Abstract
The enzymatic ribonucleoprotein telomerase maintains telomeres in many eukaryotes, including humans, and plays a central role in aging and cancer. Saccharomyces cerevisiae telomerase RNA, TLC1, is a flexible scaffold that tethers telomerase holoenzyme protein subunits to the complex. Here we test the hypothesis that a lengthy conserved region of the Est1-binding TLC1 arm contributes more than simply Est1-binding function. We separated Est1 binding from potential other functions by tethering TLC1 to Est1 via a heterologous RNA-protein binding module. We find that Est1-tethering rescues in vivo function of telomerase RNA alleles missing nucleotides specifically required for Est1 binding, but not those missing the entire conserved region. Notably, however, telomerase function is restored for this condition by expressing the arm of TLC1 in trans. Mutational analysis shows that the Second Essential Est1-arm Domain (SEED) maps to an internal loop of the arm, which SHAPE chemical mapping and 3D modeling suggest could be regulated by conformational change. Finally, we find that the SEED has an essential, Est1-independent role in telomerase function after telomerase recruitment to the telomere. The SEED may be required for establishing telomere extendibility or promoting telomerase RNP holoenzyme activity.
Keywords: telomerase, TLC1, Est1, RNA, ncRNA, lncRNA, RNP, SEED, yeast, S. cerevisiae
INTRODUCTION
Linear chromosomes cannot be fully replicated by DNA polymerases, resulting in shortening from their ends with successive cell divisions (Blackburn 2006). If left unchecked, this end-replication problem will result in a failure in telomere capping and a cellular G2/M arrest known as senescence (Weinert and Hartwell 1988; Lundblad and Szostak 1989; Abdallah et al. 2009). To counteract this problem, most eukaryotes use the specialized ribonucleoprotein (RNP) complex telomerase to synthesize telomeric DNA. The telomerase core enzyme consists of the telomerase reverse transcriptase (TERT) and the RNA subunit, which contains the stretch of template nucleotides used for reverse transcription and also acts as a scaffold for holoenyzme subunits (Greider and Blackburn 1989; Lingner et al. 1997b; Zappulla and Cech 2004).
The budding yeast Saccharomyces cerevisiae telomerase RNA, TLC1, is 1157 nt and has a secondary structure composed of three long arms, which radiate from a central catalytic core (see Fig. 1A; Dandjinou et al. 2004; Zappulla and Cech 2004). Yeast TERT, Ever-Shorter Telomeres 2 (Est2), binds to the central core where the RNA template is located (Lingner et al. 1997b; Livengood et al. 2002). Est2 and the core of TLC1 are sufficient for telomerase enzymatic activity in vitro (Cohn and Blackburn 1995; Lingner et al. 1997a; Zappulla et al. 2005), but several accessory protein subunits are also required in vivo (Lendvay et al. 1996). The essential Est1 protein binds within the distal half of one arm, while the important Ku heterodimer and Sm7 complexes bind to sites on the other arms (see Fig. 1A; Seto et al. 1999, 2002; Peterson et al. 2001; Stellwagen et al. 2003; Lubin et al. 2012). The Est3 protein has been reported to bind Est2 (Talley et al. 2011) as well as Est1 (Tuzon et al. 2011). Also, TLC1 contains a conserved three-way junction near its 3′ end, which has been shown in other species’ telomerase RNAs to interact with TERT and to be essential, although not in S. cerevisiae (see TWJ in Fig. 1A; Tesmer et al. 1999; Mitchell and Collins 2000; Livengood et al. 2002; Zappulla et al. 2005; Brown et al. 2007).
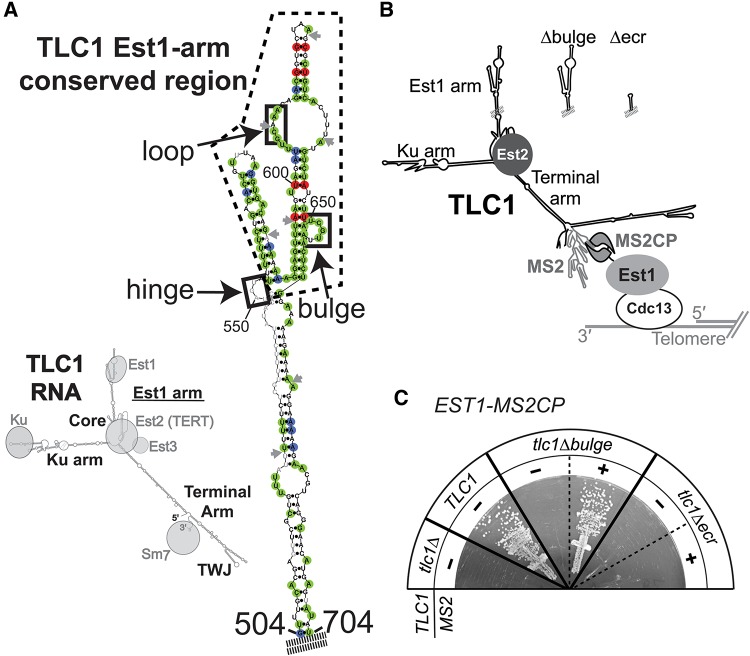
FIGURE 1.
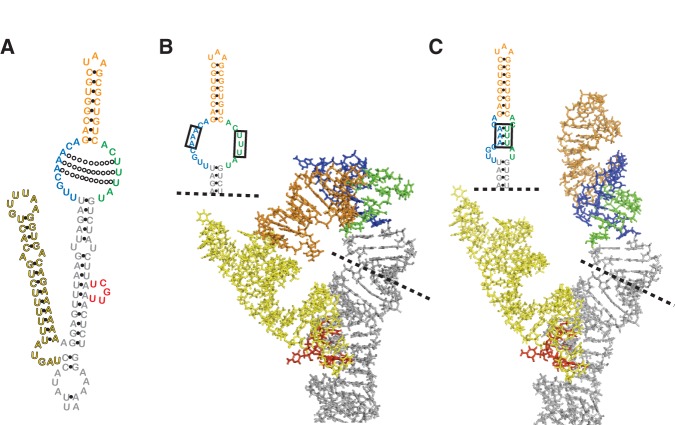
The Est1-binding arm of TLC1 has a second essential function in telomere maintenance. (A) The Est1-arm conserved region of TLC1 telomerase RNA. (Left) Phylogenetically supported secondary structure for TLC1 (Zappulla and Cech 2004) bound to protein subunits. (Right) The phylogenetically supported secondary structure of the S. cerevisiae Est1-binding arm of TLC1, with nucleotides highlighted based on the sequence alignment of 36 telomerase RNAs from seven Saccharomyces species (see also Supplemental Fig. S1). Green = 100% conservation, red = covarying, blue = varying while maintaining base-pairing. Gray nucleotides, absent in one or more sequences. Gray arrows, locus of ≥1 nt insertion in one or more sequences. Dashed line indicates 108-nt Est1-arm conserved region (nucleotides 554–661). Solid black boxes indicate elements proposed to be required for Est1 association. (B) Schematic of MS2-tethered Est1 mediating telomerase RNP recruitment to the telomere. (C) MS2-tethering Est1 to tlc1Δbulge, but not tlc1Δecr, is sufficient for telomerase function. TLC1 alleles with or without MS2 hairpins were expressed in EST1-MS2CP cells. Streaks are shown at 125 generations of growth.
Although EST1 was the first telomerase subunit-encoding gene identified (Lundblad and Szostak 1989), many details of its function are still unknown. The primary role for Est1 protein is recruitment of telomerase to the telomere through an interaction with Cdc13, which also binds single-stranded telomeric DNA (Evans and Lundblad 1999; Mitton-Fry et al. 2002; Wu and Zakian 2011; Tucey and Lundblad 2013). In addition, Est1 has a poorly understood role in activating telomerase function (Taggart et al. 2002), and deletion of EST1 results in telomere shortening even when Est1-mediated recruitment is bypassed by fusing Est2 directly to Cdc13 (Evans and Lundblad 1999, 2002).
Mutations in three discrete sets of nucleotides of the Est1-binding arm of TLC1 RNA have been shown to adversely affect association with Est1 protein in vivo (boxes in Fig. 1A; Seto et al. 2003; Lubin et al. 2012). The first of these regions comprises 5 nt predicted to form a bulge in the arm (Seto et al. 2002). The second is within an internal loop closer to the tip of the Est1 arm; nucleotide sequence of the 5′ side of the loop is important and single-stranded RNA in the loop is critical for Est1 association with TLC1 (Lubin et al. 2012). Two phylogenetically supported secondary structure models have been proposed for this internal loop; a single, large loop (Zappulla and Cech 2004) or two smaller loops separated by three A–U base pairs (Dandjinou et al. 2004). Finally, the third region of TLC1 implicated in Est1 association is a single-stranded junction at the base of the conserved region (see “hinge” in Fig. 1A). TLC1 mutants lacking single-stranded RNA in this region have largely disrupted association with Est1 protein and exhibit short but stable telomeres (Lubin et al. 2012). Together, these three structural elements provide the first essential function of the Est1 arm to be identified; namely, tethering Est1 protein to telomerase RNA.
The TLC1 long noncoding RNA has now been shown to act as a flexible scaffold for each of the three RNA-binding accessory protein subunits. The essential Est1 and important Ku and Sm7-binding sites on the RNA can be relocated to different locations within TLC1 while still functioning in vivo (Zappulla and Cech 2004; Zappulla et al. 2011; Mefford et al. 2013). In addition, telomerase tolerates miniaturization achieved by deleting the bulk of the arms (i.e., two-thirds of the entire RNA) between the central core and distal regions of the protein-binding arms in the secondary structure model (Zappulla et al. 2005). The arms can also be stiffened by converting them into double-stranded RNA struts, holding the accessory proteins away from the catalytic core, with retention of telomerase function in vivo (Lebo and Zappulla 2012). Taken together, these findings indicate that the yeast telomerase RNP does not require the RNA-tethered subunits to occupy precise positions or orientations in the RNP for them to function. However, it remains unclear whether the arms of the RNA function only as tethers for binding the accessory subunits or if they have additional roles in telomerase function.
