Abstract
Environmental RNAi (eRNAi) is a sequence-specific regulation of endogenous gene expression in a receptive organism by exogenous double-stranded RNA (dsRNA). Although demonstrated under artificial dietary conditions and via transgenic plant presentations in several herbivorous insects, the magnitude and consequence of exogenous dsRNA uptake and the role of eRNAi remains unknown under natural insect living conditions. Our analysis of coleopteran insects sensitive to eRNAi fed on wild-type plants revealed uptake of plant endogenous long dsRNAs, but not small RNAs. Subsequently, the dsRNAs were processed into 21 nt siRNAs by insects and accumulated in high quantities in insect cells. No accumulation of host plant-derived siRNAs was observed in lepidopteran larvae that are recalcitrant to eRNAi. Stability of ingested dsRNA in coleopteran larval gut followed by uptake and transport from the gut to distal tissues appeared to be enabling factors for eRNAi. Although a relatively large number of distinct coleopteran insect-processed plant-derived siRNAs had sequence complementarity to insect transcripts, the vast majority of the siRNAs were present in relatively low abundance, and RNA-seq analysis did not detect a significant effect of plant-derived siRNAs on insect transcriptome. In summary, we observed a broad genome-wide uptake of plant endogenous dsRNA and subsequent processing of ingested dsRNA into 21 nt siRNAs in eRNAi-sensitive insects under natural feeding conditions. In addition to dsRNA stability in gut lumen and uptake, dosage of siRNAs targeting a given insect transcript is likely an important factor in order to achieve measurable eRNAi-based regulation in eRNAi-competent insects that lack an apparent silencing amplification mechanism.
Keywords: RNAi, eRNAi, dsRNA uptake, corn rootworm, siRNAs
INTRODUCTION
One of the first observations of sequence-specific knockdown of gene expression via dsRNA ingestion in animals came from experiments where Caenorhabditis elegans was presented dsRNA in solution or through dsRNA-expressing bacteria (Timmons et al. 2001) and was subsequently termed environmental RNA interference or eRNAi (Whangbo and Hunter 2008). In this process, dsRNA enters from the intestinal lumen and is distributed systemically to distal cells in the organism where it down-regulates gene expression in a sequence-specific manner. While the details of intracellular RNAi pathways have become increasingly well understood, elucidation of the mechanism, prevalence, and ecological role of RNA uptake from the environment lags behind. Currently, most knowledge in this field has been generated in nematode model systems. Although insightful, even in nematodes of the same genus, the capacity for eRNAi is quite variable indicating a potential complex interplay between function and selective pressures in the environment (Nuez and Félix 2012). Other species that have demonstrated eRNAi (e.g., insects) do not show clear mechanistic orthologs to key components such as dsRNA transporter SID-2 (Winston et al. 2007; Huvenne and Smagghe 2010), or do have some homologous genes known to be essential to the process in C. elegans (e.g., SID-1), yet with differential contribution in the examined insects’ ability to uptake and distribute dsRNA (Miyata et al. 2014).
With the observed variation in eRNAi, there is an outstanding question as to what natural function this process plays in insects and other invertebrates. RNAi has a role in viral immunity and its uptake and cell-to-cell movement likely play into viral responses at an organismal level (Saleh et al. 2009). At the same time, there is evidence that eRNAi might function in processes other than antiviral defense in C. elegans. It has been demonstrated that Escherichia coli noncoding RNAs OxyS and DsrA can regulate gene expression and physiological condition of C. elegans, and this points to the potential for a broader ecological role eRNAi may have (Liu et al. 2012). In addition to viral sources, plant tissues possess several mechanisms to produce long dsRNA and represent a rich source of natural dsRNAs and as such, could provide regulatory input to responsive species that consume them. Numerous endogenous long dsRNAs have been reported in the model plant, Arabidopsis (Zheng et al. 2010). Likewise, genome-wide analysis of the transcriptome from several major plant crops, including corn (Zea mays), found that a broad portion of the crop genomes produce long dsRNAs (Jensen et al. 2013). Thus, expression of a species-centric spectrum of dsRNAs could represent a common feature in higher plants. Are there defensive capacities in this background of plant dsRNA expression? Alternatively, do the herbivorous species use cues in the dsRNAs to productively modify their own gene expression?
Several insect species have been reported to be susceptible to eRNAi under laboratory conditions, but the degree of susceptibility varies dramatically and often very large doses of synthesized dsRNA must be ingested to induce any measurable effect on target gene expression (Baum and Roberts 2014). Large doses of singular dsRNAs are not likely to be available under natural feeding conditions, and consequently, dsRNA uptake under laboratory conditions is not reflective of uptake and eRNAi effect under realistic levels of natural exposure. In contrast, some insect species, including important agricultural insect pests such as Diabrotica virgifera virgifera LeConte, western corn rootworm (WCR, one of the Diabrotica spp. that feeds on corn roots as larvae), are highly responsive to dsRNA through feeding and at doses that are biologically relevant in plant presentation scenarios. The RNAi response in WCR has been noted in artificial diet and transgenic corn plants expressing WCR-targeted dsRNA (Baum et al. 2007). Recent studies have increased the understanding of details of mode of action of eRNAi in Diabrotica ssp. (Bolognesi et al. 2012). It has been shown that dsRNA longer than 50–60 base pairs (bp) is required to trigger eRNAi in larvae via a bioassay feeding experiment (Bolognesi et al. 2012). Consistent with this observation, endogenous plant miRNAs showed no significant accumulation in WCR larvae fed on corn roots (Zhang et al. 2012), further supporting the observation that exogenous small RNAs are not taken up efficiently or are less stable via oral delivery. Interestingly, in C. elegans, SID-2-dependent dsRNA transport is required for eRNAi and exhibits size dependency by facilitating the uptake of dsRNA longer than 50 bp (McEwan et al. 2012). In spite of the obvious similarity in length requirement for the uptake of dsRNA in WCR and C. elegans, the SID-2 ortholog has not been identified in WCR (Miyata et al. 2014).
We report that WCR larvae took up numerous, endogenous corn long dsRNAs during feeding on host plant tissues. This result was confirmed in another coleopteran species susceptible to eRNAi, Colorado potato beetle (CPB, Leptinotarsa decemlineata Say), when its larvae fed on tomato plants. In these insects, exogenous dsRNA was stable in the gut lumen and plant endogenous long dsRNAs were taken up by the insect and processed into 21 nucleotide (nt) siRNAs, similar to transgenically expressed long dsRNA. In contrast, exogenous dsRNA was less stable in lepidopteran insects recalcitrant to eRNAi, such as fall armyworm (FAW, Spodoptera frugiperda J.E. Smith) and corn earworm (CEW, Helicoverpa zea Boddie). Concomitantly, we have not detected comparable accumulation of host plant-derived RNAs in these insects, revealing a significant difference in eRNAi susceptibility in herbivorous insects with similar feeding behavior. Finally, using genome-wide transcriptome analysis of WCR larvae fed on a diet supplemented with total RNA from different plant species, we evaluated the potential for an effect of eRNAi on the WCR transcriptome.
