Abstract
Quality control systems monitor and stop translation at some ribosomal stalls, but it is unknown if halting translation at such stalls actually prevents synthesis of abnormal polypeptides. In yeast, ribosome stalling occurs at Arg CGA codon repeats, with even two consecutive CGA codons able to reduce translation by up to 50%. The conserved eukaryotic Asc1 protein limits translation through internal Arg CGA codon repeats. We show that, in the absence of Asc1 protein, ribosomes continue translating at CGA codons, but undergo substantial frameshifting with dramatically higher levels of frameshifting occurring with additional repeats of CGA codons. Frameshifting depends upon the slow or inefficient decoding of these codons, since frameshifting is suppressed by increased expression of the native tRNAArg(ICG) that decodes CGA codons by wobble decoding. Moreover, the extent of frameshifting is modulated by the position of the CGA codon repeat relative to the translation start site. Thus, translation fidelity depends upon Asc1-mediated quality control.
Keywords: translation stalls, reading frame, ribosome, frameshift, Asc1, Rack1
INTRODUCTION
Ribosomes stall due to a variety of translational impediments, including structures in the mRNA (Doma and Parker 2006), the absence of stop codons (Frischmeyer et al. 2002; van Hoof et al. 2002), the presence of inhibitory amino acid combinations (Gutierrez et al. 2013; Woolstenhulme et al. 2013), or the presence of inhibitory codon combinations (Letzring et al. 2010; Seidman et al. 2011). Such sequences may substantially impact downstream translation. For instance, in the yeast Saccharomyces cerevisiae, a single CGA–CGA codon pair is translated inefficiently, which depending on its location can cause a 30%–50% reduction in expression of downstream sequences (Letzring et al. 2010).
In many cases, cells follow a kingdom-dependent program to escape the bottleneck caused by the stalled ribosome. In bacteria, such as Escherichia coli, stalled ribosomes are sometimes rescued by the trans-acting tmRNA·SmpB system (Moore and Sauer 2007), but may also undergo translational recoding, in particular +1 frameshifting (Seidman et al. 2011). Errors in which ribosomes switch reading frames generally occur at extremely low rates, 3 × 10−5 (Kurland 1992), and are almost certain to result in a nonfunctional polypeptide. However, particular codon combinations that create a slippery sequence are one of the key elements in programmed frameshift sites, at which ribosomes transfer reading frames at much higher frequencies (Dinman 2012).
In eukaryotes, quality control systems stop translation at the stall site, recycle the ribosomes and often cause degradation of the nascent peptide and mRNA (Brandman et al. 2012; Shoemaker and Green 2012). In S. cerevisiae, the Asc1 protein, which binds to the small ribosomal subunit (Gerbasi et al. 2004; Sengupta et al. 2004; Coyle et al. 2009; Rabl et al. 2011), affects the response to some stalled ribosomes, since deletion of ASC1 allows read-through of particular inhibitory sequences (Kuroha et al. 2010; Brandman et al. 2012) including internal CGA codon repeats (Letzring et al. 2013). Asc1, the highly conserved yeast ortholog of human Receptor of Activated Kinase 1 (RACK1), binds tightly to the eukaryotic 40S ribosome (Gerbasi et al. 2004) near the mRNA exit site (Sengupta et al. 2004; Rabl et al. 2011) contacting ribosomal proteins, rpS16e, rpS17e, and rpS3e (Rabl et al. 2011). Human RACK1, a member of the Trp-Asp (WD) repeat family, integrates numerous signaling pathways with translation regulation (Adams et al. 2011), for instance, by mediating phosphorylation of eIF6 by protein kinase C to promote joining of the ribosomal subunits (Ceci et al. 2003). Yeast Asc1 likewise has pleiotropic effects on the cell (Rachfall et al. 2013), at least some of which are caused by its interaction with the ribosome (Coyle et al. 2009); asc1-Δ mutants de-repress the general amino acid control genes in rich media (Hoffmann et al. 1999), alter expression of several signaling pathways (Rachfall et al. 2013), and fail to recruit the mRNA binding protein Scp160 to the ribosome (Baum et al. 2004). Moreover, Asc1 protein levels are modulated, decreasing substantially during hypoxic stress (Bruckmann et al. 2009), perhaps indicating that the response to ribosome stalls is regulated. Despite this body of knowledge, it is unknown what happens to translation elongation if Asc1 fails to stop the stalled ribosome.
There is substantial evidence that quality control and its components are important for the health of the cell. In yeast, lack of Dom34, which cooperates with Hbs1 and Rli1 to rescue arrested ribosomes (Shoemaker and Green 2011; Tsuboi et al. 2012), results in an accumulation of nontranslating ribosomes in the 3′ UTRs of mRNAs (Guydosh and Green 2014) and in synthetic genetic interactions indicative of a limiting ribosome supply (Bhattacharya et al. 2010; Balagopal and Parker 2011). In mice, mutations in Ltn1, an E3 ubiquitin ligase (Wilson et al. 2007; Bengtson and Joazeiro 2010) and a member of a ribosome quality control complex (RQC) (Brandman et al. 2012), result in neurodegeneration (Chu et al. 2009). However, we do not know if the activity of the quality control systems to end translation is important to prevent aberrant translation by the ribosome.
Since stalled ribosomes in Escherichia coli sometimes frameshift (Seidman et al. 2011), we considered that Asc1 might not only restrict continued translation by ribosomes paused at internal CGA codon repeats (Letzring et al. 2013), but might also prevent recoding events. We provide evidence here that, in the absence of Asc1 protein, ribosomes stalled at an internal CGA codon repeat undergo substantial frameshifting. Nearly 40% frameshifting occurs in an asc1-Δ strain during translation of an artificial construct with six adjacent CGA codons, a level observed only in a fraction of programmed frameshift events (Dinman 2012). Furthermore, the length of the nascent polypeptide modulates the extent of frameshifting, as well as CGA-mediated translation inhibition. We conclude that the Asc1 protein (or proteins recruited by Asc1) constrains read-through by ribosomes stalled at internal CGA repeats in order to inhibit promiscuous changes in reading frame at this stall. Thus, translation fidelity is dependent upon Asc1-mediated quality control.
RESULTS
Ribosomes lacking Asc1 frameshift at CGA codon repeats
We assessed frameshifting at a (CGA)4 codon repeat in yeast, using Renilla luciferase-codon insert-GFP fusion reporters (Fig. 1A), in which both the codon insert and the reading frame of GFP were varied. We refer to these reporters throughout the manuscript by the codon repeat and reading frame insert. For example, a Renilla luciferase-(CGA)4+1-GFP reporter containing four CGA codons and an extra nucleotide to place the downstream GFP out of frame relative to the Renilla luciferase is called a (CGA)4+1 reporter. In wild-type yeast, ribosomes paused at internal CGA repeats (Letzring et al. 2013) do not frameshift at an appreciable level, based on measurements in this construct (Fig. 1B). Thus, we asked if reading frame maintenance might be controlled by quality control components, such as Asc1.
FIGURE 1.