A recent publication reported an important role for portions of the apical hairpin and internal loop of the Est1-binding arm distinct from Est1-binding function (Laterreur et al. 2013). The reported mutants resulted in stably shorter telomeres, although not quite senescence. These alleles did not greatly affect RNA levels and the authors found these mutant TLC1 RNAs remained nuclear by microscopy, which permitted them to conclude Est1 was still binding. This publication provides evidence that the region includes a “telomerase-stimulating structure” that plays a nonessential role in telomerase enzyme activity. These findings provide further support for the hypothesis that the Est1 arm of TLC1 has additional functions in telomere maintenance beyond simply tethering Est1 to the RNP.
Here we demonstrate that, in addition to tethering Est1 protein to the RNP, the Est1 arm of TLC1 has a second essential function in telomere maintenance in vivo. Using a heterologous RNA-protein binding system, we were able to separate the Est1-binding role of the Est1 arm of TLC1 from other functions. We find that a Second Essential Est1-arm Domain (SEED) is located in an internal loop in the Est1-binding arm. SHAPE and mutagenesis suggest that the structure of the internal loop, rather than its sequence, is required. Overall, these data reveal an additional essential function for the Est1-binding arm of TLC1 and begin to elucidate Est1-arm structural conformations that could regulate telomerase action in vivo.
RESULTS
Although telomerase RNA is evolving very rapidly, the nucleotides and secondary structure in the distal half of the Est1 arm of TLC1 RNA—which includes three regions reported to be required for the essential Est1 association (see Fig. 1A; Seto et al. 2002; Lubin et al. 2012)—is highly conserved among budding yeasts (Dandjinou et al. 2004; Lin et al. 2004; Zappulla and Cech 2004). To further examine conservation in this region, we aligned all 36 publicly available distinct Saccharomyces TLC1 sequences (from seven different Saccharomyces species; Supplemental Fig. S1), and mapped sequence and base-pairing conservation onto the secondary structure model of the Est1 arm (Fig. 1A). This analysis revealed that the distal half of the arm is more than twice as conserved as the core-proximal region, with 67% of nucleotides being identical or varying while maintaining secondary structure compared with only 28% in the core-proximal region (Fig. 1A; Supplemental Fig. S1). This 108-nt Est1-arm conserved region (ECR) includes two elements reported to be important for Est1 binding, while the third feature resides just outside of it (Seto et al. 2002; Lubin et al. 2012).
The Est1-binding arm of TLC1 has a second essential function
In order to test if the 108-nt Est1-arm conserved region of TLC1 has additional functions in telomere maintenance, we set up a system to tether Est1 to TLC1 using a heterologous protein-RNA interaction domain. We added DNA sequences encoding two tandem bacteriophage MS2 coat protein domains downstream from genomic EST1, using eight glycines as a linker. This EST1-MS2CP allele was functional, although it maintained telomeres slightly shorter than wild type (see Fig. 3D, below). We also used a TLC1 RNA with 10 MS2 hairpins inserted just before the 3′ end of the gene, upstream of the Sm7-binding site (Supplemental Fig. S2A). This TLC1-MS2 construct has been shown to function similarly to wild-type TLC1 in vivo (Gallardo et al. 2011), although we observed moderately reduced RNA abundance and slightly shorter telomeres (see Fig. 3, below). With this experimental design, the MS2CP tag on Est1 should bind to the MS2 RNA hairpins in TLC1, thereby tethering Est1 to TLC1 even if the endogenous Est1-TLC1 binding interaction is disrupted (Fig. 1B).
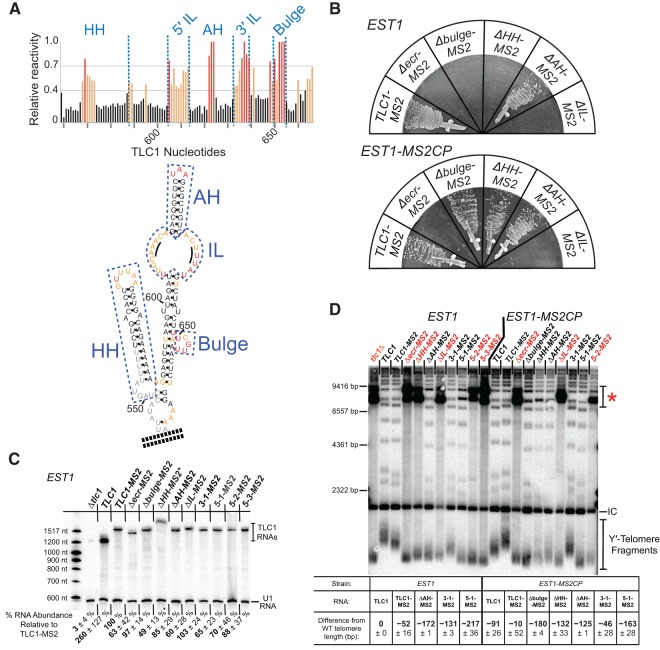
FIGURE 3.
The Second Essential Est1-arm Domain (SEED) maps to a large internal loop of the Est1 arm. (A) SHAPE-reactivity results corroborate the secondary structure model of the Est1-binding arm of TLC1 RNA. SHAPE analysis was performed on a 181-nt section of the Est1 arm (nucleotides 514–694), with a CG-clamp at its base. (Top) Graph showing relative normalized SHAPE reactivity of Est1-arm RNA nucleotides. Red lines, highly reactive (>70% relative reactivity). Orange lines, moderately reactive (40%–70% relative reactivity). Black lines, low reactivity (<40% relative reactivity). (Bottom) Est1-arm conserved region secondary structure, colored based on nucleotide reactivity to SHAPE as indicated above. Structural elements of the Est1 arm are boxed: (HH) hinge-hairpin (nucleotides 549–588), (AH) apical hairpin (nucleotides 614–632), (IL) internal loop (nucleotides 605–613 and 633–639). Black lines in the internal loop highlight nucleotides 609–611 and 635–637, which have been proposed to form base pairs. (B) MS2-tethering Est1 to TLC1 alleles with deletions of structural elements of the Est1-arm maps the second function to the internal loop. All TLC1 alleles are tagged with MS2 hairpins, and expressed in cells with EST1 (top) or EST1-MS2CP (bottom). Streaks shown represent 150 generations of growth. (C) Northern blot of Est1-arm mutants in an EST1 strain. TLC1 RNAs were probed for the shared 3′ end, and normalized to U1 RNA. Average relative RNA abundance from two independent isolates at 25 generations of growth is indicated ± SD. (*) Note that tlc1ΔHH migration is impeded in Urea-PAGE due to the stiffened arm, and the RNA abundance is likely underestimated (Lebo and Zappulla 2012). (D) Telomere Southern blot for Est1-binding arm mutants in EST1 and EST1-MS2CP strains at 250 generations of growth. Red mutant names indicate survivor colonies that arose from senescent samples. A telomere pattern typical of Type 1 survivors can be observed in the senescent samples lanes (red asterisk; Teng and Zakian 1999), confirming that telomerase-mediated telomere lengthening was disrupted. Average change in Y′ telomere length ± sample SD is shown relative to wild type (TLC1 in EST1) from two independent isolates. (IC) internal control, a nontelomeric Xho I fragment from chromosome IV.
Tethering Est1-MS2CP protein to TLC1-MS2 RNA resulted in telomere length indistinguishable from wild type, a net increase compared with either individual fusion construct (see Fig. 3D, below). We next MS2-tethered Est1 to two different mutant TLC1 RNAs: (1) tlc1Δbulge, which lacks the 5-nt bulge in the Est1 arm and has greatly reduced binding to Est1 protein and loss of telomere maintenance (Seto et al. 2002; Lubin et al. 2012) or (2) tlc1Δecr, which is missing the entire 108-nt Est1-arm conserved region of TLC1 (see Fig. 1A; Supplemental Fig. S1). The tlc1Δbulge and tlc1Δecr RNAs without MS2 hairpins did not functionally complement a tlc1Δ strain, and failed to maintain telomeres, leading to senescence (Fig. 1C). However, MS2-tethering tlc1Δbulge to Est1 restored telomere maintenance, allowing cells to grow without senescing (Fig. 1C) and supporting stably shorter telomeres (see Fig. 3D, below). This indicates that the loss of telomerase function caused by the Δbulge mutation is primarily a failure of Est1 to physically associate with the RNP, and that establishing direct binding through the MS2 tether is sufficient to rescue function. Furthermore, these results extend the flexible scaffold model for TLC1 by demonstrating that an essential Est1-binding interface in TLC1 can be functionally replaced with a heterologous RNA-protein direct-binding system.
Whereas MS2-tethering tlc1Δbulge RNA to Est1 protein rescued telomerase function in vivo, tethering tlc1Δecr to Est1 did not permit telomerase-mediated telomere maintenance, and cells senesced by 125 generations (Fig. 1C). Senescence was confirmed by telomere Southern blots, which showed that the few cells that occasionally survived were post-senescence survivors, which maintain telomeres telomerase-independently via recombination (see Fig. 3D, below; Lundblad and Blackburn 1993; Teng and Zakian 1999). The abundance of tlc1Δecr-MS2 RNA was ∼60% of TLC1-MS2 RNA levels (see Fig. 3C, below), showing that RNA abundance is not responsible for abolished telomerase function. The fact that MS2-tethering Est1 to tlc1Δecr does not allow telomerase function strongly suggests that a portion the conserved region of the Est1-binding arm (Fig. 1A; Supplemental Fig. S1) retained in tlc1Δbulge, but missing in tlc1Δecr, is essential for an Est1-binding-independent telomerase process. We therefore propose that this region contains a Second Essential Est1-arm Domain (SEED) functionally distinct from the reported Est1-binding sites.
