Abstract
Plants have evolved sophisticated genetic and epigenetic regulatory systems to respond quickly to unfavorable environmental conditions such as heat, cold, drought, and pathogen infections. In particular, heat greatly affects plant growth and development, immunity and circadian rhythm, and poses a serious threat to the global food supply. According to temperatures exposing, heat can be usually classified as warm ambient temperature (about 22–27°C), high temperature (27–30°C) and extremely high temperature (37–42°C, also known as heat stress) for the model plant Arabidopsis thaliana. The genetic mechanisms of plant responses to heat have been well studied, mainly focusing on elevated ambient temperature-mediated morphological acclimation and acceleration of flowering, modulation of circadian clock and plant immunity by high temperatures, and thermotolerance to heat stress. Recently, great progress has been achieved on epigenetic regulation of heat responses, including DNA methylation, histone modifications, histone variants, ATP-dependent chromatin remodeling, histone chaperones, small RNAs, long non-coding RNAs and other undefined epigenetic mechanisms. These epigenetic modifications regulate the expression of heat-responsive genes and function to prevent heat-related damages. This review focuses on recent progresses regarding the genetic and epigenetic control of heat responses in plants, and pays more attention to the role of the major epigenetic mechanisms in plant heat responses. Further research perspectives are also discussed.
Keywords: heat, genetic mechanism, epigenetic regulation, small RNAs, transgenerational memory
Introduction
Owing to the global warming, the annual mean maximum and minimum temperatures have been reported to increase by 0.35 and 1.13°C, respectively, for the period 1979–2003 (Peng et al., 2004). It is probable that the growing season temperatures in the tropics and subtropics by the end of the 21st century will exceed the most extreme seasonal temperatures recorded from 1900 to 2006 (Battisti and Naylor, 2009). Global warming has profound and diverse effects on plants. Warmer temperature has advanced the average first flowering date of 385 British plant species by 4.5 days (Fitter and Fitter, 2002). Besides, recent climate warming (2001–2008) has shifted vascular plant species’ ranges to higher altitudes in European mountainous regions (Pauli et al., 2012). Furthermore, it is noteworthy that climate warming poses a serious threat to the global crop yields. Over the past three decades (1980–2008), heat has caused a decrease of 3.8 and 5.5% in the global maize and wheat production (Lobell et al., 2011). It is estimated that global yields of the six most widely grown crops (wheat, rice, maize, soybeans, barley, and sorghum) drop by 0.6 ∼ 8.9% for every 1°C the temperature increases (Lobell and Field, 2007). In the dry season, global warming has been estimated to cause a ∼10% reduction in rice yield for every 1°C increase in growing-season minimum temperature (Peng et al., 2004). Thus, it is critical to dissect the heat sensing and signal transduction pathways in plants.
Genetic knowledge of plant responses to heat stress has been accumulating, including several putative heat sensors, HSFs and HSPs (heat shock factors and proteins) response pathways, and the network of phytohormones, chaperones, and secondary metabolites (Bokszczanin and Fragkostefanakis, 2013; Qu et al., 2013). However, our understanding of plant responses to warm temperature is limited, despite recent discoveries implicating the central role of the basic helix-loop-helix (bHLH) transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4) in warmth-mediated morphological acclimation and acceleration of flowering (Proveniers and van Zanten, 2013). The modulation of circadian clock and immunity by high temperature also remains largely unknown. Recently, epigenetic regulations of heat responses have attracted increasing interests. The epigenetic mechanisms in response to heat include covalent modifications of DNA and histones, histone variants, ATP-dependent chromatin remodeling, histone chaperones, small RNAs, long non-coding RNAs (lncRNAs), and other undefined mechanisms (Table 1). This review briefly introduces the genetic mechanisms of plant responses to heat and highlights recent progresses regarding the underlying epigenetic regulations mainly in the Arabidopsis model, with aspects of some important physiological processes.
Table 1.
Different epigenetic regulations involved in different heat responses.
| Plants | Heat treatment | Major effects | Major epigenetic regulations | Reference |
|---|---|---|---|---|
| Saccharina japonica | 20°C for 3 h | Regulation of tolerance to heat stress | miRNAs | Liu et al. (2014) |
| Arabidopsis thaliana | Grown at 26°C | Elevated survival of Turnip Crinkle Virus-infected Plants | siRNAs | Zhang et al. (2012) |
| A. thaliana | Grown at 27°C | Early flowering, hypocotyl and petiole elongation | H2A.Z | Kumar and Wigge (2010), Kumar et al. (2012) |
| Nicotiana benthamiana | Grown at 27°C | Enhanced antiviral defense | siRNAs | Qu et al. (2005) |
| N. benthamiana Manihot esculenta | Grown at 25–30°C | Enhanced antiviral defense | siRNAs | Chellappan et al. (2005), Andika et al. (2013) |
| A. thaliana | Grown at 30°C | Warmth-induced PTGS release with transgenerational memory | miRNAs and siRNAs | Zhong et al. (2013) |
| Oryza sativa | 34°C for 48 h | Smaller seed size | DNA methylation, H3K9me2 | Folsom et al. (2014) |
| Hordeum vulgare | 35.5°C for 24 h | Regulation of tolerance to heat stress | miRNAs | Kruszka et al. (2014) |
| Gossypium hirsutum | 35–39°C /29–31°C day/night for 7 days | Regulation of anther development | DNA methylation, histone modifications | Min et al. (2014) |
| A. thaliana | 37°C for 3 h/day during the day for 1 week | Increase in homologous recombination frequency with transgenerational memory | DNA methylation, small RNAs | Boyko et al. (2010) |
| A. thaliana | 37°C for 1–4 h | Regulation of tolerance to heat stress | Ta-siRNAs and miRNAs | Guan et al. (2013), Li et al. (2014b) |
| Populus tomentosa | 37°C for 8 h | Regulation of tolerance to heat stress | miRNAs | Chen et al. (2012) |
| A. thaliana | 37°C for 12 h | Regulation of tolerance to heat stress | long non-coding RNAs | (Di et al., 2014) |
| A. thaliana | 37°C for 12 h | Heat stress-induced alternative splicing of miR400 | MiR400 | Yan et al. (2012) |
| A. thaliana | 37°C for 16 h | Mediating the temporary growth arrest | ATP-dependent chromatin remodeling | Mlynarova et al. (2007) |
| A. thaliana | 37°C for 24 h | Transgenerational retrotransposition of ONSEN | siRNAs | Ito et al. (2011) |
| Helianthus annuus | 37°C for 24 h | Regulation of tolerance to heat stress | miRNA396 | Giacomelli et al. (2012) |
| P. trichocarpa | 37°C for 24 h | Regulation of tolerance to heat stress | miRNAs | Lu et al. (2008) |
| M. esculenta | 37°C for 24 h | Regulation of tolerance to heat stress | miRNAs | Ballen-Taborda et al. (2013) |
| A. thaliana | 37°C for 30 h | Release of TGS with reduced nucleosome occupancy and loss of chromocenter organization | CAF-1-dependent chromatin assembly complex | Pecinka et al. (2010) |
| A. thaliana | 4°C for 1 week and then 37°C for 15 h | Release of TGS | Unorthodox and potentially new mechanisms | Tittel-Elmer et al. (2010) |
| A. thaliana | 37°C for 4 days or 44°C for 30 min | Reorganization of chromatin and release of transcriptional gene silencing, | HIT4-dependent TGS regulation pathway | Wang et al. (2014) |
| A. thaliana | 38°C for 1 h | Down regulation of HSFB2a involved in gametophyte development | Long non-coding antisense RNA asHSFB2a | Wunderlich et al. (2014) |
| Apium graveolens | 38°C for 1 h | Regulation of tolerance to heat stress | miRNAs | Li et al. (2014a) |
| A. thaliana | 38°C for 1.5 h per day in the dark and then returned to normal growth conditions; for 7 consecutive days | Modulation of pattern-triggered immunity | Histone modifications | Singh et al. (2014) |
| A. thaliana | Several heat cycles (37°C for 12 h in the light and 21°C for 12 h in the dark) | Activation of the imprinted gene SDC | An undefined epigenetic mechanism | Sanchez and Paszkowski (2014) |
| Triticum aestivum | 40°C for 1 h | Regulation of tolerance to heat stress | miRNAs and long non-coding RNAs | Xin et al. (2010), Xin et al. (2011) |
| Chlamydomonas reinhardtii | 40°C for 1 h, for three times | Regulation of tolerance to heat stress | Histone modifications and chromatin remodeling | Strenkert et al. (2011) |
| O. sativa | 42°C day/36 °C night for 24 h | Regulation of tolerance to heat stress | miRNAs | Sailaja et al. (2014) |
| T. aestivum | 42°C for 2 h | Regulation of tolerance to heat stress | miRNAs | Kumar et al. (2014) |
| Brassica rapa | 42°C for 3 h per day for 7 days | Stress-induced transgenerational inheritance | miR168 and braAGO1 | Bilichak et al. (2015) |
| A. thaliana | 42°C for 16 h | Transcriptional reprogramming | DNA methylation, histone acetylation | Popova et al. (2013) |
| A. thaliana | 42°C for 48 h | Stress-induced release of GUS silencing | H3K9ac1 and H3K9/14ac2 | Lang-Mladek et al. (2010) |
| A. thaliana | BT and ATa | Gene transcription activation | Histone chaperone ASF1 | Weng et al. (2014) |
| A. thaliana | BT and ATa | Regulation of tolerance to recurring environmental stress | The miR156-SPL module | Stief et al. (2014) |
| Brassica rapa | 46°C for 1 h | Regulation of tolerance to heat stress | miRNAs, nat-siRNAs, chloroplast small RNAs | Wang et al. (2011), Yu et al. 2012b, (2013) |
| A. thaliana | 50°C for 3 h/day for 5 day | Transgenerational phenotypic and epigenetic changes | H3K9 methylation and DNA methylation | Migicovsky et al. (2014) |
| Quercus suber | Temperature increases by 10°C every 3 days from 25–55°C | Acclimation to high temperature | DNA methylation, histone acetylation | Correia et al. (2013) |
aBT, basal thermotolerance, 45°C for 2 h; AT, acquired thermotolerance, pretreated at 37°C for 1–1.5 h and returned to 22°C for several hours or days for recovery, treated at 45°C for 2 h.
Genetic Mechanisms of Plant Responses to Heat
Warm Temperature-Mediated Morphological Acclimation and Acceleration of Flowering
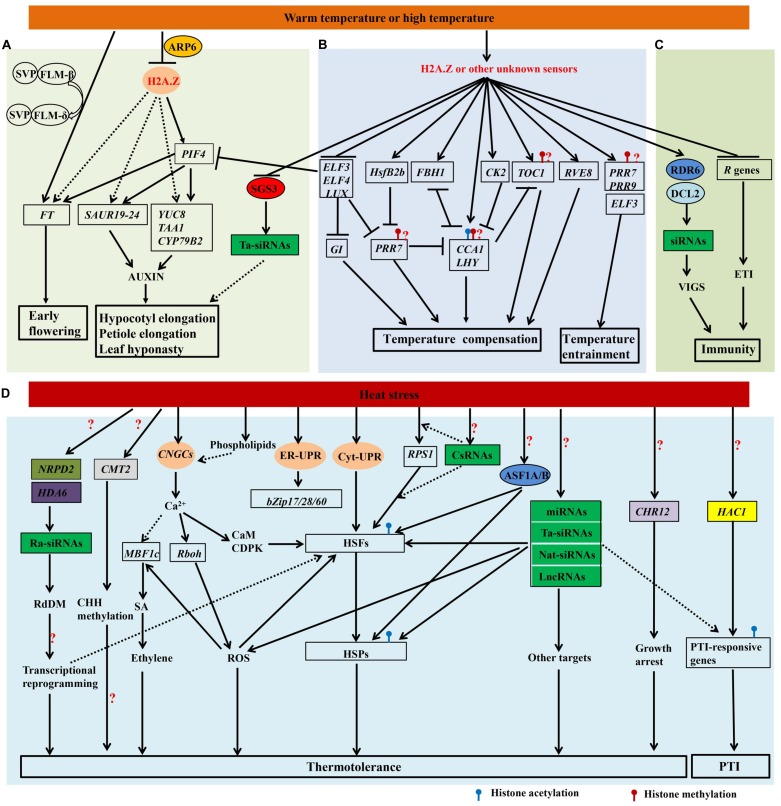
The responses of Arabidopsis plants to warm temperature include hypocotyl and petiole elongation, leaf hyponasty, and early flowering (Gray et al., 1998; Balasubramanian et al., 2006; Koini et al., 2009). Warm temperature promotes auxin accumulation and activate the gibberellin (GA) and brassinosteroids (BRs) pathway resulting in hypocotyl elongation (Gray et al., 1998; Stavang et al., 2009). PIF4 plays a central positive role in the acclimation to increased ambient temperature (Figure 1A; Proveniers and van Zanten, 2013). Warm temperature induces transient expression of PIF4 (Koini et al., 2009; Kumar et al., 2012). PIF4 has been demonstrated to control morphological acclimation to warm temperature via auxin. PIF4 binds to the promoters of the key auxin biosynthesis genes in a temperature-dependent manner (Franklin et al., 2011; Sun et al., 2012). PIF4 may also target the auxin-responsive gene INDOLE-3-ACETIC ACID INDUCIBLE 29 (IAA29) at warm temperature (Koini et al., 2009). Moreover, PIF4 directly or indirectly stimulates the expression of auxin target genes SMALL AUXIN UP RNA (SAUR) 19–24, which drive warmth-induced hypocotyl elongation and probably petiole elongation and leaf hyponasty (Franklin et al., 2011). In addition to morphological acclimation, PIF4 controls warm temperature-mediated floral induction through direct activation of the floral pathway integrator gene FLOWERING LOCUS T (FT) by binding its promoter (Kumar et al., 2012). A receptor-like kinase SCRAMBLED/STRUBBELIG (SCM/SUB) also plays a role in coordinating cell proliferation and differentiation during leaf development under increased ambient temperature (Lin et al., 2012).
FIGURE 1.
