Abstract
Sjögren’s syndrome (SS) of humans and SS-like (SjS-like) diseases in mouse models are characterized by chronic immune attacks against the salivary and lacrimal glands leading to exocrine dysfunction. One characteristic of SS and SjS-like diseases repeatedly observed is a strong upregulated expression of both the type I (α/β) and type II (γ) interferons (IFNs). In addition, recent global transcriptome studies have identified a variety of IFN-stimulated gene (ISG) transcripts differentially expressed in tissues of SS patients and mouse models exhibiting SjS-like disease. Analyses of these transcriptome databases indicate that the sets of differentially expressed genes are highly restricted, suggesting that there is a unique specificity in ISGs activated (or suppressed) during development and onset of disease. As a result, these observations have led to both SS and SjS-like diseases being designated as ‘interferon-signature’ diseases. While SS and SjS-like diseases may be designated as such, very little effort has been made to determine what an interferon-signature might signify relative to autoinflammation and whether it might point directly to an underlying etiopathological mechanism. Here, we review these limited data and provide a model of how the products of these genes interact molecularly and biologically to define critical details of SS pathology.
Introduction
Sjögren’s syndrome (SS) of humans and SS-like (SjS-like) diseases in various mouse models are best classified as systemic, hypersensitive Type II inflammatory-based rheumatic diseases characterized by chronic progressive immune attacks primarily against the salivary and lacrimal glands, resulting, respectively, in dry mouth (stomatitis sicca /xerostomia) and dry eye (keratoconjunctivitis sic-ca /xerophthalmia) diseases [1–5]. In humans, the vast majority of SS patients are post-menopausal women, implicating a strong hormone influence. In addition to the apparent primary sites of autoimmunity, multiple tissues may become involved including the GI tract, skin, lungs, vasculature, kidneys, bladder and vagina. Furthermore, perhaps 20% of SS patients exhibit various neuropathies, including sensory, peripheral, cranial and myelopathic complications [6]. Cognitive impairments such as dementia, lack of concentration, memory loss and various psychiatric disorders (ranging from depression to anxiety) are also noted in patients during clinic visits [7–9], a condition often referred to as ‘mental fogginess’. Involvement of the musculature can lead to fibromyalgia-like symptoms and chronic fatigue [3, 4], the latter being one of the most prevalent complaints. Many of these secondary complaints are thought to be a consequence of circulating interferons and autoantibodies. Although SS is generally not considered a lethal disease unless the patient develops a high-grade B cell lymphoma [10], patients have an increasingly diminished quality of life as the disease progresses [11, 12]. To date, intervention therapies have been ineffective, especially long term, but this may reflect the extreme heterogeneous nature of the clinical disease once diagnosis is confirmed.
One fascinating feature of SS autoimmunity in both humans and animal models is the upregulated expression of interferon (IFN) proteins, both IFN-α/β (type I) and IFN-γ (type II), as well as multiple IFN-regulated genes, referred to collectively as interferon-stimulated genes, or IFN-stimulated genes (ISGs) [13–20]. Few studies of SS /SjS-like diseases are published today that do not mention the role of IFN. No doubt, IFNs represent one of the most vibrant cell-autonomous immune mechanisms effecting antimicrobial defences, as recently reviewed by MacMicking [21], but carries with it a heavy genetic burden involving more than 1900 genes [22]. While elevated levels of the interferons are often associated with viral infections, the IFN-virus connection is also one of the main focuses of SS /SjS-like pathology. However, to date there is only circumstantial evidence suggesting that SS might be a viral-based disease, and this is generally founded on recent observations derived from global tran-scriptome data analyses, showing that genes encoding TLR3, TLR7, TLR9 and virtually all factors in both the toll-like receptor (TLR) and IFN-signalling pathways are markedly upregulated prior to disease onset [15, 23–25]. In addition, we recently presented several lines of evidence that indirectly point to the possible involvement of a dsRNA virus in SjS-like disease of the C57BL /6.NOD-Aec1Aec2 mouse model of primary SS [26]. These include the following: (1) an upregulated co-expression of Tlr3 and Tlr4, two genes encoding pathogen recognition receptors (PRRs) that signal through Traf3 via Trif and Traf6 via a Trif–Trim23 complex to activate NF-κβ and Irf3 /Irf7 transcription of pro-inflammatory cytokines including IFN (2) the upregulated expression of Ifih1, encoding the cytoplasmic dsRNA PRR Mda-5, in the absence of any concomitant upregulated expression of Ddx58, encoding the cytoplasmic ssRNA PRR Rig-1 and (3) an upregulation of the interferon-responsive factor genes Irf1, Irf3, Irf7 and Irf9, plus the interferon-inducible transmembrane factor genes Ifitm2 and Ifitm3 and numerous tripartite motif (Trim) genes whose products have antiviral functions.
As stated, then, based on the observations that both SS patients and SjS-susceptible (SjSS) mice exhibiting spontaneous disease can present with elevated blood interferon levels (although not routinely measured) [13, 14, 16], together with activation of multiple ISGs by both genetic and microarray studies [15, 27–32], SS and SjS-like diseases, similar to systemic lupus erythematosus (SLE) [33]and now rheumatoid arthritis (RA) [34], have been designated autoimmune diseases with an ‘interferon-signature’. Furthermore, high levels of circulating IFN is implicated as one of perhaps several likely underlying molecular causes for the frequent incidence of chronic fatigue and lethargy. Nevertheless, as pointed out in our recent article published in the Journal of Clinical Rheumatology and Musculoskeletal Medicine [26], ISGs differentially expressed within the exocrine glands represent highly select subsets within the multiple subfamilies of the ISGs. Thus, it seems imperative to question whether the ‘interferon-signature’ merely reflects the fact that type I and type II IFNs are ubiquitous cytokines involved in most cell-autonomous immune responses, thereby a consequence of general immunity, or whether the subsets of differentially expressed genes comprising the ‘interferon-signature’ define unique biological processes and molecular mechanisms that are the key to development and onset of a specific autoimmune disease, that is, SLE, RA or SS. In this abbreviated topical perspective, we focus on sets of specific genes defining the IFN-signature of SS /SjS-like diseases to generate a model that might explain development and onset of the initial inflammatory response that metamorphs to autoimmunity.
