Summary
Apoptosis is typically considered an anti-oncogenic process since caspase activation can promote the elimination of genetically unstable or damaged cells. We report that a central effector of apoptosis, caspase 3, facilitates, rather than suppresses, chemical and radiation-induced genetic instability and carcinogenesis. We found that a significant fraction of mammalian cells treated with ionizing radiation can survive, despite caspase 3 activation. Moreover, this sublethal activation of caspase 3 promoted persistent DNA damage and oncogenic transformation. In addition, chemically-induced skin carcinogenesis was significantly reduced in mice genetically deficient in caspase 3. Furthermore, attenuation of Endo G activity significantly reduced radiation-induced DNA damage and oncogenic transformation, identifying Endo G as a downstream effector of caspase 3 in this pathway. Our findings suggest that rather than acting as a broad inhibitor of carcinogenesis, caspase 3 activation may contribute to genome instability and play a pivotal role in tumor formation following damage.
Introduction
Apoptosis, or programmed cell death, is the most well-defined mode of cell death in multicellular organisms (Horvitz, 2003; Horvitz et al., 1994; Yuan et al., 1993). A major physiological function of apoptosis is to get rid of damaged or unwanted cells in early development or to maintain somatic tissue homeostasis at later stages. As such it is generally assumed that apoptosis is an anti-carcinogenic process due to its essential role in removing cells that have suffered DNA damage (Hanahan and Weinberg, 2000; Reed, 1999). DNA damage and subsequent mutations in key oncogenes and tumor suppressor genes is a key process leading to cancer (Lengauer et al., 1997). Therefore, the current paradigm is that apoptosis is a barrier for carcinogenesis. For example, many tumor suppressor genes mutated in cancer often have apoptosis-promoting functions. Examples of these include p53 (Lowe et al., 1994), PTEN (Weng et al., 2001), bax (Wang et al., 1996), and INK4a/ARF (Kim et al., 2000). Mutations in these genes often allow damaged cells to survive when they should die. On the other hand, many oncogenes whose expressions are often enhanced in cancer cells possess anti-apoptotic functions. Examples of these include bcl2(Cory and Adams, 2002), bcl-xL(Cory and Adams, 2002), Akt/PKB(Sabbatini and McCormick, 1999), and mTOR(Panner et al., 2005).
However, there is increasing recognition that the involvement of apoptosis in carcinogenesis may be more complicated. Some oncogenes appear to sensitize cells to apoptosis. For example, the apoptosis-promoting functions of c-Myc and E1A are well-documented in different studies (Debbas and White, 1993; Evan et al., 1992; Fanidi et al., 1992; Rao et al., 1992). However, the role of apoptosis in carcinogenesis induced by these genes is not well-understood. For both c-Myc and E1A, their pro-apoptotic activities appear coupled to their hyperproliferative activities. Thus it was suggested that apoptosis acts as a fail-safe mechanism to limit the consequences of aberrant mitogenic signaling (Lowe and Lin, 2000). There is also a suggestion that oncogene-mediated excessive apoptosis create a selection pressure to overcome apoptosis to allow cancer cells to become more malignant (Lowe and Lin, 2000). However, these hypotheses have not been experimentally evaluated.
Evidence is emerging that some factors in the apoptotic machinery may play facilitative roles in carcinogenesis. One example is Fas Ligand (CD95), which is a major player in mediating the extrinsic pathway of cellular apoptosis. Genetic knockout of Fas ligand attenuated tumorigenesis in mice instead of promoting it. Fas Ligand was shown to promote carcinogenesis by activating downstream c-Jun and JNK pathway (Chen et al., 2010). Fas ligand has also been shown to promote cancer cell invasiveness and metastasis by interacting and activating c-Met oncogene(Lin et al., 2012). Another recent study showed that higher levels of activated caspase 3 in head and neck cancer or breast cancer were correlated increased post-therapy tumor recurrence or patient death, contrary to conventional wisdom (Huang et al., 2011).
In this study, we designed experiments to examine the counter-intuitive hypothesis that caspase 3 facilitates carcinogenesis by inducing genetic instability in cells that survived cytotoxic stress. We provided strong evidence that activated caspase 3 can indeed promote oncogenic transformation in human cells and in mice by inducing persistent genetic instability through its downstream effector, endonuclease G, whose normal function is to fragment genomic DNA in apoptotic cells.
Results
Non-lethal activation of caspase 3 in irradiated cells
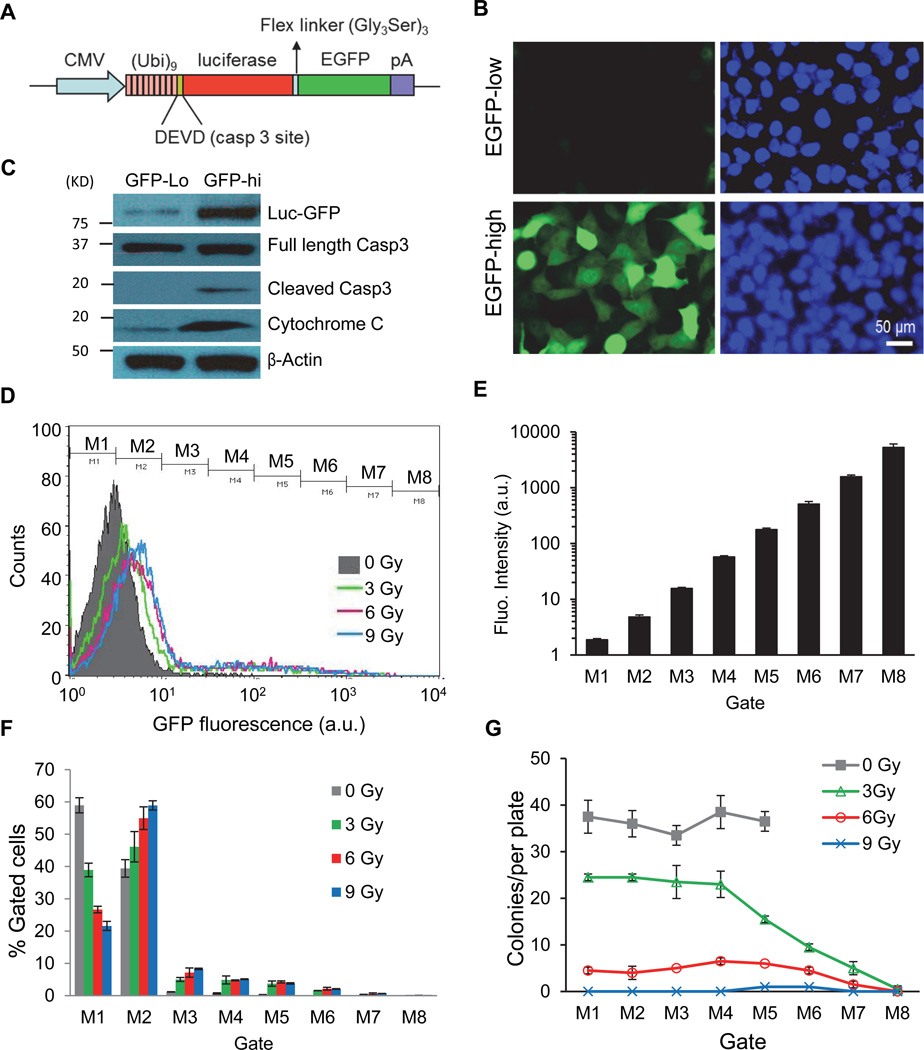
Caspases are crucial enzymes in apoptosis through which damaged or unwanted cells systematically destroy their own infrastructure to commit suicide. Activation of caspases in cells exposed to cytotoxic stress (e.g. radiation) is usually considered an irreversible process that will result in the elimination of host cells. Thus it has been generally assumed that all cells with apoptotic caspase activation will die. However, data in this respect are lacking for mammalian cells. We decided to carry out a detailed analysis of the relationship between caspase 3 activation and the fate of host cells because caspase 3 is involved at the end stage of apoptosis (Nicholson et al., 1995; Taylor et al., 2008). To do this we used a non-invasive caspase 3 reporter recently developed in our laboratory (Fig. 1A)(Huang et al., 2011; Li et al., 2010). In this reporter, the EGFP is fused with firefly luciferase reporter through a flexible linker. The fusion protein is further linked up with a polyubiquitin domain, which renders the fusion reporter protein very unstable because it is recognized by the proteasome system as an ubiquitylated protein and rapidly degraded. In between the EGFP-luc and polyubiquitin domains a caspase 3 cleavage site is inserted. Under normal circumstances steady-state reporter protein level will be very low in host cells. However, when caspase 3 is activated, the polyubiquitin domain will be cleaved off and the EGFP-Luc fusion protein will be stabilized.
