Abstract
Xanthomonas albilineans causes leaf scald, a lethal disease of sugarcane. Compared to other species of Xanthomonas, X. albilineans exhibits distinctive pathogenic mechanisms, ecology and taxonomy. Its genome, which has experienced significant erosion, has unique genomic features. It lacks two loci required for pathogenicity in other plant pathogenic species of Xanthomonas: the xanthan gum biosynthesis and the Hrp-T3SS (hypersensitive response and pathogenicity-type three secretion system) gene clusters. Instead, X. albilineans harbors in its genome an SPI-1 (Salmonella pathogenicity island-1) T3SS gene cluster usually found in animal pathogens. X. albilineans produces a potent DNA gyrase inhibitor called albicidin, which blocks chloroplast differentiation, resulting in the characteristic white foliar stripe symptoms. The antibacterial activity of albicidin also confers on X. albilineans a competitive advantage against rival bacteria during sugarcane colonization. Recent chemical studies have uncovered the unique structure of albicidin and allowed us to partially elucidate its fascinating biosynthesis apparatus, which involves an enigmatic hybrid PKS/NRPS (polyketide synthase/non-ribosomal peptide synthetase) machinery.
Keywords: Xanthomonas albilineans, leaf scald disease of sugarcane, genomic features, albicidin, NRPS and PKS genes
Introduction
Xanthomonas albilineans (Ashby) Dowson is known to invade the xylem of sugarcane and to cause leaf scald disease (Rott and Davis, 2000; Birch, 2001). Symptoms of this disease vary from a single, white, narrow, sharply defined stripe to complete wilting and necrosis of infected leaves, leading to plant death. Dissemination of X. albilineans occurs mainly mechanically through use of contaminated harvesting tools and by distribution and planting of infected cuttings. However, aerial transmission and potential for epiphytic survival have also been reported for this pathogen (Autrey et al., 1995; Daugrois et al., 2003; Champoiseau et al., 2009).
Xanthomonas albilineans is a representative of the genus Xanthomonas, members of which are exclusively Gram-negative plant-associated bacteria that collectively cause dramatic damage to hundreds of plant species of ornamental or agronomical interest. Indeed, both monocotyledonous (e.g., rice, sugarcane, or banana) and dicotyledonous (e.g., citrus, cauliflower, bean, pepper, cabbage, and tomato) plants are targeted worldwide by various Xanthomonas species. While sharing numerous phenotypic characteristics, at least 27 species and over 120 pathovars (variants of pathogeny) of the genus Xanthomonas are currently recognized. Each pathovar individually exhibits a very restricted host range and/or tissue-specificity and this leads to clustering of bacterial strains causing similar symptoms on the same host.
Multilocus sequence analysis (MLSA) with four housekeeping genes resulted in the distribution of Xanthomonas species in two clades. The main one contains the majority of species whereas the secondary clade contains X. albilineans, Xanthomonas sacchari, Xanthomonas theicola, Xanthomonas hyacinthi, and Xanthomonas translucens (Young et al., 2008). Phylogenetic analyses with the gyrB sequence indicate that this secondary group also contains several uncharacterized species of Xanthomonas isolated mainly on rice, banana or sugarcane (Studholme et al., 2011, 2012). Intriguingly, two multiMLSA studies with 28 genes and 228 genes, respectively, in which X. albilineans is the only representative of this secondary clade, resulted in the branching of Xylella fastidiosa between X. albilineans and the main clade (Rodriguez-R et al., 2012; Naushad and Gupta, 2013). X. fastidiosa is a xylem-limited bacterium which is insect-vectored to a variety of diverse hosts, has a reduced genome and lacks the Hrp-T3SS (hypersensitive response and pathogenicity–type III secretion system; Simpson et al., 2000).
Analysis of the X. albilineans genome has revealed unusual features compared to other xanthomonads, the most prominent being the absence of the Hrp-T3SS gene cluster and the occurrence of genome erosion. Furthermore, to our knowledge, X. albilineans is the only xanthomonad that produces the phytotoxin albicidin. This mini-review aims to summarize the characteristics that, taken together, make X. albilineans so unique.
Genome Erosion
The genome of X. albilineans strain GPE PC73 has been fully sequenced and annotated. It consists of a 3,768,695-bp circular chromosome with a G+C content of 63%, and three plasmids of 31,555-bp, 27,212-bp and 24,837-bp, respectively (Pieretti et al., 2009). This genome size is much smaller than that of any other xanthomonad sequenced to date (commonly ∼5 Mb). Examination of the genome of strain GPE PC73 together with OrthoMCL comparative analyses performed with other sequenced xanthomonads highlights several genomic features that distinguish X. albilineans from its near relatives (Pieretti et al., 2009, 2012; Marguerettaz et al., 2011; Royer et al., 2013).