Proposed model integrating genetic and epigenetic controls of heat responses. Genes and proteins are represented in boxes and circles, respectively. The genes and proteins in color are involved in the epigenetic regulation of heat responses. The four putative heat sensors, H2A.Z, the calcium channel in the plasma membrane (CNGCs), two unfolded protein sensors in ER (ER-UPR) and the cytosol (Cyt-UPR), are indicated. The speculative regulatory paths are indicated with broken arrows. (A) Warm temperature mediates the morphological acclimation and acceleration of flowering. Under warm temperatures, the expression of PIF4 could be induced by the eviction of H2A.Z at its promoter. PIF4 binds to the promoters of target genes and plays a central role in the morphological acclimation and acceleration of flowering. Warm temperature also induces the transition from SVP-FLM-β to the competing SVP-FLM-δ complex, the latter is then released from the promoter of FT. The inhibition of ta-siRNAs (green box) through the down-regulation of SGS3 protein (red circle) by warm temperature may be also involved in the morphological acclimation. (B) The genetic mechanisms of temperature entrainment and temperature compensation are proposed. ELF3, PRR7 and PRR9 are involved in temperature entrainment, while CCA1, LHY, PRR7, PRR9, GI, CK2, RVE8, FBH1 and HsfB2b are proved to play roles in temperature compensation. Note that histone modifications of LHY, CCA1, TOC1, PRR7 and PRR9, such as H3K56ac, H3K9/14ac, H3K4me3 and H3K4me2, may (question mark) be regulated by high temperatures. (C) High temperature inhibits R genes-mediated ETI and enhances RNA-silencing mediated resistance. Reduced H2A.Z-containing nucleosome occupancy or other unknown mechanism are likely involved in the modulation of clock (B) and immunity (C). (D) Heat sensors and main signal transduction pathways in heat stress responses (HSR) are shown. Heat stress activates CNGCs, ER-UPR, and Cyt-UPR, and triggers signaling through multiple kinases as well as transcriptional regulators of the HSR, such as HSFs, MBF1c, and Rboh. RPS1 in the chloroplast also responds to heat stress, and generates a retrograde signal to activate HsfA2-dependent heat-responsive genes in the nucleus. Some csRNAs are highly sensitive to heat stress and may regulate RPS1-mediated heat stress responses. Heat stress also affects the production of some ra-siRNAs, miRNAs, ta-siRNAs, nat-siRNAs, and lncRNAs. These non-coding RNAs may regulate HSFs, HSPs, and other target genes that function in heat acclimation. The NRPD2 (olive box) and HDA6 (purple box)-dependent RdDM pathway and the CMT2 (gray box)-dependent CHH methylation may be required for thermotolerance. AtASF1A/B proteins (blue circle) are recruited onto chromatin and facilitate H3K56ac, which promotes the activation of some HSFs and HSPs. The chromatin-remodeling gene CHR12 (light purple box) plays a vital role in mediating the temporary growth arrest of Arabidopsis under heat stress. Repetitive heat stress has also been reported to modulate PTI in a HAC1 (yellow box)-dependent manner. Many unknown steps (?) remain to be recognized in this model.
Besides PIF4, MADS-box genes SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING (MAF) 1–5 modulate flowering time in response to ambient temperature changes (Balasubramanian et al., 2006; Lee et al., 2007; Gu et al., 2013). These genes act as flowering repressors and loss of their function leads to accelerated flowering independent of the photoperiod pathway. Interestingly, the RNA processing-related gene products are enriched upon thermal induction, suggesting that temperature might affect RNA processing in Arabidopsis. For instance, FLM is subject to temperature-dependent alternative splicing (Balasubramanian et al., 2006). The SVP-FLM-β complex is predominately formed at 17°C and prevents precocious flowering. By contrast, the competing SVP-FLM-δ complex is impaired in DNA binding and acts as a dominant-negative activator of flowering at 27°C (Figure 1A; Lee et al., 2013; Pose et al., 2013). Therefore, PIF4 is an activator of FT while the MADS-box genes are repressors. However, how the two antagonistic pathways are integrated to modulate flowering time at warm conditions still needs to be genetically dissected.
The Effect of High Temperature on Circadian Clock
As a cellular time-keeper mechanism, the circadian clock allows plants to coordinate environmental time cues, such as photocycles (light/dark) and thermocycles (warm/cold), with endogenous biological rhythms with a period of ∼24 h. Recent studies have suggested that plant circadian clock consists of three interlocked transcriptional feedback loops, i.e., a core oscillator loop, a morning loop, and an evening loop (Hsu and Harmer, 2014). Key players in this interconnected network are two MYB transcription factors CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION1/PSEUDO-RESPONSE REGULATOR1 (TOC1/PRR1). These three components repress the activity of each other and direct temporal regulation of most other clock components (Hsu and Harmer, 2014).
The two key responses of circadian clock to high temperatures are temperature entrainment and temperature compensation (Figure 1B). Thermocycles are able to entrain the clock in constant light with shorter periods than photocycles in Arabidopsis (Boikoglou et al., 2011). The genes governing temperature entrainment remain largely unknown, except for the evening loop component EARLY FLOWERING 3 (ELF3), and the morning loop components PRR7 and PRR9. The etiolated elf3-1 seedlings are unable to exhibit classic indicators of entrainment by temperature cycles in darkness (Thines and Harmon, 2010). The prr7-3 prr9-1 double mutants fail to entrain to thermocycles of 22/12°C, but can entrain to 28/22°C thermocycles without a robust oscillation (Salome and McClung, 2005; Salome et al., 2010). Temperature compensation refers to the ability of maintaining a relatively constant period over a range of environmental temperatures. As temperature increases from 12 to 27°C, the periodicity has no significant changes in Arabidopsis (Gould et al., 2006). High temperature enhances the CCA1 binding affinity to the promoters of the oscillator genes, which is precisely antagonized by protein kinase CASEIN KINASE2 (CK2; Portoles and Mas, 2010) and transcription factor FLOWERING BASIC HELIX-LOOP-HELIX 1 (FBH1; Nagel et al., 2014) to maintain the circadian period. At high temperatures, the activities of CCA1 and LHY are counterbalanced by the temperature-dependent regulation of TOC1 and GI (Gould et al., 2006) as well as PRR7 and PRR9 (Salome et al., 2010). REVEILLE8 (RVE8), a homolog of CCA1 and LHY, is also required for temperature compensation, as rve8 mutants have long-period and its overexpression lines have short-period phenotypes under high temperature (Rawat et al., 2011). Moreover, the activity of PRR7 is regulated by HEAT SHOCK FACTOR B2b (HsfB2b; Kolmos et al., 2014) and the evening complex night-time repressor consisting of ELF3, ELF4, and LUX ARRHYTHMO (LUX; Mizuno et al., 2014). The night-time repressor also mediate temperature responses of the clock transcriptional circuitry by regulating other targets GI, LUX and PIF4/5. This activity of night-time repressor is antagonized by warm temperature, suggesting that the PIF4-mediated morphological acclimation may be regulated by the night-time repressor under warm temperature (Mizuno et al., 2014). It is noteworthy that heat-induced alternative splicing of clock components such as CCA1, PRR7, TOC1, and ELF3 may be an important mechanism in temperature compensation (Kwon et al., 2014; Filichkin et al., 2015). Overall, our knowledge on the response of circadian clock to high temperature is rather limited. How plants integrate circadian clock with immunity under high temperature remains elusive.
Modulation of Plant Immunity by High Temperature
The effect of high temperatures on plant immunity has been well summarized recently (Hua, 2013). Two major influences of high temperature on plant immunity are that high temperature often inhibits the effector triggered immunity (ETI) and enhances RNA-silencing mediated resistance (Figure 1C). In ETI, pathogen effectors are recognized by the host proteins encoded by resistance (R) genes, of which most are nucleotide binding-leucine rich repeat (NB-LRR) class of proteins (Martin et al., 2003). SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 (SNC1) is the first identified R gene mediating high temperature inhibition of resistance (Yang and Hua, 2004), which is negatively regulated by BONZAI1 (BON1; Zhu et al., 2010). At 22°C, the bon1-1 loss-of-function mutation activates SNC1, which induces constitutive salicylic acid (SA)-mediated defense responses and inhibits plant growth. While at 28°C, the nuclear accumulation of SNC1 protein is reduced by high temperature, which may inhibit the activity of SNC1 protein and suppress the defense responses (Zhu et al., 2010). Besides BON1, other negative regulators of SNC1 have been identified, such as BON1-ASSOCIATED PROTEIN 1(BAP1), BAK1-INTERACTING RECEPTOR-LIKE KINASE 1(BIR1), SUPPRESSOR OF rps4-RLD (SRFR1), CONSTITUTIVE EXPRESSER OF PR GENES 1(CPR1) and MAP KINASE PHOSPHATASE 1 (MKP1; Gou and Hua, 2012). These genes tightly control SNC1 activities at both the transcriptional and posttranscriptional levels. Besides, abscisic acid (ABA) plays a positive role in the high temperature-mediated inhibition of disease resistance, as ABA deficiency promotes nuclear accumulation of SNC1 and potentiates defense responses at 28°C (Mang et al., 2012; Zhu et al., 2012b). In contrast, nitric oxide (NO) may act as a negative regulator in the high temperature-mediated inhibition of disease resistance (Wang et al., 2012a). It will be interesting to further dissect the interplay between ABA, NO, and SA-mediated defense responses under high temperature. Another major effect of high temperature in plant immunity is the enhancement of RNA-silencing mediated resistance. The underlying mechanisms will be discussed later in this review (see Small Interfering RNAs).
Thermotolerance in Plants
The mechanisms of thermotolerance to heat stress in plants have been elaborated, including the HSFs and HSPs, ROS, phospholipids and calcium signaling pathways, and the network of hormones (Figure 1D; Qu et al., 2013). The thermotolerance in Arabidopsis consists of basal and acquired thermotolerance. The basal thermotolerance is an inherent ability for plants to survive in exposure to temperatures above the optimal for growth, while acquired thermotolerance refers to the ability to cope with lethal high temperatures after acclimatization to mild high temperatures (Clarke et al., 2004). It is reported that SA, jasmonic acid (JA) and ethylene signaling pathways and ROS scavenging are required for basal thermotolerance (Miller et al., 2008; Clarke et al., 2009). The transcriptional co-activator MULTIPROTEIN BRIDGING FACTOR 1C (MBF1c) is required for basal thermotolerance and functions upstream of SA, trehalose and ethylene signaling pathways during heat stress (Suzuki et al., 2008). Moreover, NADPH oxidases RESPIRATORY BURST OXIDASE HOMOLOGUE (Rboh) enhances the production and maintenance of ROS, which is important for basal thermotolerance (Miller et al., 2008). HSFs and HSPs play central roles in the acquired thermotolerance in plants. HSFs are the central regulators responsible for the expression of HSP genes. The Arabidopsis genome contains 21 HSF members that can be sorted into classes A, B, and C (Baniwal et al., 2004). HsfA1a is a master regulator for acquired thermotolerance that triggers the heat stress response through the induction of HsfA1b and HsfA2 expression, while HsfA2 is a major heat stress factor and induces the expression of HSPs under heat stress. HsfB1 acts as a co-regulator enhancing the activity of HsfA1a and HsfA2 (Baniwal et al., 2004). HSPs are categorized into five classes based on their approximate molecular masses: Hsp100, Hsp90, Hsp70, Hsp60, and small Hsps (sHsps). These HSPs function as molecular chaperones and play complementary and sometimes overlapping roles in stabilizing proteins and membranes and assisting in protein refolding under heat stress (Wang et al., 2004). A variety of signaling molecules, such as ABA, H2O2, ethylene, SA, calcium, and phospholipids, are also involved in acquired thermotolerance as well. These signaling molecules regulate the expression of HSFs and HSPs and protect cells against heat stress-induced oxidative damage (Song et al., 2012).
Although great progress has been achieved in the elucidation of molecular mechanisms of thermotolerance, how plants sense and transduce the signal of heat stress is still an important topic to be addressed. It is hard to define the primary heat sensor(s) as heat stress simultaneously poses a threat to almost all macromolecules and all organelles in the cells. Several putative heat sensors have been proposed, including a plasma membrane cyclic nucleotide gated calcium channel (CNGC) and two unfolded protein sensors in the endoplasmic reticulum (ER) and the cytosol (Figure 1D). The CNGC2 gene in Arabidopsis and its ortholog CNGCb from Physcomitrella patens act as the primary heat sensors of land plant cells (Saidi et al., 2009; Finka et al., 2012). Heat shock impairs the protein stability and activates the unfolded protein response (UPR) in the ER and the cytosol. The cytosolic UPR is triggered by unfolded proteins in the cytosol and is notably regulated by HsfA2 (Sugio et al., 2009). Heat promotes the translocation of two basic leucine-zipper domain-containing transcription factors bZIP17 and bZIP28 to the nucleus. The nuclear-localized bZIPs not only activate ER chaperone genes and induce the ER-UPR, but also activate BR signaling, which is required for heat stress acclimation and growth (Che et al., 2010). Heat-induced cleavage of bZIP60 by the RNA splicing factor IRE1b also triggers the ER-UPR (Deng et al., 2011). Taken together, these results suggest that the primary heat sensor may lie in the plasma membrane, ER or cytosol. However, a heat-responsive retrograde pathway in chloroplast has recently been reported (Yu et al., 2012a). The photosynthetic apparatus in the chloroplast are the primary susceptible targets of heat stress. Through proteomic screening, the chloroplast ribosomal protein S1 (RPS1) is also identified as a heat-responsive protein. Under heat stress, RPS1 plays a critical role in modulating the translational efficiency of thylakoid proteins to maintain the stability and integrity of thylakoid membranes. The capacity of protein translation in chloroplasts generates the retrograde signals to activate HsfA2-dependent heat-responsive genes in the nucleus (Yu et al., 2012a). Thus, the chloroplasts are proposed as heat sensors as well.
