Abstract
Precursor mRNA (pre-mRNA) splicing is a critical step in gene expression that results in the removal of intronic sequences from immature mRNA, leading to the production of mature mRNA that can be translated into protein. Alternative pre-mRNA splicing is the process whereby alternative exons and/or introns are selectively included or excluded, generating mature mRNAs that encode proteins that may differ in function. The resulting alterations in the pattern of protein isoform expression can result in changes in protein–protein interaction, subcellular localization, and flux through metabolic pathways. Although basic mechanisms of pre-mRNA splicing of introns and exons are reasonably well characterized, how these mechanisms are regulated remains poorly understood. The goal of this review is to highlight selected recent advances in our understanding of the regulation of pre-mRNA splicing by nutrients and modulation of nutrient metabolism that result from changes in pre-mRNA splicing.
Keywords: skeletal muscle, alternative splicing, mRNA, macronutrient, micronutrient, splice factors
Introduction
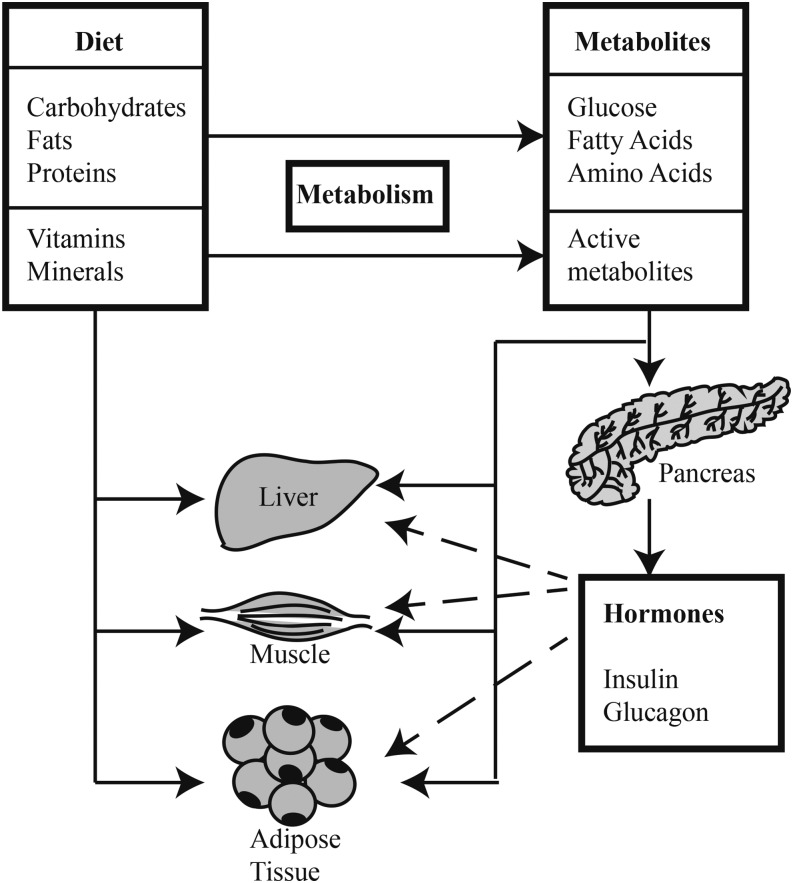
Classically, nutrients have been defined as the dietary components needed for survival and growth of organisms. Macronutrients are metabolized to provide energy and the precursors for the synthesis of more complex molecules (e.g., amino acids are used in protein synthesis), whereas micronutrients provide essential cofactors involved in various enzymatic reactions. More recent studies have shown that, in addition to these classic functions, nutrients play important roles in regulating gene expression. In mammals, gene expression involves a number of steps starting with the transcription of a gene into precursor mRNA (pre-mRNA)6, which is usually processed into a mature mRNA molecule that is exported from the nucleus for translation by cytoplasmic ribosomes. Many studies have focused on the regulation of the first and last steps in gene expression, i.e., DNA transcription and mRNA translation. However, both macro- and micronutrients are important regulators of mRNA splicing, a critical step in the conversion of pre-mRNA into mature mRNA. In addition, recent studies highlight the importance of nutrients in modulating alternative splicing of pre-mRNA, a process that is important to the production of proteins with diverse functions and/or subcellular localization by generating multiple mature mRNAs from single genes. These splicing events can be regulated in various tissues by signaling pathways activated by nutrients and/or their metabolites or by hormones secreted in response to dietary nutrients (Figure 1). The goal of this review article is to highlight the role of both macro- and micronutrients in regulating pre-mRNA splicing and the control of nutrient metabolism through alterations in the splicing of the pre-mRNA encoding enzymes involved in various metabolic pathways. In both cases, the focus is on recent studies that serve as examples of this type of regulation. Unfortunately, page limits prevent us from citing many important works, and we apologize in advance to the authors of those studies not referenced herein.
