Dylan Clevenger, Tiffany Clinton, Benjamin J Cody, Rainna Coelho, Sarah Coffee, Gayle Coggins, Kali Coghlan, Karen Cohen, Lianne B Cohen, Joanna Katherine Claire Coker, Tiffany Colburn, Allison Cole, Kris Cole, Arlixer Coleman, Maggie Colicchio, Eric Collin, Carol M Collins, Joseph M Collins, Justin Collins, Theresa Collins, Kimberly Colombini, Jennifer Colquhoun, Jessica E Colunga, Kevin Colvin, Ronald Comeaux, Brian Conahan, Destinee Cone, Erik Cone, Robert Connolly, Ashley Connors, Sandra Connors, Paul Consiglio, Kathrina Consing, Daniel Conti, Troy Coody, Stephanie Cook, Jamika Cookson, Charles Coomer, Crystal Cooper, Jacob Cooper, Cianna E Corbacio, Gabriel Cordero-Bernard, Sarah Corley, Kathleen Cornely, Eduardo Correa, Earl Cosby, Michael Costello, Catherine Cota, Idalid Cotto-Berríos, Alexis Cotto-Rosario, Mariele Courtois, Ashley Cox, Michelle Cox, Madeline Cox, Elizabeth Craig, Lara Crawford, Michael Crawford, Cheryl Creed, Victor A Crespo-Vega, Charlotte Cronenweth, Trevor Cross, Cody Crossley, Luis Cruz-Garcia, Daniel Cui Zhou, Ryan Cullen, Nicole Cullen, Andrew Cullett, Samantha Culpepper, John Culver, Nico Cunanan, Christina Cunha, Richard Cunningham, Taylor Cunningham, Michael Cuoco, Chiara P Curcillo, Esmeralda Curiel, James Curlin, Renee Curry, Matt Cusmiani, Christie L Cutting, Lauren Czaja, Karolina W Czarnecki, Emila Czyszczon, Thomas D'Addario, Satish Dahal, Alysan Dahl, Katherine Daily, Tiffany Damiani, Kyle Dammann, Jolene SP Damoiseaux, Duy Dang, Samantha Danguilan, Chuck Daniels, Richard L Daniels, Benjamin Danner, Michael A Darcy, Marija Dargyte, Rachel Darko, Adam Darwiche, Aditya Das, Thressa DaSilva, Amina Dasin, Ekaterina Dasiuk, Cierra Dauenhauer, Sarahanne Davidson, H Lane Davis, Katie Davis, Dillon Davis, Jeremy S Davis, Kimberly Davis, Marshall Davis, William B Davis, Ariangela J Davis-Kozik, Zazil-xa Davis-Vazquez, Allie Day, Jelani Days, Nydia De La Cruz, Jessica De La Luz, Carla De Los Santos, Melissa De Mattos, Alexander J DeBernardo, LaJoyce Debro, Samuel DeCero, Sarah DeCou, Kimberly DeGlopper, Ari Dehn, Morgan Deihs, Randall J DeJong, Rafaelle Delaney, Alex Delenko, Veronique A Delesalle, Hilda Delgadillo, Zachary DeLong, Maggie DelPonte, Katherine E Deming, Cassondra Demming, Renee M Deneweth, Lisa Deng, Danielle M DeNigris, John J Dennehy, Shannon Denny, Jonquil Dent, Dee R Denver, Armelle DeRiso, Payal P Desai, Dana DeSantis, Ria Deshpande, Alana Deutsch, Veena Devaraju, Bethany DeVault, Sarah Devine, Elizabeth B DeVore, Jodie DeVries, Brittney DeWald, Jaskirat Dhanoa, Molly Diamond, Crystal Diaz, Felix Diaz-Medero, Mike DiCandia, Leon Dickson, Jr, Lauren Dieleman, Joshua T Dimmick, Sasha DiNitto, Luke E Diorio-Toth, Anthony Disteso, Nikita Divekar, Michael DiVito, Ravi Dixit, Andrea Doak, Joanne Dobbins, Pamela Dockstader, Shea Dolan, Ebony Domingo, Bianca Dominguez, Evaristo Dominguez-Rodriguez, Julie Donna, John Paul Donohue, Melindy Dorcin, Emilio Doring, Robert Dorit, Stanna Dorn, Viviana Doros-Bonciu, Amrit Dosanjh, Meredith Doughty, Jasmine Douglas, Erin Doyle, Matthew Doyle, Nicolette Driscoll, Karlee Driver, Bayless E Drum, Alan Yicong Du, Heloise Dubois, Madison Duckworth, Emily Duex, Mary Duff, Jackie Duffy, Jacob Dums, David Dunbar, Courtney Dunkerley, Azaline Dunlap-Smith, Matthew Dunn, Matthew Dunworth, Quoc-viet Duong, Stephen Duong, Ben Duplechain, Mackenzie Durham, Ryan J Durham, Mieke Dykhouse, Maciej Dzikowski, Keith Earley, Brian J Early, Nicole A Ebalo, Annelle Eben, Erich Eberts, Eric Edewaard, Nicholas P Edgington, Jessica Edwards, Maria Eguiluz, Bernadette M Eichman, Rachel Ekdahl, Ashuvinee Elangovan, Sarah CR Elgin, Shelby A Ellis, Catherine E Elorette, Moustafa ElSayed, Shannon Ely, Abby Emanuel, Nicholas Emard, Jan A Enabore, Pauline Encarnacion, Nicole Enciso, Rachel Ende, Abby Engelkes, Angela Engelsen, Jessica M Engle, Belle V English, Sandy Enriquez, Elizabeth Ensink, Marcella L Erb, Gereltuya Erdenejargal, Jessica Erlich, Dana Escareno-Linger, Dulce Escobar, Joshua Esguerra, Militza Espada-Ramos, Kathryn Esposito, Lauren A Esposito, Maria Esquinca, Paige Estave, Amanda Estes, Crystal Estrada, Yesenia Estrada-Rivera, Ann-Scott Ettinger, Nicole Deanne Evangelista, Jared Evans, Mikala Evans, Tom Everding, Mitchell Eyerman, Daniel R Ezzo, Deborah Fadoju, Mohammed Fahad, J Grant Fahey, Michael Falahat, Emily Falch, Alexandra L Falk, Yiwen Fang, Michael Fangman, Jennifer Farina, Charles Newton Farmer, Amal Farooq, Summer Farooq, Kanhai Farrakhan, Elias Farran, Joseph Farrell, Tolulope Fasoranti, Mokunfope Fatukasi, Jonathan Faughn, Emilio Feal, Cameron Feathers, Melissa Feeney, Joel Feldhake, Zachery Feldker, Juan Feliciano-Figueroa, Celia Feng, Chelsea L Ferguson, Jacquelyn R Ferguson, Asia Fernandes, Alyka Glor Fernandez, Mariceli Fernandez-Martinez, Pilar Fernandez-Rodriguez, Michelle Fernando, Aisha Ferrazares, Gregory J Ferroni, Kyra Feuer, Alex Fields, Rachel Fieman, Laura Z Filliger, Christy Fillman, Jared Filut, Ann M Findley, Adam W Fine, Joseph Fiorenza, Marlie Fisher, Joshua NB Fisher, Jodi Fitzgerald, Nicholas M Flaherty, Brandon Flatgard, Taylor Fleet, Robert Fleming, Alexandru Florea, Desirey Flores, Izamar Flores Castillo, Shikira Flounory, Caroline E Flowers, Matthew Flowers, Kelsey Focht, Rose Fogliano, Chase Fong, Lindsey M Fong, Amy Fontenot, Lauren Ford, Jacquelyn Ford, Berencia Fore, Rebecca Foreman, Kathryn M Forman, Steven Forrester, Katherine S Forsyth, Mark H Forsyth, Gloria Foster, Lisa Anne Foster, Deitrick Fowler, Kristen Fowler, Courtney Fox, Tyler M Fox, Janey Foxe, Ethan Fram, Sarah Francisco, Rene D Francolini, Samantha Frangos, Shanah Frankel, Regina Frawley, Ryan Frazier, Christina M Freeman, Carlyn Freeman, Vanessa Freitas, Stanislav Fridland, Iddo Friedberg, Shawn Friel, Gabriel Michael Frischer, Chadley D Froes, Julia Froud, Aubree Frownfelter, Megan Fruchte, Katherine Fu, Kayci Fudge, Ana Lucia Fuentes, Chelsea Fulmore, Ho Yee Joyce Fung, Kaitlin Fusco, Deanna Fyffe, Jamal Gaddis, Christopher Gager, Eliot Gagne, Miranda Gagnon, Jasmine Gajeton, Maria E Galassi, Ruth Galatowitsch, Chanah Gallagher, Jordan Gallardo, Isaura Gallegos, Jenna Galletta, Tyler Galvelis, Aakash Y Gandhi, Ryan Gandy, Danielle Gannon, Alejandra Garcia, Carlos Garcia, Karla Garcia, Oscar Garcia, Samuel Garcia, Karla Garcia-Delgado, Hernan Garcia-Ruiz, Adam V Gardner, Jeremy Garner, Jacqualyn Garrett, Logan Garthe, Samantha M Gatt, Brianna Gaytan, Abraham Gebreselassie, Kaitlyn Geffen, Sarah Geiger, Katelyn Geleynse, Ethan Gelke, William Gendron, Jessica Genkil, Cody Gensen, Katia George, Kara Geraci, Shaunt Gharabegian, Kamalini Ghosh, Bryan C Gibbon, Zane Gibbs, Allison Gibson, Katherine Giddens, Dometria Gilbert, Neil