Abstract
Protein turnover is an important regulatory mechanism that facilitates cellular adaptation to changing environmental conditions. Previous studies have shown that ribosome abundance is reduced during nitrogen starvation by a selective autophagy mechanism termed ribophagy, which is dependent upon the deubiquitinase Ubp3p. In this study, we asked whether the abundance of various translation and RNA turnover factors are reduced following the onset of nitrogen starvation in Saccharomyces cerevisiae. We found distinct differences in the abundance of the proteins tested following nitrogen starvation: (1) The level of some did not change; (2) others were reduced with kinetics similar to ribophagy, and (3) a few proteins were rapidly depleted. Furthermore, different pathways differentially degraded the various proteins upon nitrogen starvation. The translation factors eRF3 and eIF4GI, and the decapping enhancer Pat1p, required an intact autophagy pathway for their depletion. In contrast, the deadenylase subunit Pop2p and the decapping enzyme Dcp2p were rapidly depleted by a proteasome-dependent mechanism. The proteasome-dependent depletion of Dcp2p and Pop2p was also induced by rapamycin, suggesting that the TOR1 pathway influences this pathway. Like ribophagy, depletion of eIF4GI, eRF3, Dcp2p, and Pop2p was dependent upon Ubp3p to varying extents. Together, our results suggest that the autophagy and proteasomal pathways degrade distinct translation and RNA turnover factors in a Ubp3p-dependent manner during nitrogen starvation. While ribophagy is thought to mediate the reutilization of scarce resources during nutrient limitation, our results suggest that the selective degradation of specific proteins could also facilitate a broader reprogramming of the post-transcriptional control of gene expression.
Keywords: autophagy, proteasome, nitrogen starvation, translation factors, RNA turnover factors
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is an important mechanism used by eukaryotic cells to degrade cytosolic contents and recycle the resulting building blocks for the synthesis of new macromolecules during stress conditions. Autophagy in yeast occurs mainly in response to nutrient limitation (Takeshige et al. 1992). In this process, portions of the subcellular environment are sequestered into de novo formed double membrane vesicles and routed to the yeast vacuole (or mammalian lysosome) where the contents are degraded. The resulting degradative products are transported back into the cytoplasm through vacuolar permeases to facilitate their reuse in biosynthetic pathways. In this way, autophagy maintains cytoplasmic amino acid levels and basal protein synthesis during starvation conditions (Onodera and Ohsumi 2005; Yang et al. 2006; Yang and Klionsky 2007).
The target of rapamycin (TOR1) kinase functions as an important sensor of nitrogen and amino acid availability in eukaryotic cells. Addition of the TOR1 inhibitor, rapamycin, induces autophagy during nutrient-rich growth (Noda and Ohsumi 1998). TOR1 regulates the autophagy pathway by direct phosphorylation of Atg13p (Kamada et al. 2010). TOR1 kinase activity is rapidly inhibited under starvation conditions, allowing dephosphorylated Atg13p to accumulate and induction of autophagy to occur. However, TOR1 function is gradually reactivated in an autophagy-dependent manner during prolonged starvation in both yeast and mammalian cells, suggesting that TOR1 reactivation may play a role in the attenuation of autophagy (Yu et al. 2010; Shin and Huh 2011).
Until recently, autophagy was thought to facilitate random degradation of cytoplasmic macromolecules and organelles. However, the involvement of ubiquitin as a specificity factor for selective autophagy is emerging, and recent evidence suggests that crosstalk exists between proteasome-mediated degradation and selective autophagy (Kraft et al. 2010). Many examples of selective autophagy have been reported. One of these, ribophagy, specifically degrades ribosomes upon nitrogen starvation. Both 40S and 60S ribosomal subunits are degraded by ribophagy, but only 60S ribosomal subunit decay requires the activity of the deubiquitinase Ubp3p (Kraft et al. 2008). Kraft et al. (2008) found that Rpl25p and other 60S ribosomal subunit proteins were enriched with ubiquitin conjugates in a ubp3Δ strain during nitrogen starvation, suggesting that deubiquitination of ribosomal proteins may facilitate degradation of 60S subunits during nitrogen starvation. In addition, Ubp3p has been shown to physically interact with Cdc48p, a key component of the ubiquitin–proteasome system. Cdc48p binds to multiple cofactors, including the deubiquitinase Ubp3p and the ubiquitin ligase Ufd3p (Rumpf and Jentsch 2006; Dargemont and Ossareh-Nazari 2012). The interaction of these distinct classes of ubiquitin-processing enzymes with Cdc48p is thought to make it a “decision platform” for substrate ubiquitination. Finally, the ribosome-associated E3 ubiquitin ligase Ltn1p/Rkr1p was recently found to antagonize Ubp3p activity and act as an inhibitor of ribophagy during growth in nutrient-rich conditions (Ossareh-Nazari et al. 2014).
Besides ribosomes, a small subset of cytoplasmic proteins, including eIF4GI, have been shown to undergo selective degradation during nitrogen starvation in Saccharomyces cerevisiae (Berset et al. 1998; Gelperin et al. 2002; Onodera and Ohsumi 2004; Shimobayashi et al. 2010). eIF4GI is also degraded upon rapamycin addition, while other translation initiation factors such as eIF4E and eIF4A remain stable under these conditions in both mammalian and yeast cells (Berset et al. 1998; Powers and Walter 1999; Kuruvilla et al. 2001; Ramirez-Valle et al. 2008). The selective degradation of eIF4GI during nitrogen starvation is interesting from the gene regulation standpoint since depletion of eIF4GI in mammalian cells by shRNA knockdown not only decreases the translation of mRNAs involved in cell proliferation, but also promotes the induction of autophagy (Ramirez-Valle et al. 2008). Deletion of the yeast TIF4631 gene encoding eIF4GI impairs global translation initiation rates and cell growth (Clarkson et al. 2010). These results suggest that the abundance of eIF4GI broadly influences the translation of many classes of mRNAs.
Given the previous findings that ribosomes and eIF4GI undergo selective degradation during nitrogen starvation, we examined the fate of 14 translation and mRNA decay factors under these conditions. We found that two translation factors, eRF3 and eIF4GI, are rapidly degraded by autophagy in a manner that requires the ribophagy deubiquitinase Ubp3p. Furthermore, two mRNA turnover factors, Dcp2p and Pop2p, were depleted during nitrogen starvation at a faster rate than eRF3 or eIF4GI. The decreased abundance of Dcp2p and Pop2p was Ubp3p-dependent, but also required the proteasome pathway in a manner that was repressed by TOR1 during nutrient-rich conditions. We also found that Ubp3p itself is depleted during nitrogen starvation. Taken together, our data show that Ubp3p mediates the depletion of a subset of translation and RNA turnover factors by both the proteasome and autophagy pathways during nitrogen starvation.
RESULTS
A subset of translation and RNA turnover factors are degraded by autophagy during nitrogen starvation
Previous studies have shown that nitrogen starvation causes the selective depletion of ribosomes through a process called ribophagy (Kraft et al. 2008; Ossareh-Nazari et al. 2010). To initially explore ribophagy in our strains, we monitored the abundance of the endogenous large ribosomal subunit protein Rpl3p, as well as the 18S and 25S rRNAs during nitrogen starvation. We found that the initial abundance of Rpl3p and both rRNAs was reduced by twofold within 24 h of the onset of nitrogen starvation (Fig. 1A); no further decrease was observed after 48 h of starvation. Thus, ribophagy decreased ribosome content by up to twofold in our strains following the onset of nitrogen starvation. Thereafter, ribosome abundance reequilibrated at a new steady-state level.
FIGURE 1.