Orthologous analyses show that X. albilineans and X. fastidiosa have experienced a convergent genome reduction during their respective speciation, with a more extensive genome reduction for X. fastidiosa (Pieretti et al., 2009). Based on these analyses, X. albilineans has lost at least 592 genes that were present in the last common ancestor of the xanthomonads. Interestingly, most of these ancestral genes are conserved in the genome of X. sacchari strains NCPPB4393 and LMG 476 and Xanthomonas spp. strains NCPPB1131 and NCPPB1132, which are the sequenced strains phylogenetically closest to X. albilineans (Studholme et al., 2011, 2012; Pieretti et al., 2015). This indicates that genome erosion is specific to X. albilineans. Convergent genome erosion of X. albilineans and X. fastidiosa could be linked to a similar adaptation to a xylem-invading lifestyle in which interactions with living plant tissues are minimal (Pieretti et al., 2009). More recently, a study of the somewhat reduced genome of Xanthomonas fragariae (4.2 Mb) led to the hypothesis that the convergent genome reduction observed in some xanthomonads could be linked to their endophytic lifestyle and typically to their commitment to a single host (Vandroemme et al., 2013).
Compared to other xanthomonads, a low number of insertion sequences (IS) has been found in the genome of X. albilineans. Taken together with a limited recombination of the chromosome and a GC skew pattern containing a low number of distortions, it was postulated that genome erosion of X. albilineans was mainly not due to IS and other mechanisms were proposed for this erosion (Pieretti et al., 2009). The low number of IS could be linked to the activity of CRISPR (clustered regularly interspaced short palindromic repeats) systems. Strain GPE PC73 of X. albilineans possesses two CRISPR loci. The first one, CRISPR-1, is conserved in X. oryzae pv. oryzae, X. axonopodis pv. citri, X. campestris pv. vasculorum, and X. campestris pv. musacearum. The second, CRISPR-2, is present in X. campestris pv. raphani (Pieretti et al., 2012). Interestingly, many spacers of CRISPR-1 and CRISPR-2 of strain GPE PC73 are identical to IS or phage-related DNA sequences present on the chromosome of this strain (Pieretti et al., 2012).
Specific Genes Linked to a Xylem-Invading Lifestyle
Although determinants for host- or tissue-specificity of X. albilineans remain unclear, the presence in its genome of genes encoding cell-wall-degrading enzymes (CWDEs) with specific features is probably important for its ability to spread in xylem and for pathogenicity. Indeed, all CWDEs from X. albilineans harbor a cellulose-binding domain (CBD) and a long linker region both adapted to the utilization of cell-wall breakdown products as carbon source and to the ability to spread in sugarcane xylem vessels (Pieretti et al., 2012). These enzymes may also be required to disrupt pit membranes in sugarcane, thereby promoting propagation of the bacteria in the plant. Interestingly, X. fastidiosa also encodes two CWDEs containing a long linker and a CBD. It has been shown that one of these two CWDEs is involved in the spread of X. fastidiosa in the xylem by increasing the pore size of pit membranes. CWDEs are therefore considered as virulence factors (Roper et al., 2007; Chatterjee et al., 2008; Pérez-Donoso et al., 2010). TonB-dependent transporters (TBDTs) may be used by X. albilineans to transport cell-wall-degrading products resulting from the activity of CWDEs, and thus may facilitate spread of the organism in the nutrient-poor conditions prevailing in the xylem of sugarcane. In the genome of X. albilineans, 35 TBDT genes have been identified, including one specific to this species and two others that are functionally associated to pathogenicity of the bacterium (Rott et al., 2011; Pieretti et al., 2012).