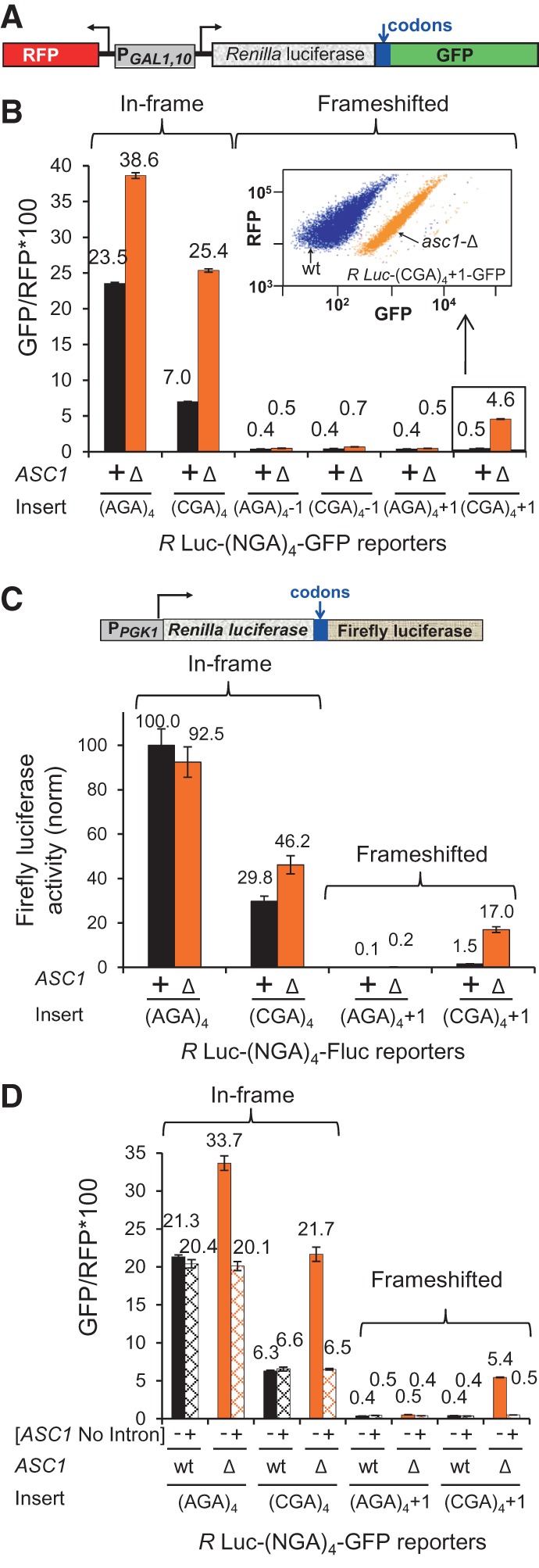
Ribosomes lacking Asc1 frameshift at CGA codon repeats. (A) Schematic of GFP Frameshifting Reporter. In this variant of the RNA-ID reporter (Dean and Grayhack 2012; Letzring et al. 2013), the reading frame of GFP is varied by insertion of 1 or 2 nt immediately downstream from the codon insert. Noise is minimized by independent measurement of RFP to obtain GFP/RFP and by integrating the reporter into the chromosome. (B) Expression of GFP in the +1 frame is observed in the asc1-Δ strain with a (CGA)4+1 codon insert but not with an (AGA)4+1 insert. Quantification of median GFP/RFP from in-frame and frameshifted reporters with (CGA)4 and (AGA)4 inserts in wild-type and asc1-Δ strains. A scatter plot of flow cytometry of GFP versus RFP for (CGA)4+1 reporters in wild-type (blue) and asc1-Δ (orange) strains is shown. (NGA)4 indicates either four Arg AGA or four Arg CGA codons inserted in-frame with Renilla luciferase. Values are derived from flow cytometry of four individual strains, and standard deviations are indicated. (C) Expression of firefly luciferase in the +1 frame is observed from a Renilla luciferase-firefly luciferase fusion protein with a (CGA)4+1 insert in an asc1-Δ strain. A diagram of the Renilla luciferase-firefly luciferase fusion reporter is shown. Firefly luciferase activity is normalized to firefly luciferase activity of the R Luc-(AGA)4-F Luc reporter in the wild-type. (D) A plasmid-borne copy of the ASC1 gene, lacking the intron encoding snR24, complements the effects of the ASC1 deletion on translational frameshifting and on suppression of CGA inhibition.
We find that, in the absence of Asc1, ribosomes translating a (CGA)4+1 reporter undergo ∼11% frameshifting at CGA codon repeats (Fig. 1B), which is estimated as the percentage of GFP/RFP from the (CGA)4+1 construct (4.6 ± 0.04) minus GFP/RFP background (0.4) relative to the GFP/RFP from the in-frame (AGA)4 construct (38.6 ± 0.4) minus GFP/RFP background (0.4). This 11% estimate of frameshifting is a minimal estimate of the percentage of ribosomes that frameshift, since it is relative to the GFP/RFP from the in-frame (AGA)4 construct, which represents the maximum translation efficiency for this polypeptide. Frameshifting is not observed from either the (AGA)4+1 construct, or from any construct in the −1 frame (Fig. 1B), since GFP/RFP from all of these constructs is at or near background levels of 0.4. Similarly, we observe ∼18% frameshifting in an asc1-Δ mutant with a (CGA)4+1 insertion in a Renilla luciferase-firefly luciferase reporter (Fig. 1C); with this reporter, no expression is detected with an (AGA)4+1 insert in either the wild-type or the asc1-Δ strain, and little expression is detected with the (CGA)4+1 insert in the wild-type strain. Furthermore, frameshifting in the (CGA)4+1 construct is due to lack of Asc1, since it is completely suppressed by a plasmid-borne copy of the ASC1 gene, even one without the ASC1 intron, which encodes snR24 (Fig. 1D; Kiss-László et al. 1996).
Improved CGA decoding inhibits frameshifting
We find that frameshifting depends upon inefficient decoding of the CGA codons since tRNAs that suppress the expression defect of CGA codon repeats (Letzring et al. 2010) also suppress frameshifting. CGA-mediated frameshifting is eliminated by expressing the anticodon mutated tRNAArg(UCG) (which forms W:C base pairs with CGA) (Fig. 2A), and is markedly reduced by overproducing the native tRNAArg(ICG) on a multicopy plasmid (Fig. 2B), since GFP/RFP from the (CGA)4+1 construct is reduced from 5.1 without tRNA to 0.9 with additional copies of tRNAArg(ICG). Increasing the quantity of the native tRNA does not alter its function in the ribosome (either the codon–anticodon interactions or translocation efficiency of the tRNA), but is likely to increase the rate of translation elongation by increasing the rate of A site acceptance (due to tRNA availability). Thus, we infer that frameshifting in the asc1-Δ mutant depends critically on a slow rate of translation through the CGA codon repeat.
FIGURE 2.
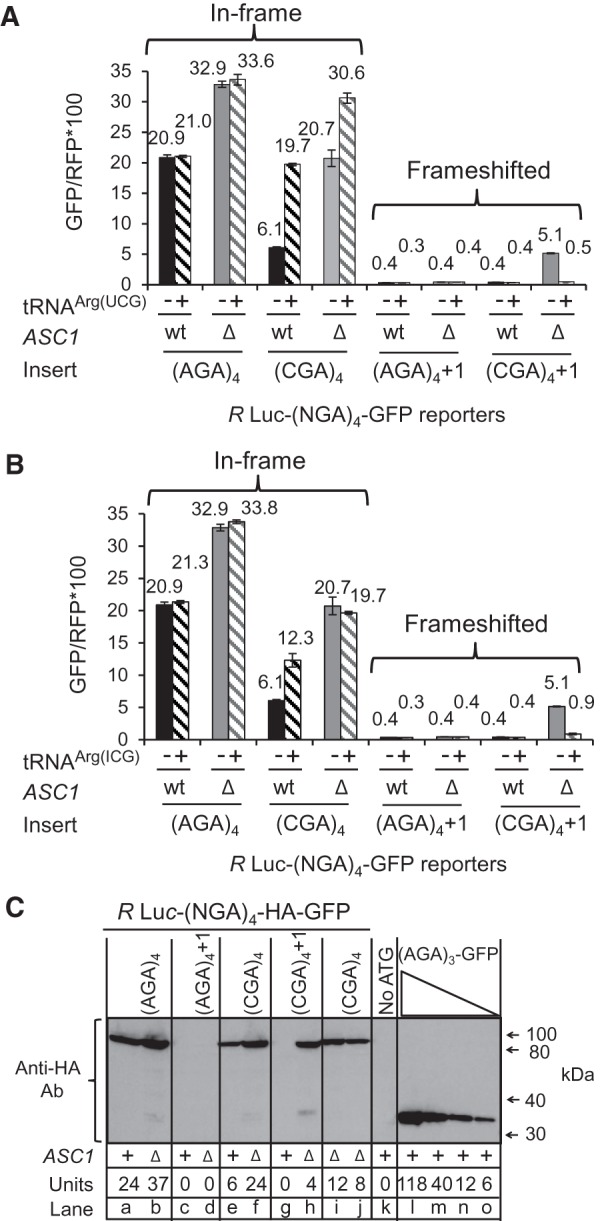
Frameshifting depends upon inefficient decoding of CGA codons and yields a full-length polypeptide. Expression of the nonnative anticodon mutated tRNAArg(UCG) (A) or the native tRNAArg(ICG) (B) on a 2μ plasmid suppresses frameshifting in the (CGA)4+1 construct in an asc1-Δ mutant. Median GFP/RFP expression of the indicated R Luc-(NGA)4-GFP constructs in the indicated wild-type and asc1-Δ strains. (C) CGA-mediated frameshifting yields a full-length fusion protein. Yeast extracts containing indicated units of GFP/RFP were resolved by SDS-PAGE and probed with an anti-HA antibody to detect GFP; the HA epitope is encoded just downstream from the frameshift site in-frame with GFP. The sample in lane k is derived from a GFP construct lacking an initiating methionine, while samples in lanes l–o are derived from yeast extracts expressing a 3C-HA-His6-superfolder GFP protein (lacking Renilla luciferase), derived from pEKD1024 (Dean and Grayhack 2012). Relative amounts of input protein were varied in lanes l–o and lanes f, i, and j, as indicated.