A subset of translation and RNA turnover factors are rapidly degraded by autophagy following the onset of nitrogen starvation. (A) Western blots of Rpl3p and Pgk1p, and an RNA gel of rRNA from cells harvested at various times following the onset of nitrogen starvation. Quantitation of Rpl3p (center) and rRNA (right) abundance is also shown. (B) Western blots of translation and RNA turnover factors from a wild-type strain whose abundance did not decrease following a shift to nitrogen starvation (−N). (C) Western blots of translation and RNA turnover factors in wild-type and atg7Δ strains that showed a modest decrease in abundance following a shift to nitrogen starvation (−N). (D) Western blots of translation and RNA turnover factors in wild-type and atg7Δ strains that showed a significant decrease in abundance following a shift to nitrogen starvation (−N). A shift to glucose starvation (−Glu) for the indicated times was also examined. (E) Quantitation of Western blots from D for proteins showing the largest changes in abundance after exposure to nitrogen starvation (−N) for the indicated times (compared with the ribophagy control, Rpl3p). (F) Western blots of eRF3 and eIF4GI in an atg19Δ strain after exposure to nitrogen starvation (−N) for the indicated times. Quantitation is shown to the right. Protein abundance was normalized to Pgk1p from the same extract as an internal control. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation.
This decrease in ribosome content upon nitrogen starvation led us to inquire about the fate of other proteins associated with the post-transcriptional control of gene expression. To do this, we examined the fate of nine translation factors and five RNA decay factors following the onset of nitrogen starvation. The choice of these proteins was primarily based on the availability of specific antibodies or epitope-tagged expression clones of each. Changes in the abundance of these proteins were compared with the ribophagy control protein Rpl3p, whose abundance routinely decreased 10%–25% in the first 6 h following nitrogen depletion. The cytosolic protein Pgk1p and the mitochondrial protein Tom70p were used as controls and found to be stable under the conditions used (Fig. 1B–D).
We found that the abundance of three translation factors (eIF2α, eIF5B, and eEF3) and two RNA turnover factors (Lsm1p and Tpa1p) did not change appreciably during the first 6 h of nitrogen starvation (Fig. 1B). Five other proteins (the translation factors eIF5A, eEF1A, eEF2, eRF1, and the poly(A) binding protein Pab1p) exhibited modest (20%–40%) decreases in abundance following the onset of nitrogen starvation, roughly similar to the ribophagy marker Rpl3p (Fig. 1C). To examine the role of autophagy in reducing the abundance of these latter proteins upon nitrogen starvation, we utilized an atg7Δ strain. Atg7p shares homology with E1 activating enzymes and mediates the conjugation of Atg12p with Atg5p, and Atg8p with phosphatidylethanolamine. The absence of Atg7p blocks autophagosome formation, an essential early step of autophagy (Feng et al. 2014). We found that the decrease in the abundance of these proteins (as well as the appearance of a degradation product for eIF5A) was blocked in the atg7Δ strain in a manner similar to Rpl3p (Fig. 1C). These results suggest that the reduced abundance of these proteins is due to degradation by the autophagy pathway.
Interestingly, we found that the abundance of three other proteins, the translation factors eIF4GI and eRF3, as well as the decapping enhancer Pat1p, was reduced much more rapidly than Rpl3p, with decreases of four- to fivefold within 4 h of nitrogen starvation (Fig. 1D,E). The decreased abundance of these proteins was also mediated primarily by autophagy, since they were stabilized in an atg7Δ strain under the same conditions. However, the inability of the atg7Δ mutation to completely block the decrease of these proteins may suggest that another mechanism, such as reduced synthesis, may also contribute to their decrease. The degradation of these proteins is specific for nitrogen starvation, since the onset of glucose starvation did not induce eIF4GI degradation as previously reported (Gelperin et al. 2002) or eRF3 degradation (Fig. 1D). These results demonstrate that the abundance of a subset of translation and mRNA turnover factors is rapidly reduced following the onset of nitrogen starvation, and much of this decrease is dependent upon the autophagy pathway.
The cytoplasm to vacuole (Cvt) pathway is a specialized form of autophagy that delivers at least two hydrolases, α-mannosidase (Ams1) and aminopeptidase I (Ape1), to the vacuole. During assembly of the Cvt complex, Atg19p serves as a cargo receptor (Lynch-Day and Klionsky 2010). To test whether the Cvt pathway also participates in the preferential degradation of eIF4GI and eRF3 during nitrogen starvation, we examined the effect of an atg19Δ mutation on their depletion. We found that the decreased abundance of both proteins was unaffected by the atg19Δ mutation (Fig. 1F), indicating that the Cvt pathway is not responsible for their turnover.
To confirm that the reduced abundance of eRF3 and eIF4GI was mediated (at least in part) by protein degradation, we carried out metabolic pulse-chase experiments using antibodies to these proteins (Fig. 2). Two identical cultures were labeled for 15 min with [35S]-methionine in nitrogen rich conditions. A chase period was then initiated by the addition of excess unlabeled methionine and cells from both cultures were rapidly harvested. Subsequently, one culture was resuspended in nitrogen rich medium, while the other was shifted to nitrogen-free medium. Aliquots of each were harvested at time points thereafter to quantitate the amount of [35S]-labeled eRF3 and eIF4GI remaining. We found that the half-life of both proteins decreased significantly following the onset of nitrogen starvation (Fig. 2A,B). These results confirm that an increase in the rate of degradation by autophagy plays a significant role in the reduced abundance of eRF3 and eIF4GI following the onset of nitrogen starvation.
FIGURE 2.
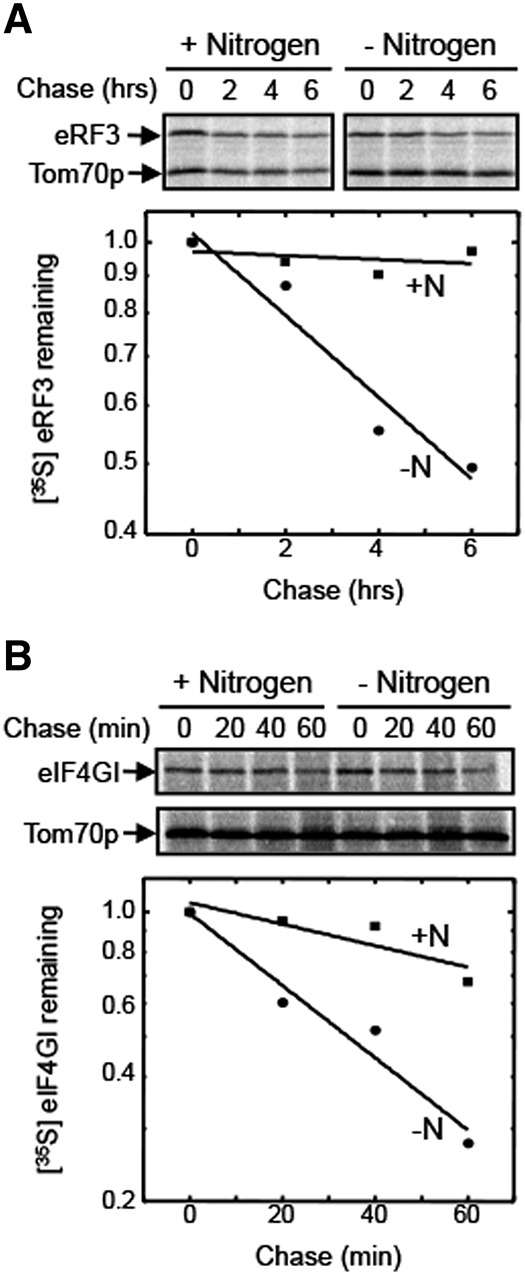
The stabilities of eRF3 and eIF4GI are decreased following the onset of nitrogen starvation. Pulse-chase measurements of protein half-life were carried out for (A) eRF3 and (B) eIF4GI. Measurements were carried out twice for each protein and condition with similar results.
The abundance of Dcp2p and Pop2p is rapidly reduced in a proteasome-dependent manner following the onset of nitrogen starvation
We also examined the abundance of Dcp2p and Pop2p following the onset of nitrogen starvation. Dcp2p is the catalytic subunit of the Dcp2p/Dcp1p decapping complex, while Pop2p (also known as Caf1p) is a component of the Ccr4p–Pop2p deadenylase complex. We found that the steady-state level of both proteins was reduced in a rapid manner following the onset of nitrogen starvation (Fig. 3A,B), with their levels decreasing by four- to fivefold within 2 h. Intriguingly, their degradation was not blocked in the atg7Δ strain, indicating that their depletion was not due to degradation by autophagy.
FIGURE 3.