Defining the ‘interferon-signature’ of SjS-like disease
Previous studies reported by Cha et al. [19, 20, 35]revealed that high levels of IFN-γ are detected in mice of the NOD /ShiLtJ and NOD-derived congenic C57BL /6.NOD-Aec1Aec2 lines as early as time of birth. In contrast, if these SjSS mice expressed a non-functional Ifnγ (Ifng) or Ifnγ-receptor (Ifngr)-encoding gene, they failed to develop any aspect of SjS-like disease, revealing an absolute requirement for Ifnγ in the development and onset of SjS. However, why Ifnγ plays such an important role in promoting disease in these mice remains only speculative. Therefore, in an attempt to define an interferon-signature for SjS-like disease, and thereby for SS, follow-up studies were carried out to analyse temporal gene expression profiles generated for both salivary and lacrimal glands isolated from C57BL /6.NOD-Aec1Aec2 mice for known interferon-encoding, interferon-stimulating and interferon-regulatory genes [see 21]. More recently, follow-up analyses have focused on genes primarily belonging to a limited number of ISG families, including Tlr, Irf, Ifi, Ifr and Trim genes [26]. The most obvious observation drawn from these new analyses is the fact that about half of the genes in each ISG subfamily are upregulated, while the other half of these genes are either neutral or downregulated. A second observation is the fact that, of the genes whose expressions were upregulated, one subset showed optimal expression during the innate immune stage of disease, while a second subset showed optimal expression during the phase of adaptive immunity [26]. On rare occasion, individual ISG subfamily genes exhibited a biphasic response correlating to both the innate and adaptive immune responses. There were no direct correlations identified between time of optimal gene expression and type of interferon, but this may be due in part to the fact that multiple ISGs are activated by both type I and type II IFN.
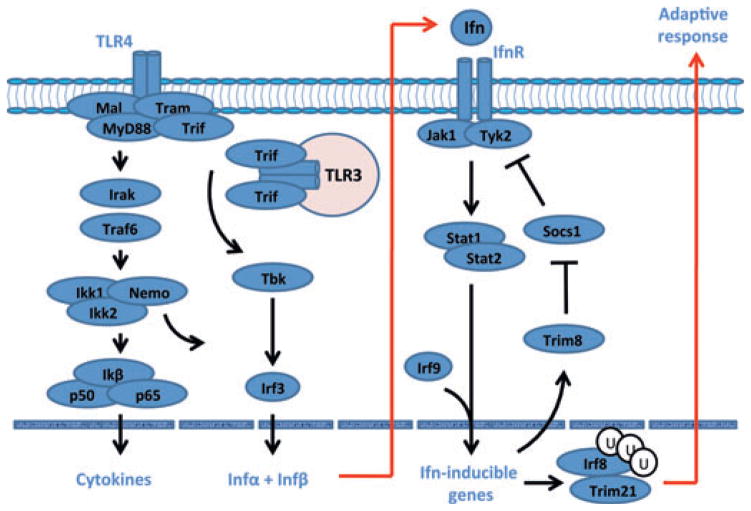
With the expansion of the number of genes being added to the Interferome database ([22]), we now know to examine even larger sets of ISGs that not only are activated or suppressed during development and onset of SS /SjS-like disease, but that actually function as both positive and negative-feedback regulatory molecules to ensure maximum host defences against microbial infections at all levels while preventing hyper-reactivity leading to unwanted host injury. One example of this type of network is depicted in Fig. 1, in which the feedback loop to suppress interferon signalling is a balance between Socs1 (suppressor of cytokine synthesis, factor 1) that negatively regulates interferon signalling by blocking phosphorylation of the signal transducer and activator of transcription factor 1 (Stat1) molecules and Trim8, which in turn negatively regulates Socs1. In SjS-like disease of C57BL /6.NOD-Aec1Aec2 mice, Trim8 is highly upregulated while Socs1 is downregulated, and this can lead to subsequent transcription of Trim21 (Ro52) and Irf8 that tandemly act as transcription factors to activate multiple initiators of adaptive immune responses, for example IL12 and CD40 [36, 37], two molecules shown to be highly upregulated in salivary glands of SS patients [38, 39]. At the same time, Trim21 has been shown to stabilize the function of Irf3 through blocking its interaction with Pin1, thereby also promoting Ifn signalling [40], and most likely adaptive immunity.
Figure 1.
An example of positive and negative regulation of the Ifn-signalling pathway. Activation of toll-like receptors, Tlr3 and Tlr4, results in the well-described cytokine storm of SS and SjS, including the synthesis of type 1 interferons. Signal transduction following activation of the Ifnα/β receptor involves the Jak /Tyk-Stat1 /Stat2 pathway. Irf9 acts as a transcription factor that is involved in the activation of Trim molecules, many of which are known to interact at multiple points of viral infections as antimicrobial factors. A major regulator of the Infαβ signalling pathway is Socs, a molecule that interferes with the activation loop of Jak kinase, thereby preventing phosphorylation of Stat molecules. Trim8 functions as an inhibitor of Socs1, promoting continuation of Ifn signalling. Similarly, Trim21 stabilizes the function of Irf3 through blocking its interaction with Pin1, thereby promoting Ifn signalling.
Does the IFN-signature define a biological process?