Gilbert, Claire Gillette, Brooke Gillispie, Sinead Gilmore, Meghana Ginjpalli, Chris R Gissendanner, Jennifer Giulietti, Felipe Giuste, John Givler, Mitchell Go, Grayland W Godfrey, Erich Goebel, Eric A Goethe, Amber Goins, Urszula P Golebiewska, Pawel Golyski, Norma Gomez-Fuentes, Germarie Gomez-Garcia, Jessmarie Gonzales, Alfredo Gonzalez, Gabriela Gonzalez, Bridget Gonzalez, Elizabeth D Gonzalez, Joshua Gonzalez, Stephanie Gonzalez, Yvonne Gonzalez, Joshua Gonzalez-Berrios, Leanabel Gonzalez-Menendez, Karla Gonzalez-Pagan, Benjamin Goodwin, Emma Goodwin, Matthew Goodwin, Sean Goralski, Sonja Gorman, Alexander Goss, Maya Gotsatsenko, Emily Gough, Jayalakshmi A Govindan, Hannah M Gowaty, Jamie Gradishar, Neshaun Grady, Hannah Graff, Allison Graine, Jordan M Grainger, Melanie Grajales, Jose Grajeda, Marcela Grajeda, Brittany Grandaw, John Grant, Deborah Grant, Racheal Granville, Kaitlynn Graven, Austin Graves, Andrea Graves, Abby Grawe, Angela Gray, Ken Gray, Kevin Gray, Veronica C Gray, Eric Greene, Kyla L Greenfield, Stephanie Gregory, Janelle Grendler, Jacob Gries, Phyllis Griffard, Andrew Griffin, Alishia K Griffin, Maura Griffith, Phoebe Grijalva, Wendy Grillo, Melvin Grimes, Kimberly Grome, Julianne H Grose, Oleg Gross, Adam Groth, Halle Grove, Clayton Gruber, Katherine Zhen H Guan, Hebe Guardiola-Diaz, Brittany Guay, Genevieve Guerra, Stephanie L Guerra, Stuart W Guertin, Nicole Guevara, Ayele Gugssa, Nancy Guild, Nathaniel Guilford, Desire Guillory, Quentin Guillory, Bridget Guiza, Shan Gulamani, Naomi D Gunawardena, Jinny Guo, Stella Guo, Auroni Gupta, Swati Gupta, Eric L Gustafson, Nicholas Guthrie, Alexandra Gutierrez, Jesus Gutierrez, Omar Gutierrez Ruiz, Ivan Guzman, Soo Jung Ha, Brandon Haake, Jake Hackel, Alexander Hadik, Kaylee Hagen, Ritika Halder, Richard H Hale, Emily Hall, Jeremy Hall, Michelle Hall, Andrew Halleran, Mitchell Hallman, Alexander Hallwachs, Catherine Halpern, Peter Hamar, Jameel Hamdan, Ariel Hamil, Elizabeth Hamilton, Kaitlin Hamilton, Taylor Hammock, Latanya P Hammonds-Odie, Maxwell Hampton, Stephanie A Haney, Adam W Hansen, Brent Harbaugh, Emily Hardison, Charles Hardnett, Shyla L Hardwick, Raven Hardy, Victoria Hare, Raven Hargrove, Nyema Harmon, Jeremy Harmson, Jesse Haro, Jourdan Harper, Ravyne Harper, Donyelle Harrigan, Alex Harris, Celina Harris, Jamie Harris, Katrina Harris, Jenna N Harrison, Jon Harrison, Melinda Harrison, Andrew Hart, Matthew Hartman, Grant A Hartzog, Robert Harvell, Jayla Harvey, Samuel E Harvey, Vivian Harvey, Patrick Hashiguchi, Hina Hashmi, Selena Hasircoglu, Kyler J Haskell, Rachel Hastert, Paige Hasty, J Rob Hatherill, Sarah Hausmann, Tyler Hawk, Christina B Hawkins, Harry Hawthorne, Samantha Hawtrey, Kendra Hayden, Joseph Haydock, Mallorie Hayes, Michael Hayes, Camilla Haynes, Michaela Haynie, Sarah Hays, Diana He, Zezhong He, Kevin He, Siping He, Kaitlin E Healy, Stacey Heaver, Emily Heckman, Eric Hederstedt, Morgan Hefner, Erin M Hegarty, Neal Hegarty, Cally Hein, Bryce Henderson, Melissa S Henderson, Hope Hendricks, Blake Henley, Colleen Henry, Lanese Henry, Robert Henry, Shana-kay Henry-Grant, Ellen Hensle, Ryan M Hensleigh, Vanessa Hernandez, Emma Herold, April Hester, Sara A Heyn, Henry Higby, Charles Highfield, Gabriela Hilario, Heather Hill, Rose Zabel Hill, Sarah Hillson, Brenda E Hinojoza, Daniel Hinson, Traci-Lynn Hirai, Aspen Hirsch, Hana Hoang, Malayna Hocker, Kimberly Hodgson, Krista Hoevemeyer, Taylor Hoff, Hilary Hoffman, Gina Hogan, Sara E Hoge, Ryan Holden, Alexis Holder, Cameron Holder, Courtney Hollingsworth, Johnathon Hollis, Gail Hollowell, Georgeanna M Holmes, Le'nica Holmes, Bethany Holtz, Nathan Holz, David Homan, Joris Hoogendoorn, Stacy Hooker, James Horsfall, David Daniel Horstman, Ellen Hostert, Katherine Hotze, Laken C Houser, Catherine M Howard, Lauren A Howell, Sunnie Hsiung, G Jason Huang, Tina Huang, Vincent J Huang, Jasmine Hubley, Erica Hufford, Raya Hughes, Lee E Hughes, Emily Huizenga, Nathaniel Hunnewell, Kellie Hunnicutt, Gregory Hunt, Taylor Hunt, Kessler Hurd, Mackenzie Hurlbert, Kayla Hurst, Arturo Husein, Keith W Hutchison, Sydney Hutton, Huyen Huynh, Stephanie Huynh, Vicky Hwang, Joshua Hynes, Chikodi Ibe, Aubrey Ibele, Catherine Ibrahim, Amanda Icazatti-Burtell, Torri Igou, Jasmine Ikejiani, Mary Illback, Emily Illingworth, Ariel Imler, Joshua Imperial, Emily J Interrante, John Ion, Andra Ionescu, Khristina Ipapo, Shubha K Ireland, Camille E Irwin, Sharon Isern, Akira Issac, Tambi F Issac, Kristina Ivanova, Varun Iyengar, Osai Ize-Iyamu, Antonio Jackson, Charity Jackson, Stephen Jackson, Jelissa Jackson, Naveen Jain, Marilyn Jalal, Aleksandar Jamborcic, Brett James, Wendy Jamison, Alicia Jancevski, Elaina Jandourek, Emily Jankowski, Gary Janssen, Jonathan W Jarvik, Paul G Jasinto, Rahul Jaswaney, Rohit Jaswaney, Kishore L Jayakumar, Alec Jeffers, Myleka Jefferson, Sharese Jefferson, Tori Jefferson, Sabrina Jen, Timothy Jen, Adam C Jenkins, Meagan M Jenkins, Valerie R Jenkins, Stephen Jensen, Chandler Jensen-Cody, Ji-il Jeon, Huiyi Jiang, Wenxuan Lilith Jiang, Xuexia Jiang, Yi Jiang, Allison A Johnson, Anna Johnson, Ashley Marie Johnson, Brett Johnson, Cody Johnson, Christopher Johnson, Jessica Johnson, Joseph E Johnson, Ember Johnson, Benjamin K Johnson, Katie Johnson, Laura Johnson, Mark Johnson, Nicholas Johnson, Rakiyah Johnson, Rachel Johnson, Alexander Johnson, Emily Johnston, Rachel Johnston, Kevin Johnston, Netherland Joiner, Alana Jones, George Jones, Isabel Jones, Jackson Jones, Nancy L Jones, J Derek Jones, Keara Jones, Latoya Jones, Mackenzie Jones, Margaret Jones, Morgan Jones, Laura Joo, Taylor Jordan, Caelan Jordan-Ferrer, Michelle Juarez, Billy Judd, Mark Juel, Ryan Juhring, Harsha Jujjavarapu, John F Julian, Nickolas W Julian, Erin Jung, Jennifer Jung, Peter Jung, Jesus Jurado, Ziomara Jurado, Nicholas Justus, Kyle Kaczynski, Day Nahm Kagy, Maria Kakonikos, Cody Kamstra, Thalia Kanani-Hendijani, Karen Kantor, Aaron James Kappe, Alexandra Karacozoff, Ann Karam, Lakshmi Karamsetty, Semir Karic, Srilakshmi Karuturi, Surabhi Kasera, Woderyelesh Kassa, Manuel Kassardjian, Tomas Kasza, Dylan Katon, Balpreet Kaur, Kevin Kaurich, Mousa Kawwa, Katelynn Kazane, Michelle Keag, Sean Kearney, Alex Kearns, Michael G Kearse, Gabrielle Keeler, Nigel Keen, Ayleen Keeton, Meghan Keleher, Chandler Keller, Paige Keller, Aubrey Kelley, Brianna Kelley, Tess Kelley, Bobbi Kelling, Evan Kelly, Karla Kelly, Jessica Kelsey, Margaret A Kenna, Emma Kennedy, Erin Kennedy, Kendall Kennedy, Lacey Kennedy, Emily J Kenney, Samuel Kerk, Matthew Kern, McKenzie Kerr, Mark Kerry, Anna C Kesaris, Evan Kesinger, Stecey B Kessel, Yousif Kettoola, Joshua Keyes, Zenab Khan, Sher Adam Khan, Rohan