Lack of Hrp-T3SS
Most phytopathogenic bacteria rely on the type III secretion system (T3SS) of the hypersensitive response and pathogenicity family (Hrp1 and Hrp2, respectively). This syringe-like apparatus allows pathogens to deliver, into their host cells, proteins (type III effectors) that modulate plant physiology and immunity for the benefit of the pathogen. Interestingly, genes encoding the injectisome and associated effectors of the Hrp-T3SS are missing in the genome of X. albilineans, as is also the case in the genomes of X. sacchari strains NCPPB4393 and LMG 476 and Xanthomonas spp. strains NCPPB1131 and NCPPB1132 (Studholme et al., 2011, 2012; Pieretti et al., 2015). Yet, an Hrp system is present in other close neighbor species of X. albilineans, such as X. translucens pv. graminis strain 29, X. translucens pv. translucens strain DSM18974, and X. translucens strain DAR 61454 (Wichmann et al., 2013; Gardiner et al., 2014). Although the Hrp-T3SS is described as a crucial key component in plant–host interactions for most Xanthomonas spp, it seems not to be essential in X. translucens pv. graminis strain 29 for xylem colonization, even though it is involved in symptom development (Ryan et al., 2011; Wichmann et al., 2013). Similarly, despite being devoid of any Hrp T3SS, X. albilineans displays pathogenicity and is able to cause serious damage to sugarcane.
Acquisition of a SPI-1 T3SS
The annotated sequence of the genome of X. albilineans strain GPE PC73 reveals the presence of a T3SS belonging to the Salmonella pathogenicity island-1 (SPI-1) injectisome family. Genes encoding this system are located near the terminus of the replication site of the chromosome and were probably acquired by lateral gene transfer. This secretion system, found mainly in mammals and insects bacterial pathogens or symbionts, exhibits high similarity to that described in Burkholderia pseudomallei—a human pathogen causing melioidosis (Stevens et al., 2002). The SPI-1 needle-like assemblies of X. albilineans strain GPE PC73 and B. pseudomallei strain K96243 are homologous. Both species share all but two genes—orgA and orgB, encoding putative oxygen-regulated invasion proteins involved in type three secretion that are not conserved in B. pseudomallei. The genome composition of the SPI-1 T3SS in X. albilineans additionally includes genes encoding translocon components (xipB, xipC, and xipD), injectisome components (xsaJ to xsaS and xsaV to xsaZ) and a chaperone (xicA). Furthermore, the locus contains 15 additional genes referred to as xapA–xapO, encoding hypothetical proteins. These genes, which show homology neither to sequences from B. pseudomallei nor to sequences available from protein sequence databases, are specific to X. albilineans and their products represent good candidates to be considered as effectors for this SPI-1 T3SS (Marguerettaz et al., 2011). Interestingly, this SPI-1 T3SS is conserved in Xanthomonas axonopodis pv. phaseoli strains CFBP 2534, CFBP 6164 and CFBP 6982, which moreover possess a second T3SS belonging to the Hrp2 family (Alavi et al., 2008; Marguerettaz et al., 2011). Pathogenicity of X. albilineans strains seems not to be linked to the presence of the SPI-1 T3SS in their genome; besides, no SPI-1 T3SS locus has been identified in strain PNG130 of X. albilineans even though it is able to spread in sugarcane. Functional analyses showed that, in planta, multiplication of a SPI-1 T3SS knockout mutant of X. albilineans was not impaired when compared to the wild-type, indicating that the SPI-1 T3SS is not required for spread in sugarcane vessels or for development of leaf scald symptoms. The role of the SPI-1 T3SS of X. albilineans remains unclear, although it has been conserved during its evolution in X. albilineans without frame-shifting indels or nonsense mutations (Marguerettaz et al., 2011). It remains possible, in conditions other than those tested with our knockout mutant, that the SPI-1 T3SS system may be required for interaction with sugarcane, as in the case of SPI-1 of Salmonella, which is involved in interactions with Arabidopsis thaliana (Schikora et al., 2011). The SPI-1 T3SS system may also be associated with other aspects of the X. albilineans lifestyle, e.g., an involvement in adherence as reported for Erwinia tasmaniensis (Kube et al., 2008) or in formation of pellicle or biofilm-like structures (Jennings et al., 2012), which could be related to epiphytic survival on sugarcane leaves. Although no insect vector has been identified for X. albilineans to date, we cannot rule out that the SPI-1 T3SS could be involved in insect association or might mediate persistence of the bacterium in an insect vector as was shown for Pantoea stewartii (Correa et al., 2012).
Lack of T6SS and the Xanthan Gum Gene Cluster
Xanthomonas albilineans lacks two other major pathogenicity factors that are common features of most xanthomonads. First, it lacks the gum gene cluster for extracellular polysaccharide (EPS) synthesis. This gene cluster is responsible for biofilm and xanthan gum formation, and is associated with pathogenesis in xanthomonads (Katzen et al., 1998; Kim et al., 2009; Galván et al., 2012). Exceptions are X. fragariae, which lacks the gumN, gumO and gumP genes, and X. albilineans, which lacks the complete set of gum genes, indicating those are not essential for virulence of both these pathogens (Pieretti et al., 2012; Vandroemme et al., 2013).