RESULTS
Exogenous long dsRNA uptake and movement in insects
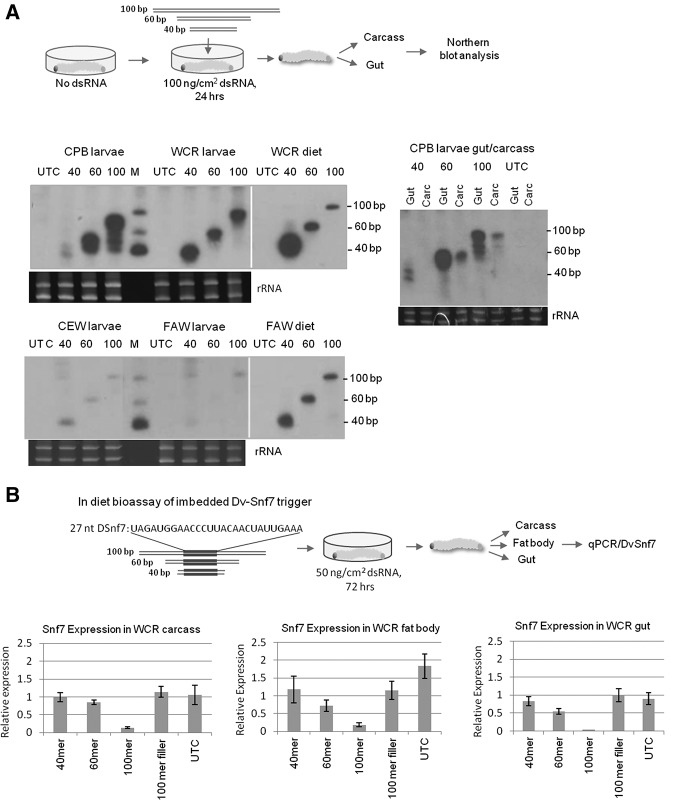
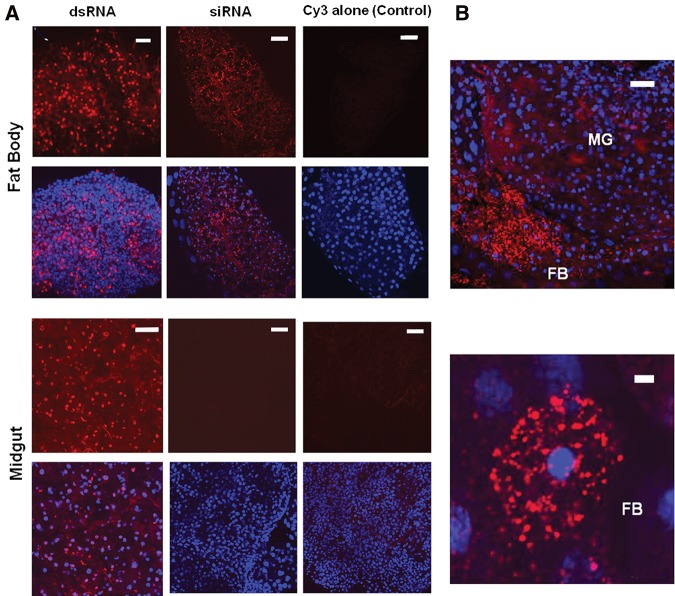
We have previously reported that dsRNA longer than 50–60 bp in size is required to induce eRNAi in southern corn rootworm (SCR, Diabrotica undecimpunctata howardi Barber) by feeding larvae with a size series of long dsRNAs targeting an essential gene and evaluating the effect on larvae mortality (Bolognesi et al. 2012). To understand the details of long dsRNA uptake and spread in herbivorous insects, we conducted molecular analysis of uptake and processing of exogenous dsRNA in two coleopteran insects sensitive to eRNAi, WCR and CPB and two lepidopteran insects, FAW and CEW which are recalcitrant to eRNAi. Recalcitrance was defined by no significant stunting or mortality in a diet bioassay when lepidopteran neonates were fed on diets with dsRNAs at high dose (e.g., feeding the respective species-specific dsRNA for V-ATPase A subunit at >1.82 μg/cm2 in diet overlay, which is >1000-fold higher than the LC50 of the orthologous dsRNA in WCR) (Baum et al. 2007). Larvae were fed for 2 d on an artificial diet supplemented with an absorbed overlay of 104 ng/cm2 of 40, 60, and 100 bp long dsRNA, respectively, of a “neutral” sequence with no homology with any insect genes used in these studies, followed by Northern blot analysis (Fig. 1A). Analysis of the applied dsRNA size series triggers revealed that dsRNAs were stable in CPB and WCR larvae when ingested with diet (Fig. 1A). In contrast, dsRNAs were much less stable in the gut of the two tested lepidopteran insects consistent with the general lack of robust eRNAi response in these species (Terenius et al. 2011). Dissection of gut and carcass tissues of CPB fed on long dsRNA revealed that dsRNAs of the size series 60 bp or longer were detected in tissues other than gut (Fig. 1A). Detection of long dsRNA in tissues other than primary recipient gut cells suggests that long dsRNA is capable of moving from the recipient gut cells to distal carcass tissues. This observation is interesting because WCR and CPB do not posses any known RNA-dependent RNA polymerase activity and apparently lack secondary siRNA amplification mechanisms, suggesting a role for the primary trigger in systemic silencing in these insects, although comovement of derived siRNAs cannot be ruled out. qPCR analysis of WCR larvae fed on the 40, 60 and 100 bp size series of long dsRNA with an embedded common 27 nt fragment of WCR Snf7 gene (DvSnf7) (as in Bolognesi et al. 2012) confirmed down-regulation of the target gene with dsRNAs longer than 60 bp in gut, fat body and carcass tissues (Fig. 1B). This indicated uptake and systemic spread of a silencing signal, but not with shorter sequences, thus corroborating the requirement of longer dsRNAs for eRNAi in susceptible insects. To further evaluate the ability of long dsRNA and siRNA to be taken up by primary recipient midgut cells and other cells in eRNAi-sensitive insects, we used Cy3-labeled 240 bp long dsRNA and 21 bp siRNA in a WCR tissue culture assay. The labeled RNAs were incubated with WCR midgut tissue or fat body cultures followed by microscopic visualization. As expected, long dsRNA, but not siRNA, uptake was observed in midgut cells (Fig. 2A,B). In contrast, both long dsRNAs and siRNAs were taken up by fat body cells. This result suggests that while the primary uptake of exogenous RNA from the gut lumen is limited to long dsRNA, internal cells may have a less stringent threshold for cellular import. We also observed uptake of long dsDNA used as a control by fat body but not by gut tissues and competition between dsDNA and dsRNA uptake in fat body (Supplemental Fig. 1A,B) suggesting a difference in mechanism of nucleic acid uptake in fat body and gut tissues at least under in vitro conditions tested in this study. Further studies would be needed to validate this result in whole larvae.