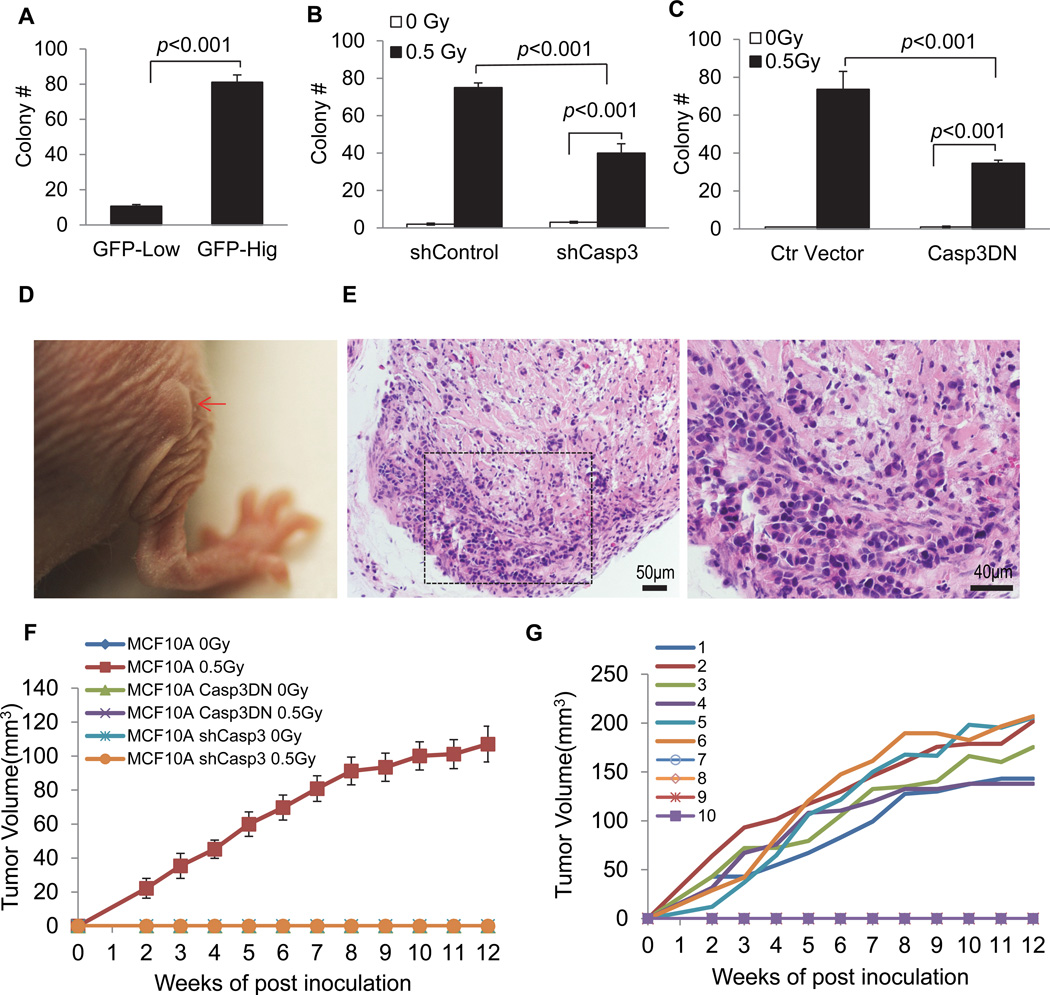
Figure 1. Non-lethal activation of caspases 3 in MCF10A cells exposed to ionizing radiation.
A). Diagram of the caspase 3 reporter gene. (Ub)9, a nine-ubiquitin polyubiqutin domain that serves as the proteasome recognition signal that causes the rapid degradation of the reporter protein, which consists of EGFP and firefly luciferase linked by a flexible linker.
B). Irradiated (0.5 Gy 600MeV 56Fe ions) MCF10A cells with low (top panels) and high (lower panels) caspase 3 reporter activities after separation by a FACS sorter based on Luc-GFP expression levels 7 days after cellular exposure to radiation.
C). Western blot analysis of caspase 3 cleavage and activation in cells with high and low Casp3Luc-EGFP reporter activities 14 days after irradiation. Cytochrome c was probed using cytosolic extracts while other proteins were probed with whole cell lysates.
D). Flow cytometry profiles of MCF10A-Casp3EGFP reporter activities in cells exposed to different doses of x-rays. Cells were analyzed 4 days after irradiation. Cells were gated into 8 different groups for colony forming assays according to their fluorescence intensity levels (M1-M8).
E). The mean (geometric) GFP fluorescence intensities of gated MCF10A cells.
F). Distribution of MCF10A cells in each gate after different irradiation doses. For each radiation dose, M1+M2+…+M8=100%.
G). Colony forming abilities of cells with different Casp3EGFP reporter activities. Cell from varying levels of reporter activities (M1-M8) were flow-sorted into individual wells of 96-well plates at 1 cell/well. Two weeks later, the numbers of MCF10A colonies on each 96-well plate were counted and plotted.
Error bars in 1E, 1F, and 1G represent standard deviation. All values are derived from the average of three independent experiments.
See also Figure S1.
To study the fate of cells with caspase 3 activation, we transduced the Casp3EGFP-Luc reporter gene into the MCF10A human mammary epithelial cells. We then subjected the cells to a moderate dose (0.5 Gy) of high energy (600 MeV/µ) 56Fe ion irradiation. High energy particle radiation has been shown to be more potent in inducing DNA damage and carcinogenesis compared with lower energy radiation (x-rays and γ-rays)(Durante and Cucinotta, 2008). We show that radiation induced significant caspase 3 reporter activation in a persistent manner, with some cells expressing high EGFP at 2 weeks post irradiation. To characterize reporter activation, the irradiated cells were sorted by use of FACS according to their cellular GFP levels. Those with high GFP expression were separated from those with low GFP expression (Fig. 1B). Interestingly, many cells with high EGFP levels appeared normal morphologically. This was despite robust cleaved caspase 3 cleavage and cytochrome C release status in irradiated cells in general (Fig. S1A, see Table S1 for a list of antibodies used in this study) and in cells with high EGFP levels (Fig. 1C). Those data therefore contradict with the currently established view that damaged cells with caspase 3 activation are destined to die. We further carried out experiments to observe the growth of the cells with high Casp3EGFP reporter activities. Our data suggest that both the EGFP-hi and EGFP-low groups of cells maintained continuous cellular growth and irradiated cells with high or low GFP expression grew only slightly slower than untreated parental cells (Fig.S1B). Because caspase 3 activation is often accompanied by cytochrome c leakage in apoptotic cells, we carried out immunofluorescence analysis of cytochrome c in parental and Casp3-EGFP-high cells. Our data (Fig. S1C & D) showed that many irradiated GFP-hi cells showed a distinct cytochrome c staining pattern with a small but significant percentage of cytochrome c located in the extra-mitochondrial regions (Fig. S1C, lower panels). In contrast, the vast majority of non-irradiated cells showed only mitochondrial cytochrome c staining (Fig. S1C, top panels). Quantitatively, the percentage of GFP-hi cells with the “leaky” extra-mitochondrial cytochrome c staining pattern was about 23% at 14 days after irradiation. This is compared with about 0.5% with the “leaky” pattern in the non-irradiated cells (Fig. S1D). The persistence of cells with cytochrome c leakage suggests that the leakage was at levels that did not lead to cell death. An analysis of mitochondrial membrane potential by use of the TMRE-flow cytometry assay (Fig. S1E) indicated that compared with parental MCF10A cells, those with high caspase3EGFP reporter activities showed higher degree of mitochondrial membrane depolarization, indicating persistent and incomplete mitochondrial membrane leakage, consistent with the leaky cytochrome c pattern shown in Fig. S1C & D.
To further characterize the fate of individual cells with caspase 3 activation, MCF10A cells transduced with caspase 3 reporters were irradiated with different doses of radiation and analyzed through flow cytometry (Fig. 1D). The cells were then sorted through 8 different gates according to caspase 3 reporter activation levels (Fig. 1D, E). Higher radiation doses increased the fraction of cells with higher levels of caspase 3 activation (Fig. 1D, F). Cells from different gates were then sorted 1 cell/well into different wells of 96-well plates. EGFP expression in the wells was further confirmed though fluorescence microscopy (data not shown). All the individual wells examined showed positive identification of 1 cell/well. In addition, EGFP expression was seen clearly in cells from M4-M8 gates. Colony formation from the individually plated cells with different GFP expression status (M1-M8) was then analyzed after 2 weeks in the 96-well plates. As expected, there was a clear radiation dose-dependent decrease in colony forming abilities (Fig. 1G). However, the relationship between caspase 3 activation and colony forming abilities was more complicated. It appeared that cells could tolerate a wide range of caspase 3 activation levels (Fig. 1G, M1-M4 gates), especially at lower doses of radiation. At a moderate radiation dose (3Gy), the relationship between caspase 3 activation and clonogenic survival become linear, with M4 as the threshold. At higher levels (>3Gy), most of the cells could not form colonies irrespective of caspase 3 reporter activities, consistent with the ability of radiation to kill cells in an apoptosis-independent manner. Our data thus clearly established that radiation could activate caspase 3 in a non-lethal manner at moderate radiation doses (≤6 Gy).