Prolonged activation of interferon signalling is critical in dealing with chronic infections, not only for activating an adaptive response, but also for orchestrating cooperative antimicrobial processes between ISG and autophagic factors that opsonize cytosolic pathogens or disrupt compartmentalized pathogens to facilitate efficient killing in autophagolysosomes [21]. Unique inducible molecular mechanisms have evolved in mammalian hosts to counter the many schemes used by microorganisms to gain a foothold in cells. Considering the multitude of functions that gene products associated with the various ISG families exhibit, one can hypothesize that global transcriptome data would appear to distinguish between the different interferon-induced cell-autonomous effector biological processes used to kill and /or clear specific pathogens. The first consideration in this analysis is whether the pathogen is compartmentalized, for example, in phagocytic vacuoles (or pathogen-containing inclusion bodies) or residing freely as a cytosolic pathogen. The subsequent obvious consideration is whether the make-up of interferon-signature profiles at the transcription level can identify first a specific molecular mechanism, then a specific pathogen. Currently, however, any conclusion must be tempered by the fact that the function(s) of many ISGs remain unknown.
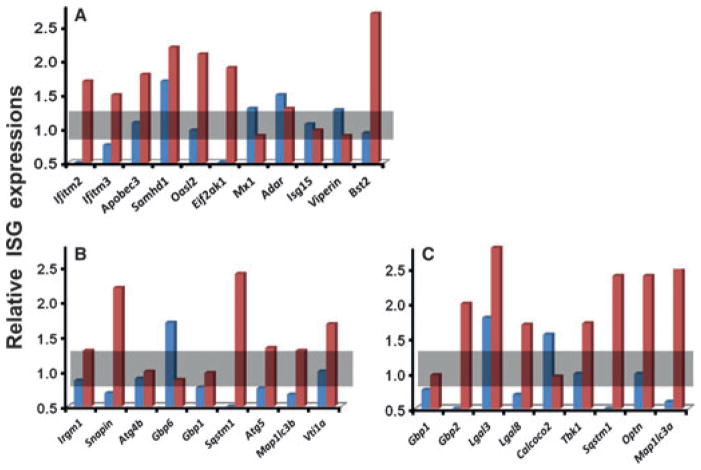
Assuming that a SS /SjS-like disease profile represents a specific host response, one or more of three basic host molecular mechanisms are likely to be identifiable: in the first mechanism, generally involving responses to viruses, genes from multiple ISG families, including Ifitm, Trim and /or Mx, are activated to block viral entry into the cytoplasm, while apolipoprotein-B mRNA-editing enzyme (encoded by Apobec3), SAM-domain- and HD-domain-containing protein (encoded by Samhd1), adenosine deaminase RNA-specific enzyme (encoded by Adar1 or Adarb1), and nitric oxide synthetase (encoded by Nos2) inhibit viral transcription and translation. In addition, the molecules viperin (encoded by Rsad2) and tetherin (encoded by Bst2) prevent viral assembly and release. In the second mechanism, generally involved in responses to bacteria present in phagosomes, molecular complexes of various gene products, for example Irgm1 (encoded by Irgm1), Snapin (encoded by Snapin), Sequestosome-1 (encoded by Sqstm1), Atg4b (encoded by Atg4b) and LC3 (encoded by Map1lc3a), plus members of the Gbp and Irg families, opsonize phagosomes that then interact with snare proteins to mediate fusion with lysosomes, resulting in the formation of autophagolysosomes with subsequent pathogen degradation. In a third mechanism, usually associated with responses to cytosolic bacterial pathogens, host defence begins with ubiquinylation of the pathogen followed by attraction of molecules such as sequestosome-1, optineurin (encoded by Optn), calcium-binding and coiled-coil domain-2 (or Ndp52 encoded by Calcoco2) and various galectins (encoded by Lgal genes). These complexes aggregate with Map1lc3a-encoded LC3-containing autophagophores that subsequently fuse with lysosomes again forming autophagolysosomes in which the pathogen is degraded. Unfortunately, profiles of the various genes being differentially expressed in the salivary glands of SjSS C57BL /6.NOD-Aec1Aec2 compared to SjS non-susceptible (SjSNS) C57BL /6J mice at 12 weeks of age (a time of active innate immunity) relative to 4 weeks of age (a time considered predisease), presented in Fig. 2, have yet to reveal any clear decisive patterns pointing to a single process, based on this set of genes. Nevertheless, the overall response profile, together with previously published results [26], continues to support a viral aetiology, but perhaps one that involves a cytoplasmic dsRNA virus.
Figure 2.
Relative expression of genes associated with cell-autonomous responses to infection. (A) Genes encoding factors known to function as cellular antiviral agents. (B) Genes encoding factors known to be involved with the formation of autophagolysosomes in compartmentalized microbial infections. (C) Genes encoding factors involved in the formation of autophagolysosomes in cytosolic microbial infections. Gene expressions observed in SjSNS C57BL /6J mice are indicated in blue, and gene expressions observed in SjSS C57BL /6.NOD-Aec1Aec2 mice are indicated in red.
Are the human and mouse IFN-signatures of SS /SjS diseases comparable?
One encouraging aspect of the transcriptome data sets that have been published for SS and SjS-like diseases, although still limited, is the fact that the genes used to establish the IFN-signature in both species overlap. First, the IFN-responsive genes that have thus far been reported as differentially expressed in human SS patients by several groups [15, 27–32]include IRF7, Mx1, GIP2, GIP3, OAS1, OAS2, PKR, IFI16, IFI27, IFI30, IFI35, IFI44, ISG20, ISG56K, IFIT1, IFIT2, IFIT4, IFITM1, IFITM3, IP10 /CXCL10 and STAT1a. More than half of these genes, or their equivalents, are also represented in the differentially expressed genes upregulated in the exocrine glands of the C57BL /6.NOD-Aec1Aec2 mice [26]. Second, the numbers of IFN-responsive genes upregulated and differentially expressed in SS patients again represented only a fraction of the total ISGs. Nevertheless, this set of differentially expressed genes must be considered an important subset of responsive genes that we would hypothesize point to specific etiopathological processes. Third, despite the fact that a significant number of SS patients, when tested for IFN levels during clinic visits, can exhibit an elevated levels of plasma IFN, the human transcriptome data indicate that few, if any, genes encoding an IFN per se were noted as being upregulated, compared to normal, healthy individuals [12–19]. In contrast, transcriptome data indicate many ISGs are upregulated. Thus, similar to the lack of differentially expressed IFN-encoding genes in the exocrine glands of SjSS C57BL /6.NOD-Aec1Aec2 mice [19], results published first by Hjelmervik et al. [27]using human minor salivary glands (huMSGs), then by Emamian et al. [15]using peripheral blood mononuclear cells, once again point to a lack of plasmacytoid dendritic cells (pDCs) in the starting tissue. We have interpreted these results to suggest that the earliest disease stage(s) actually occur(s) outside of the targeted exocrine glands where pDCs are likely to occur and in higher numbers, and /or IFN-α/β expression highly relevant for innate immunity is quickly replaced by IFN-γ expression strongly associated with adaptive immunity.
