Abstract
Group II introns are some of the largest ribozymes in nature, and they are a major source of information about RNA assembly and tertiary structural organization. These introns are of biological significance because they are self-splicing mobile elements that have migrated into diverse genomes and played a major role in the genomic organization and metabolism of most life forms. The tertiary structure of group II introns has been the subject of many phylogenetic, genetic, biochemical and biophysical investigations, all of which are consistent with the recent crystal structure of an intact group IIC intron from the alkaliphilic eubacterium Oceanobacillus iheyensis. The crystal structure reveals that catalytic intron domain V is enfolded within the other intronic domains through an elaborate network of diverse tertiary interactions. Within the folded core, DV adopts an activated conformation that readily binds catalytic metal ions and positions them in a manner appropriate for reaction with nucleic acid targets. The tertiary structure of the group II intron reveals new information on motifs for RNA architectural organization, mechanisms of group II intron catalysis, and the evolutionary relationships among RNA processing systems. Guided by the structure and the wealth of previous genetic and biochemical work, it is now possible to deduce the probable location of DVI and the site of additional domains that contribute to the function of the highly derived group IIB and IIA introns.
Keywords: RNA splicing, transposition, ribozyme, enzymology, crystallography
General introduction
Group II introns are a large and diverse family of self-splicing retroelements that have played a major role in shaping the genomic organization of terrestrial life forms (Mattick, 1994; Mattick and Gagen, 2001; Rest and Mindell, 2003), from bacteria (Martinez-Abarca and Toro, 2000) to multicellular eukaryotes (Bonen and Vogel, 2001; Lehmann and Schmidt, 2003; Robart and Zimmerly, 2005; Pyle and Lambowitz, 2006). Splicing by group II introns occurs through several different pathways, and, although splicing chemistry is catalyzed by intron RNA, splicing and reverse-splicing reactions are facilitated by host and intron-encoded proteins (Belfort et al., 2002; Lambowitz and Zimmerly, 2004; Solem et al., 2009).
Group II introns are not extensively conserved in primary sequence, but they share a common secondary structure (Figure 1) (Michel et al., 1989; Qin and Pyle, 1998; Toor et al., 2001), and it is likely that their basic tertiary structures are also quite similar (Michel et al., 2009; Toor et al., 2009). Understanding the group II intron tertiary structure has been the goal of many investigations, employing powerful phylogenetic, genetic, biochemical, spectroscopic and crystallographic methods (Dayie and Padgett, 2008; Pyle, 2008; Toor et al., 2009; Michel et al., 2009). Together, these complementary approaches have resulted in a detailed picture of group II intron architecture that has yielded new insights into molecular evolution and splicing mechanism. Perhaps more broadly, the work has expanded our understanding of RNA folding and of tertiary structure stabilization, helping to define the building blocks by which large, complex RNA molecules assemble into unique structures.
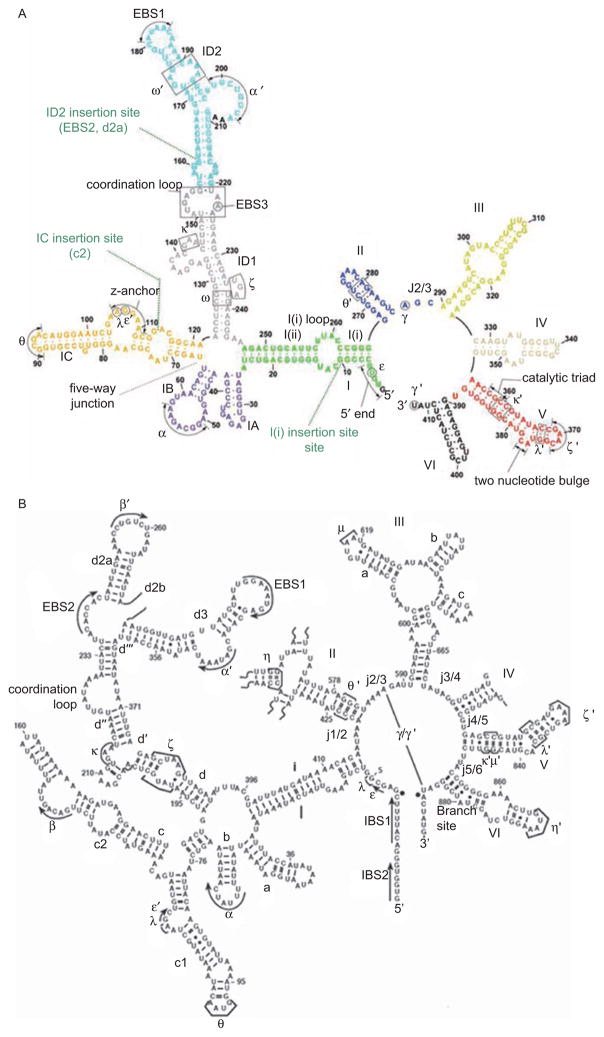
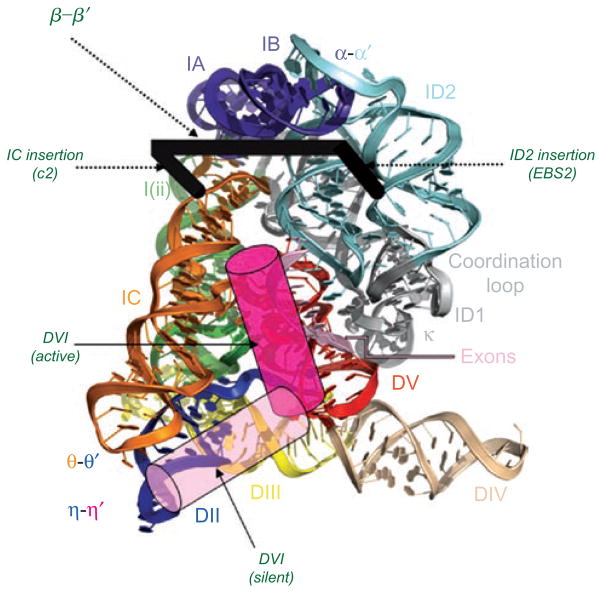
Figure 1.
Secondary structures of smaller (Group IIC) and larger (Group IIB) intron variants. (A) Secondary structure of the crystallized group IIC intron construct from Oceanobacillus iheyensis (O.i.). The sequence of the full-length O.i. intron is provided in Figure S2 of Toor et al. (2008). Domains are indicated with roman numerals and long-range interactions are indicated by Greek letters. Color coding is the same as shown in Figure 6, for the three-dimensional structure. Probable sites of the ID2, IC and I(i) insertions are indicated with green dotted lines, as indicated. (B) Secondary structure of the ai5γ Group IIB intron from the mitochondrial genome of Saccharomyces cerevisiae. Domains are indicated with roman numerals and subdomains with lower case letters. Long-range tertiary interaction partners are indicated with Greek letters. Note that EBS1 and EBS2 form Watson–Crick base pairs with IBS1 and IBS2 of the 5′-exon, respectively. Exon/intron boundaries are indicated by black dots.
A. The evolution, biological function and reactions of group II introns
Modern forms of group II introns are common in bacteria (Ferat and Michel, 1993; Martinez-Abarca and Toro, 2000; Toro, 2003), archaea (Toro, 2003), and they are abundant in fungi and plants (Bonen, 2008), where they are found in the mitochondrial and chloroplast genomes (Michel et al., 1989; Fedorova and Zingler, 2007). They have also recently been observed in the genomes of certain animals (Dellaporta et al., 2006; Valles et al., 2008; Burger et al., 2009). Ancient forms of group II introns are believed to have entered the eukaryal lineage from primitive bacteria, during endosymbiont events that led to the major organelle structures (Rest and Mindell, 2003; Koonin, 2006). Once absorbed into eukaryotic genomes, group II introns are believed to have become fragmented, and to have developed into ribonucleoprotein machines such as the eukaryotic nuclear spliceosome (Wheelan et al., 2005; Koonin, 2006), which processes eukaryotic pre-mRNA molecules and which contains RNA components that are closely related to group II intron domains in structure and function. Indeed, it has been hypothesized that the spliceosome (Keating et al., 2010), most nuclear introns, components of telomerase, and the abundant LINE-element form of transposons all descend from group II introns, thereby constituting a significant portion of the human genome (Boeke, 2003).
Group II introns continue to have a major impact on the metabolism and genomic structure of modern organisms, as they are mobile elements that continue to diversify and bring new information when they migrate (Dai and Zimmerly, 2002; Simon et al., 2008). Some organisms have succeeded in “taming” group II introns, incorporating them as regulatory elements for controlling gene expression (Michel et al., 2007) or using them as a framework for new RNA processing machines, such as the unusual ribonucleoproteins (RNPs) that are essential for splicing of certain gene families in plants (Lambowitz and Perlman, 1990; Ostersetzer et al., 2005; Solem et al., 2009). But the major influence of group II introns stems from their retrotransposition in all of these organisms (Beauregard et al., 2008), and on the plasticity this confers to genomic organization (Lambowitz and Zimmerly, 2004). Indeed, fast mutation and genomic flux can lead to rapid diversification during periods of environmental stress, resulting from mutations that may increase fitness of an organism or species. Environmental factors and stress have been specifically shown to influence the mobility of group II introns (Coros et al., 2005; 2008; 2009), and the implications of these processes are soberly underscored by the isolation of a group II intron insertion within a multi-drug resistant clinical isolate of the pathogen Serratia marcescens (Centron and Roy, 2002).
There are three major families of group II introns – groups IIA, IIB and IIC (Toor et al., 2001) – and several more classes that appear, at least so far, to be restricted to bacteria (bacterial classes D, E and F) (Simon et al., 2008). The group IIA and IIB families are large introns that can undergo both site-specific homing into intronless alleles, as well as nonspecific retrotransposition into other sites (Lambowitz and Zimmerly, 2004; Pyle and Lambowitz, 2006). Introns of the group IIC class are almost half the size of its cousins (~400 nucleotides (nts) rather than 900 nts, exclusive of open reading frames (ORFs)), and yet these introns contain all of the major active-site components (Granlund et al., 2001, Toor et al., 2001). The IIC introns contain only a short, single 5′-exon recognition element (EBS1-IBS1, see below), and as a result they retrotranspose in a sequence-nonspecific manner, behaving purely as retroelements (Dai and Zimmerly, 2002; Simon et al., 2008). These introns are believed to be the most ancient lineage, and they are likely to have evolved into the more derived IIA and IIB classes (Rest and Mindell, 2003). Consistent with divergence before the endosymbiont event, group IIC introns are observed only in bacteria (Simon et al., 2008). Because group IIC introns are small and compact, they are excellent systems for biophysical investigation of the group II intron structure (Toor et al., 2008a; 2009).