To determine if GFP measured from frameshifted Renilla luciferase-GFP constructs is due to translation of a Renilla luciferase-codons-HA epitope-GFP fusion protein or to internal initiation at the HA epitope-GFP, we examined the size of the frameshifted polypeptide by SDS-PAGE and antibody detection. A full-length fusion protein is the primary polypeptide detected from all constructs, including the (CGA)4+1 construct in the asc1-Δ strain (Fig. 2C). Furthermore, the intensity of the Western signal from the frameshifted polypeptide in the asc1-Δ strain (Fig. 2C, lane h) is similar to that in lanes with comparable GFP/RFP activity units (Fig. 2C, lanes e and j). Thus, we conclude that the Asc1 protein directly or indirectly prevents a stalled ribosome from moving to another reading frame. We also observe trace amounts of a smaller polypeptide that migrates similarly to the 3C-HA-His6-superfolder GFP protein (lacking Renilla luciferase) in extracts from the asc1-Δ strain expressing the (CGA)4+1 construct (Fig. 2C, lane h); this is most likely due to internal initiation at an AUG in the +1 frame located 10 codons upstream of the start of the CGA repeat.
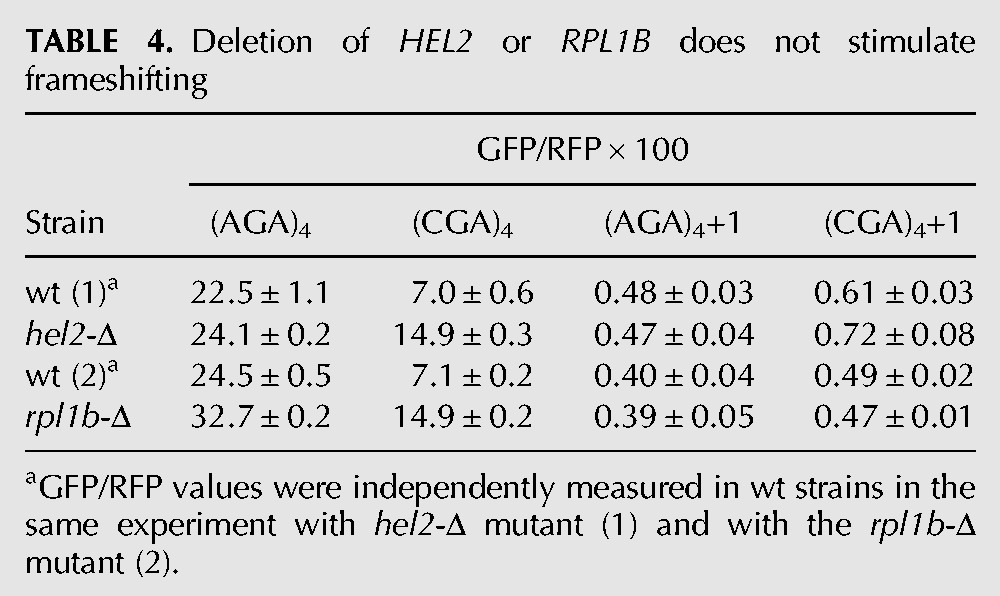
We considered that the frameshifting observed in the asc1-Δ mutant might actually be due to a failure of other known quality control systems, caused by the absence of ASC1. For instance, Asc1 is implicated in recruitment of the ribosome quality control (RQC) complex, in which the E3 ubiquitin ligase Ltn1 is not only a member but is required for assembly of the complex (Bengtson and Joazeiro 2010; Brandman et al. 2012). If recruitment of the RQC complex actually stops translation and prevents frameshifting, we would expect to observe frameshifting in an ltn1-Δ mutant; however, we do not observe any detectable frameshifting in an otherwise wild-type strain bearing a deletion in LTN1 (Fig. 3A). No direct evidence exists that Asc1 recruits other quality control proteins, such as Dom34 and Hbs1, which are involved in recycling stalled ribosomes (Doma and Parker 2006; Shoemaker et al. 2010), or Nam7 and Nmd2, which are involved in nonsense-mediated decay (Isken and Maquat 2007), but we also tested these mutants and did not find evidence of significant frameshifting in any of these single mutants (Tables 1, 2). We noted that GFP/RFP from the in-frame Renilla luciferase-(AGA)4-GFP was increased in the nam7-Δ and nmd2-Δ strains, suggesting that our Renilla luciferase-GFP construct might be a natural target of the NMD pathway (Table 2). To determine if mRNA stabilization played a role in (CGA)4+1 frameshifting in the asc1-Δ mutant, we examined GFP/RFP expression and GFP/actin mRNA levels (by qPCR) from (AGA)4 and (CGA)4+1 constructs in nmd2-Δ single mutant and in the nmd2-Δ asc1-Δ double mutant. The differences in GFP mRNA levels between the (CGA)4+1 constructs in the nmd2-Δ mutant and the nmd2-Δ asc1-Δ mutant strains are relatively small (approximately twofold) and cannot account for the differences in frameshifted GFP expression (GFP/RFP of 0.6 in the nmd2-Δ mutant versus 8.1 in the nmd2-Δ asc1-Δ mutant [Table 3]). Furthermore, no frameshifting is observed in strains with deletions in either of two other genes implicated in CGA codon read-through: HEL2, a putative E3 ubiquitin ligase (Brandman et al. 2012; Letzring et al. 2013), or RPL1B, encoding one of two copies of ribosomal L1 protein (Table 4; Letzring et al. 2013).
FIGURE 3.
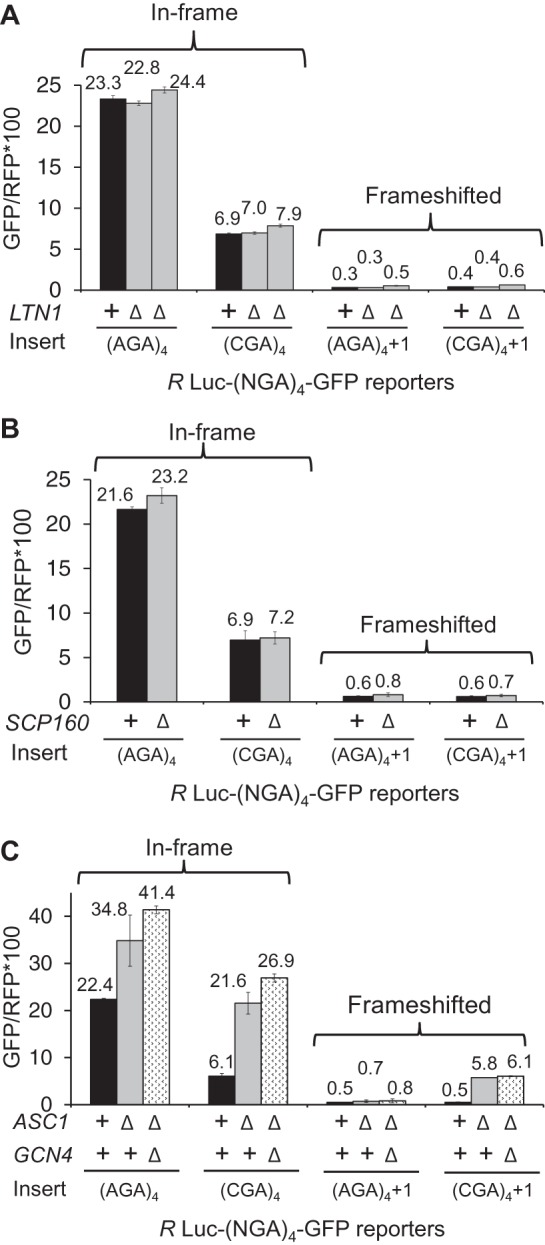
Frameshifting is not observed in ltn1-Δ or scp160-Δ mutants and is not impaired in an asc1-Δ gcn4-Δ mutant. (A) Deletion of LTN1 does not stimulate frameshifting. GFP/RFP expression from in-frame and frameshifted reporters with (CGA)4 and (AGA)4 inserts in wild-type, and two independently constructed ltn1-Δ strains. (B) Deletion of SCP160 does not stimulate frameshifting. GFP/RFP expression from in-frame and frameshifted reporters with (CGA)4 and (AGA)4 inserts in wild-type, and scp160-Δ strains. (C) Frameshifting is not dependent upon activation of the GCN4 pathway. GFP/RFP expression from in-frame and frameshifted reporters with (CGA)4 and (AGA)4 inserts in wild-type, asc1-Δ and double mutant asc1-Δ gcn4-Δ strains.