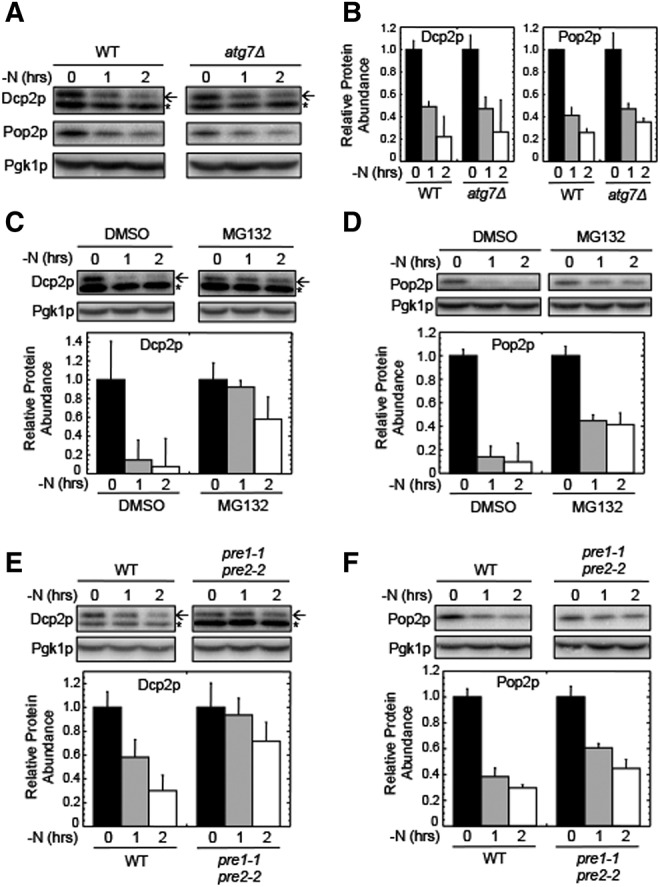
Dcp2p and Pop2p are rapidly degraded by the proteasome following the onset of nitrogen starvation. (A) Western blots of Dcp2p and Pop2p abundance in wild-type and atg7Δ strains harvested at the indicated times following the onset of nitrogen starvation (−N). (B) Quantitation of Western blots from panel A. (C) Western blots and quantitation of Dcp2p in a wild-type strain harvested at various times following a shift to nitrogen starvation (−N). (D) Western blots and quantitation of Pop2p in a wild-type strain harvested at various times following a shift to nitrogen starvation (−N). In panels C and D, MG132 (100 µM) was added at the onset of nitrogen starvation as indicated. (E) Western blots and quantitation of Dcp2p in wild-type and pre1-1, pre2-2 strains harvested at various times following a shift to nitrogen starvation (−N). (F) Western blots and quantitation of Pop2p in wild-type and pre1-1, pre2-2 strains harvested at various times following a shift to nitrogen starvation (−N). The experiments shown in panels E and F were carried out at 30°C. For Dcp2p blots, the arrow indicates the Dcp2p band, while the small star indicates a nonspecific band recognized by the HA antibody. For quantitation, protein abundance was normalized to Pgk1p from the same extract as an internal control. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation.
Since autophagy did not mediate the decreased abundance of Dcp2p and Pop2p, we next asked if the proteasome was responsible. We found that addition of the proteasome inhibitor MG132 to the culture medium partially stabilized both proteins during nitrogen starvation (Fig. 3C,D). These results suggest that both Dcp2p and Pop2p become substrates for proteasomal degradation following the onset of nitrogen starvation. To confirm the role of the proteasome, we examined the levels of these proteins in a pre1-1/pre2-2 strain, which carries mutations in two of the three catalytic subunits of the 20S proteasome (Heinemeyer et al. 1991, 1993). We found that both Dcp2p and Pop2p were partially stabilized in a pre1-1/pre2-2 strain when grown at 30°C (a semi-restrictive temperature) (Fig. 3E,F). These results confirm that the rapid decrease in abundance of Dcp2p and Pop2p following the onset of nitrogen starvation is at least partially mediated by the proteasome.
To confirm that the Dcp2p and Pop2p proteins are subject to direct proteasomal degradation under these conditions, we again attempted pulse-chase experiments as described above. However, we were unable to detect either of these proteins by immunoprecipitation. Therefore, while direct proteasomal degradation of Dcp2p and Pop2p is the most direct interpretation of these results, we cannot yet exclude an indirect role of the proteasome in their decreased steady-state abundance following the onset of nitrogen starvation.
Ubp3p is required for selective degradation of translation and RNA turnover factors by the autophagy and proteasomal pathways during nitrogen starvation
Activation of the ribophagy pathway following the onset of nitrogen starvation requires the deubiquitinase Ubp3p and its associated cofactor, Bre5p (Cohen et al. 2003; Kraft et al. 2010). Since our results showed that eRF3, eIF4GI, and Pat1p are rapidly degraded by autophagy following the onset of nitrogen starvation, we next asked whether their turnover was also Ubp3p-dependent. We found that the rapid decrease in both eRF3 and eIF4GI abundance was moderated in both ubp3Δ and bre5Δ strains (Fig. 4A,B), suggesting that deubiquitinase activity of the Ubp3p–Bre5p complex facilitates degradation of these proteins. In contrast, the ubp3Δ mutation did not alter the degradation of Pat1p. We also examined whether Ubp3p is required for the proteasomal degradation of Dcp2p and Pop2p during nitrogen starvation. We found that both proteins were strongly stabilized in the ubp3Δ strain (Fig. 4A,C). These results indicate that Ubp3p participates in the rapid and selective reduction of a subset of translation and RNA turnover factors by both the autophagy and the proteasomal pathways during nitrogen starvation.
FIGURE 4.
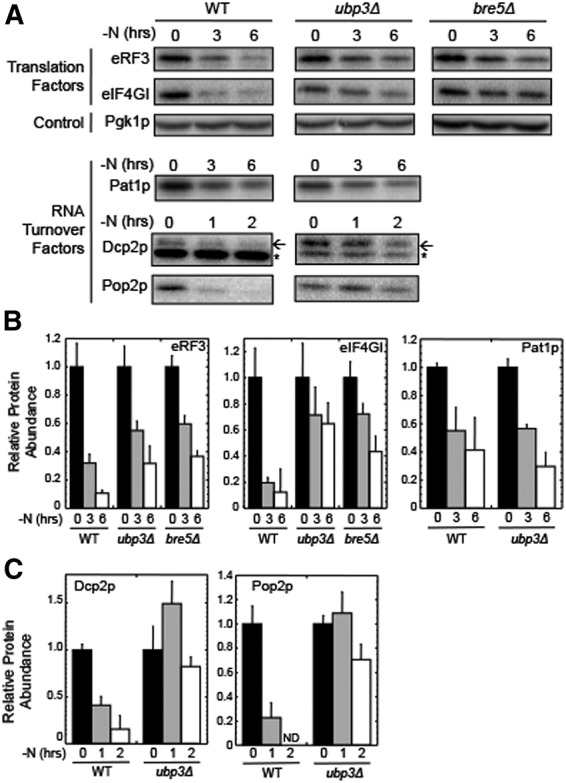
The ribophagy factor Ubp3p is required for selective protein degradation by both the autophagy and proteasomal pathways during nitrogen starvation. (A) Western blots of eRF3, eIF4GI, Pat1p, Dcp2p, and Pop2p abundance in wild-type, ubp3Δ, and bre5Δ strains harvested at the indicated times following the onset of nitrogen starvation (−N). (B) Quantitation of results from panel A for eRF3, eIF4GI, and Pat1p. (C) Quantitation of results from panel A for Dcp2p and Pop2p. For Dcp2p blots, the arrow indicates the Dcp2p band, while the small star indicates a nonspecific band recognized by the HA antibody. For quantitation, protein abundance was normalized to Pgk1p from the same extract as an internal control. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation. (ND) not detectable.