The profound impact of group II introns on the evolution of terrestrial genomes stems from their diverse mechanisms for self-splicing and retrotransposition. Group II introns catalyze phosphodiester cleavage and ligation reactions on both DNA and RNA molecules, resulting in several possible mechanisms for excision and migration into new sites (Pyle and Lambowitz, 2006). The basic chemical reaction catalyzed by group II introns is the in-line, nucleophilic attack of an alcohol moiety or a water molecule on an activated phosphodiester linkage, resulting in release of a 3′-hydroxyl leaving group and a 5′-phosphate or new phosphodiester linkage (Figure 2) (Pyle, 2008). As expected for a typical SN2 reaction, this results in the inversion of configuration at phosphorus (Podar et al., 1995b). A two-metal ion mechanism for catalysis has been implicated by functional group mutagenesis studies showing that both the 3′-hydroxyl leaving group and the attacking nucleophile are coordinated to divalent metal ions (Figure 2) (Gordon et al., 2007, Gordon and Piccirilli, 2001, Sontheimer et al., 1999). Thus, group II introns have long been thought to be metalloenzymes, with a catalytic mechanism that is likely to resemble other phosphotransferase enzymes. This is consistent with the group II intron crystal structure, in which two metal ions are observed within the conserved active-site (Toor et al., 2008a; 2009).
Figure 2.
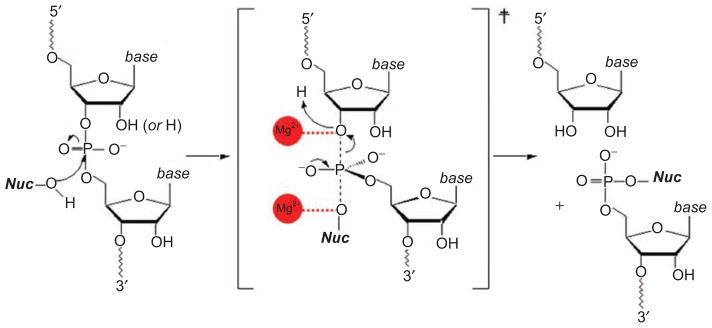
The chemical mechanism of catalysis by group II introns. This is the basic reaction underlying both steps of splicing and the reverse-splicing reactions involved in intron mobility. In this scheme, “Nuc” can represent the 2′-OH group of the branch-site, the 3′-OH group of the 5′-exon or the terminus of the intron, and it can also represent water. Note that the 2′-group on the departing sugar moiety can be a 2′-hydroxyl group (for RNA) or a 2′-hydrogen (for DNA), as the intron can react with either type of substrate. Metal ions are indicated with red balls, and they are in the positions indicated through biochemical work, and implicated by the position of metal ions in the crystal structure, as shown in Figure 11.
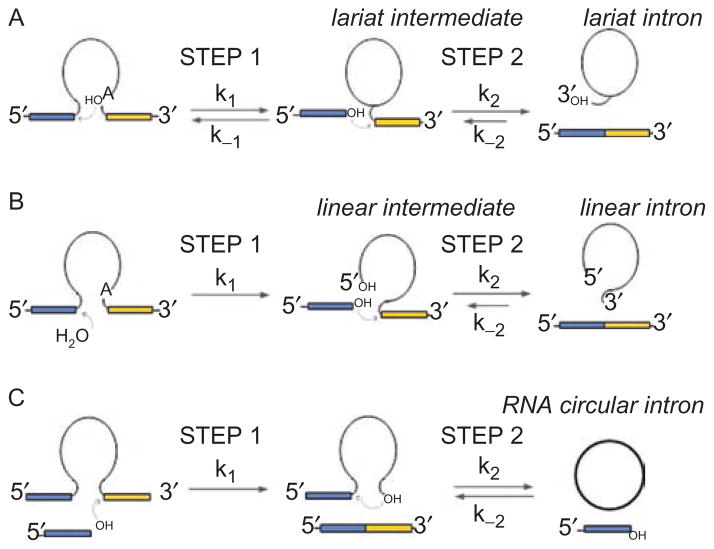
By employing the chemical reaction described in Figure 2, group II introns are excised through one of three pathways (Figure 3), resulting in free introns that can subsequently attack new target sequences through reverse-splicing, leading to intron mobility. A prevalent pathway for self-splicing and intron release is lariat formation (Figure 3A) (Perlman and Butow, 1989; Perlman and Podar, 1996), whereby the 2′-hydroxyl group on a specific adenosine within intron domain VI (the branch-point A) attacks the 5′-splice site, resulting in a lariat-3′-exon intermediate. This is followed by the second step of splicing, in which the 3′-hydroxyl group on the 5′-exon attacks the 3′-splice site, resulting in ligated exons and free lariat intron molecules. Both steps of this reaction are highly reversible (Pyle and Lambowitz, 2006), and free intron molecules can utilize reverse-splicing to insert themselves into DNA and RNA target molecules that contain sequences similar to the original 5′-exon. While the chemical steps of splicing and reverse-splicing are catalyzed by intron RNA, proteins play essential roles in the function of group II introns. Host and intron-encoded proteins facilitate formation of active RNA structures, and they are required for most stages of retrotransposition and mobility (Belfort et al., 2002; Lambowitz and Zimmerly, 2004; Pyle and Lambowitz, 2006; Beauregard et al., 2008; Solem et al., 2009).
Figure 3.
Three pathways for group II intron splicing and intron excision. (A) The branching pathway that leads to lariat formation. (B) The hydrolytic pathway that leads to release of linear intron. Note that the reverse reactions shown for (A) and (B) are the basis for the intron mobility reaction, whereby group II introns insert themselves into the sense-strand of target DNA (Pyle and Lambowitz, 2006). (C) A possible circle formation pathway. This reaction may be initiated by a free 5′-exon that has been liberated through the spliced-exon reopening reaction on a different intron boundary. Note that 5′-exons are shown in blue and 3′-exons are shown in yellow. The intron is indicated by a thin black line.
A second common pathway for self-splicing involves the attack of water as the nucleophile during the first step of splicing (van der Veen et al., 1987; Jarrell et al., 1988; Daniels et al., 1996; Podar et al., 1998; Vogel and Borner, 2002), which leads to the release of ligated exons and free linear introns (Figure 3B). This process readily occurs in most introns that have been biochemically studied, and it represents a major competitive pathway with lariat formation both in vitro and in vivo (Pyle and Lambowitz, 2006). The first step of reverse-splicing by linear intron is highly efficient (Roitzsch and Pyle, 2009) and it has been shown to represent a viable pathway for subsequent intron mobility (Zhuang et al., 2009). While branching is generally believed to be the most common pathway by which group II introns self-splice and retrotranspose, it is clear that hydrolytic splicing represents a potential mechanistic alternative that may confer significant advantages under specific environmental conditions. Indeed, there are entire families of group II introns that self-splice only through hydrolysis (Bonen and Vogel, 2001; Vogel and Borner, 2002).
A third pathway for intron release involves circle formation (Jarrell, 1993; Murray et al., 2001; Li-Pook-Than and Bonen, 2006; Molina-Sanchez et al., 2006), through a pathway reported for several group IIB introns (Figure 3C). This mechanism results in the release of intron circles, rather than the short 3′-tails that typify lariat molecules. Circular forms of RNA (including lariats of group II introns and group I-like ribozymes (Nielsen et al., 2005)) are highly resistant to cellular nucleases, thereby providing significant advantages to selfish, mobile RNA elements that must persist in stable forms within host organisms (Pyle, 2005). Although there are several possible models for the formation of intron circles, it has been suggested that circle formation is initiated by the attack of free 5′-exon molecules, which are an abundant product of spliced-exon reopening (a side-reaction catalyzed by linear and lariat group II introns in which the ligated exons are hydrolytically cleaved into free 5′ and 3′ exons). Free 5′-exon is thought to attack the 3′-splice site, liberating the downstream intron terminus while splicing to the 3′-exon (Jarrell, 1993; Murray et al., 2001; Molina-Sanchez et al., 2006). In the second step of circle formation, the terminal 3′-hydroxyl (or 2′-hydroxyl) group of the intron may attack the 5′-splice site, releasing intron circles and regenerating free 5′-exon. Although it is not yet well characterized, it is believed that the liberated intron circle can join with maturase proteins to catalyze intron mobility through pathways that have not yet been described.
B. Major functional domains and their architectural organization
The domain structure of group II introns derives from the earliest phylogenetic analysis of their secondary structures (Michel et al., 1989). This domain organization, along with knowledge of numerous tertiary interactions and structural constraints, has allowed for the construction of hypothetical three-dimensional models for group II introns (DeLencastre et al., 2005; Hamill and Pyle, 2006; Dai, 2008). Despite new information on the functional tertiary structure (Toor et al., 2008; Toor et al., 2008a), the original domain designations and intron models remain robust and useful (Figure 1). Of the two exons that flank group II introns, the 5′-exon displays extensive base complementarity with recognition sequences within D1 (the EBS1-IBS1 and EBS2-IBS2 pairings, Figure 1B) (Jacquier and Michel, 1987), while 3′-exon recognition is limited to the single EBS3-IBS3 base pair (Costa et al., 2000). Domain I (DI) is the largest region of the intron and it is divided into various sectors that control RNA folding, exon recognition, and the docking of catalytic domains. RNA folding studies conducted in vitro indicate that DI folds first and it serves as a scaffold for subsequent assembly of the other domains (Fedorova and Zingler, 2007; Pyle et al., 2007). The overall architecture of DI is dictated by proper formation of the conserved five-way junction (Toor et al., 2008a) and by a set of internal loops in the κ–ζ region of stem DId (Figure 1) (Waldsich and Pyle, 2007). Domain II contains motifs important for architectural assembly (Costa et al., 1997), and it can function as a site for encoding ORFs. The junction between DII and DIII (J2/3) is among the most conserved regions of the intron, and it plays a key role in construction of the active-site (Toor et al., 2008a). DIII contains a semi-conserved internal loop that is important for structural assembly and this domain forms a section of the active-site cleft (Toor et al., 2010). DIV functions primarily as the site of ORF insertion, and it encodes “maturase” proteins that are obligate partners in splicing and intron mobility (Zimmerly et al., 2001). The most phylogenetically conserved and functionally critically region of the intron is domain V (Toor et al., 2009) (Figure 4). Together with J2/3, DV forms the active site for ribozyme activity by the intron and almost every atom on this domain serves a specific function (Figures 4 and 5). DVI is not highly conserved, but it contains the location of the branch-point adenosine, which is usually a bulged nucleotide that is located near the base of the hairpin (Figure 1B) (Chu et al., 2001). These various domains assemble into an elaborate three-dimensional structure that finally explains patterns of conservation and covariation that were apparent, but unexplained, for many years.