Khazanchi, Elizaveta Khenner, Farzana Kibria, Joshua Kiehl, Michael Kiflezghi, Albert Kiladjian, Chad Killen, Bradley W Killingsworth, Lauren Killough, Myles S Killpatrick, Bryan Kim, Justine S Kim, Deborah Kim, Nelly Kim, Young Kim, Jonathan Kindberg, Meghan Kinder, Allyson King, Katie King, Tierra King, Rodney A King, Christina King Smith, Britni Kiosse, Jonathan Kirk, Ericka Kirkpatrick, Joe Kirsch, Taylor Kiskamp, Evan Kittaka, Amy Kivela, Beth Klein, Rachel Klein, Holly Klepek, Christine Kline, Josh Klinger, Linus Klingler, Tyler Klobucher, Karen Klyczek, Rachel Knapp, Anna Knight, Jacob Knol, Joshua Knopf, Andrew Knutson, Kevin Ko, Tae Wuk Ko, Staci Koberstein, Ann P Koga, Anna Kogler, William Kohlway, IV, Kristen Kokkonen, Christopher R Kolar, Ljuvica Kolich, Meghan Konda, Brett Konzek, Joseph Koon, Timothy Kopper, Christopher A Korey, Kristen Kornsey, Michal Kozdronkiewicz, Aleksey Kozlov, Marie-Morella Kponou, Allison Kraft, Lauren Kraft, Zachary Kramer, Arcadia Kratkiewicz, Taylor Kratochvil, Aurora Kraus, Amy Krause, Jessica Kraut, Amanda Kreger, Megan Kreitzman, Olivia Krejcarek, Jacqueline Kremer, Laura Krings, Grace Mahony Kroner, Greg P Krukonis, Alex Kryger, Lawrence Chihyung Ku, James L Kuhn, Logan Kuhn, Gary Kuleck, Emil Kurian, Meghan Kurz, Thomas Kwak, Samantha Kwok, Kaitlin Gee-Mei Kwong, Shelby Labe, Fabiani Laboy-De-Jesus, Holly LaFerriere, Maryanne LaFollette, Caitlin Lahey, Lindsay A Lahoda, Peggy Lai, Gilbert Laim, Christian Laing, Kelly Laird, Conner Lajoie, Zachery Lake, Dieter Ka Yeung Lam, Yuwan Lam, Nicole Lamas, Evan Lambert, Nicholas Lambrecht, Jessica Lambright, Anne Lamsa, Ignacio Landaverde, Joseph F Lang, Jennifer Lang, Jessica Lang, Tyler Langenbrunner, Allison Langone, Elizabeth Lanum, Jonathan Lapin, Anthony Lapsansky, Erin Lapsansky, Kristina Lardner, Lisa Lark, Tori Larsen, Victoria Larsen, Emily Larson, Victoria Larson, Julie Lau, Charlotte Laven, Joseph Lawhead, Zachary Lawrence, Stephen Layfield, Manuel Lazos, Derek Le, Mimi Le, John Leatherman, Eric Lebel, Janine M LeBlanc-Straceski, Dave Lee, HeaGie Lee, Ling-Ling Y Lee, Paul Lee, Stacey Lee, Taewoo Lee, Lee Ying-Chiang Jeffrey, Julia Y Lee-Soety, Laquisha Leemow, Taylor Leet, Julie H Lench, Donicia Lenori, Toinette Lenori, Justina Leo, Brittany Leonard, Dean M Leonard, Jonathan Leonard, Felecia Leslie, Samantha Leslie, Aden Lessiak, Robert J Lester, Nobel LeTendre, Robin Levy, Azlin Lewis, Demi Lewis, Jennifer Lewis, Lynn O Lewis, Nakia Lewis, Elizabeth Lewis Roberts, Amber Li, Diana Li, Frances Li, Ardy Li, Yi Li, Amy J Li, Vicky Li, Wei Li, Wendy F Li, Benjamin Lichtenfels, Lauren Liddy, Gretchen Lidicker, Mitchell Lienemann, Morgan Light, Jonathan Lin, Tiffany Lin, Sawyer E Linke, Amanda Links, Leah C Liston, Megan Little, Kunyao Liu, Mingchun Liu, Lisa Lizarraga, Manuel Llano, Maria Llanos, Ivan Llavona-Cartagena, Ashley Lloyd, Roxanne Lockhart, Paola Lockwood, Kathryn E Loesser-Casey, Kathleen R Logan, Lebron Logan, Alexandra Lombard, Laricca London, Shawn C London, Angelina Londono-Joshi, Christopher B Long, Michael Longmire, Luther Loose, Carlos Lopez, A Javier Lopez, Juan Lopez, Naomy Lopez-Cuevas, Todd Lorenz, Brian Lott, Kevin K Lou, Lize Loubser, Dustin Lovas, Emily Love, Ryan Loviza, Abigail Lowery, Katelyn Lowery, Paola Lozada, Alexander J Lu, Jacky W Lu, Alexis Lubbers, Jenna M Luczka, Catherine M Ludwig, Sean Joseph Lund, Emily Lundberg, David Lunderberg, Bryce L Lunt, Yan Luo, Lena N Lupey, Danielle Lutyk, Brittany Lynch, Tatiana Lyons, Lena Ma, Angela Maccarrone, Ana L Maccarrone, Margaret MacGibeny, Roger Machin-Rivera, Maxwell Machurick, Heyward B Mack, Stephanie Mack, Anne MacKenzie, Lara Madison, Tanya Maestas, John Kevin Magarian, Lauren Magee, Catherine M Mageeney, Samantha Magier, Michael Majekodunmi, Juan Maldonado-Rodriguez, Natalia Maldonado-Vazquez, Sarah Mam, Richa D Manglorkar, Coreen Manley, Janet Mansaray, Diane Marcillo, Joseph E Marcus, Matthew Mardis, Cody J Marfizo, Alyssa Marian, Emil Maric, Diane Marin, Joseph Marin, Raven Mark, Bryan Marone, Anahi Marquez, Nicholas Marquis, Paige Marsolek, Jordyn Mart, Tehja Martin, Alexander Martinez, Katherine Martinez, Matthew Martinez, Yanilda Martinez-Vega, Ryan Martinie, Olivia A Martino, Victoria L Martino, Olumide A Martins, Jutta Y Marzillier, John Mason, Siiri M Mason, Benjamin Massat, Jorge Mata, James Matern, Reny M Mathew, Oliver Mathrani, Hedieh Matinrad, Andrea Matthew, Danielle Mauch, Kimberly Maurer, Jadae A Maxton, Jessica Maya, Christine Mayer, Eric Mayer, Luke Maynard, Rebecca Catherine Mazahreh, Katherine McArthur, Jim Mcburney-lin, Kirk McCall, Christina McChesney, Casey A McConnell, Shanika McCray, Kyle McCurdy, Taashaylaray McDuffie, Reshard McElrath, Kyren McGary, Earyn McGee, Breann McGee, Hayley McGinnis, Erik McGuire, Dustin McHugh, Kristina McInnes, Emily McKee, Melanie McKell, Angela L McKinney, Kelly E McKinnon, Lisa McLellan, Connor W McMahon, Sean McMahon, Thomas McNamara, Lauren McNulty, Abigail McPherson, Erin E McPherson, Briana McRae, Jacqueline Measer, Andrew J Medenbach, Stephanie Medina-Delgado, Theodore Medling, Anna M Meese, Joseph Meggs, Haley Mehalik, Christopher Meier, Trevor Meindertsma, Riley Meister, Nisa Melendez-Rodriguez, Brett Mellbye, Kimberly Menendez, Monica Menkis, Theresa Menna, Akshay Mentreddy, Seth Menzer, Alvin Mercado, Raysean Mercer, Paul Merlau, Bryan D Merrill, Brenden Merriman, Robert Merritt, Nairobi Mesa, Adriana Messyasz, Maeva Metz, Eli Metzler-Prieb, Danielle Meyer, Jordan Meyer, Malcolm Mezue, Scott F Michael, Steven Micheletti, Nicole Michmerhuizen, Michael Miera, Olivia Miess, John Mike, Devon Miles, Adela Miller, Barrett Miller, Brenda Miller, Dustin Miller, Eric S Miller, Lindsay Miller, Brittany Miller, Savannah Miller, Mistie Miller, Meredith Millman, Shelby Mills, Taylor Mills, Mariah Minder, Thomas Minkiewitz, Briaunna Minor, Jessica Minor, Crystal Miranda-Mendoza, Lauren Misel, Brianna Mishler, Nikita Mishra, Neil Mistry, Andreas Mitchell, Casey Mitchell, Dan Mize, Jordan Moberg Parker, Samantha Moffat, Kim Mogen, Jay Mohan, Tal Mohn, Zaffiro Molina-Rivera, Molly Mollica, Sally D Molloy, Kirsten Monsen-Collar, Erin Montgomery, Greg Montgomery, Denise Monti, Steven Montiel-Melgar, Nina Montoya, Austin L Moody, Joshua Moore, Justin Moore, Tiffany Moore, Christina Moore, Jose Morales, Marjorie Morales, Abimelec Morales-Rivera, Deborah Moran, Lauren Mordukhaev, Robert Morefield, Benji Morehead, Jonathan Morena, Cameron Morford, Alexandra Morgan, Brandon Morgan, Miranda Nicole Morgan, Gabrielle Morgan, Yolanda Morgan, John D Morrell, Carlye Morris, Lailonny Y Morris, Tessa Morris, Tara Morton, Jordan Mosier, Allegra Mosley, Jonah Moss, Sarah Mosteller, Maroutcha