Another area of importance but less well-studied in SS is the role of complement. In two recent papers from Nordmark et al. [41, 42], the authors advance the hypothesis that complement may be a factor that prolongs the immune response in SS patients. In fact, genes encoding complement components are identified as being upregulated in transcriptome data set analyses. Of special interest relative to this point are the roles of complement in phagocytosis and cell lysis. The role of complement in SjS disease of C57BL /6.NOD-Aec1Aec2 mice has been clearly demonstrated in studies by Nguyen et al. [43, 44]in which blocking C’3 function suppressed development of disease. These data have been further supported by the fact that the genes encoding C’1q, C’3 and each of the components of the alternate pathway (but not the classical pathway) are strongly upregulated [45]. Studies of the relationship between complement and the IFN-signature certainly deserve further attention.
While it is natural to focus on the many similarities in the gene sets differentially expressed in human SS patients and SjSSC57BL /6.NOD-Aec1Aec2 mice, there are also important major differences. A few that stand out include expression profiles for Mx1, Irf8, Isg15 (or G1p2), Ifi202b–Ifi205 encoding the p200 molecules, and the three interferon-inducible genes, Ifi27, Ifi30 and Ifi44. One might expect a difference in Mx1 expression as laboratory mouse strains, especially C57BL /6J, carry a non-coding Mx1 gene [46], thus Mx1 is not upregulated in our SSS mice. On the other hand, the Irf8 gene, which encodes a factor that is involved in myeloid differentiation and Fas-mediated apoptosis, as well as B cell development and transcriptional regulation of germinal centre formation [37], deserves special attention and one of the highest upregulated expressions in our SjSS mice. Our earlier studies postulated that myeloid cells enter the exocrine glands during the early innate response (8-12 weeks of age) in response to Fas-FasL-mediated apoptosis of acinar tissue, while B cells enter the salivary glands transiently during the adaptive immune phase (after 16 weeks of age). Interestingly, the temporal expression profile of Irf8 showed a bimodal profile, in line with this hypothesis. The inability to detect an upregulated expression of IRF8 in SS patients is an interesting aspect to examine further, as binding of the transcriptional factor PU.1 to IRF8 leads to upregulation of OAS1 and /or OAS2, two molecules that can bind and degrade dsRNA viral RNA [47], and are highly upregulated in SS patients. In contrast, the p200 molecules, encoded by the Ifi200 family of genes, are known to sense cytoplasmic DNA, leading to the formation and activation of inflammasomes with subsequent production of antinuclear antibodies [48]. Although there was an Ifi202b upregulated gene expression in the exocrine glands of C57BL /6.NOD-Aec1Aec2 mice, we have not found evidence for the activation of inflammasomes in these mice, in contrast to their comparative SjSNS C57BL /6J partners (ABP, unpublished data). Lastly, whereas IFI27, IFI30 and IFI44 have been consistently found to be upregulated in SS patients [15, 27, 30, 32], these three Ifi genes with distinct functions were not found to be differentially expressed in the exocrine glands of C57BL /6.NOD-Aec1Aec2 mice. Considering IFI44 is associated with hepatitis c virus (HCV) and respiratory syncytial virus (RSV) infections, we would contend that this difference between humans and mice lies in the fact that the underlying aetiological agents of SS and SjS-like diseases are different and such differences indicate different environmental triggers in the two species. In contrast, common genes probably indicate activation of similar downstream immunopathological processes. Interestingly, IFI44L was identified as a marker gene in RA [34]. Thus, both similarities and differences in the interferon-signatures are critical to understanding SS.
Is the ‘interferon-signature’ sculpted by regulators of the interferon signalling pathways?