Epigenetic Regulation of Heat Responses in Plants
DNA Methylation
DNA methylation is a biological process by which a methyl group is added to the cytosine bases of DNA to form 5-methylcytosine. In plants, DNA methylation occurs frequently in all three sequence contexts: the symmetric CG and CHG contexts (where H = A, T or C) and the asymmetric CHH context. In Arabidopsis, overall genome-wide levels of 24% CG, 6.7% CHG and 1.7% CHH methylation are observed (Cokus et al., 2008). DNA methylation in plants predominantly occurs on transposons and other repetitive DNA elements (Zhang et al., 2006). Different proteins are involved in the establishment, maintenance and removal of DNA methylation. De novo methylation in all sequence contexts is catalyzed by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), and DNA methylation is maintained by three different pathways: CG methylation is maintained by METHYLTRANSFERASE 1 (MET1) and DECREASE IN DNA METHYLATION 1 (DDM1); CHG methylation is maintained by CHROMOMETHYLASE 3 (CMT3), a plant-specific DNA methyltransferase; and asymmetric CHH methylation is maintained by DRM2 (Chan et al., 2005). The DRM2 activity is regulated by the RNA-directed DNA methylation (RdDM) pathway (Matzke and Mosher, 2014). The plant-specific RNA polymerase IV (Pol IV) transcribes heterochromatic regions to generate single-stranded RNA (ssRNAs). RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) then synthesizes double-stranded RNA intermediates (dsRNAs) as precursors for RNase III-class DICER-LIKE 3 (DCL3) to process into 24-nt small interfering RNAs (siRNAs). Following incorporation into ARGONAUTE 4 (AGO4), the 24-nt siRNAs base-pair with Pol V scaffold transcripts, which results in DRM2 recruitment and DNA methylation at the source loci (Matzke and Mosher, 2014). In plants, four bifunctional 5-methylcytosine glycosylases, REPRESSOR OF SILENCING 1 (ROS1), DEMETER (DME), DME-like 2 (DML2) and DML3, have been implicated in the active removal of 5-methylcytosine from DNA through the base excision repair pathway (Zhang and Zhu, 2012). DNA methylation has two main roles in plants: defending the genome against selfish DNA elements and regulating gene expression. DNA methylation induces the transcriptional gene silencing (TGS) of transgene as well as endogenous transposons and retrotransposons to maintain genome stability (Chan et al., 2005). DNA methylation of promoter regions usually inhibits transcription initiation, while methylation within the gene body quantitatively impedes transcript elongation in Arabidopsis (Zilberman et al., 2007).
The global methylation can be differently affected by heat in different species. Exposure of Arabidopsis plants to heat stress results in an increased global methylation and a higher homologous recombination frequency (HRF; Boyko et al., 2010). The up-regulation of DRM2, NUCLEAR RNA POLYMERASE D 1(NRPD1) and NRPE1 in response to heat stress may contribute to the increased genome methylation in Arabidopsis (Naydenov et al., 2015). An increase in global methylation is also observed in Cork oak (Quercus suber L.) grown at 55°C (Correia et al., 2013). In Brassica napus, the DNA methylation levels increase more in the heat-sensitive genotype than in the heat-tolerant genotype under heat treatment (Gao et al., 2014). However, in cotton (Gossypium hirsutum) anthers, high temperature significantly decreases the expression of S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE1 (SAHH1) and DNA methyltransferases (DRM1 and DRM3), resulting in the genome-wide hypomethylation at the tetrad stage and the tapetal degradation stage (Min et al., 2014). It appears that there is no consistent trend in the changes of DNA methylation under heat in different species. The methylation status of certain loci may be affected by heat stress. In developing rice seeds, the DNA methylation level of Fertilization-Independent Endosperm1 (OsFIE1), a member of Polycomb Repressive Complex 2 (PRC2), is reduced and the transcript abundance of OsCMT3 is repressed by a moderate heat stress (34°C) at 48 h after fertilization, which may lead to the misregulation of OsFIE1 (Folsom et al., 2014).
Heat stress induces transcriptional activation of various transgenes that are previously silenced via TGS, such as the 35S promoter of Cauliflower Mosaic Virus and β-glucuronidase (GUS), which occurs without detectable changes in the levels of DNA methylation (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010). Warm temperature slightly increases the methylation level in some regions of GUS but decreases it in other regions (Zhong et al., 2013). Thus, it seems that DNA methylation is not involved in the regulation of heat responses. However, an analysis of the heat tolerance of mutants defective in DNA methylation reveals that the RdDM pathway is required for basal thermotolerance (Popova et al., 2013). Plants deficient in NRPD2, the second-largest subunit of Pol IV and V, are hypersensitive to heat stress, while rdr2, dcl3 and ago4 mutants are less sensitive. In nrpd2 mutants recovering from heat, the misexpression of some protein-coding genes is associated with the epigenetic regulation of adjacent transposon remnants (transposons and retrotransposons; Popova et al., 2013). For example, the expression of the COPIA-like transposon At1g29475 is induced by heat but not decreases during recovery in nrpd2 plants, which may repress the adjacent six highly homologous auxin-responsive genes during recovery (Popova et al., 2013). Another study has also reported that the expression of Calmodulin-like 41 (CML41) gene is up-regulated by high temperature with reduced DNA methylation level in the TE insertion very closely to the transcriptional start site (Naydenov et al., 2015). However, the activation of ONSEN, an LTR-copia type retrotransposon, could not be explained by the reduction of DNA methylation at the promoter upon heat stress (Cavrak et al., 2014). Whether the changes in DNA methylation of TEs play a causal role in the heat-induced activation of nearby genes needs to be explored.
Interestingly, in a genome-wide association analysis to detect loci with plastic response to climate, CMT2 has been found to be associated with temperature seasonality in Arabidopsis (Shen et al., 2014). The accessions with CMT2STOP allele, which contains a premature stop codon, have broader geographic distribution than accessions with the wild-type allele. The CMT2STOP allele can alter the genome-wide CHH-methylation pattern and cmt2 mutant plants have an improved heat-stress tolerance, suggesting that CMT2-dependent CHH methylation may act as an important alleviator of heat stress responses. Moreover, the CMT2STOP allele is associated with increased leaf serration and higher disease presence after bacterial inoculation (Shen et al., 2014). In summary, the above studies have not elaborated on the role of DNA methylation in heat responses. cmt2 mutant plants have an improved heat-stress tolerance while nrpd2 mutant plants are hypersensitive to heat stress, suggesting the different roles of CMT2-dependent CHH methylation and the RdDM pathway in response to heat (Figure 1D). Whether DNA methylation regulate plant immunity and circadian clock under heat needs to be investigated.
Histone Covalent Modification
In eukaryotic cells, genomic DNA is packaged into chromatin. The fundamental unit of chromatin is the nucleosome composed of ∼147 bp- DNA wrapped around a histone octamer consisting of two copies of H2A, H2B, H3, and H4. Histone tails can be covalently modified at various amino acids and via different types, such as acetylation, mono/di/trimethylation, phosphorylation, ubiquitination, glycosylation, ADP ribosylation, carbonylation, sumoylation, and biotinylation. These modifications can activate or repress transcription by generating either ‘open’ or ‘closed’ chromatin configurations, respectively, thereby regulating the accessibility of chromatin to transcriptional regulators (Li et al., 2007). In plants, histone methylation and acetylation have been well characterized. As one of the most complex modifications, histone methylation not only occurs at distinct sites of lysine and arginine residues but also differs in the number of methyl groups added. In Arabidopsis, histone methylation mainly occurs at Lys4 (K4), Lys9 (K9), Lys27 (K27), Lys36 (K36), and Arg17 (R17) of histone H3, and Arg3 (R3) of histone H4 (Liu et al., 2010). These methylation types have different roles. H3K4me and H3K36me mainly generate ‘open’ chromatin configurations and activate transcription, whereas H3K9me and H3K27me create a “closed” chromatin and transcriptional repression. H3K9me2 functions as a silencing mark linked to DNA methylation, while H3K27me3 represses the expression of many genes targeted by PRC2. The effects of histone methylation on genome management, transcriptional regulation, and development in plants have been well reviewed (Liu et al., 2010; He et al., 2011). Histone acetylation and deacetylation are catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. Histone acetylation are directly connected with transcriptional activation and affect a variety of biological processes in plant growth and development as well as biotic and abiotic stress responses (Chen and Tian, 2007).
Similar to DNA methylation, the histone modifications can be differently affected by heat in different species. In the unicellular green alga Chlamydomonas reinhardtii, there are higher levels of histone H3/4 acetylation and lower levels of H3K4me1 at promoter regions of active genes compared with inactive promoters and transcribed and intergenic regions after heat stress (Strenkert et al., 2011). The transcription factor HSF1 may mediate the acetylation of histones H3/4, remodeling of the H3K4 methylation, and transcription initiation/elongation upon heat stress (Strenkert et al., 2011). However, temperature shift from 25 to 45°C decreases the acetylated histone H3 levels in the forest tree Cork oak (Correia et al., 2013). The deacetylated H3 may be responsible for repressive chromatin in gene promoters and repression of gene transcription. Histone modifications are also involved in rice seed and cotton anther development at high temperature. The H3K9me2 level of OsFIE1 is sensitive to moderate heat stress and may be an important component involved in regulating OsFIE1 when developing rice seeds are exposed to a moderate heat stress (Folsom et al., 2014). In cotton anthers, one histone methyltransferase, one histone monoubiquitination gene and two jumonji C (jmjC) domain-containing genes are down-regulated upon high temperature (Min et al., 2014). The roles of the differently regulated histone modifications in heat responses remain unknown.
After heat stress, the levels of H3K9me2, H3K27me1 and H3K4me3 at a transcriptionally silenced GUS transgene (TS-GUS) and a non-LTR retrotransposon LINE039 showed only minor changes or remain unchanged (Lang-Mladek et al., 2010). But the amounts of H3K9ac1 and H3K9/14ac2 significantly increased in response to heat. The histone deacetylase HDA6 may be involved in this process as the TS-GUS activity showed a pronounced increase in hda6 mutants (Lang-Mladek et al., 2010). Similarly, another study also demonstrates that levels of H3K4me3, H3K9me2, H3K27me2, and H3K27me3 were unaffected by temperature shift from 4 to 37°C for 15 h while a modest enrichment in H3K9ac-K14ac was detected at 5S rDNA, 106B long terminal-like dispersed repeats and a Mutator-like transposable element related locus MULE-F19G14 (Tittel-Elmer et al., 2010). Thus, the heat-induced release of silencing seems to be associated with histone acetylation but not histone methylation. However, similar to the wild-type control, the transcripts from these three targets over-accumulated in hda6 mutants exposed to temperature shift, but reverted to the initial level after 2 days of recovery. These results exclude the possibility that HDA6 activity is required for the release of gene silencing (Tittel-Elmer et al., 2010). Interestingly, both the levels of repressive H3K9me2 and active H3K4me3 significantly reduced directly after long heat stress (37°C for 30 h) and returned to the initial level after 7 days of recovery (Pecinka et al., 2010). After long heat stress, nucleosomes and all the histone modifications on them were partially removed through unknown mechanisms, and then reloaded to the chromatin upon returned to ambient temperatures, while the levels of histone modifications on the remaining histones remained relatively unchanged (Pecinka et al., 2010). Thus, histone modifications may not play an important role in the heat-induced release of silencing.
Recently, environmental history of repetitive heat stress has been reported to modulate Arabidopsis pattern-triggered immunity (PTI) in a HISTONE ACETYLTRANSFERASE1 (HAC1)-dependent manner (Figure 1D; Singh et al., 2014). Arabidopsis plants exposed to repetitive heat stress were more resistant to virulent bacteria than plants grown in a more stable environment. The enhanced resistance in repetitively stress-challenged plants occurred with priming of PTI-responsive genes and the potentiation of PTI-mediated callose deposition. The transcriptional activation of PTI-responsive genes was associated with enrichment of H3K9/14ac, H3K4me2 and H3K4me3, indicating a positive relationship between the bacterial resistance and histone modifications after heat stress. In hac1-1 mutants, repetitively heat stress failed to induce enhanced resistance to bacteria, priming of PTI, and enrichment of H3K9/14ac, H3K4me2 and H3K4me3. These findings reveal that HAC1 is a necessary component for bacterial resistance, priming of PTI, and open chromatin configurations mediated by repetitive heat stress exposure. Whether H3K4 methylation have a similar role needs to be further analyzed (Singh et al., 2014). Multiple histone modifications, such as H3K56ac, H3K9/14ac, H3K4me3, and H3K4me2, have been found to closely correlate with the rhythmic expression of LHY, CCA1, TOC1, PRR7, and PRR9 in Arabidopsis (Seo and Mas, 2014). Histone acetylation may contribute to the circadian peak of expression by influencing transcription factor accessibility under different temperature conditions, while H3K4me3 may antagonize clock repressor binding, ensuring a proper timing and duration of gene activation (Figure 1B). Overall, the roles of histone modifications in response to heat stress are largely obscure and need to be further recognized.
Histone Chaperones
Histone chaperones are a group of proteins that bind histones and prevent non-productive aggregation between highly positive charged histones and highly negative charged DNA without using the energy of ATP (Zhu et al., 2012a). They play a crucial role in nucleosome assembly during different processes such as DNA replication, repair, and transcription. In general, histone chaperones can be classified as either H3–H4 or H2A–H2B chaperones on the basis of their preferential histone binding. In plants, the well-studied chaperones include H3–H4 chaperones ANTI-SILENCING FUNCTION 1 (ASF1), CHROMATIN ASSEMBLY FACTOR-1 (CAF-1) and HISTONE REGULATORY HOMOLOG A (HIRA), and the H2A–H2B chaperones NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1), NAP1-RELATED PROTEIN (NRP) and FACILITATES CHROMATIN TRANSCRIPTION (FACT; Zhu et al., 2012a). Only a few studies have reported the role of histone chaperones in heat responses. The reload of nucleosome, whose occupancy is reduced by long heat stress, requires the CAF-1-dependent chromatin assembly complex (Pecinka et al., 2010). Wild-type plants lost nucleosomes immediately after heat stress and restored the original level during recovery. By contrast, the fasciata1 (fas1) and fas2 mutants that lack different subunits of CAF-1, had the already reduced nucleosome occupancy before heat treatment. The nucleosome occupancy was further reduced by long heat stress in the mutants, and there was no restoration even after 7 days of recovery. The heat stress-induced loss of nucleosomes and heterochromatin decondensation led to the activation of transcriptionally silenced repetitive elements. The CAF-1-dependent chromatin assembly complex may provide a safeguarding mechanism to minimize the heat-induced epigenetic damage in the germ line (Pecinka et al., 2010). AtASF1A and AtASF1B have also been reported to participate in basal and acquired thermotolerance (Figure 1D; Weng et al., 2014). Upon heat stress, AtASF1A/B proteins were recruited onto chromatin, and their enrichment was correlated with nucleosome removal and RNA polymerase II accumulation at the promoter and coding regions of some HSF and HSP genes. Moreover, AtASF1A/B facilitated H3K56ac, which also promotes the activation of some HSFs and HSPs (Weng et al., 2014).