FIGURE 1.
Nutrient regulation of alternative splicing. The metabolites of dietary macronutrients and micronutrients act directly and indirectly (e.g., through hormones produced by the pancreas) to modulate alternative splicing events in target tissues. This process is regulated by signaling pathways downstream of nutrients, metabolites, and hormones.
Pre-mRNA Splicing
The vast majority of mammalian pre-mRNAs contain introns that are removed by 2 trans-esterification reactions catalyzed by the spliceosome and associated auxiliary proteins that result in the removal of an intron and the joining of the 2 exons that border the intron (1) (Figure 2). The core of the spliceosome is composed of a complex of 5 small nuclear ribonucleoprotein particles (snRNPs) that are recruited to the pre-mRNA in a sequence-dependent manner. The splicing process is under robust regulation to ensure temporal and cell- and tissue-specific expression of splice variants. Part of this regulation is due to the utilization of distinct snRNPs and auxiliary proteins for different types of splicing events. At least 45 snRNPs have been identified as components of the spliceosome and >170 auxiliary proteins are known to regulate the process. Splicing is dynamic and remodeling of RNA–protein and protein–protein interactions mediates progression of the machinery along the pre-mRNA.
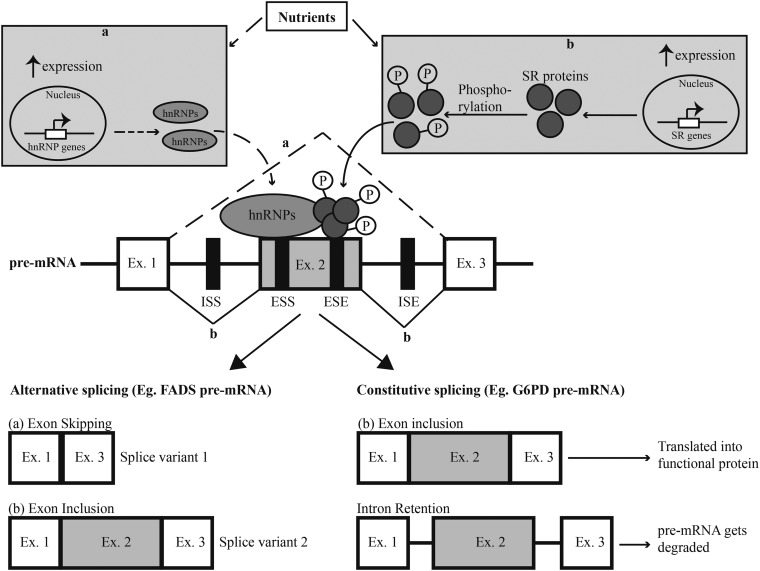
FIGURE 2.
Mechanisms for regulation of alternative splicing by cis and trans factors. The pre-mRNA depicted in this diagram consists of 3 exons, represented by white and gray boxes. The lines connecting the exons denote the introns that are removed during the splicing process. There are 2 types of splicing processes. Alternative splicing leads to different types of splice variants, depending on which exons are included or excluded. In the example depicted here, Ex. 2 can be alternatively spliced and the outcome of splicing events is regulated by both cis elements (denoted by black boxes) and trans factors. (a) Nutrients and/or their metabolites can mediate increased expression of hnRNPs that bind ESSs in Ex. 2 and inhibit the assembly of splicing machinery at the intron–exon junction. As a result, Ex. 2 is excluded from the mature mRNA and splice variant 1 is formed. (b) Alternatively, nutrients can also mediate increased expression and phosphorylation of SR proteins that bind to ESEs within an exon to promote its inclusion. The mature mRNA-produced splice variant 2 includes all 3 exons. Constitutive splicing mediates the removal of introns and inclusion of all exons in the mature mRNA that can be translated into a functional protein. Defective regulation of this process results in intron retention and the unspliced transcript cannot be translated into protein and is degraded. ISEs and ISSs function similarly to exonic elements. ESE, exonic splicing enhancer; ESS, exonic splicing silencer; Ex., exon; FADS, fatty acid desaturase; G6PD, glucose-6-phosphate dehydrogenase; hnRNP, heterogeneous nuclear ribonucleoprotein; ISE, intronic splicing enhancer; ISS, intronic splicing silencer; P, phosphate; pre-mRNA, precursor mRNA; SR protein, serine/arginine–rich protein.