As suggested earlier, and depicted in Fig. 1, the IFN-signalling pathway, after activation by PRRs such as Tlr3, Tlr4 and Mda5, is regulated by at least one negative-feedback mechanism, specifically, the balance between the Trim8 and Socs1 molecules. In recent years, it has become generally accepted that, during normal homoeostasis, the clearance of apoptotic cells by macrophages, DCs and neighbouring tissue generally initiates an immunosuppressive and anti-inflammatory activity to regulate or suppress inflammation, whereas defective clearance of apoptotic cells can activate inflammatory responses and autoimmunity [49–52]. Clearance of apoptotic cells is a highly complex and sophisticated biological process involving multiple molecular sequences. Apoptotic cellular debris tends to express phosphatidylserine (PS). PS can be recognized directly by the PS receptor (PtdSerR) plus several additional receptors present on phagocytic cells via bridging molecules. These latter include the following: the TAM receptors (Tyro, Axl and Mer) via Gas6 or Protein S, the integrin molecule αvβ5 via thrombospondin-1, the integrin molecule αvβ3 via the Megf8 molecule and even the interferon receptor IfnαR via Ifnα. In addition, phagocytic cells can bind apoptotic debris via CD31–CD31, CD36-oxidized low-density lipoprotein (OxLDL) and ICAM3–ICAM3 interactions. Our transcriptome data from C57BL /6.NOD-Aec1Aec2 mice indicate that the genes for each of these processes, except PtdSerR, are highly upregulated (unpublished data). The intrinsic inhibition of inflammation following phagocytosis of apoptotic cells is mediated, in large part, by these TAM receptor protein tyrosine kinases (RTKs) present on phagocytic and antigen-presenting cells by interacting with the IfnαRs, and represents a critical mechanism for regulating inflammation and self-recognition, primarily during the innate immune response, but also in adaptive immunity. The effectors of this regulation are molecules belonging to the SOCS family of proteins, especially Socs1 and Socs3. Deficiencies in TAM RTK signalling or synthesis of the Socs molecules has been shown to lead to autoimmunity [52]. Not surprising, then, that transcriptome data from the salivary glands of SjSS C57BL /6.NOD-Aec1Aec2 mice indicate that neither Socs1 nor Socs3 expressions are upregulated (ABP, unpublished data), and this fact clearly plays into modelling the innate immune response promoted by ISGs, discussed below. Interestingly, silencing the throm-bospondin-1 gene (Thbs1) in C57BL /6J mice results in a SjS-like disease in this otherwise SjSNS mouse [53]. Activation of the αvβ5-thrombospondin-1 results in downstream signalling of the Ras-Raf-Erk and Rac1-CDC42 pathways, critical for cell migration and phagocytosis.
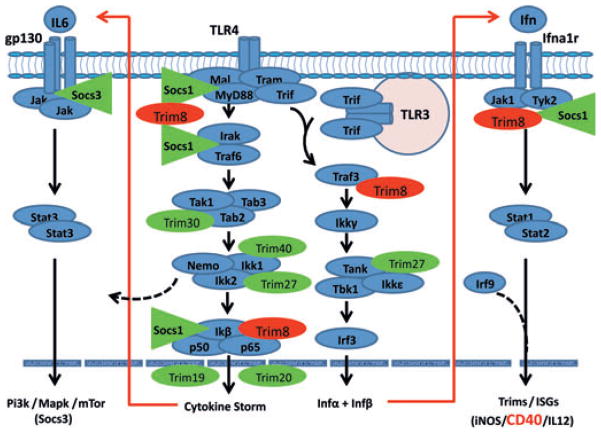
To determine whether the chronic activation of the IFN-signalling pathway results from signals associated with ISGs, initial studies were carried out in SjSS C57BL /6.NOD-Aec1Aec2 mice [23]looking at gene expressions of various Trim genes reportedly regulating the TLR-and IFN-signalling pathways during viral infections. Results of this analysis, summarized in Fig. 3, revealed that Trim8 (encoding Trim8), whose gene product functions to suppress the action of the Socs molecules as depicted in Fig. 1, is strongly upregulated, while the expression of genes encoding Socs1 and Socs3 is suppressed. This would prevent Socs1 from interfering with phosphorylation of appropriate signalling pathway molecules, thus permitting persistent signal transduction and synthesis of ISGs. As both TRIM and SOCS molecules are progressively becoming realized as critical factors in viral infections, and innate immune responses in general [recently reviewed in [54–57], their expression profiles during infections clearly help define the interferon-signature, and this is true for both SS and SjS-like diseases. Consistent with this, in SjSS mice, the genes encoding four molecules (Trim21, Trim23, Trim25 and Trim56) whose functions are to upregulate the Tlr3, Tlr4 and Mda5 pathways at different signalling steps, are each upregulated, while the genes of three additional Trim molecules (Trim27, Trim30 and Trim40) whose functions are to downregulate the signal transductions of Tlr4, Tlr3 and Mda5 at different signalling points, were downregulated. Considered as a whole, this profile would be expected to promote upregulation of pathways leading to strong transcription of pro-inflammatory cytokines, interferons and molecules that are known activators of adaptive responses (e.g. IL6, IL12p40, Rantes, CD40, CD80 and CD56), and genes that provide a CD40-signature are differentially expressed in C57BL /6.NOD-Aec1Aec2 mice (ABP, unpublished data). This precise gene expression pattern is reversed in SjSNS C57BL /6J mouse. Not surprising, then, that the innate phase of SS /SjS-like diseases transitions to a chronic adaptive inflammatory immune phase that, in genetically predisposed individuals, can progress to autoimmunity. However, whether these data result from the environmental trigger or the nature of an intrinsic host response is still unresolved. This question requires further study.
Figure 3.
Scheme depicting the interactive roles of Trim and Socs molecules regulating the innate response in SjSS C57BL /6.NOD-Aec1Aec2 mice. Genes encoding molecules that function to inhibit the innate response (Socs1, Trim27, Trim30 and Trim40) are shown to be downregulated (green), while genes encoding factors that function to generally activate innate responses are shown to be upregulated (red). This scheme is consistent with the strong interferon-signature observed in SS /SjS-like diseases.