FIGURE 1.
Exogenous dsRNA stability and silencing signal spread in herbivorous insects. (A) Outline of diet feeding experiment. WCR, CPB, CEW, or FAW larvae were fed for 24 h on diet supplemented with of 40, 60, or 100 bp dsRNA of “neutral” sequence at 100 ng/cm2, followed by Northern blot analysis of whole larvae or dissected gut and carcass tissues. (UTC) Untreated (non-dsRNA) control; (M) molecular weight markers. (B) qPCR analysis of WCR Snf7 gene (DvSnf7) expression in WCR larvae fed on the 40, 60, and 100 bp size series of long dsRNA with an embedded 27 nt fragment of DvSnf7. Three larvae were pooled for each of the three replicates. Relative expression and standard deviation are shown.
FIGURE 2.
Exogenous RNA uptake in WCR. (A) WCR midgut and fat body tissue cultures incubated with labeled long dsRNA and siRNA for 15 h. Both Cy3-labeled long dsRNA (240 bp) and siRNA (21 bp) were taken up by fat body cells, but only Cy3-labeled long dsRNA and not siRNA were internalized by midgut tissue (scale bar, 50 μM). (B) WCR midgut (MG) and fat body (FB) co-cultures Cy3-labeled siRNA were taken up by only fat body tissue but not by midgut tissue (upper panel) (scale bar, 50 μM). Higher magnification of Cy3-labeled siRNA internalization in fat body tissue (lower panel) (scale bar, 5 μM). In both panels, DAPI (blue signal) reveals nuclear DNA staining.
Endogenous dsRNAs in plants
Most data for eRNAi in WCR and other insects have been generated by feeding synthesized or transgenically expressed dsRNA to insects, but there is a clear need for connecting laboratory data with an understanding of the eRNAi process in the more complex biological context that exists in nature. Analysis of plant transcriptomes suggests that endogenously produced dsRNAs are abundant in plants (Zheng et al. 2010), a primary food source for herbivorous insect species. Long dsRNA molecules in plants are typically produced by several RNA-dependent RNA polymerases (Baulcombe 2004; Carthew and Sontheimer 2009), but can also be formed by intermolecular base-pairing from highly repetitive regions of plant genomes or due to convergent transcription (Borsani et al. 2005). Because dsRNA precursor-independent production of siRNAs has not been described in plants, it is assumed that most siRNAs in plants are derived from long dsRNA precursors through the activity of several Dicer-like, dsRNA RNAase III-type nucleases (Brodersen and Voinnet 2006). Based on this assumption, we exploited genome-wide small RNA sequencing to predict dsRNA-producing loci in corn and tomato (Jensen et al. 2013). While it is considered that many long dsRNAs are relatively unstable and short lived in plant cells, we hypothesized that the actual steady-state abundance of dsRNA in a particular cell depends on the rate of dsRNA synthesis and processing, creating a dynamic pool of long dsRNAs. Mapping and analysis of strand-specific RNA-seq reads and small RNA (sRNA) reads to individual loci in the corn genome identified many instances with overlap between RNA-seq and sRNA reads that mapped to both strands of DNA (Supplemental Fig. 2A). This was confirmed on a genome-wide level (Supplemental Fig. 2B), thus validating the accumulation of endogenous dsRNA transcripts. Northern blot analysis combined with RNase treatment of total RNA samples from transgenic corn expressing long dsRNAs transcribed from an inverted repeat cassette with sequence derived from the WCR DvSnf7 gene also revealed accumulation of long dsRNA, indicating that only a portion of the dsRNA was processed into siRNAs in corn tissues (Supplemental Fig. 2C). Accumulation of long WCR DvSnf7 dsRNA in corn is in agreement with down-regulation of DvSnf7 gene expression in WCR larvae fed on this transgenic corn. These data indicate that long dsRNA molecules can accumulate in corn roots, a natural food source for WCR larvae.
Uptake of ingested plant long dsRNA by insects
Considering that dietary long dsRNA can induce gene silencing in WCR when ingested with either artificial diet or transgenic plant tissues expressing long dsRNA (Fig. 1B; Baum et al. 2007), we speculated that a steady-state pool of endogenous corn long dsRNAs meeting the minimal size threshold should also be taken up by WCR larvae. To test this hypothesis, we allowed WCR and CPB neonates to feed on host plants, corn roots or tomato leaves, for 12 and 11 d, respectively, to evaluate uptake of plant dsRNA under natural feeding conditions. Similarly, neonates of a lepidopteran insect recalcitrant to eRNAi, FAW, were fed on corn leaves for 11 d. Larvae were collected from host plants, and midguts were removed to eliminate unprocessed plant material. The remaining carcass and fat body tissues were used for sRNA sequencing. We also conducted sRNA sequencing of all food sources (corn roots and leaves, and tomato leaves) to be used as reference controls.sRNAs from plant food sources in the study were dominated by 24 nt sRNAs while the major classes of sRNAs detected in WCR, CPB, and FAW were 21–23 nt (Supplemental Fig. 3). To identify plant-derived siRNAs accumulated in insects, we mapped sRNA reads generated in this set of experiments to the host plant genomes, corn and tomato, as well as to insect genome and transcriptome sequence data. Sequence analysis revealed a significant accumulation of host plant-derived 21 nt siRNAs in WCR and CPB, while we did not observe comparable accumulation of 21 nt plant-derived siRNAs in FAW (Fig. 3A). We also did not detect corn-derived siRNAs in freshly hatched, unfed WCR neonates used as control (Fig. 3A). Overall, corn and tomato-derived 21 nt long siRNAs represented up to 1.71% ± 0.46 and 0.6% ± 0.31 of all sRNAs detected in WCR and CPB, respectively (Fig. 3A). Considering that exogenous dsRNAs are processed by insects primarily into 21 bp siRNAs, this result suggests that this is a major class of siRNAs involved in eRNAi in WCR and CPB, and that they are likely processed by the same RNAi pathway responsible for the generation of insect endogenous 21 nt siRNAs. Interestingly, plant-derived 21 nt siRNAs can represent up to 12% and 3.9% of all 21 nt siRNAs identified in carcass tissues of WCR and CPB larvae, fed on corn roots or tomato leaves, respectively (Fig. 3B). In contrast, only 0.78% of all 21 nt siRNAs identified in FAW were plant-derived; however, we did not observe any enrichment for the 21 nt siRNA size class in plant-derived sRNAs identified in FAW (Fig. 3A) suggesting that detected siRNAs are contaminating RNA degradation products rather than functional siRNAs.