Xanthomonas albilineans is also devoid of any type VI secretion system (T6SS) described in other xanthomonads, as for example in Xanthomonas fuscans pv. fuscans strain 4834-R and Xanthomonas citri subsp. citri strain 306, which each contain a single T6SS (Potnis et al., 2011; Darrasse et al., 2013) or X. translucens strain DAR61454, which encodes two distinct T6SS (Gardiner et al., 2014). Structurally, the T6SS looks like an inverted bacteriophage. Functionally, this system is able to interact with both eukaryotic and prokaryotic cells by delivering effectors or toxins into host cells to subvert the signaling process to its own advantage, but also into other bacteria from the same habitat to outcompete them during infection (Filloux, 2013; Russell et al., 2014). Despite its multifunctional roles during host–pathogen interactions, the lack of T6SS in Xanthomonas campestris pv. campestris strain 8004, Xanthomonas gardneri strain 101, and X. albilineans seems to have no effect on pathogenesis of these xanthomonads.
Albicidin and Other Non-Ribosomally Synthesized Peptides
A unique feature of X. albilineans is the production of albicidin—a phytotoxin causing the white foliar stripe symptoms characteristic of leaf scald disease of sugarcane (Birch and Patil, 1985). Albicidin is a potent DNA gyrase inhibitor that blocks the differentiation of chloroplasts (Figure 1). It also targets bacterial gyrase by a mechanism different from that of other DNA gyrase inhibitors like coumarins and quinolones (Hashimi et al., 2007). This mode of action accounts for the potent antibacterial activity of albicidin, which inhibits the growth of Gram-positive and Gram-negative pathogenic bacteria at nanomolar concentrations (Birch and Patil, 1985). Albicidin gives a competitive advantage to X. albilineans against other bacteria within the xylem vessels of sugarcane (Magnani et al., 2013). Interestingly, two sugarcane-living bacteria harbor an albicidin resistance gene: Leifsonia xyli (Monteiro-Vitorello et al., 2004) and Pantoea dispersa (Zhang and Birch, 1997).
FIGURE 1.
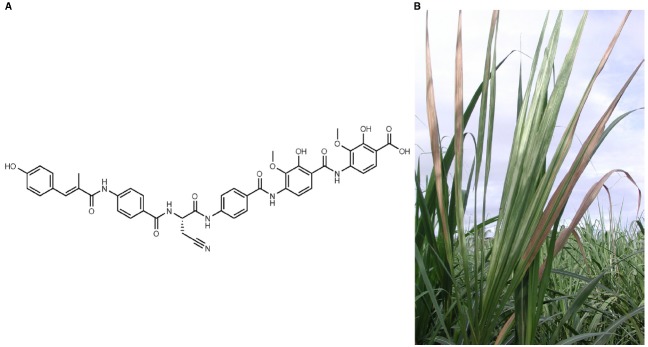
Xanthomonas albilineans produces the phytotoxin albicidin—a potent gyrase inhibitor that blocks chloroplast differentiation, resulting in sugarcane leaf scald disease symptoms. (A) Structure of albicidin, a hybrid PKS/NRPS compound with unique composition including p-aminobenzoic acid and cyanoalanine. (B) Diseased sugarcane plant with characteristic leaf scald symptoms: white foliar bleaching and necrosis of infected leaves (© J. H. Daugrois/Cirad).
Albicidin is produced by a hybrid polyketide synthase (PKS)/non-ribosomal peptide synthetase (NRPS) enzyme complex. PKS and NRPS genes are often clustered together with a large set of regulatory, transport or modification (tailoring) genes, as well as genes involved in the biosynthesis of non-proteinogenic amino acids. In addition to a phosphopantetheinyl transferase required for activation of the PKS/NRPS system and a HtpG chaperone, the role of which remains unclear, a locus (alb cluster) containing 20 genes is required for albicidin biosynthesis. Among these 20 genes, 3 encode the PKS/NRPS system; 15 others act as transport, regulatory, modification or resistance genes (Royer et al., 2004).