Figure 4.
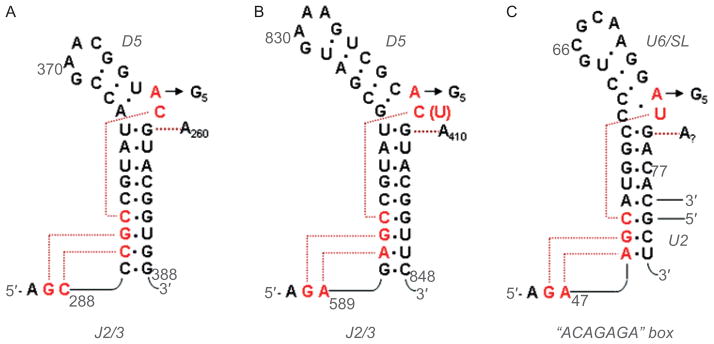
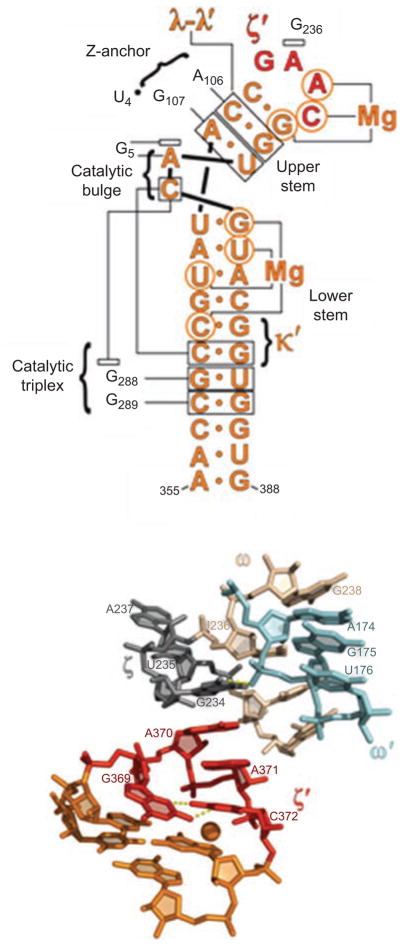
Secondary structures of DV and the U6 snRNA, together with anticipated contacts within the active-site. (A) The DV and J2/3 sequences from group IIC intron Oceanobacillus iheyensis, with active-site tertiary interactions determined from the crystal structure (Keating et al., 2010). The J2/3 nucleotides form a major groove triplex with the “catalytic CGC triad” of conserved nucleotides at the base of DV, and with C259 from the bulge. (B) The DV and J2/3 sequences from group IIB intron ai5γ. Anticipated long-range interactions are shown, by analogy to the O.i. crystal structure. These can be readily modeled into the active-site of the solved structure (Keating et al., 2010). (C) The U6 snRNA from the human spliceosome. The ACAGAGA box is hypothesized to function like J2/3, forming a triple helix that supports the metal-binding, actives-site bulge at positions 73 and 74. The U6 bulge region is hypothesized to form tertiary interactions similar to those observed within the IIC intron (Keating et al., 2010). Reprinted with permission from RNA (Keating et al., 2010).
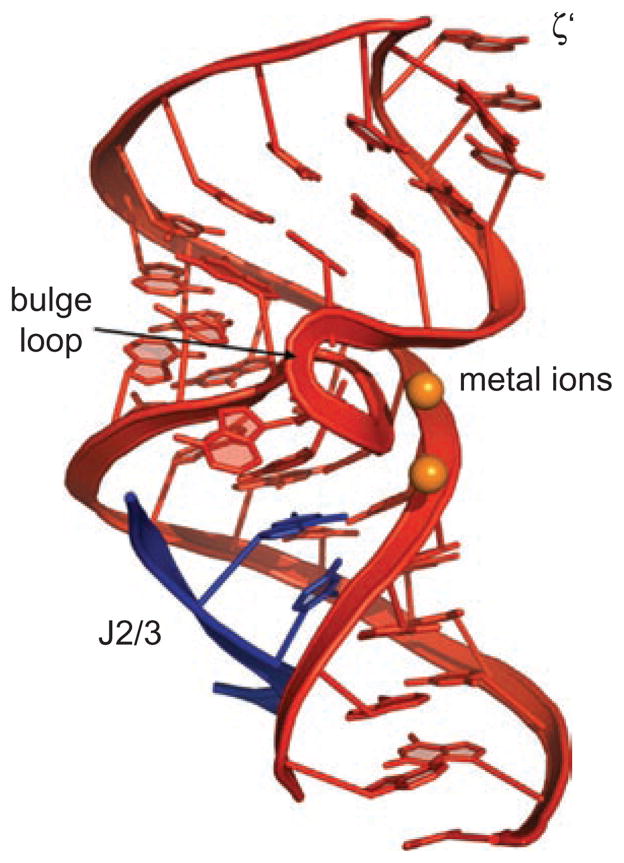
Figure 5.
The structure of DV within the catalytic core of the folded group IIC intron. DV nucleotides are shown in red. Two metal ions (orange spheres) are bound near the tightly twisted bulge loop. The J2/3 nucleotides (blue ribbon) form a triple helix with the major groove at the base of DV. The site for ζ–ζ′ interaction with DI is indicated. This image was rendered from pdb code 3IGI.
Domain V and the reactive center of the intron
The group II intron is constructed like a seed, which is apropos given its role in disseminating genetic information and generating genomic complexity. The kernel of this seed is the small, highly conserved hairpin loop structure (DV) that contains a small “elbow”, or bulge of two bases (Schmidt et al., 1996), in the middle of the stem (Figure 4). DV has long been known to be the most important component for the catalytic reactivity of group II introns (Lehmann and Schmidt, 2003; Pyle and Lambowitz, 2006; Toor et al., 2009). If one believes that the U6 snRNA is related to DV (Figure 4), which is likely given their many common elements (Keating et al., 2010), then DV has changed very little from bacteria to man.
When bound within the center of the intron, which enfolds DV like a protective shell (Toor et al., 2008a, 2010, Keating et al., 2010), DV adopts a conformation that is highly conducive to catalytic reactivity (Figure 5). The asymmetric internal loop twists tightly upon itself, concentrating the backbone phosphates in space and thereby creating a site of extremely negative electrostatic potential. Perhaps as a result, two divalent cations bind tightly at this site, and they are spaced 4Å apart, much like the catalytic metal ions that are common in other phosphodiesterase protein enzymes and in group I introns (Steitz, 1993; Stahley and Strobel, 2006; Stahley et al., 2007). Based on their spacing, their attachment to specific RNA ligands that are required for reaction chemistry, and by their location relative to scissile phosphodiester linkages on bound substrates (Toor et al., 2008b), it is highly likely that these two metal ions play a key role in the catalytic mechanism of group II introns. The metal ion binding site is supported from below by a triple-helical structure that results from binding of a conserved junction region (J2/3) to an invariant region within the major groove of the DV lower stem (Keating et al., 2010). This major-groove triple helix is a new type of RNA motif that appears to be of general importance in biologically important RNAs, including the pseudoknot from telomerase RNA (Theimer et al., 2005; Keating et al., 2010).
The surrounding intronic substructure appears to be essential for maintaining this catalytically reactive conformation of DV (Toor et al., 2008a) (Figure 6). This is achieved through the formation of the triple helix with J2/3 and through interactions such as λ–λ′ within the Z-anchor region in D1 (Boudvillain et al., 2000; Toor et al., 2008a), which enforce the unique base conformations within the DV bulge region and enable the negative charge on the backbone to become highly focused, like a lens of catalytic potential.
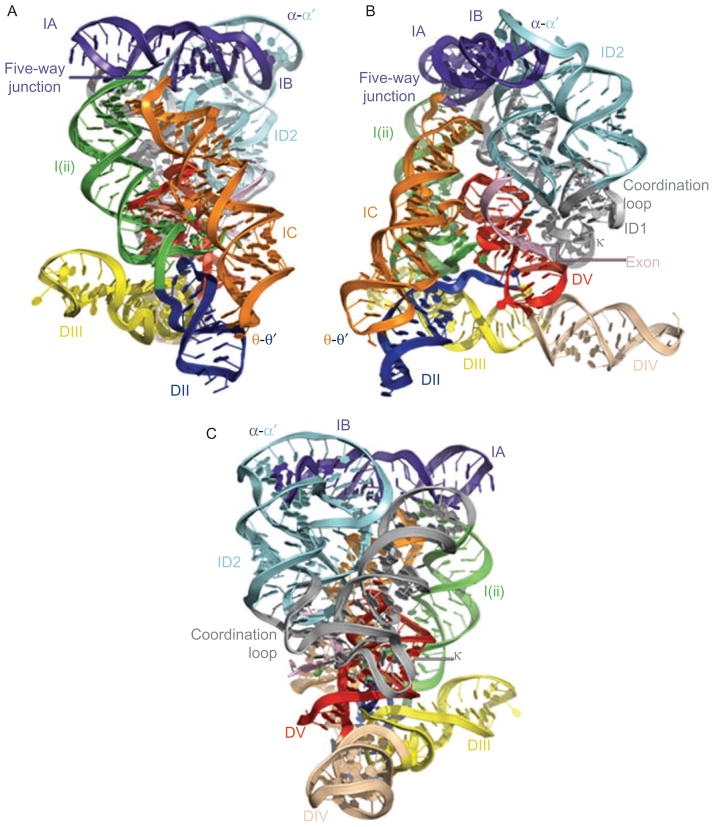
Figure 6.