FIGURE 3.
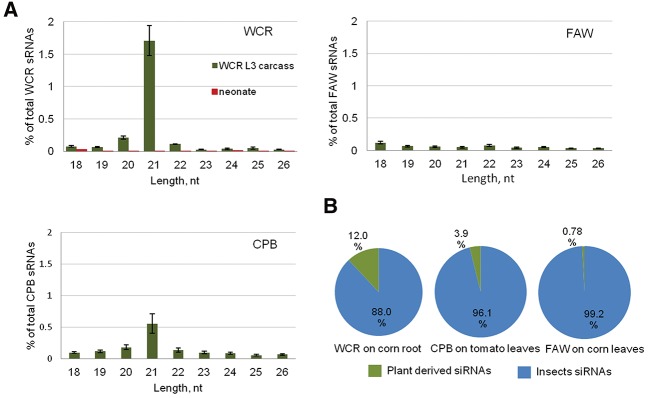
Plant-derived siRNA accumulation in insects fed on host plants. (A) Size distribution of host plant sRNAs detected in WCR, CPB, and FAW fed on host plant organs of corn roots, tomato leaves, and corn leaves, respectively. Individual bars represent the percentage of plant-derived siRNA of a certain size to all siRNAs (18–26 nt range) detected in insect larvae. Larvae were fed 11–12 d until third–fourth instars, followed by gut dissection, sRNA sequencing of RNA of carcass tissues and mapping of sRNA reads to corresponding plant genomes. WCR neonates (red) which were not exposed to corn roots were sequenced as a control. sRNA sequencing was done in three replicates and standard deviation bars are shown. (B) Abundance of plant-derived (exogenous) and insect (endogenous) 21-nt long siRNAs in WCR, CPB, and FAW larvae fed on host plants. Error bars show standard deviation.
In general, 21-nt long siRNAs were one of the less abundant size classes of siRNAs out of the three major classes of corn and tomato sRNAs (21, 22, and 24 nt) (Supplemental Fig. 3). Thus, accumulation of 21 nt long plant-derived siRNAs in WCR and CPB, and lack of accumulation of abundant 22 and 24-nt long plant siRNAs, indicate that plant RNAs are likely taken up as long dsRNA by WCR and CPB and then processed into 21 nt siRNAs by the insects’ RNAi machinery. This result, obtained under feeding on whole plant tissues, is consistent with the requirement for long dsRNA for eRNAi as demonstrated in artificial diet feeding experiments for WCR and CPB (Fig. 1A) and as well as for SCR (Bolognesi et al. 2012), and the lack of siRNA uptake in WCR midgut tissues (Fig. 2A,B). To validate that plant-derived 21 nt siRNAs found in the carcass tissues of WCR originated primarily from predicted plant dsRNA loci, we mapped them to the corn genome. Our analysis confirmed that the vast majority of plant-derived 21 nt siRNAs detected in WCR are from predicted dsRNA loci in corn (Supplemental Fig. 4A,B). Interestingly, the majority of predicted dsRNA loci in corn are hotspots for siRNA size classes other than 21 nt (e.g., 24 nt sRNAs), while siRNAs found in WCR are primarily 21 nt (Fig. 3A), supporting the conclusion that ingested plant long dsRNAs are processed by insect machinery. From a technical point of view, these results also indicate minimal cross contamination of insect sRNA libraries with plant siRNAs from the food source. If cross contamination was prevalent, we would expect a similar ratio of different classes of plant sRNAs (21, 22, and 24 nt) in insect and plant sRNA libraries. Genome-wide mapping of corn-derived siRNAs detected in WCR larvae carcass revealed a distribution across all 10 corn chromosomes (Supplemental Fig. 4C). Thus, our data indicate that WCR and CPB larvae uptake a variety of long dsRNAs from plants during natural feeding scenarios and exogenous long dsRNAs present in the food sources are processed into RNAi-capable 21 nt siRNAs by consuming insects.
Effect of exogenous dsRNA on the insect transcriptome
The uptake of numerous plant long dsRNAs of diverse sequence and processing into 21 nt siRNAs by insects susceptible to eRNAi raises an intriguing question of whether these siRNAs are capable of regulating gene expression in insects. Short regions of sequence identity among organisms of different taxa, are not uncommon given the size and complexity of genomes (Ivashuta et al. 2009; Jensen et al. 2013). Genome-wide transcriptome sequence comparison between corn and WCR revealed multiple regions with sequence homology longer than 21 nt (Jensen et al. 2013), thus indicating a possibility of naturally occurring RNAi-based gene regulation in WCR feeding on corn.
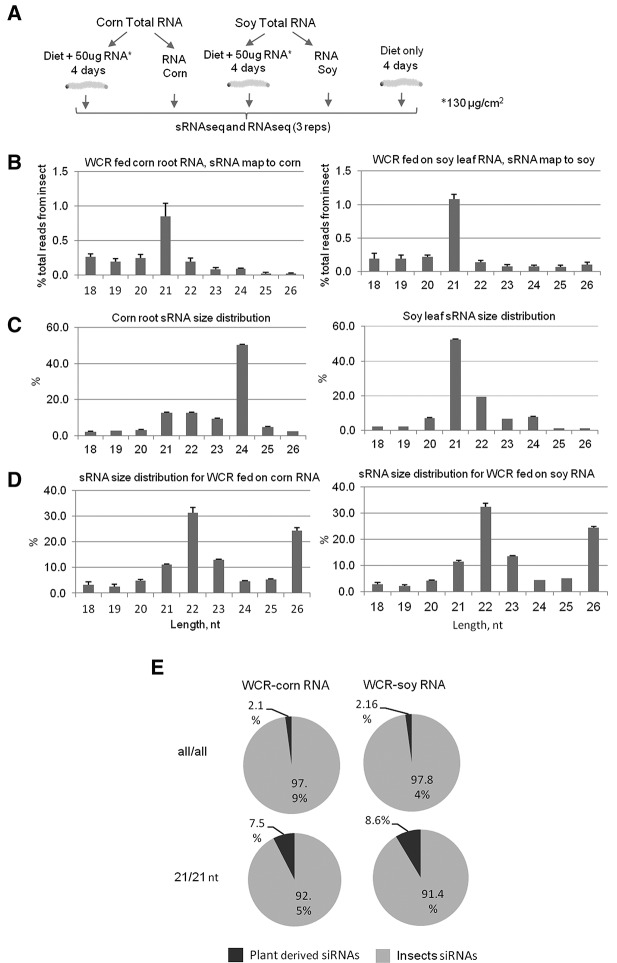
One way to evaluate natural eRNAi-mediated effects on insects is by genome-wide expression analysis to identify genes differentially expressed in insects fed on plants with drastically different genomic compositions, and correspondingly, dsRNA transcriptomes. Unfortunately, such an approach has technical limitations. Species such as WCR will not readily feed on nonhost species and there is the challenge to differentiate genes regulated by eRNAi in a sequence-specific manner from genes regulated indirectly or by other components of the plant food such as secondary metabolites, enzymes, nutrients, etc. In an attempt to overcome this limitation, we developed an approach that would allow for the assessment of the global impact of the plant transcriptome on gene expression in WCR larvae. In this approach, we fed WCR on artificial diet supplemented with total RNA isolated from host and nonhost plants with distinct genomes. To validate the method, we compared the accumulation of plant-derived 21 nt siRNAs in WCR neonate larvae fed on corn roots or on artificial diet supplemented with total RNA from corn roots for 4 d. The pilot experiment revealed a similar pattern and distribution of plant-derived 21 nt siRNA accumulation in WCR larvae when 50 µg per well (130 µg/cm2) of total RNA was presented in artificial diet or larvae were fed on corn roots. The gut was not removed from the WCR neonates used for sequencing due to small larvae size in this sequencing experiment, but plant miRNAs and 22 or 24 nt plant siRNAs were still underrepresented in this whole larvae sequence data set, suggesting that plant sRNAs might be less stable in the WCR larval gut environment as compared with long dsRNA that may be stabilized through uptake and distribution. Feeding WCR larvae on diet supplemented with 50 µg of total RNA extracted from transgenic corn expressing 426 bp long dsRNA targeting DvSnf7 (as shown in Supplemental Fig. 2C) resulted in WCR mortality as expected and similar to what has been observed when WCR were fed on transgenic roots. Mapping of plant-derived siRNAs identified in WCR fed on corn roots or on RNA extracted from corn roots revealed similar patterns (Supplemental Fig. 5), reconfirming the similarity in exogenous dsRNA uptake by WCR from corn roots and from total RNA preparation.