Non-ribosomal peptide synthetases are multimodular megasynthetases used by bacteria and fungi to produce peptides in a ribosome-independent manner (Strieker et al., 2010). Each module governs the specific incorporation of an amino acid substrate based on signature sequences in the adenylation (A) domains (Stachelhaus and Marahiel, 1995), which are loaded onto peptidyl carrier protein (PCP) domains. Elongation of the peptide is mediated by condensation (C) domains present within each module. PKSs function according to the principles of fatty acid biosynthesis (Weissman and Leadlay, 2005).
For decades, the structure elucidation of albicidin was impeded by its extremely low production yield by X. albilineans. A first step to overcome this bottleneck was achieved by transferring the biosynthetic genes into a heterologous host, namely X. axonopodis pv. vesicatoria, resulting in a significant increase in albicidin production (Vivien et al., 2007). Extensive HPLC purification of albicidin and thorough analysis of the purified compound by means of mass spectrometry and nuclear magnetic resonance spectroscopy then allowed us to unravel its unique structure (Figure 1). Albicidin proved to be a linear pentapeptide composed of cyanoalanine and p-amino benzoic acids N-terminally linked to a p-coumaric acid derivative (Cociancich et al., 2015). Although over 500 different monomers (amino acid substrates) have been identified to date as being incorporated by NRPS systems, elucidation of the structure of albicidin revealed for the first time the incorporation by NRPSs of cyanoalanine and p-amino benzoic acids. Moreover, the incorporation of p-amino benzoic acids is the first example of incorporation of a δ-aminoacid by NRPSs, since all NRPSs described to date incorporate only α or β aminoacids. The use of unusual amino acid substrates is linked to unique features that were identified in silico 10 years ago within the albicidin NRPS modules sequence (Royer et al., 2004). The formation and incorporation of cyanoalanine most likely occurs in situ through an additional module present in the PKS-NRPS assembly line that was investigated in one of our present studies (Cociancich et al., 2015).
Chemical synthesis of albicidin is now available, allowing both production of high quantities of the compound for further study of its mode of action and activity spectrum, and the synthesis of analogs (Kretz et al., 2015). The uniqueness of its structure and the specific mode of action of this compound make albicidin a strong lead structure for antibiotic development.
Data mining of the genome of X. albilineans strain GPE PC73 has led to the identification, in addition to the albicidin biosynthesis locus, of five other NRPS loci (Pieretti et al., 2012; Royer et al., 2013). The first, named Meta-B, encodes megasynthases performing peptidic elongation of a 16-amino acid lipopeptide. This locus also encodes a transcription regulator belonging to the AraC family, a cyclic peptide transporter, and enzymes involved in biosynthesis of the non-proteinogenic amino acids di-amino butyric acid and dihydroxyphenylglycine. Interestingly, the NRPS locus Meta-B has been identified in the genome of strains of three other Xanthomonas species, namely Xanthomonas oryzae pv. oryzae strains BAI3 and X11-5A, X. translucens strain DAR61454 and Xanthomonas spp. strain XaS3 (Royer et al., 2013). Despite a similar organization of the genes within these loci, the in silico prediction of the sequences of the peptides produced indicates that each strain produces a different lipopeptide.
Two other NRPS gene clusters, Meta-A and Meta-C, have been identified in the genome of X. albilineans strain GPE PC73. They encode megasynthases that perform the biosynthesis of peptides of 12 and 7 amino acids, respectively. A partial sequence has been predicted for each of these peptides (Royer et al., 2013).
Finally, two short NRPS genes have also been identified on the chromosome of X. albilineans: they both encode only one NRPS module. Interestingly, there is an overlap between both these genes and a gene encoding a glycosyltransferase. It has been hypothesized that these genes encode glycosylated amino acids, to which, however, no precise function could yet be attributed (Royer et al., 2013).
Conclusion
Although most xanthomonads require pathogenicity factors such as gum genes, T3SS Hrp and T6SS for survival, growth and spread within host plants, X. albilineans lacks these pathogenicity factors, de facto reducing its artillery to circumvent sugarcane defense mechanisms and innate immunity. While being “disarmed” could be disadvantageous for a vascular plant pathogen, X. albilineans remains able to invade and spread in sugarcane, suggesting that it uses other strategies, such as stealth, i.e., being unobtrusive in planta, to minimize inducible host defense responses. On the other hand, the reduced genome of X. albilineans has specific features that may be involved in the adaptation of the bacterium to live and spread in sugarcane xylem vessels. For example, specific CWDEs and TBDTs appear to be optimized for life in the nutrient-poor sugarcane xylem environment. The uniqueness of X. albilineans resides also in the production of the phytotoxin and antibiotic albicidin. The recently unraveled structure and concomitant development of a chemical synthesis protocol for this compound leads to additional prospects for its use in the antibiotherapy field. According to the specificities deriving from the biological, biochemical, phylogenetic and genomic analyses described in this review, one can truly say that X. albilineans is quite unique amongst the genus Xanthomonas.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work on albicidin was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG SU239/11-1; SU 18-1), by the Cluster of Excellence “Unifying Concepts in Catalysis (UniCat)” (DFG) and by a grant from the Agence Nationale de la Recherche (ANR-09-BLAN-0413-01). The authors are indebted to Helen Rothnie for English editing.