Three views of the molecular structure of the Oceanobacillus iheyensis group IIC intron (Toor et al., 2010), color-coded and labeled as shown in Figure 1A. This image was rendered from pdb code 3IGI. Reprinted with permission from RN (Toor et al., 2010).
In addition to these interactions that support catalytic reactivity of the intron, there are several sets of interactions between DI and the “back” side of DV that are necessary for presenting and orienting catalytic groups properly within the active site. Based on functional group mutagenesis studies of bases and backbone residues of DV, it has long been known that the D5 hairpin has a “catalytic face” and a “binding face” on its opposite side (Abramovitz et al., 1996, Konforti et al., 1998). All of these have now been visualized crystallographically in their anticipated locations (Toor et al., 2008a). An essential interaction for docking DV is the ζ–ζ′ interaction that occurs between the tetraloop at the DV tip and a tetraloop-receptor in DI (Keating et al., 2008). While this is usually a cannonical tetraloop-receptor interaction in group IIA and IIB introns, it has evolved into an idiosynchratic form in group IIC introns that has only been observed in that unique context (Figure 7), suggesting that the motif has evolved to fit a particular structural niche (Keating et al., 2008). A second interaction is κ–κ′, which braces the lower helix of DV through a network of contacts with base and backbone moieties (Boudvillain and Pyle, 1998; Toor et al., 2010).
Figure 7.
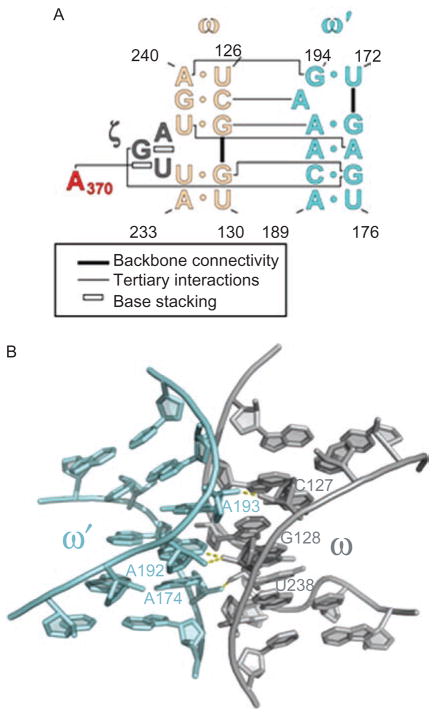
The network of interactions involving DV. (A) A secondary structural map of the interactions observed between DV and the rest of the intron. Brackets indicate sites of long-range interaction and contacts with metals, open rectangles indicate stacked nucleobases, labels indicate identity of tertiary interactions. Red nucleotides are involved in the ζ–ζ′ interaction. (B) The molecular structure of the ζ–ζ′ interaction, determined from the crystal structure of the Oceanobacillus iheyensis group IIC intron. Unlike most tetraloop receptors, this interaction is dictated only by a single stack between A370 of DV and G236 in DI. The unusual tetraloop contains a bound metal ion. The receptor is buttressed by the ω–ω′ interaction that is idiosynchratic to IIC introns (Keating et al., 2008). First published in J Mol Biol (2008) 283, 475–481; Figures 1a and 2a, pages 476 and 478 respectively.
Given these many activating interactions between DI and the DV helix (Figure 7), it is tempting to speculate that the intact intron is absolutely required for catalytic potential of DV. While this appears to be true for DV in group II introns (DV displays no phosphodiesterase reactivity when completely by itself), it is not necessarily true for the U6 snRNA, which has been shown to have a small amount of phosphodiesterase activity when presented with RNA substrates (Valadkhan et al., 2007; 2009). In isolation, DV and U6 may occasionally sample the active conformation and be capable of reaction. This will be particularly true for U6, which is more “self-sufficient” in that it carries the equivalent of the J2/3 triple helical strand along with it, in the form of the appended ACAGAGA box upstream of the helical region (Keating et al., 2010) (Figure 4).
Domain I and the intron shell
Domain I (DI) is the largest intronic domain and it is transcribed first, in its entirety, when the intron is being synthesized (Figure 1). Not surprisingly, this domain also folds first (Figure 8), forming the basic scaffold for subsequent docking by downstream catalytic and ORF-containing domains (Qin and Pyle, 1997; Pyle et al., 2007; Fedorova et al., 2007; Steiner et al., 2008). When examining the secondary structure, DI can be viewed as a bipartite domain, with two major lobes that are connected by a narrow neck region (helix ID or Id). The center of this neck contains a structurally conserved internal loop region that is located at the approximate position of κ and ζ (the “folding control element”, see Figure 1B), and it appears to be very important for conferring the proper shape of D1 (Waldsich and Pyle, 2007; 2008).
Figure 8.
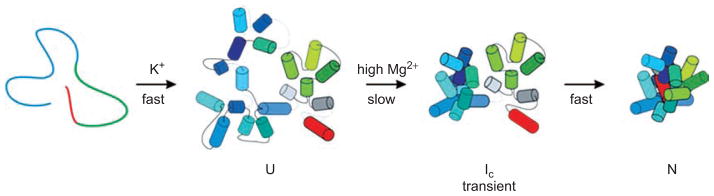
Generalized tertiary folding pathway of the group IIB intron ai5γ. After forming the basic secondary structure (defined here as U), domain I (blue cylinders) folds first, forming an intermediate (Ic) in which the catalytic domains are not docked within the core. The Ic–N transition involves the docking of D5 and D3. First published in TIBS (2007) 32, 138–145; Figure 3a, page 142.
Two folding events appear to dictate the architectural organization of DI (both of which lead to the U → I transition shown in Figure 8). These include a solidification of the folding control element near κ and ζ, which is rate-limiting in the ai5γ group IIB intron (Waldsich and Pyle, 2007; 2008). Folding of this region may bend the neck of DI and enable the two lobes of the domain to interact, thereby forming α–α′ and other features. A second folding event that is essential for proper placement of the D1 helices is the local folding and coaxial stacking network of the five-way junction (Toor et al., 2008a, 2010), from which all the DI subdomains radiate.
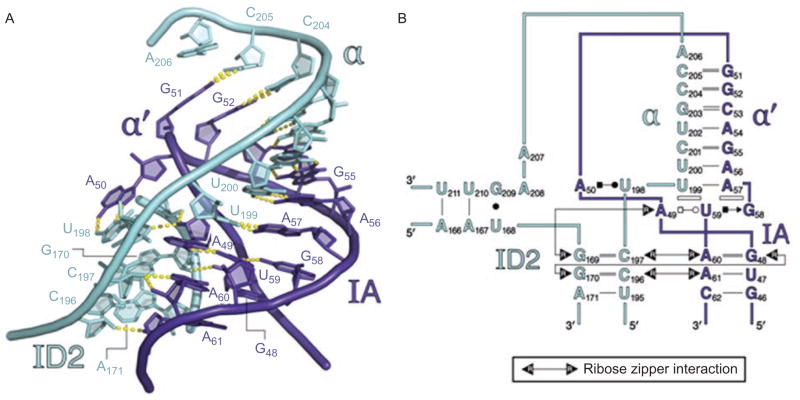
Once DI has folded, one can imagine that it is shaped like a square pillow that has been twisted slightly around its middle (Toor et al., 2010) (Figure 6). This visualization allows one to define four basic “corners” of the domain, each of which is terminated by a specific tertiary interaction motif. The interaction motifs at each vertex define the specificity of each fold, but they are unlikely to be the only thermodynamic determinants for local stability in each region. Indeed, each of these motifs (discussed below) is flanked on all sides by extensive ribose zippers that effectively anneal adjacent strands and helices together, thereby cementing the local structure in a fixed configuration (Figure 9) (Toor et al., 2010).
Figure 9.
The α–α′ interaction and its supporting ribose zipper network. (A) Molecular structure of α–α′, showing that the strands beneath it are joined through sequential contacts between 2′-hydroxyl groups (ribose zippers). (B) Secondary structural representation of the same region. Open rectangles indicate base stacking interactions. Types of base-pairing are represented using the Leontis–Westhof symbolism. Color coding is as shown in Figure 1A and 6. Reprinted with permission from RNA (Toor et al., 2010).
Along the “top” edge of the DI scaffold, there are two vertices, or “corners”, formed by the five-way junction and α–α′ motifs, respectively (Figure 6). The five-way junction region is comprised of a T-loop (formed by the terminus of helix IA), that is intercalated by an adenosine from stem ID1 (A245). This motif forms the top of an extended stacking network that brings numerous strands together through an array of interdigitated bases and buttressing hydrogen bonds. On the opposite side of the square, the second vertex is provided by the α–α′ region (Figures 6 and 9) (Michel et al., 1989; Harris-Kerr et al., 1993), which is a kissing loop interaction that is reinforced from below by a new interaction, ω–ω′, that was identified only through analysis of the crystal structure and was not apparent from previous analyses (Toor et al., 2008a). The ω–ω′ interaction is a type of ribose zipper that occurs in a sequence-specific context (Figure 10) (Tamura, 2002). Like the idiosynchratic ζ–ζ′ (Figure 7), it appears to be unique to group IIC introns (Keating et al., 2008; Toor et al., 2008a).
Figure 10.
The ω–ω′ interaction of group IIC introns. (A) Secondary structural representation of this sequence-specific ribose zipper motif. Color coding is as shown in Figures 1A and 6. First published in J Mol Biol (2008) 283, 475–481; Figure 1b, page 476. (B) Molecular structure of the ω–ω′ region within the Oceanobacillus iheyensis group IIC intron, showing the ribose zipper hydrogen bonds in yellow. First published in Science (2008) 320, 77–82; Figure 3c, page 79.
Along the “base” of the DI scaffold, the two major “corners” are comprised of the θ–θ′ region and the κ-coordination loop region. The θ–θ′ interaction, which is a cannonical tetraloop-receptor interaction that was phylogenetically predicted (Costa et al., 1997), joins the tip of the IC helix with the stem of DII (Figures 1 and 6). This important interaction serves to brace the IC helix and to govern the ultimate orientation of the DII and DIII stems. The κ-coordination loop region is a highly complex RNA substructure that cradles the back side of DV and provides the binding interface for the 5′ and 3′-exons (and the branch-site, if it were present) (Boudvillain and Pyle, 1998; Costa et al., 2000; Hamill and Pyle, 2006). This region of the molecule is highly elaborate and does not readily lend itself to description using conventional motif nomenclature. A detailed analysis of its multi-lobed architecture has revealed networks of purine–purine base stacks, ribose interactions and multi-stranded structures (Costa et al., 2000; Toor et al., 2010).