To identify genes potentially regulated by accumulated host plant-derived siRNAs in WCR, we fed larvae on an artificial diet supplemented with total RNA from host and nonhost plants, corn roots and soy leaves, respectively (Fig. 4A). Previous analysis indicated that both corn and soy contain a comparable complexity of predicted dsRNA transcriptome (Jensen et al. 2013) while having a significantly different transcriptome sequence. As described above in the experimental design, we expected to exclude non-sequence-specific effects of various components of plant diet on gene expression in WCR larvae and expected the major variable between experimental samples to be due to differences in nucleotide sequence from corn and soy total RNA. sRNA sequence analysis in WCR revealed an expected enrichment of plant-derived siRNAs for the 21nt class in WCR larvae (Fig. 4B) fed on diet supplemented with total RNA from corn root or soy leaves relative to the food sources (Fig. 4C) and endogenous insect sRNAs (Fig. 4D). Plant-derived 21 nt siRNAs in larvae fed on diet supplemented with total RNA from corn root or soy leaves comprised 7.64% ± 1.69, and 9.52% ± 0.62 of all 21 nt siRNAs in WCR larvae that mapped to corn and soy genomes, respectively (Fig. 4E).
FIGURE 4.
Effect of ingested plant dsRNA on WCR transcriptome. (A) Outline of experiments to identify genes regulated by plant-derived siRNAs accumulated in WCR larvae fed on artificial diet supplemented with total RNA from corn roots and soy leaves, respectively. All graphs show % of total reads from the respective organism and standard deviation. (B) Plant-derived siRNA size distribution in WCR larvae fed on total RNA from corn or soy. (C) Plant siRNA size distribution found in total RNA fed to WCR. (D) Endogenous sRNA size distribution in WCR fed on total RNA from corn or soy. (E) Plant-derived siRNA accumulation in WCR larvae fed total RNA from corn roots or soy leaves.
A comparison of RNA-seq profiling sets of larvae fed on corn or soy total RNA identified 22 genes (out of 95,910 unigenes) with at least 1.5-fold change in expression. We next looked at how many of these differentially regulated genes have sequence homology with either corn or soy-derived siRNAs detected in larvae after feeding. Surprisingly, we found that only a few siRNAs match differentially regulated WCR transcripts (up to 3 mismatches out of 21 allowed, Supplemental Table 1), supporting a lack of detectable impact of eRNAi on the steady-state WCR transcriptome under tested conditions. This is consistent with the need for high identity matches across the 21 nt RNA to significantly regulate gene expression in vivo (Bachman et al. 2013). We identified a relatively large number of unique plant-derived siRNAs in WCR larvae matching thousands of genes in WCR (Supplemental Table 2) with up to 66 (22.4 reads per million, rpm) perfectly matched plant-derived siRNAs to some WCR genes, but the overall number of siRNAs per predicted targeted WCR gene was low and the vast majority had just one plant-derived siRNA identified (Fig. 5). To support our conclusion obtained from global gene expression analysis, we evaluated if corn or soy dsRNA taken up by larvae has an impact on larval development. We did not detect any visible effect on WCR larvae development or any change in larvae body mass when they were fed for 8 d on diet supplemented with 50 µg/well (130 µg/cm2) of total RNA from either corn or soy (Supplemental Fig. 6) during a period in the life cycle where rapid growth is occurring and large quantities of food/diet is consumed (≈45-fold mass gain starting out as a neonate at 0.05 mg and developing to 2.25 mg during the course of the experiment on control larvae).
FIGURE 5.
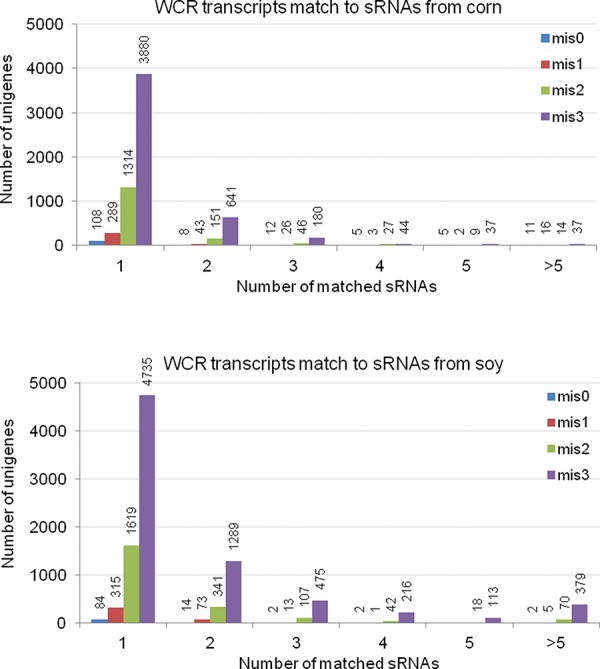
Number of WCR unigenes match to plant-derived siRNAs. Plant-derived siRNAs identified in WCR larvae fed on the diet supplemented with total RNA from corn roots or soy leaves. “mis0” indicates perfect identity between siRNA and putative WCR target transcript; “mis1,” “mis2,” and “mis3” indicate 1, 2, or 3 allowed mismatches, respectively, between pairings.