The SEED can function as part of an Est1-arm RNA expressed in trans
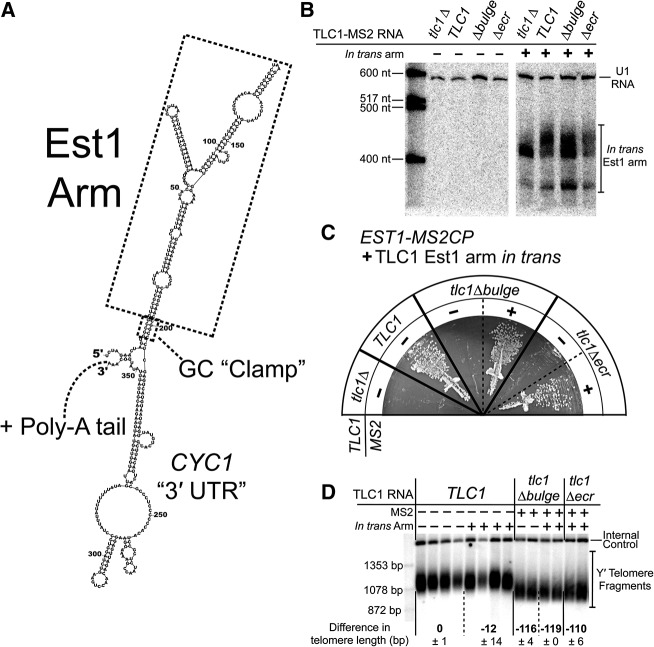
If the second essential function of the Est1 arm of TLC1 is distinct from binding Est1 protein to tether it to the telomerase RNP, then the SEED could perhaps even function in trans. We tested this by expressing the Est1 arm from a constitutive TEF2 promoter harbored on a multiple-copy 2µ plasmid (Fig. 2A; Mumberg et al. 1995). Although the Est1 arm expressed in trans is expected to bind to Est1 protein, it should not help tether Est1 to the telomerase RNP since it is a separate RNA molecule. Northern blots confirm the expression of the in trans Est1-binding arm in cells, show it is the expected size, and suggest it is also polyadenlyated (Fig. 2B).
FIGURE 2.
Expression of the Est1 arm in trans restores telomerase activity to the condition where Est1 is MS2-tethered to tlc1Δecr. (A) Lowest energy Mfold secondary structure prediction for the in trans arm RNA transcript. The Est1 arm is predicted to fold as in TLC1. A GC “clamp” at the base of the Est1 arm helps to ensure proper folding. Upstream and downstream sequences are derived from the TEF2 promoter and CYC1 terminator, respectively. Like wild-type TLC1, the in trans RNA is a Pol II transcript, and is expected to be polyadenylated. (B) Northern blot probed for Est1-arm expression in Est1-MS2CP cells. The two Est1-arm bands correspond to predicted sizes for a nonpolyadenylated arm (∼389 nt; lower band) and a polyadenylated arm (∼400–450 nt; upper band). (C) TLC1 alleles with or without MS2 hairpins expressed in EST1-MS2CP cells along with the Est1 arm in trans. Streaks shown at 125 generations of growth. Presence of the in trans Est1 arm rescues cells with tlc1Δecr only when the TLC1 mutant is tethered to Est1 protein. (D) Telomere Southern blot for Est1-MS2CP cells with or without the in trans Est1 arm. Two or four independent transformants are shown for each condition after 250 generations of growth. Average changes in Y′ telomere length relative to TLC1 are indicated ± SD. Internal control = nontelomeric Xho I fragment from chromosome IV.
Strikingly, expression of the Est1 arm of TLC1 in trans while Est1 protein was MS2-tethered to the tlc1Δecr RNA rescued cells from senescence (Fig. 2C), maintaining telomeres at a similar length to cells with Est1 tethered to tlc1Δbulge RNA (Fig. 2D). This indicates that the SEED can function in telomere maintenance as a separate RNA molecule. However, in the absence of MS2-tethering Est1 to tlc1Δecr RNA, the in trans Est1 arm did not prevent senescence (Fig. 2B), confirming that Est1 protein binding to full-length TLC1 is still required for the ectopic arm to provide its other function. Expressing the Est1 arm in trans with wild-type TLC1 did not have a significant effect on telomere length (Fig. 2D). In summary, these results show that the SEED can function in trans when Est1 is MS2-tethered to the telomerase RNA, indicating that the Est1 arm of TLC1 has a second essential function beyond simply scaffolding proteins in the telomerase RNP.
SHAPE analysis of the Est1 arm of TLC1 RNA
We set out to identify which nucleotides within the 108-nt Est1-arm conserved region of TLC1 are required for the identified second essential function. To accurately target structural elements proposed to exist in the conserved region for mutational analysis, we first tested the secondary structure model of the entire 181-nt Est1 arm of TLC1 (nucleotides 514–694) in vitro. We used selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to chemically interrogate physical flexibility at each position of purified in vitro-transcribed RNA (Wilkinson et al. 2006) using methods described for the catalytic core of yeast and human telomerase RNAs (Niederer and Zappulla 2015). Nucleotides predicted to comprise apical loops of the two hairpins and the 5-nt bulge in the secondary structure model were highly reactive, showing that they are flexible and suggesting that they are indeed unpaired (Fig. 3A; Supplemental Fig. S3). Correspondingly, nucleotides within predicted helices were nearly all unreactive. These data strongly support the secondary structure model for the Est1 arm, which was based on phylogenetic information as well as computational modeling that maximizes free energy (Seto et al. 2002; Dandjinou et al. 2004; Zappulla and Cech 2004). Interestingly, SHAPE reactivity revealed flexibility at every nucleotide in the internal loop of the Est1 arm, suggesting they are not paired and, therefore, that this conformation is favored (see “IL” in Fig. 3A).
A conserved internal loop of the TLC1 Est1 arm is required for SEED function
To determine more precisely the region containing the SEED, we designed a series of deletions in the Est1 arm of TLC1 to test specific RNA elements: the apical hairpin (AH), the hinge-hairpin structure (HH), and the internal loop (IL) (Fig. 3A; Supplemental Fig. S2A). Deleting the 19-nt AH from the Est1 arm did not cause senescence (Fig. 3B), in agreement with previously reported deletions (Lubin et al. 2012; Laterreur et al. 2013).
Next, we deleted the hinge-hairpin (HH) domain (Fig. 3A). Previously published results and our own findings indicate that single-stranded nucleotides are required at this locus for telomerase function (Fig. 4; Lubin et al. 2012). We tested a series of Est1-arm mutants with single-stranded nucleotides deleted, in the context of wild-type TLC1 or Triple-Stiff-Arm TLC1 (TSA-T, with stiffened Ku and Est1 arms; Lebo and Zappulla 2012). Deleting just the HH domain from TLC1 does not cause senescence (see TLC1Δ550–590 in Fig. 4). However, deleting HH from the stiffened Est1 arm (tsa-tΔHH or tlc1ΔHH) causes loss of telomere maintenance. Reintroducing the 8-nt hinge from wild-type or a random 8-nt sequence at the native HH locus restores telomerase function (TLC1(ssH) and TLC1(randH) in Fig. 4). But adding the 8-nt hinge into the middle of the stiffened arm (tlc1(midH)) does not rescue telomere maintenance, showing that a physically flexible point simply anywhere in the arm is insufficient. Together, these data indicate that unpaired nucleotides of nonspecific sequence are required near the HH locus for telomerase function in the cell.
FIGURE 4.
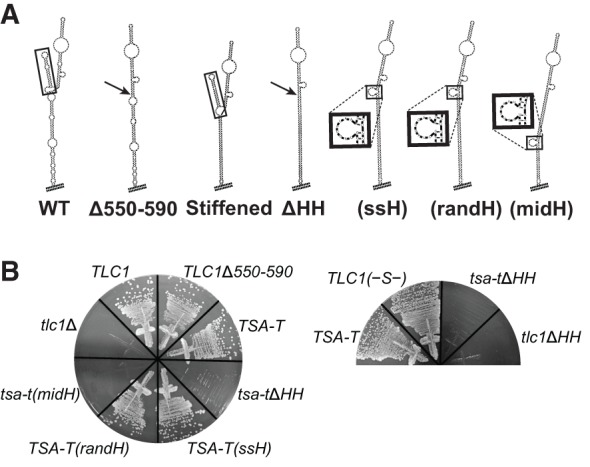
Single-stranded nucleotides in the hinge-hairpin (HH) domain are required for telomerase function in vivo. (A) Lowest energy Mfold-predicted secondary structures for Est1-arm mutants, from left to right, with hinge domains boxed, or sites of HH deletion indicated by an arrow: wild-type Est1 arm; Δ550-590; stiffened Est1 arm, referred to as TLC1(–S–) when in wild-type context; ΔHH; (ssH) with wild-type 8-nt single-stranded hinge restored at the HH locus (UAGUAUAA; see box); (randH) with 8-nt single-stranded hinge of random sequence at the HH locus (CUAGUCAG; see box); (midH), with wild-type 8-nt hinge added into the middle of the stiffened Est1 arm (see box). (B) TLC1 HH mutant alleles expressed from CEN plasmids in yeast, shown at 250 generations of growth.