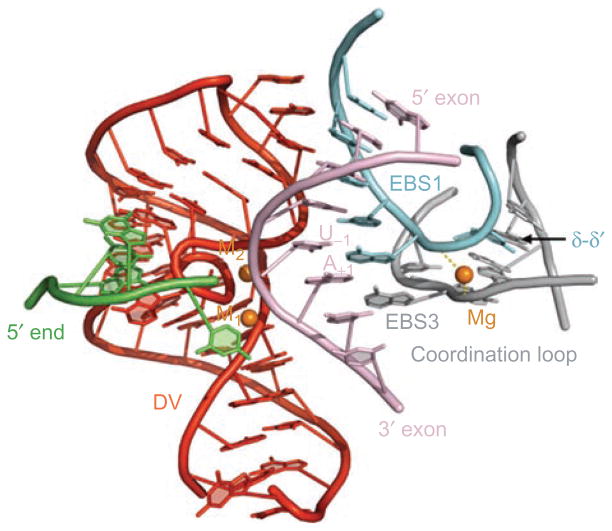
DI recognizes both the 5′ and 3′ exons simultaneously through base-pairing with a contiguous recognition surface that is composed of the EBS1 and EBS3 elements (Figure 11). These two substructures are stacked on one another and melded together through formation of the phylogenetically-predicted δ–δ′ interaction (Costa et al., 2000) within the coordination loop (so named because it coordinates the approach of all reactants necessary for splicing: 5′-splice site, 3′-splice site and branch-site; see Costa et al., 2000; Hamill and Pyle, 2006). Furthermore, the backbones of strands extending from EBS1 and EBS3 are brought into close proximity through interactions with a bridging magnesium ion (Toor et al., 2009, Toor et al., 2008b). The existence of this continuous binding interface for both exons explains how the two splice sites are brought together before the first step of splicing, and how their relative positions are maintained through the second step (Toor et al., 2008b; 2009).
Figure 11.
Exon recognition surface within the Oceanobacillus iheyensis group IIC intron. Bound target oligonucleotide (pink) interacts with the intron core through base-pairing interactions with EBS1 (blue) and EBS3 (gray), which are joined together and form one structural unit that is stabilized through the δ–δ′ interaction and a bridging metal ion. Note that the result is a duplex in which EBS3 stacks beneath EBS1, as they both base pair simultaneously with the oligonucleotide. Rendered from pdb code 3IGI.
Domain II: a scaffolding element that provides sites for external projections
Domain II contains key anchor points for the DIC helix (through the θ–θ′ interaction (Costa et al., 1997) as described above) and for a “silent” conformation of DVI (through the η–η′ interaction (Costa et al., 1997), as described below). Once DII is locked into position through θ–θ′ and a second network of contacts with the S-turn in DIII (see next section), the DII terminus projects away from the intron core and out into solution (Figure 12) (Toor et al., 2008a; 2010). In this location, DII can serve as site for additional elaborations to the intron structure that do not interfere with the function of the ribozyme core. It is therefore not surprising that DII may contain insertions of additional RNA which, in some cases, form ORFs for encoded proteins (Simon et al., 2008). Thus, DII might serve as a potential vehicle for encoding additional genes that are brought into new host sites when the intron retrotransposes. However, ORFs in DII are much more uncommon than the ORFs that are found within intron domain IV (DIV), which almost always carry genes for encoded proteins(Simon et al., 2008).
Figure 12.
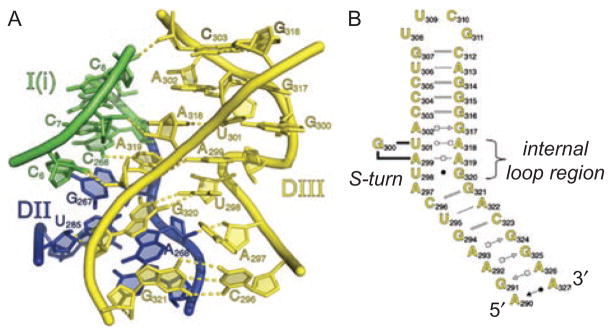
The internal loop in DIII dictates placement of the DI and DII stems. (A) The molecular structure of the DIII loop region (yellow), showing that it forms extensive interactions with the bottom of the DI (blue) and DII (green) stems, helping the junction of these coaxially stacked stems to become properly oriented within the structure. Rendered from pdb code 3IGI. (B) Secondary structure of DIII from Oceanobacillus iheyensis, as determined from the crystal structure. Color coding is the same as in Figures 1A and 6. Reprinted with permission from RNA (Toor et al., 2010).
Domain III and the adjacent J2/3 linker: elements for active-site construction
The appearance of DIII differs among the various types of group II intron, although if one looks closely, there is an important common element among them all (Toor et al., 2001). In each case, the basal DIII stem projects for almost a helical turn before it is punctuated by an internal loop that displays high levels of phylogenetic conservation, particularly within intronic subgroups (Michel et al., 1989; Toor et al., 2001; Simon et al., 2008). Biochemical studies have shown that the internal loop plays a major role in stimulating catalytic activity of group II introns (Podar et al., 1995a; Jestin et al., 1997; Fedorova and Pyle, 2008). The function of the DIII internal loop has been made clear by the crystal structure of the Oceanobacillus iheyensis (O.i.) intron, which reveals it to adopt a nearly helical structure that is composed of numerous purine–purine pairs, resulting in a very wide major groove that is made further accessible by a canonical S-turn motif (Leontis, 1998; Toor et al., 2010). This flat platform, which is rich in molecular recognition determinants projecting from the bases, is a receptor for binding the basal stems of both DI and DII, which are coaxially stacked upon one another (Figure 12). This unique junction between stems of DI, DII and DIII results in a multi-stranded structure that sets the three major intron structural domains into their proper orientation.
More derived introns, such as the IIB family, contain additional domains that extend from this junction (Michel et al., 1989; Toor et al., 2001), potentially supporting the intron superstructure in additional ways (Fedorova and Pyle, 2008). For example, the μ–μ′ interaction between the DIIIa terminal loop of group IIB intron ai5γ and the κ′ region of DV appears to result in a more elaborate κ–κ′ interaction in group IIB introns (Figure 1B) (Fedorova and Pyle, 2005), potentially strengthening the binding interface between DV and the outer intron shell. In addition, the single-stranded region between helices DIIIa and DIIIb is involved in functional communication with the basal stem of DII (Fedorova et al., 2003), which is part of the θ–θ′ tertiary contact. This suggests that, in group IIB introns, the θ–θ′ interaction site may include elements of DIII. Biochemical studies suggest that in group IIB introns, the internal loop of DIII is proximal to the ε–ε′ interaction within in D1, and that it may allosterically influence the bulge of DV (Jestin et al., 1997; Fedorova and Pyle, 2008). Thus, the positioning of the DIII internal loop in IIB introns may be slightly different than that in IIA and IIC introns, in that it has shifted into a position that facilitates interactions with active site elements ε–ε′, J2/3 and the bulge of DV (Fedorova and Pyle, 2008).
Domain IV and the ORF region
DIV and DIII coaxially stack upon each other (Toor et al., 2008a) (Figure 8), and this interaction sends the DIV helix away from the intron core and into peripheral space. Given this position away from the catalytic center, DIV is located in an ideal site for encoding ORFs and additional genetic structures that contribute to mechanisms of intron mobility. Because DIV is a peripheral domain that projects away from the catalytic center, it is free to balloon out, to evolve and develop without disturbing basic intronic function. Thus, the maturase proteins that facilitate splicing and retrotransposition are encoded by ORFs in DIV, and other genes are also found inserted at this region (Zimmerly et al., 2001; Lambowitz and Zimmerly, 2004; Simon et al., 2008). One can therefore visualize DIV as a vehicle for carrying extra parts, allowing them to be disseminated when the intron migrates. In addition, this domain often contains stem-loop regions that contribute to intron recognition and binding by maturase proteins that are translated from the ORF(Wank et al., 1999).
C. The location of the branching region: domain 6
Although the crystal structure of the O.i. group IIC intron has provided complete information on the structure and placement of DI–DV, there is a disappointing lack of electron density attributable to the DVI hairpin (Toor et al., 2010). The O.i. intron was crystallized from a full-length, self-splicing construct that contained an intact DVI region and it was therefore puzzling when this substructure could not be visualized. The result was not entirely surprising because DVI has long been known to flip in and out of the intron core, having few tertiary contacts with which it can form a stable network of interactions (Dib-Hajj et al., 1993; Chin and Pyle, 1995; Chu et al., 1998; 2001). The dynamic nature of DVI may be important for its function, as described below, but it makes crystallization very challenging. In order to determine whether DVI was simply disordered in the crystal or whether it had somehow become degraded, a biochemical investigation was conducted on the O.i. construct before and after crystallization (Toor et al., 2010). These studies revealed that, after splicing in vitro, the O.i. construct “bites off its own tail”, cleaving RNA from the 3′-end of the intron and ultimately excising DVI completely. Thus the dynamic behavior of D6 renders it vulnerable to attack by the highly reactive intron core, which chews it to bits under certain reaction conditions (Toor et al., 2010).
It is possible that one role of the encoded maturase protein (which was not included in the crystallization construct) is to control the fate and dynamics of DVI, enabling its functions to be controlled and preserving the intact intron for subsequent retrotransposition. This is supported by the fact that group IIC introns cannot undergo branching in vitro in the absence of the maturase protein, and they splice exclusively through the hydrolytic pathway (Granlund et al., 2001; Toor et al., 2006). However, group IIC introns in vivo have been observed to excise themselves as lariats, indicating that branching is viable under more native conditions that include the intron-encoded protein (Centron and Roy, 2002).
Although DVI cannot be directly visualized from the crystal structure, its location and even the path of its dynamic transitions can be deduced by combining our knowledge of the group II intron structural scaffold with biochemical studies on the architectural placement of DVI. When it docks within the core and participates in the branching reaction, there is only one probable location for the DVI hairpin: the domain is expected to lie within a crevice that is formed between the IC helix and κ-coordination loop substructure (dark pink cylinder, Figure 13). Based on site-directed crosslinking studies, DVI should be oriented along side the DIC helix (DeLencastre et al., 2005; Hamill and Pyle, 2006). In this position, DVI will form well-characterized interactions with the coordination loop and other regions of the intron (DeLencastre et al., 2005; Hamill and Pyle, 2006). In addition, the bulged adenosine branch-site will be poised over the scissile phosphodiester linkage of the 5′-splice site. Indeed, substrate (or exon, pink strand, Figure 13) strands will be necessarily sandwiched between DVI and DV, which are expected to lie side-by-side, as indicated by photocrosslinking experiments (Podar and Perlman, 1999).