Transgenically expressed dsRNA with 100% sequence identity to a target WCR transcript induces robust gene silencing when consumed by WCR larvae (Baum et al. 2007). We wondered how the transgenic contribution of dsRNA fares in competition with the background of endogenous long dsRNA coming from the corn genome and if the net abundance of plant-derived siRNAs accounts for observed gene silencing through plant presentation of long dsRNA. For this investigation, we introduced an inverted repeat cassette into corn plants to express transgenic long dsRNA of a similar size and expression pattern as described in Baum et al. (2007). The sequence of this transgenic inverted repeat had no similarity to known WCR transcripts and should therefore be a neutral way to monitor accumulation of transgenic and endogenous plant-derived siRNAs without inducing negative impacts on larval physiology while feeding on corn roots. sRNA sequencing of larvae fed on wild type or transgenic corn plants revealed a similar accumulation of net sRNAs derived from the host plant, regardless of origin status (i.e., transgenic vs. not transgene-derived) with 2.5% of all sRNAs being derived from the native corn genome, or 8.4% and 10.0% of all 21 nt siRNA coming from the native corn genome in nontransgenic or transgenic lines, respectively (Supplemental Fig. 7A). The sRNAs derived from transgenic long dsRNA represented 0.4% of all siRNAs or 1.8% of 21 nt siRNAs in WCR consuming transgenic roots. Thus, ingestion of long dsRNA expressed in plant leads to accumulation of hundreds of unique siRNAs in WCR larvae with perfect matches to a potential target. A number of these unique siRNAs have been found at very high abundance (up to 300 rpm) in WCR tissues (Supplemental Fig. 7B). To evaluate if processing of ingested transgenic dsRNA in WCR tissues and accumulation of large quantities of siRNAs of certain sequence has any nonspecific effect and whether they interfere with endogenous sRNA biogenesis in WCR, we compared endogenous sRNA abundance and distribution in WCR fed on wild type and transgenic plants. No significant perturbations of endogenous WCR sRNAs were observed indicating the insect RNAi machinery is capable of coping with at least small variations in plant dsRNA inputs or that specific sequences presented do not have a perturbing influence on the homeostatic sRNA balance (Supplemental Fig. 8A,B).
DISCUSSION
RNA uptake through diet and its possible impact on the consuming animals is an intriguing phenomenon and highly discussed topic of interaction with the environment (Witwer and Hirschi 2014). A number of factors that complicate investigations of eRNAi under natural feeding conditions include heterogeneity of primary food source, feeding behavior, significant variation in sensitivity to eRNAi, and a limited spectrum of species examined. Our data represent one of the first detailed examinations of RNA movement between host plants and herbivorous insects and indicates significant uptake of plant endogenous dsRNAs into species responsive to eRNAi. In agreement with previous observations on SCR (Bolognesi et al. 2012), dsRNA smaller than 50–60 bp long was insufficient to engage eRNAi within species such as WCR or CPB. We demonstrated that only long dsRNAs were selectively accumulated in insects receptive to eRNAi while abundant plant small RNAs and smaller RNA fragments were not taken up or were unstable in the gut lumen environment.
We also found that plant-derived long dsRNA was processed by receptive larvae predominantly into 21 nt long siRNAs by the insect endogenous RNAi machinery. This process occurs in gut epithelial cells—a primary dsRNA receptive tissue in insects. At the same time, detection of exogenous long dsRNA in CPB carcass suggests that at least some long dsRNA can be transported from tissues where primary uptake occurred to other insect organs. Thus, while only long dsRNA can be efficiently taken up from gut lumen, it is possible that both long dsRNA and processed siRNAs are transported to other insect organs and tissues. WCR tissue culture assays suggests that at least some cell types such as fat bodies can uptake both long dsRNA and siRNA further supporting a hypothesis that both exogenous long dsRNA and processed siRNAs can be a systemic silencing signal in insects (Fig. 2A). The surprising result of the inherent dsRNA uptake process by gut tissues was the magnitude of accumulated plant-derived RNAs in a responsive species. We observed that up to 12% of all 21 nt siRNAs found in the WCR third instar larval body (excluding midgut tissues) were not of insect origin but originated from the host corn roots (Fig. 3B). Mapping of plant-derived siRNAs found in WCR larvae to the corn genome revealed a broad distribution in their origin suggesting a lack of obvious sequence specificity for dsRNA uptake, processing and transport within the larval body. This distribution was colocalized with loci previously identified as candidates for endogenous long dsRNA production in corn (Jensen et al. 2013). The primary long dsRNA transcripts produced within corn are subject to the activity of Dicer-like enzymes and processed into 24, 22 and 21 nt long siRNAs. Even if this processing is rapid, the resulting steady-state level of transcripts above 50–60 bp in size (the uptake threshold), must be efficiently recognized and imported by the responsive insect species. Conversely, inefficient uptake/accumulation of the more abundant plant sRNAs and other small degraded RNA fragments may serve as a protection mechanism to avoid overloading of the insect RNAi machinery and to prevent compromising the function of endogenous RNAi pathways. Size, but not sequence-selective uptake of long dsRNA from the intestinal lumen would fit the hypothesis of an antiviral role for eRNAi. The import of long plant dsRNA may be the result of a system unable to differentiate the origin of dsRNA and as a consequence, such species have learned to manage endogenous sRNA-based processes amidst a background of nonregulatory sRNAs. In this light, the breadth of ecological roles eRNAi can play in invertebrates needs further investigation.
One of those roles could be to regulate the level of endogenous transcripts leading to suppression that could drive developmental cues, etc. in the recipient species. Alternatively, the faculty of eRNAi could be exploited by a plant against an invertebrate herbivore, i.e., an insect system that is otherwise designed for non-sequence-specific uptake as part of a viral defense system could be a means for a plant to deliver dsRNAs that antagonize herbivory. Either of these scenarios is conceivable given the numerous short, high identity matches that are observed in the predicted long dsRNA sequences in corn (Jensen et al. 2013). To help guide future investigation, we examined the response of the WCR larval transcriptomes to dsRNAs from distinct plant sources. Unexpectedly, genome-wide transcriptome analysis of WCR fed on total RNA from host and nonhost plant on artificial diet did not reveal evidence of significant gene regulation by endogenous plant dsRNA at the level of transcript accumulation. sRNA sequencing has revealed multiple plant-derived siRNAs in WCR with perfect or near-perfect complementarity to WCR genes, but the overall number of plant-derived siRNAs per gene was low suggesting very limited if any direct effect of exogenous siRNAs on WCR gene expression. A high degree of suppression via transcript turnover may depend on multiple exact matches. This was observable in the LC50 values of SCR fed a transcript containing only a single embedded 21 nt match to DvSnf7 relative to the equivalent, fully complementary sequence (Bolognesi et al. 2012). In these SCR bioassays, LC50 values of comparably sized dsRNAs bearing only a single embedded 21 nt match to the DvSnf7 gene ranged from 8.0 to 20.3 ng/mL of diet while an 240 nt match had an LC50 of 1.2 ng/mL, thus the amount of dsRNA required jumped an order of magnitude to achieve the same phenotypic response when there was less sequence identity present.