The pre-mRNA includes 4 important regions that serve as sites for recognition by the spliceosome (1). The 3′- and 5′-splice sites are located at exon–intron junctions and are defined by consensus sequences consisting minimally of the dinucleotides GU at the 5′-end of the intron and AG at the 3′-end. In addition, introns can contain branch sites and/or polypyrimidine tracts that are critical in defining the location of the splice site. These sites recruit specific proteins of the spliceosome by base pairing with the snRNPs. Splice site recognition is further mediated by cis- and trans-regulatory elements (Figure 2). The cis-regulatory elements are present within the pre-mRNA in regions proximal to the splice site and include intronic or exonic sequences that recruit auxiliary proteins (i.e., trans factors) that regulate spliceosomal assembly. Trans factors are broadly classified as splicing enhancers, e.g., serine/arginine–rich proteins (SR proteins), that facilitate splice site recognition by the spliceosome, or splicing repressors, e.g., heterogeneous nuclear ribonucleoproteins (hnRNPs) that inhibit splice site recognition (2). The efficiency of the splicing process depends on the competitive and mutually exclusive binding of the splicing factors to the cis elements. However, some splicing factors, such as hnRNP L and hnRNP H, can act as both enhancers and repressors depending on the type of cis element to which they are bound or the location of the cis element in the pre-mRNA. Importantly, the binding of trans factors to the cis elements can be modulated in response to environmental cues (e.g., hormones and nutrients) through phosphorylation of trans factors such as the SR proteins (2, 3).
The process of alternative pre-mRNA splicing is similar to constitutive splicing. Indeed, many of the proteins involved in the former also mediate constitutive splicing reactions (2). However, alternative pre-mRNA splicing differs from constitutive splicing in that the former can lead to intron retention, exon skipping, mutually exclusive exon inclusion/exclusion, and alternative 5′- or 3′-splice site usage. Whether constitutive or alternative splicing occurs depends on both how well the sequences in the regulatory elements described above correspond to the ideal, and the binding of trans factors to cis regulatory domains. Thus, it has been proposed that constitutive splicing takes place at strong splice sites that closely match the consensus, whereas alternative splicing occurs at weak splice sites in which the sequence elements diverge from the consensus and are recognized less efficiently by the spliceosome (4). Moreover, alternative splicing at a particular site depends upon the abundance and activity of trans factors, such as the SR proteins and hnRNPs. Recent estimates suggest that 95% or more of human pre-mRNAs are subject to alternative splicing (4), whereas in other organisms this number is much lower (e.g., ∼40% in Drosophila). The number of mature mRNAs that can be generated from a single pre-mRNA varies widely, with some pre-mRNAs predicted to generate as many as several thousand mature mRNAs. However, the abundance of some alternatively spliced mature mRNAs is low (i.e., <1% of the total) and the number of functionally distinct proteins produced by alternative pre-mRNA splicing is undoubtedly much smaller than the number of possible alternative splicing events predicted solely by splice-site identification.
In addition to the role they play in pre-mRNA splicing, a number of splice factors have additional roles in controlling post-transcriptional gene expression, including nuclear export of mRNA, initiation of mRNA translation, and mRNA degradation (5). For example, the SR protein splice factor 2/alternative splice factor 1 (SF2/ASF) promotes phosphorylation of the eukaryotic initiation factor 4E binding protein 1, thereby upregulating cap-dependent translation of specific mRNAs (6). Moreover, some splice factors have additional roles outside of mRNA processing and translation. A particularly interesting example of such a role is the recent identification of hnRNP E1 as a possible sensor of folate deficiency (7).
Regulation of Pre-mRNA Splicing by Macronutrients
Carbohydrates.
Carbohydrate ingestion leads to the secretion of a variety of hormones, including the incretins (e.g., glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide 1), insulin, and glucagon, which act to promote the uptake and storage of carbohydrates as glycogen as well as the modulation of carbohydrate metabolism (8). The pre-mRNAs for a number of proteins involved in carbohydrate metabolism [e.g., glucose-6-phosphate dehydrogenase (G6PD) (9) and fructose-6-phosphate 2-kinase (10)], as well as those encoding monosaccharide transporters [e.g., glucose transporters (11)], are alternatively spliced. The following genes represent a few examples of pre-mRNA splicing that occur in direct response to changes in dietary carbohydrates or to hormones secreted as a result of carbohydrate consumption.