A key facilitative role of caspase 3 in radiation induced DNA damage foci formation in surviving cells
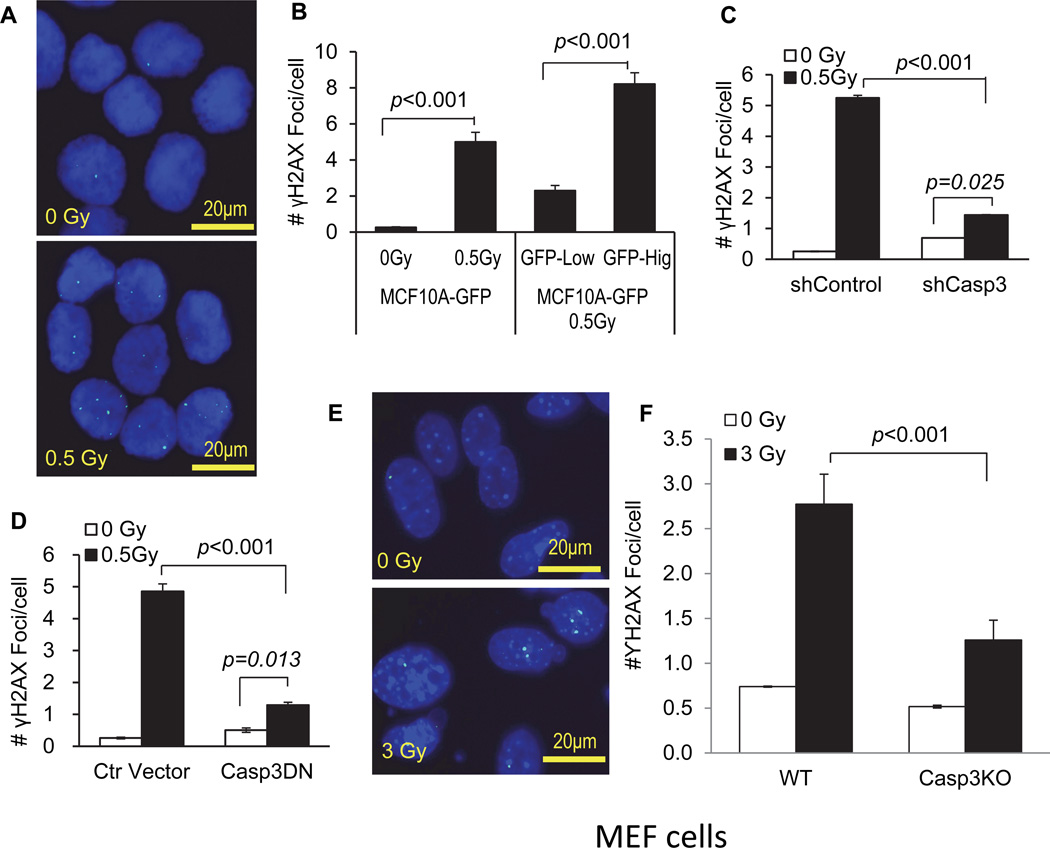
Our observation of non-lethal caspase 3 activation in MCF10A cells points to an important question: what happens to the genome of the cells that survive caspase 3 activation? This question is important since caspase 3 activation in those cells could lead to downstream nuclease activation and DNA fragmentation, similar to what occurs in apoptotic cells, which can potentially wreak havoc in the genomes of cells experiencing “abortive apoptosis”. To answer this question, we carried out experiments to examine DNA damage in cells that have survived caspase 3 activation. Our data show that radiation exposure induced significantly higher levels of γH2AX foci, an indication of DNA double strand breaks (DSBs) (Rogakou et al., 1998), when compared with sham-irradiated cells (Fig. 2A, and Fig. 2B (left side)) at 14 days after irradiation. There was a dose-dependent increase in the number of γH2AX foci in irradiated cells (Fig. S1F). Furthermore, cells with higher levels of Casp3EGFP reporter activation had significantly more γH2AX foci, when compared with those with low levels of reporter activation (Fig. 2B, right side). Importantly, higher numbers of γH2AX foci persist in the irradiated MCF10A cells for more than three months in the irradiated cells (Fig. S1G).
Figure 2. A key role for activated caspase 3 in radiation induced foci formation.
A). Typical micrographs of γH2AX foci in control (0 Gy) and 0.5 Gy 56Fe ions irradiated MCF10A cells.
B). The average number of γH2AX foci in caspase 3 reporter-transduced MCF10A cells exposed 0.5 Gy of 56Fe ions 14 days after 56Fe ions irradiation. The left two bars shows the numbers for non-irradiated vs 0.5 Gy irradiated cells while the right two bars show the numbers for irradiated MCF10A cells with high and low caspase 3 reporter activities based on GFP expression levels.
C). The effect of caspase 3 expression attenuation on γH2AX foci formation. MCF10A cells transduced with an shRNA gene against the CASP3 gene (shCASP3) or control (shControl) were evaluated for γH2AX foci formation at 10 days after 0.5 Gy or sham irradiation.
D). The effect of caspase 3 inhibition on radiation induced γH2AX foci formation in MCF10A cells. Cells transduced with a control lentiviral vector or those transduced with a dominant negative caspase 3 gene (CASP3DN) were evaluated for γH2AX foci formation with sham or 0.5 Gy of irradiation at 14 days after irradiation.
E). Typical micrographs of γH2AX foci in control (0 Gy) and 3 Gy x-Ray irradiated MEF cells at 4 days after irradiation.
F). The numbers of γH2AX foci in irradiated caspase 3 deficiency MEFs were significantly lower than those of caspase 3 proficiency MEFs at 4 days after irradiation.
Error bars in 2B, 2C, 2D and 2F represent standard error of the mean (SEM). All values are derived from the average of three independent experiments.
See also Figure S1–S3.
To determine if activation of caspase 3 plays a causative role in the observed DNA strand breaks, we established MCF10A cells that express a small hairpin RNA (shRNA) gene against the caspase 3 gene (see Fig. S2A, top panels for confirmation of knockdown). In those cells, the numbers of γH2AX foci were significantly reduced after exposure to 56Fe ions radiation when compared with those transduced with a control shRNA gene (Fig. 2C).
The causative role of caspases 3 was further confirmed by use of MCF10A cells transduced with a dominant negative version of caspase 3 (Casp3DN, Fig. S2A, lower panels). The dominant negative version of the caspase 3 has one single-base pair mutation that inactivates its cleavage activity (Stennicke and Salvesen, 1997). As shown in Fig. 2D, the number of γH2AX foci was significantly lower in irradiated MCF10A cells expressing Casp3DN at 14 days after irradiation.
In addition to the above data implicating caspase 3 in mediating radiation induced DNA damage in human cells, we also examined radiation induced γH2AX foci formation using wild type and Casp3KO mouse embryonic fibroblast (MEF) cells. We observed significantly attenuated radiation induced foci formation in Casp3KO, than in wild type MEF cells (Fig.2E & F). Thus our data support a key role for caspase 3 in mediating radiation induced γH2AX foci formation in murine cells as well.
Previous studies have shown that caspase 3 and caspase 7 shared similar cleavage site but are functionally distinct in their effect on host mice when genetically ablated (Lakhani et al., 2006; Walsh et al., 2008). In order to compare the relative contribution of caspases 3 &7, we also examined γH2AX foci formation in WT, Casp3KO, Casp7KO, and caspase 3&7 double knockout (DKO) cells (Fig. S2B). Our data show that Casp3 knockout had a more pronounced effect than Casp7 knockout. In addition, double knockout cells did not exhibit less radiation induced DNA damage than caspase 3 knockout cells alone, indicating that caspase 3 and caspase 7 have mostly overlapping functions in terms of DNA damage with caspase 3 playing a much more dominant role.
We next examined dynamic 53BP1 foci formation in irradiated cells. 53BP1 is a protein shown to be actively involved in DSB repair (Dimitrova et al., 2008; Wang et al., 2002). It forms foci with many other DNA DSB repair proteins such as γH2AX at sites of DNA double strand breaks. Using a non-invasive red fluorescent protein (mCherry) based 53BP1 foci reporter (Dimitrova et al., 2008), we monitored 53BP1 foci formation in the nuclei of irradiated wild type and Casp3-deficeint MEF cells. Our results show a significantly higher fraction of cells with 53BP1 foci in irradiated wild type MEF cells when compared with sham irradiated controls (Fig. S2C & D). In addition, deficiency in caspase 3 caused a significant reduction in 53BP1 foci (Fig. S2D), similar to γH2AX foci. By time-lapse video, we were also able to observe dynamic and persistent 53BP1 foci formation in irradiated MCF10A cells and their progeny (Fig. S2E).