To help ensure that no unpaired nucleotides remained at the base of the conserved region in the tlc1ΔHH deletion mutant, we also deleted all bulges and loops from the core-proximal portion of the Est1-arm conserved region (Fig. 4; Supplemental Fig. S2A). This tlc1ΔHH RNA was unable to maintain telomeres, resulting in senescence (Fig. 3B). This phenotype was not due to decreased RNA abundance, since Northern blots showed this RNA (as well as each of the other TLC1 Est1-arm mutants) was substantially above the threshold level for function (Fig. 3C; Seto et al. 1999; Mefford et al. 2013). Furthermore, the addition of MS2 hairpins to this or any of the Est1-binding arm mutants reported here was not responsible for the senescence phenotype, since each also senesced in the absence of both MS2 hairpins and the MS2CP tag on Est1 protein (Supplemental Fig. S2B). Consistent with tlc1ΔHH having a defect in Est1 binding, when Est1 was MS2-tethered to tlc1ΔHH, it rescued this telomerase RNA and cells did not senesce (Fig. 3B). After being propagated for 250 generations to reach equilibrium, telomeres from these cells were 135 bp shorter than TLC1-MS2, similar to the 134-bp shortening for the condition with Est1 tethered to tlc1Δbulge (Fig. 3D). This suggests that, like the 5-nt bulge, the hinge-hairpin RNA element in the Est1 arm is important for proper Est1 protein association with TLC1, and not for SEED function.
Finally, we deleted the Est1-arm internal loop (IL) (Fig. 3A; Supplemental Fig. S2A) and found that it caused senescence. However, in contrast to the Δbulge and ΔHH deletion mutants, MS2-tethering Est1 to tlc1ΔIL did not rescue telomerase function (Fig. 3B). This unique result provides compelling evidence that the Second Essential Est1-arm Domain of TLC1 is located within this internal loop.
Mutations that disrupt structure of the internal loop also disrupt SEED function
To understand how structure of the internal loop relates to SEED function, we made a series of nucleotide substitution mutations within the internal loop. We first altered the sequence of the entire 3′ side of the loop, while maintaining internal loop secondary structure based on Mfold predictions (Fig. 5A; Supplemental Fig. S2A). Mutant TLC1(3-1) did not exhibit senescence (Fig. 5B), maintaining telomeres slightly shorter than wild type (Fig. 3D), and indicating that the sequence of the 3′ side of the internal loop is not essential for binding Est1 or for SEED function.
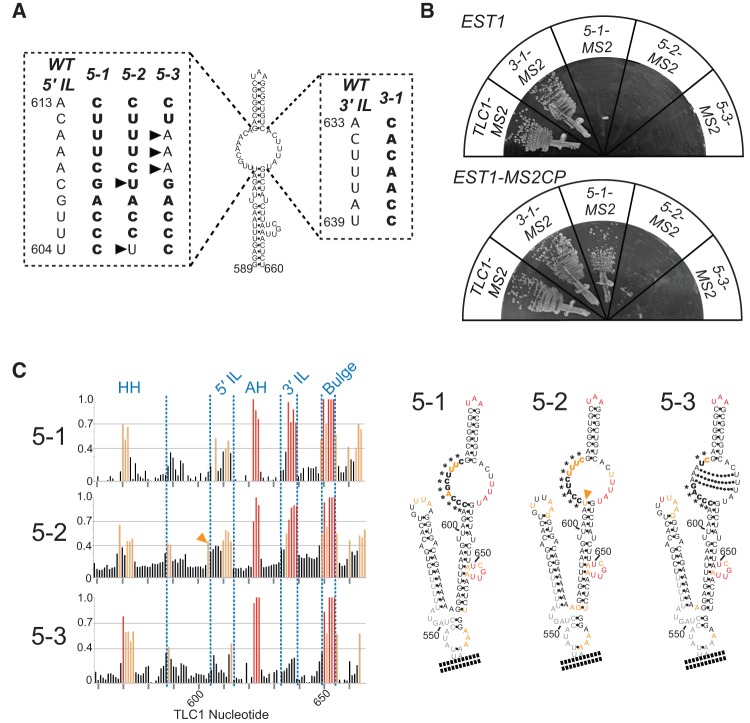
FIGURE 5.
SEED function is dependent on the structure of the Est1-arm internal loop. (A) Schematic of nucleotide substitution mutants in the large internal-loop of the Est1-arm conserved region. Mutant nucleotides are boldface. Arrowheads indicate nucleotides that differ from mutant 5-1. (B) Mutants tlc1(5-2) and tlc1(5-3) have lost SEED function since they cannot be rescued by MS2-tethering Est1. All TLC1 alleles were tagged with MS2 hairpins, and expressed in cells expressing EST1 (top) or EST1-MS2CP (bottom). Streaks shown represent 150 generations of growth. (C) SHAPE analysis of internal-loop nucleotide sequence-substitution mutants shows that mutants 5-2 and 5-3 have altered internal loop structures. (Left) Graphs showing relative normalized SHAPE reactivity of TLC1 Est1-arm nucleotides. Red lines correspond to highly reactive positions, orange lines indicate moderately reactive positions, as in Figure 3A. Blue text above indicates Est1-arm structural elements. (Right) Internal-loop-mutant Est1-arm secondary structures, colored based on reactivity to NMIA. Asterisks indicate non-wild-type nucleotides in the mutants. Orange arrow indicates residue U604, with high SHAPE reactivity in mutant 5-2. Dotted lines indicate base pairs predicted to form by Mfold in mutant 5-3.
We next made sequence mutations in the 5′ side of the internal loop. Mutant tlc1(5-1) has all 5′-side nucleotides substituted as shown in Figure 5A, and is still predicted by Mfold to form the single internal loop (Supplemental Fig. S2A). SHAPE analysis demonstrated that there is a moderate reduction in nucleotide reactivity in the internal loop compared to wild type, demonstrating a loss of flexibility in portions of the structure, whereas the lower half of the 3′ side of the loop remained highly reactive (Fig. 5C). Cells expressing tlc1(5-1) grew poorly and occasionally displayed a delayed senescence phenotype in the absence of tethering (Fig. 5B, top), which is consistent with this allele exhibiting extremely short telomeres (Fig. 3D).
MS2-tethering Est1 protein to tlc1(5-1) prevented senescence and restored telomere maintenance to levels similar to the condition of Est1 MS2-tethered to tlc1Δbulge (Figs. 5B, 3D). Since tethering Est1 to tlc1(5-1) suppresses its phenotype, we conclude that mutations in the 5′ side of the internal loop cause a loss of Est1 association with the RNA, similar to previously reported internal loop mutants (Lubin et al. 2012), and do not abolish SEED function.
While nucleotide substitutions in the internal loop are tolerated, mutants that alter the structure of the loop disrupt SEED function. We designed a second IL 5′-side mutant, tlc1(5-2), with two different nucleotide substitutions than 5-1 (indicated by black arrows in Fig. 5A), including restoring nucleotide 604 to the wild-type U. Tlc1(5-2) resulted in a senescence phenotype not suppressible by MS2-tethering Est1 protein, indicating lost SEED function (Fig. 5B). It is unlikely that the sequence of tlc1(5-2) per se is responsible for disruption of the SEED, given that tlc1(5-1)—which has all nucleotides in the 5′ side of the internal loop changed—retains SEED function.
SHAPE reactivity was similar for both 5-1 and 5-2 in the 3′-most quadrant of the internal loop (Fig. 5C; Supplemental Fig. 3), but there was alternate folding in the basal part of the internal loop exclusively in SEED-disrupting 5-2 allele. In particular, U604, which is essentially unreactive in wild-type TLC1, is strongly reactive in tlc1(5-2), suggesting it is not forming the wild-type base pair (see orange arrow in Fig. 5C). Phylogenetic analysis indicates that this base pair forms in all Saccharomyces yeast species (see Fig. 1A; Supplemental Fig. S1). A possible explanation for the SHAPE results for 5-2 is G640 pairing with C606 or C605, in turn causing U604 to be unpaired and significantly changing the structure in the base of the internal loop. The SHAPE analysis suggests an altered internal loop structure in tlc1(5-2) results in SEED dysfunction.
To test the hypothesis that the structure of the internal loop is more important than its sequence for SEED function, we designed a modification to tlc1(5-1) that disrupts the structure with fewer mutated residues. This mutant, 5-3, has 7 of the same 10-nt substitutions as 5-1, but retains the wild-type AAA sequence at positions 609–611 (Fig. 5A). Mfold predicts that the internal loop of mutant 5-3 will contain three A–U base pairs, resulting in the formation of two small loops separated by this 3-bp helix (Fig. 5C; Supplemental Fig. S2A). This structure may be biologically relevant and has been suggested previously to form in wild-type TLC1 (Dandjinou et al. 2004). SHAPE analysis of tlc1(5-3) indicated a clear reduction in reactivity of essentially all nucleotides in the internal loop (Fig. 5C; Supplemental Fig. 3), consistent with the expected increase in base-pairing between the two sides. The greatest reduction in reactivity was in the 3′-most quadrant of the loop (Fig. 5C), a region we found earlier to be highly reactive by SHAPE in wild-type TLC1 and the 5-1 allele with its functional SEED. Cells expressing tlc1(5-3) exhibited loss of telomere maintenance leading to senescence, which could not be bypassed by MS2-tethering Est1 (Fig. 5B). Low RNA abundance or the presence of MS2 hairpins is unlikely to contribute substantially to this loss-of-function phenotype for tlc1(5-3), because even in the absence of MS2 hairpins, when RNA abundance is 78% of wild-type TLC1, cells expressing tlc1(5-3) senesced (Supplemental Fig. S2B). Thus, these data show that disrupting the structure of the internal loop results in loss of SEED function.