Histone Variants
In addition to the conventional histones, which are deposited mostly during the S phase of the cell cycle, all eukaryotes have non-allelic histone variants that can be incorporated into nucleosomes in a DNA replication-independent manner during the entire cell cycle. Histone variants can alter the properties of the nucleosomes they occupy and play important roles in maintenance of genome stability, transcriptional activation and repression (Kamakaka and Biggins, 2005). There are 15 histone H3 genes in the Arabidopsis genome, including six canonical H3.1 or H3.1-like genes, eight H3.3 or H3.3-like genes and one centromeric histone H3 gene (Okada et al., 2005). The roles of H3 variants in heat responses have not been reported hitherto. Thirteen H2A-encoding genes have been identified in Arabidopsis, including four canonical H2A genes, two H2A.X genes, three H2A.Z genes and other four less categorized genes (March-Diaz and Reyes, 2009). Recently, an important study revealed that H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis (Kumar and Wigge, 2010). In a genetic screen of mutants defective in heat sensing, the ARP6 gene was identified to mediate the response to increased temperature. The APR6 protein is an essential component of the SWR1 complex required for H2A.Z incorporation into chromatin (March-Diaz and Reyes, 2009). When grown at 22°C, the arp6 mutants display phenotypes similar to wild-type plants grown at 27°C, such as hypocotyl and petiole elongation, leaf hyponasty, and early flowering. It is proposed that H2A.Z occupancy represses gene expression by creating a physical block to transcription or by preventing the binding of transcription activators at cooler temperatures, and eviction of H2A.Z at higher temperatures would thereby facilitate transcription of target genes. This temperature-induced H2A.Z nucleosome dynamics has been proved to regulate the binding of PIF4 to the FT promoter, thereby controlling the thermosensory activation of flowering. Based on these results, H2A.Z-containing nucleosomes are recognized as temperature sensors in the nucleus (Figure 1A; Kumar et al., 2012). However, it is unclear whether this mechanism is also responsible for the regulation of other heat-induced genes, such as auxin biosynthesis genes required for warm temperature-mediated morphological acclimation, HSF and HSP genes in acquired thermotolerance, and genes involved in the modulation of plant immunity and circadian clock by high temperature.
ATP-Dependent Chromatin Remodeling
ATP-dependent chromatin remodeling complexes use the energy of ATP hydrolysis to alter the structure of chromatin by destabilizing histone–DNA interactions, moving histone octamers or catalyzing the incorporation of histone variants. According to ATPases used, the complexes can be grouped into four main classes: the SWItch/Sucrose Non-Fermentable (SWI/SNF) class, the imitation switch (ISWI) class, the inositol requiring 80 (INO80) class, and the chromodomain and helicase-like domain (CHD) class (Clapier and Cairns, 2009). The SWI/SNF complex is the first ATP-dependent chromatin remodeling complex identified, and 41 SNF2 proteins in Arabidopsis have yet been identified. Functional analysis indicates that many of these proteins play important roles in plant development and stress response. Moreover, some of these proteins are involved in epigenetic regulation, such as TGS (dependent or independent of DNA methylation), H2A.Z deposition and histone modifications (Knizewski et al., 2008).
The Swi2/Snf2-related (SWR1) complex regulates transcription by replacing the H2A–H2B histone dimers in nucleosome with dimers containing the H2A.Z variant. As mentioned above, the ARP6 protein, which is an essential component of the SWR1 complex, plays an important role in temperature sensing (Figure 1A; Kumar and Wigge, 2010). The SNF2/Brahma-type chromatin-remodeling gene CHROMATIN REMODELING (CHR12) also plays a vital role in mediating the temporary growth arrest of Arabidopsis under heat, drought and salinity stresses (Figure 1D; Mlynarova et al., 2007). When exposed to stress conditions, a gain-of-function mutant overexpressing AtCHR12 showed growth arrest of normally active primary buds and reduced growth of the primary stem. In contrast, the loss-of-function mutant showed less growth arrest than the wild-type when exposed to moderate stress (Mlynarova et al., 2007). In Chlamydomonas reinhardtii, heat stress induces low nucleosome occupancy at promoter regions of active genes, which is mediated by HSF1 and other unknown chromatin remodeling complexes (Strenkert et al., 2011). However, the heat stress-mediated release of TGS is at least partly independent of the activity of MORPHEUS’ MOLECULE 1 (MOM1)/CHR15, a well-known DNA methylation-independent transcriptional silencer, and DECREASED DNA METHYLATION 1 (DDM1)/CHR1, a component required for DNA methylation and H3K9me2 (Tittel-Elmer et al., 2010).Whether other ATP-dependent chromatin remodeling complexes play roles in heat responses remains elusive.
Small RNAs
Small RNAs are 18–30 nt non-protein-coding RNAs, which have emerged as key guide molecules in the control of gene expression. Two major types of small RNAs in plants, microRNAs (miRNAs) and siRNAs, are distinguished by the different proteins involved in their biogenesis and the modes of regulation (Ghildiyal and Zamore, 2009).
microRNAs
Plant miRNAs are a class of 20–24 nt endogenous small RNAs that derive from the miRNA genes (MIR; Rogers and Chen, 2013). MIR genes are transcribed by Pol II to generate primary miRNA transcripts called pri-miRNAs. The pri-miRNAs are processed into stem–loop precursor pre-miRNA and further excised as miRNA/miRNA∗ duplex by the endonuclease activity of the DCL1 protein complex in the nucleus. The mature miRNAs are exported to the cytoplasm and incorporated into AGO proteins, mediating posttranscriptional gene silencing (PTGS) through slicing or translational inhibition, or TGS by targeting chromatin for cytosine methylation (Rogers and Chen, 2013).
A diversity of conserved and non-conserved heat-responsive miRNAs have been identified by small RNA deep-sequencing in different species, but few of them have been validated by either northern blots or real time PCR. As listed in Table 2, most of the conserved heat-responsive miRNAs are differently regulated in various species, except for miR159, 166 and 472 families. miR159 has been found to be down-regulated by heat in Arabidopsis (Zhong et al., 2013), wheat (Triticum aestivum; Wang et al., 2012b; Kumar et al., 2014) and cassava (Manihot esculenta; Ballen-Taborda et al., 2013). The main targets of miR159 are MYB transcription factors. Tae-miR159 has been demonstrated to direct the cleavage of TaGAMYB1 and TaGAMYB2 (Wang et al., 2012b). The tae-miR159 overexpression rice lines and Arabidopsis myb33myb65 double mutants are more sensitive to heat stress relative to the wild-types, indicating that the down-regulation of miR159 and up-regulation of its targets after heat stress might participate in a heat stress-related signaling pathway and contribute to heat stress tolerance (Wang et al., 2012b). MiR166, which targets HD-Zip transcription factors, is up-regulated by heat in Arabidopsis (Zhong et al., 2013), wheat (Xin et al., 2010), and barley (Hordeum vulgare; Kruszka et al., 2014). The heat-induced up-regulation of hvu-miR166a and down-regulation of its targets, PHAVOLUTA (PHV), REVOLUTA (REV) and a homeobox-leucine zipper protein HOX9-like gene, might influence the leaf morphology (Kruszka et al., 2014). miR472 may be down-regulated by heat in Arabidopsis (Zhong et al., 2013) and Chinese white poplar (Populus tomentosa; Chen et al., 2012), but need to be further validated.
Table 2.
The conserved heat-responsive miRNAs in different plant species.
| Family | miRNAa | Heat treatment | Regulation | Validationc | Target proteins |
|---|---|---|---|---|---|
| 156 | ath-miR156h | ATb | Up-regulation | Yes | SPL transcription factor |
| ath-miR156g,h | Grown at 30°C | Up-regulation | No | ||
| bra-miR156g,h | 46°C for 1 h | Up-regulation | Yes | BracSPL2 | |
| osa-miR156a,g,h | 42°C day/36°C night for 24 h | Down-regulation in roots and shoots | Yes | SPL transcription factor | |
| tae-miR156a-g | 40°C for 2 h | Up-regulation | Yes | SPL transcription factor | |
| tae-miR156 | 42°C for 2 h | Up-regulation | Yes | Heat shock protein 90 | |
| mes-miR156a | 37°C for 24 h | Down-regulation | Yes | SPL transcription factor | |
| 159 | ath-miR159a,b | Grown at 30°C | Down-regulation | No | MYB and TCP transcription factors |
| tae-miR159 | 40°C for 2 h | Down-regulation | Yes | GAMYB1 and GAMYB2 | |
| tae-miR159a,b | 42°C for 2 h | Down-regulation | Yes | WRKY transcription factor; MYB3; alkaline phosphatase family protein, cytochrome P450, cobalamine adenosyl transferase, Mob1-like protein and TLD family protein | |
| mes-miR159a | 37°C for 24 h | Down-regulation | Yes | myb-like HTH transcriptional regulators | |
| 160 | ath-miR160a-c | Grown at 30°C | Up-regulation | No | Auxin response factors |
| osa-miR160a | 42°C day/36°C night for 24 h | Down-regulation in roots, up-regulation in shoots | Yes | ||
| pto-miR160a-d | 37°C for 8 h | Down-regulation | No | ||
| tae-miR160 | 40°C for 2 h | Up-regulation | No | Heat shock protein 70; ARF; tetratricopeptide repeat (TPR) | |
| tae-miR160 | 42°C for 2 h | Down-regulation | Yes | ||
| hvu-miR160a | 35.5°C for 24 h | Up-regulation | Yes | ARF17 and ARF13 | |
| mes-miR160a | 37°C for 24 h | Down-regulation | Yes | Auxin response factor | |
| celery-miR160 | 38°C for 1 h | Up-regulation | Yes | Auxin response factor | |
| 162 | osa-miR162a | 42°C day/36°C night for 24 h | Down-regulation in both roots and shoots | Yes | Endoribonuclease DCL1 |
| 164 | ath-miR164a-c | Grown at 30°C | Up-regulation | No | NAC domain containing transcription factors |
| tae-miR164 | 42°C for 2 h | Down-regulation | Yes | Small heat shock proteins 17; NAC transcription factor; target genes involved in mitogen-activated protein kinase (MAPK) signaling pathways | |
| celery-miR164 | 38°C for 1 h | Up-regulation | Yes | NAC domain containing transcription factors | |
| 166 | ath-miR166a | Grown at 30°C | Up-regulation | No | HD-Zip transcription factors including PHV and REVOLUTA |
| tae-miR166a-d | 40°C for 2 h | Up-regulation | Yes | Unknown | |
| hvu-miR166a | 35.5°C for 24 h | Up-regulation | Yes | HD-Zip transcription factors including PHV and REVOLUTA; a homeoboxleucine zipper protein HOX9-like gene | |
| 167 | ath-miR167c,d | Grown at 30°C | Up-regulation | No | HD-Zip transcription factors including PHV and PHB |
| ath-miR167d | ATb | Down-regulation | No | ||
| bra-miR167 | 42°C for 3 h per day for 7 days | Up-regulation | No | TOM1-like protein 2; Tudor domain-containing protein 3 | |
| bra-miR167∗ | 42°C for 3 h per day for 7 days | Up-regulation | No | GDSL esterase/lipase; Ribulose bisphosphate carboxylase/oxygenase activase | |
| bra-miR167 | 46°C for 1 h | Up-regulation | No | BracARF6 | |
| osa-miR167a,c,d | 42°C day/36°C night for 24 h | Down-regulation in both roots and shoots | Yes | Class III HD-Zip protein 4; heat repeat family protein | |
| sja-miR167a | 20°C for 3h | Down-regulation | No | Unknown | |
| pto-miR167c,d,f,g | 37°C for 8 h | Up-regulation | Yes | Unknown | |
| tae-miR167 | 42°C for 2 h | Up-regulation | Yes | Dnaj heat shock n-terminal domain-containing protein | |
| hvu-miR167h | 35.5°C for 24 h | Up-regulation | Yes | ARF8 and a serine/threonine-protein kinase Nek5-like gene | |
| 168 | bra-miR168 | 42°C for 3h per day for 7 days | Up-regulation | Yes | BraAGO1 |
| osa-miR168a | 42°C day /36°C night for 24 h | Down-regulation in shoots, no expression in roots | Yes | AGO1 | |
| sja-miR168a | 20°C for 3 h | Down-regulation | No | Unknown | |
| pto-miR168a,b | 37°C for 8 h | Down-regulation | Yes | Unknown | |
| tae-miR168 | 40°C for 2 h | Up-regulation | No | Unknown | |
| celery-miR168 | 38°C for 1 h | Up-regulation | Yes | Unknown | |
| 169 | ath-miR169a,d-n | Grown at 30°C | Down-regulation | No | CCAAT Binding Factor (CBF) and HAP2-like transcription factors |
| ath-miR169d,e,k,i,m | ATb | Down-regulation | No | ||
| ath-miR169b,c | Grown at 30°C | Up-regulation | No | ||
| osa-miR169a,b,g | 42°C day/36°C night for 24 h | Down-regulation in roots, up-regulation in shoots | Yes | Nuclear transcription factor Y subunit | |
| pto-miR169j-m | 37°C for 8 h | Up-regulation | No | Unknown | |
| pto-miR169n-t | 37°C for 8 h | Down-regulation | No | Unknown | |
| tae-miR169a-d | 40°C for 2 h | Up-regulation | No | Unknown | |
| 171 | ath-miR171a-c | Grown at 30°C | Up-regulation | No | Scarecrow-like transcription factors |
| ath-miR171b,c | ATb | Up-regulation | No | ||
| bra-miR171a-1 | 42°C for 3 h per day for 7 days | Up-regulation | No | 26S protease regulatory subunit 6A homolog | |
| pto-miR171a-i | 37°C for 8 h | Down-regulation | No | Unknown | |