Another important question is whether the observed caspase 3-dependent foci formation requires initial cellular exposure to ionizing radiation. In order to examine this issue, we transduced an artificially inducible, dimerizable caspase 3 gene (iCasp3) into MCF10A cells. The iCasp3 gene is engineered by fusing caspase3 gene with FK506-binding sites (FKBPs) so that the engineered iCasp3 gene can dimerize and be activated when exposed to an FK506 analogue in the absence of upstream apoptotic signaling (Fujioka et al., 2011; MacCorkle et al., 1998). We show that the addition of the chemical dimerizer (AP20187, an FK506 analogue) was sufficient to induce caspase 3 activation and γH2AX foci formation (Fig. S3A–C). These results indicate that caspase 3 activation alone is sufficient to induce DNA damage in host cells.
In another experiment, we evaluated γH2AX foci formation in wild-type and Casp3KO MEF cells exposed to staurosporine, an apoptosis inducer not known to cause direct DNA damage like ionizing radiation. Our results show that staurosporine was able to induced γH2AX foci formation three days after exposure in wild type MEF cells. However, the induction was almost abolished in Casp3KO cells (Fig. S3D & E). These results provide further support for activated caspase 3 in inducing DNA damage.
Evidence for involvement of caspase 3 in radiation-induced persistent DNA strand breaks as determined by use of the comet assay
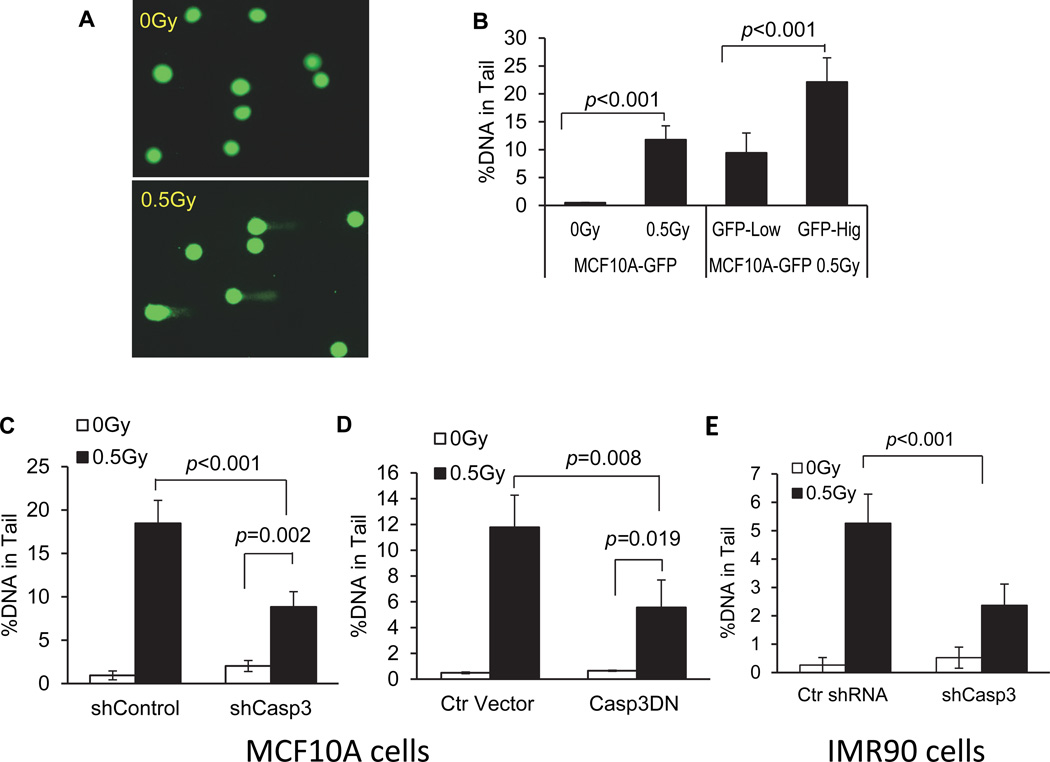
We next determined the roles of caspase 3 in radiation induced persistent DNA damage in MCF10A cells using the comet assay, another approach to measure DNA strand breaks (Olive et al., 1990; Ostling and Johanson, 1984; Singh et al., 1988). Our experiments show that in MCF10A cells exposed to 56Fe ions irradiation, the “comet tails’ of the cells whose sizes reflect the extent of DNA damage in host cells, were significantly bigger than those of the control cells at 14 days after irradiation (Fig. 3A & B), indicating significantly higher levels of persistent DNA strand breaks in the irradiated cells. Furthermore, those cells with higher levels of caspase 3 activities possessed significantly larger DNA tails when examined by use of the comet assay (Fig. 3B). In MCF10A cells transduced with an shRNA gene against caspase 3, comet assay analyses detected significantly reduced levels of radiation-induced DNA damage when compared with wild type cells at 10 days after irradiation (Fig. 3C). Similar results were also obtained with cells transduced with CASP3DN (Fig. 3D). Thus results obtained with comet assay are very consistent with results obtained with the γH2AX assay (Fig. 2).
Figure 3. A key role for caspase 3 in radiation-induced DNA damage as determined by the comet assay.
A). Typical examples of control and irradiated cells when run through electrophoresis during the comet assay.
B). The fraction of DNA in the comet tail in caspase 3 reporter-transduced MCF10A cells exposed 0.5 Gy of 56Fe ions 14 days after 56Fe ions irradiation. The left two bars shows fractions for non-irradiated vs 0.5 Gy irradiated cells while the right two bars show the fractions for irradiated MCF10A cells with high and low caspase 3 reporter activities based on GFP expression levels.
C). The effect of caspase 3 expression attenuation on the fraction of DNA in the comet tails. MCF10A cells transduced with an shRNA gene against the CASP3 gene (shCASP3) or control (shControl) were evaluated for the fraction of DNA in the comet tails at 10 days after 0.5 Gy or sham irradiation.
D). The effect of caspase 3 inhibition on radiation induced DNA strand breaks as quantified by the amount of cellular DNA in the comet tail in MCF10A cells. Cells transduced with a control lentiviral vector or those transduced with a dominant negative caspase 3 gene (CASP3DN) were evaluated for the amount of DNA in the comet tails with sham or 0.5 Gy of irradiation at 14 days after irradiation.
E). The effect of caspase 3 attenuation on the fraction of DNA in the comet tails in IMR90 cells. IMR90 cells transduced transduced with an shRNA gene against the CASP3 gene (shCASP3) or control (shControl) were evaluated for the fraction of DNA in the comet tails at 10 days after 0.5 Gy or sham irradiation.
Error bars in 3B, 3C, 3D and 3E represent standard deviation. All values are derived from the average of three different samples within one experimental group. For each group, at least 50 cells from randomly chosen fields were scored by use of the Image J software (NIH) for DNA distribution. Two sided Student’s t-test was used to calculate the p-values.
See also Fig. S3&S4.
In addition to immortalized MCF10A cells, we examined the roles of caspase 3 in 56Fe ions radiation induced comet tails in IMR90 cells, a primary human fibroblast cell strain. Our data show that caspase 3 attenuation significantly down-regulated radiation induced DNA damage in terms of comet assays in those cells as well (Fig. 3E). The role of caspase 3 in radiation induced DNA damage in IMR90 cells were further examined by use of the γH2AX foci assay (Fig. S3F) and chromosome aberration assay (Fig. S3G). Our results show that shCasp3-mediated caspase 3 attenuation reduced 56Fe ions induced DNA damage in both assays in IMR90 cells.
We also carried out experiments to determine if the p53 protein, the “guardian of the genome”, plays any role in radiation induced, persistent DNA damage as measured by γH2AX foci formation. We show that in MCF10A cells, radiation induced p53 and its downstream p21 protein expression at 24 hrs post exposure, similar to IMR90 cells (Fig. S4A). By 10 days post irradiation, both p53 and p21 expression mostly went back to control levels (Fig. S4A). In MCF10A cells transduced with p53DN, a dominant-negative version of p53 (Kendall et al., 2005), radiation-induced p21 induction is significantly reduced compared with parental MCF10 cells. However, when γH2AX foci formation was measured in these cells at different time points post-irradiation, we show that no statistical differences exist between these two cells in terms both the total number of foci and the kinetics of the foci induction (Fig. S4B), indicating the p53 did not play a significant role in this process.