TABLE 1.
Deletion of DOM34 or HBS1 does not stimulate frameshifting
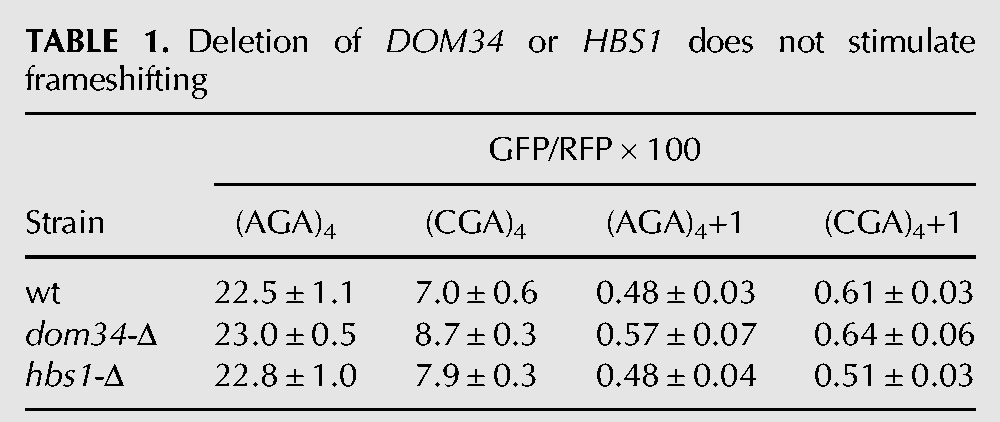
TABLE 2.
Inactivation of nonsense-mediated decay does not stimulate frameshifting
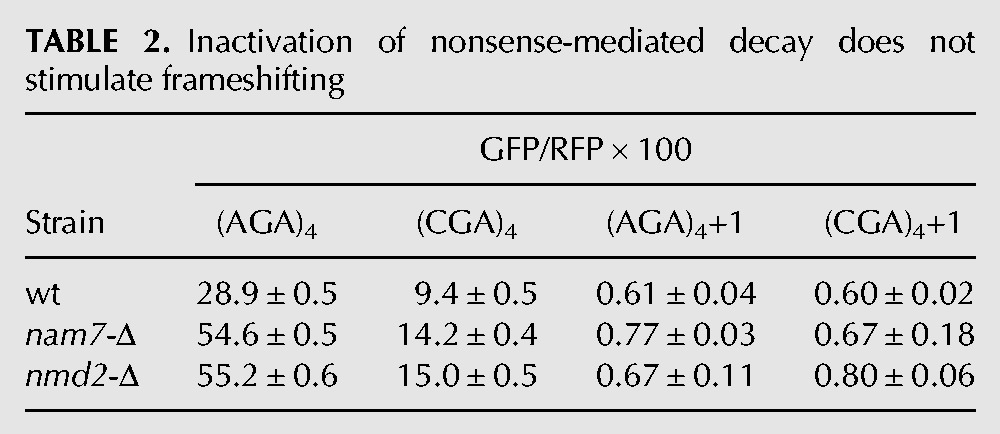
TABLE 3.
GFP/RFP and GFP/actin mRNA in nmd2-Δ and nmd2-Δ asc1-Δ strains
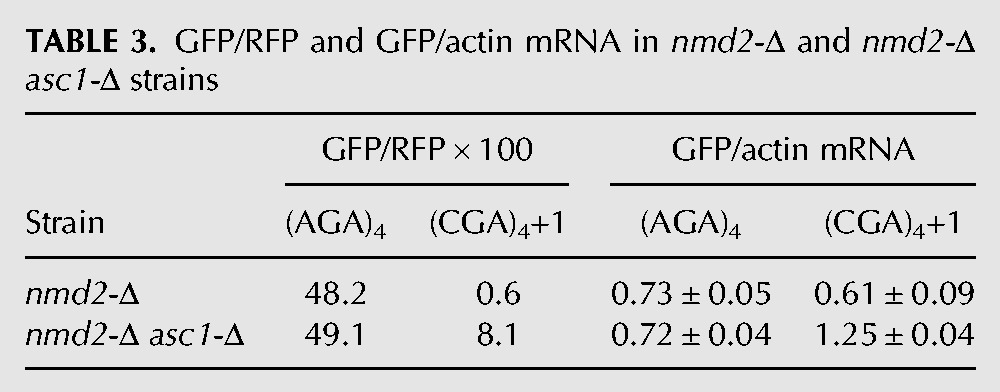
TABLE 4.
Deletion of HEL2 or RPL1B does not stimulate frameshifting
We considered that frameshifting might be dependent upon other known interactions of the Asc1 protein or known effects caused by mutating ASC1. However, deletion of SCP160, an mRNA binding protein recruited to the ribosome by Asc1 (Baum et al. 2004), is not sufficient to cause frameshifting (Fig. 3B). Furthermore, frameshifting does not require the function of the Gcn4 pathway, which is induced in rich media in the asc1-Δ mutant, since frameshifting is not reduced in the double asc1-Δ gcn4-Δ mutant (Fig. 3C; Hoffmann et al. 1999). Thus, frameshifting in the asc1-Δ strain is not simply explained by known quality control systems, or altered CGA decoding, or known Asc1 interactions.
Frameshifting depends upon CGA codon dosage
To determine if frameshifting is primarily dependent upon the number of CGA codons, or if there are additional constraints upon frameshifting, we examined the effects of increasing numbers of adjacent CGA codons on both in-frame and frameshifted expression of GFP/RFP in the wild-type and asc1-Δ strains (Fig. 4). We have previously noted that adjacent CGA–CGA codons (CGA codon pairs) are far more efficient inhibitors of expression than individual CGA codons. Thus, we increased CGA codon pairs by adding adjacent CGA codons (Fig. 4A). (Note each additional CGA codon [after two CGA codons] adds another CGA codon pair: a CGA1–CGA2–CGA3 sequence includes two CGA codon pairs since the ribosome first translates CGA1–CGA2 and then CGA2–CGA3.)
FIGURE 4.