Proteins depleted by the proteasome during nitrogen starvation are also reduced following rapamycin addition
Rapamycin is a macrolide antibiotic that mediates rapid TOR1 inhibition, which results in the induction of autophagy (Noda and Ohsumi 1998). Since our results indicated that Dcp2p and Pop2p are rapidly decreased in a proteasome-dependent manner following the onset of nitrogen starvation, we next asked whether rapamycin also induced a similar response. We found that the abundance of Dcp2p and Pop2p also rapidly decreased upon rapamycin treatment (Fig. 5A,B). This turnover was blocked by MG132, indicating that their depletion under these conditions was again dependent upon proteasome function. To confirm that TOR1 inhibition was required for this decrease, we transformed strains with a plasmid expressing the rapamycin resistant tor1-1 allele. The tor1-1 mutation prevents TOR1 binding by the FKBP-rapamycin complex (Schmelzle and Hall 2000). Thus, expression of this mutant allele should prevent degradation of Dcp2p and Pop2p upon rapamycin addition if TOR1 regulates this proteasomal process. Indeed, we found that degradation of both Dcp2p and Pop2p was blocked in strains expressing the tor1-1 allele (Fig. 5C,D). These results indicate that TOR1 activity represses the proteasomal depletion of Dcp2p and Pop2p during growth in nutrient-rich conditions, while TOR1 inactivation following nitrogen starvation or rapamycin exposure induces selective, rapid depletion of these proteins by the proteasome. We note that the smaller decrease in Pop2p abundance observed following rapamycin addition as compared with nitrogen starvation could be due to the fact that rapamycin is a relatively specific inhibitor of the TOR1 pathway, while nitrogen starvation perturbs not only the TOR1 pathway, but also other physiological processes.
FIGURE 5.
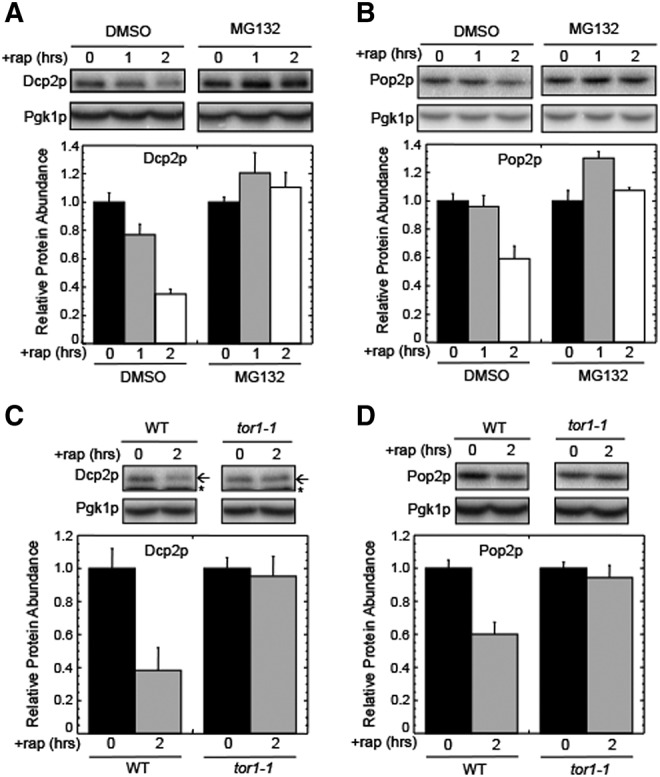
TOR1 inhibition by rapamycin induces proteasomal degradation of Dcp2p and Pop2p. (A) Western blots and quantitation of Dcp2p abundance following the addition of 200 nM rapamycin (+rap) to a wild-type strain. (B) Western blots and quantitation of Pop2p abundance following the addition of 200 nM rapamycin (+rap) to a wild-type strain. For panels A and B, MG132 (100 µM) was added where indicated. (C) Western blots and quantitation of Dcp2p abundance following the addition of 200 nM rapamycin (+rap) to wild-type and tor1-1 strains. (D) Western blots and quantitation of Pop2p abundance following the addition of 200 nM rapamycin (+rap) to wild-type and tor1-1 strains. For Dcp2p blots, the arrow indicates the Dcp2p band, while the small star indicates a nonspecific band recognized by the HA antibody. For quantitation, protein abundance was normalized to Pgk1p from the same extract as an internal control. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation.
Ubp3p is depleted during nitrogen starvation and has a genetic interaction with the proteasome
We next examined the abundance of Ubp3p during nitrogen starvation. We found that the level of Ubp3p is rapidly decreased during nitrogen starvation (Fig. 6A), and the decrease of full-length Ubp3p coincided with the appearance of a fragment that was ∼30 kDa smaller. Since our Ubp3p construct carried a carboxy-terminal HA epitope tag, this indicated that the smaller species lacked the amino-terminal region that includes the Bre5p binding site (Li et al. 2007). We found that Ubp3p turnover was reduced in the atg7Δ strain (Fig. 6B), as well as following the addition of MG132 (Fig. 6C). Together, these results indicate that both the autophagy and proteasomal pathways participate in Ubp3p depletion during nitrogen starvation.
FIGURE 6.
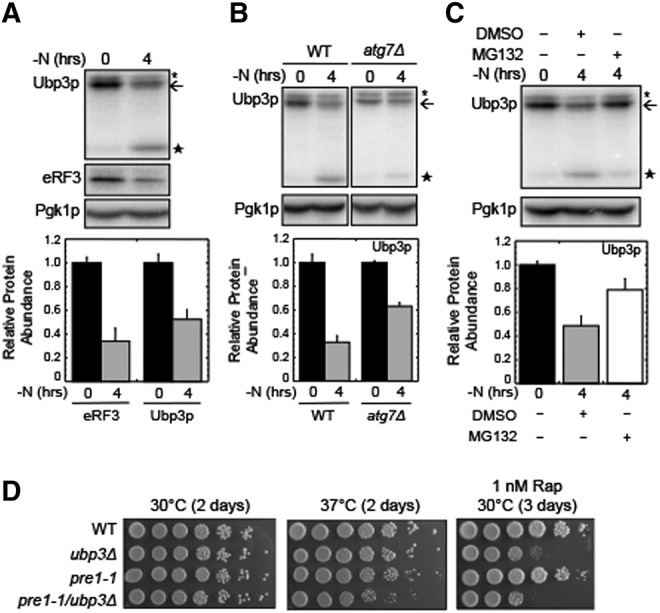
Ubp3p is degraded during nitrogen starvation. (A) Western blots and quantitation of Ubp3p following the onset of nitrogen starvation (−N) in a wild-type strain. eRF3 is included as a control. (B) Western blots and quantitation of Ubp3p following the onset of nitrogen starvation (−N) in wild-type and atg7Δ strains. (C) Western blots and quantitation of Ubp3p following the onset of nitrogen starvation (−N) in a wild-type strain. MG132 (100 µM) was added where indicated. For panels A–C, the arrow indicates the location of full-length Ubp3p. The large star is a stable Ubp3p degradation product, while the small star indicates a nonspecific band recognized by the HA antibody. (D) Evidence of a genetic interaction between the ubp3Δ and pre1-1 proteasomal mutations. Wild-type, ubp3Δ, pre1-1, and pre1-1/ubp3Δ cells were adjusted to a cell density of one A600 unit/mL and spotted on YPD plates, or YPD plates with 1 ng/mL rapamycin using fivefold serial dilutions at the indicated temperatures. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation.
Since our results suggest that Ubp3p participates in the proteasomal degradation of Dcp2p and Pop2p, we tested for a genetic interaction between the ubp3Δ mutation and the pre1-1 mutation of the 20S proteasome. Serial dilutions of WT, ubp3Δ, pre1-1, and ubp3Δ/pre1-1 strains were plated on YPD plates in the absence or presence of 1 nM rapamycin. Colony size and cell viability of these strains were similar on YPD plates grown at 30°C (Fig. 6D, left panel). When YPD plates were incubated at 37°C, we found that the colony size of the pre1-1 strain was reduced, while the ubp3Δ/pre1-1 double mutant grew more slowly and exhibited poorer viability than either of the single mutants (Fig. 6D, center panel). When these strains were tested on plates containing 1 nM rapamycin at 30°C, the pre1-1 strain exhibited only a slight growth defect (Fig. 6D, right panel). The ubp3Δ strain exhibited both reduced viability and slow growth, indicating an enhanced sensitivity to TOR1 inhibition as observed previously (Kraft et al. 2008; Ossareh-Nazari et al. 2010). Finally, the ubp3Δ/pre1-1 double mutant exhibited both slow growth and a greater loss of viability than the ubp3Δ strain. These results demonstrate that rapamycin sensitivity of the ubp3Δ strain is exacerbated by the pre1-1 mutation, further implicating TOR1 in the regulation of proteasomal function.