| tae-miR171a | 42°C for 2 h | Down-regulation | Yes | Scarecrow-like protein | |
| ptc-miR171l-n | 37°C for 24 h | Down-regulation | Yes | SCL, clathrin assembly protein | |
| 172 | ath-miR172b∗ | Grown at 30°C | Down-regulation | No | Eukaryotic translation initiation factor 5, putative; calcium-transporting ATPase |
| ath-miR172c,d,e | Grown at 30°C | Up-regulation | No | AP2 transcription factors | |
| ath-miR172a,e | ATb | Up-regulation | No | ||
| tae-miR172a,b | 40°C for 2 h | Down-regulation | Yes | AP2 transcription factors; floral homeotic protein | |
| 319 | ath-miR319a,b | Grown at 30°C | Up-regulation | No | MYB and TCP transcription factors |
| ath-miR319a,c | ATb | Up-regulation | No | ||
| ath-miR319c | Grown at 30°C | Down-regulation | No | ||
| tae-miR319 | 42°C for 2 h | Down-regulation | Yes | MYB3; histone protein-associated genes | |
| 393 | ath-miR393a,b | Grown at 30°C | Down-regulation | No | F-box proteins and bHLH transcription factors |
| tae-miR393 | 40°C for 2 h | Up-regulation | Yes | Genes involved in auxin signaling pathway and basal defense | |
| 394 | ath-miR394a,b | Grown at 30°C | Down-regulation | No | F-box proteins |
| celery-miR394 | 38°C for 1 h | Up-regulation | Yes | Unknown | |
| pto-miR394a,b | 37°C for 8 h | Down-regulation | No | Unknown | |
| 395 | ath-miR395a-f | Grown at 30°C | Up-regulation | No | ATP sulphurylases; leucine-rich repeat family protein |
| ath-miR395d | ATb | Up-regulation | No | ||
| pto-miR395a-j | 37°C for 8 h | Down-regulation | Yes | Unknown | |
| sja-miR395x | 20°C for 3 h | Up-regulation | No | Unknown | |
| tae-miR395b | 42°C for 2 h | Up-regulation | No | sulfur transporters and ATP sulphurylases | |
| celery-miR395 | 38°C for 1 h | Up-regulation | Yes | Unknown | |
| 396 | ath-miR396a | Grown at 30°C | Up-regulation | No | Growth-regulating factor (GRF) transcription factors; rhodenase-like proteins; kinesin-like protein B |
| han-miR396 | 37°C for 24 h | Down-regulation | Yes | HaWRKY6 | |
| 397 | ath-miR397a,b | Grown at 30°C | Up-regulation | No | Laccases and beta-6 tubulin |
| osa-miR397b.2 | 42°C for 8 h | Up-regulation | Yes | L-ascorbate oxidase | |
| osa-miR397 | 42°C day/36°C night for 24 h | Down-regulation in shoots, no epression in roots | Yes | L-ascorbate oxidase precursor; F-box domain containing protein | |
| mes-miR397a | 37°C for 24 h | Down-regulation | Yes | Laccase/Diphenol oxidase family protein | |
| 398 | ath-miR398b | Grown at 30°C | Up-regulation | No | CSD1, CSD2 and CCS |
| ath-miR398 | 37°C for 4 h | Up-regulation | Yes | ||
| bra-miR398a,b | 46°C for 1 h | Down-regulation | Yes | BracCSD1 | |
| osa-miR398 | 42°C day/36°C night for 24 h | Down-regulation in both roots and shoots | Yes | Superoxide dismutase (SOD) gene family | |
| sja-miR398a-5p | 20°C for 3 h | Up-regulation | No | Unknown | |
| tae-miR398 | 42°C for 2 h | Up-regulation | Yes | Superoxide dismutase (SOD) gene family | |
| 399 | ath-miR399b-d, f | Grown at 30°C | Up-regulation | No | Phosphatase transporter |
| ath-miR399c,d | ATb | Down-regulation | No | ||
| bra-miR399b | 46°C for 1 h | Down-regulation | No | BracPHO2 | |
| 400 | ath-miR400 | 37°C for 12 h | Down-regulation | Yes | Pentatricopeptide (PPR) repeat-containing protein |
| 408 | ath-miR408 | Grown at 30°C | Up-regulation | No | Peptide chain release factor; plantacyanin |
| sja-miR408b-5p | 20°C for 3 h | Down-regulation | Yes | An unknown conserved protein | |
| pto-miR408 | 37°C for 8 h | Down-regulation | Yes | Unknown | |
| mes-miR408 | 37°C for 24 h | Down-regulation | Yes | Plantacyanin | |
| celery-miR408 | 38°C for 1 h | Up-regulation | Yes | Unknown | |
| 472 | ath-miR472 | Grown at 30°C | Down-regulation | No | RFL1 (RPS5-LIKE 1) |
| pto-miR472a,b | 37°C for 8 h | Down-regulation | No | Disease resistance protein; F-box protein | |
| 482 | pto-miR482 | 37°C for 8 h | Down-regulation | Yes | Unknown |
| 827 | ath-miR827 | Grown at 30°C | Down-regulation | No | SPX (SYG1/Pho81/XPR1) domain-containing protein; DNA-binding storekeeper protein-related |
| ath-miR827 | ATb | Down-regulation | No | ||
| bra-miR827 | 46°C for 1 h | Down-regulation | Yes | Unknown | |
| tae-miR827 | 40°C for 2 h | Up-regulation | No | Unknown | |
| ptc-miR827 | 37°C for 24 h | Down-regulation | Yes | Sec14 cytosolic factor family protein | |
| 1117 | tae-miR1117 | 42°C for 2 h | Down-regulation | Yes | Calcium dependent protein kinase 1 |
| 1450 | pto-miR1450 | 37°C for 8 h | Up-regulation | No | Unknown |
| ptc-miR1450 | 37°C for 24 h | Down-regulation | Yes | Leucine-rich repeat transmembrane protein kinase |
The miRNA family, kinds of heat treatment, up or down-regulation, validated by either northern blot or realtime PCR, and their target proteins are presented. aath: Arabidopsis thaliana (Yan et al., 2012; Guan et al., 2013; Zhong et al., 2013; Stief et al., 2014); bra: Brassica rapa (Yu et al., 2012b; Bilichak et al., 2015); celery: Apium graveolens (Li et al., 2014a); han:Helianthus annuus (Giacomelli et al., 2012); hvu: Hordeum vulgare (Kruszka et al., 2014); mes: Manihot esculenta (Ballen-Taborda et al., 2013); osa: Oryza sativa (Jeong et al., 2011; Sailaja et al., 2014); ptc: Populus trichocarpa (Lu et al., 2008); pto: P. tomentosa (Chen et al., 2012); sja: Saccharina japonica (Liu et al., 2014); tae: Triticum aestivum (Xin et al., 2010; Wang et al., 2012b; Kumar et al., 2014). bAT: acquired thermotolerance, pretreated at 37°C for 1 h and returned to 22°C for 1.5 h for recovery, treated at 44°C for 45 min. cvalidated by either northern blot or realtime PCR.
Several miRNA families seem to be responsive to heat in at least four species, including miR156, 160, 167, 168, 169, 171, 395, 398, 408, and 827 families (Table 2). Some members of the miR156 family are induced by heat in Arabidopsis (Zhong et al., 2013; Stief et al., 2014), Brassica rapa (Yu et al., 2012b), and wheat (Xin et al., 2010; Kumar et al., 2014), but are repressed by heat in rice (Sailaja et al., 2014) and cassava (Ballen-Taborda et al., 2013). In Arabidopsis, miR156 isoforms are highly induced after heat stress and target SQUAMOSA-PROMOTER BINDING-LIKE (SPL) transcription factor genes (especially SPL2 and SPL11) that are master regulators of developmental transitions (Stief et al., 2014). AGO1 acting through miR156 and its target SPLs appears to mediate the adaptation to recurring heat stress (HS memory) by inducing the expression of HS memory–related genes (Stief et al., 2014). Bra-miR156h and bra-miR156g were also heat-induced and their putative target BracSPL2 was down-regulated (Yu et al., 2012b). The up-regulation of tae-miR156 and down-regulation of its putative target SPL genes Ta3711 and Ta7012 were also validated in wheat (Xin et al., 2010). However, the roles of the down-regulated miR156 in rice (Sailaja et al., 2014) and cassava (Ballen-Taborda et al., 2013) remain unknown. The regulation of miR160 family by heat is quite different in various species, although they all target auxin response factors (ARFs). After heat stress, miR160 was up-regulated in Arabidopsis (Zhong et al., 2013), barley (Kruszka et al., 2014) and celery (Apium graveolens; Li et al., 2014a), but down-regulated in cassava (Ballen-Taborda et al., 2013) and Chinese white poplar (Chen et al., 2012). An increase in barley miR160a during heat stress down-regulated the expression level of ARF17 and ARF13, which might affect shoot morphology and root growth (Kruszka et al., 2014). miR160 as well as miR169 in rice showed differential expression in roots and shoots under heat stress, suggesting the different regulation of the target genes by heat in this two different tissues (Sailaja et al., 2014). It is also amazing that miR160 in wheat was up-regulated by 40°C for 2 h in the heat tolerant genotype TAM107 (Xin et al., 2010), but down-regulated with the up-regulation of its putative target HSP70 by 42°C for 2 h in another heat tolerant cultivar HD2985 (Kumar et al., 2014). miR167 was proved to be up-regulated in Chinese white poplar (Chen et al., 2012), wheat (Kumar et al., 2014) and barley (Kruszka et al., 2014) but down-regulated in rice (Sailaja et al., 2014). Heat stress enhanced the miR167h-guided cleavage of the ARF8 and NEK5 transcript in barley (Kruszka et al., 2014). miR168 has also been shown to be up-regulated by heat in Brassica rapa (Bilichak et al., 2015) and celery (Li et al., 2014a), but down-regulated in Chinese white poplar (Chen et al., 2012) and rice (Sailaja et al., 2014). A differential expression of bra-miR168 following heat shock in the parental tissues was observed to be negatively correlated with transcript levels of its putative target braAGO1 in the corresponding tissues, suggesting the important role of bra-miR168 in heat responses (Bilichak et al., 2015).
The miR398 family have been validated to be up-regulated in Arabidopsis (Guan et al., 2013) and wheat (Kumar et al., 2014), but down-regulated in rice (Sailaja et al., 2014) and Brassica rapa (Yu et al., 2012b). Heat stress rapidly induced ath-miR398 and reduced transcripts of its target genes COPPER/ZINC SUPEROXIDE DISMUTASE 1(CSD1), CSD2 and COPPER CHAPERONE FOR SOD 1 (CCS) that control ROS accumulation (Guan et al., 2013). The altered redox status contributed to the consequent accumulation of HSFs and HSPs that are critical for thermotolerance. Transgenic plants overexpressing miR398-resistant versions of CSD1, CSD2, or CCS under the control of their native promoters were hypersensitive to heat stress, and the expression of many HSF and HSP genes under heat stress was reduced in these plants. In contrast, csd1, csd2, and ccs plants were more tolerant to heat stress than wild-type plants with the increased expression levels of HSF and HSP genes. Moreover, HSFA1b and HSFA7b were found to be responsible for heat induction of miR398. Thus, HSFs, miR398 and its target genes CSD1, CSD2, and CCS form an essential regulatory loop for thermotolerance in Arabidopsis (Guan et al., 2013). However, in Brassica rapa, heat stress reduced the expression of the conserved miRNAs bra-miR398a and bra-miR398b, which guides heat response of their target gene BracCSD1 (Yu et al., 2012b).The expression of most members in miR169, 171, 395, and 827 families have not been experimentally validated, and their targets remain largely unknown (Table 2). In addition to the above miRNA families, a lot of conserved miRNA families response to heat only in 1–3 species (Table 2). Among them, han-miR396 in sunflower (Helianthus annuus) was found to be repressed by high temperature, which results in the up-regulation of the putative target HaWRKY6 (Giacomelli et al., 2012). But plants overexpressing miR396-resistant versions of HaWRKY6 were hypersensitive to heat shock, indicating that HaWRKY6 is involved in a fine modulation in response to heat (Giacomelli et al., 2012).
In addition to the conserved miRNAs, many non-conserved and novel heat-responsive miRNAs have been validated. For instance, ptc-miR1445, 1446a-e and 1447 were down-regulated by heat in P. trichocarpa (Lu et al., 2008); pto-smR7, 8, and 9 were down-regulated by heat in P. tomentosa (Chen et al., 2012); osa-miR1884 was down-regulated in roots but up-regulated in shoots by heat (Sailaja et al., 2014); tae-candidate_3466 and 5064 were up-regulated in wheat by heat (Kumar et al., 2014). Interestingly, the splicing of introns hosting miR160a and miR5175a in barley was heat induced, but the roles of these spliced isoforms in response to heat stress are unclear (Kruszka et al., 2014). Such heat stress-induced alternative splicing also regulates the miR400 expression in Arabidopsis (Yan et al., 2012). The intronic MIR400 is co-transcribed with its host gene At1g32583. Upon heat stress, a specific alternative splicing occurred at the first intron of At1g32583 containing the miR400 hairpin, which led to a decrease of mature miR400, but did not affect the host gene expression. This alternative splicing event may be favorable for thermotolerance, as overexpression of MIR400 made the plants more sensitive to heat stress (Yan et al., 2012). These findings extend our view about the regulatory mechanism linking miRNAs and heat stress.
It is worth noting that some heat-responsive miRNAs also function in other biotic and abiotic stresses. The miR156-SPL pathway in rice also functions in other stresses such as cold, salt and drought stress, suggesting a vital role of miR156 in modulating plant development and responses to abiotic stress (Cui et al., 2014). tae-miR827 and 2005 were up-regulated in wheat by both powdery mildew infection and heat stress (Xin et al., 2010). ptc-miR 171l-n, 530a, 1445, 1446a-e, and 1447 were down-regulated in response to heat as well as cold, salt and dehydration in P. trichocarpa (Lu et al., 2008). mes-miR156a, 159a, 160a, 397a, and 408 were down-regulated by heat and drought stresses in cassava (Ballen-Taborda et al., 2013). Thus, miRNAs may integrate the regulatory networks of heat stress with that of other biotic and abiotic stresses.