Perspective
Despite efforts to define an environmental, genetic and /or immunopathological basis for SS, the underlying aetiology remains poorly defined with little consensus in the field, in part, due to the fact that patients are currently diagnosed only after onset of overt clinical disease. Furthermore, patients present with multiple disease phenotypes, and this important fact clearly suggests a need to consider each phenotype separately when analysing large data sets such as global transcriptome or proteome results. In this regard, transcriptome studies that are beginning to define a ‘disease-specific interferon-signature profile’ appear to offer a viable approach, if not an absolute answer, at least a basis for developing disease models for testing. To this end, we hypothesize that for the C57BL /6.NOD-Aec1Aec2 mouse model virtually all data are beginning to point to a cytoplasmic RNA viral aetiology and a dysregulated innate immune response giving rise to an autoimmune inflammatory pathology. Support for these concepts lie in the observations that: (1) the three activated PRRs in our model (Tlr3, Tlr4 and Mda-5) are receptors involved in activating the interferon-based innate response against dsRNA viruses. No other PRR (or class of PRRs) has been identified to be activated, including Nod, Nalp, Ipaf, Naip, Rage, Rxfp1 and Dai receptors (unpublished data) (2) the genes associated with cell-autonomous immune effector mechanisms exhibiting upregulated expressions generally defined an anticytoplasmic viral response and (3) the expression of specific Trim and Socs molecules known to regulate the IFN pathway remains in a balance favourable for activating innate immunity, not in favour of suppression. It should be reiterated, of course, that the aetiological agents underlying SS and SjS-like diseases are probably different in the two species, despite the fact that both exhibit upregulation of TLR3 and TLR4. Assuming that the concomitant upregulation of Mda-5 in C57BL /6.NOD-Aec1Aec2 mice is an important clue to a possible aetiological agent, one might surmise the involvement of picorna-virus, murine norovirus, murine hepatitis virus or reovirus. In any event, the downstream outcome is the activation and dysregulation of an immune response strongly mediated by the interferons and a wide range of regulatory molecules of the Ifn pathways. Taken as a whole, these overall profiles indicate upregulation of pathways leading to strong transcription of pro-inflammatory cytokines, interferons and molecules that are known activators of adaptive responses (e.g. IL6, IL12p40, Rantes, CD40, CD80 and CD56). Not surprising, then, that the innate phase of SS transitions to an adaptive immune phase; unfortunately, in genetically predisposed individuals this adaptive phase remains dysregulated. Perpetuation of disease in SS patients has recently been discussed by Voulgarelis and Tzioufas [58].
Overall, results of gene profiling derived from transcriptome data provide a strong basis for both modelling and investigating molecular events and their biological processes underlying SS and SjS disease. At the same time, the unique temporal changes exhibited by IFN-responsive genes and genes involved in molecular signalling and biological processes reveal differential expressions of select sets of genes, indicating that detection of differentially expressed genes that are crucial to specific disease-associated molecular processes and those genes that are merely bystanders remains complicated. Any measurement at one time point of disease development, a serious weakness of applying transcriptome data analysis to human autoimmune diseases, no doubt represents an incomplete picture, yet still defines critical elements of the etiopathological processes. Studies using the C57BL /6.NOD-Aec1Aec2 mouse model of primary SS document the fact that there are multiple ISGs that are never differentially expressed and that this lack of gene expression is not because of disease phase-restricted expression, thus invoking a need to consider both up-regulated and downregulated genes in defining an interferon-signature. With respect to the transcriptome studies thus far completed, the dichotomy between gene expressions and levels of plasma interferon raises questions as to how best to define a biological theme such as the IFN-signature for SS. Is it merely that an IFN-signature should be defined as a high number of IFN-related genes found to be upregulated and that this fact may eventually be extrapolated to a potential underlying cause of autoimmunity, or does the pattern of IFN-responsive gene activation indicate a need to sort out the relationship between the activated and non-activated IFN-related genes to piece together how an environmental trigger can circumvent all the naturally built-in mechanisms (perhaps even distinct in different tissues) that are meant to prevent autoimmunity? On the other hand, because the IFNs represent a ubiquitous cytokine involved in most, if not all, innate and adaptive immune responses, will each IFN-signature be specifically reflective of a particular immune response, and thereby be a diagnostic marker?
Acknowledgments
This study was supported financially in part by PHS grants DE014344 (ABP), AI081952 (ABP, CQN) and DE018958 (CQN) from the National Institutes of Health (NIH), a research grant from the Sjögren’s Syndrome Foundation (CQN) and funds from the University of Florida’s Center for Orphaned Autoimmune Disorders.
Footnotes
The authors state that they have no financial or other conflicts of interest with the content of this study.
References
- 1.Jonsson R, Haga HJ, Gordon TP. Current concepts on diagnosis, autoantibodies and therapy in Sjogren’s syndrome. Scand J Rheumatol. 2000;29:341–8. doi: 10.1080/030097400447525. [DOI] [PubMed] [Google Scholar]
- 2.Hansen A, Lipsky PE, Dorner T. New concepts in the pathogenesis of Sjogren syndrome: many questions, fewer answers. Curr Opin Rheumatol. 2003;15:563–70. doi: 10.1097/00002281-200309000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Fox RI. Sjogren’s syndrome. Lancet. 2005;366:321–31. doi: 10.1016/S0140-6736(05)66990-5. [DOI] [PubMed] [Google Scholar]