Evidence implicating an important role for caspase 3 activation in radiation induced chromosome aberrations
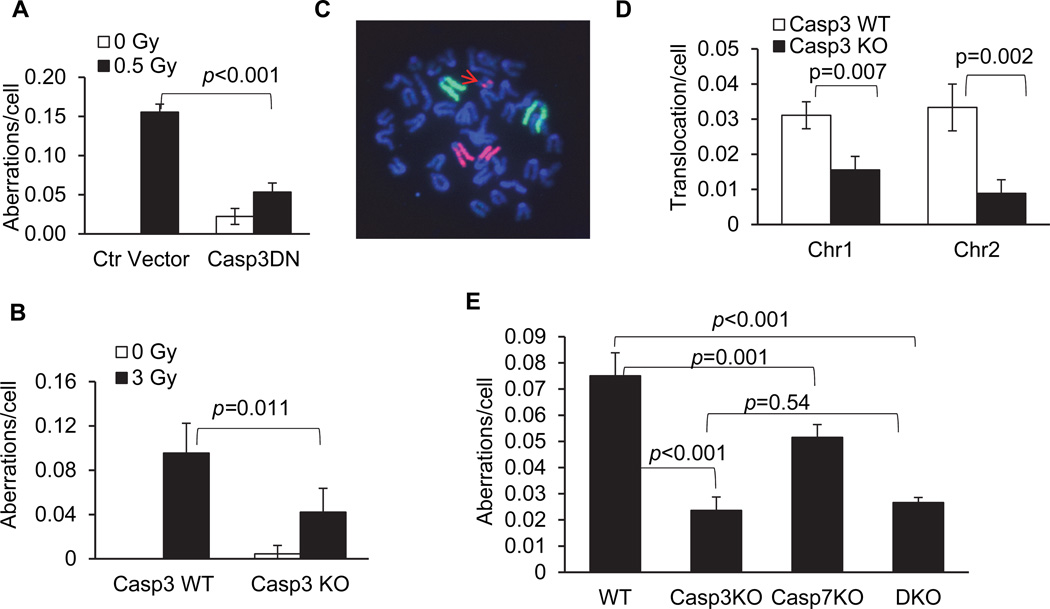
We conducted chromosome aberration analyses to further determine the roles of caspase3 in radiation-induced chromosome instability. Chromosome instability is key characteristic of cancer cells (Lengauer et al., 1997, 1998). We carried out chromosome aberration analysis in MCF10A cells. Our results show that inhibition of caspase 3 by use of CASP3DN significantly reduced radiation induced chromosomal aberrations in MCF10A cells (Fig. 4A, Fig. S4C–F, Table S2).
Figure 4. A key role for caspase 3 activities in radiation induced chromosome aberrations.
A). The effect of caspase 3 inhibition on radiation induced chromosome aberrations in MCF10A cells. Cells transduced with a control lentiviral vector or a dominant negative caspase 3 gene (casp3DN) were evaluated for chromosome aberrations with sham or 0.5 Gy of 600MeV 56Fe ions irradiation at 14 days after irradiation.
B). Radiation-induced chromosome aberrations in wild type or CASP3 gene knockout (CASP3KO) C57BL/6 mice. Results were obtained from bone marrow cells harvested from irradiated mice 3 days after 3 Gy x-ray irradiation.
C). Whole chromosome painting results for chromosomes 1 (FITC) & 2 (rhodamine). A cellular chromosome spread showing a translocation of part of chromosome 2 (arrow in red) in irradiated wild type C57BL/6 mice.
D). Quantitative summary of radiation-induced chromosome 1 &2 translocations in wild type or CASP3 gene knockout (CASP3KO) C57BL/6 mice. Results were obtained from bone marrow cells harvested from irradiated mice 3 days after. No translocations were identified in cells from either wild type or CASP3KO mice without irradiation.
E). Radiation induced chromosomal aberrations in WT, Casp3KO, Casp7KO, and Casp3KOCasp7KO (DKO) mouse embryonic fibroblast cells 5 days post irradiation.
Error bars in 4A, 4B, 4D, and 4E represent standard deviations. All values are derived from the average of three separate experiments. In each experiment, 150 cells for each cell type were counted without knowledge of the cell types to enumerate chromosome aberrations. Two-tailed Student’s t-test was used to calculate the p-values.
See also Figure S4 and Tables S2&S3.
We also assessed radiation-induced chromosome aberrations in vivo in wild type or caspase 3 deficient (Casp3KO) C57BL/6 mice (Kuida et al., 1996). Radiation induced a significant amount of chromosome aberrations in both wild type and Casp3KO bone marrow cells (Fig. 4B, also see Table S3). On the other hand, bone marrow cells in the caspase 3 knockout mice showed significantly less chromosome aberration after exposure to radiation when compared with wild type mice.
In additional experiments, we examined radiation-induced chromosome translocations in mouse bone marrow cells by use of whole chromosome painting to evaluate the frequencies of chromosomes 1&2 translocations. Bone marrow cells in the caspase 3 knockout mice showed significantly less chromosome 1&2 translocations after exposure to radiation when compared with wild type mice (Fig. 4C & D).
In further experiments, we examined the relative contribution of caspase 3 and caspase 7 in radiation induced chromosome aberrations by use of mouse embryonic fibroblasts from wild type, Casp3KO, and Casp7KO cells. Our results (Fig. 4E) show that both caspase 3 knockout and caspase 7 knockout cells exhibit significantly reduced, radiation-induced chromosome aberrations with caspase 3 playing a more prominent role. On the other hand, the caspase 3 and caspase 7 double knockout MEF cells (DKO), did not show additional reduction in chromosome aberrations, indicating that Casp3 plays a more dominant role in facilitating radiation induced chromosome aberrations.
Caspase 3 plays a facilitating role in oncogenic transformation of MCF10A cells induced by ionizing radiation
Our results so far clearly established a causative role for caspase 3 in radiation induced genetic instability at both the DNA and chromosome levels. We next explored the relationship between non-lethal caspase 3 activation and oncogenic transformation because genetic instability has been closely linked with carcinogenesis. We first use FACS to sort out cells with low or high caspase 3 activities. About 20% of the cells showed significantly increased caspase 3 reporter activities after 0.5 Gy 600 MeV 56Fe ions irradiation (data not shown). Those cells were then cultured for two weeks and then plated into soft agar. The ability to grow in an anchorage-independent manner in soft agar is a hallmark of transformed cells (Cifone and Fidler, 1980; Weinberg, 2007). Our data show that MCF10A cells exposed to 56Fe ions readily formed colonies in soft agar (Fig. 5A–C, also see Fig. S5A–C for actual photos of soft agar colonies). Interestingly, those with high Casp3EGFP activities formed soft agar colonies at significantly higher frequencies than those with low Caspa3EGFP caspase 3 activities (Fig. 5A). Those data suggest that higher caspase 3 activities correlated with significantly higher frequencies of oncogenic transformation despite minimal effect of caspase 3 activation on the proliferation of the cells in culture (Fig. S1B). We further examined if a causative relationship exist between caspase 3 activation and soft agar growth in irradiated MCF10A cells by use of small hairpin RNA against caspase 3 (shCasp3) and casp3DN to block caspase 3 activities. We observed significantly lower rates of soft agar colony formation in irradiated MCF10A-shCASP3 cells (Fig. 5B). In further experiments, we showed that cells with Casp3DN expression also had attenuated radiation induced soft agar colony formation significantly (Fig. 5C). Our results thus confirmed a significant facilitative role for capases 3 in radiation-induced oncogenic transformation of MCF10A cells. We further attempted to confirm the tumorigenic nature of the irradiated cells in nude mice. Non-irradiated parental MCF10A cells did not form any tumors in nude mice (0/10) 12 weeks post subcutaneous injection into nude nice. On the other hand, irradiated (0.5 Gy 56Fe ions) MCF10A cells readily formed tumors in nude mice (Fig. 5D–F). In fact, 6 out of 10 mice injected with irradiated MCF10A cells formed tumors (Fig. 5G). In contrast, neither control nor irradiated MCF10A-shCasp3 or MCF10A-CaspDN cells formed any tumors (0/10 for each of the two experimental groups, Fig. 5F). Similar results were also obtained for MCF10A-Casp3DN cells. None of the mice injected with either non-irradiated or irradiated cells formed any tumor in 12 weeks of observation (0/10 for each group, Fig. 5F). These data indicate that caspase 3 activities are required for tumorigenicity in 56Fe ions irradiated MCF10A cells.
Figure 5. A facilitative role for caspase 3 in radiation-induced oncogenic transformation of MCF10A cells.
A). Soft agar colony formation in MCF10A cells with high and low caspase 3 reporter activities. Irradiated (0.5 Gy of 600MeV 56Fe ions) cells were sorted for high and low caspase 3 reporter expression by use of an FACS sorter. They were then cultured for 14 days and seeded in soft agar plates.
B). The effects of small hairpin caspase 3 (shCASP3) gene expression on soft agar colony formation in irradiated MCF10A cells.
C). The effects of dominant-negative caspase 3 (CASP3DN) expression on soft agar colony formation ability of irradiated MCF10A cells.
Error bars in 5A, 5B, and 5C represent standard error of the mean (SEM). All values are derived from the average of three independent experiments. Student’s t-test was used to calculate the p-values in A–C.
D). A xenograft tumor in nude mice after 5 weeks inoculation of irradiated MCF10A cells (indicated by arrow in red).
E). H&E staining of tissues derived from a xenograft tumor formed from irradiated MCF10A cells in nude mice. The right panel shows an amplified image of part of the left panel.
F). Tumorigenic abilities of different MCF10A cells. Only the wild type MCF10A cells irradiated with 0.5 Gy can form tumors in nude mice. For each group of tumor cells, 10 mice were used.