The regulation of G6PD activity by carbohydrates is a classic example of nutrient-mediated constitutive exon splicing and intron retention (9). G6PD catalyzes the first and rate-limiting step in the oxidative branch of the pentose phosphate pathway that leads to the production of NAD(P)H, which can be utilized for FA biosynthesis in the liver and adipose tissue. Starvation and refeeding studies highlight the role of post-transcriptional, rather than transcriptional mechanisms in regulating G6PD activity. Thus, G6PD activity is repressed in the fasted state as a result of decreased splicing of the G6PD transcript, without a change in the rate of transcription. Upon refeeding a high carbohydrate diet, an increase in the splicing of the G6PD pre-mRNA leads to an increased abundance of the fully spliced mRNA and enhanced production of the active protein. An analysis of the G6PD transcript revealed the presence of a cis-acting element in exon 12 that can function as an exon splicing silencer and enhancer. The efficiency of splicing depends on the mutually exclusive binding of trans-acting factors to the cis elements (12). For example, starvation leads to increased hnRNP K expression and increased binding of the protein to exon 12 of the G6PD pre-mRNA, thereby leading to inhibition of the splicing of nearby introns (13). In contrast, refeeding causes increased expression and phosphorylation of SR proteins such as serine/arginine–rich splice factor (SRSF) 3 that recruit the spliceosome to the G6PD transcript and result in increased splicing (9, 14). The effect of dietary carbohydrates on G6PD pre-mRNA splicing is primarily mediated by insulin that is released in response to the increased circulating glucose concentrations (9). Protein kinases downstream of insulin, e.g., Akt, phosphorylate and activate SR proteins and thereby promote increased G6PD pre-mRNA splicing (3). In addition, intermediates of carbohydrate metabolism, such as glucose and fructose and other hormones like glucocorticoids, have been predicted to influence G6PD pre-mRNA splicing (9).
X-box binding protein 1 (XBP1) is a transcription factor that plays an important role in activating the unfolded protein response (UPR) pathway to protect cells after the induction of endoplasmic reticulum (ER) stress (15). The XBP1 pre-mRNA has 2 overlapping open reading frames (ORFs), ORF1 and ORF2. In response to ER stress that occurs after consumption of a high carbohydrate diet (16) or the incubation of cells in culture under hyperglycemic conditions (17), an endoribonuclease, inositol-requiring, ER-to-nucleus signaling protein 1α (IRE1α) is activated and removes 26 nucleotides from ORF1 of the XBP1 pre-mRNA. This event results in a frame shift in the processed mRNA that replaces a portion of the C-terminus of ORF1 with ORF2, resulting in a spliced mRNA that encodes the active transcription factor (15). The resulting increase in active XBP1 abundance leads to the enhanced transcription of genes involved in FA synthesis (18). Together, the altered splicing of the G6PD and XBP1 pre-mRNAs are important facets in the carbohydrate-induced upregulation of FA synthesis.
FAs.
In addition to being regulated by carbohydrates, splicing of the pre-mRNAs encoding G6PD and XBP1 is also modulated by FAs. For example, in primary cultures of rat hepatocytes, incubation with the PUFA arachidonic acid (20:4n–6) leads to decreased phosphorylation of SR proteins, resulting in decreased binding of SRSF3 to the cis-acting element in exon 12 of the G6PD pre-mRNA and decreased splicing efficiency (14). The effect of dietary PUFAs on G6PD pre-mRNA splicing can also be mediated by an inhibition of insulin signaling in the liver (19). Also, similar to carbohydrates, dietary SFAs induce ER stress in several tissues, including liver (20) and pancreas (21), leading to increased splicing of the XBP1 pre-mRNA by IRE1α and subsequently to increased transcription of genes involved in the UPR pathway (18).
Long-chain PUFAs (LCPUFAs) can be synthesized by fatty acid desaturases (FADSs), including FADS1, FADS2, and FADS3, in the liver. The genes encoding these enzymes have been linked to hyperlipidemia (22), as well as cardiovascular and neuronal diseases (23). Increased dietary consumption of LCPUFAs leads to decreased activity of FADS1 and FADS2 because of a change in the pattern of alternative splicing of these genes, thereby decreasing biosynthesis of LCPUFAs (22). Wijendran et al. (24) observed that consumption of a diet high in LCPUFAs results in decreased expression of the FADS2 AT1 transcript in the liver of suckling piglets because of the binding of hnRNP I to the exon splicing silencer in the FADS2 pre-mRNA. Altered FADS expression leads to an imbalance in the ratio of intracellular PUFAs and intermediate metabolites involved in inflammatory pathways (25).