Overall, SHAPE and functional analyses of the three 5′ IL mutants and wild-type TLC1 suggests that the base of the internal loop is of particular structural importance to the second essential function of the Est1 arm, and therefore comprises the SEED.
The SEED has an Est1-independent function in telomerase mechanism
To explore the mechanism of SEED function in telomerase, we further examined the functional relationships between TLC1 and Est1 in vivo. The essential role of Est1 protein in recruiting telomerase to the telomere can be bypassed by fusing the telomere-binding protein Cdc13 to the telomerase catalytic subunit Est2 (Fig. 6A; Evans and Lundblad 1999). We used this CDC13-EST2 system to test (1) if the TLC1 SEED function is separable from Est1-mediated telomerase recruitment and (2) if SEED function is dependent on the presence of Est1 protein. We integrated a DNA sequence of eight glycine-encoding codons followed by the EST2 gene in place of the stop codon of CDC13 to generate CDC13-Gly8-EST2. In an EST1 genetic background, after 500 generations of passaging cells the Cdc13-Est2 fusion protein had caused extensive hyperlengthening of telomeres (see bracket “a” in Fig. 6B, lanes 1,2), as observed for the originally reported Cdc13-Est2 fusion (Evans and Lundblad 1999). Due to the highly elongated telomeres, cells were able to grow for over 600 generations after TLC1 was deleted before senescing (∼6 times longer than wild-type cells survive without telomerase). The decrease in telomere length in tlc1Δ cells was apparent by Southern blotting; as telomeres shortened, distinct bands became apparent on the Southern blot due to differences in the size of subtelomeric regions in the XhoI-digestion products (Fig. 6B, lanes 17–20). As previously reported, deletion of EST1 in the CDC13-EST2 fusion strain caused relative telomere shortening, but not senescence (Evans and Lundblad 1999). Although telomeres in the est1Δ CDC13-EST2 strain (lanes 3,4) became shorter, even after 500 generations they appeared stably longer than standard wild-type telomeres.
FIGURE 6.
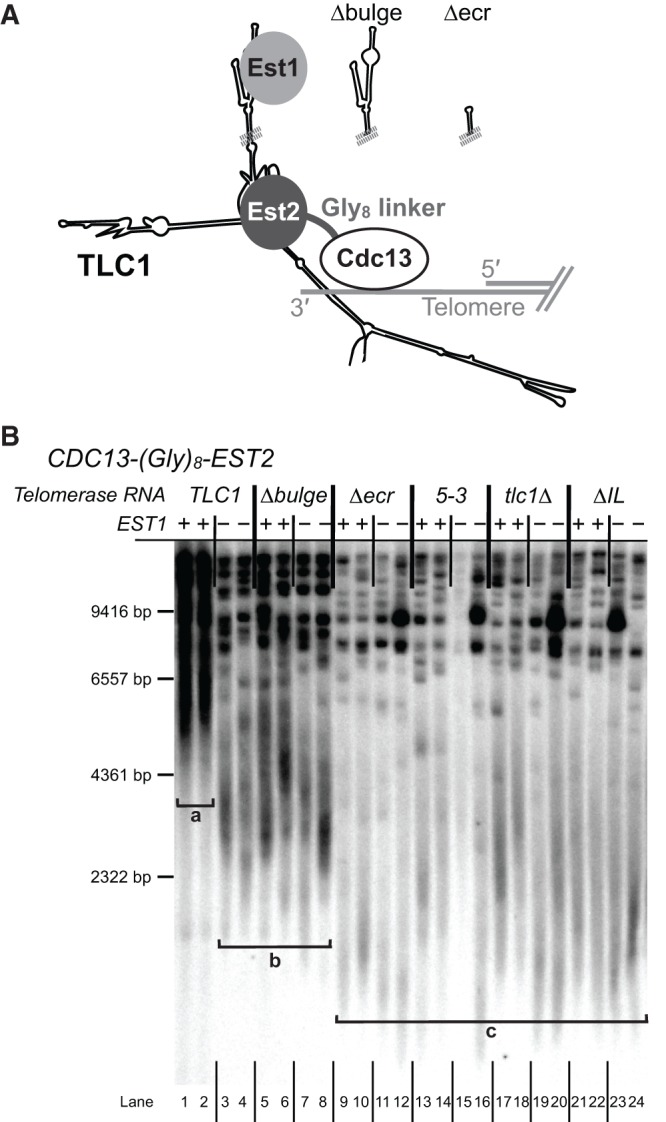
The SEED is essential in a Cdc13-Est2 fusion-protein strain. (A) Schematic showing Est1-bypassing recruitment of telomerase to the telomere in a Cdc13-Est2 protein fusion strain. CDC13-Gly8-EST2 is genomically encoded in either an EST1 or est1Δ background. TLC1 alleles are expressed from centromeric plasmids. (B) Telomere Southern blot from CDC13-EST2 cells from two biological replicates at 500 generations. Brackets indicate different observed classes of telomere clusters: bracket “a,” extremely hyperlengthened telomeres seen in lanes 1 and 2; bracket “b,” stably shorter but still hyperlengthened telomeres seen in lanes 3–8; bracket “c,” telomeres shortening like a tlc1Δ strain, seen in lanes 9–24. Marker sizes are based on [γ-32P]-labeled λHindIII DNA ladder.
We next examined TLC1 alleles with Est1-arm mutations in the CDC13-EST2 strain to test if the Est1-binding or SEED functions are required after recruitment of telomerase to the telomere (see Fig. 6A). Expressing tlc1Δbulge (which disrupts Est1 binding to the RNA but not SEED function) in the CDC13-EST2 EST1 strain resulted in telomere length similar to TLC1 est1Δ CDC13-EST2 cells (see bracket “b” in Fig. 6B; lanes 5 and 6 versus 3 and 4), consistent with the reported activation function of Est1 in telomerase requiring TLC1 binding (Taggart et al. 2002). Furthermore, simultaneous loss of EST1 and the bulge from TLC1 in tlc1Δbulge est1Δ CDC13-EST2 cells resulted in a very similar telomere length phenotype (lanes 7,8). This lack of a genetic interaction provides evidence that these mutations disrupt the same nonessential secondary function of Est1 protein, resulting in stably shorter telomeres (bracket “b” in Fig. 6B).
Having demonstrated that disrupting the Est1-tethering function of the TLC1 Est1 arm results in moderate telomere shortening, we next tested whether the SEED plays a role in telomere maintenance when telomerase recruitment to telomeres via Est1 is bypassed. We expressed tlc1Δecr—which lacks the Est1-binding sites as well as the SEED—in CDC13-EST2 cells, and found that it resulted in loss of telomere maintenance in both EST1 and est1Δ strains (Fig. 6B, lanes 9–12). Telomeres shortened similarly to those of tlc1Δ CDC13-EST2 cells (lanes 17–18), and senescence was apparent in several samples after ∼25 restreaks despite being a RAD52+ strain. Additionally, expression of tlc1ΔIL in CDC13-EST2 cells caused a similar loss of telomere maintenance in both the presence and absence of EST1 (lanes 21–24). Finally, tlc1(5-3), with only 7 nt substitutions, caused tlc1Δ-like telomere shortening in the CDC13-EST2 fusion strains (lanes 13–16). The fact that all of the SEED mutants tested result in a telomere pattern similar to a complete loss of telomerase TLC1 RNA (see bracket “c” in Fig. 6B) indicates that the SEED is essential even when Est2 is covalently fused to Cdc13, suggesting that SEED function is required after recruitment of telomerase to the telomere. In addition, the results indicate that the mechanism of SEED function is independent of Est1 protein once telomerase has been recruited to the telomere.
DISCUSSION
The 1157-nt yeast telomerase RNA TLC1 acts as a flexible scaffold that tethers essential and important accessory subunits to the RNP complex (Zappulla and Cech 2004, 2006; Zappulla et al. 2011; Lebo and Zappulla 2012; Mefford et al. 2013). As additional long noncoding RNAs (lncRNAs) have subsequently been proposed to function through flexible scaffold mechanisms like TLC1 (Chu et al. 2011; Guttman et al. 2011; Wang et al. 2011), this class of RNPs defined by TLC1 (Zappulla and Cech 2004) is being increasingly recognized as an important archetype (Zappulla and Cech 2006; Wang and Chang 2011). While TLC1 has been shown to tether each of the three known accessory subunits—Est1, Ku, and Sm7—to the telomerase RNP without fixing the subunits into precise positions in the RNP (Zappulla and Cech 2004; Zappulla et al. 2011; Lebo and Zappulla 2012; Mefford et al. 2013), it was not known if any of these protein-binding sites on TLC1 RNA had additional roles in RNP function. Although only 33% of TLC1 nucleotides are conserved among species of the same genus, sequence preservation is clearly highest around proposed protein-interacting regions in the catalytic core and at the ends of the three long arms (Dandjinou et al. 2004; Zappulla and Cech 2004; Mefford et al. 2013). In this regard, we have been intrigued by alignments of TLC1 from known Saccharomyces sequences in the Est1-interacting region of TLC1 that show it to be an expansive 108-nt stretch of highly conserved nucleotides (Fig. 1A; Supplemental Fig. S1). This contrasts markedly with the conserved 25 nt shown to bind the ∼152-kDa Ku complex (Peterson et al. 2001; Dalby et al. 2013) and 13-nt conserved nucleotides that bind the ∼91-kDa Sm7 complex (Jones and Guthrie 1990; Seto et al. 1999; Zappulla and Cech 2004; Mefford et al. 2013). The expansive conservation in the Est1-arm of TLC1 and reports of mutations in different positions over this region that disrupt Est1 binding led us to hypothesize that the Est1-arm conserved region comprises a domain with sophisticated structure that is important for functions beyond simply binding Est1 to tether it to the RNP.