Effect of Ubp3 and autophagy on translational efficiency
Translation is severely reduced during nitrogen starvation (Schmelzle and Hall 2000). This is a consequence of TOR1 inactivation, which greatly reduces translation initiation on most mRNAs and allows the expression pattern of specific mRNAs to increase in response to nutrient limitation. However, selective autophagy of proteins, including eIF4G and eRF3, could possibly fine-tune translation during nitrogen starvation. To address this point, we first asked whether autophagy had any influence on the overall translational down-regulation that occurs upon nitrogen starvation by carrying out polysome analysis in wild-type and atg7Δ strains. We observed a robust level of polysomes during nutrient-rich growth in both strains (Fig. 7A) (polysome/monosome ratios of 2.0). Following nitrogen starvation for 2 h, we observed a severe reduction in the polysome pools in both strains (polysome/monosome ratios of <0.2). These results demonstrate that the TOR1-mediated down-regulation of translation initiation during nitrogen starvation is the primary means of controlling bulk translation during nitrogen starvation, and this mechanism was not adversely affected by the loss of autophagy in the atg7Δ strain. However, we cannot exclude the possibility that the reduction in ribosome content or the depletion of specific translation factors may alter the translation of specific mRNAs.
FIGURE 7.
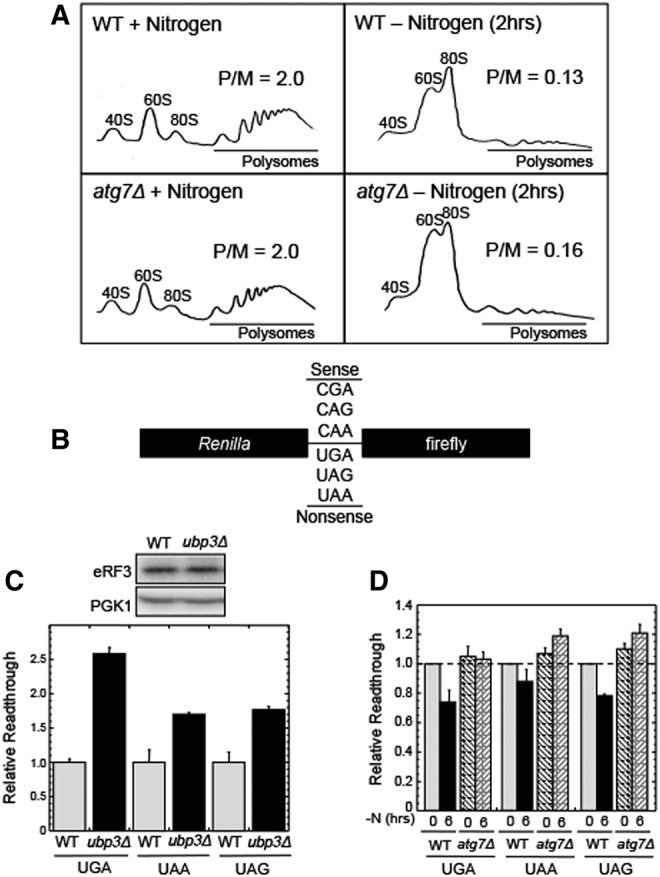
Effects of autophagy and Ubp3p on translation following the onset of nitrogen starvation. (A) Polysome analysis on wild-type and atg7Δ strains under conditions of nitrogen excess (left) and nitrogen starvation (right). (P/M) Polysome-to-monosome ratio (where monosome includes the sum of the 40S, 60S, and 80S peaks). (B) Schematic of the dual luciferase readthrough reporters. Each reporter contains either an in-frame nonsense or sense codon between the Renilla and firefly ORFs. (C) Effect of the ubp3Δ mutation on the relative efficiency of translation termination and eRF3 abundance during growth in nitrogen excess. (D) Effect of nitrogen starvation (−N) on the relative efficiency of translation termination in wild-type and atg7Δ strains. All experiments were carried out two or more times with similar results. Bar graphs are plotted as mean ± standard deviation.
We next asked whether the deubiquitinase Ubp3p influences translation termination during growth in nutrient-rich conditions using dual luciferase readthrough reporters. These reporter constructs contain a nonsense codon (or a sense codon as control) inserted in-frame between the Renilla and firefly reading frames (Fig. 7B; Grentzmann et al. 1998; Keeling et al. 2004). This configuration allows us to use the firefly/Renilla ratio as an indicator of the efficiency of translation termination. We found that the ubp3Δ strain exhibited 1.5 to 2.5-fold more readthrough at nonsense codons than the wild-type strain, although the steady-state level of eRF3 did not change (Fig. 7C). This suppressor phenotype suggests that Ubp3p (and by inference, the ubiquitin–proteasome system) influences the efficiency of translation termination during normal growth conditions.
Finally, we explored the effect of autophagy on translation termination during nitrogen starvation. The efficiency of translation termination is determined by the relative abundance of release factors, aminoacyl tRNAs, and terminating ribosomes (Betney et al. 2010). During nitrogen starvation, the steady-state levels of both ribosomes and eRF3 are reduced. Accordingly, the reduction in eRF3 may modulate the efficiency of translation termination during these conditions. To test this possibility, we used dual luciferase readthrough reporters to compare the relative stop codon readthrough levels in wild-type and atg7Δ strains in nutrient-rich and nitrogen-starved conditions. In the wild-type strain, the readthrough observed was modestly reduced (10%–25%) at the three stop codons during nitrogen starvation (Fig. 7D), indicating that the efficiency of translation termination had marginally increased. This anti-suppressor effect was not observed in the atg7Δ strain, indicating that autophagy (and possibly the reduced level of eRF3) may fine-tune the efficiency of translation termination during nitrogen starvation.
DISCUSSION
In this study, we found that the translation factors eIF4GI and eRF3 undergo rapid and selective Ubp3p-dependent degradation by autophagy following the onset of nitrogen starvation (Figs. 1D,E, 2). eIF4GI was previously shown to be degraded during either nitrogen starvation or rapamycin exposure, although the mechanism of degradation was not known (Powers and Walter 1999; Kuruvilla et al. 2001; Gelperin et al. 2002; Ramirez-Valle et al. 2008). We found that the turnover of eIF4GI and eRF3 is highly selective during nitrogen starvation, since no decrease in abundance of the initiation factors eIF2α and eIF5B, or the elongation factor eEF3 were observed under these conditions. Previous studies found that two other initiation factors, eIF4E and eIF4A, also remain stable in both yeast and mammalian cells following nutrient limitation or rapamycin addition (Berset et al. 1998; Powers and Walter 1999; Kuruvilla et al. 2001; Ramirez-Valle et al. 2008). While the degradation of eIF4GI and eRF3 shares some components of the ribophagy machinery, the turnover rate of these proteins is much faster than ribophagy, and the overall extent of protein turnover is also much greater. These results indicate that selective autophagy of specific translation factors occurs following the onset of nitrogen starvation in a manner that complements the ribophagy process.
The balance between mRNA synthesis and degradation controls the steady-state levels of cellular mRNAs. Regulation of mRNA synthesis rates has long been appreciated as a key mechanism that controls gene expression, while the importance of regulated mRNA decay is generally less appreciated. We found that the degradation of Dcp2p (an mRNA decapping factor) and Pop2p (a component of the CCR4/Pop2p mRNA deadenylase complex) during nitrogen starvation was extremely rapid and dependent upon proteasome function (Fig. 3) and the ribophagy deubiquitinase, Ubp3p (Fig. 4). Similarly, a previous study found that Ubp3p interacts with the proteasome to stimulate the selective degradation of a protein (Rad4p) to suppress nucleotide excision repair (Mao and Smerdon 2010). The degradation we observed was selective, as other proteins involved in mRNA decay, such as Tpa1p and Lsm1p, were stable under the same conditions (Fig. 1B). Two other proteins, the poly(A) binding protein Pab1p and decapping enhancer, Pat1p, were also depleted by autophagy at different rates (Fig. 1C–E). When taken together, these results indicate that specific translation and RNA metabolism factors are selectively degraded by either the proteasome or autophagy during nitrogen starvation. It is possible that the differential degradation of these proteins helps cells alter their gene expression profile to adapt to starvation conditions.