Mouawad, Christopher Mulhern, Stefanie E Mundhenk, Trapper Munn, David Munoz, Miranda J Munoz, Ernesto Munoz-Pena, Chris Murdock, Meghan M Muretta, Alexandra Murphy, Ashley Murphy, Brendan Murphy, Jason Murphy, Zachery Murphy, Cara Murphy, Ryan Murphy, Jessica Murray, Rachel Murray, Brandon C Mus, Munia M Mustafa, Justin Muste, Rebecca Muthalaly, Mallory E Myers, Samantha Midori Nadeau, Colby Nance, Joshua Napial, Alberto Napuli, Ryan Narbutas, Courtney Navarro, Mercedes Navarro-Ohara, Saba Nawaz, Ann Nduati, Kevin Ndukwe, Kelli Neal, Robert Neblett, Mekala Kavya Neelakantan, Om Neelay, Fathom Neft, Nicholas Negretti, Casey Neisius, James J Neitzel, Brock Nelson, Matthew Nelson, Peter R Nelson, Samantha Nelson, Victoria A Nelson, Rahmi Nemri, Julie Nessler, Paul Nestor, Elizabeth K Neumann, Ann Neumeyer, Jordan Newhof, Alysha Newsom, Octavius Newsome, Carmen Ng, Aundrea Nguyen, Minh Nguyen, Emilie Nguyen, Emily Dao Anh Nguyen, Josephina Nguyen, Katrina Nguyen, Katrina Nguyen, Lida K Nguyen, Christine Nguyen, Thai Nguyen, Catherine Nicholas, Conor P Nichols, Janet Nickels, Rachel E Nicoletto, Natalie Nigg, Yana A Nikolayeva, Ryan Nintzel, Emily Noble, Ron Noble, Emily Nolting, Poochit Nonejuie, Heather Nootbar, Megan North, Gina Notaro, Adam Novajovsky, Peter Novick, Daniel Andre Novoa, Ian Noyes, Amy Nusbaum, Brenna Nye, James O'Brien, Heather O'Connell, Delaney O'Malley, Lauren P O'Neil, Kayla O'Sullivan, Stacey A Oakes, Nadeem Obaydou, Kasey A Ober, Trevor Obrinsky, Natalia Ocasio-Ramos, Raul Ochoa, Richard Ode, Nwamara Ogbonna, Marcus Oglesby, Kasopefoluwa Oguntuyo, David Ohnstad, Kristina Ok, Oreoluwa M Olaniyan, Margot Oliver, Olivia Oliver, Helena Olivieri, Alishe Olmo-Colon, Heather Olney, Morenike Olu, Uche Onoh, Judy Oranika, Kathryn Orban, Kristina A Orbe, Javier Ordonez, Alexander S Orenstein, Iris Orion, Chelsea M Orlando, Ryan Orloski, Beu Oropeza, Andrew M Orr, Kiara Ortiz-Camacho, Eric Ortiz-Lopez, Luis Ortiz-Munoz, Erin Osborne, Gina Osburn, Brandon P Ossont, Elijah Ostalza, Larissa Osterbaan, Taylor Oswald, Hidalisse Otero-Ruiz, Nathaly Otero-Ruiz, Cassandra Ott, Nakeya Owens, Sarah Owens, Oreoluwa Oyetan, Oluwaseun Oyewole, Jose Pabon-Lopez, Joanna Padolina, Shallee Page, Josue Paico, Jillian Pailin, Ramya Palaniappan, Jamie L Palmer, Caressie Palomino, Haoyu Pan, Marianne Pan, Lianette M Pappaterra, Mili Parikh, Min Jin Park, Minyoung Park, Peter J Park, Jacob Parker, Lindsay Parnell, Laura E Parrella, Prasanna Parthasarathy, Natasha Pascal, Valerie Paschalis, Jacob Pascual, Anjali P Patel, Mira Patel, Krupa Patel, Nikita Patel, Shivani Patel, Pooja M Patel, Pooja B Patel, Pooja Patel, Rikita Patel, Ritesh Patel, Ruchik Patel, Shivum Patel, Sarah Patno, Briawn Patrick, Emma Patschorke, Shejuti Paul, David E Payne, II, Yogitha Pazhani, Jacelyn Peabody, Bria Peacock, Katelyn C Peasley, Eric N Pederson, Marisa L Pedulla, Luke Peebles, Amber Peek-Simpson, Alexandra Peister, Fei Peng, Katelyn Pennington, Laura Penrod, Kathleen Peoples, Gretchen Peppers, Luigi Peracchi, Natalia Perecki, Ariane N Pereira, Paola Pereira-Rivera, Varahenage R Perera, Dahiana Perez, Ricardo Perez, Eliezer Perez-Colomba, Gabriel Perez-Lopez, Joseph Perez-Otero, Abbey Perl, Christina Perri, Benjamin M Perrin, Christine Elise Peters, Caroline Petersen, Christian Peterson, Kraig Peterson, Thalia Peterson Galvez, Mitchell Petredis, David Peyton, Brandon Pezley, Patrick Pfaffle, Shawna Pfeif, Rachel E Pferdehirt, Elizabeth Pflugradt, Anthony Pham, Danny Pham, Michael Pham, Mai-Trinh Pham, Alex Phillips, Kaysi Phillips, Melissa Phillips, Olivia Phillips, Connie Phung, Carlie Pickrel, Anya Pierson, Britta Pihl, Nathan Pihl, Jessamine Pilcher, Indiren Pillay, Robert Pinches, Megan Pineda, Jonathan Pinney, Matthew Piotrowiak, Yasaman Camelia Pirahanchi, Brianna Piro, Kaknika Pisith, Andrew Pita, Lauren Pittak, Thomas J Pittman, Amanda Pitts, Sonja Pizzini, Marie Pizzorno, Anna Plantinga, Andryus Planutis, Melanie Plastini, Ruth Plymale, Adam Poff, Joe Pogliano, Kit Pogliano, Zeeshan Polani, Rachel Polet, Galen Polise, Elliot Pollack, Tatianna Pollak, Aja Pollard, Georgia Pollard, Mariela Ponce-Torres, Sita Pongthunyaviriya, Amanda Porter, Chritiane J Porter, Lindsey Porter, Michelle Portillo, Abigail J Porzucek, Lindsay M Poss, Matthew Postolowski, Joseph Potesta, Michael Potter, Destinee Powell, Elijah Powell, Emily Powell, Jordan Powell, Taylor R Power, Marianne Poxleitner, Riemer Praamsma, Ashley Prasad, Smrithi Prem, Anthony Preston, Jordan Price, Kirsten Price, Kimberly Prince, Arturo Principe-Cartagena, Amber Prins, Stephenie Prinsen, Sara Privatt, Daysi Proano, Atticus M Proctor, Lora Prosser, Ashley Prout, Brooke Pruitt, Alyssa Prush, Seamus Pugh, Sandra Pumper, Taylor Purks, Kendyl B Pyfer, Zhijie Qi, Hong Qin, Michael Qiu, Conrad Quade, Ariana Quattlebaum, Ashley Queen, Thaina Quiles, Alexandra Quinn, Christopher Quintanal-Segarra, Hector Quintero-Alvarez, Hannah S Rabinowitz, Hiram Rabri-Diaz, Aleksandar Radakovic, Brendan Radel, Casey Radice, Nicholas J Radigan, Jennifer Radke, Paige Radtke, Miranda Raevsky, Haley Rafferty, Prartana Ramachandran, Eloy Ramirez, Juan Ramirez, Valeria Ramirez, Jose Ramirez-Diaz, Mauro Ramos, Kareliz Ramos-Aponte, Laura Ramos-Flores, Taylor Randell, Alex Randoph, Alexa Raney, Surabhi Rao, Dustin Raper, Rehnuma Rashid, Amir Rastegari, Suchita Rastogi, Rebecca Ratusnik, Luke Raudaskoski, Tiffany Ravelomanantsoa, Rawnok Rayeka, John R Raymond, Ulugbek Razakov, David C Ream, Krisanavane Reddi, Sherry Reddix, Kayla Reed, Abigayle Reed, Jeremiah Reenders, Lukas Rees, Brooke Reese, Jordan Rego, Amanda Reicherter, Margaret Reilly, Meagan Reinecke, Emily Reinhart, Mickey J Reiss, Leah Relko, Daniel Renner, Piper Replogle, Tessa Retzlaff, Daniel Reyes, Angel Reyes-Aponte, Adriana Reyes-DeLeon, Nathan Reyna, Brady Reys, Cortney Rhoades, Christopher Rhodes, Christopher Rhodes, Emily Rhude, Corwin N Rhyan, Ricci-tam Chiara Jeun-Ning Elena, Jeremy Rice, Benjamin James Rich, Erin Morris Richard, Mitchell Richards, Megan Richters, Jenna Rickus, Kendall Riddelle, Tanya Riddick, Benjamin C Riley, Claire A Rinehart, Sarah Ring, Alyssa Riordan, Luis Rivera-Arce, Yoliveliz Rivera-Bernier, Myrielis Rivera-Burgos, Denise Rivera-Cordero, Asdrubal Rivera-Dones, Shalimar Rivera-Gonzalez, Marlene Rivera-Martinez, Yamil Rivera-Ortiz, Carlivette Rivera-Rivera, Alejandra Rivera-Rodriguez, Miriam Rivera-Rolon, Angelica Rivera-Rosa, Melanie Rivera-Rosario, Rivera-San Antonio Jennifer, Antonella Rivezzi, Erin Rizzo, Haleigh Roach, Darcy Roberson, Megan Robertson, Molly Robertson, Laura Robey, Courtney Robinson, Danielle Robinson, Jana Robinson, Kayla Robinson, Tamara S Robinson, Joseph Robinson, Sarah