Using the MS2 bacteriophage-based RNA-protein interaction system allowed us to separate the primary essential function of the Est1-binding arm—recruitment of Est1 to the RNP (Lundblad and Szostak 1989)—from other roles essential for telomere maintenance (Fig. 1B). This allowed testing TLC1 mutants that would otherwise disrupt Est1 binding, while still promoting Est1 recruitment to the telomerase RNP and led to identification and structural mapping of the Second Essential Est1-arm Domain (SEED).
Our findings expand the flexible scaffold model for yeast telomerase RNA. MS2-tethering Est1 to the tlc1Δbulge RNA rescues telomere maintenance, demonstrating that an essential Est1-binding interface in TLC1 can be functionally replaced with a heterologous RNA-protein direct-binding system (Fig. 1C). Furthermore, like the protein-binding sites on TLC1, the SEED appears to be flexibly scaffolded in the RNP; it retains function when relocated on TLC1 (Zappulla and Cech 2004), and even functions in trans as a separate RNA (Fig. 2). This in trans functionality strongly suggests that the SEED does not act to recruit protein subunits to the RNA scaffold. The presence of the SEED indicates that the accessory-protein-binding arms of TLC1 can have roles in telomere maintenance in addition to flexibly scaffolding the holoenzyme protein components.
Our results indicate that the SEED mechanism of action is Est1-independent, and does not require Est1 protein once telomerase has been recruited to the telomere-binding protein Cdc13. We hypothesize that Est1 first binds to TLC1 to promote telomerase recruitment to the telomere by also binding Cdc13, and then the SEED functions as part of the recruited telomerase RNP•telomere complex.
We find it interesting that Est1 association with the RNA and SEED function are each dependent on the internal loop structure in the Est1-binding arm, despite being at least partially functionally independent. The existence of an internal loop structure below the apical hairpin is conserved across many species of yeast within the Saccharomyces, Kluyveromyces, and Candida genera (Gunisova et al. 2009). Even reported models for the distantly related fission yeast S. pombe appears to have a large internal loop in the Est1-binding arm, although nucleotide sequence in the region is not conserved (Webb and Zakian 2012; DC Zappulla, unpubl.). In fact, the conserved “CS2a” region of TLC1, which includes the 5′ side of the internal loop, is even more conserved among budding yeast than the essential bulge-containing “CS2” region, with relatively high conservation even through the Candida genus (Gunisova et al. 2009).
Previously, phylogenetic models predicted two possible structures for the internal loop: one large loop (Zappulla and Cech 2004) or two smaller loops separated by a 3-bp helix (Fig. 4; Dandjinou et al. 2004). Our SHAPE analysis on purified Est1-arm RNA suggests the former conformation dominates in vitro in the absence of protein (Fig. 3A; Supplemental Fig. 3). In addition, phylogenetic analysis showed that just two of the A residues, and none of the pair-partnering U residues, were conserved among TLC1 sequences, and there is no covariation evident that would suggest conserved base-pairing (Fig. 1A; Supplemental Fig. S1). Together, these data support a single, large internal loop secondary structure as the most favorable conformation for the naked RNA. However, it remains quite possible that limited pairing across this internal loop occurs in the ribonucleoprotein complex, either transiently or in equilibrium with the unpaired loop in the cell. Consistent with this, we and other groups have shown that pairing across the internal loop can have potent effects on telomerase function (Fig. 5; Lubin et al. 2012; Laterreur et al. 2013).
The seemingly small secondary structure difference between the two possible internal loop folding states may have a large effect on the tertiary RNA structure in the region. We used bioinformatic software to model the 3D structure of the 193-bp Est1 arm (Fig. 7; Popenda et al. 2012). We first input the nucleotide sequence along with secondary structure information based on the model for the wild-type TLC1 Est1 arm with the large, single internal loop (Fig. 7B). The 3D structure prediction shows that the bulge and hinge domains come into close proximity; since both of these domains have been implicated in Est1 association with the RNA, this tertiary structure may represent an important Est1-binding site. As for the internal loop containing the SEED, it is predicted to bend sharply, forming a kink that imparts a large-scale conformational change, orienting the apical hairpin back toward the hinge-hairpin and exposing nucleotides along the internal loop required for SEED function.
FIGURE 7.
Three-dimensional computational modeling of the TLC1 Est1-arm conserved region. (A) TLC1 Est1-arm conserved region secondary structure, with sub-elements colored. (Blue) 5′ side of the internal loop, (green) 3′ side of internal loop, (orange) apical hairpin, (yellow) hinge-hairpin, and (red) bulge. Open circles indicate the potential A–U pairs across the internal loop. (B) Three-dimensional structure prediction by RNAComposer of the Est1-arm conserved region with unpaired internal loop. Inset shows internal loop secondary structure, with the three unpaired A and U residues boxed. The structure of the entire 193-nt Est1 arm (nucleotides 508–700) was modeled. (C) Three A–U base pairs in the internal loop lead to prediction of a very different tertiary structure with coaxially stacked RNA helices. Inset shows the corresponding secondary structure, with the A–U pairs boxed.
Next, based on our results with tlc1(5-3) (Fig. 5C) and the slightly different secondary structure proposed for this region previously (Dandjinou et al. 2004), we computationally modeled the same wild-type Est1-arm conserved region, but this time with the three A–U pairs across the internal loop (Fig. 7C). The modeling shows the three base pairs in the internal loop dramatically switching the overall 3D conformation of the Est1-arm conserved region; it is no longer predicted to be bent, but rather the apical hairpin coaxially stacks with the helix between the internal loop and the bulge, thus projecting away from the catalytic core. This tertiary structure is consistent with our SHAPE results that showed the internal loop nucleotides being less exposed in this conformation (Fig. 5C). Thus, 3D modeling helps explain how the secondary structure within the internal loop could cause large-scale conformational changes in the tertiary structure of the Est1 arm of yeast telomerase RNA.
We hypothesize that modulating the secondary structure of the internal loop may act as a regulatory mechanism, switching the 3D structure between active and inactive SEED states. It is not yet known precisely where Est1 protein binds the internal loop, although binding does require at least some unpaired nucleotides in the internal loop and is also at least partially affected by nucleotide substitutions in the 5′ part of the loop (Lubin et al. 2012). The SEED, however, appears to be inactive in the partially paired internal-loop conformation (see mutant 5-3 in Fig. 5). Instead, the SEED is functional when the internal loop is more flexible (see mutant 5-1 in Fig. 5), which may allow for a tertiary structure that exposes nucleotides along the loop (Fig. 7B). Although the essential SEED function is Est1 independent, it remains possible that Est1 protein binding has a role in modulating the conformational state of the SEED. Since both Est1-binding and SEED nucleotides are essential, future studies examining the relationship of these functions will be facilitated by identification of more conditional and partial loss-of-function alleles that individually affect these processes. These studies will extend our results based on MS2-tethering Est1 to telomerase as a means to parse Est1-binding and SEED functions of the Est1 arm.
The trans functionality is evidence that the SEED does not function in RNP assembly or recruiting protein components, but rather affects telomerase action in a different way. We do not favor the hypothesis that the SEED activates the yeast telomerase reverse transcriptase (TERT), Est2. Although in organisms other than Saccharomyces it has been shown that telomerase activity in vivo and in vitro requires a portion of telomerase RNA in addition to the central core that can function in trans (Tesmer et al. 1999; Mitchell and Collins 2000; Mason et al. 2003; Qi et al. 2013), simply the central core of TLC1—entirely without the Est1 arm—is sufficient to reconstitute robust yeast telomerase activity with Est2 in vitro (Qiao and Cech 2008; Mefford et al. 2013). This suggests the SEED is not analogous to the human CR4/5, the ciliate stem-terminus element, or the three-way junction in other yeasts (Tesmer et al. 1999; Mitchell and Collins 2000; Theimer and Feigon 2006; Brown et al. 2007). Instead, we favor the hypothesis that SEED mechanism involves a telomerase-regulating protein such as Cdc13 or the also-essential telomerase RNP component Est3. Since the SEED appears to function after telomerase has been recruited to the telomere by way of Est1-Cdc13 interaction, it is more likely that it promotes the telomerase-extendible state of the telomere and/or permits multiple rounds of template-directed telomerase activity (Fig. 8).
FIGURE 8.
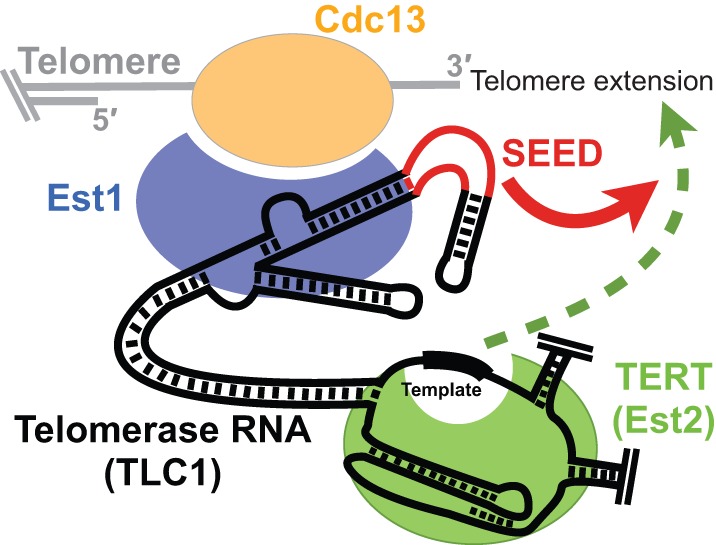
Model for SEED function in telomerase. The SEED plays a role in telomere synthesis downstream from Est1-Cdc13-mediated telomerase recruitment to the telomere. It is unlikely to directly activate TERT enzymatic function, but may be required to establish the extendable state of the telomere and/or increase telomerase holoenzyme activity.