References
- Alavi S. M., Sanjari S., Durand F., Brin C., Manceau C., Poussier S. (2008). Assessment of the genetic diversity of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans as a basis to identify putative pathogenicity genes and a type III secretion system of the SPI-1 family by multiple suppression subtractive hybridizations. Appl. Environ. Microbiol. 74, 3295–3301. 10.1128/aem.02507-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autrey L., Saumtally S., Dookun A., Sullivan S., Dhayan S. (1995). Aerial transmission of the leaf scald pathogen, Xanthomonas albilineans. Proc. Int. Soc. Sug. Cane Technol. 21, 508–526. [Google Scholar]
- Birch R. G. (2001). Xanthomonas albilineans and the antipathogenesis approach to disease control. Mol. Plant Pathol. 2, 1–11. 10.1046/j.1364-3703.2001.00046.x [DOI] [PubMed] [Google Scholar]
- Birch R. G., Patil S. S. (1985). Preliminary characterization of an antibiotic produced by Xanthomonas albilineans which inhibits DNA synthesis in Escherichia coli. J. Gen. Microbiol. 131, 1069–1075. 10.1099/00221287-131-5-1069 [DOI] [PubMed] [Google Scholar]
- Champoiseau P., Rott P., Daugrois J. H. (2009). Epiphytic populations of Xanthomonas albilineans and subsequent sugarcane stalk infection are linked to rainfall in Guadeloupe. Plant Dis. 93, 339–346 10.1094/pdis-93-4-0339 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Almeida R. P. P., Lindow S. (2008). Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 46, 243–271. 10.1146/annurev.phyto.45.062806.094342 [DOI] [PubMed] [Google Scholar]
- Cociancich S., Pesic A., Petras D., Uhlmann S., Kretz J., Schubert V., et al. (2015). The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat. Chem. Biol. 11, 195–197. 10.1038/nchembio.1734 [DOI] [PubMed] [Google Scholar]
- Correa V. R., Majerczak D. R., Ammar E.-D., Merighi M., Pratt R. C., Hogenhout S. A., et al. (2012). The bacterium Pantoea stewartii uses two different type III secretion systems to colonize its plant host and insect vector. Appl. Environ. Microbiol. 78, 6327–6336. 10.1128/aem.00892-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse A., Carrere S., Barbe V., Boureau T., Arrieta-Ortiz M., Bonneau S., et al. (2013). Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genom. 14:761. 10.1186/1471-2164-14-761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugrois J. H., Dumont V., Champoiseau P., Costet L., Boisne-Noc R., Rott P. (2003). Aerial contamination of sugarcane in Guadeloupe by two strains of Xanthomonas albilineans. Eur. J. Plant Pathol. 109, 445–458 10.1023/a:1024259606468 [DOI] [Google Scholar]
- Filloux A. (2013). The rise of the type VI secretion system. F1000Prime Rep. 5:52. 10.12703/p5-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván E. M., Ielmini M. V., Patel Y. N., Bianco M. I., Franceschini E. A., Schneider J. C., et al. (2012). Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology 23, 259–272. 10.1093/glycob/cws146 [DOI] [PubMed] [Google Scholar]
- Gardiner D. M., Upadhyaya N. M., Stiller J., Ellis J. G., Dodds P. N., Kazan K., et al. (2014). Genomic analysis of Xanthomonas translucens pathogenic on wheat and barley reveals cross-kingdom gene transfer events and diverse protein delivery systems. PLoS ONE 9:e84995. 10.1371/journal.pone.0084995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi S. M., Wall M. K., Smith A. B., Maxwell A., Birch R. G. (2007). The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 51, 181–187. 10.1128/aac.00918-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. E., Quick L. N., Ubol N., Shrom S., Dollahon N., Wilson J. W. (2012). Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS ONE 7:e33080. 10.1371/journal.pone.0033080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen F., Ferreiro D. U., Oddo C. G., Ielmini M. V., Becker A., Pühler A., et al. (1998). Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J. Bacteriol. 180, 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-G., Li X., Roden J. A., Taylor K. W., Aakre C. D., Su B., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21, 1305–1323. 10.1105/tpc.108.063123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz J., Kerwat D., Schubert V., Grätz S., Pesic A., Semsary S., et al. (2015). Total synthesis of albicidin: a lead structure from Xanthomonas albilineans for potent antibacterial gyrase inhibitors. Angew. Chem. Int. Ed. 54, 1969–1973. 10.1002/anie.201409584 [DOI] [PubMed] [Google Scholar]