Small Interfering RNAs
Plant siRNAs are processed by DCL2-4 from long dsRNAs, which are generated directly from virus replication and inverted repeats (IRs), or converted from ssRNAs by RDRs, or by annealing of two complementary and separately transcribed RNA strands (Bologna and Voinnet, 2014). SiRNAs guide a silencing effector complex to homologous DNA loci to trigger TGS or target mRNAs for transcript cleavage. Several exogenously triggered PTGS pathways resulting in transcript cleavage have been reported. These PTGS pathways can be induced by sense transgenes (S-PTGS), antisense transgenes (A-PTGS), inverted-repeat transgenes (IR-PTGS) and virus replication (VIGS). The diverse PTGS pathways play important roles in plant immunity and silencing of transgenes (Brodersen and Voinnet, 2006). In Arabidopsis, several distinct classes of endogenous siRNAs have also been uncovered, including repeat-associated siRNAs (ra-siRNAs), trans-acting siRNAs (ta-siRNAs), natural antisense transcript-derived siRNAs (nat-siRNAs), endogenous IR-derived siRNAs, and double-strand-break-induced RNAs (diRNAs; Bologna and Voinnet, 2014). Ra-siRNAs are typically 24-nt small RNAs that are derived from genomic repetitive sequences, which usually direct DNA methylation through the RdDM pathway (Matzke and Mosher, 2014). The ta-siRNAs arise from eight recognized Arabidopsis TAS loci (TAS1a-c, TAS2, TAS3a-c, and TAS4) through a miRNA-dependent biogenesis pathway (Fei et al., 2013). Ta-siRNAs are distinguished for the ability to function in trans to suppress the expression of target genes, such as disease resistance genes and transcription factors. Nat-siRNAs, which originate from the overlapping region of a pair of natural antisense transcripts (NAT), have been found a role in stress responses. In the case of two published nat-siRNAs, one transcript of the NAT pair is constitutively expressed and the other is induced by salt or bacterial pathogen, which induce the production of nat-siRNAs (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). Nat-siRNAs target the constitutive expressed transcript for cleavage, which confers tolerance to the inductive stress. Endogenous IR-derived siRNAs are processed by DCLs from genomic loci rearranged to form extended IRs that produce perfect or near-perfect dsRNA molecules (Dunoyer et al., 2010). IR-derived siRNAs can drive non-cell-autonomous silencing at both transcriptional and posttranscriptional levels and may have adaptive value by integrating temporally and/or spatially restricted stresses or environmental signals at the whole-plant level and perhaps in progenies (Dunoyer et al., 2010). DiRNAs derive from both sense and antisense strands around double-strand-break sites and may function as guide molecules directing chromatin modifications or the recruitment of protein complexes to source sites to facilitate repair (Wei et al., 2012).
In addition to miRNAs, the diverse exogenous and endogenous siRNAs are affected by heat. An increase in growth temperature from 22 to 30°C effectively inhibited S-PTGS and A-PTGS but not IR-PTGS in Arabidopsis (Zhong et al., 2013). The warmth-induced PTGS release most likely occured during a critical step that leads to the formation of stable dsRNAs involving RDR6 and SUPPRESSOR OF GENE SILENCING 3 (SGS3). The abundance of many endogenous tasiRNAs was also significantly reduced by the 30°C growth, consistent with increased transcript levels of TAS and tasiRNA-target genes, which may affect the morphological acclimation (Figure 1A). The temperature increase reduced the protein abundance of SGS3, as a consequence, attenuating the formation of stable dsRNAs. Overexpression of SGS3 could release such warmth-triggered inhibition of siRNA biogenesis (Zhong et al., 2013). Heat shock (37°C for 1 h) also decreased the accumulation of TAS1-derived siRNAs, whereas their target genes HEAT-INDUCED TAS1 TARGET1 (HTT1) and HTT2 were highly up-regulated by heat shock (Li et al., 2014b). Meanwhile, HTT1 and HTT2 were directly activated by HsfA1a through binding to their promoters. HTT1 mediated thermotolerance by acting as a cofactor of Hsp70-14 complexes (Li et al., 2014b; Figure 1D). Some nat-siRNAs are responsive to heat stress in Brassica rapa and Arabidopsis. For example, nat-siRNAs derived from Bra018216/Bra018217 and its homologous NAT pair AT3G46230/AT3G46220 were induced by heat, leading to the induction of the former gene (Bra018216 and AT3G46230) and the repression of the latter gene (Bra018217 and AT3G46220; Yu et al., 2013; Figure 1D). A novel class of heat-responsive small RNAs derived from the chloroplast genome of Brassica rapa has been reported (Wang et al., 2011). Many members of chloroplast small RNAs (csRNAs) families are highly sensitive to heat stress, and some csRNAs respond to heat stress by silencing target genes (Wang et al., 2011). It will be interesting to investigate the role of these csRNAs in RPS1-mediated heat-responsive retrograde pathway (Figure 1D). Although ra-siRNAs-mediated RdDM pathway is required for basal thermotolerance (Popova et al., 2013), the underlying mechanism is still not clear (Figure 1D). The study of ONSEN reveals a novel regulation mechanism via siRNAs in heat responses (Ito et al., 2011). In Arabidopsis seedlings, ONSEN is transiently activated by heat stress and re-silenced during the recovery period. A surprisingly high frequency of retrotransposition, which produces new ONSEN insertions, is observed in the progeny of stressed nrpd1 plants but not of the wild-type plants, suggesting a crucial role of the ra-siRNA pathway in restricting transgenerational retrotransposition triggered by heat stress. Moreover, natural and experimentally induced variants in endogenous loci harboring new ONSEN insertions confer heat responsiveness to nearby genes. Therefore, heat-induced mobility bursts of ONSEN may generate novel, stress-responsive regulatory gene networks (Ito et al., 2011). A recent study in maize has also demonstrated that allelic variation for insertions of the TEs associated with heat stress-responsive expression can contribute to variation in the regulation of nearby genes, probably by providing binding sites for transcription factors or influencing chromatin (Makarevitch et al., 2015). The roles of endogenous IR-derived siRNAs and diRNAs in heat responses remain to be investigated.
High temperatures often enhance the VIGS-mediated disease resistance (Figure 1C). The temperature shift from 25 to 30°C induced the accumulation of siRNAs and increases the cassava geminivirus-induced RNA silencing in plants (Chellappan et al., 2005). Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires DCL2, AGO2, and HEN1 (Zhang et al., 2012). The activity of DCL2 was up-regulated by high temperature, suggesting that DCL2 protein may be a temperature-sensitive component responsible for modulation of RNA silencing pathway (Zhang et al., 2012). In addition, RDR6 may be closely related to the temperature sensitivity of the silencing pathway in Nicotiana benthamiana (Qu et al., 2005). Plants with reduced expression of NbRDR6 were more susceptible to various viruses and this effect was more pronounced at higher growth temperatures (Qu et al., 2005). Moreover, NbRDR6 plays a root-specific role in the inhibition of Chinese wheat mosaic virus (CWMV) accumulation and biogenesis of CWMV siRNAs at higher temperatures (Andika et al., 2013). It will be interesting to investigate whether other components of RNA silencing also affect virus resistance under high temperatures.
Other Epigenetic Regulation of Heat Responses
In recent years, new epigenetic mechanisms have been revealed to regulate the heat responses, including lncRNAs, HEAT INTOLERANT 4 (HIT4)-mediated non-canonical TGS regulation and other two unorthodox pathways. LncRNAs, with size larger than 200 nt, are precursors for siRNA biogenesis and act as scaffolds for the establishment of DNA methylation and histone modifications (Wierzbicki, 2012). In Arabidopsis, the expression of HSFB2a was counteracted by a natural and heat-inducible long non-coding antisense RNA, asHSFB2a (Wunderlich et al., 2014). In leaves, the antisense RNA gene was only expressed after heat stress and dependent on the activity of HSFA1a/HSFA1b. HSFB2a and asHSFB2a RNAs were also present in the absence of heat stress in the female gametophyte. HSFB2a activity temporarily repressed vegetative growth during development and after heat stress, the antisense regulation by asHSFB2a counteracted this effect to restore growth and further development (Figure 1D; Wunderlich et al., 2014). Other 15 heat-responsive lncRNAs have been found in Arabidopsis, but their functions are still unknown (Di et al., 2014). Seventy-seven putative heat-responsive lncRNAs, which are not conserved among plant species, have been identified in wheat (Xin et al., 2011). Among them, TahlnRNA27 and TalnRNA5 are the precursors of tae-miR2010 and tae-miR2004, respectively. These two lnRNAs and miRNAs were up-regulated after heat stress in heat sensitive genotype Chinese Spring (CS) and heat tolerant genotype TAM107. Nine heat-responsive lncRNAs such as TalnRNA21 may be precursors of siRNAs. TalnRNA9 and TalnRNA12 are identified as signal recognition particle (SRP) 7S RNA variants and can be regulated by siRNAs. Besides, three lncRNAs (TahlnRNA12, TahlnRNA23, and TahlnRNA29) are characterized as U3 snoRNAs (Xin et al., 2011). Interestingly, lnc-508 was down-regulated by heat and cold, while lnc-168 was down-regulated by heat and salt in Arabidopsis (Di et al., 2014). Twenty-three lncRNAs respond to both powdery mildew infection and heat stress in wheat (Xin et al., 2011). These results suggest that like miRNAs, lncRNAs may also integrate the regulatory networks of heat stress with that of other biotic and abiotic stresses.
HIT4 has been reported to mediate heat-induced decondensation of chromocenters and release from TGS with no change in the level of DNA methylation (Wang et al., 2013, 2014). HIT4 acts independent of MOM1 at the level of heterochromatin organization and this activity is essential for basal thermotolerance in plants. Thus, HIT4 delineates a novel TGS regulation pathway, involving a currently unidentified component that links HIT4 relocation and the large-scale reorganization of chromatin (Wang et al., 2013, 2014). A special inductive temperature shift released the heterochromatin-associated silencing in Arabidopsis plants in a genome-wide manner (Tittel-Elmer et al., 2010). This occurred without alteration of repressive epigenetic modifications and did not involve common epigenetic mechanisms. Such induced release of silencing was rapidly restored, without the involvement of factors known to be required for silencing initiation. Therefore, stress-induced destabilization of heterochromatic TGS and its re-establishment may involve novel mechanisms that repress transcription (Tittel-Elmer et al., 2010). In a recent study, long-term heat stress activated the Arabidopsis imprinted gene SUPPRESSOR OF DRM1 DRM2 CMT3 (SDC), which encodes a putative F-Box protein and is silent during vegetative growth due to DNA methylation (Sanchez and Paszkowski, 2014). The heat-mediated transcriptional induction of SDC occurred only above a particular window of absolute temperature and was proportional to the level of stress. After heat stress, SDC was slowly re-silenced, allowing a temporal extension of SDC activity to contribute to the recovery of plant biomass. The SDC activation seems to occur independently and in parallel to canonical heat-shock perception and signaling, but rely on a yet undefined epigenetic mechanism (Sanchez and Paszkowski, 2014).
Transgenerational Memory and Evolutionary Adaptation
Transgenerational memory, also known as epigenetic inheritance, refers to the transmittance of epigenetic states and certain environmental responses from one generation to the next. These transgenerational effects may offer the offspring an adaptive advantage or genomic flexibility for better fitness. Recent evidence suggests that some abiotic and biotic stress responses are transgenerational in plants. For example, exposure of Arabidopsis plants to UV-C and flagellin can induce transgenerational increases in HRF (Molinier et al., 2006). Heat responses also exhibit transgenerational epigenetic inheritance (Boyko et al., 2010; Lang-Mladek et al., 2010; Ito et al., 2011; Zhong et al., 2013; Iwasaki and Paszkowski, 2014; Migicovsky et al., 2014). The immediate progeny of heat-stressed Arabidopsis plants have fewer, but larger leaves, and tend to bolt earlier (Migicovsky et al., 2014). These plants have increased expression of HSFA2, but reduced expression of ROS1 and several Su(var)3–9 homologs (SUVH) genes involved in H3K9 methylation and DNA methylation. These phenotypic and epigenetic changes are partially deficient in the offspring of heat-stressed dcl2 and dcl3 mutants (Migicovsky et al., 2014). It is also reported that transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins (Boyko et al., 2010). However, the transgenerational retrotransposition of ONSEN is prevented by the siRNAs pathway (Matsunaga et al., 2012), while SGS3 overexpression could decrease the warmth-induced transgenerational memory (Zhong et al., 2013). Thus, the role of siRNAs pathway in transgenerational memory of heat responses remains controversial. The involvement of AGO1 and the miR156-SPL pathway has been demonstrated to maintain the short memory of acquired thermotolerance in the adaptation to recurring heat stress at the physiological and molecular level in Arabidopsis (Stief et al., 2014). Bra-miR168 and its target braAGO1 are also suggested to be putative messengers that mediate meiotic epigenetic inheritance in Brassica rapa (Bilichak et al., 2015). Further experiments on transgenerational heat stress in the hypomorphic ago1 mutants may shade a new light on the contribution of AGO1 and the miRNA pathway to epigenetic inheritance in plants.
The role of DNA methylation in transgenerational memory is also obscure. It is reported that there seems no consistent correlation between DNA methylation changes of transgene and the warmth-induced transgenerational release of PTGS (Zhong et al., 2013). Similarly, DNA methylation may be also not involved in the release of transgene TGS by heat stress (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010). However, it remains a possibility that changes in DNA methylation at certain sites of a silenced target gene or at certain loci of the genome are responsible for the transgenerational memory, as CG methylation plays a central role in transgenerational stability of the Arabidopsis epigenome (Mathieu et al., 2007). Loss of CG methylation triggers genome-wide aberrant de novo non-CG methylation by interfering with the RdDM process and expression of DNA demethylases, as well as progressive H3K9 remethylation of heterochromatin. It is proposed that immediate, non-heritable stress responses may be associated with alteration of non-CG methylation patterns mediated by siRNAs/RdDM and ROS1/DME, while long-term, heritable adaptation to a changing environment would require modulation of CG patterns (Mathieu et al., 2007). Recently, a forward genetic screen revealed that DDM1 and MOM1 act redundantly in preventing the transmission of stress-induced transcriptional changes to progeny of the stressed plants (Iwasaki and Paszkowski, 2014). Such DDM1- and MOM1-mediated or other mechanisms of chromatin resetting could prevent the transgenerational transmission of environmentally induced epigenetic traits. The roles of other epigenetic mechanisms in transgenerational memory, such as H2A.Z and H3K27me3, are worth further investigation.