While this work was under review, a publication reported a “telomerase-stimulating structure” (TeSS) in the conserved region of Est1 (Laterreur et al. 2013), mapping to the apical hairpin/upper portion of the internal loop of the Est1-binding arm. The mutants analyzed showed a nonessential function for this region, causing shortened telomeres but not senescence. The TeSS and SEED reported here may represent distinct functional domains. Alternatively, the TeSS mutants may be altering the structure of the internal loop, partially disrupting the SEED without fully abolishing SEED function.
In summary, we have demonstrated that a second essential function exists for the Est1-binding arm of yeast telomerase RNA, in addition to tethering the protein to the RNP. This Second Essential Est1-arm Domain is located in the base of the internal loop of the Est1-arm conserved region. Despite also mapping to a region of TLC1 implicated in binding Est1 protein, the SEED can function in trans and its role in telomere synthesis does not require Est1. Dynamic tertiary structural conformations of the Est1 arm may regulate SEED function. In summary, the SEED represents a key, nonscaffolding domain of TLC1 RNA required for telomerase-mediated telomere extension.
MATERIALS AND METHODS
Phylogenetic analysis of TLC1
To analyze the Est1-binding arm in the context of an alignment of entire TLC1 sequences, we aligned 36 unique TLC1 sequences from NCBI ascribed to seven Saccharomyces species: cerevisiae, paradoxus, pastorianus, cariocanus, kudriavzevii, mikatae, and bayanus. The alignment was viewed using Jalview (Waterhouse et al. 2009) to generate what is shown in Supplemental Figure S1. Percent conservation values were calculated by comparing the number of 100% conserved residues (green in Fig. 1A; Supplemental Fig. S1) or total number of conserved and covarying nucleotides (green, red, and blue) by the length of the alignment for each section.
Creation of a heterologous binding interaction system for tethering Est1 protein to TLC1 RNA in vivo
We used the MS2 bacteriophage coat protein-RNA interaction system to tether Est1 to TLC1 RNA. For the MS2 hairpin-tagged TLC1 RNA, a version of the gene with 10 MS2 hairpins inserted at position 1135 was used (Gallardo et al. 2011). In order to tag various TLC1 alleles with the MS2 hairpins in the same vector backbones, we first cloned the BclI to NsiI fragment, including the MS2 sites, into a pRS314-based centromere-containing (CEN) vector harboring TLC1, including ∼518 bp of genomic sequence upstream of and 795 bp of genomic sequence downstream from TLC1 (pSD107), to make pDZ641. This fragment was also cloned into tlc1Δbulge vector pAS558 (Seto et al. 2002) to make tlc1Δbulge-MS2 (pDZ353) and tlc1Δecr vector pDZ428 to make tlc1Δecr-MS2 (pDZ432). See Supplemental Figure S2A for TLC1-MS2 Mfold-predicted secondary structure.
In order to carboxy-terminally tag Est1 with two tandem copies of the bacteriophage MS2 coat protein domain (MS2CP), we PCR amplified the MS2CP2 coding region using an MS2CP-containing plasmid, pRS426-TAP-MS2CP2 (Gallardo et al. 2011), using primers that insert eight glycine residues (Gly8) upstream, which has been shown to maintain function of carboxy-terminally tagged Est1 (Sabourin et al. 2007). This fragment was cloned into PacI/AscI-digested pFA6a-3HA-His3MX6 vector (Longtine et al. 1998). We next PCR amplified Gly8-MS2CP::His3MX6 and integrated it in place of the stop codon of EST1 in the strain TCy127 (MATa, ADE2, his3Δ, leu2Δ0, lys2Δ0, trp1Δ63, ura3Δ0, RAD52, tlc1::KanR, pTLC1-URA3) (Mozdy and Cech 2006) to make EST1-Gly8-MS2CP2 strain yDZ385. Correct integration was confirmed by colony PCR.
In trans Est1-arm expression in yeast
The Est1 arm of TLC1 (nucleotides 514–694) was amplified using PCR primers that add a five C or G residues to either side, respectively, designed to form G–C base pairs at the base of the RNA's secondary structure to help promote proper folding. The product was cloned into SpeI/XhoI-digested 2µ vector p425TEF, harboring the TEF2 promoter and a CYC1 terminator (Russo and Sherman 1989; Mumberg et al. 1995). The in trans Est1-arm transcript is predicted to include additional nucleotides from the promoter and terminator regions; Mfold predicts proper folding of the Est1 arm in the transcript. See Figure 2 for in trans Est1-arm Mfold-predicted secondary structure.
Est1-arm mutants
We deleted the TLC1 Est1-arm conserved region by ligating two PCR products, resulting in tlc1Δecr (TLC1(552–662::C); pDZ428 without MS2s, pDZ432 with MS2s). Est1-arm conserved region sub-element deletions TLC1ΔAH (Δ614–632; pDZ767 without MS2s, pDZ760 with MS2s) and tlc1ΔIL (Δ605–613, 633–639; pDZ768 without MS2s, pDZ761 with MS2s) were also made using PCR fragments. For tlc1ΔHH, deletion of the hinge-hairpin domain alone was insufficient for fully disrupting hinge function, presumably due to remaining unpaired nucleotides near the native HH locus (Fig. 4). Therefore, we used a stiffened Est1 arm, with all unpaired nucleotides deleted (Lebo and Zappulla 2012). NcoI–HpaI and HpaI–MluI gene fragments were synthesized, sequence-verified, and subcloned by GenScript (Piscataway). Then, the two fragments were cloned into NcoI/MluI-digested vector pDZ386 (harboring TLC1 with a stiffened Est1 arm) (Lebo and Zappulla 2012), creating tlc1ΔHH (pDZ389 without MS2s, pDZ794 with MS2s). We created the internal loop sequence mutations in TLC1-MS2 using PCR fragments (3-1 = pDZ766, 5-1 = pDZ791, 5-2 = pDZ765, 5-3 = pDZ788). All Est1-arm mutants were cloned into CEN plasmids expressing TLC1 from its endogenous promoter, either in pSD107 (TLC1) or pDZ641 (TLC1-MS2). Mfold software secondary structure predictions were used to guide the creation of all mutants, to reduce the likelihood of off-target effects on folding (Zuker and Jacobson 1998; Zuker 2003). See Supplemental Figure S2A for Mfold lowest-free-energy structure models of each mutant.
Senescence experiments in yeast
We transformed TRP1-marked CEN plasmids harboring TLC1 alleles into haploid S. cerevisiae strain TCy127 (Mozdy and Cech 2006) for testing in the absence of MS2-tethering, or yDZ385 (MATa, ADE2, his3Δ, leu2Δ0, lys2Δ0, trp1Δ63, ura3Δ0, RAD52, tlc1::KanR, pTLC1-URA3, EST1-Gly8-MS2CP2::HIS3) for testing in the presence of Est1-MS2CP. The pTLC1-URA3 cover plasmid was shuffled out using counter-selection on 5-fluoroacetic acid (5-FOA), and colonies were restreaked sequentially 10 times on synthetic-complete medium lacking tryptophan. Each colony-forming unit was estimated to represent 25 generations. Colonies were visually monitored for signs of senescence. Due to the presence of RAD52 in the strains, “survivor” colonies sometimes occur; these cells maintain telomeres through a telomerase-independent recombination-based pathway (Lundblad and Blackburn 1993). Senescence was indicated by an obvious decrease in colony viability before the onset of any survivors. Additionally, telomeric Southern blotting was used to verify telomerase-independent survivorship by the characteristic telomeric restriction fragment patterns (see Fig. 3D; Lundblad and Blackburn 1993; Teng and Zakian 1999). Cells were restreaked a minimum of 10 times (∼250 generations) to test for late-senescence phenotypes.
For the in trans Est1-arm tests (Fig. 2), we co-transformed the LEU2-marked 2µ plasmid into cells along with the TRP1-marked TLC1-allele-harboring plasmids. After shuffling out pTLC1WT-URA3 using 5-FOA-containing medium, cells were restreaked on synthetic complete medium lacking tryptophan and leucine, while carefully monitoring for senescence.
All in vivo experiments were performed multiple times.
Cdc13-Est2 fusion-protein experiments
We created a strain expressing CDC13-Gly8-EST2 from the genomic CDC13 locus. To do this, we amplified EST2 from the genome by PCR using primers to add eight glycine residues (Gly8) to the 5′ end of the open reading frame. The product was cloned into PacI/AscI-digested pFA6a-3HA-His3MX6 (Longtine et al. 1998) to make pDZ699. Gly8-EST2::HIS3 was PCR-amplified off of the plasmid using primers containing 50 bp of homology with the 3′ end of CDC13 on both sites of the product. The insertion cassette was transformed into TCy127 for integration in place of the CDC13 stop codon, and was confirmed by PCR to have created CDC13-Gly8-EST2 (yDZ453). The est1Δ CDC13-EST2 strain was made by knocking out EST1 in yDZ453. This was achieved using Candida glabrata LEU2 (a gift from Aaron Neiman, Stony Brook University), PCR-amplified with primers adding 50 bp of homology upstream of and downstream from EST1. The product was transformed into yDZ453 cells and the est1Δ genotype was then confirmed by PCR (yDZ485). Telomeres in the CDC13-EST2 and est1Δ CDC13-EST2 strains were longer than previously reported (Evans and Lundblad 1999); this is likely due to genomic expression of Cdc13-Est2, instead of plasmid-based expression.