Figure 13.
The hypothetical position of DVI, EBS2 and β–β′. Superimposed on a model of the group II intron crystal structure (rendered from pdb file 3IGI), the proposed position of various derived domains is shown. DVI is likely to have two functional conformations. The “active” conformation that participates in branching is likely to be in the position indicated by the dark pink cylinder. The “silent” conformation is stabilized by the η–η′ interaction between DVI and DII is likely to be located in the position indicated by the light pink cylinder. DVI probably transits between these two sites in order to toggle between branching and other aspects of intron function (see text). The likely sites of EBS2, β–β′ and the d2a and c2 stems are shown by the thick black bracket. The ID2 insertion (which evolved into EBS2 and the d2a stem) occurred approximately between nucleotides 114 and 115 (Oceanobacillus iheyensis numbering), placing EBS2 on the right hand side of the bracket. The IC insertion occurred approximately between nucleotides 158 and 159, placing helix c2 on the left hand side of the bracket. The β–β′ interaction between loops of c2 and d2a is shown as the connecting bar of the bracket, spanning structural domains of the core.
However, DVI has two mutually exclusive orientations, and both of them are likely to play an important role in the function of the intron. In its secondary conformation, DVI appears to be pulled completely out of the intron core and it is fastened along the intron periphery through a tetraloop-receptor interaction with DII (Figure 13, light pink cylinder) (Costa et al., 1997). This conserved interaction (known as η–η′), joins the tip of the DVI tetraloop with a distal region of DII (Costa et al., 1997). In this orientation, DVI is not expected to be capable of branching, as the branch-site is located at least 20 Å from the bulge of DV and from 5′-splice site bound within the core (Figure 13). Intriguingly, the crystal structure reveals a clear path for the DVI helix to traverse the distance between its primary conformation in the core and its secondary conformation joined to DII. Using the J5/6 linker region as a hinge, the DVI hairpin can swing ~90° in order to move between its two binding sites within and outside of the core (Figure 13).
It has been suggested that η–η′ might allow DVI to adopt two different conformations in the first and second steps of splicing (Chanfreau and Jacquier, 1996). Based on the existing crystal structure, it is possible that DVI might occupy the “active” conformation during the first step of splicing and then toggle to the “silent” conformation for the second step of splicing. This transition would require some adjustment in the position of DII for the O.i. intron, and it might be more probable in the more derived introns where DIII subdomains play a bigger role in the structure. Indeed, it has been shown that branch-site choice by the ai5γ group IIB intron is decoupled from 3′-splice site selection, which is consistent with the notion that DVI can exit the active-site during the second step (Chu et al., 2001). However, other evidence contraindicates this large conformation change between the two steps of splicing. For example, crosslinking studies indicate that DV and DVI residues are located in approximately the same architectural positions before, during and immediately after splicing (DeLencastre et al., 2005; Hamill and Pyle, 2006). Nonetheless, the DVI toggle remains an interesting way to conceptualize the active-site as it might switch between steps of splicing.
Alternatively, the η–η′ interaction may play a key regulatory role in the function of DVI, potentially by protecting this domain from degradation by the active site. It is possible that DVI is “stored” at η–η′, and released only when the 5′-splice site is inserted within the core and properly configured for the first step of splicing. However, the events that mediate this transition are presently unknown and await structural characterization of other intron isoforms.
Another regulatory role for the η–η′ interaction and the silent conformation of DVI lies with the fact that group II introns perform several reactions other than branching (see section A). Indeed, branching and lariat formation may not always be advantageous reactions during the splicing and transposition of group II introns, and these effects may be particularly pronounced under certain reaction conditions or environmental stresses. Group II introns commonly splice through an alternative hydrolytic pathway, which releases linear introns (van der Veen et al., 1987; Jarrell et al., 1988; Daniels et al., 1996; Podar et al., 1998; Bonen and Vogel, 2001; Vogel and Borner, 2002; Molina-Sanchez et al., 2006; Toor et al., 2006), and they can also undergo a set of transesterification reactions that release intron circles (Molina-Sanchez et al., 2006). All of these reactions are now known to be prevalent pathways for intron function and they have now both been shown to be relevant for specialized retrotransposition pathways (Lambowitz and Zimmerly, 2004; Molina-Sanchez et al., 2006; Roitzsch and Pyle, 2009; Zhuang et al., 2009). Thus, the η–η′ interaction may confer inherent mechanistic flexibility to group II introns, enabling them to sample one of several reaction pathways by shutting down the branching reaction.
D. The location of derived subdomains that have conferred new functions
The recent crystal structure of the O.i. group IIC intron has obviously provided a wealth of information on the spatial locations and catalytic functions of the major intronic substructures. The features revealed by the crystal structure represent those elements that are common among all group II introns, and they are likely to represent the most ancient and basic components of the intron construction (Rest and Mindell, 2003). Ironically, the crystal structure has also revealed the probable location of domains that are not within group IIC introns and are found within the more derived and highly evolved group IIA and IIB introns. These derived domains are indispensable to the function of other intron classes and it is therefore very important to know their architectural placement and biological role within the intron superstructure. It is possible to deduce the location of these domains with reasonable precision because they appear to derive from “insertions” of RNA into discrete locations within the group IIC helical framework (Figure 13, black bracket), which is well established from the crystal structure. This information, together with the extensive phylogenetic and biochemical characterization of the secondary and tertiary structure of the derived domains, allows one to model their approximate location upon the intron surface and to speculate on their role in elaborating group II intron function.
Like the insertion of many sequences into genomes, it is not clear how the group II intron insertions occurred, what mobile elements were involved and why these particular substructures persisted through time. However, it is notable that all the known insertions (ID2, IC, I(i) and 3′-terminal, described below) are observed at positions where IIC introns contain an internal loop (see Figure 1A). This may imply that insertions occurred on the RNA level, or it may imply that modern IIC introns are actually degenerated forms of introns that once contained the same substructures as IIA and IIB introns. However, phylogenetic analysis of group II intron lineages is inconsistent with the latter model (Rest and Mindell, 2003; Simon et al., 2008).
The ID2 insertion
The EBS2 region and the d2a helix (which are essential to the function of IIB introns (Figure 1B)) appear to have arisen from the insertion of an RNA stem-loop structure into the ID2 strand (Figures 1A and 13). Based on the typical location of these regions in secondary structure, the insertion occurred on the exterior of the ID2 helix, at a position that is almost exactly between α–α′ and the coordination loop, approximately 1/2 turn from each (Figure 13). From this location, the d2a hairpin loop is likely to extend away from the intron core, allowing it to present the EBS2 strand and the β′ loop strand for molecular recognition by their corresponding pairing partners. The ID2 insertion and the EBS2/β′ elements that evolved from it are found in all IIA and IIB introns (Michel et al., 1989), suggesting that this substructure developed prior to the divergence of IIA and IIB introns. It is notable that EBS2, but not β–β′ and its associated stems, is found in bacterial class E and F introns (Simon et al., 2008), implying that the evolution of EBS2 and β–β′ are not necessarily linked.
The EBS2 element
At the base of the ID2 insertion, the EBS2 region is situated immediately next to EBS1 in three-dimensional space (Figures 1B and 13). It is therefore located at an ideal site for recognition of the 5′-exon, with which it forms an extended set of Watson–Crick base-pairs in group IIA and IIB introns. The evolution of the EBS2-IBS2 recognition element allowed IIA and IIB introns to achieve their extraordinary levels of sequence-selectivity and to develop from sequence-nonselective retroelements (as in the IIC introns) to highly selective homing introns that target specific genes (as in the IIB introns). Many group IIB introns, for example, have extended EBS1/EBS2 recognition surfaces that enable them to form more than 13 sequence-specific base-pairs with target DNA or RNA elements (Xiang et al., 1998; Su et al., 2001). This attribute endows group IIB (and probably IIA) introns with sufficient specificity to potentially target a single gene in a human being (Xiang et al., 1998). Group II intron gene therapy, and indeed any targeted reactivity within eukaryotes, is in the earliest stages of development (Jones et al., 2005; Yao and Lambowitz, 2007). However, the extraordinary specificity of these introns may render them medically useful (Nazari and Joshi, 2008).
Although the spatial location of EBS2 can readily be deduced from the crystal structure of IIC introns and from the highly conserved secondary structures of IIA and IIB introns, its exact location and orientation relative to EBS1 awaits a crystal structure of the other group II intron families. For example, EBS2/IBS2 may be coaxial with EBS1, but biochemical work on target recognition by group IIB introns suggests that this is unlikely (Qin and Pyle, 1997; 1999; Xiang et al., 1998). Fluorescence studies suggest a bend, or discontinuity between EBS1 and 2, and the effects of mutations at the EBS1/EBS2 interface are inconsistent with a continuous network of stacked bases (Qin and Pyle, 1997; Xiang et al., 1998). By calculating the energetic contribution of each base-pair within the EBS1/IBS1 and EBS2/IBS2 helices, it has been shown that these two helices are independent of one another and that there is an energetic penalty that is incurred when they both form (Qin and Pyle, 1999). This penalty serves to preserve the information content of each base pair while reducing the overall binding affinity that might cause the intron to become too “sticky” and to bind with targets that contain mispairs (Qin and Pyle, 1999). This finding, together with the fact that EBS2 mutations influence the chemical step of group II intron catalysis, causes group IIB introns to display inordinately high levels of sequence-specificity (relative kcat/Km of 106). (Xiang et al., 1998).
In addition to lengthening the target sequence, the EBS2 structure (together with β–β′) is located in a region that is likely to sterically block the active site and it should reduce the number of directions from which a target oligonucleotide can approach DV. In a IIC intron such as the one that was recently crystallized, EBS1 is completely accessible to solvent and it is expected to experience many more collisions with ligands and potential targets. By forming sequence-specific interactions and serving as a steric “filter” for the approach of RNA into the active-site, EBS2 is a derivation that may have improved the control of intron function, and contributed to changes in the biological function of the IIB and IIA classes.