FAs also regulate splicing of the pre-mRNA encoding the GIP receptor (GIPR). GIP is an incretin secreted by intestinal cells in response to dietary glucose that has distinctive effects on pancreatic β cells (26) and adipocytes (27). In pancreatic β cells, a splice variant of GIPR that retains intron 8 has been identified (26). This splice variant encodes an inactive form of GIPR with a truncated C-terminal domain. In response to a high fat diet, there is a change in the pattern of GIPR pre-mRNA splicing, resulting in a decrease in the ratio of inactive to active isoform expression. As a result, there is an increase in the sensitivity of β cells to GIP, leading to high fat diet–induced hyperinsulinemia. GIP also has been shown to promote inflammation and insulin resistance in the adipose tissues of obese individuals (27). Exon (Ex) 9 of the GIPR pre-mRNA encodes the transmembrane domain and decreased expression of the exon 9–containing splice variant (Ex9+ GIPR) results in decreased GIPR function in adipose tissue. Ahlqvist et al. (27) reported that decreased Ex9+ GIPR expression has a protective effect and corresponds to lower BMI and improved insulin sensitivity in obese individuals.
Protein.
Reduced dietary availability of one or more essential amino acids leads to induction of the amino acid response (AAR) that promotes activation of several signaling pathways controlling transcription, processing, and turnover of mRNA (28). Expression of several genes in the AAR, e.g., Asn synthetase, is regulated by activating transcription factor 3 (ATF3), and different splice variants of ATF3 have been identified in cells in culture deprived of histidine, serum, or glucose (29). For example, Pan et al. (30) observed that there was a change in the ratio of ATF3 mRNA splice form expression in response to histidine deprivation in HepG2 cells. The full length ATF3 isoform forms complexes with corepressors and inhibits AAR gene expression. The other ATF3 splice variants encode truncated isoforms that lack the DNA binding domain for AAR genes. These truncated isoforms sequester the corepressors associated with ATF3 and thereby mediate activation of AAR gene transcription. Unfortunately, the mechanisms involved in amino acid–induced regulation of ATF3 pre-mRNA splicing have not yet been elucidated.
Regulation of Pre-mRNA Splicing by Micronutrients
Vitamins.
Vitamins are obtained in trace quantities from the diet and metabolized into their active forms that can bind to vitamin response receptors and activate downstream signaling pathways. The direct effects of vitamins on pre-mRNA splicing are mediated by the active metabolites and the indirect effects are mediated by oxidative stress in response to vitamin deficiency. Most vitamins, including the vitamin B complex and vitamin C, regulate the alternative splicing of their own transporters and vitamin binding proteins. For example, 3 splice variants of Met synthase (vitamin B-12 dependent) have been identified in the human cerebral cortex (31). Vitamins also can regulate expression of splicing factors. For example, a vitamin E–deficient diet has been shown to decrease the expression of splicing factors such as splicing component 35kDa (SC35) in the liver (32).
Vitamin A plays an important role in differentiation of neuronal cells and is involved in both prosurvival (33) and apoptotic (34) pathways. Retinoic acid (RA) is the biologically active metabolite of vitamin A that acts through the retinoic acid receptor (RAR) to regulate the expression of genes containing the retinoic acid response element (RARE) (33). Laserna et al. (35) observed that RA-induced differentiation of neurons resulted in phosphorylation of several trans-acting splicing factors such as the SR proteins via the phosphatidyl-inositol-3-kinase signaling pathway, leading to their activation and increased splicing efficiency. Consequently, RA alters the pattern of pre-mRNA splicing in neuronal cells. For example, RA treatment leads to increased expression of the protein kinase C (PKC) δ-VIII isoform because of the utilization of an alternative 5′-splice site located downstream of exon 10 (33). The SR protein SC35 binds to the cis-acting element in the PKC δ pre-mRNA to mediate this splicing event. In addition, the PKC δ-II isoform that has antiapoptotic functions is also regulated during neuronal differentiation (34). Recent studies (36) have established the role of apoptotic chromatin condensation inducer in the nucleus (Acinus) in regulating RA-induced splicing. Acinus has been shown to bind to the spliceosome and the exon junction complex. The C-terminal region has an RNA recognition motif, similar to SR proteins. Acinus targets RARE-containing pre-mRNAs by interacting with the RAR. The RNA recognition motif domain of Acinus can bind to splicing enhancers in the alternative 5′-splice site, followed by recruitment of the spliceosomal machinery and other SR splicing factors to the pre-mRNA.