Previous studies found that depletion of either eIF4GI or eRF3 reduces growth rates and inhibits TOR signaling in mammalian cells (Chauvin et al. 2007; Ramirez-Valle et al. 2008). These similar effects suggest that these proteins may share a common function, such as a shared role in translation. In this regard, both eIF4GI and eRF3 participate in the formation of mRNA closed-loop structures required for efficient initiation of cap-dependent translation (Amrani et al. 2008). Based on our finding that the deubiquitinase Ubp3p and its binding partner Bre5p are required for efficient degradation of eIF4GI and eRF3 (Fig. 4A,B), the autophagy machinery and the Ubp3p complex may facilitate eRF3 and eIF4GI degradation in order to reprogram translation initiation during adaptation to nitrogen starvation conditions.
Previous studies have shown that selective autophagy exists in organisms ranging from yeast to mammals. Targets of this process can range from specific proteins to protein aggregates, ribosomes, various organelles, and bacteria. While the mechanism of selective autophagy remains poorly understood, a role for ubiquitin is a common theme. For example, degradation of various substrates by autophagy in mammalian cells requires ubiquitination, as well as ubiquitin-binding receptors such as p62 (Kraft et al. 2010). Similarly, the selective degradation of ribosomes following nitrogen starvation requires the deubiquitinase activity of Ubp3p and its partner, Bre5p (Kraft et al. 2008). Recently, the Ubp3p/Bre5p complex was shown to interact with both Cdc48p, which plays a role in the proteasomal pathway, as well as Ufd3p, a ubiquitin-binding adaptor of Cdc48 (Ossareh-Nazari et al. 2010). These Ubp3p partners are required for efficient ribophagy but are not required for bulk autophagy. Since inhibition of proteasomal activity does not block ribophagy, it was proposed that Cdc48p functions as a molecular platform on which ubiquitinated ribosomes are recognized and deubiquitinated before delivery to the vacuolar for degradation.
In the current study, we found that Ubp3p itself is degraded during nitrogen starvation. The reduced steady-state abundance of full-length Ubp3p is accompanied by the accumulation of a carboxy-terminal fragment that corresponds to the UCH catalytic domain of Ubp3p that lacks the Bre5p binding domain (Fig. 6; Li et al. 2007). The loss of full-length Ubp3p and appearance of the Ubp3p fragment are influenced by both the proteasome and autophagy, since inhibition of either pathway moderates the decrease in full-length Ubp3p and the appearance of the degradation fragment (Fig. 6B,C). These results suggest that Ubp3p, possibly in complex with known binding partners such as Cdc48p and Ufd3p (Ossareh-Nazari et al. 2010), may be regulated by the activities of the proteasome and autophagy during nitrogen starvation to limit these degradative processes. Previous studies have described a process termed Regulated Ubiquitin Proteasome-dependent processing (RUP) to yield polypeptide fragments with biological activity (Rape and Jentsch 2004). It is possible that accumulation of a Ubp3p fragment that retains the UCH catalytic domain but lacks the Bre5p binding domain could down-regulate its deubiquitinase activity and possibly allow it to switch functions (Cohen et al. 2003; Li et al. 2007).
Based on this information and our current results, we propose the model presented in Figure 8. During nitrogen excess (Fig. 8A), ubiquitination of Dcp2p, Pop2p, eRF3, and eIF4GI may be blocked by TOR1 activity (step 1). Alternatively, these factors may become ubiquitinated, but their Ubp3p-dependent turnover is inhibited by TOR1 activity (step 2). Following the onset of nitrogen starvation (Fig. 8B), TOR1 activity is repressed. This allows Dcp2p, Pop2p, eRF3, and eIF4GI to be ubiquitinated and subsequently processed by the Ubp3p complex (consisting of Ubp3p–Cdc48p–Ufd3p) (Ossareh-Nazari et al. 2010). Engagement of the Ubp3p complex leads to the transport of Dcp2p and Pop2p to the proteasome, where their degradation occurs (step 3). Similarly, the Ubp3p complex recruits eRF3 and eIF4GI into early autophagosomes, leading to their subsequent degradation by autophagy (step 4). The selective degradation of Dcp2p, Pop2p, eRF3, and eIF4GI may gradually become limiting as processing and/or turnover depletes Ubp3p (step 5) during prolonged nitrogen starvation.
FIGURE 8.
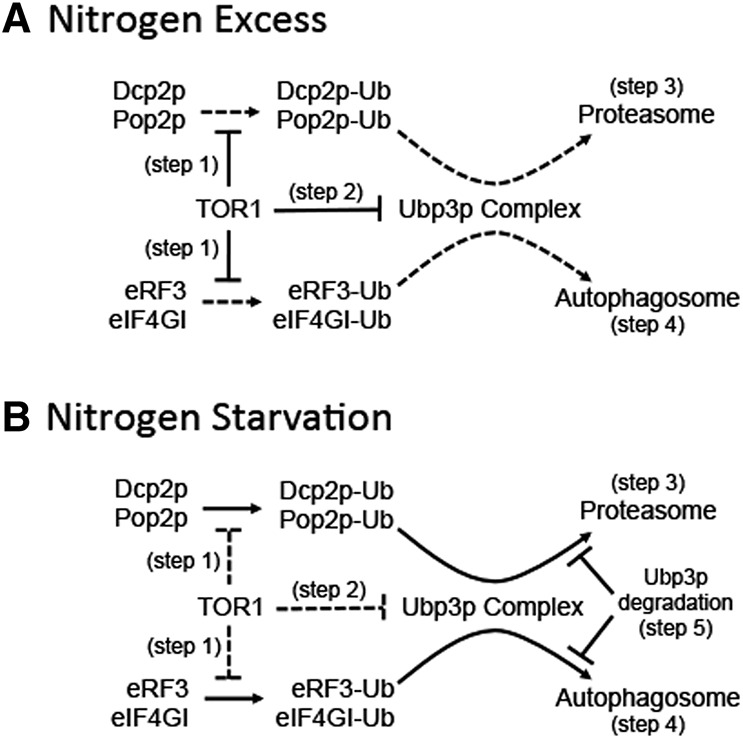
Model for selective turnover of translation and mRNA decay factors during nitrogen starvation by the autophagy and proteasomal pathways. (A) Nitrogen excess conditions. (B) Nitrogen starvation conditions. See text for details.
The ubiquitin–proteasome system was previously shown to be dispensable for both ribophagy and autophagy in yeast (Krick et al. 2010; Ossareh-Nazari et al. 2010). However, our results suggest that the ubiquitin–proteasome system mediates the regulated degradation of selected proteins during nitrogen starvation. We also found that proteasomal degradation of Dcp2p and Pop2p is dependent on TOR1 inhibition, suggesting that the TOR1 pathway regulates a branch of the ubiquitin–proteasome system (Fig. 5). Previous results have also suggested interplay between the proteasome and the TOR1 signaling pathway. Chan et al. (2000) found that some proteasome mutants are hypersensitive to rapamycin. Furthermore, rapamycin addition promotes proteasome activation, while proteasome inhibition decreases TOR function in mammalian cells (Jin et al. 2009; Ko et al. 2011). These results provide strong evidence of crosstalk between the ubiquitin–proteasome and TOR1 pathways, and that TOR1 activity inversely correlates with proteasome function under some conditions.