G). Tumor growth kinetics from each of the 10 mice injected with the wild type MCF10A cells irradiated with 0.5 Gy of 56Fe ions. Six out of ten mice showed tumor growth.
See also Figure S5.
Significantly reduced skin carcinogenesis in Casp3 (−/−) mice after two-stage chemical carcinogenesis
We further conducted chemically induced carcinogenesis experiments in wild type and caspase 3 deficient (Casp3 (−/−)) C57BL/6 mice. We used a long established 7,12-Dimethylbenz(a)anthracene (DMBA) + 12-O-tetradecanoyphobol-13-acetate (TPA) two-stage carcinogenesis protocol following published procedures (Abel et al., 2009). In our induction regimen, an initial application of DMBA was followed by 20 weeks of TPA administration.
When the skins from C57/BL6 mice that were treated with DMBA+TPA were analyzed for caspase 3 activation, we observed a gradual increase in caspase 3 activation that peaked at around 3 weeks post treatment and attenuated afterwards (Fig. 6A & B). In wild type mice, DMBA+TPA induced numerous tumors, as expected. Tumor incidence was significantly earlier and higher in wild type mice than in Casp3 (−/−) mice (Fig. 6C & D). In addition, in Casp3-knockout (Casp3−/−) mice, the numbers of chemically induced tumors were significantly reduced (Fig. 6E). Furthermore, the aggregate tumor sizes in wild type mice were significantly larger than those in the Casp3(−/−) mice (Fig. 6F). Supplementary Fig. S5D shows typical H&E staining of two tumor sections from wild type and Casp3(−/−) mice, respectively.
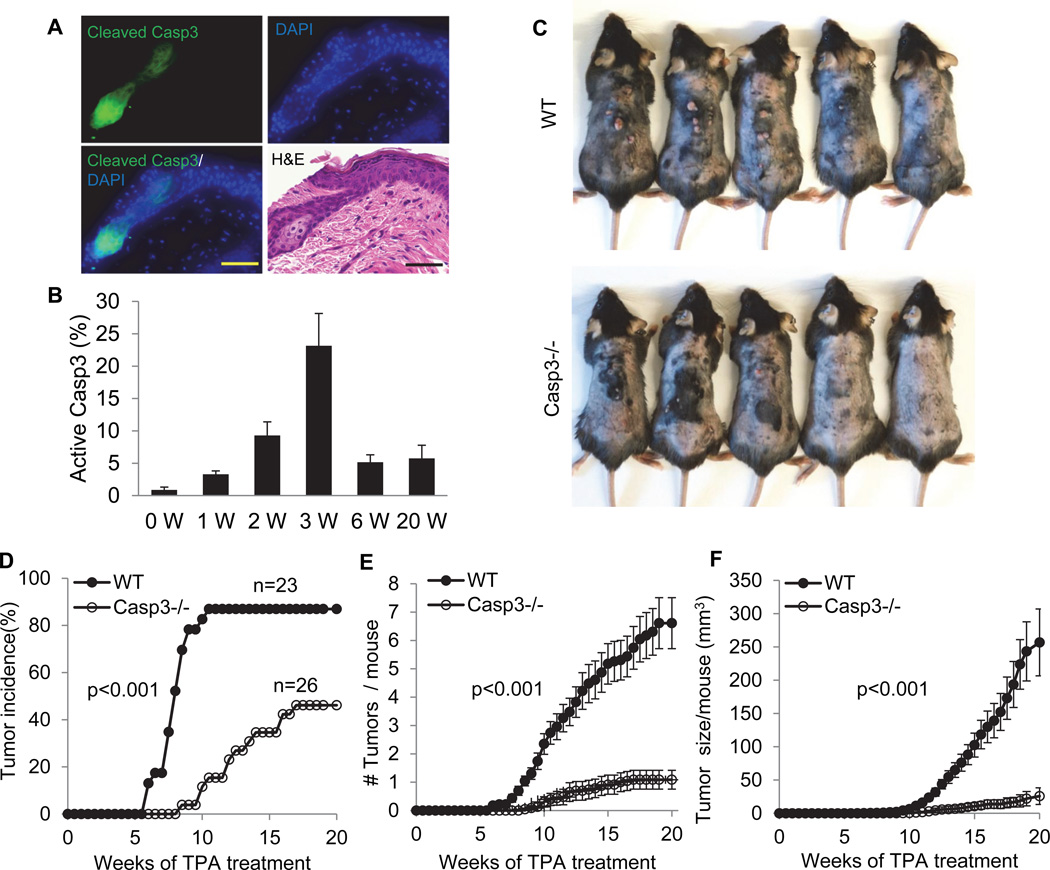
Figure 6. A facilitative role for caspase 3 in two-stage chemically induced skin carcinogenesis in wild type and Casp3 (−/−) C57BL/6 mice.
A). Activated caspase 3 in DMBA+TPA treated pre-malignant mouse skin. Scale bars represent 50 µm.
B). Quantitative measurement of caspase 3 activation in DMBA+TPA treated C57BL/6 mouse skin. Error bars show standard deviation (n=3).
C). Photographs representative of skin tumor formation in wild type and Casp3 (−/−) C57BL/6 mice at 20 weeks after the initiation of two-stage chemical treatment.
D). Tumor incidence in DMBA+TPA treated wild type (WT) (n=23) and Casp3 (−/−) (n=26) mice. p<0.001, logrank test.
E). Average number of tumors per mouse in DMBA+TPA treated wild type and Casp3 (−/−) mice at 20 weeks after initiation of chemical treatment. The difference between the two groups is statistically significant (p <0.001, two-sided ANOVA test).
F). Average tumor volume per mouse during the course of two-stage chemical induction. For each mouse, tumor burden represent the aggregate of all tumors in the mouse. The difference between the two groups is statistically significant (p <0.001, two-sided ANOVA test).
Error bars in 6E&F represent standard error of the mean (SEM).
See also Fig. S5&S6.
A careful analysis further reveals that there were clear sex differences in DMBA+TPA tumor induction in wild type (WT) and Casp3KO mice (supplementary Fig. S6A–C). Female mice appeared to be more susceptible to DMBA+TPA induced tumors. In both sexes, caspase 3 knockout caused profound reductions in tumorigenesis. However, female mice showed earlier and higher tumor incidence (Fig. S6A), higher numbers of tumors per mouse (Fig. S6B), and bigger tumor sizes (Fig. S6C) in wild type as well as Casp3KO mice.
We further compared the role of caspase 7 with that of caspase 3 in DMBA+TPA induced carcinogenesis. Our data suggest that caspase 7 knockout also led to significantly attenuated carcinogenesis (Fig. S6D–F). However, compared to caspase 3, knocking out caspase 7 had a more moderate effect in terms of attenuating DMBA+TPA induced carcinogenesis, a result consistent with results from radiation induced DNA damage experiments obtained with Casp7−/−mouse embryonic fibroblasts (supplementary Fig. S2B & Fig. 4E).
Endonuclease G as a major downstream effector of caspase 3-mediated DNA strand breaks
We next attempted to determine the factors downstream of caspase 3 that are responsible for generating DNA damage in cells with non-lethal activation of caspases 3. We focused our attention on endonuclease G (endoG), because it normally resides within the mitochondria and can migrate into the nucleus to fragment nuclear DNA in the event of caspase 3 activation(Li et al., 2001; Parrish et al., 2001).
We first examined endoG status through immunofluorescence staining. Our data indicated that in non-irradiated cells, most of cells exhibited faint staining in the cytoplasmic region with a small fraction of cells showed nuclear staining. Cytoplasmic endoG were localized mostly in the perinuclear regions that correlated with mitochrondria staining (Fig. 7A, top panels), consistent with published literature. In irradiated cells, there is a significant increase in the fraction of cells with nuclear endoG staining (Fig. 7A, lower panels), consistent with endoG movement from the mitochondria into the nucleus. Caspase 3 activity appears to be a major regulator of endonuclease G’s cytoplasmic to nuclear movement as attenuation of Casp3 through either Casp3DN or shCasp3 expression in irradiated MCF10A cells significantly reduced the fraction of cells with nuclear endoG staining (Fig. 7B). Additional data using MEF cells with Casp3 gene knockout further confirmed the role of caspase 3 in mediating radiation induced nuclear migration of endoG (supplementary Fig. S7A & B). Importantly, when DMBA+TPA induced skin tumors were analyzed, tumor tissues showed significantly more nuclear endoG staining than adjacent normal skin tissue. In comparison, nuclear migration was significantly reduced in tumors induced in Casp3−/− mice (supplementary Fig. S7C & D), consistent with the hypothesis that endoG nuclear migration was involved in DMBA+TPA induced carcinogenesis.