Vitamin D is another important modulator of pre-mRNA splicing (37). For example, in colon cancer cells, the expression of different splice variants of vitamin D metabolizing enzymes, e.g., cytochrome p450 24A (CYP24A), is controlled by the active form of the vitamin, 1α,25-dihydroxyvitamin D [1,25(OH)2D]. Several mechanisms have been proposed for the regulation of pre-mRNA splicing by vitamin D. For example, 1,25(OH)2D can bind to the vitamin D receptor (VDR) and couple the regulation of transcription and processing of vitamin D response element–containing genes and pre-mRNAs. Zhang et al. (38) showed that the VDR coregulator nuclear receptor coactivator/ski-interacting protein (NCoA/SKIP) is a component of the spliceosomal complex and its recruitment to VDR is essential for vitamin D–dependent splicing events. In addition to its role in cancer cells, 1,25(OH)2D also mediates the increased expression of the short splice variant of the plasma membrane calcium pump (PMCA) 1 in osteoblasts (39).
Minerals.
Many of the mineral ion transporters have splice variants with distinctive functions. For example, 3 splice variants of the sodium/potassium/chloride cotransporter (NKCC2) have been identified, and a low-salt diet increases the expression of the high affinity isoform NKCC2B in the renal cortex (40). Other examples include salt-sensitive hypertension-induced changes in the alternative splicing pattern of the pre-mRNA for the epithelial sodium channel (41), dietary iodide-induced alterations in the expression of different splice variants of the intestinal sodium/iodide symporter that result in increased absorption activity (42), and zinc-induced alterations in phosphorylation of the SR protein SRSF6 that lead to changes in splicing of the Bcl-2-interacting mediator of cell death (Bim) pre-mRNA and modulation of apoptosis in neuroblastoma cells (43).
Conclusion
The above examples demonstrate that nutrients can both directly and indirectly affect gene expression through the modulation of alternative splicing of pre-mRNAs. Such events have the potential to alter not only the processes involved in nutrient metabolism, but other events required for cell homeostasis. Because the area is still relatively unexplored, the available information likely represents the tip of the iceberg in regard to nutrient control of gene expression. However, given that pre-mRNAs encoding key regulatory proteins (e.g., the insulin receptor, leptin receptor, etc.) associated with diseases such as obesity undergo important splicing events, and the fairly limited splicing machinery proteins involved, in-depth studies of the effects of nutrients on the regulation of constitutive and alternative splicing processes likely will be a fertile endeavor, and could lead to the identification of potential targets for therapeutic intervention.
Acknowledgments
We thank Dr. Leonard S Jefferson for helpful comments during preparation of the manuscript. SR, RJS, and SRK wrote the manuscript; SRK had responsibility for the final content. All authors read and approved the final version of the manuscript.
Footnotes
Abbreviations: AAR, amino acid response; ATF3, activating transcription factor 3; BIM, Bcl-2-interacting mediator of cell death; CYP24A, cytochrome p450 24A; ER, endoplasmic reticulum; FADS, fatty acid desaturase; GIP, glucose-dependent insulinotropic polypeptide; GIPR, glucose-dependent insulinotropic polypeptide receptor; G6PD, glucose-6-phosphate dehydrogenase; hnRNP, heterogeneous nuclear ribonucleoprotein; IRE1α, inositol-requiring, ER-to-nucleus signaling protein 1α; LCPUFA, long-chain PUFA; NCoA/SKIP, nuclear receptor coactivator/ski-interacting protein; NKCC2, sodium/potassium/chloride cotransporter; ORF, open reading frame; PKC, protein kinase C; PMCA, plasma membrane calcium pump; pre-mRNA, precursor mRNA; RA, retinoic acid; RAR, retinoic acid receptor; RARE, retinoic acid response element; SC35, splicing component 35 kDa; SF2/ASF, splice factor 2/alternative splice factor 1; snRNP, small nuclear ribonucleoprotein particles; SR protein, serine/arginine–rich protein; SRSF, serine/arginine–rich splice factor; UPR, unfolded protein response; VDR, vitamin D receptor; XBP1, X-box binding protein 1; 1,25(OH)2D, 1α,25-dihydroxyvitamin D.