Although the mechanisms remain to be elucidated, the transgenerational memory of heat responses may contribute to evolutionary adaptation. The warmth-induced epigenetic memory was maintained for at least three generations with gradually declining (Zhong et al., 2013). Heat stress also induced transgenerational phenotypic changes over three generations (Suter and Widmer, 2013a,b). Ancestral exposure to elevated temperatures over P and F1 generations resulted in increased fitness in the F3 heat-treated Arabidopsis plants (Whittle et al., 2009). Thus, the transgenerational memory of heat responses may allow potentially long-term adaptation and rapid evolution, as chromatin modifications can be mitotically or meiotically heritable. Stress-induced epigenetic changes may lead to the formation of heritable epialleles and transcriptional activation of TEs (Mirouze and Paszkowski, 2011). The epialleles and transposon-driven variation in gene expression may contribute to the phenotypic diversity of different individuals in a population or a species that can be subject to natural selection (Mirouze and Paszkowski, 2011). The transgenerational retrotransposition of ONSEN may reshape gene regulatory networks and potentially create novel traits for adaptation to heat stress, as genes in the vicinity of ONSEN neo-insertions aquired heat-responsiveness (Ito et al., 2011). Given that the activity of transposable element is important for adaptive plant evolution (Lisch, 2013), and ONSEN is evolutionary conserved and transcriptionally activated by environmental heat stress in some Brassicaceae species (Ito et al., 2013), it will be interesting to explore the possible role of ONSEN and other TEs in plant evolution. Whether heat mediates the formation of heritable epialleles still needs to be investigated. Random RdDM-mediated epiallele formation is suggested to play a greater role in evolution than genetic variation (Matzke and Mosher, 2014). Epigenetic variation in DNA methylation among epigenetic recombinant inbred lines (epiRILs) that are nearly isogenic but highly variable at the level of DNA methylation, can cause substantial heritable variation of drought tolerance and nutrient plasticity (Zhang et al., 2013). It will be worth investigating whether heat-induced changes of global methylation creates potential for the evolution of phenotypic plasticity.
Concluding Remarks
Heat greatly affects the growth, development, and productivity of plants. Several heat sensors have been reported, including the calcium channel in the plasma membrane, H2A.Z-containing nucleosomes in the nucleus, and two unfolded protein sensors in ER and the cytosol. Importantly, different epigenetic regulations may also be involved in the responses to different heat treatments (Table 1). The epigenetic regulation of warm and high temperatures mainly involves warmth-induced PTGS release, enhanced VIGS-mediated resistance, and H2A.Z-mediated morphological acclimation and acceleration of flowering. Various epigenetic mechanisms (known or unknown) are involved in response to heat stress. It is notable that different lengths (from 1 h to 4 days) of heat treatment at 37°C have diverse effects on the epigenome, suggesting the complexity in the epigenetic regulation of heat stress.
Despite recent advances in our understanding of the genetic and epigenetic mechanisms involved in heat stress sensing in plants, many questions remain to be answered by future research (Box 1). Perhaps the most important questions in the genetic mechanisms of heat responses are: what are the primary heat sensors? In addition to the four heat sensors mentioned above, a list of other components like mRNAs, miRNAs and hormonal import and antiport channels may also be plausible thermometers based on physical capacities (McClung and Davis, 2010). Systematic analysis of the changes in the genome, transcriptome, microme, and proteome by omic approaches may help to identify novel transcriptional, translational, and posttranslational regulation components and underlying mechanisms in plant heat responses (Bokszczanin and Fragkostefanakis, 2013; Hasanuzzaman et al., 2013). Perhaps plants sense heat through different organelles in different phases of the response, and then these signaling pathways are integrated and work synergisticly or differentially to defend plants from heat-induced deleterious effects.
Box 1. Proposals of future researches.
• What are the primary heat sensors? Are the CNGCs, H2A.Z or the unfolded protein sensors the true heat sensor?
• How are the different heat-sensing pathways integrated in plant cells?
• What are the roles of the RdDM pathway in response to heat in crop plants?
• How do the different histone modifications regulate the heat responses in different plants?
• Do histone modifications regulate the response of circadian clock to high temperature and heat stress?
• By which precise mechanism is the H2A.Z occupancy regulated by high temperature? Is H2A.Z occupancy regulated by heat stress? Is H2A.Z occupancy involved in the modulation of plant immunity and circadian clock by high temperature?
• How are the heat-responsive miRNAs, siRNAs and lncRNAs regulated by heat? What are the regulatory networks of their targets in plants?
• How is the epigenetic regulation of heat responses integrated with the epigenetic regulation of other biotic and abiotic responses?
• How is the transgenerational memory of heat responses controlled?
• How to improve the thermotolerance of crops without sacrificing growth?
As listed in Box 1, a set of questions concerning the epigenetic regulations of heat responses are proposed. One of the major challenge ahead may be to discover the mechanisms of transgenerational memory heat responses. Systemic screening for factors regulating transgenerational memory of heat may address the long-term controversial issue. Besides, the roles of DNA methylation and histone modifications in response to heat need to be defined. miRNAs, lncRNAs and the chromatin-remodeling gene AtCHR12 have been suggested to integrate the epigenetic regulation of heat stress with the regulation of other biotic and abiotic stresses. Other epigenetic regulations may also have similar functions. Further investigations should be focused on the epigenetic regulatory networks between heat stress and other biotic and abiotic stresses. We should note that most experiments on the role of genetic and epigenetic regulation in heat responses are limited to the model Arabidopsis plants in laboratory conditions with short-term heat treatment. As temperatures in the field change seasonally and fluctuate daily, further studies should also be centered on the genetic and epigenetic regulations of heat responses in crop plants in the field, which may produce practical approaches to develop crop plants with improved thermotolerance.
Acknowledgment
This work was supported by grants from the National Research Program of China (2011CB100700).
References
- Andika I. B., Sun L., Xiang R., Li J., Chen J. (2013). Root-specific role for Nicotiana benthamiana RDR6 in the inhibition of Chinese wheat mosaic virus accumulation at higher temperatures. Mol. Plant Microbe Interact. 26 1165–1175 10.1094/mpmi-05-13-0137-r [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2:e106 10.1371/journal.pgen.0020106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballen-Taborda C., Plata G., Ayling S., Rodriguez-Zapata F., Becerra Lopez-Lavalle L. A., Duitama J., et al. (2013). Identification of cassava microRNAs under abiotic stress. Int. J. Genomics 2013:857986 10.1155/2013/857986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal S. K., Bharti K., Chan K. Y., Fauth M., Ganguli A., Kotak S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29 471–487 10.1007/BF02712120 [DOI] [PubMed] [Google Scholar]
- Battisti D. S., Naylor R. L. (2009). Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323 240–244 10.1126/science.1164363 [DOI] [PubMed] [Google Scholar]
- Bilichak A., Ilnytskyy Y., Woycicki R., Kepeshchuk N., Fogen D., Kovalchuk I. (2015). The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 6:5 10.3389/fpls.2015.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boikoglou E., Ma Z., Von Korff M., Davis A. M., Nagy F., Davis S. J. (2011). Environmental memory from a circadian oscillator: the Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics 189 655–664 10.1534/genetics.111.131417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokszczanin K. L., Fragkostefanakis S. (2013). Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 4:315 10.3389/fpls.2013.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna N. G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65 473–503 10.1146/annurev-arplant-050213-035728 [DOI] [PubMed] [Google Scholar]
- Borsani O., Zhu J. H., Verslues P. E., Sunkar R., Zhu J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 10.1016/j.cell.2005.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., et al. (2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5:e9514 10.1371/journal.pone.0009514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280 10.1016/j.tig.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Cavrak V. V., Lettner N., Jamge S., Kosarewicz A., Bayer L. M., Mittelsten Scheid O. (2014). How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet 10:e1004115 10.1371/journal.pgen.1004115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W., Henderson I. R., Jacobsen S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360 10.1038/nrg1601 [DOI] [PubMed] [Google Scholar]
- Che P., Bussell J. D., Zhou W., Estavillo G. M., Pogson B. J., Smith S. M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 3:ra69 10.1126/scisignal.2001140 [DOI] [PubMed] [Google Scholar]
- Chellappan P., Vanitharani R., Ogbe F., Fauquet C. M. (2005). Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 138 1828–1841 10.1104/pp.105.066563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ren Y., Zhang Y., Xu J., Sun F., Zhang Z., et al. (2012). Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 504 160–165 10.1016/j.gene.2012.05.034 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., Tian L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769 295–307 10.1016/j.bbaexp.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78 273–304 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Cristescu S. M., Miersch O., Harren F. J., Wasternack C., Mur L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182 175–187 10.1111/j.1469-8137.2008.02735.x [DOI] [PubMed] [Google Scholar]
- Clarke S. M., Mur L. A., Wood J. E., Scott I. M. (2004). Salicylic acid dependent signaling promotes basal thermotolerance but is not essential for acquired thermotolerance in Arabidopsis thaliana. Plant J. 38 432–447 10.1111/j.1365-313X.2004.02054.x [DOI] [PubMed] [Google Scholar]
- Cokus S. J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C. D., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215–219 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B., Valledor L., Meijon M., Rodriguez J. L., Dias M. C., Santos C., et al. (2013). Is the interplay between epigenetic markers related to the acclimation of cork oak plants to high temperatures? PLoS ONE 8:e53543 10.1371/journal.pone.0053543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. G., Shan J. X., Shi M., Gao J. P., Lin H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80 1108–1117 10.1111/tpj.12712 [DOI] [PubMed] [Google Scholar]
- Deng Y., Humbert S., Liu J. X., Srivastava R., Rothstein S. J., Howell S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 7247–7252 10.1073/pnas.1102117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C., Yuan J., Wu Y., Li J., Lin H., Hu L., et al. (2014). Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 80 848–861 10.1111/tpj.12679 [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Brosnan C. A., Schott G., Wang Y., Jay F., Alioua A., et al. (2010). An endogenous, systemic RNAi pathway in plants. EMBO J. 29 1699–1712 10.1038/emboj.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fei Q., Xia R., Meyers B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25 2400–2415 10.1105/tpc.113.114652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Cumbie J. S., Dharmawardhana J. P., Jaiswal P., Chang J. H., Palusa S. G., et al. (2015). Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 8 207–227 10.1016/j.molp.2014.10.011 [DOI] [PubMed] [Google Scholar]
- Finka A., Cuendet A. F., Maathuis F. J., Saidi Y., Goloubinoff P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24 3333–3348 10.1105/tpc.112.095844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter A. H., Fitter R. S. (2002). Rapid changes in flowering time in British plants. Science 296 1689–1691 10.1126/science.1071617 [DOI] [PubMed] [Google Scholar]
- Folsom J. J., Begcy K., Hao X., Wang D., Walia H. (2014). Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165 238–248 10.1104/pp.113.232413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Lee S. H., Patel D., Kumar S. V., Spartz A. K., Gu C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108 20231–20235 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Li J., Li H., Li F., Xu K., Yan G., et al. (2014). Comparison of the heat stress induced variations in DNA methylation between heat-tolerant and heat-sensitive rapeseed seedlings. Breed Sci. 64 125–133 10.1270/jsbbs.64.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10 94–108 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli J. I., Weigel D., Chan R. L., Manavella P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195 766–773 10.1111/j.1469-8137.2012.04259.x [DOI] [PubMed] [Google Scholar]
- Gou M., Hua J. (2012). Complex regulation of an R gene SNC1 revealed by auto-immune mutants. Plant Signal. Behav. 7 213–216 10.4161/psb.18884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P. D., Locke J. C., Larue C., Southern M. M., Davis S. J., Hanano S., et al. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 10.1105/tpc.105.039990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W. M., Ostin A., Sandberg G., Romano C. P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95 7197–7202 10.1073/pnas.95.12.7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Le C., Wang Y., Li Z., Jiang D., Wang Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4:1947 10.1038/ncomms2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Lu X., Zeng H., Zhang Y., Zhu J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74 840–851 10.1111/tpj.12169 [DOI] [PubMed] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M. M., Roychowdhury R., Fujita M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14 9643–9684 10.3390/ijms14059643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Elling A. A., Deng X. W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62 411–435 10.1146/annurev-arplant-042110-103806 [DOI] [PubMed] [Google Scholar]
- Hsu P. Y., Harmer S. L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19 240–249 10.1016/j.tplants.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J. (2013). Modulation of plant immunity by light, circadian rhythm, and temperature. Curr. Opin. Plant Biol. 16 406–413 10.1016/j.pbi.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472 115–119 10.1038/nature09861 [DOI] [PubMed] [Google Scholar]