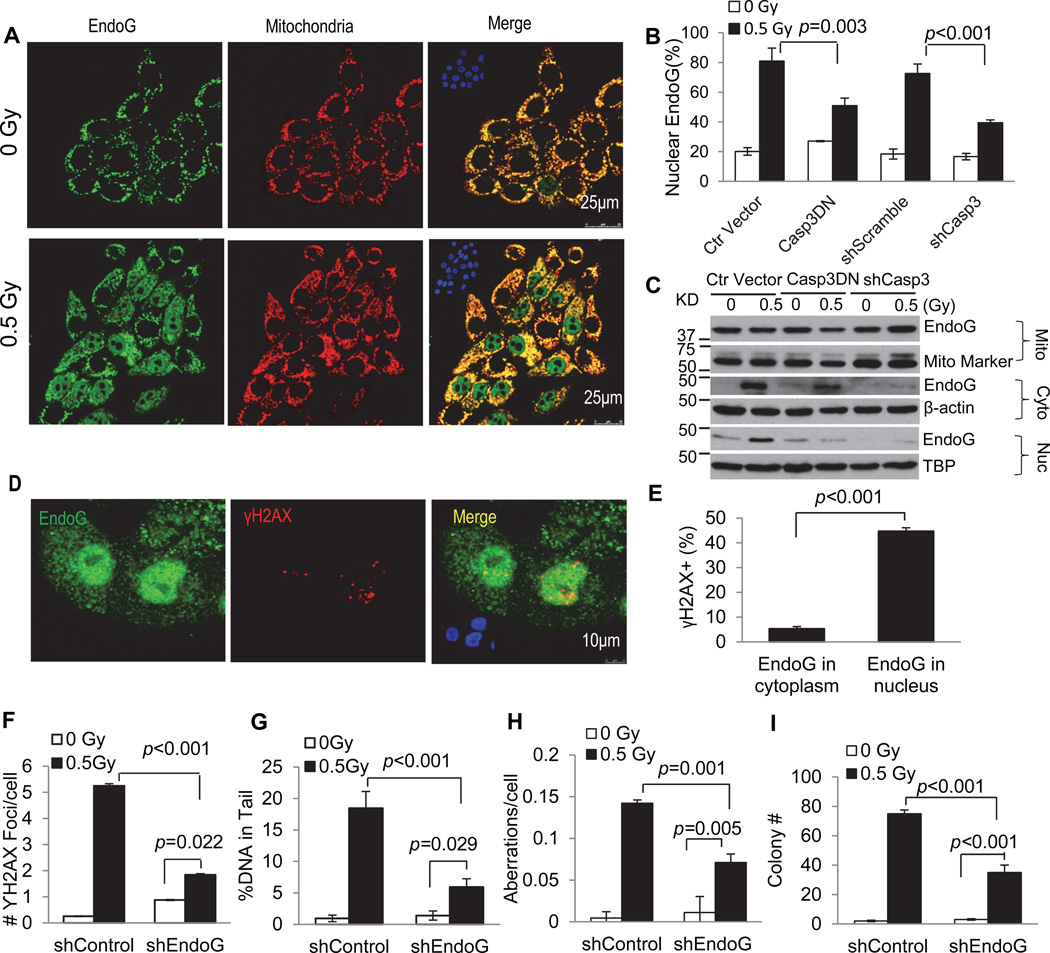
Figure 7. A significant role for endonuclease G as a downstream factor of caspase 3 in mediating radiation induced DNA damage and transformation.
A). Immunofluorescence co-staining of endonuclease G (green) and mitochondria (red) in control (top panels) and irradiated (lower panels) MCF10A cells. Insets: DAPI staining of the same slides.
B). Effect of caspase 3 inhibition (through CASP3DN or shCasp3 expression) on radiation-induced endoG migration from cytoplasmic to nuclear locations.
C). Western blot analysis of endoG in the mitochondria, cytoplasmic and nuclear fractions of MCF10A cells with or without Casp3DN or shCasp3 expression. Mito marker, TBP (TATA binding protein) and β-actin were used as mitochondria, nuclear and cytoplasmic loading controls, respectively.
D). Immunofluorescence co-staining of endoG (green) and γH2AX foci (red) in irradiated MCF10A cells. Insets: DAPI staining of the same slides.
E). Fraction of cells with γH2AX foci among cells with endoG staining in the cytoplasm only or those in both the cytoplasm and nucleus.
F). Effect of attenuating endoG expression on radiation induced γH2AX foci in MCF10A cells.
G). Effects of attenuating endoG expression on radiation induced comet tail DNA amount (an estimate of DNA strand breaks) in MCF10A cells.
H). Effects of attenuating endoG expression on the fraction of cells with radiation-induced chromosome aberrations in MCF10A cells.
I). Effects of attenuating endoG expression on the soft agar colony forming abilities of irradiated MCF10A cells.
In F–I, shEndoG represents shRNA2 from supplementary Fig.S7F.
Error bars in 7B, 7E–I represent standard error of the mean (SEM). The values in 7B, 7E, 7G and 7H are derived from the average of three samples within of each cell group. The values in 7F and 7I are derived from the average of three separate experiments. For γH2AX foci, at least 200 cells were counted for each measurement. For comet assay, a minimum of randomly chosen 50 cells was measured automatically by Image J software (NIH) for DNA distribution. For chromosome aberration analysis, a minimum of 150 cells was counted without knowledge of cell identities.
Student’s t-test was used to calculate the p-values.
See also Figure S7.
We also carried western blot analysis of endoG location in MCF10A cell before and after irradiation (Fig. 7C). Our results indicate that radiation induced a clear migration of endoG to the cytoplasm as well as the nucleus. However, down-regulation of caspase 3 activity through Casp3DN or shCasp3 expression significantly reduced the migration.
In further analysis, we show that irradiated cells with nuclear endoG staining contained significantly higher proportion of cells with γH2AX foci (~45%) when compared with those without nuclear endoG staining (~5%, Fig. 7D, E) at 10 days after irradiation, consistent with an important role for endoG in inducing persistent DNA strand breaks.
In additional experiments, we examined whether endoG was required for radiation induced persistent genetic instability by use of MCF10A cells stably transduced with endoG targeted shRNAs (Fig. S7E). Our results indicated that attenuation of endoG expression significantly lowered the fraction of cells with persistent γH2AX foci among irradiated MCF10A cells (Fig. 7F), thereby establishing endoG as the key downstream effector of caspase 3 in causing DNA damage. Comet assay showed reduced DNA strand breaks in irradiated MCF10A cells with endoG knockdown (Fig. 7G). These results were further supported by chromosome aberration analyses of MCF10A cells with shRNA knockdown (Fig. 7H, supplementary Table S2). Finally, we show that endoG knockdown also reduced the number of radiation induced soft agar colonies from the MCF10A cells (Fig. 7I). The results of down-regulating endoG were demonstrated with 3 independent shRNAs against endoG (supplementary Fig. S7E–G), suggesting that they were not likely from off-target effects.
Discussion
Apoptosis and factors involved in the apoptotic machinery are generally considered tumor suppressive (Hanahan and Weinberg, 2011). However, the absence of mutations in the apoptosis inducing factors (Casp3, Casp9, or APAF) in patient-derived tumor samples suggests that these factors are not major obstacles for carcinogenesis in mammalian cells. The most striking aspect of the present study is that our results demonstrate that caspase 3 can play an active role in promoting genetic instability and carcinogenesis.
How do we reconcile the fact that most previous studies support the notion of apoptosis-inducing factors being tumor suppressive with our finding that caspase 3 activation promotes carcinogenesis? To really understand these conflicting results, we need to re-examine the fundamental premise of the apoptosis- tumor suppressive hypothesis. At first glance it is a very straightforward scenario: cells are exposed to stress and activate the apoptosis program, they die, and get scavenged. The whole process should be anti-oncogenic because there is no inflammation and no-leftover DNA damage. While this scenario is certainly true for cells that do die, an implied but critical assumption is that all cells that initiate the apoptotic process will die. While it is a reasonable assumption, it has not been carefully examined except in C. elegans. It was shown that some C. elegans cells with caspase activation can actually survive if they are not engulfed by surrounding cells (Hoeppner et al., 2001; Reddien et al., 2001). In addition, previous studies have unveiled non-apoptotic roles of caspase 3 and its downstream caspase-dependent DNase (CAD) in muscle differentiation (Fernando et al., 2002; Larsen et al., 2010). In the present study, we show clear evidence that many irradiated mammalian cells with caspase 3 activation can survive. It is in these surviving cells with caspase 3 activation that one sees significantly elevated levels of genetic instability and oncogenic transformation.
Our results are consistent with a recent report that indicates treatment of glioma or MEF cells with TRAIL or FasL, an apoptosis-inducing agent, causes increased DNA damage and mutagenesis that is caspase 8-dependent(Lovric and Hawkins, 2010). It is also consistent with another study which shows that prolonged mitotic arrest triggers partial caspase activation that causes increased DNA damage(Orth et al., 2012).
In conclusion, in this study we find that many cells can survive caspase 3 activation after exposure to ionizing radiation. We further reveal a surprising and unconventional function for caspase 3 in mammalian cells exposed to radiation: causing and sustaining DNA damage, and facilitating oncogenic transformation.