References
- 1.Coelho MB, Smith CW. Regulation of alternative pre-mRNA splicing. Methods Mol Biol 2014;1126:55–82. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 2009;10:741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013;122:191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 2013;14:153–65. [DOI] [PubMed] [Google Scholar]
- 5.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009;136:688–700. [DOI] [PubMed] [Google Scholar]
- 6.Michlewski G, Sanford JR, , . Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E–BP1. Mol Cell 2008;30:179–89. [DOI] [PubMed] [Google Scholar]
- 7.Tang YS, Khan RA, Zhang Y, Xiao S, Wang M, Hansen DK, Jayaram HN, Antony A. Incrimination of heterogeneous nuclear ribonucleoprotein E1 (hnRNP-E1) as a candidate sensor of physiological folate deficiency. J Biol Chem 2011;286:39100–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin Biochem 2013;46:1339–52. [DOI] [PubMed] [Google Scholar]
- 9.Salati LM, Szeszel-Fedorowicz W, Tao H, Gibson MA, Amir-Ahmady B, Stabile LP, Hodge DL. Nutritional regulation of mRNA processing. J Nutr 2004;134:2437S–43S. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe F, Furuya E. Alternative splicing of novel exons of rat heart-type fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase gene. Biochem Biophys Res Commun 2001;282:803–10. [DOI] [PubMed] [Google Scholar]
- 11.Augustin R. The protein family of glucose transport facilitators: It's not only about glucose after all. IUBMB Life 2010;62:315–33. [DOI] [PubMed] [Google Scholar]
- 12.Szeszel-Fedorowicz W, Talukdar I, Griffith BN, Walsh CM, Salati LM. An exonic splicing silencer is involved in the regulated splicing of glucose 6-phosphate dehydrogenase mRNA. J Biol Chem 2006;281:34146–58. [DOI] [PubMed] [Google Scholar]
- 13.Cyphert TJ, Suchanek AL, Griffith BN, Salati LM. Starvation actively inhibits splicing of glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS element bound by hnRNP K. Biochim Biophys Acta 2013;1829:905–15. [DOI] [PMC free article] [PubMed]
- 14.Walsh CM, Suchanek AL, Cyphert TJ, Kohan AB, Szeszel-Fedorowicz W, Salati LM. Serine arginine splicing factor 3 is involved in enhanced splicing of glucose-6-phosphate dehydrogenase RNA in response to nutrients and hormones in liver. J Biol Chem 2013;288:2816–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001;107:881–91. [DOI] [PubMed] [Google Scholar]
- 16.Pfaffenbach KT, Nivala AM, Reese L, Ellis F, Wang D, Wei Y, Pagliassotti MJ. Rapamycin inhibits postprandial-mediated X-box-binding protein-1 splicing in rat liver. J Nutr 2010;140:879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao D, Liu J, Ni J, Wang Z, Shen Y, Zhou L, Huang Y, Wang J, Xue H, Zhang W, et al. Suppression of XBP1S mediates high glucose-induced oxidative stress and extracellular matrix synthesis in renal mesangial cell and kidney of diabetic rats. PLoS ONE 2013;8:e56124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glimcher LH, Lee AH. From sugar to fat: How the transcription factor XBP1 regulates hepatic lipogenesis. Ann N Y Acad Sci 2009;1173: Suppl 1:E2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao H, Szeszel-Fedorowicz W, Amir-Ahmady B, Gibson MA, Stabile LP, Salati LM. Inhibition of the splicing of glucose-6-phosphate dehydrogenase precursor mRNA by polyunsaturated fatty acids. J Biol Chem 2002;277:31270–8. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Wei Y, Pagliassotti MJ. Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology 2006;147:943–51. [DOI] [PubMed] [Google Scholar]
- 21.Xu T, Yang L, Yan C, Wang X, Huang P, Zhao F, Zhao L, Zhang M, Jia W, Liu Y. The IRE1α-XBP1 pathway regulates metabolic stress-induced compensatory proliferation of pancreatic β-cells. Cell Res 2014;24:1137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reardon HT, Hsieh AT, Park WJ, Kothapalli KS, Anthony JC, Nathanielsz PW, Brenna JT. Dietary long-chain polyunsaturated fatty acids upregulate expression of FADS3 transcripts. Prostaglandins Leukot Essent Fatty Acids 2013;88:15–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park WJ, Kothapalli KS, Reardon HT, Lawrence P, Qian SB, Brenna JT. A novel FADS1 isoform potentiates FADS2-mediated production of eicosanoid precursor fatty acids. J Lipid Res 2012;53:1502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wijendran V, Downs I, Srigley CT, Kothapalli KS, Park WJ, Blank BS, Zimmer JP, Butt CM, Salem N, Brenna JT. Dietary arachidonic acid and docosahexaenoic acid regulate liver fatty acid desaturase (FADS) alternative transcript expression in suckling piglets. Prostaglandins Leukot Essent Fatty Acids 2013;89:345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reardon HT, Park WJ, Zhang J, Lawrence P, Kothapalli KS, Brenna JT. The polypyrimidine tract binding protein regulates desaturase alternative splicing and PUFA composition. J Lipid Res 2011;52:2279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada N, Yamada Y, Tsukiyama K, Yamada C, Nakamura Y, Mukai E, Hamasaki A, Liu X, Toyoda K, Seino Y, et al. A novel GIP receptor splice variant influences GIP sensitivity of pancreatic beta-cells in obese mice. Am J Physiol Endocrinol Metab 2008;294:E61–8. [DOI] [PubMed] [Google Scholar]