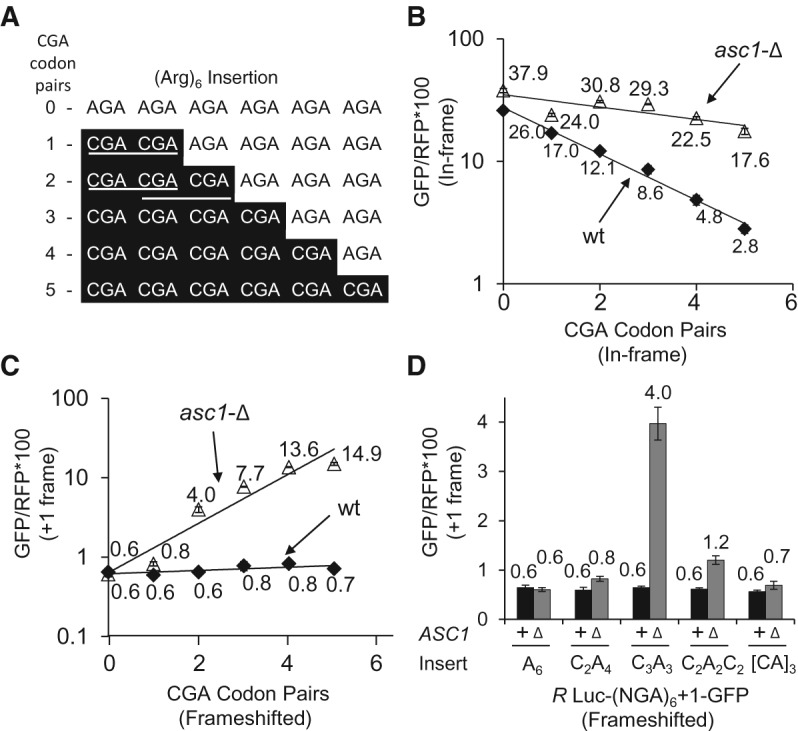
Frameshifting efficiency rises with increased CGA–CGA codon pair dosage. (A) Schematic of constructs made to measure the dose dependence of CGA codon pairs on expression and frameshifting. CGA codon pairs are denoted with the white lines for the (CGA)2(AGA)4 and (CGA)3(AGA)3 constructs, illustrating that addition of a third CGA codon results in two CGA–CGA codon pairs. (B) Exponential decrease in GFP/RFP as function of CGA–CGA codon pair dosage for the in-frame Renilla Luciferase-GFP reporter in wild-type and asc1-Δ strains. (C) Exponential increase in GFP/RFP from the +1 frameshifted Renilla Luciferase-GFP reporter as function of CGA–CGA codon pair dosage in the asc1-Δ strain. Frameshifting is not observed in the wild-type strain. (D) Comparison of frameshifting with a single (CGA)3 triple, with one or two separated (CGA)2 pairs. AGA codons are designated as A and CGA codons are designated as C.
In-frame expression of GFP/RFP decreases progressively with increasing numbers of CGA codon pairs in both strains (Fig. 4B) and fits a single exponential with exponential coefficients of inhibition (kTI) (Letzring et al. 2010) that are consistent with differences in the effectiveness of the CGA inhibition in the wild-type (kTI = −0.44) and asc1-Δ (kTI = −0.12) strains. We note that the best fit occurs if the exponential is fit to CGA codon pairs. Frameshifted GFP/RFP increases progressively with increasing numbers of CGA codon pairs in the asc1-Δ strain (and also fits a single exponential), although there is little or no indication of frameshifting with this construct in the wild-type strain (Fig. 4C). Frameshifted GFP/RFP reaches a maximum of 14.9 with six adjacent CGA codons, equivalent to ∼38% frameshifted product (relative to the in-frame AGA control). Thus, CGA-mediated frameshifting is limited primarily by the number of adjacent CGA codons.
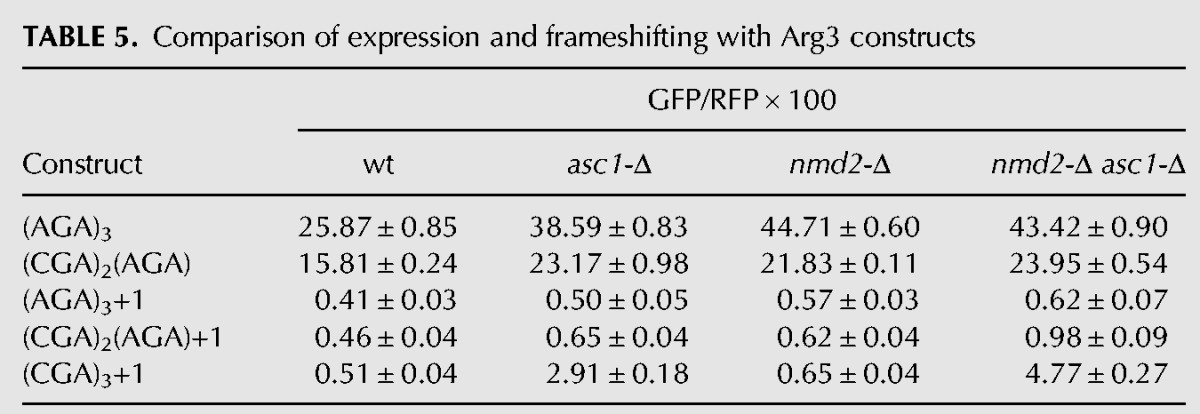
We noted that frameshifting with two adjacent CGA codons, a single CGA–CGA pair, is relatively low (GFP/RFP is 0.8 ± 0.05), but is greater than GFP/RFP observed from either the (AGA)6+1 construct or the identical (CGA)2(AGA)4+1 construct in the wild-type strain (GFP/RFP is 0.6 ± 0.04) (Fig. 4C). We also noted that GFP/RFP was higher from a construct with two CGA–CGA pairs that are separated from each other by two AGA codons (GFP/RFP is 1.2) (Fig. 4D), although even more frameshifting is seen with three adjacent CGA codons (GFP/RFP is 4.0). To establish whether or not frameshifting occurs at a single CGA–CGA codon pair, we compared GFP/RFP expression in a set of Arg3 constructs encoded with (AGA)3+1, [(CGA)2(AGA)+1] and (CGA)3+1 in both an asc1-Δ mutant and in an nmd2-Δ asc1-Δ mutant, subjecting the resulting values to Welch's 1-tailed t-test. In both strains, frameshifted GFP/RFP from the [(CGA)2(AGA)+1] construct was significantly greater than the corresponding frameshifted GFP/RFP from the (AGA)3+1 construct (Table 5) with P values <1 × 10−5. Based on these results, we conclude that ribosomes lacking Asc1 do frameshift at a CGA–CGA pair, albeit inefficiently, and thus that frameshifting could occur in the 26 yeast genes with CGA–CGA codon pairs (Tumu et al. 2012).
TABLE 5.
Comparison of expression and frameshifting with Arg3 constructs
The nascent polypeptide length modulates frameshifting, as well as CGA-mediated inhibition
We considered that the effects of Asc1 on frameshifting, as well as on suppression of CGA-mediated inhibition, might depend upon the distance between the CGA codon repeat and the translation start site, since we had previously observed that deletion of ASC1 improved read-through of CGA codon repeats at amino acid 318, but not at amino acid 4 (Letzring et al. 2013). To investigate this, we altered the location of the codon insert by encoding distinct segments of the amino-terminal domain (NTD) of the GLN4 gene upstream of these codons. The GLN4 NTD is highly expressed in yeast and forms a well-structured domain with several distinct α helices (Grant et al. 2011), allowing us to position the CGA codons between α helices (Fig. 5A). Control experiments show similar levels of expression of each of the GLN4-(AGA)4-GFP fusion proteins (Table 6).
FIGURE 5.
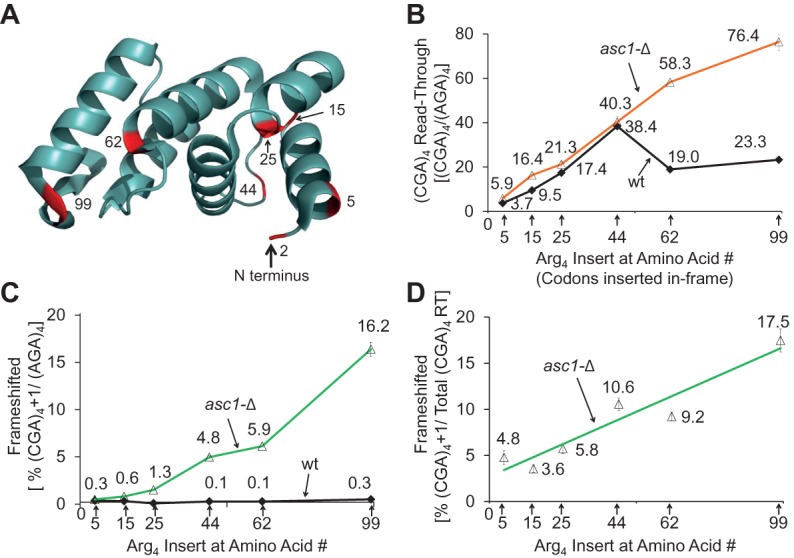
The length of coding sequence upstream of the CGA codon insert modulates CGA-mediated frameshifting and inhibition. (A) Structure of the Gln4 NTD (amino acids 1–187) based on the crystal structure (PDB ID: 3TL4) (Grant et al. 2011), indicating the location of the C termini used in constructs in B–D. Effects of the length of the nascent polypeptide preceding the CGA codons on CGA-mediated inhibition (B) and frameshifting (C,D) in wild-type and asc1-Δ yeast strains. (CGA)4-GFP and (AGA)4-GFP were inserted downstream from the indicated number of amino acids derived from the GLN4 NTD, starting at the native GLN4 AUG. In B, read-through for each in-frame (CGA)4 construct is reported as the percentage of the expression of the in-frame (AGA)4 construct with the identical leader ([GFP/RFP from (CGA)4 construct/GFP/RFP from (AGA)4 construct] × 100). In C, expression of the frameshifted (CGA)4+1 constructs are reported as the percentage of the in-frame (AGA)4 construct with the identical leader. In D, expression of the frameshifted (CGA)4+1 constructs in the asc1-Δ strain are reported as the percentage of total GFP/RFP from read-through (RT) at CGA codons, that is GFP/RFP from the in-frame (CGA)4 construct plus GFP/RFP from the frameshifted (CGA)4+1. GFP/RFP expression of each construct is reported in Table 6.