- Kube M., Migdoll A. M., Müller I., Kuhl H., Beck A., Reinhardt R., et al. (2008). The genome of Erwinia tasmaniensis strain Et1/99, a non-pathogenic bacterium in the genus Erwinia. Environ. Microbiol. 10, 2211–2222. 10.1111/j.1462-2920.2008.01639.x [DOI] [PubMed] [Google Scholar]
- Magnani G. S., Cruz L. M., Weber H., Bespalhok J. C., Daros E., Baura V., et al. (2013). Culture-independent analysis of endophytic bacterial communities associated with Brazilian sugarcane. Genet. Mol. Res. 12, 4549–4558. 10.4238/2013.October.15.3 [DOI] [PubMed] [Google Scholar]
- Marguerettaz M., Pieretti I., Gayral P., Puig J., Brin C., Cociancich S., et al. (2011). Genomic and evolutionary features of the SPI-1 type III secretion system that is present in Xanthomonas albilineans but is not essential for xylem colonization and symptom development of sugarcane leaf scald. Mol. Plant Microbe Interact. 24, 246–259. 10.1094/mpmi-08-10-0188 [DOI] [PubMed] [Google Scholar]
- Monteiro-Vitorello C. B., Camargo L. A., Van Sluys M. A., Kitajima J. P., Truffi D., do Amaral A. M., et al. (2004). The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant Microbe Interact. 17, 827–836. 10.1094/mpmi.2004.17.8.827 [DOI] [PubMed] [Google Scholar]
- Naushad H. S., Gupta R. S. (2013). Phylogenomics and molecular signatures for species from the plant pathogen-containing order Xanthomonadales. PLoS ONE 8:e55216. 10.1371/journal.pone.0055216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Donoso A. G., Sun Q., Roper M. C., Greve L. C., Kirkpatrick B., Labavitch J. M. (2010). Cell wall-degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa-infected grapevines. Plant Physiol. 152, 1748–1759. 10.1104/pp.109.148791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti I., Bolot S., Carrère S., Barbe V., Cociancich S., Rott P., et al. (2015). Draft genome sequence of Xanthomonas sacchari LMG 476. Genome Announc. 3, e00146-15. 10.1128/genomeA.00146-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti I., Royer M., Barbe V., Carrere S., Koebnik R., Cociancich S., et al. (2009). The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genom. 10:616. 10.1186/1471-2164-10-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti I., Royer M., Barbe V., Carrere S., Koebnik R., Couloux A., et al. (2012). Genomic insights into strategies used by Xanthomonas albilineans with its reduced artillery to spread within sugarcane xylem vessels. BMC Genom. 13:658. 10.1186/1471-2164-13-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis N., Krasileva K., Chow V., Almeida N., Patil P., Ryan R., et al. (2011). Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genom. 12:146. 10.1186/1471-2164-12-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-R L., Grajales A., Arrieta-Ortiz M., Salazar C., Restrepo S., Bernal A. (2012). Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 12:43. 10.1186/1471-2180-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper M. C., Greve L. C., Warren J. G., Labavitch J. M., Kirkpatrick B. C. (2007). Xylella fastidiosa requires polygalacturonase for colonization and pathogenicity in Vitis vinifera grapevines. Mol. Plant Microbe Interact. 20, 411–419. 10.1094/mpmi-20-4-0411 [DOI] [PubMed] [Google Scholar]
- Rott P., Davis M. (2000). “Leaf scald,” in A Guide to Sugarcane Diseases, eds Rott P., Bailey R., Comstock J., Croft B., Saumtally A. (Montpellier: CIRAD-ISSCT press; ), 339. [Google Scholar]
- Rott P., Fleites L., Marlow G., Royer M., Gabriel D. W. (2011). Identification of new candidate pathogenicity factors in the xylem-invading pathogen Xanthomonas albilineans by transposon mutagenesis. Mol. Plant Microbe Interact. 24, 594–605. 10.1094/mpmi-07-10-0156 [DOI] [PubMed] [Google Scholar]