Rock, Kathy Rodogiannis, Christina Rodriguez, Jasmin Rodriguez, Katherine M Rodriguez, Lauren Rodriguez, Neudalis Rodriguez-Burgos, Frances Rodriguez-Diaz, Ashley Rodriguez-Gonzalez, Claribel Rodriguez-Lopez, Stephen Rogers, Thomas Rogers, Kelvin J Rojas, Lucero Rojas Gallegos, David Rokhinson, Lauren E Rollman, Alfredo Roman, Will Romero, Kimberly A Rosales, German Rosas-Acosta, Elizabeth L Rosenthal, Meredith K Rosenthal, Joseph Ross, Jessica Rossi, Lindsay Roth, Kasey Roush, Aislinn Rowan, Ansley T Royal, Michael R Rubin, Jeffrey Rubin, Jessica Ruby, Leanna Rucker, Maxwell Sung Ruckstuhl, Xiang Rui, Michael Ruiz, Sarafina Rush, Jeanette Russell, Alfredo Ruvalcaba, Joseph Ryan, Nathan Ryan, Ryan Amelia-Frances M, Jessica L Sabo, Daniel Sabzghabaei, Lindsey Sacco, Quang V Sack, Rachna Sadana, Margaret S Saha, Katrina Sahawneh, Kenny Salamea, Samaher M Saleh, Mary Salim, Angelica Salinas, Brianna Salverda, Andria Sammon, Jorrel Sampana, Shaterrica Sampson, Jessica Sanchez, Marieli Sanchez-Collazo, Erin Sanders, James Sandoz, Eric Sanford, Michael Santana, Alexander Santiago, Jan Clement Santiago, David Santiago-Ochoa, Jose Santiago-Vazquez, Natanael Santillana, Eddy Santos, Christy Saquin, Heba Sarhan, Richard Sater, Lori Saunders, Erin Sauve, Judith G Savitskaya, Lauren E Sawyer, Aatif Sayeed, Heidi Sayre, Tyler Scaff, Allysan Scatterday, Claire Schaar, Amy Schade, Claire Schafer, Lauren B Schellenberger, Anne Scherer, Alexander Schierbeek, Isaac B Schiller, Catherine Schilling, Jessica Schipper, Jennifer Schlegel, Elizabeth Schleh, Joshua Schmidt, Theresa Schmidt, Carson Schneider, Seth Schneider, Sydney Schneider, Christine E Schnitzler, Morgan B Schoer, Leo R Scholl, Amanda Schorr, Sarah Schrader, Stephanie Schramm, Ariel L Schroeder, Katherine Schroeder, Allison Schroeder, Monica Schroll, Karyssa Schrouder, Jacob Schrull, Kaitlyn Schuberth, Olivia Schuele, Thomas Schulte, Ellen Schultz, Lisa Schultz, Michael B Schultz, Morgan Schultz, Megan Schulz, AJ Schumacher, Victoria Schwartz, Amanda Schwarz, Shelby Scola, Amanda Scott, Taylor D Scott, Vincent Scuttaro, Corey D Seacrist, Sabe Sears, Amanda Seaton, Hailey N Seaver, Ethan Sebasco, M Esa Seegulam, J Bradley Segal, Gabriel C Segarra, Ana Segura Lerma, Reuben Seidl, Robert Semler, Sageanne Senneff, Jiwon Seo, Bijan Sepheri, Erica Sewell, Amy Shafer, Rachel A Shaffer, Christopher D Shaffer, Madeena Shafiq, Harsh Shah, Zalak Shah, Sohum C Shah, Lindsey Shain, Peter Shank, Devon Shannonhouse-Wilde, Ananya Sharma, Sanskriti Sharma, Shaylen Sharp, Spencer Sharp, Nadia Sheen, Marshall G Sheide, Kathryn E Sheldon, Michael A Shelfo, Laura Shellooe, Matthew Sheltra, Adrie Shen, Jean Shen, Morgan Sherer, Andrea Sherod, Chringma Sherpa, Eileen Shi, Rani Shiao, Kelly S Shibuya, Hyo Jung Shin, Marla Shipton, Katharine Shively, Breanne Short, Rahul Shrikanth, Michael Shultz, Shruthi Shyamala, Zia Siddiqui, Hannah Sides, Aziz Sidra, Talles Sidronio, Christina Godfried Sie, Satchel Siegel, Mary Siki, Jeremy Silva, Abigail Silva, Ethan Sim, Jacqueline Simeon, Nicholas J Simitzi, Abigail R Simmons, Jacob Simon, Stephanie E Simon, Zachary Simon, Anita Simonian, Nathan Simpson, Erika F Sims, Danielle Sin, Ramya Singireddy, Samantha Siomko, Benjamin Siranosian, Emily Sirek, Jordan Skinner, Brittany Sklenar, Tyler Slade, Lucas Slivicke, Katherine Smart, Megan Smeets, Thomas J Smith, Abigail J Smith, Damien Smith, Erica Smith, Elliott G Smith, Joanna Smith, Jason P Smith, Katherine A Smith, Kyle M Smith, Logan Smith, Luke D Smith, Savanna K Smith, Colby Smith, Dennis Smith, Jalen Smith, Kyle C Smith, Veronica Smith, Brett Snyder, Sarah L Sokol, Divyakshi Solanki, Vincent Sonderby, Robert Soohey, Stephen Soohey, Talia K Sopp, Samantha Sorenson, Erin Sorge, Jesus Sotelo, Subada Soti, Rebecca Soto, Kara Soucek, Hannah Souers, Maura J Southwell, Hannah Space, Alexia L Sparrow, Blaire Spaulding, Kayla Spears, Michael Clayton Speed, Shannon B Spencer, Lauren Spicer, Preethy S Sridharan, Meghan K St. Cyr, Erika Stairs, Katelyn J Stanley, Julie Stanton, John Starner, John Starnes, Beth Statler, Richard Stauffer, Hernando Steidel, Jackson Steinberg, Ivy J Stejskal, Oleg Stens, Rachel Sternberg, Leah Stetzel, McKayla Stevens, Joseph Charles Steward, Damion Stewart, Eric Stewart, Shawntavia Stewart, Julianne J Sticha, Jennifer Stiles (Beane), Julia Stimpfl, Jonathan Stites, Timothy Stoddard, Kaitlyn Stoddart, Molly Storer, Elizabeth K Storm, Emily Stowe, Andrew Straszewski, Clark Straub, Zachary Streeter, William Strober, Alyssa Stubblefield, Joseph Stukey, Rachel Sturge, Trevor Sughrue, Niles Sulkko, Matthew Sullivan, Ryan Sullivan, David Sullivan, Cassandra Sulski, Grace Sundeen, Nandini Surendranathan, Minnu H Suresh, Jacob A Surges, Theresia Sutherlin, Sarah Swalley, David Swartout, Elliot Swartz, Ramata Sy, Najah Syed, Cole Sylvester, Charity Sylvester, Jacqueline Synder, Mary R Szurgot, Marta Szyszka, Shandee Tachick, Kayla Taggard, Michael Taguiam, Kareen J Taha, Ahmed Tahseen, Katherine Tai, George Taishin, Shiori Takashima, Jordan Takasugi, Jaee Tamhane, Kathleen Tan, Tin-Yun Tang, Natalie Tanke, Lucas Tans, Brian Tarbox, Lubaba Tasnim, Devon Taylor, Rebecca Taylor, Sarah Taylor, Barbara J Taylor, Warren Taylor, Kristina Taynor, Laura Teal, Gavin L Teichman, Shreya Tekumalla, David W Temme, Louise Temple, Autumn Tendler, Mayra Terres, Treyc Terry, Michael Teti, Patricia M Thang, Franklin L Thelmo, Anapaula Themann, Monique Theriault, Sarah Thibault, Arsema A Thomas, Ditte C Thomas, Sherwin Thomas, Athena Thomas, Jennifer Thomas, Katherine Thomas, Jasper V Thompson, Juri Thompson, Justin Thompson, Kayla Thompson, Sara Thompson, Christopher Thompson, Elyse Thompson, Tasha Elizabeth Thompson, Cheng Tian, Taylor Tibbs, Melissa Tighe, Ainsley Timmel, Marsha W Timmermanm, Jasmine Ming Jing Ting, Vishnu Tirumala, Margaret Tish, Mehrgol Tiv, Andrew Tobias, Deborah Tobiason, Benjamin P Todd, Audrey Tolbert, Marc Tollis, Skyler Tomisato, Brianne Tomko, Alex Tonthat, Michael Torello, Melissa Torres, Sarah Torres, Javier Torresdey, Elizabeth Toscan, Raymond Totah, Mariama Tounkara, Monica Towler, Colby Townsend, Cody Townsend, Khue Vi Tran, Quy T Tran, Brandon Tran, Connie Tran, Alexandra Treadaway, Marissa A Tremoglie, Kathryn Trentadue, Mason Trieu, Clayton Trisler, Daryl R Trumbo, Quang Truong, Brendan Tsai, Edwin Tsay, Jonathan Tsay, Chung Him Tse, Lanya Tseng, Smaragdi Tsourapa, Jonathan Tucker, Joshua Tull, Jacqueline C Tully, Jessica Turen, Chelsey Turner, Eric Turner, Paul Turner, Sydnie Turner, Beth Tuttle, Cassandra Tuttman, Joy L Twentyman, Dionne Ubungen, Dhruva Rajesh Udani, Neha Udayakumar, Yakntoro Udoumoh, William Ueckermann, Sharon Uhder, Kristin Ullberg, Hannah