TABLE 6.
Expression of the GLN4-(NGA)4-GFP fusion proteins
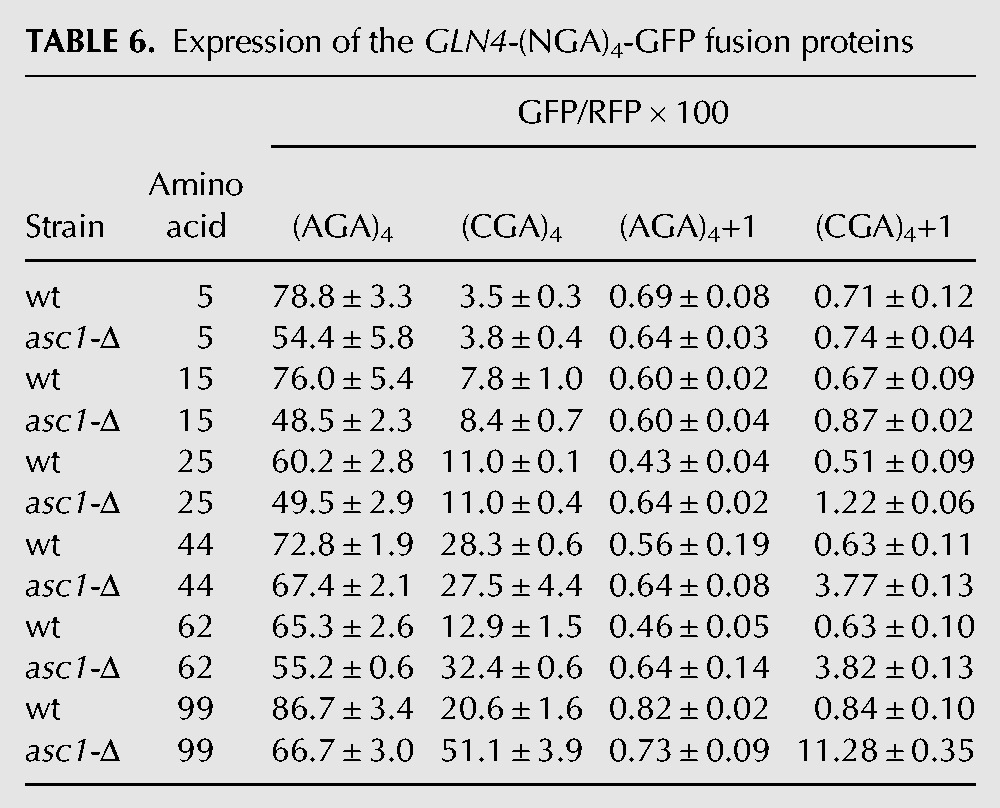
We find that the distance between the initiating methionine and the CGA codons influences three aspects of CGA-mediated effects on translation. First, although four adjacent CGA codons are inhibitory at every position in a wild-type strain, in-frame translation of GFP downstream from four CGA codons (relative to the identical construct with four AGA codons) increases approximately linearly as the insertion position is increased, up to GLN4 amino acid 44, in both the wild-type and asc1-Δ strains. Each construct is designated by the amino acid position in GLN4 after which the CGA or AGA codons are inserted. Expression of the (CGA)4 construct with insertion beginning after amino acid 5 results in 2.9 units GFP/RFP (3.2 units in the asc1-Δ strain) (after subtraction of 0.58 units background) and is 3.7% (5.9% in the asc1-Δ) that of the (AGA)4 construct (Fig. 5B). Expression of the (CGA)4 construct in which the CGA codons are inserted after GLN4 amino acid 44 increases to GFP/RFP of 27.8 (26.9), which is 38.4% (40.3%) that of the corresponding (AGA)4 construct (Fig. 5B). Second, suppression of CGA-mediated inhibitory effects by the asc1-Δ mutation is not apparent until CGA codons are positioned after amino acid 62 (31.8 GFP/RFP [58.3%] in asc1-Δ versus 12.3 [19.0%] in the wild-type strain) (Fig. 5B). The observation that suppression by asc1-Δ is not observed until amino acid 62 is consistent with the idea that CGA codons near the initiating methionine inhibit expression in a manner that is independent of Asc1-mediated effects. Thus, there are likely two modes of codon-mediated inhibition.
Third, frameshifting in the asc1-Δ strain is detectable (albeit minimally) with (CGA)4+1 inserted after amino acid 15, and increases steadily with (CGA)4+1 inserts after amino acid 25, 44, 62, and 99 (Fig. 5C). GFP/RFP levels rise from 0.16 GFP/RFP (0.3%) at amino acid 5 to 10.7 GFP/RFP (16.2%) (Table 6). Controls reveal very little, if any, (<0.26 GFP/RFP) frameshifting with any construct in the wild-type strain or with the (AGA)4 codon insert in either strain (Table 6). Moreover, if we consider the fraction of GFP/RFP that derives from frameshifting at CGA codons relative to the total GFP/RFP from both in-frame and frameshifted (CGA)4 constructs, we find an increase in the percentage of frameshifted GFP/RFP with distance from the initiating methionine (Fig. 5D), at least over this range of amino acids. We presume that frameshifting has reached a maximum by amino acid 99 since frameshifting is similar at amino acid 318 in the Renilla luciferase-GFP constructs. Thus, among ribosomes that continue translation past CGA codons, frameshifting appears to be the more favored outcome as the distance from the initiating methionine increases.
DISCUSSION
We have provided evidence that the Asc1/RACK1 protein plays a role in reading frame maintenance, a critical feature of translation, and infer that Asc1-mediated cessation of translation is important to avoid synthesis of aberrant proteins through frameshifting, although the precise nature of the frameshifting event (+1 or −2) (Fang et al. 2012) that occurs in the asc1-Δ mutant is unknown. Moreover, the effects of the asc1-Δ mutant may be physiologically relevant, since Asc1 protein levels are decreased substantially during hypoxic stress (Bruckmann et al. 2009), and since frameshifting is detectable (albeit minimally) at CGA–CGA codon pairs, which are found in 26 yeast genes, with two occurrences of CGA–CGA pairs in one gene (BUD8). In most cases, the CGA codons are located well into the coding sequence, median position beginning at codon 209 and in only five genes are the CGA–CGA codon pairs found in the first 50 codons. Furthermore, the finding that frameshifting occurs at CGA–CGA codon pairs leads to speculation that such frameshifting occurs at some other inhibitory codon combinations.