The d2a and β–β′ elements
Adjacent to the EBS2 strand, the d2a helix will extend outward and project away from the intron core (Figure 13). At its tip is a loop sequence (β′) that is complementary to a loop in the c2 stem (β), as described below. When these two elements pair, they form the β–β′ kissing loop interaction (Figure 1B). Based on the pitch and length of the connecting helices, the β–β′ duplex will bridge the ID2 and IC helices, which are located on opposite sides of the core. The β–β′ interaction will lie just beneath (and potentially parallel with) the critical α–α′ helix, buttressing it and creating a stronger framework for the core (Figure 13, center of bracket). Like EBS2, this substructure will also obstruct the active site and reduce the possible paths for entry of small molecules and RNA ligands, potentially facilitating the high sequence-specificity observed for IIA and IIB introns.
The β–β′ interaction is likely to have provided a selective advantage by significantly rigidifying the D1 scaffold and creating a more protected shell around the active site that is centered over the DV bulge. This innovation would have reduced the exchange of water and substrates into the active site, potentially increasing transesterification activity and reducing hydrolysis. Indeed, hydrolysis is a more prevalent form of splicing by the group IIC introns (Granlund et al., 2001; Toor et al., 2006), which lack c2, d2a or β–β′ substructures. Based on all these factors, the β–β′ innovation is likely to have conferred multiple, significant advantages for derived introns that contain this substructure.
The IC insertion
The long IC helix is like an extended strut which, together with the I(ii) helix (Figures 6 and 8), provides much of the rigid frame for the intron core. The exterior of the IC helix was the site of an insertion at a position that is located 1/2 helical turn from the five-way junction. This inserted hairpin-loop resulted in the c2 subdomain, which is common among the IIA and IIB classes of intron (Figure 1B), suggesting that the insertion event occurred before these two groups diverged from one another.
In IIA and IIB introns, the c2 subdomain contains a loop (β) that is complementary to the loop within the d2a subdomain (β′), as described above (Figure 1B). In three-dimensional space, the c2 and d2a insertions are very close together (Figure 13), and the β–β′ kissing-loop interaction between these elements may have evolved in response to their close proximity.
E. The location of unusual subdomains within specialized intron classes
The I(i) insertion
A small but interesting set of group II introns contain an insertion that is located in the very first stem of domain I (Figure 1), in helix I(i), adjacent to a semi-conserved internal loop that forms part of the Z-anchor region visualized in the group IIC crystal structure (Michel et al., 2007). This insertion was first observed upon characterization of the Azotobacter vinlandii (Avi) intron, which self-splices at high temperatures and is located adjacent to the stop codon for a bacterial heat shock gene (Adamidi et al., 2003; Ferat et al., 2003). Since the initial identification of this specialized intron, many related variants have been identified and they have been classified as a distinct lineage within class CL1 (Michel et al., 2007). These introns, all of which contain the I(i) insertion at approximately the same position, tend to invade sites at the boundaries of genes, although it is not yet clear how or whether the inserted sequences confer this novel specialization (Michel et al., 2007; Simon et al., 2008).
It has been proposed that the inserted sequences contain alternative 5′-splice sites and IBS1 sequences, which might provide some form of useful splicing regulation (Ferat et al., 2003; Michel et al., 2007). The insertion of this large substructure immediately next to the Z-anchor region (Toor et al., 2008a) (it would invade at intron position 10 of the O.i. intron, Figure 1A), which is essential for holding the intron frame together and for providing an interface with the catalytic face of DV, is expected to profoundly influence intron behavior and to modulate it in unpredictable ways that are likely to depend on the local structure of the inserted substructure.
The 3′-terminal insertion, or domain 7
Although they are not highly conserved in sequence, group II introns have a highly distinctive secondary structure that is composed of six domains that radiate from a central junction (Michel et al., 1989). It was therefore quite surprising when a new group of IIB introns was recently reported, containing an apparently seventh domain that is located between DVI and the 3′-splice site (Tourasse et al., 2005; Stabell et al., 2007; 2009). While perhaps not a “domain” as such, this insertion has now been identified within several group IIB introns that are phylogenetically related. In some cases, the inserted stem-loop structure influences splicing of the adjacent intron (Stabell et al., 2009), and structure–function analyses of this interesting intron class are underway. These introns may add to our knowledge of adaptation by group II introns, and they may also provide insights into mechanisms for 3′-splice site recognition by group II introns in general.
Concluding remarks
The group II intron tertiary structure has been an important challenge because it promised to provide new information on RNA molecular recognition and architectural motifs. While these expectations have certainly been met, it is now clear that the structure provides much more than we anticipated. It provides a map of potential interactions that can be tested within the eukaryotic spliceosome (Keating et al., 2010), in order to finally examine the question of common ancestry between these systems. In addition, the structure reveals hints of dynamic change within group II introns as they carry out different biochemical tasks and as they evolve through time. Most importantly, the structure points the way to important new structural and biochemical experiments that are designed to explore the chemical basis for RNA metabolism and genomic plasticity in terrestrial life.
Acknowledgments
I thank Kevin Keating for stimulating discussions on the probable location of derived domains, for critical reading of the manuscript and for assistance with the preparation of figures. I also thank Olga Fedorova and Amelia Johnson for assistance with figures and editing of the manuscript. I thank Philip Perlman and Marlene Belfort for stimulating discussions and suggestions.
Footnotes
Declaration of interest
I am an Investigator with the Howard Hughes Medical Institute, which supported this work. This work was also supported by a generous grant from the National Institutes of Health (GM50313). The author report no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Abramovitz DL, Friedman RA, Pyle AM. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science. 1996;271:1410–1413. doi: 10.1126/science.271.5254.1410. [DOI] [PubMed] [Google Scholar]
- Adamidi C, Fedorova O, Pyle AM. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M, Derbyshire V, Parker M, Cousineau B, Lambowitz A. Mobile introns: pathways and proteins. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 761–783. [Google Scholar]
- Boeke JD. The unusual phylogenetic distribution of retrotransposons: a hypothesis. Genome Res. 2003;13:1975–1983. doi: 10.1101/gr.1392003. [DOI] [PubMed] [Google Scholar]
- Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudvillain M, Delencastre A, Pyle AM. A new RNA tertiary interaction that links active-site domains of a group II intron and anchors them at the site of catalysis. Nature. 2000;406:315–318. doi: 10.1038/35018589. [DOI] [PubMed] [Google Scholar]
- Burger G, Yan Y, Javadi P, Lang BF. Group I-intron trans-splicing and mRNA editing in the mitochondria of placozoan animals. Trends Genet. 2009;25:381–386. doi: 10.1016/j.tig.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Centron D, Roy P. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO. 1996;15:3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Chin K, Pyle AM. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- Chu V-T, Liu Q, Podar M, Perlman PS, Pyle AM. More than one way to splice an RNA: Branching without a bulge and splicing without branching in group II introns. RNA. 1998;4:1186–1202. doi: 10.1017/s1355838298980724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Adamidi C, Liu Q, Perlman PS, Pyle A. Control of branch-site choice by a group II intron. EMBO J. 2001;20:6866–6876. doi: 10.1093/emboj/20.23.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros CJ, Landthaler M, Piazza CL, Beauregard A, Esposito D, Perutka J, Lambowitz AM, Belfort M. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol. 2005;56:509–524. doi: 10.1111/j.1365-2958.2005.04554.x. [DOI] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Belfort M. A mutant screen reveals RNase E as a silencer of group II intron retromobility in Escherichia coli. RNA. 2008;14:2634–2644. doi: 10.1261/rna.1247608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M. Global regulators orchestrate group II intron retromobility. Mol Cell. 2009;34:250–256. doi: 10.1016/j.molcel.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Deme E, Jacquier A, Michel F. Multiple tertiary interactions involving domain II of group II self- splicing introns. J Mol Biol. 1997;267:520–536. doi: 10.1006/jmbi.1996.0882. [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F, Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–5018. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Zimmerly S. The dispersal of five group II introns among natural populations of Escherichia coli. RNA. 2002;8:1294–1307. doi: 10.1017/s1355838202023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Chai D, Gu SQ, Gabel J, Noskov SY, Blocker FJ, Lambowitz AM, Zimmerly S. A three-dimensional model of a group II intron RNA and its interaction with the intron-encoded reverse transcriptase. Molecular Cell. 2008;30:472–485. doi: 10.1016/j.molcel.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Michels WJ, Jr, Pyle AM. Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J Mol Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- Dayie KT, Padgett RA. A glimpse into the active site of a group II intron and maybe the spliceosome, too. RNA. 2008;14:1697–1703. doi: 10.1261/rna.1154408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLencastre A, Hamill S, Pyle AM. A single active-site region for a group II intron. Nat Struct Mol Biol. 2005;12:626–627. doi: 10.1038/nsmb957. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Xu A, Sagasser S, Jakob W, Moreno MA, Buss LW, Schierwater B. Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci USA. 2006;103:8751–8756. doi: 10.1073/pnas.0602076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD, Boulanger SC, Hebbar SK, Peebles CL, Franzen JS, Perlman PS. Domain 5 interacts with Domain 6 and influences the second transesterification reaction of group II intron self-splicing. NAR. 1993;21:1797–1804. doi: 10.1093/nar/21.8.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM. Linking the group II intron catalytic domains: tertiary contacts and structural features of domain 3. Embo J. 2005;24:3906–3916. doi: 10.1038/sj.emboj.7600852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Pyle AM. A conserved element that stabilizes the group II intron active site. RNA. 2008;14:1048–1056. doi: 10.1261/rna.942308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol Chem. 2007;388:665–678. doi: 10.1515/BC.2007.090. [DOI] [PubMed] [Google Scholar]
- Fedorova O, Mitros T, Pyle AM. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J Mol Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferat JL, Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- Ferat JL, Le Gouar M, Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- Gordon PM, Piccirilli JA. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol. 2001;8:893–898. doi: 10.1038/nsb1001-893. [DOI] [PubMed] [Google Scholar]