A previous study found that proteasome activity plays an important role in maintaining amino acid availability during the early stages of acute amino acid starvation in mammalian cells, while the autophagy system provides amino acids at later times (Vabulas and Hartl 2005). Here, we observed a similar temporal sequence of events for selective protein degradation following nitrogen starvation in yeast. Dcp2p and Pop2p were rapidly and selectively depleted by the proteasome, while eIF4GI, eRF3, and Pat1p were degraded more slowly by autophagy. Other translation and mRNA turnover factors were degraded by autophagy at even slower rates that were comparable to the rate of ribosome turnover by autophagy. Finally, some factors were completely stable over the same time period. The selective, temporally controlled nature of this process suggests that the turnover of these proteins is not a bulk process that simply occurs to provide amino acids to the starving cell. Instead, the rapid loss of Dcp2p and Pop2p may increase the stability of a specific subset of mRNAs, as previously shown for strains lacking the RNA turnover factors Xrn1p and Dcp1p (He et al. 2003). Similarly, depletion of eIF4GI and eRF3 could reprogram the translation machinery to facilitate the synthesis of proteins required for survival during nitrogen starvation conditions. Further studies will be required to test these predictions.
MATERIALS AND METHODS
Nitrogen starvation assays
Cells were grown to log phase in synthetic dextrose (SD) media and diluted overnight. When cultures reached a cell density of 0.5 A600 units/mL, 5 A600 units of cells were harvested and flash frozen as the time zero sample. The rest of the culture was centrifuged, washed with nitrogen starvation media (SD medium without amino acids or nitrogen) (Difco) and resuspended in the same volume of nitrogen starvation media. Cells were subsequently harvested at the indicated times. Samples were flash frozen and kept at −80°C until they were processed for Western blotting. In some experiments cells were washed in nitrogen starvation media with 100 μM MG132 (Caymen Chemical) and resuspended in the same. In other experiments, cells were grown to log, a time = 0 time point was taken, then 200 nM rapamycin (LC Laboratories) was added and aliquots taken at the indicated time-points.
Western blotting and antibodies
To harvest proteins for Western blots, 5 A600 units of cells were added to 100% trichloroacetic acid (TCA) to achieve a final TCA concentration of 5%. Cells were incubated on ice for 30 min, harvested by centrifugation, washed four times with 100% acetone, and dried under vacuum. Cells were then resuspended in 100 μL of sodium dodecyl sulfate (SDS) boiling buffer (1% SDS, 50 mM Tris, pH 7.5, 1 mM EDTA), and lysed by agitation with glass beads (four cycles, 1 min each) with 1 min on ice between each cycle. Protein concentrations were determined by the Lowry method (Lowry et al. 1951). Fifteen to twenty-five micrograms of protein was loaded into each lane of an 8% SDS polyacrylamide gel. Protein was then transferred to an Immobilon-P transfer membrane (Millipore) using a Genie Electrophoretic Transfer system (Idea Scientific). The membranes were blocked in 0.3% Tween 20–phosphate-buffered saline buffer containing 5% nonfat milk. Membranes were incubated in the same buffer with the indicated antibodies or antiserum at their recommended concentrations for 2 h at room temperature. When a secondary antibody was required, either Rabbit anti-mouse (MP Biomedical, 55436) or Rabbit anti-rat (Abcam, ab6703) sera were used. Bound antibodies were detected using [125I]-protein A (PerkinElmer, NEX146L100UC), and results were visualized using a Storm PhosphorImager (GE Healthcare). A mouse monoclonal antibody to S. cerevisiae Pgk1p (22C5D8; Invitrogen, cat. No. 459250) was used as an internal control. Rabbit antiserum to eIF4GI was a gift from Michael Altmann (Berset et al. 1998). Rabbit antisera to S. cerevisiae eEF3, eEF1A, eEF2 were gifts from Terry Kinzy, and rabbit antisera to eIF2α, eIF5B, and eIF5A were gifts from Tom Dever. Rabbit antiserum to Tom70p (Mas70p) was published previously (Koh et al. 2001), as were rabbit antisera to eRF3 or eRF1 (Salas-Marco and Bedwell 2004). Mouse monoclonal antibodies to S. cerevisiae Rpl3p (ScRPL3) were obtained from the Developmental Studies Hybridoma Bank. HA antibodies (Covance, MMS-101R) were used to detect HA-tagged versions of Lsm1p, Pat1p, Dcp2p, Pop2p, and Ubp3p. Pab1p antibodies are available commercially (EnCor Biotech, MCA-1G1). In some cases, nonspecific bands were consistently observed in Western blots using HA antibodies. When these were near proteins of interest (e.g., Dcp2p or Ubp3p), the correct band was confirmed by its absence in extracts from the same strain lacking the HA-tagged protein.
Pulse-chase experiments
To measure protein half-life, 16 A600 units of cells were grown to log (0.5 A600) in synthetic dextrose (SD) medium overnight at 30°C. The culture was then split into two equal portions. Cells were spun down and resuspended in the same prewarmed media to an OD600 of 4 OD/mL and incubated at 30°C for 5 min in a shaking water bath. EasyTag [35S] protein labeling mix (PerkinElmer) was added to each tube at 200 μCi/mL final volume and incubated for 15 min at 30°C with shaking. After 15 min a 150× chase mix (1 mg/mL methionine, 1 mg/mL cysteine, 15% yeast extract) was added to 1× final concentration and cells were incubated at 30°C for 2 min in a shaking water bath. Cells were then rapidly harvested and washed. One tube was washed in SD medium without methionine, while the other was washed in SD medium without nitrogen or amino acids. Cells were spun down once more and resuspended in the same wash medium. Time-points were taken at the indicated times and flash frozen. Samples were incubated with 5% TCA and washed twice with ice-cold acetone. The samples were then dried, resuspended in 50 μL SDS boiling buffer, disrupted by glass bead lysis, and subjected to immunoprecipitation as previously described (Bedwell et al. 1987). A rabbit antiserum to Tom70p (Mas70p) was used as an internal control for immunoprecipitation samples (Koh et al. 2001). The final samples were subjected to 8% SDS-PAGE, and the gel was dried onto filter paper. Radiolabeled proteins were detected using a Storm PhosphorImager (GE Healthcare).
Polysome profiles
Polysome profiles were conducted as described (Landry et al. 2009). Briefly, strains were grown in SD medium to a cell density of 0.5 A600 units/mL. Cycloheximide was added to a final concentration of 0.1 mg/mL, and cells were harvested by centrifugation (6000 rpm, 4 min at 4°C) and washed twice with lysis buffer (20 mM Tris–HCl at pH 8.0, 140 mM KCl 1.5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 0.1 mg/mL cycloheximide, 1 mg/mL heparin). After centrifugation (7000 rpm for 4 min, 4°C), pellets were resuspended in lysis buffer and cells were lysed by glass bead beating. Lysates were cleared by centrifugation at 9500 rpm for 5 min at 4°C and layered over a 20%–50% sucrose gradient (containing 20 mM Tris–HCl at pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.1 mg/mL cycloheximide, 1 mg/mL heparin). Gradients were centrifuged in a Beckman SW41 rotor at 35K rpm for 160 min at 4°C. Fractions were collected, and absorbance at 254 nm was recorded using an ISCO UA-5 absorbance monitor (Teledyne).
Strains
The Saccharomyces cerevisiae strains used in this study are described in Table 1. Strains YDB0665, YDB0657, and YDB0707 were derived from strain MHY501 using standard genetic techniques. Strains YDB0681, YDB0680, YDB0682, YDB0688, YDB0695, YDB0683, YDB0687, YDB0689, and YDB0717 were derived from strains MHY501, MHY659, or YDB0665 by integrating carboxy-terminal tags into the appropriate genomic locus as described (Longtine et al. 1998). YDB0686 and YDB0679 were made using standard genetic techniques starting with strain YDB0646/pDB1227. Strains YDB0708 and YDB0709 were derived from YH129/w and YH129/1, respectively. Strains YDB0694 and YDB0695 were derived from YWO607 and YWO612 as described (Longtine et al. 1998). yJC750 was a gift from Dr. Jeff Coller. YJW615 was a gift from Dr. Jonathan Weissman and YH129/w, YH129/1, YWO607, and YWO612 were gifts from Dr. Dieter Wolf.
TABLE 1.