A high number of siRNAs of perfect complementarity helps account for successful gene suppression in plant-expressed dsRNA-expressing transgenes targeting insect pests (Baum et al. 2007; Mao et al. 2007). Taking into account the lack of obvious secondary siRNA amplification components in insects, such as RdRP (Tomoyasu et al. 2008), the abundance of primary siRNAs processed from an exogenous long dsRNA trigger is likely a requirement of robust gene silencing in insects. To evaluate the level of plant-derived siRNA accumulation in WCR fed on transgenically produced dsRNA, we expressed a long dsRNA sequence and driven by the same promoter as described in (Baum et al. 2007). We chose to transgenically present a dsRNA sequence with no known homology with WCR transcripts in order to minimize any confounding effect on larvae physiology that could impact processing, accumulation, and systemic spread. In WCR fed on transgenic roots expressing the “neutral” long dsRNA sequence, transgenic siRNA accounted for ∼1.8% of all 21 nt siRNA in larva body, thus providing a large number of perfectly complementary siRNAs for a hypothetical target. Transgenic sRNA accumulation still represents a fraction of the total load of sRNAs in the WCR as a percentage of total plant sRNAs accumulated in the insect (0.4%). The high background of dsRNA/sRNA originating from the corn genome relative to transgene contribution indicates that successful pest targeting with transgenes expressing dsRNA is due to high identity on-target effects (Baum et al. 2007; Bolognesi et al. 2012; Bachman et al. 2013) versus nonspecific impacts such as saturation of RNAi machinery that has been reported in some siRNA-based oligonucleotide therapeutic drug studies (for review, see Jackson and Linsley 2010).
In summary, our data demonstrates that several insects with similar feeding behavior (chewing herbivores) and consuming a common plant food source manifest a dramatically different capability to take up exogenous dsRNA. Unlike coleopterans examined in this study, the recalcitrant lepidopterans showed enhanced dsRNA degradation in the gut lumen and a lack of stable accumulation of plant-derived sRNAs, indicating a significant variation in the potential ability to participate in eRNAi. The threshold for “long dsRNA” may be a broad prerequisite and signatures of eRNAi-competency can be found by comparing the small RNA populations of potentially receptive invertebrate species to those of the food source that generates the long dsRNAs (a priori this could be from any defined source: plant, animal, fungal, microbial, etc.). This type of analysis could also permit assessment when the invertebrate of interest is not amenable to rearing and experimentation under laboratory conditions if the observations can be substantiated outside of the coleopterans and lepidopterans examined here. While dsRNA stability in gut lumen and uptake appeared to be key factors that differentiate between eRNAi-competent and recalcitrant insects, the insect transcriptome analysis also highlighted a requirement for a high dose of exogenous trigger in order to achieve measurable eRNAi-based regulation even in eRNAi-competent insects. Such a requirement for high dose of exogenous trigger likely reflects the lack of RdRP and silencing signal amplification mechanism in insects and represents a key difference with the most studied animal model, C. elegans that possess a robust silencing amplification mechanism. While we did not observe significant difference in WCR gene regulation at the level of transcript abundance under feeding conditions tested in this work (corn vs. soy total RNA feeding), that does not obviate a potential role of eRNAi in gene regulation under other environmental conditions considering the variety of factors that may affect certain dsRNAs accumulation in host plants. It is clear that the resident insect RNA-based regulatory system at least in WCR and CPB has adapted to this food borne “background” of exogenous dsRNA influx and functions despite its presence or functionally uses it in ways that are yet to be determined.
MATERIALS AND METHODS
Insect testing and RNA analysis
For the dsRNA feeding experiments, dsRNA was made using the MEGAscript kit (Ambion), as previously described (Bolognesi et al. 2012). For the dsRNA stability experiment, neonates were transferred to artificial diet containing dsRNA treatments (100 ng/cm2) administered via diet overlay as in Baum et al. (2007) for 1 d. Larvae were removed from the diet, rinsed in phosphate buffered saline to remove residual diet and dsRNA treatment, and collected. Sixteen WCR, 3 CPB, and 24 FAW and CEW larvae were pooled for each of the three replications. The remaining diet was also collected. For experiments with CPB and WCR testing dissected material, neonates were grown on artificial diet until they reached the first (CPB) or third (WCR) instar and then transferred to artificial diet containing dsRNA treatments (100 ng/cm2 for CPB, 50 ng/cm2 for WCR). After 2 (CPB) or 3 (WCR) days, larvae were rinsed as previously described, then dissected. Three larvae were pooled for each of the three replicates. In real-time RT-PCR analyses, RNA was treated with TURBO DNA-free DNase (Ambion) and used as a template for real-time RT-PCR using iTaq Universal SYBR Green One-Step Kit (Bio-Rad) as previously described (Bolognesi et al. 2012).
For the insect feeding study on whole plant tissues, WCR neonates were applied to corn roots (Zea mays) and allowed to feed until the third instar CPB neonates were applied to tomato leaves (Lycopersicon esculentum) and allowed to feed until they reached the fourth instar. FAW neonates were placed on freshly detached corn leaves which were replenished as needed until insects reached the fifth instar. For all insects, carcass material (gut dissected and removed) from 10 (WCR) or 3 (CPB, FAW) insects was harvested and pooled for each replicate.
Evaluation of isolated plant total RNA was performed on artificial diet as for other purified RNA samples. Neonates were infested onto the diet overlaid with the respective RNA treatments. Single WCR neonates were fed on 50 µg of total corn RNA per artificial diet well (130 µg/cm2) for 8 d and then measured for larval mass. Individual WCR collected for RNA-seq and sRNA sequencing were allowed to feed on 50 µg total plant RNA (130 µg/cm2) (corn or soy, Glycine max) for 4 d, after which 20 whole larvae per each of the three replicates were collected. WCR neonates were also allowed to feed on corn roots for 4 d, after which 10 whole larvae for each of the three replicates were collected.
In vitro tissue culture assay for uptake of labeled dsRNA
An in vitro tissue culture assay was performed following the methods described in (Bolognesi et al. 2012). Both midguts and fat bodies dissected from second instar WCR were used in this system. Labeling of long dsRNAs, siRNA, and dsDNA, processing of tissues after exposure to labeled nucleic acids, and documentation were done as described in (Bolognesi et al. 2012).