Experimental Procedures
Cell lines and tissue culture
Early passage, immortalized, non-transformed human breast epithelial cell line, MCF10A, was a kind gift from Dr. Hatsumi Nagasawa of Colorado State University (Fort Collins, CO). MCF10A growth medium was composed of DMEM/F12 (Sigma, St. Louis, MO) supplemented with 5% donor horse serum (Sigma), 20 ng/ml epidermal growth factor (EGF; R&D, Minneapolis, MN), 0.5 µg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 µg/ml insulin (Invitrogen, Grand Island, NY), and 100 unites/ml penicillin and 100 µg/ml streptomycin. For γH2AX foci assays, the cells were cultured in growth medium with only 2% horse serum and without EGF.
Exposure to higher energy 56Fe ions
To conduct irradiation with 56Fe ions, cells to be irradiated or sham-irradiated were shipped by FedEx to the National Aeronautics and Space Administration (NASA)-sponsored Space Radiation Laboratory at Brookhaven National Laboratory (BNL; Brookhaven, Long Island, NY) in sealed T-25 flasks. The iron beam energy used was 600 MeV/μ. The dose rate for exposure was 0.5 Gy/min. After irradiation, the cells were immediately shipped back to our laboratory in Durham, NC for further analysis.
γ2AX foci assay
To detect the radiation-induced DNA double strand breaks, γH2AX foci in the irradiated cells was examined through immunofluorescence by use of an established protocol (Paull et al., 2000; Rogakou et al., 1998).
Alkaline comet assay
We also used alkaline comet assay (Olive et al., 1990; Ostling and Johanson, 1984; Singh et al., 1988) to detect DNA double strand breaks. To do this we used a commercial kit following the manufacturer (Trevigen, Gaithersburg, MD)’s instructions.
Chromosome aberration analysis
We carried out chromosome aberration analysis in cultured cells and in bone marrow cells irradiated in mice. We also analyzed chromosome translocations analysis by use of FISH analysis in bone marrow cells from sham and irradiated mice.
Two-stage carcinogenesis
An established protocol (Abel et al., 2009) was used to induce skin carcinogenesis in mice by use of combined DMBA+TPA administration onto shaved mouse skin. One initial DMBA treatment and periodic (2× weekly) TPA treatments were carried. At the end of 20 weeks, the number of tumor per mice and tumor sizes were enumerated and quantified, respectively.
Statistical analysis
Specific statistical methods are mostly described in the figure legends. Where it is not stated, two-tailed Student’s t-test was used to compare differences between two groups. In other instances, ANOVA or log rank tests were also used and described in the figure legends.
Supplementary Material
Acknowledgements
This study was supported in part by grants CA131408, CA136748, CA155270, ES024015 from the National Institutes of Health (to C-Y Li), and grant NNX12AB88G (to C-Y Li) from NASA Space Radiation Biology Research Program; the Duke Skin Disease Research Core Center grant (AR066527) from the NIH to R. P. Hall; and grants 30428015, 30325043 from the National Science Foundation of China, grant 2004CB518804 from Ministry of Science of China “973 project” to Q. Huang;
We thank Dr. Sally Kornbluth (Duke University) for reading our manuscript and providing insightful suggestions. We also thank Dr. Richard A. Flavell (Yale University) for depositing the CASP3−/− mouse to Jackson Laboratory for research use; and Drs. Guy Salveseen (Sanford Burnham Institute), David M Spencer (Baylor college of medicine), Dr. Chris Counter (Duke University ), and Titia de Lange (Rockefeller University) for sharing their CASPDN, iCasp, p53DN, and 53BP1-mCherry plasmids, respectively, through Addgene.
We thank Drs. Adam Rusek and Peter Guida for their help in carrying out 56Fe ions irradiation of our cells at the Brookhaven National Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Please also see Supplemental Experimental Procedures for additional details on various experimental methods used in this study.
Author contributions:
X.L., Y. H., and C.L. conceived of the study and designed the experiments. X.L. carried out most of the experiments. Y.H. carried out some of the initial studies and key initial carcinogenesis experiment. F.L. and Q.H. carried out some of the soft agar studies. T.K. carried out some of the chromosome aberrations analysis. R.P.L. provided intellectual input in designing some of the experiments and helped in writing the manuscript. X.L. and C.L. wrote the manuscript.
References
- Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4:1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Park SM, Tumanov AV, Hau A, Sawada K, Feig C, Turner JR, Fu YX, Romero IL, Lengyel E, et al. CD95 promotes tumour growth. Nature. 2010;465:492–496. doi: 10.1038/nature09075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifone MA, Fidler IJ. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci U S A. 1980;77:1039–1043. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Debbas M, White E. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer. 2008;8:465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Fanidi A, Harrington EA, Evan GI. Cooperative interaction between c-myc and bcl-2 proto-oncogenes. Nature. 1992;359:554–556. doi: 10.1038/359554a0. [DOI] [PubMed] [Google Scholar]
- Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA. Caspase 3 activity is required for skeletal muscle differentiation. Proc Natl Acad Sci U S A. 2002;99:11025–11030. doi: 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Tokano H, Fujioka KS, Okano H, Edge AS. Generating mouse models of degenerative diseases using Cre/lox-mediated in vivo mosaic cell ablation. J Clin Invest. 2011;121:2462–2469. doi: 10.1172/JCI45081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 2001;412:202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- Horvitz HR. Worms, life, and death (Nobel lecture) Chembiochem. 2003;4:697–711. doi: 10.1002/cbic.200300614. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Shaham S, Hengartner MO. The genetics of programmed cell death in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1994;59:377–385. doi: 10.1101/sqb.1994.059.01.042. [DOI] [PubMed] [Google Scholar]
- Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, O'Sullivan B, He Z, Peng Y, Tan AC, et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall SD, Linardic CM, Adam SJ, Counter CM. A network of genetic events sufficient to convert normal human cells to a tumorigenic state. Cancer Res. 2005;65:9824–9828. doi: 10.1158/0008-5472.CAN-05-1543. [DOI] [PubMed] [Google Scholar]
- Kim M, Katayose Y, Rojanala L, Shah S, Sgagias M, Jang L, Jung YJ, Lee SH, Hwang SG, Cowan KH. Induction of apoptosis in p16INK4A mutant cell lines by adenovirus-mediated overexpression of p16INK4A protein. Cell Death Differ. 2000;7:706–711. doi: 10.1038/sj.cdd.4400703. [DOI] [PubMed] [Google Scholar]
- Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- Lakhani SA, Masud A, Kuida K, Porter GA, Jr, Booth CJ, Mehal WZ, Inayat I, Flavell RA. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci U S A. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li F, He Z, Shen J, Huang Q, Li W, Liu X, He Y, Wolf F, Li CY. Apoptotic caspases regulate induction of iPSCs from human fibroblasts. Cell Stem Cell. 2010;7:508–520. doi: 10.1016/j.stem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Lin HC, Lai PY, Lin YP, Huang JY, Yang BC. Fas ligand enhances malignant behavior of tumor cells through interaction with Met, hepatocyte growth factor receptor, in lipid rafts. J Biol Chem. 2012;287:20664–20673. doi: 10.1074/jbc.M111.326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovric MM, Hawkins CJ. TRAIL treatment provokes mutations in surviving cells. Oncogene. 2010;29:5048–5060. doi: 10.1038/onc.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Bodis S, McClatchey A, Remington L, Ruley HE, Fisher DE, Housman DE, Jacks T. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- MacCorkle RA, Freeman KW, Spencer DM. Synthetic activation of caspases: artificial death switches. Proc Natl Acad Sci U S A. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banath JP, Durand RE. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the "comet" assay. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell. 2012;23:567–576. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- Panner A, James CD, Berger MS, Pieper RO. mTOR controls FLIPS translation and TRAIL sensitivity in glioblastoma multiforme cells. Mol Cell Biol. 2005;25:8809–8823. doi: 10.1128/MCB.25.20.8809-8823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci U S A. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- Reed JC. Dysregulation of apoptosis in cancer. J Clin Oncol. 1999;17:2941–2953. doi: 10.1200/JCO.1999.17.9.2941. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sabbatini P, McCormick F. Phosphoinositide 3-OH kinase (PI3K) and PKB/Akt delay the onset of p53-mediated, transcriptionally dependent apoptosis. J Biol Chem. 1999;274:24263–24269. doi: 10.1074/jbc.274.34.24263. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Stennicke HR, Salvesen GS. Biochemical characteristics of caspases-3, −6, −7, and −8. J Biol Chem. 1997;272:25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Walsh JG, Cullen SP, Sheridan C, Luthi AU, Gerner C, Martin SJ. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci U S A. 2008;105:12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. BID: a novel BH3 domain-only death agonist. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- Weinberg R. The Biology of Cancer. New York: Garland Science; 2007. [Google Scholar]
- Weng L, Brown J, Eng C. PTEN induces apoptosis and cell cycle arrest through phosphoinositol-3-kinase/Akt-dependent and -independent pathways. Hum Mol Genet. 2001;10:237–242. doi: 10.1093/hmg/10.3.237. [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.