- Royer M., Costet L., Vivien E., Bes M., Cousin A., Damais A., et al. (2004). Albicidin pathotoxin produced by Xanthomonas albilineans is encoded by three large PKS and NRPS genes present in a gene cluster also containing several putative modifying, regulatory, and resistance genes. Mol. Plant Microbe Interact. 17, 414–427. 10.1094/mpmi.2004.17.4.414 [DOI] [PubMed] [Google Scholar]
- Royer M., Koebnik R., Marguerettaz M., Barbe V., Robin G., Brin C., et al. (2013). Genome mining reveals the genus Xanthomonas to be a promising reservoir for new bioactive non-ribosomally synthesized peptides. BMC Genom. 14:658. 10.1186/1471-2164-14-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Peterson S. B., Mougous J. D. (2014). Type VI secretion effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Vorhölter F.-J., Potnis N., Jones J. B., Van Sluys M.-A., Bogdanove A. J., et al. (2011). Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat. Rev. Microbiol. 9, 344–355. 10.1038/nrmicro2558 [DOI] [PubMed] [Google Scholar]
- Schikora A., Virlogeux-Payant I., Bueso E., Garcia A. V., Nilau T., Charrier A., et al. (2011). Conservation of Salmonella infection mechanisms in plants and animals. PLoS ONE 6:e24112. 10.1371/journal.pone.0024112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A., Reinach F., Arruda P. (2000). The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406, 151–157. 10.1038/35018003 [DOI] [PubMed] [Google Scholar]
- Stachelhaus T., Marahiel M. A. (1995). Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol. Lett. 125, 3–14. 10.1111/j.1574-6968.1995.tb07328.x [DOI] [PubMed] [Google Scholar]
- Stevens M. P., Wood M. W., Taylor L. A., Monaghan P., Hawes P., Jones P. W., et al. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659. 10.1046/j.1365-2958.2002.03190.x [DOI] [PubMed] [Google Scholar]
- Strieker M., Tanovic A., Marahiel M. A. (2010). Non-ribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 20, 234–240. 10.1016/j.sbi.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Studholme D. J., Wasukira A., Paszkiewicz K., Aritua V., Thwaites R., Smith J., et al. (2011). Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas Group 1 Clade. Genes 2, 1050–1065. 10.3390/genes2041050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme D. J., Wasukira A., Paszkiewicz K., Aritua V., Thwaites R., Smith J., et al. (2012). Correction: Studholme et al., Draft genome sequences of Xanthomonas sacchari and two banana-associated xanthomonads reveal insights into the Xanthomonas Group 1 Clade. Genes 2011, 2, 1050–1065. Genes 3, 88–89 10.3390/genes3010088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandroemme J., Cottyn B., Baeyen S., De Vos P., Maes M. (2013). Draft genome sequence of Xanthomonas fragariae reveals reductive evolution and distinct virulence-related gene content. BMC Genom. 14:829. 10.1186/1471-2164-14-829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivien E., Pitorre D., Cociancich S., Pieretti I., Gabriel D. W., Rott P. C., et al. (2007). Heterologous production of albicidin: a promising approach to overproducing and characterizing this potent inhibitor of DNA gyrase. Antimicrob. Agents Chemother. 51, 1549–1552. 10.1128/aac.01450-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman K. J., Leadlay P. F. (2005). Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Micro. 3, 925–936. 10.1038/nrmicro1287 [DOI] [PubMed] [Google Scholar]
- Wichmann F., Vorhölter F.-J., Hersemann L., Widmer F., Blom J., Niehaus K., et al. (2013). The noncanonical type III secretion system of Xanthomonas translucens pv. graminis is essential for forage grass infection. Mol. Plant Pathol. 14, 576–588. 10.1111/mpp.12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. M., Park D. C., Shearman H. M., Fargier E. (2008). A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 31, 366–377. 10.1016/j.syapm.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Zhang L., Birch R. (1997). The gene for albicidin detoxification from Pantoea dispersa encodes an esterase and attenuates pathogenicity of Xanthomonas albilineans. Proc. Natl. Acad. Sci. U.S.A. 94, 9984–9989. 10.1073/pnas.94.18.9984 [DOI] [PMC free article] [PubMed] [Google Scholar]