Ullery, Miranda Ulmer, Kendrick Underwood, Tyler J Uppstrom, Carl Urbinati, Eduardo Urias, Akif Uzman, Steven Valentino, Lourdes Valenzuela, Andrew Valesano, Braeden Van Deynze, David Van Doren, Dylan van Krieken, Vanessa Van Tongel, Rebecca Van Zanen, Alexis Vance, Peter VandeHaar, Amy VanderStoep, Zoe Vann, Cassie VanWynen, Corina Vasquez, Josue Vasquez, Gwendolyn S Vasquez, Tremain Vass, Anastasia Dmitrievna Vavilina, Edwin Vazquez, Alyssa Vecchio, Quinn C Vega, Rahulsimham Vegesna, Paola Vela, Rebecca Velasco, Kelsey Veldkamp, Mariano Velez, Kara Venema, Marissa Venero, Yaamini Ranjani Venkataraman, Vaidehi Venkataraman, Kristen Verdoorn, Jake Verduzco, Daniel Vessells, Breeze Victor, McElroy Victoria, Steven Vigil Roach, Janahan Vijanderan, Ammu Vijayakumar, Aishwarya Vijayan, Albert Vill, Erika Villa, Sabra Villa, Maria Virginia Villadiego-Punto, Jose Villagomez, Ana Villanueva-Pereira, Rachel Villegas, William Villella, Yvette Villero Martinez, Kelly Vining, Meredith Virk, Bhula Vishal, Nikita Vispute, Victoria Viveen, Melody Vo, Quynh Vo, Stephanie Vu, John Vu, Jamie Vulgamore, Chiraag Vyas, Kristen Wade, Marisa H Wagner, Shalia Wagner, Ryan Wagner, Abigail Waidelich, Andrew Wakefield, Isaac Wakiro, Tyler Walburn, Erin Walch, Gretchen Walch, Angie Waldron, Loni Walker, Savannah Walker, Brenden Wall, Jasmine C Wall, Lauren Beth Waller, Rachel Walstead, Rachel S Walter, Dustin Walter, Andy Yixun Wang, Da Wang, Jay-Shing Wang, Judy Jingxuan Wang, Hao-Yi Wang, Yuqi Wang, Vassie C Ware, John Robert Warner, David Warren, Samantha Warwar, Jacqueline M Washington, William Waterstreet, Colby Watkins, Laura C Watkins, Erin L Waugaman, Brent Webb, Jessica Webb, Marc Webb, Laurence Webb, Jeffrey Wei, Christine Weir, Emilie Weisser, Holly Welfley, Marie Wells, Joshua Welsch, Braden Wenndt, John T Wertz, Aliah S West, Daniel Westholm, Kathryn Weston, Kathleen A Weston-Hafer, Victoria Westra, Abigail Whalen, Ellen Wheeler, James Wherley, Emily Whitaker, Dakota White, Laura L White, Lamanuel White, Rebekah K White, Xander White, Stephen Whitfield, Carmen Wickware, Peter Widitz, Allison MD Wiedemeier, Sophia R Wienbar, Dion Wigfall, Katherine Wikholm, Luke Wilde, William Wilde, Adrienne Wilen, Abigail Wilhelm, Garrett Wilkerson, Kellyn Wilkes, Chantelle Willette, Chandler Williams, Ciera Williams, Devin M Williams, Drake Williams, Kristina Williams, Lauren H Williams, Madelaine Williams, Richard Williams, Tyler Williams, Sara Williams, Kurt E Williamson, Sarah M Williamson, Whitney Willis, Monique Willoughby, Alix C Wilson, Christine R Wilson, Elisa Wilson, John Wilson, Trevor Wilson, Lauren Wilson, Tyriana Wilson, Justin Wimberly, Danielle Winders, Shane Wing, Sarah Winokur, Victoria Marie Winslow, Hannah S Wirtshafter, Eric Witherspoon, Evan Witt, Donna Wodarski, Meredith Wojcik, Victoria Wolf, Brooke Wolff, Cody Wolterman, Michael J Wolyniak, Adrienne Evelyn Wong, Chung Ki Wong, Stephanie Wood, Mitchell Woodford, Andrew Woodruff, Avery Woods, Dayton Wooldridge, Michael Woolford, Cassandra Worner, Rebecka Worrell, Michael J Wozny, Nina Wren, Brigham A Wright, Bianca Wright, Heather E Wright, Spencer Wright, Shelby Wright, Kathryn Wrobel, Cynthia E Wu, Hao Wu, Kit Wu, Xiangying Wu, Jiewei Wu, Yi Shuan Wu, Jalyn Wurm, Shacaria Wyke, Kevin Wyllie, Kristen Wymore, Christian J Xander, Xiao Xiao, Helen Xun, Anastaciya Yakovenko, Keianne Dale Yamada, Kyoko Yamaguchi, Keying Yan, Jonathan Yang, Sophia Yang, Tsering Yangzom, Rayce D Yanney, Tyler Yates, Chen Ye, Brandon Yee, Sarah Yeend, Daniel Yehdego, Justin Yen, Benjamin Yoder, Amber Yohn, Priscilla Yong, Santiago Yori, Alicia Young, Elizabeth Young, Lauren K Young, Hannah Marie Youngwirth, Hussain Yousaf, Jullie Yu, Victor Yu, Eric Yu, Stefan Yu, Han Yuan, Christine Zabel, Joleen Zackowski, David A Zaidins, Alice Zalan, Stephanie Zamora, Rachel Zarchy, Michael V Zavorski, Mariam Zayed, Franck Zeba, Gerard Zegers, Michael Zehner, Lucas Zellmer, Manhao Zeng, Bruce H Zhang, Bo Zhang, Carolyn Min Zhang, Daiyuan Zhang, Hairong Zhang, James Zhang, Junhao Zhang, Mitchell Jia Zhao, Alec Zimmer, Zachary Zimmer, Anastasia M Zimmerman, Sarah Zimmermann, Tai Zollars, and Melina Y Zuniga
Phage Hunters Integrating Research and Education:
Amma Ababio, Jamil Alhassan, Zohair Azmi, Michelle Boyle, April Burch, Stephen Canton, Alexandra Cathcart, Brittany Dey, CourtneyBeth Dohl, Samantha Eppinger, Emma Fisher, Rodrigo Gonzalez, Forrest Guilfoile, David Hauser, Christina Hwang, Saikrishna Kothapalli, Darwin Leuba, Kohana Leuba, Sequoia Leuba, Austin Li, Edeline Loh, Matthew Luchansky, Nathaniel MacKenzie, Kaitlin Mitchell, Matthew Olm, Yein Park, Terence Parker, Kaitlin Price, Elina Roine, Rachel Rush, Paul Salamanca, Jennifer Schaub, Lauren Schmidt, Victoria Schneider, Ana Sencilo, Emilee Shine, Kailey Slavik, Shahwar Tariq, Waleed Tariq, Enoch Tse, Kathy Van Hoeck, Alex Waldherr, Ana Wan, Brett Weingart, Albin Wells, Jakob Wells, Philip Williams, Randi Wilson, Gabriel Winbush, and Amanda Yurick
Mycobacterial Genetics Course:
Naazneen Adam, Ashmita Arjoon, Lugani Bengani, Rooksana Carim, Nafiisah Chotun, Navisha Dookie, Nabila Essack, Karnishree Govender, Viveshree Govender, Nandini Gramoney, Jessica Hunter, Chernoh Jalloh, Afsana Kajee, Nathan Kieswetter, Michelle H Larsen, Jared Mackenzie, Fiona Maiyo, Gugulethu Masondo, Mpilwenhle Mbanjwa, Yanga Mdleleni, Khanyisile Mngomezulu, Katherine Moccia, Chantal Molechan, Odessa Moodley, Zama Msibi, Tessa Naido, Anand Naranbhai, Vivek Naranbhai, Nomfundo Ncobeni, Fortunate Ndlandla, Bridget Nduna, Silindile Ngobese, Nokonwaba Nkondlo, Shirwin Pillay, Yathisha Ramlakhan, Nicole Reddy, Eric J Rubin, Neo Sehloko, Shilisha Shanmugam, Sarisha Singh, Melisha Sukkhu, and Po-Cheng Tang
Funding Information
This paper was supported by the following grants:
Howard Hughes Medical Institute (HHMI) 54308198 to Graham F Hatfull.
National Institutes of Health (NIH) GM51975 to Graham F Hatfull.
Howard Hughes Medical Institute (HHMI) 52007054 to Graham F Hatfull.
Brigham Young University to Sandra Burnett.
Cabrini College to David Dunbar.
National Institutes of Health—INBRE GM103408 to R Luke Daniels.
National Science Foundation (NSF) to John Dennehy.
Queens College to John Dennehy.
Lehigh University to Vassie Ware.
Merrimack College to Janine LeBlanc-Straceski.
National Institutes of Health (NIH) GM094712 to Nicanor Austriaco.
National Institutes of Health—INBRE GM103430 to Kathleen Cornely.
Davis Foundational Grant to Kathleen Cornely.
Providence College to Kathleen Cornely.
St. Joseph's University to Christina King Smith.
University of Houston, Downtown to Rachna Sadana.
University of Maine, Honors College to Keith Hutchinson.