Est1-arm mutants on TRP1-CEN plasmids were transformed into yDZ453 or yDZ485 cells. After pTLC1-URA3 was shuffled out on solid synthetic medium containing 5-FOA, cells were restreaked on solid synthetic complete media lacking tryptophan for 30 restreaks (∼750 generations).
Southern blots
Southern blots were performed as previously described (Zappulla and Cech 2004; Zappulla et al. 2011). Cell pellets were prepared from liquid cultures grown from serially restreaked plates, and genomic DNA was isolated (Gentra Puregene system). Equal amounts of DNA were digested with XhoI, and electrophoresed through a 1.1% agarose gel for 17 h at 70 V. DNA was transferred by capillary action to a Hybond-N+ Nylon membrane (GE), and probed for telomeric sequence. A nontelomeric 1627-bp fragment probe of chromosome IV was also included as an internal control. Average Y′ telomere fragment length was quantified using the weighted average mobility (WAM) assay (Zappulla et al. 2011).
Northern blots
Northern blots were performed as previously described (Zappulla et al. 2005). Briefly, total cellular RNA was isolated from yeast cultures using a modified hot-phenol RNA isolation method (Köhrer and Domdey 1991). After boiling, ∼10 µg of total RNA was separated by Urea-PAGE, transferred to a Hybond-N+ Nylon Membrane (GE), UV-crosslinked (Spectrolinker XL-1500 UV Crosslinker, “Optimal Crosslink” setting), and prehybridized in Church buffer for 10 min at 55°C. The membrane was then probed for the 3′ end of TLC1 (nucleotides 906–1140; Lebo and Zappulla 2012) or for the Est1 arm (nucleotides 504–704), and for the U1 snRNA (Friedman and Cech 1999). Relative abundances were determined by normalized TLC1 or Est1-arm levels to U1 snRNA. Since U1 is far more abundant, 100-fold less U1 probe was used relative to TLC1 probe.
SHAPE analysis of purified TLC1 Est1-arm RNAs
A 181-nt TLC1 Est1-arm RNA (nucleotides 514–694) was T7-transcribed along with 5 C–G base pairs added to the base of the arm to drive folding at the core-proximal side (as performed with the in trans arm construct; see Fig. 2A). SHAPE was performed as recently described (Niederer and Zappulla 2015). Of note, 2 pmol of purified TLC1 Est1-arm RNA suspended in 8 μL 0.5× TE was denatured at 95°C for 2 min, then cooled on ice for 3 min. On ice, samples were treated with 6 μL 3.3× folding buffer (333 mM HEPES pH 8.0, 333 mM NaCl, 20 mM MgCl2). After folding, RNAs were treated with 1 μL 65 mM N-methylisatoic anhydride (NMIA) in anhydrous DMSO and incubated at 37°C for 22 min (Steen et al. 2012). No-NMIA control reactions were treated with 1 μL of DMSO. RNAs were recovered by ethanol precipitation and resuspended in 9 μL 0.5× TE.
Primer extensions were performed as described in the SuperScript III reverse transcriptase protocol (Invitrogen) with the following minor adjustments. Of note, 1 pmol of each RNA was denatured at 95°C for 1 min, then cooled on ice for 2 min. Three microliters of [γγ−32P]-end-labeled primer DZ1348 (CCAACAGGATCCGGATGGCCTATCCTGGGCCGTTCCCTGACGTTCTTTTTCCTTTTTC) was added and the mixture was incubated at 65°C for 2 min, followed by annealing at 35°C for 10 min. The supplied reverse transcription buffer was added along with 5 mM DTT (final concentration), 1 mM dNTPs (final total concentration) and 100 units of SuperScript III. The mixture was incubated at 52°C for 10 min prior to analysis on a 10% polyacrylamide sequencing gel (Supplemental Fig. 3).
Modification intensities at each position were quantified using SAFA (Das et al. 2005). Results were adjusted by selecting a reference band with the least variability between lanes, calculating the factor by which the experimental band varied from the reference, and adjusting each lane by the calculated scaling factor necessary to make the reference band equal across all lanes. Normalized SHAPE reactivity was calculated by subtracting the intensity of the corresponding DMSO-only control band from the +NMIA band. To determine the SHAPE-reactivity pattern the average of the 10 most reactive nucleotides was set to 100% (i.e., the maximum reactivity). Relative reactivity was determined for each remaining nucleotide in relation to the maximum (Figs. 3A, 5C). Relative reactivity values >70% were considered highly reactive, while values between 40% and 70% were considered moderately reactive.
RNA three-dimensional structure modeling
We generated dot-bracket notations for the phylogenetically supported Est1-arm secondary structure model (i.e., nucleotides 508–700). We then also made a modified version of the dot-bracket structure, with three A–U pairs in the internal loop, formed between nucleotides 609–611 and 635–637. The wild-type Est1-arm sequence was submitted alongside each dot-bracket structure to the RNAComposer Automated RNA Structure 3D Modeling Server (Popenda et al. 2012).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Zappulla laboratory for comments on the manuscript and Sarah Woodson for help with SHAPE. We also thank Raymund Wellinger (Université de Sherbrooke) for sharing TLC1-(MS2)10 and data prior to publication. This work was supported by US National Institutes of Health (National Institute of General Medical Sciences) funding from R00 GM80400 to D.C.Z. and startup funds from The Johns Hopkins University. K.J.L. and R.O.N. have been supported by National Institutes of Health Cellular and Molecular Biology graduate student training grant 2T32 GM007231.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.049379.114.
REFERENCES
- Abdallah P, Luciano P, Runge KW, Lisby M, Géli V, Gilson E, Teixeira MT 2009. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol 11: 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH 2006. A history of telomere biology. In Telomeres (ed. de Lange T, Lundblad V, Blackburn EH), pp. 1–19 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y 2007. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol Cell 44: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH 1995. Telomerase in yeast. Science 269: 396–400. [DOI] [PubMed] [Google Scholar]
- Dalby AB, Goodrich KJ, Pfingsten JS, Cech TR 2013. RNA recognition by the DNA end-binding Ku heterodimer. RNA 19: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandjinou AT, Lévesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ 2004. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V 2002. The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Cech TR 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev 13: 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo F, Laterreur N, Cusanelli E, Ouenzar F, Querido E, Wellinger RJ, Chartrand P 2011. Live cell imaging of telomerase RNA dynamics reveals cell cycle-dependent clustering of telomerase at elongating telomeres. Mol Cell 44: 819–827. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337. [DOI] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L 2009. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Guthrie C 1990. Unexpected flexibility in an evolutionarily conserved protein–RNA interaction: genetic analysis of the Sm binding site. EMBO J 9: 2555–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhrer K, Domdey H 1991. Preparation of high molecular weight RNA. Methods Enzymol 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Laterreur N, Eschbach SH, Lafontaine DA, Wellinger RJ 2013. A new telomerase RNA element that is critical for telomere elongation. Nucleic Acids Res 41: 7713–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo KJ, Zappulla DC 2012. Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA 18: 1666–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V 1996. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH 2004. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci 101: 14713–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V 1997a. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci 94: 11190–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR 1997b. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR 2002. Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lubin JW, Tucey TM, Lundblad V 2012. The interaction between the yeast telomerase RNA and the Est1 protein requires three structural elements. RNA 18: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73: 347–360. [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643. [DOI] [PubMed] [Google Scholar]
- Mason DX, Goneska E, Greider CW 2003. Stem-loop IV of Tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol 23: 5606–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford MA, Rafiq Q, Zappulla DC 2013. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. EMBO J 32: 2980–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell 6: 361–371. [DOI] [PubMed] [Google Scholar]
- Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS 2002. Conserved structure for single-stranded telomeric DNA recognition. Science 296: 145–147. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Niederer RO, Zappulla DC 2015. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 21: 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67. [DOI] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW 2012. Automated 3D structure composition for large RNAs. Nucleic Acids Res 40: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li Y, Honda S, Hoffmann S, Marz M, Mosig A, Podlevsky JD, Stadler PF, Selker EU, Chen JJ 2013. The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res 41: 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P, Sherman F 1989. Transcription terminates near the poly(A) site in the CYC1 gene of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 86: 8348–8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Fisher TS, Zakian VA 2007. A flexible protein linker improves the function of epitope-tagged proteins in Saccharomyces cerevisiae. Yeast 24: 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR 2003. A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9: 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen KA, Rice GM, Weeks KM 2012. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J Am Chem Soc 134: 13160–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Teng SC, Zakian VA 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297: 1023–1026. [DOI] [PubMed] [Google Scholar]
- Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem 286: 26431–26439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Zakian VA 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE 1999. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol 19: 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J 2006. Structure and function of telomerase RNA. Curr Opin Struct Biol 16: 307–318. [DOI] [PubMed] [Google Scholar]
- Tucey TM, Lundblad V 2013. A yeast telomerase complex containing the Est1 recruitment protein is assembled early in the cell cycle. Biochemistry 52: 1131–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Wu Y, Chan A, Zakian VA 2011. The Saccharomyces cerevisiae telomerase subunit Est3 binds telomeres in a cell cycle- and Est1-dependent manner and interacts directly with Est1 in vitro. PLoS Genet 7: e1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. 2011. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA 2012. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase–telomere association. Genes Dev 26: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zakian VA 2011. The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc Natl Acad Sci 108: 20362–20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2004. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR 2006. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077. [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich KJ, Arthur JR, Gurski LA, Denham EM, Stellwagen AE, Cech TR 2011. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA 17: 298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Jacobson AB 1998. Using reliability information to annotate RNA secondary structures. RNA 4: 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.