- 4.Manthorpe R, Bredberg A, Henriksson G, Larsson A. Progress and regression within primary Sjogren’s syndrome. Scand J Rheumatol. 2006;35:1–6. doi: 10.1080/03009740500537945. [DOI] [PubMed] [Google Scholar]
- 5.Fox PC, Bowman SJ, Segal B, et al. Oral involvement in primary Sjogren syndrome. J Am Dent Assoc. 2008;139:1592–601. doi: 10.14219/jada.archive.2008.0101. [DOI] [PubMed] [Google Scholar]
- 6.Delalande S, de Seze J, Fauchais AL, et al. Neurologic manifestations in primary Sjogren syndrome: a study of 82 patients. Medicine (Baltimore) 2004;83:280–91. doi: 10.1097/01.md.0000141099.53742.16. [DOI] [PubMed] [Google Scholar]
- 7.Malinow KL, Molina R, Gordon B, Selnes OA, Provost TT, Alexander EL. Neuropsychiatric dysfunction in primary Sjogren’s syndrome. Ann Intern Med. 1985;103:344–50. doi: 10.7326/0003-4819-103-3-344. [DOI] [PubMed] [Google Scholar]
- 8.Belin C, Moroni C, Caillat-Vigneron N, et al. Central nervous system involvement in Sjogren’s syndrome: evidence from neuropsychological testing and HMPAO-SPECT. Ann Med Interne (Paris) 1999;150:598–604. [PubMed] [Google Scholar]
- 9.Valtysdottir ST, Gudbjornsson B, Lindqvist U, Hallgren R, Hetta J. Anxiety and depression in patients with primary Sjogren’s syndrome. J Rheumatol. 2000;27:165–9. [PubMed] [Google Scholar]
- 10.Theander E, Manthorpe R, Jacobsson LT. Mortality and causes of death in primary Sjogren’s syndrome: a prospective cohort study. Arthritis Rheum. 2004;50:1262–9. doi: 10.1002/art.20176. [DOI] [PubMed] [Google Scholar]
- 11.Voulgarelis M, Moutsopoulos HM. Lymphoproliferation in autoimmunity and Sjogren’s syndrome. Curr Rheumatol Rep. 2003;5:317–23. doi: 10.1007/s11926-003-0011-y. [DOI] [PubMed] [Google Scholar]
- 12.Ansell P, Simpson J, Lightfoot T, et al. Non-Hodgkin lymphoma and autoimmunity: does gender matter? Int J Cancer. 2011;129:460–6. doi: 10.1002/ijc.25680. [DOI] [PubMed] [Google Scholar]
- 13.Bave U, Nordmark G, Lovgren T, et al. Activation of the type I interferon system in primary Sjogren’s syndrome: a possible etiopathogenic mechanism. Arthritis Rheum. 2005;52:1185–95. doi: 10.1002/art.20998. [DOI] [PubMed] [Google Scholar]
- 14.Wildenberg ME, van Helden-Meeuwsen CG, van de Merwe JP, Drexhage HA, Versnel MA. Systemic increase in type I interferon activity in Sjogren’s syndrome: a putative role for plasmacytoid dendritic cells. Eur J Immunol. 2008;38:2024–33. doi: 10.1002/eji.200738008. [DOI] [PubMed] [Google Scholar]
- 15.Emamian ES, Leon JM, Lessard CJ, et al. Peripheral blood gene expression profiling in Sjogren’s syndrome. Genes Immun. 2009;10:285–96. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L, Zhang Z, Yu C, Tu L, Zhong L, Yang C. Association between IFN-alpha and primary Sjogren’s syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:e12–8. doi: 10.1016/j.tripleo.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Vakaloglou KM, Mavragani CP. Activation of the type I interferon pathway in primary Sjogren’s syndrome: an update. Curr Opin Rheumatol. 2011;23:459–64. doi: 10.1097/BOR.0b013e328349fd30. [DOI] [PubMed] [Google Scholar]
- 18.Mavragani CP, Crow MK. Activation of the type I interferon pathway in primary Sjogren’s syndrome. J Autoimmun. 2010;35:225–31. doi: 10.1016/j.jaut.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Cha S, Brayer J, Gao J, et al. A dual role for interferon-gamma in the pathogenesis of Sjogren’s syndrome-like autoimmune exocrinopathy in the nonobese diabetic mouse. Scand J Immunol. 2004;60:552–65. doi: 10.1111/j.0300-9475.2004.01508.x. [DOI] [PubMed] [Google Scholar]
- 20.Cha S, van Blockland SC, Versnel MA, et al. Abnormal organogenesis in salivary gland development may initiate adult onset of auto-immune exocrinopathy. Exp Clin Immunogenet. 2001;18:143–60. doi: 10.1159/000049194. [DOI] [PubMed] [Google Scholar]
- 21.Macmicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 2012;12:367–82. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertzog P, Forster S, Samarajiwa S. Systems biology of interferon responses. J Interferon Cytokine Res. 2011;31:5–11. doi: 10.1089/jir.2010.0126. [DOI] [PubMed] [Google Scholar]
- 23.Killedar SY, Eckenrode SE, McIndoe RA, et al. Early pathogenic events associated with Sjogren’s syndrome (SjS)-like disease of the nod mouse using microarray analysis. Lab Invest. 2006;86:1243–60. doi: 10.1038/labinvest.3700487. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami A, Nakashima K, Tamai M, et al. Toll-like receptor in salivary glands from patients with Sjogren’s syndrome: functional analysis by human salivary gland cell line. J Rheumatol. 2007;34:1019–26. [PubMed] [Google Scholar]
- 25.Spachidou MP, Bourazopoulou E, Maratheftis CI, et al. Expression of functional Toll-like receptors by salivary gland epithelial cells: increased mRNA expression in cells derived from patients with primary Sjogren’s syndrome. Clin Exp Immunol. 2007;147:497–503. doi: 10.1111/j.1365-2249.2006.03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peck AB, Nguyen CQ, Sharma A, McIndoe RA, She JX. The interferon-signature of Sjögren’s syndrome: what does it say about the etiopathology of autoimmunity. J Clin Rheum & Musculoskeletal Med. 2011;1:1–17. [Google Scholar]
- 27.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjogren’s syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–44. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 28.Gottenberg JE, Cagnard N, Lucchesi C, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjogren’s syndrome. Proc Nat Acad Sci USA. 2006;103:2770–5. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez P, Anaya JM, Aguilera S, et al. Gene expression and chromosomal location for susceptibility to Sjogren’s syndrome. J Autoimmun. 2009;33:99–108. doi: 10.1016/j.jaut.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Kimoto O, Sawada J, Shimoyama K, et al. Activation of the interferon pathway in peripheral blood of patients with Sjogren’s syndrome. J Rheumatol. 2011;38:310–6. doi: 10.3899/jrheum.100486. [DOI] [PubMed] [Google Scholar]