- Ito H., Yoshida T., Tsukahara S., Kawabe A. (2013). Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 518 256–261 10.1016/j.gene.2013.01.034 [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Paszkowski J. (2014). Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl. Acad. Sci. U.S.A. 111 8547–8552 10.1073/pnas.1402275111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.-H., Park S., Zhai J., Gurazada S. G., De Paoli E., Meyers B. C., et al. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23 4185–4207 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Biggins S. (2005). Histone variants: deviants? Genes Dev. 19 295–310 10.1101/gad.1272805 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Morgan R., Dahlbeck D., Borsani O., Villegas A., Jr., Zhu J. K., et al. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U.S.A. 103 18002–18007 10.1073/pnas.0608258103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizewski L., Ginalski K., Jerzmanowski A. (2008). Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13 557–565 10.1016/j.tplants.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Koini M. A., Alvey L., Allen T., Tilley C. A., Harberd N. P., Whitelam G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19 408–413 10.1016/j.cub.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Kolmos E., Chow B. Y., Pruneda-Paz J. L., Kay S. A. (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111 16172–16177 10.1073/pnas.1418483111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K., Pacak A., Swida-Barteczka A., Nuc P., Alaba S., Wroblewska Z., et al. (2014). Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 65 6123–6135 10.1093/jxb/eru353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. R., Pathak H., Sharma S. K., Kala Y. K., Nirjal M. K., Singh G. P., et al. (2014). Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genomics 10.1007/s10142-014-0421-0 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Kumar S. V., Lucyshyn D., Jaeger K. E., Alos E., Alvey E., Harberd N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484 242–245 10.1038/nature10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. V., Wigge P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140 136–147 10.1016/j.cell.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Kwon Y. J., Park M. J., Kim S. G., Baldwin I. T., Park C. M. (2014). Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14:136 10.1186/1471-2229-14-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Mladek C., Popova O., Kiok K., Berlinger M., Rakic B., Aufsatz W., et al. (2010). Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 3 594–602 10.1093/mp/ssq014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Ryu H.-S., Chung K. S., Posé D., Kim S., Schmid M., et al. (2013). Regulation of temperature-responsive flowering by MADS-Box transcription factor repressors. Science 342 628–632 10.1126/science.1241097 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yoo S. J., Park S. H., Hwang I., Lee J. S., Ahn J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21 397–402 10.1101/gad.1518407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L. (2007). The role of chromatin during transcription. Cell 128 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- Li M. Y., Wang F., Xu Z. S., Jiang Q., Ma J., Tan G. F., et al. (2014a). High throughput sequencing of two celery varieties small RNAs identifies microRNAs involved in temperature stress response. BMC Genomics 15:242 10.1186/1471-2164-15-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liu J., Liu Z., Li X., Wu F., He Y. (2014b). HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in Arabidopsis. Plant Cell 26 1764–1780 10.1105/tpc.114.124883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Zhong S. H., Cui X. F., Li J., He Z. H. (2012). Characterization of temperature-sensitive mutants reveals a role for receptor-like kinase SCRAMBLED/STRUBBELIG in coordinating cell proliferation and differentiation during Arabidopsis leaf development. Plant J. 72 707–720 10.1111/j.1365-313X.2012.05109.x [DOI] [PubMed] [Google Scholar]
- Lisch D. (2013). How important are transposons for plant evolution? Nat. Rev. Genet. 14 49–61 10.1038/nrg3374 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61 395–420 10.1146/annurev.arplant.043008.091939 [DOI] [PubMed] [Google Scholar]
- Liu F., Wang W., Sun X., Liang Z., Wang F. (2014). Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ. 10.1111/pce.12484 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Lobell D. B., Field C. B. (2007). Global scale climate-crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2 1–7 10.1088/1748-9326/2/1/014002 [DOI] [Google Scholar]
- Lobell D. B., Schlenker W., Costa-Roberts J. (2011). Climate trends and global crop production since 1980. Science 333 616–620 10.1126/science.1204531 [DOI] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Chiang V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55 131–151 10.1111/j.1365-313X.2008.03497.x [DOI] [PubMed] [Google Scholar]
- Makarevitch I., Waters A. J., West P. T., Stitzer M., Hirsch C. N., Ross-Ibarra J., et al. (2015). Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11:e1004915 10.1371/journal.pgen.1004915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang H. G., Qian W., Zhu Y., Qian J., Kang H. G., Klessig D. F., et al. (2012). Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24 1271–1284 10.1105/tpc.112.096198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Diaz R., Reyes J. C. (2009). The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2 565–577 10.1093/mp/ssp019 [DOI] [PubMed] [Google Scholar]
- Martin G. B., Bogdanove A. J., Sessa G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54 23–61 10.1146/annurev.arplant.54.031902.135035 [DOI] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130 851–862 10.1016/j.cell.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Matsunaga W., Kobayashi A., Kato A., Ito H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol. 53 824–833 10.1093/pcp/pcr179 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 394–408 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- McClung C. R., Davis S. J. (2010). Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 20 R1086–R1092 10.1016/j.cub.2010.10.035 [DOI] [PubMed] [Google Scholar]
- Migicovsky Z., Yao Y., Kovalchuk I. (2014). Transgenerational phenotypic and epigenetic changes in response to heat stress in Arabidopsis thaliana. Plant Signal. Behav. 9:e27971 10.4161/psb.27971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133 481–489 10.1111/j.1399-3054.2008.01090.x [DOI] [PubMed] [Google Scholar]
- Min L., Li Y., Hu Q., Zhu L., Gao W., Wu Y., et al. (2014). Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164 1293–1308 10.1104/pp.113.232314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M., Paszkowski J. (2011). Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14 267–274 10.1016/j.pbi.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Mizuno T., Nomoto Y., Oka H., Kitayama M., Takeuchi A., Tsubouchi M., et al. (2014). Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 55 958–976 10.1093/pcp/pcu030 [DOI] [PubMed] [Google Scholar]
- Mlynarova L., Nap J. P., Bisseling T. (2007). The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 51 874–885 10.1111/j.1365-313X.2007.03185.x [DOI] [PubMed] [Google Scholar]
- Molinier J., Ries G., Zipfel C., Hohn B. (2006). Transgeneration memory of stress in plants. Nature 442 1046–1049 10.1038/nature05022 [DOI] [PubMed] [Google Scholar]
- Nagel D. H., Pruneda-Paz J. L., Kay S. A. (2014). FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111 14595–14600 10.1073/pnas.1416666111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov M., Baev V., Apostolova E., Gospodinova N., Sablok G., Gozmanova M., et al. (2015). High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 87 102–108 10.1016/j.plaphy.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Okada T., Endo M., Singh M. B., Bhalla P. L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44 557–568 10.1111/j.1365-313X.2005.02554.x [DOI] [PubMed] [Google Scholar]
- Pauli H., Gottfried M., Dullinger S., Abdaladze O., Akhalkatsi M., Benito Alonso J. L., et al. (2012). Recent plant diversity changes on Europe’s mountain summits. Science 336 353–355 10.1126/science.1219033 [DOI] [PubMed] [Google Scholar]
- Pecinka A., Dinh H. Q., Baubec T., Rosa M., Lettner N., Mittelsten Scheid O. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22 3118–3129 10.1105/tpc.110.078493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Huang J., Sheehy J. E., Laza R. C., Visperas R. M., Zhong X., et al. (2004). Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. U.S.A. 101 9971–9975 10.1073/pnas.0403720101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova O. V., Dinh H. Q., Aufsatz W., Jonak C. (2013). The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol. Plant 6 396–410 10.1093/mp/sst023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoles S., Mas P. (2010). The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6:e1001201 10.1371/journal.pgen.1001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pose D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503 414–417 10.1038/nature12633 [DOI] [PubMed] [Google Scholar]
- Proveniers M. C. G., van Zanten M. (2013). High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18 59–64 10.1016/j.tplants.2012.09.002 [DOI] [PubMed] [Google Scholar]
- Qu A. L., Ding Y. F., Jiang Q., Zhu C. (2013). Molecular mechanisms of the plant heat stress response. Biochem. Biophys. Res. Commun. 432 203–207 10.1016/j.bbrc.2013.01.104 [DOI] [PubMed] [Google Scholar]
- Qu F., Ye X., Hou G., Sato S., Clemente T. E., Morris T. J. (2005). RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79 15209–15217 10.1128/jvi.79.24.15209-15217.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P. Y., Jones M. A., Schwartz J., Salemi M. R., et al. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7:e1001350 10.1371/journal.pgen.1001350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25 2383–2399 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi Y., Finka A., Muriset M., Bromberg Z., Weiss Y. G., Maathuis F. J., et al. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21 2829–2843 10.1105/tpc.108.065318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B., Voleti S. R., Subrahmanyam D., Sarla N., Prasanth V. V., Bhadana V. P., et al. (2014). Prediction and expression analysis of miRNAs associated with heat stress in Oryza sativa. Rice Science 21 3–12 10.1016/S1672-6308(13)60164-X [DOI] [Google Scholar]
- Salome P. A., McClung C. R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17 791–803 10.1105/tpc.104.029504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome P. A., Weigel D., Mcclung C. R. (2010). The role of the Arabidopsis morning loop components CCA1 LHY, PRR7 and PRR9 in temperature compensation. Plant Cell 22 3650–3661 10.1105/tpc.110.079087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D. H., Paszkowski J. (2014). Heat-induced release of epigenetic silencing reveals the concealed role of an imprinted plant gene. PLoS Genet. 10:e1004806 10.1371/journal.pgen.1004806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Mas P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26 79–87 10.1105/tpc.113.119842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., De Jonge J., Forsberg S. K., Pettersson M. E., Sheng Z., Hennig L., et al. (2014). Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 10:e1004842 10.1371/journal.pgen.1004842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Yekondi S., Chen P. W., Tsai C. H., Yu C. W., Wu K., et al. (2014). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26 2676–2688 10.1105/tpc.114.123356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L. L., Jiang Y. L., Zhao H. Q., Hou M. F. (2012). Acquired thermotolerance in plants. Plant Cell Tiss Organ Cult. 111 265–276 10.1007/s11240-012-0198-6 [DOI] [Google Scholar]
- Stavang J. A., Gallego-Bartolome J., Gomez M. D., Yoshida S., Asami T., Olsen J. E., et al. (2009). Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 60 589–601 10.1111/j.1365-313X.2009.03983.x [DOI] [PubMed] [Google Scholar]
- Stief A., Altmann S., Hoffmann K., Pant B. D., Scheible W.-R., Bäurle I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26 1792–1807 10.1105/tpc.114.123851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D., Schmollinger S., Sommer F., Schulz-Raffelt M., Schroda M. (2011). Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23 2285–2301 10.1105/tpc.111.085266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Dreos R., Aparicio F., Maule A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21 642–654 10.1105/tpc.108.062596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8:e1002594 10.1371/journal.pgen.1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter L., Widmer A. (2013a). Environmental heat and salt stress induce transgenerational phenotypic changes in Arabidopsis thaliana. PLoS ONE 8:e60364 10.1371/journal.pone.0060364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter L., Widmer A. (2013b). Phenotypic effects of salt and heat stress over three generations in Arabidopsis thaliana. PLoS ONE 8:e80819 10.1371/journal.pone.0080819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Bajad S., Shuman J., Shulaev V., Mittler R. (2008). The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J. Biol. Chem. 283 9269–9275 10.1074/jbc.M709187200 [DOI] [PubMed] [Google Scholar]
- Thines B., Harmon F. G. (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 107 3257–3262 10.1073/pnas.0911006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M., Bucher E., Broger L., Mathieu O., Paszkowski J., Vaillant I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genet. 6:e1001175 10.1371/journal.pgen.1001175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lu Y., Liu P., Wen W., Zhang J., Ge X., et al. (2012a). The ammonium/nitrate ratio is an input signal in the temperature-modulated, SNC1-mediated and EDS1-dependent autoimmunity of nudt6-2 nudt7. Plant J. 10.1111/tpj.12032 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun F., Cao H., Peng H., Ni Z., Sun Q., et al. (2012b). TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 7:e48445 10.1371/journal.pone.0048445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. C., Wu J. R., Chang W. L., Yeh C. H., Ke Y. T., Lu C. A., et al. (2013). Arabidopsis HIT4 encodes a novel chromocentre-localized protein involved in the heat reactivation of transcriptionally silent loci and is essential for heat tolerance in plants. J. Exp. Bot. 64 1689–1701 10.1093/jxb/ert030 [DOI] [PubMed] [Google Scholar]
- Wang L. C., Wu J. R., Hsu Y. J., Wu S. J. (2014). Arabidopsis HIT4, a regulator involved in heat-triggered reorganization of chromatin and release of transcriptional gene silencing, relocates from chromocenters to the nucleolus in response to heat stress. New Phytol. 205 544–554 10.1111/nph.13088 [DOI] [PubMed] [Google Scholar]
- Wang L., Yu X., Wang H., Lu Y. Z., De Ruiter M., Prins M., et al. (2011). A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genomics 12:289 10.1186/1471-2164-12-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9 244–252 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., et al. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149 101–112 10.1016/j.cell.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Weng M., Yang Y., Feng H., Pan Z., Shen W. H., Zhu Y., et al. (2014). Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environ. 37 2128–2138 10.1111/pce.12299 [DOI] [PubMed] [Google Scholar]
- Whittle C. A., Otto S. P., Johnston M. O., Krochko J. E. (2009). Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 87 650–657 10.1139/B09-030 [DOI] [Google Scholar]
- Wierzbicki A. T. (2012). The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 15 517–522 10.1016/j.pbi.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Wunderlich M., Gross-Hardt R., Schoffl F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85 541–550 10.1007/s11103-014-0202-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D., et al. (2011). Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61 10.1186/1471-2229-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Xie C., Peng H., Ni Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Liu P., Wu C. A., Yang G. D., Xu R., Guo Q. H., et al. (2012). Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 48 521–531 10.1016/j.molcel.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Yang S. H., Hua J. (2004). A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 1060–1071 10.1105/tpc.020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. D., Yang X. F., Chen S. T., Wang Y. T., Li J. K., Shen Q., et al. (2012a). Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genet. 8:e1002669 10.1371/journal.pgen.1002669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang H., Lu Y., De Ruiter M., Cariaso M., Prins M., et al. (2012b). Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 63 1025–1038 10.1093/jxb/err337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Yang J., Li X., Liu X., Sun C., Wu F., et al. (2013). Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 13:208 10.1186/1471-2229-13-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu J. K. (2012). Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 77 161–173 10.1101/sqb.2012.77.014936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S. W., Chen H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang X., Singh J., Li D., Qu F. (2012). Temperature-dependent survival of Turnip crinkle virus-infected Arabidopsis plants relies on an RNA silencing-based defense that requires DCL2 AGO2 and HEN1. J. Virol. 86 6847–6854 10.1128/jvi.00497-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Y., Fischer M., Colot V., Bossdorf O. (2013). Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol. 197 314–322 10.1111/nph.12010 [DOI] [PubMed] [Google Scholar]
- Zhong S. H., Liu J. Z., Jin H., Lin L., Li Q., Chen Y., et al. (2013). Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 9171–9176 10.1073/pnas.1219655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Shen W. H. (2012a). Histone variants and chromatin assembly in plant abiotic stress responses. Biochim. Biophys. Acta 1819 343–348 10.1016/j.bbagrm.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Mang H. G., Sun Q., Qian J., Hipps A., Hua J. (2012b). Gene discovery using mutagen-induced polymorphisms and deep sequencing: application to plant disease resistance. Genetics 192 139–146 10.1534/genetics.112.141986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qian W., Hua J. (2010). Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6:e1000844 10.1371/journal.ppat.1000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69 10.1038/ng1929 [DOI] [PubMed] [Google Scholar]