National Institutes of Health (NIH) GM1003423 to Keith Hutchinson.
Howard Hughes Medical Institute (HHMI) to Michael Rubin, Kirk Anders.
University of Puerto Rico to Michael Rubin.
University of Wisconsin, River Falls to Karen Klyczek.
Western Kentucky University to Claire Rinehart.
Gatton Academy of Science and Mathematics to Rodney King.
Georgia College to Indiren Pillay.
Del Mar College to John Hatherill.
Miami University to Iddo Friedberg.
National Science Foundation (NSF) DUE-1205059 to John Hatherill.
National Science Foundation (NSF) ABI-1146960 to Iddo Friedberg.
Howard Hughes Medical Institute (HHMI) 52007572 to SK Ireland.
Doris Duke Charitable Foundation to Michelle Larsen.
Gonzaga University to Kirk Anders.
National Science Foundation (NSF) DUE-1245778 to Kirk Anders.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
WHP, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
DJ-S, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
CAB, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
DAR, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
DJA, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
WRJ, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
RWH, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
JGL, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
GFH, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
SGC, Acquisition of data, Analysis and interpretation of data, Contributed unpublished essential data or reagents.
Additional files
List of 627 sequenced mycobacteriophages and cluster designations.
Full list of group author details.
Major datasets
The following datasets were generated:
Russell D, Hatfull G, 2015, Mycobacteriophage database, http://phagesdb.org, Publicly available at the Mycobacteriophage Database (http://phagesdb.org).
Bowman C, Cresawn S, Hatfull G, 2014, Mykobacteriophage_627, http://phamerator.webfactional.com/databases_Hatfull, Accessed via the program Phamerator, publicly available at http://phagesdb.org/Phamerator/faq/.
References
- Brown KL, Sarkis GJ, Wadsworth C, Hatfull GF. Transcriptional silencing by the mycobacteriophage L5 repressor. The EMBO Journal. 1997;16:5914–5921. doi: 10.1093/emboj/16.19.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Brockhurst M. Bacteria-virus coevolution. Advances in Experimental Medicine and Biology. 2012;751:347–370. doi: 10.1007/978-1-4614-3567-9_16. [DOI] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Pope WH, Jacobs-Sera D, Bowman CA, Russell DA, Dedrick RA, Adair T, Anders KR, Ball S, Bollivar D, Breitenberger C, Burnett SH, Butela K, Byrnes D, Carzo S, Cornely KA, Cross T, Daniels RL, Dunbar D, Findley AM, Gissendanner CR, Golebiewska UP, Hartzog GA, Hatherill JR, Hughes LE, Jalloh CS, De Los Santos C, Ekanam K, Khambule SL, King RA, King-Smith C, Klyczek K, Krukonis GP, Laing C, Lapin JS, Lopez AJ, Mkhwanazi SM, Molloy SD, Moran D, Munsamy V, Pacey E, Plymale R, Poxleitner M, Reyna N, Schildbach JF, Stukey J, Taylor SE, Ware VC, Wellmann AL, Westholm D, Wodarski D, Zajko M, Zikalala TS, Hendrix RW, Hatfull GF. Comparative genomics of cluster O mycobacteriophages. PLOS ONE. 2015;10:e0118725. doi: 10.1371/journal.pone.0118725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Ignacio-Espinoza JC, Gregory AC, Poulos BT, Weitz JS, Hugenholtz P, Sullivan MB. Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. 2014;513:242–245. doi: 10.1038/nature13459. [DOI] [PubMed] [Google Scholar]
- Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae. Virology. 2014;468–470:421–443. doi: 10.1016/j.virol.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose JH, Jensen GL, Burnett SH, Breakwell DP. Erratum: genomic comparison of 83 Bacillus phages reveals 12 clusters, 14 singletons and remarkable diversity. BMC Genomics. 2014;15:1184. doi: 10.1186/1471-2164-15-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Current Opinion in Microbiology. 2005;8:444–450. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. Mycobacteriophages: genes and genomes. Annual Review of Microbiology. 2010;64:331–356. doi: 10.1146/annurev.micro.112408.134233. [DOI] [PubMed] [Google Scholar]
- Hatfull GF. The secret lives of mycobacteriophages. Advances in Virus Research. 2012;82:179–288. doi: 10.1016/B978-0-12-394621-8.00015-7. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Current Opinions in Virology. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O'Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW. Comparative genomic analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. Journal of Molecular Biology. 2010;397:119–143. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WR, Jr, Lawrence JG, Hendrix RW. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLOS Genetics. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) Program. KwaZulu-Natal Research Institute for Tuberculosis and HIV (K-RITH) Mycobacterial Genetics Course. University of California—Los Angeles Research Immersion Laboratory in Virology. Phage Hunters Integrating Research and Education (PHIRE) Program Complete genome sequences of 63 mycobacteriophages. Genome Announcements. 2013;1 doi: 10.1128/genomeA.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M, Mayer CE, Soding J. kClust: fast and sensitive clustering of large protein sequence databases. BMC Bioinformatics. 2013;14:248. doi: 10.1186/1471-2105-14-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Current Opinion in Microbiology. 2003;6:506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proceedings of the National Academy of Sciences of USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskisson PA, Sumby P, Smith MC. The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology. 2015;477:100–109. doi: 10.1016/j.virol.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter AB, Laursen SL, Seymour E. Becoming a scientist: the role of undergraduate research in students' cognitive, personal, and professional development. Science Education. 2007;91:36–74. doi: 10.1002/sce.20173. [DOI] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Ignacio-Espinoza JC, Solonenko SA, Sullivan MB. The global virome: not as big as we thought? Current Opinion in Virology. 2013;3:566–571. doi: 10.1016/j.coviro.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Science Education Alliance Phage Hunters Advancing Genomics And Evolutionary Science Sea-Phages Program. Modlin RL, Hendrix RW, Hatfull GF. On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio. 2014;5:e01051–e01013. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. Journal of Molecular Biology. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- Kropinski AM, Sulakvelidze A, Konczy P, Poppe C. Salmonella phages and prophages–genomics and practical aspects. Methods in Molecular Biology. 