Strains and plasmids used

Plasmids
The plasmids used in this study are described in Table 2, while the oligonucleotides used are shown in Table 3. pDB1227 was made by first amplifying a SalI-POP2-HA-BamHI fragment from chromosomal DNA using primers DB2996 and DB2997, and inserting the resulting fragment into the SalI–BamHI sites of YCpLac33 (Gietz and Sugino 1988). A HindIII–SalI fragment corresponding to the 500 bp preceding the start codon was then amplified from chromosome DNA using primers DB2998 and DB2999 was inserted into the HindIII–SalI sites. Lastly, a fragment corresponding to 248 bp distal to the stop codon was amplified using primers DB3000 and DB3001 from chromosome DNA and inserted into the BamH1–SacI sites of the construct. The final construct was confirmed by sequencing using primers DB3023, DB3024, and DB3025. pDB1226 was made by first amplifying the TPA1 ORF 932 bp upstream start and 200 bp downstream stop codon with primers DB0873 and DB0874 and inserting this into the PstI–BamHI sites of YCpLac22 (Gietz and Sugino 1988). A SalI restriction site was then introduced at the stop codon using site-directed mutagenesis and primers DB0876 and DB0877. The amplified fragment was subcloned back into the original construct using the KpnI and BamH1 sites. Finally, a ∼250 bp SalI–BamHI fragment with a SalI site followed by an HA tag and 200 bp of the TPA1 3′ UTR followed by a BamHI site was amplified with DB0874 and DB3184 and inserted into the SalI–BamH1 sites of the construct. For pDB1287, the 5′ UTR and ORF of SCH9 was amplified and HA-tagged using primers 3933 and 3934 from chromosomal DNA giving a ∼3000 bp fragment with HindIII–BamHI sites. This was cloned into the HindIII–BamHI sites of YCplac33 (Gietz and Sugino 1988). The 500 bp 3′ UTR was amplified using primers 3935 and 3936 to introduce BamHI and SacI sites. This was then cloned into the larger construct. Finally, plasmid pYDF23, which carries the dominant-negative tor1-1 allele, was a kind gift from Dr. Ted Powers.
TABLE 2.
Plasmids used
TABLE 3.
Oligonucleotides used
Dual luciferase assays
The dual luciferase reporters used to monitor readthrough of stop codons in yeast were described previously (Keeling et al. 2004). Strains harboring the indicated plasmids were grown to log in selective synthetic media and assayed for readthrough either during nutrient excess or 6 h after the onset of nitrogen starvation. The dual luciferase assays were carried out as previously described (Grentzmann et al. 1998; Keeling et al. 2004). This system monitors readthrough of a stop codon by measuring firefly luciferase activity, and allows the normalization to the level of upstream Renilla luciferase activity expressed in the same open reading frame. Assays were also done with reporter that contained a sense codon in place of the stop codon to determine maximum (100%) readthrough.
Serial dilution assays
Cells were grown overnight in YPD. Of note, 1 A600 unit of cells was spun down and resuspended in 1 mL sterile water. Fivefold serial dilutions were made in sterile water and 3 μL of each dilution was spotted onto YPD plates in the absence or presence of 1 ng/mL rapamycin. Plates were incubated as indicated.
ACKNOWLEDGMENTS
The authors thank Jeff Coller, Jonathan Weissman, Roy Parker, Ted Powers, and Dieter Wolfe for providing strains and plasmids, and Michael Altmann, Terri Kinzy, and Tom Devor for providing antisera. The authors also thank Kim Keeling for providing critical comments on the manuscript. This work was supported by National Institutes of Health (NIH) R01 grant GM068854 (to D.M.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045211.114.
REFERENCES
- Amerik AY, Li SJ, Hochstrasser M 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem 381: 981–992. [DOI] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, Jacobson A 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453: 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedwell DM, Klionsky DJ, Emr SD 1987. The yeast F1-ATPase β subunit precursor contains functionally redundant mitochondrial protein import information. Mol Cell Biol 7: 4038–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M 1998. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci 95: 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betney R, de Silva E, Krishnan J, Stansfield I 2010. Autoregulatory systems controlling translation factor expression: thermostat-like control of translational accuracy. RNA 16: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TF, Carvalho J, Riles L, Zheng XF 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc Natl Acad Sci 97: 13227–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Salhi S, Jean-Jean O 2007. Human eukaryotic release factor 3a depletion causes cell cycle arrest at G1 phase through inhibition of the mTOR pathway. Mol Cell Biol 27: 5619–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson BK, Gilbert WV, Doudna JA 2010. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One 5: e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol 5: 661–667. [DOI] [PubMed] [Google Scholar]
- Dargemont C, Ossareh-Nazari B 2012. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim Biophys Acta 1823: 138–144. [DOI] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ 2014. The machinery of macroautophagy. Cell Res 24: 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D, Horton L, DeChant A, Hensold J, Lemmon SK 2002. Loss of Ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics 161: 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486. [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH 1991. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: Mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J 10: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Möhrle V, Mahé Y, Wolf DH 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem 268: 5115–5120. [PubMed] [Google Scholar]
- Jin HK, Ahn SH, Yoon JW, Park JW, Lee EK, Yoo JS, Lee JC, Choi WS, Han JW 2009. Rapamycin down-regulates inducible nitric oxide synthase by inducing proteasomal degradation. Biol Pharm Bull 32: 988–992. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10: 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM 2006. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 26: 5237–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JK, Choi CH, Kim YK, Kwon CH 2011. The proteasome inhibitor MG-132 induces AIF nuclear translocation through down-regulation of ERK and Akt/mTOR pathway. Neurochem Res 36: 722–731. [DOI] [PubMed] [Google Scholar]
- Koh JY, Hájek P, Bedwell DM 2001. Overproduction of PDR3 suppresses mitochondrial import defects associated with a TOM70 null mutation by increasing the expression of TOM72 in Saccharomyces cerevisiae. Mol Cell Biol 21: 7576–7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 10: 602–610. [DOI] [PubMed] [Google Scholar]
- Kraft C, Peter M, Hofmann K 2010. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 12: 836–841. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M 2010. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol 190: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruvilla FG, Shamji AF, Schreiber SL 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc Natl Acad Sci 98: 7283–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Ossareh-Nazari B, Liu X, Dargemont C, Marmorstein R 2007. Molecular basis for bre5 cofactor recognition by the ubp3 deubiquitylating enzyme. J Mol Biol 372: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275. [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ 2010. The Cvt pathway as a model for selective autophagy. FEBS Lett 584: 1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Smerdon MJ 2010. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J Biol Chem 285: 37542–37550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 273: 3963–3966. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y 2004. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem 279: 16071–16076. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y 2005. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280: 31582–31586. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, Van Dorsselaer A, Dargemont C 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep 11: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Niño CA, Bengtson MH, Lee JW, Joazeiro CA, Dargemont C 2014. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol 204: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Walter P 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell 10: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ 2008. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 181: 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Jentsch S 2004. Productive RUPture: activation of transcription factors by proteasomal processing. Biochim Biophys Acta 1695: 209–213. [DOI] [PubMed] [Google Scholar]
- Reinke A, Chen JC, Aronova S, Powers T 2006. Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem 281: 31616–31626. [DOI] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S 2006. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell 21: 261–269. [DOI] [PubMed] [Google Scholar]
- Salas-Marco J, Bedwell DM 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24: 7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN 2000. TOR, a central controller of cell growth. Cell 103: 253–262. [DOI] [PubMed] [Google Scholar]
- Shimobayashi M, Takematsu H, Eiho K, Yamane Y, Kozutsumi Y 2010. Identification of Ypk1 as a novel selective substrate for nitrogen starvation-triggered proteolysis requiring autophagy system and endosomal sorting complex required for transport (ESCRT) machinery components. J Biol Chem 285: 36984–36994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CS, Huh WK 2011. Bidirectional regulation between TORC1 and autophagy in Saccharomyces cerevisiae. Autophagy 7: 854–862. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119: 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Hartl FU 2005. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310: 1960–1963. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2007. Permeases recycle amino acids resulting from autophagy. Autophagy 3: 149–150. [DOI] [PubMed] [Google Scholar]
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ 2006. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 17: 5094–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465: 942–946. [DOI] [PMC free article] [PubMed] [Google Scholar]