- 27.Ahlqvist E, Osmark P, Kuulasmaa T, Pilgaard K, Omar B, Brøns C, Kotova O, Zetterqvist AV, Stancáková A, Jonsson A, et al. Link between GIP and osteopontin in adipose tissue and insulin resistance. Diabetes 2013;62:2088–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilberg MS, Balasubramanian M, Fu L, Shan J. The transcription factor network associated with the amino acid response in mammalian cells. Adv Nutr 2012;3:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu Rev Nutr 2005;25:59–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Y, Chen H, Siu F, Kilberg MS. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple activating transcription factor-3 mRNA species that, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J Biol Chem 2003;278:38402–12. [DOI] [PubMed] [Google Scholar]
- 31.Muratore CR, Hodgson NW, Trivedi MS, Abdolmaleky HM, Persico AM, Lintas C, De la Monte S, Deth RC. Age-dependent decrease and alternative splicing of methionine synthase mRNA in human cerebral cortex and an accelerated decrease in autism. PLoS ONE 2013;8:e56927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malatesta M, Bertoni-Freddari C, Fattoretti P, Baldelli B, Fakan S, Gazzanelli G. Aging and vitamin E deficiency are responsible for altered RNA pathways. Ann N Y Acad Sci 2004;1019:379–82. [DOI] [PubMed] [Google Scholar]
- 33.Apostolatos H, Apostolatos A, Vickers T, Watson JE, Song S, Vale F, Cooper DR, Sanchez-Ramos J, Patel NA. Vitamin A metabolite, all-trans-retinoic acid, mediates alternative splicing of protein kinase C deltaVIII (PKCdeltaVIII) isoform via splicing factor SC35. J Biol Chem 2010;285:25987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel NA, Song SS, Cooper DR. PKCdelta alternatively spliced isoforms modulate cellular apoptosis in retinoic acid-induced differentiation of human NT2 cells and mouse embryonic stem cells. Gene Expr 2006;13:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laserna EJ, Valero ML, Sanz L, del Pino MM, Calvete JJ, Barettino D. Proteomic analysis of phosphorylated nuclear proteins underscores novel roles for rapid actions of retinoic acid in the regulation of mRNA splicing and translation. Mol Endocrinol 2009;23:1799–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Soprano KJ, Soprano DR. Role of Acinus in Regulating Retinoic Acid-responsive Gene Pre-mRNA Splicing. J Cell Physiol 2015;230:791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Chun RF, Lisse TS, Garcia AJ, Xu J, Adams JS, Hewison M. Vitamin D and alternative splicing of RNA. J Steroid Biochem Mol Biol 2014. Oct 16 (Epub ahead of print; DOI:10.1016/j.jsbmb.2014.09.025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Dowd DR, Staal A, Gu C, Lian JB, van Wijnen AJ, Stein GS, MacDonald PN. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem 2003;278:35325–36. [DOI] [PubMed] [Google Scholar]
- 39.Glendenning P, Ratajczak T, Dick IM, Prince RL. Regulation of the 1b isoform of the plasma membrane calcium pump by 1,25-dihydroxyvitamin D3 in rat osteoblast-like cells. J Bone Miner Res 2001;16:525–34. [DOI] [PubMed] [Google Scholar]
- 40.Schiessl IM, Rosenauer A, Kattler V, Minuth WW, Oppermann M, Castrop H. Dietary salt intake modulates differential splicing of the Na-K-2Cl cotransporter NKCC2. Am J Physiol Renal Physiol 2013;305:F1139–48. [DOI] [PubMed] [Google Scholar]
- 41.Shehata MF. Regulation of the epithelial sodium channel [ENaC] in kidneys of salt-sensitive Dahl rats: insights on alternative splicing. Int Arch Med 2009;2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicola JP, Reyna-Neyra A, Carrasco N, Masini-Repiso AM. Dietary iodide controls its own absorption through post-transcriptional regulation of the intestinal Na+/I- symporter. J Physiol 2012;590:6013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hara H, Takeda T, Yamamoto N, Furuya K, Hirose K, Kamiya T, Adachi T. Zinc-induced modulation of SRSF6 activity alters Bim splicing to promote generation of the most potent apoptotic isoform BimS. FEBS J 2013;280:3313–27. [DOI] [PubMed] [Google Scholar]