2007;394:133–175. doi: 10.1007/978-1-59745-512-1_9. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH. Order to the viral universe. Journal of Virology. 2010;84:12476–12479. doi: 10.1128/JVI.01489-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Liu J, Dubow M, Gros P, Pelletier J. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. Journal of Bacteriology. 2006;188:1184–1187. doi: 10.1128/JB.188.3.1184-1187.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proceedings of the National Academy of Sciences of USA. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. Journal of Bacteriology. 2002;184:4891–4905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Shin H, Ryu S. Characterization and comparative genomic analysis of bacteriophages infecting members of the Bacillus cereus group. Archives of Virology. 2014;159:871–884. doi: 10.1007/s00705-013-1920-3. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G, Toussaint A, Leplae R. Analysis of the phage sequence space: the benefit of structured information. Virology. 2007;365:241–249. doi: 10.1016/j.virol.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, Kim J, Miller JF, Hatfull GF, Modlin RL. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio. 2012;3 doi: 10.1128/mBio.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Current Opinion in Virology. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PCAST, PsCoSaT . Executive Office of the President. Washington, DC: 2012. Report to the president. Engage to excel: producing one million additional college graduates with degrees in science, technology, engineering, and mathematics. [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virology Journal. 2010;7:292. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Anders KR, Baird M, Bowman CA, Boyle MM, Broussard GW, Chow T, Clase KL, Cooper S, Cornely KA, Dejong RJ, Delesalle VA, Deng L, Dunbar D, Edgington NP, Ferreira CM, Weston Hafer K, Hartzog GA, Hatherill JR, Hughes LE, Ipapo K, Krukonis GP, Meier CG, Monti DL, Olm MR, Page ST, Peebles CL, Rinehart CA, Rubin MR, Russell DA, Sanders ER, Schoer M, Shaffer CD, Wherley J, Vazquez E, Yuan H, Zhang D, Cresawn SG, Jacobs-Sera D, Hendrix RW, Hatfull GF. Cluster M mycobacteriophages Bongo, PegLeg, and Rey with unusually large repertoires of tRNA isotypes. Journal of Virology. 2014a;88:2461–2480. doi: 10.1128/JVI.03363-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Ferreira CM, Jacobs-Sera D, Benjamin RC, Davis AJ, DeJong RJ, Elgin SCR, Guilfoile FR, Forsyth MH, Harris AD, Harvey SE, Hughes LE, Hynes PM, Jackson AS, Jalal MD, MacMurray EA, Manley CM, McDonough MJ, Mosier JL, Osterbann LJ, Rabinowitz HS, Rhyan CN, Russell DA, Saha MS, Shaffer CD, Simon SE, Sims EF, Tovar IG, Weisser EG, Wertz JT, Weston-Hafer KA, Williamson KE, Zhang B, Cresawn SG, Jain P, Piuri M, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4. PLOS ONE. 2011a;6:e26750. doi: 10.1371/journal.pone.0026750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Best AA, Broussard GW, Connerly PL, Dedrick RM, Kremer TA, Offner S, Ogiefo AH, Pizzorno MC, Rockenbach K, Russell DA, Stowe EL, Stukey J, Thibault SA, Conway JF, Hendrix RW, Hatfull GF. Cluster J mycobacteriophages: intron splicing in capsid and tail genes. PLOS ONE. 2013;8:e69273. doi: 10.1371/journal.pone.0069273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al-Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD, Anderson AG, Ang AAS, Ares M, Jr, Barber AJ, Barker LP, Barrett JM, Barshop WD, Bauerle CM, Bayles IM, Belfield KL, Best AA, Borjon A, Jr, Bowman CA, Boyer CA, Bradley KW, Bradley VA, Broadway LN, Budwal K, Busby KN, Campbell IW, Campbell AM, Carey A, Caruso SM, Chew RD, Cockburn CL, Cohen LB, Corajod JM, Cresawn SG, Davis KR, Deng L, Denver DR, Dixon BR, Ekram S, Elgin SCR, Engelsen AE, English BEV, Erb ML, Estrada C, Filliger LZ, Findley AM, Forbes L, Forsyth MH, Fox TM, Fritz MJ, Garcia R, George ZD, Georges AE, Gissendanner CR, Goff S, Goldstein R, Gordon KC, Green RD, Guerra SL, Guiney-Olsen KR, Guiza BG, Haghighat L, Hagopian GV, Harmon CJ, Harmson JS, Hartzog GA, Harvey SE, He S, He KJ, Healy KE, Higinbotham ER, Hildebrandt EN, Ho JH, Hogan GM, Hohenstein VG, Holz NA, Huang VJ, Hufford EL, Hynes PM, Jackson AS, Jansen EC, Jarvik J, Jasinto PG, Jordan TC, Kasza T, Katelyn MA, Kelsey JS, Kerrigan LA, Khaw D, Kim J, Knutter JZ, Ko CC, Larkin GV, Laroche JR, Latif A, Leuba KD, Leuba SI, Lewis LO, Loesser-Casey KE, Long CA, Lopez AJ, Lowery N, Lu TQ, Mac V, Masters IR, McCloud JJ, McDonough MJ, Medenbach AJ, Menon A, Miller R, Morgan BK, Ng PC, Nguyen E, Nguyen KT, Nguyen ET, Nicholson KM, Parnell LA, Peirce CE, Perz AM, Peterson LJ, Pferdehirt RE, Philip SV, Pogliano K, Pogliano J, Polley T, Puopolo EJ, Rabinowitz HS, Resiss MJ, Rhyan CN, Robinson YM, Rodriguez LL, Rose AC, Rubin JD, Ruby JA, Saha MS, Sandoz JW, Savitskaya J, Schipper DJ, Schnitzler CE, Schott AR, Segal JB, Shaffer CD, Sheldon KE, Shepard EM, Shepardson JW, Shroff MK, Simmons JM, Simms EF, Simpson BM, Sinclair KM, Sjoholm RL, Slette IJ, Spaulding BC, Straub CL, Stukey J, Sughrue T, Tang TY, Tatyana LM, Taylor SB, Taylor BJ, Temple LM, Thompson JV, Tokarz MP, Trapani SE, Troum AP, Tsay J, Tubbs AT, Walton JM, Wang DH, Wang H, Warner JR, Weisser EG, Wendler SC, Weston-Hafer KA, Whelan HM, Williamson KE, Willis AN, Wirtshafter HS, Wong TW, Wu P, Yang YJ, Yee BC, Zaidins DA, Zhang B, Zúniga MY, Hendrix RW, Hatfull GF. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLOS ONE. 2011b;6:e16329. doi: 10.1371/journal.pone.0016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope WH, Jacobs-Sera D, Russell DA, Rubin DH, Kajee A, Msibi ZN, Larsen MH, Jacobs WR, Jr, Lawrence JG, Hendrix RW, Hatfull GF. Genomics and proteomics of mycobacteriophage patience, an accidental tourist in the mycobacterium neighborhood. mBio. 2014b;5:e02145. doi: 10.1128/mBio.02145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature Methods. 2012;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, Felts B, Haynes M, Liu H, Lipson D, Mahaffy J, Martin-Cuadrado AB, Mira A, Nulton J, Pasic L, Rayhawk S, Rodriguez-Mueller J, Rodriguez-Valera F, Salamon P, Srinagesh S, Thingstad TF, Tran T, Thurber RV, Willner D, Youle M, Rohwer F. Viral and microbial community dynamics in four aquatic environments. The ISME Journal. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rohwer F. Global phage diversity. Cell. 2003;113:141. doi: 10.1016/S0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses–major players in the global ecosystem. Nature Reviews Microbiology. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]