- 31.Devauchelle-Pensec V, Cagnard N, Pers JO, Youinou P, Saraux A, Chiocchia G. Gene expression profile in the salivary glands of primary Sjogren’s syndrome patients before and after treatment with rituximab. Arthritis Rheum. 2010;62:2262–71. doi: 10.1002/art.27509. [DOI] [PubMed] [Google Scholar]
- 32.Wakamatsu E, Nakamura Y, Matsumoto I, et al. DNA microarray analysis of labial salivary glands of patients with Sjogren’s syndrome. Ann Rheum Dis. 2007;66:844–5. doi: 10.1136/ard.2006.063370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obermoser G, Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19:1012–9. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raterman HG, Vosslamber S, de Ridder S, et al. Interferon type I signature may predict non response upon rituximab in rheumatoid arthritis patients. Arthritis Res Ther. 2012;14:R95. doi: 10.1186/ar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha S, Nagashima H, Brown VB, Peck AB, Humphreys-Beher MG. Two NOD Idd-associated intervals contribute synergistically to the development of autoimmune exocrinopathy (Sjogren’s syndrome) on a healthy murine background. Arthritis Rheum. 2002;46:1390–8. doi: 10.1002/art.10258. [DOI] [PubMed] [Google Scholar]
- 36.Toniato E, Chen XP, Losman J, Flati V, Donahue L, Rothman P. TRIM8 /GERP RING finger protein interacts with SOCS-1. J Biol Chem. 2002;277:37315–22. doi: 10.1074/jbc.M205900200. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Lee CH, Qi C, et al. IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112:4028–38. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohlsson M, Szodoray P, Loro LL, Johannessen AC, Jonsson R. CD40, CD154, Bax and Bcl-2 expression in Sjogren’s syndrome salivary glands: a putative anti-apoptotic role during its effector phases. Scand J Immunol. 2002;56:561–71. doi: 10.1046/j.1365-3083.2002.01168.x. [DOI] [PubMed] [Google Scholar]
- 39.Dimitriou ID, Kapsogeorgou EK, Moutsopoulos HM, Manoussakis MN. CD40 on salivary gland epithelial cells: high constitutive expression by cultured cells from Sjogren’s syndrome patients indicating their intrinsic activation. Clin Exp Immunol. 2002;127:386–92. doi: 10.1046/j.1365-2249.2002.01752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang K, Shi HX, Liu XY, et al. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J Immunol. 2009;182:3782–92. doi: 10.4049/jimmunol.0803126. [DOI] [PubMed] [Google Scholar]
- 41.Nordmark G, Alm GV, Ronnblom L. Mechanisms of disease: primary Sjogren’s syndrome and the type I interferon system. Nat Clin Pract Rheumatol. 2006;2:262–9. doi: 10.1038/ncprheum0173. [DOI] [PubMed] [Google Scholar]
- 42.Nordmark G, Eloranta ML, Ronnblom L. Primary Sjogren’s syndrome and the type I interferon system. Curr Pharm Biotechnol. 2012 Jan 2; doi: 10.2174/138920112802273290. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen CQ, Kim H, Cornelius JG, Peck AB. Development of Sjogren’s syndrome in nonobese diabetic-derived autoimmune-prone C57BL /6. NOD-Aec1Aec2 mice is dependent on complement component-3. J Immunol. 2007;179:2318–29. doi: 10.4049/jimmunol.179.4.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen C, Cornelius J, Singson E, Killedar S, Cha S, Peck AB. Role of complement and B lymphocytes in Sjogren’s syndrome-like autoimmune exocrinopathy of NOD. B10-H2b mice. Mol Immunol. 2006;43:1332–9. doi: 10.1016/j.molimm.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen CQ, Sharma A, Lee BH, She JX, McIndoe RA, Peck AB. Differential gene expression in the salivary gland during development and onset of xerostomia in Sjogren’s syndrome-like disease of the C57BL /6. NOD-Aec1Aec2 mouse. Arthritis Res Ther. 2009;11:R56. doi: 10.1186/ar2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staeheli P, Sutcliffe JG. Identification of a second interferon-regulated murine Mx gene. Mol Cell Biol. 1988;8:4524–8. doi: 10.1128/mcb.8.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogozin IB, Aravind L, Koonin EV. Differential action of natural selection on the N and C-terminal domains of 2′-5′oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J Mol Biol. 2003;326:1449–61. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- 48.Choubey D, Duan X, Dickerson E, et al. Interferon-inducible p200-family proteins as novel sensors of cytoplasmic DNA: role in inflammation and autoimmunity. J Interferon Cytokine Res. 2010;30:371–80. doi: 10.1089/jir.2009.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2:965–75. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 50.Wu AJ, Chen ZJ, Tsokos M, O’Connell BC, Ambudkar IS, Baum BJ. Interferon-gamma induced cell death in a cultured human salivary gland cell line. J Cell Physiol. 1996;167:297–304. doi: 10.1002/(SICI)1097-4652(199605)167:2<297::AID-JCP14>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 51.Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8:327–36. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothlin CV, Lemke G. TAM receptor signaling and autoimmune disease. Curr Opin Immunol. 2010;22:740–6. doi: 10.1016/j.coi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turpie B, Yoshimura T, Gulati A, Rios JD, Dartt DA, Masli S. Sjogren’s syndrome-like ocular surface disease in thrombospondin-1 deficient mice. Am J Pathol. 2009;175:1136–47. doi: 10.2353/ajpath.2009.081058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jefferies C, Wynne C, Higgs R. Antiviral TRIMs: friend or foe in autoimmune and autoinflammatory disease? Nat Rev Immunol. 2011;11:617–25. doi: 10.1038/nri3043. [DOI] [PubMed] [Google Scholar]
- 55.McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 56.Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3:513–27. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dimitriou ID, Clemenza L, Scotter AJ, Chen G, Guerra FM, Rottapel R. Putting out the fire: coordinated suppression of the innate and adaptive immune systems by SOCS1 and SOCS3 proteins. Immunol Rev. 2008;224:265–83. doi: 10.1111/j.1600-065X.2008.00659.x. [DOI] [PubMed] [Google Scholar]
- 58.Voulgarelis M, Tzioufas AG. Pathogenetic mechanisms in the initiation and perpetuation of Sjogren’s syndrome. Nat Rev Rheumatol. 2010;6:529–37. doi: 10.1038/nrrheum.2010.118. [DOI] [PubMed] [Google Scholar]