- Gordon PM, Fong R, Piccirilli JA. A second divalent metal ion in the group II intron reaction center. Chem Biol. 2007;14:607–612. doi: 10.1016/j.chembiol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Granlund M, Michel F, Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Pyle AM. The receptor for branch-site docking within a group II intron active site. Mol Cell. 2006;23:831–840. doi: 10.1016/j.molcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Harris-Kerr CL, Zhang M, Peebles CL. The phylogenetically predicted base-pairing interaction between α and α’ is required for group II splicing in vitro. Proc Natl Acad Sci USA. 1993;90:10658–10662. doi: 10.1073/pnas.90.22.10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A, Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987;50:17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Jarrell KA. Inverse splicing of a group II intron. PNAS. 1993;90:8624–8627. doi: 10.1073/pnas.90.18.8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS. Group II intron self-splicing: Alternative reaction conditions yield novel products. J Biol Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- Jestin J-L, Deme E, Jacquier A. Identification of structural elements critical for inter-domain interactions in a group II self-splicing intron. EMBO. 1997;16:2945–2954. doi: 10.1093/emboj/16.10.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JP, 3rd, Kierlin MN, Coon RG, Perutka J, Lambowitz AM, Sullenger BA. Retargeting mobile group II introns to repair mutant genes. Mol Ther. 2005;11:687–694. doi: 10.1016/j.ymthe.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Keating KS, Toor N, Pyle AM. The GANC tetraloop: a novel motif in the group IIC intron structure. J Mol Biol. 2008;383:475–481. doi: 10.1016/j.jmb.2008.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16:1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konforti BB, Abramovitz DL, Duarte CM, Karpeisky A, Beigelman L, Pyle AM. Ribozyme catalysis from the major groove of group II intron domain 5. Mol Cell. 1998;1:433–441. doi: 10.1016/s1097-2765(00)80043-x. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus intronslate debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Perlman PS. Involvement of aminoacyltRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem Sci. 1990;15:440–444. doi: 10.1016/0968-0004(90)90283-h. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol. 1998;283:571–583. doi: 10.1006/jmbi.1998.2106. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res. 2006;34:2782–2790. doi: 10.1093/nar/gkl328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Abarca F, Toro N. Group II introns in the bacterial world. Molecular Microbiology. 2000;38:917–926. doi: 10.1046/j.1365-2958.2000.02197.x. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Introns: evolution and function. Curr Opin Genet Dev. 1994;4:823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns–a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Doucet AJ, Ferat JL. Specialized lineages of bacterial group II introns. Biochimie. 2007;89:542–553. doi: 10.1016/j.biochi.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci. 2009;34:189–199. doi: 10.1016/j.tibs.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Molina-Sanchez MD, Martinez-Abarca F, Toro N. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem. 2006;281:28737–28744. doi: 10.1074/jbc.M602695200. [DOI] [PubMed] [Google Scholar]
- Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA. Excision of group II introns as circles. Mol Cell. 2001;8:201–211. doi: 10.1016/s1097-2765(01)00300-8. [DOI] [PubMed] [Google Scholar]
- Nazari R, Joshi S. Exploring the potential of group II introns to inactivate human immunodeficiency virus type 1. J Gen Virol. 2008;89:2605–2610. doi: 10.1099/vir.0.2008/004333-0. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Westhof E, Johansen S. An mRNA is capped by a 2′, 5′ lariat catalyzed by a group I-like ribozyme. Science. 2005;309:1584–1587. doi: 10.1126/science.1113645. [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman PS, Butow RA. Mobile introns and intron-encoded proteins. Science. 1989;246:1106–1109. doi: 10.1126/science.2479980. [DOI] [PubMed] [Google Scholar]
- Perlman P, Podar M. Reactions catalyzed by group II introns in vitro. Methods Enzymol. 1996;264:66–86. doi: 10.1016/s0076-6879(96)64010-5. [DOI] [PubMed] [Google Scholar]
- Podar M, Perlman PS. Photocrosslinking of 4-thio uracil-containing RNAs supports a side-by-side arrangement of domains 5 and 6 of a group II intron. RNA. 1999;5:318–329. doi: 10.1017/s1355838299981724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Dib-Hajj S, Perlman PS. A UV-induced Mg2+-dependent cross-link traps an active form of Domain 3 of a self-splicing group II intron. RNA. 1995a;1:828–840. [PMC free article] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA. Stereochemical selectivity of group II intron splicing, reverse splicing, and hydrolysis reactions. Mol Cell Biol. 1995b;15:4466–4478. doi: 10.1128/mcb.15.8.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Chu VT, Pyle AM, Perlman PS. Group II intron splicing in-vivo by first step hydrolysis. Nature. 1998;391:915–918. doi: 10.1038/36142. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Capping by branching: a new ribozyme makes tiny lariats. Science. 2005;309:1530–1531. doi: 10.1126/science.1117957. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Group II introns: catalysts for splicing, genomic change and evolution. In: Lilley DMJ, Eckstein F, editors. Ribozymes and RNA Catalysis. New York: Springer-Verlag; 2008. pp. 201–223. [Google Scholar]
- Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland R, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor: Cold Spring Harbor Press; 2006. pp. 469–505. [Google Scholar]
- Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. Antagonistic substrate binding by a group II intron ribozyme. JMB. 1999;291:15–27. doi: 10.1006/jmbi.1999.2922. [DOI] [PubMed] [Google Scholar]
- Rest JS, Mindell DP. Retroids in archaea: phylogeny and lateral origins. Mol Biol Evol. 2003;20:1134–1142. doi: 10.1093/molbev/msg135. [DOI] [PubMed] [Google Scholar]
- Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet Genome Res. 2005;110:589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- Roitzsch M, Pyle AM. The linear form of a group II intron catalyzes efficient autocatalytic reverse splicing, establishing a potential for mobility. RNA. 2009;15:473–482. doi: 10.1261/rna.1392009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Podar M, Stahl U, Perlman PS. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: Phenotypes and suppressor mutations. RNA. 1996;2:1161–1172. [PMC free article] [PubMed] [Google Scholar]
- Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA. 2008;14:1704–1713. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM, Li-Pook-Than J. Group II introns and their protein collaborators. In: Walter NG, editor. Non-Protein Coding RNAs. Berlin: Springer-Verlag; 2009. pp. 167–182. [Google Scholar]
- Sontheimer EJ, Gordon PM, Piccirilli JA. Metal ion catalysis during group II intron self-splicing: parallels with the spliceosome. Genes Dev. 1999;13:1729–1741. doi: 10.1101/gad.13.13.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell FB, Tourasse NJ, Ravnum S, Kolsto AB. Group II intron in Bacillus cereus has an unusual 3′ extension and splices 56 nucleotides downstream of the predicted site. Nucleic Acids Res. 2007;35:1612–1623. doi: 10.1093/nar/gkm031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabell FB, Tourasse NJ, Kolsto AB. A conserved 3′ extension in unusual group II introns is important for efficient second-step splicing. Nucleic Acids Res. 2009;37:3202–3214. doi: 10.1093/nar/gkp186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley MR, Strobel SA. RNA splicing: group I intron crystal structures reveal the basis of splice site selection and metal ion catalysis. Curr Opin Struct Biol. 2006;16:319–326. doi: 10.1016/j.sbi.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Stahley MR, Adams PL, Wang J, Strobel SA. Structural metals in the group I intron: a ribozyme with a multiple metal ion core. J Mol Biol. 2007;372:89–102. doi: 10.1016/j.jmb.2007.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci USA. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. A general two-metal ion mechanism for catalytic RNA. PNAS. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Qin P, Michels W, Pyle A. Guiding ribozyme cleavage through motif recognition: the mechanism of cleavage site selection by a group II intron ribozyme. J Mol Biol. 2001;306:665–668. doi: 10.1006/jmbi.2000.4323. [DOI] [PubMed] [Google Scholar]
- Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J Mol Biol. 2002;320:455–474. doi: 10.1016/s0022-2836(02)00515-6. [DOI] [PubMed] [Google Scholar]
- Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Toor N, Hausner G, Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Robart AR, Christianson J, Zimmerly S. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006;34:6461–6471. doi: 10.1093/nar/gkl820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008a;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. 2008b;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Pyle AM. Structural insights into RNA splicing. Curr Opin Struct Biol. 2009;19:260–266. doi: 10.1016/j.sbi.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating A, Fedorova O, Rajashankar K, Wang J, Pyle AM. The tertiary architecture of a group II intron. RNA. 2010;16:57–69. doi: 10.1261/rna.1844010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro N. Bacteria and archaea group II introns: additional mobile genetic elements in the environment. Environ Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- Tourasse NJ, Stabell FB, Reiter L, Kolsto AB. Unusual group II introns in bacteria of the Bacillus cereus group. J Bacteriol. 2005;187:5437–5451. doi: 10.1128/JB.187.15.5437-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL. Proteinfree spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA. 2007;13:2300–2311. doi: 10.1261/rna.626207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Jaladat Y, Geisler S. Proteinfree small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci USA. 2009;106:11901–11906. doi: 10.1073/pnas.0902020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles Y, Halanych KM, Boore JL. Group II introns break new boundaries: presence in a bilaterian’s genome. PLoS One. 2008;3:e1488. doi: 10.1371/journal.pone.0001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Kwakman JHJM, Grivell LA. Mutations at the lariat acceptor site allow self-splicing of a Group II intron without lariat formation. EMBO Journal. 1987;6:3827–3831. doi: 10.1002/j.1460-2075.1987.tb02719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Borner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldsich C, Pyle AM. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol. 2007;14:37–44. doi: 10.1038/nsmb1181. [DOI] [PubMed] [Google Scholar]
- Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wank H, Sanfilippo J, Singh RN, Matsuura M, Lambowitz AM. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell. 1999;4:239–250. doi: 10.1016/s1097-2765(00)80371-8. [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Qin PZ, Michels WJ, Freeland K, Pyle AM. The sequence-specificity of a group II intron ribozyme: Multiple mechanisms for promoting unusually high discrimination against mismatched targets. Biochemistry. 1998;37:3839–3849. doi: 10.1021/bi972661n. [DOI] [PubMed] [Google Scholar]
- Yao J, Lambowitz AM. Gene targeting in gram-negative bacteria using a mobile group II intron (“targetron”) expressed from a broad-host-range vector. Appl Environ Microbiol. 2007;73:2735–2743. doi: 10.1128/AEM.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Mastroianni M, White TB, Lambowitz AM. Linear group II intron RNAs can retrohome in eukaryotes and may use nonhomologous end-joining for cDNA ligation. Proc Natl Acad Sci USA. 2009;106:18189–18194. doi: 10.1073/pnas.0910277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerly S, Hausner G, Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]