Northern blot analysis
Insect and plant RNA was extracted using TRIzol reagent (Invitrogen) following manufacturer's instructions. For Northern blot analysis testing the stability of dsRNA in insects, 10 µg of total RNA were resolved on a 15% polyacrylamide gel containing 7 M urea in 0.5× TBE and blotted to a positively charged nylon membrane (Hybond-XL, GE Life Sciences) with a Bio-Rad Transblot SD. Membranes were probed with complementary oligonucleotides end-labeled with 32P-ATP using OptiKinase (USB Corporation) in Sigma PerfectHyb buffer at 37°C. Final washes of the blots were performed at 37°C with 0.5× SSC, 0.1% SDS. Probe sequences are listed in Supplemental Table 3.
For Northern blot analyses shown in Supplemental Figure 2, tissue was collected from transgenic corn expressing an inverted repeat cassette that targeted the WCR DvSnf7 transcript (Supplemental Table 3). Five micrograms of total plant RNA was treated with RNase I (New England Biolabs) or RNase III (Ambion) according to manufacturers’ directions, ethanol precipitated, then resolved on denaturing gel and blotted and probed using the probe in Supplemental Table 3.
RNA-seq analysis
Transcriptome sequencing was performed using the Illumina HiSeqTM 2000 platform to generate between 15 and 35 million single-end 50 nt reads per sample. (Illumina). These single-end reads were mapped to the public B73 maize transcriptome reference, AGPv2, using Bowtie allowing for up to two mismatches in the first 28 nt “seed” sequence. Reads were filtered based on the uniqueness of their transcriptome mapping. If a read mapped to >10 transcripts, it was ignored. If it mapped to 10 or less transcripts, it was assigned equally to all of them. This was done in order to avoid underestimating the number of transcriptionally active genes. The DESeq algorithm (Simon Anders, EMBL) was then used to normalize the raw counts across all the samples in an experiment. To identify poor quality samples or sampling mislabeling, three quality control analyses were performed. Principal component analysis, hierarchical cluster analysis, and Pearson correlation analysis were performed using Partek's Genomic Suite. Total transcript counts were calculated using htseq_count with default settings (Anders et al. 2014). Differential gene expression was calculated in SAS 9.2 using the PROC GLIMMIX function to fit the read counts to an overdispersed Poisson distribution with a log link.
sRNA sequencing and analysis
sRNA library preparation, sequencing and computational analysis were as described (Dickinson et al. 2013). sRNA sequencing was done in three replicates except as indicated otherwise. In this manuscript, sRNAs of 18–26 nt long were used for analysis. sRNAs from samples of insects fed on plants were mapped back to the diet source plant genome. To get a global view of crop endogenous sRNA biogenesis and siRNAs processed by insects fed on crop dsRNAs, sRNAs from corn root and WCR fed on corn root were mapped to the corn genome public assembly v2.0 (www.maizegdb.org/). SHRiMP2 (David et al. 2009) with default settings was used as the mapping tool, and only perfect matches were allowed except as indicated otherwise. The genome was divided into 10 kilobase (kb) bins where abundance of sRNAs mapped to each bin was summed. The summed abundance was then normalized to per one million sRNA reads in the sRNA library and also to per base in the bin. Corn stranded-RNA-seq data mapping results were also binned in the same way so that comparisons could be made between corn dsRNAs predicted from the RNA-seq data and sRNA mapping results.
DATA DEPOSITION
Raw sequence data have been deposited at NCBI under BioProject accession PRJNA266439.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Emily Kraus, Jodi Beattie, Oliver Ilagan, and Michael Pleau for insect assays, and Jin Zhu and Crystal Ruth for sRNA library preparation. This work was supported by the Monsanto Company.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.048116.114.
REFERENCES
- Anders S, Pyl PT, Huber W 2014. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman PM, Bolognesi R, Moar WJ, Mueller GM, Paradise MS, Ramaseshadri P, Tan J, Uffman JP, Warren J, Wiggins BE, et al. 2013. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res 22: 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D 2004. RNA silencing in plants. Nature 431: 356–363. [DOI] [PubMed] [Google Scholar]
- Baum JA, Roberts JK 2014. Progress towards RNAi-mediated insect pest management. In Insect midgut and insecticidal proteins, Vol. 47of Advances in Insect Physiology (ed. Dhadialla TS, Gill SS), pp. 249–295 Academic Press, London. [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, et al. 2007. Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R, Ilagan O, Lawrence C, Levine S, Moar W, et al. 2012. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7: e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O 2006. The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M 2009. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics 27: 1011–1012. [DOI] [PubMed] [Google Scholar]
- Dickinson B, Zhang Y, Petrick JS, Heck G, Ivashuta S, Marshall WS 2013. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat Biotechnol 31: 965–967. [DOI] [PubMed] [Google Scholar]
- Huvenne H, Smagghe G 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol 56: 227–235. [DOI] [PubMed] [Google Scholar]
- Ivashuta SI, Petrick JS, Heisel SE, Zhang Y, Guo L, Reynolds TL, Rice JF, Allen E, Roberts JK 2009. Endogenous small RNAs in grain: semi-quantification and sequence homology to human and animal genes. Food Chem Toxicol 47: 353–360. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 9: 57–67. [DOI] [PubMed] [Google Scholar]
- Jensen PD, Zhang Y, Wiggins BE, Petrick JS, Zhu J, Kerstetter RA, Heck GR, Ivashuta SI 2013. Computational sequence analysis of predicted long dsRNA transcriptomes of major crops reveals sequence complementarity with human genes. GM Crops Food 4: 90–97. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, et al. 2012. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat Commun 3: 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25: 1307–1313. [DOI] [PubMed] [Google Scholar]
- McEwan DL, Weisman AS, Hunter CP 2012. Uptake of extracellular double-stranded RNA by SID-2. Mol Cell 47: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Ramaseshadri P, Zhang Y, Segers G, Bolognesi R, Tomoyasu Y 2014. Establishing an in vivo assay system to identify components involved in environmental RNA interference in the western corn rootworm. PLoS One 9: e101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuez I, Félix M-A 2012. Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7: e29811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, Jacquier C, Antoniewski C, Andino R 2009. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458: 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, Albrechtsen M, An C, Aymeric JL, Barthel A, et al. 2011. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57: 231–245. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Tomoyasu Y, Miller S, Tomita S, Schoppmeier M, Grossmann D, Bucher G 2008. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo JS, Hunter CP 2008. Environmental RNA interference. Trends Genet 24: 297–305. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc Nat Acad Sci 104: 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer KW, Hirschi KD 2014. Transfer and functional consequences of dietary microRNAs in vertebrates: Concepts in search of corroboration: negative results challenge the hypothesis that dietary xenomiRs cross the gut and regulate genes in ingesting vertebrates, but important questions persist. Bioessays 36: 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wiggins B, Lawrence C, Petrick J, Ivashuta S, Heck G 2012. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics 13: 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD 2010. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet 6: e1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.