Decoding at programmed frameshifts and at CGA codon repeats share two features that may be key to ribosomal frameshifting at CGA repeats. First, both types of frameshifting are associated with ribosome stalls. Thus, −1 and +1 programmed frameshifting events depend upon a ribosome stall, mediated either by codons at the frameshift site, by structures downstream from the frameshift site or by sequences upstream of the frameshift site (Dinman 2012), although stalling is not sufficient to cause frameshifting (Tu et al. 1992; Somogyi et al. 1993; Dinman 2012). Similarly, ribosomes also stall at CGA codon repeats as evidenced by mRNA cleavage at these sites (Chen et al. 2010; Letzring et al. 2010), by detection of the nascent polypeptide arrested at the CGA repeat and by destruction of the nascent polypeptide by Ltn1 (Letzring et al. 2013), a member of the RQC complex (Kuroha et al. 2010; Brandman et al. 2012). This ribosome stalling may mechanistically aid frameshifting. Ribosomes stalled at the −1 programmed frameshifting site in the E. coli dnaX gene undergo fluctuations between hybrid and classical states due to an energy barrier to translocation (Kim et al. 2014). Since it is known that yeast ribosomes can be found in two distinct conformations at different codons (Lareau et al. 2014), it is reasonable to speculate that ribosomes at CGA codon repeats might also undergo fluctuations between states.
Second, both classes of frameshifting may require a poorly decoded P site codon. In several examples, +1 programmed frameshifting depends critically on a poorly decoded P site codon, such as CUU or GCG, each decoded by wobble (Belcourt and Farabaugh 1990; Baranov et al. 2004; Dinman 2012). CGA codons, like CUU and GCG, are inefficiently decoded, and require an I·A wobble interaction, which results in distortion of the tRNA anticodon in Thermus thermophilus 30S ribosomal subunits (Murphy and Ramakrishnan 2004). Moreover, we know that I·A wobble decoding of CGA codons results in poor decoding in both the A and the P sites of the ribosome, since CGA–CGA codon pairs are synergistically inhibitory (Letzring et al. 2010), and since frameshifting requires a CGA–CGA pair. Thus poor decoding of CGA in the P site is implicated in frameshifting at CGA codon repeats. However, CGA repeats lack some sequences that facilitate frameshifting, such as the Ty1 +1 frameshift site, which has a CUU A sequence on which tRNALeu(UAG) can slip from CUU to UUA (Belcourt and Farabaugh 1990). On the other hand, there are +1 programmed frameshifting sites, such as Ty3, that lack these slippery sequences (Guarraia et al. 2007).
The specific manner in which Asc1 blocks read-through of CGA codons and prevents frameshifting is unknown. Although it is unproven that the loss of reading frame maintenance and suppression of CGA inhibitory effects are a direct consequence of translation by ribosomes lacking Asc1 protein, as compared with altered expression of another gene in the asc1-Δ mutant, this is the most parsimonious explanation of these events. Furthermore, the known interactions of Asc1 with ribosomal protein rpS3e could provide a clue to the manner in which the stall state may be transmitted to Asc1. In bacteria, rpS3p is found in the mRNA channel near a conserved base in the 16S rRNA that interacts with mRNA and undergoes conformational rearrangements during the elongation cycle; this base in 16S rRNA has been proposed to act as a pawl to hold the mRNA in place (Zhou et al. 2013). In eukaryotes, rpS3e contains additional sequences, so that rpS3e traverses the small subunit from the mRNA channel to RACK1 (near the 40S beak) (Rabl et al. 2011). Thus, changes in rpS3e might occur as the ribosome shifts between states, and such changes could affect the interaction between rpS3e and Asc1, allowing Asc1 to stop translation.
We also found that three phenomena, CGA-mediated inhibition in wild-type yeast, CGA-mediated frameshifting and suppression of CGA-mediated inhibition in asc1-Δ mutants, all depend upon distance from the initiating methionine. Since CGA codons near the initiating methionine inhibit translation in a manner independent of Asc1 protein, while much of the inhibition by CGA codon repeats downstream from amino acid 62 depends upon Asc1 protein, there are likely to be at least two mechanisms by which CGA codon repeats inhibit translation. In contrast to CGA-mediated inhibition of translation, frameshifting at the CGA codons always requires the absence of Asc1 protein.
There are at least two models that would explain the Asc1-independent inhibition by CGA codons in the first 40–60 amino acids, each of which might also explain how inhibitory effects wane with increasing distance from the initiating methionine. The first model is based on results from Chu et al. (2014) who suggest that slowly elongating ribosomes may restrict initiation by an incoming ribosome; that is, that codon-mediated effects on the translation elongation rate could result in reduced rates of translation initiation. In this model, slow translation of CGA codons closer to the initiation site (in which stalled ribosomes would directly affect the next initiating ribosome) would be more inhibitory than slow translation further from the initiation site; a feedback mechanism of this type would be independent of Asc1 protein and might only operate near the initiation site. The second model is based on the idea that the efficiency of the peptidyl transferase reaction is modulated by the nascent polypeptide within the exit tunnel, an idea for which there is precedent from nascent peptide sequences that stall the ribosome (Ramu et al. 2011; Martinez et al. 2014), and evidence for contact between the nascent polypeptide and 28S rRNA at the peptidyl transferase center (Bhushan et al. 2010). In this model, which is similar to the promoter clearance step in transcription, the ribosome becomes committed to continued elongation as the exit tunnel fills, and thus CGA codons become less inhibitory as the exit tunnel fills. The exit tunnel accommodates 30–40 amino acids in unfolded form and up to 72 amino acids in an α-helical fold (Malkin and Rich 1967; Blobel and Sabatini 1970; Kramer et al. 2001); the large effects on modulating CGA codon inhibition occur in this same range. It is intriguing that suppression caused by loss of Asc1 occurs at a point where most of the exit tunnel would be filled. Subsequent experiments are needed to shed light on the mechanism by which Asc1 mediates CGA inhibition and prevents loss of reading frame.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides
Strains, plasmids, and oligonucleotides used in these studies are listed in Supplemental Tables S1–S3, respectively. The yeast strain BY4741 (MATa his3Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0) (Open Biosystems) was the parent strain for all yeast manipulations. Yeast strains bearing deletions of NAM7, NMD2, and SCP160 were constructed by PCR amplification of the kanR cassette from the systematic deletion collection (Open Biosystems). Deletions of DOM34 and HBS1 were constructed by amplification of the bleoR cassette from pUG66 (Gueldener et al. 2002) with oligonucleotides homologous to the 5′ and 3′ ends of the genes. Yeast strains with deletions in RPL1B, HEL2, LTN1, and GCN4 have been described (Letzring et al. 2013).
The yeast strain AW768 bearing the asc1-Δ::spHIS5, and the plasmid pURA3, ASC1 was made by first creating the asc1-Δ::spHIS5 cassette using pUG27 (Gueldener et al. 2002) and primers homologous to the 5′ and 3′ ends of ASC1 (OW125, OW126), and then integrating this cassette into an asc1-Δ::kanMX strain (AW714) derived from the corresponding knockout strain in the systematic deletion collection (Open Biosystems). Then, the asc1-Δ::spHIS5 cassette with flanking regions was obtained by genomic PCR of a His+ isolate, and integrated into a strain AW765 (which is derived from BY4741 and carries the plasmid URA3 2μ ASC1 plasmid, pEAW076). Transformants were screened for colonies that retained their His+ phenotype after removal of the [ASC1] covering plasmid by FOA treatment. The resulting strain AW768 (MATa his3-Δ1, leu2-Δ0, met15-Δ0, ura3-Δ0, asc1-Δ::spHIS5, pURA3, ASC1) was verified by confirmation PCR, and a test for sensitivity to cycloheximide.
The plasmid vector pEAW012, in which Renilla luciferase is fused to GFP under PGAL1 control, was obtained by PCR amplification of the Renilla luciferase gene from pDL202 (Letzring et al. 2010) using oligonucleotides OW090 and OW091, which was inserted into the Pac1 site of pEKD1024 (Dean and Grayhack 2012) using LIC cloning; this regenerated a Pac1 site as well as a LIC site for use in cloning codon inserts as Renilla luciferase-GFP fusion proteins. Oligonucleotides carrying codon sequences were annealed and inserted in-frame or out of frame with GFP by insertion into the PacI and BbrPI sites of pEAW012 using LIC.
Plasmids containing truncated GLN4 (Grant et al. 2011) fused to GFP were obtained by cloning the GLN4-codon inserts (with or without a frameshifting nucleotide) into the PacI and BbrPI sites of pEKD1024 using LIC cloning (Dean and Grayhack 2012). The GLN4 sequences corresponding to amino acids 1–5 and 1–15 were obtained by annealing oligonucleotides with the sequences to be inserted. The GLN4 sequences encoding amino acids 1–25, 1–44, 1–62, 1–99 were obtained by PCR amplification from plasmid pJE1012a using the forward primer OJW011 and the reverse oligonucleotide corresponding to the 3′ end of GLN4 (OJY12-OJY19 and OW212-OW219).
The ASC1 2μ URA3 plasmid (pEAW076) was constructed by cloning the ASC1 gene with flanking sequences (−326 to +434) into pECB194 (derived from AVA577) (Alexandrov et al. 2004). The ASC1-Δintron URA3 Cen plasmid pEAW141 was constructed from two fragments joined by LIC methods, followed by ligation and PCR amplification of the joined fragments. Sequence from −496 to +537, directly upstream of the intron, were amplified with oligonucleotides OW119 and OW121, while sequences from 811, immediately downstream from the intron, through the coding region to +537 downstream from the TAA stop codon were amplified with oligonucleotides OW122 and OW123. The oligonucleotide OW122 had an additional 12 bases on the 5′ end, to generate homologous ends with the upstream PCR product. Twelve base single stranded homologous ends on each PCR product were generated by treatment with T4 DNA Polymerase, subsequently annealed and ligated, which enabled PCR amplification of the entire ASC1 coding sequence with flanking regions (−482 to +445) using primers OW120 and OW124. The PCR product was cloned into pAVA577 (Alexandrov et al. 2004) using LIC cloning to make pEAW141.
Flow cytometry
Yeast strains bearing the R Luc-(NGA)x-GFP, GFP or GLN4-(NGA)4-GFP reporters were constructed by integrating each Stu1-digested reporter into the ADE2 locus (Dean and Grayhack 2012) and selecting for growth on SD-Met-Ura to retain the ASC1 plasmid. Transformants were streaked onto SD-Met-Ura, grown overnight in YPD or SD-Met media, and streaked for single colonies onto SD-Met + FOA for 2–3 d. FOA treated single colonies were grown in YP + Raff + Gal + Ade at 30°C to an OD600 of 0.6–1.5 and analyzed using flow cytometry. Strains bearing LEU2 plasmids followed a similar treatment except that they were streaked onto SD-Leu + FOA plates, and grown in S-Leu + Raff + Gal + Ade at 30°C for flow cytometry. Analytical flow cytometry and subsequent data analysis were performed as previously described (Dean and Grayhack 2012). Mean GFP/RFP values and their standard deviations reported in the bar graphs are obtained from the median GFP/RFP from measurements of three (and usually four) independent yeast transformants.
Protein expression analysis
Western analysis of the R Luc-(NGA)4-GFP and (AGA)3-GFP with anti-HA antibody was performed as described previously (Gelperin et al. 2005).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eric Phizicky, Christina Brule, Stanley Fields, Gloria Culver, Anita Hopper, and Yi-Tao Yu for comments on the manuscript, Christina Brule, Jiyu Wang, and So-Yon Jang for assistance with experiments, and Jeffrey Zuber and Michael Gleghorn for help with PyMol. Support for this work was provided by National Science Foundation (NSF) grant MCB-1329545 to E.J.G.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.049080.114.
REFERENCES
- Adams DR, Ron D, Kiely PA 2011. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, et al. 2004. A facile method for high-throughput co-expression of protein pairs. Mol Cell Proteomics 3: 934–938. [DOI] [PubMed] [Google Scholar]
- Balagopal V, Parker R 2011. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17: 835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF 2004. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum S, Bittins M, Frey S, Seedorf M 2004. Asc1p, a WD40-domain containing adaptor protein, is required for the interaction of the RNA-binding protein Scp160p with polysomes. Biochem J 380: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcourt MF, Farabaugh PJ 1990. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62: 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467: 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR 2010. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol 30: 5562–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R 2010. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol 17: 313–317. [DOI] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD 1970. Controlled proteolysis of nascent polypeptides in rat liver cell fractions. I. Location of the polypeptides within ribosomes. J Cell Biol 45: 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckmann A, Hensbergen PJ, Balog CI, Deelder AM, Brandt R, Snoek IS, Steensma HY, van Heusden GP 2009. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J Proteomics 71: 662–669. [DOI] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhäuser N, Marchisio PC, Biffo S 2003. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426: 579–584. [DOI] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H 2010. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol 17: 1233–1240. [DOI] [PubMed] [Google Scholar]
- Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, et al. 2009. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci 106: 2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T 2014. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J 33: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA 2009. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 29: 1626–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean KM, Grayhack EJ 2012. RNA-ID, a highly sensitive and robust method to identify cis-regulatory sequences using superfolder GFP and a fluorescence-based assay. RNA 18: 2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD 2012. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA 3: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, et al. 2012. Efficient −2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci 109: E2920–E2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O'Donnell K, Guerrerio AL, Parker R, Dietz HC 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ 2004. Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol 24: 8276–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant TD, Snell EH, Luft JR, Quartley E, Corretore S, Wolfley JR, Snell ME, Hadd A, Perona JJ, Phizicky EM, et al. 2011. Structural conservation of an ancient tRNA sensor in eukaryotic glutaminyl-tRNA synthetase. Nucleic Acids Res 40: 3723–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarraia C, Norris L, Raman A, Farabaugh PJ 2007. Saturation mutagenesis of a +1 programmed frameshift-inducing mRNA sequence derived from a yeast retrotransposon. RNA 13: 1940–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, Green R 2014. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Mösch HU, Sattlegger E, Barthelmess IB, Hinnebusch A, Braus GH 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol Microbiol 31: 807–822. [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2007. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev 21: 1833–1856. [DOI] [PubMed] [Google Scholar]
- Kim HK, Liu F, Fei J, Bustamante C, Gonzalez RL Jr, Tinoco I Jr 2014. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci 111: 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kramer G, Ramachandiran V, Hardesty B 2001. Cotranslational folding—omnia mea mecum porto? Int J Biochem Cell Biol 33: 541–553. [DOI] [PubMed] [Google Scholar]
- Kurland CG 1992. Translational accuracy and the fitness of bacteria. Annu Rev Genet 26: 29–50. [DOI] [PubMed] [Google Scholar]
- Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T 2010. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep 11: 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO 2014. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3: e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ 2010. Control of translation efficiency in yeast by codon–anticodon interactions. RNA 16: 2516–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Wolf AS, Brule CE, Grayhack EJ 2013. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 19: 1208–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin LI, Rich A 1967. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J Mol Biol 26: 329–346. [DOI] [PubMed] [Google Scholar]
- Martinez AK, Gordon E, Sengupta A, Shirole N, Klepacki D, Martinez-Garriga B, Brown LM, Benedik MJ, Yanofsky C, Mankin AS, et al. 2014. Interactions of the TnaC nascent peptide with rRNA in the exit tunnel enable the ribosome to respond to free tryptophan. Nucleic Acids Res 42: 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem 76: 101–124. [DOI] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V 2004. Structure of a purine–purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 11: 1251–1252. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736. [DOI] [PubMed] [Google Scholar]
- Rachfall N, Schmitt K, Bandau S, Smolinski N, Ehrenreich A, Valerius O, Braus GH 2013. RACK1/Asc1p, a ribosomal node in cellular signaling. Mol Cell Proteomics 12: 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu H, Vazquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell 41: 321–330. [DOI] [PubMed] [Google Scholar]
- Seidman JS, Janssen BD, Hayes CS 2011. Alternative fates of paused ribosomes during translation termination. J Biol Chem 286: 31105–31112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11: 957–962. [DOI] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R 2011. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci 108: E1392–E1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R 2012. Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Jenner AJ, Brierley I, Inglis SC 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol 13: 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T 2012. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell 46: 518–529. [DOI] [PubMed] [Google Scholar]
- Tu C, Tzeng TH, Bruenn JA 1992. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci 89: 8636–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumu S, Patil A, Towns W, Dyavaiah M, Begley TJ 2012. The gene-specific codon counting database: a genome-based catalog of one-, two-, three-, four- and five-codon combinations present in Saccharomyces cerevisiae genes. Database (Oxford) 2012: bas002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Frischmeyer PA, Dietz HC, Parker R 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Meaux S, van Hoof A 2007. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci 110: E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF 2013. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340: 1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


