SUMMARY
Excess dormant origins bound by the minichromosome maintenance (MCM) replicative helicase complex play a critical role in preventing replication stress, chromosome instability and tumorigenesis. In response to DNA damage, replicating cells must coordinate DNA repair and dormant origin firing to ensure complete and timely replication of the genome; how cells regulate this process remains elusive. Herein, we identify a member of the Fanconi Anemia (FA) DNA repair pathway, FANCI, as a key effector of dormant origin firing in response to replication stress. Cells lacking FANCI have reduced number of origins, increased inter-origin distances and slowed proliferation rates. Intriguingly, ATR-mediated FANCI phosphorylation inhibits dormant origin firing while promoting replication fork restart/DNA repair. Using super-resolution microscopy, we show that FANCI co-localizes with MCM-bound chromatin in response to replication stress. These data reveal a unique role for FANCI as a modulator of dormant origin firing and links timely genome replication to DNA repair.
INTRODUCTION
In mammalian cells, chromosomes are replicated from multiple origins that initiate throughout the S-phase of the cell cycle (Blow et al., 2011). The regulation of DNA replication occurs in two phases: origin licensing in the G1-phase and origin firing during S-phase. Replication licensing starts as cells exit mitosis and involves the recruitment of the minichromosome maintenance proteins (MCM2-7) (Bell and Botchan, 2013) to replication origins by ORC (origin recognition complex), Cdc6 and Cdt1 proteins, to assemble the pre-replicative complex (pre-RCs) (Blow and Dutta, 2005; Diffley, 2004; O'Donnell et al., 2013). Firing of replication origins is triggered through the activation of the MCM2-7 complex by two conserved protein kinases, the Dbf4-dependent Cdc7 kinase (DDK) and the cyclin-dependent kinase (CDK).
During DNA replication, the presence of endogenous or exogenous sources of replication stress causes individual replication forks to slow or stall. How do cells overcome perturbed replication forks to finish genome replication in a timely manner? A critical response to overcome this type of replication stress is to fire additional licensed origins to complete replication within the intervening regions of the stalled forks; these backup replication origins are referred to as “dormant origins” (McIntosh and Blow, 2012). The MCM2-7 complex are loaded onto DNA in ~20-fold excess over the number of active replication origins and ORCs in the cell, presumably at dormant origins (Lei et al., 1996; Rowles et al., 1996). Studies by Blow and others showed that mild depletion of MCM5 (a subunit of MCM2-7) reduced overall chromatin-bound MCM proteins but did not affect normal rates of DNA synthesis in human cells. However, when treated with inhibitors that cause mild replication stress (stress that doesn’t activate replication checkpoint), MCM5-depleted cells experienced reduced levels of DNA synthesis and viability due to the lack of dormant origin firing (Ge and Blow, 2010; Ge et al., 2007; Ibarra et al., 2008). Furthermore, mice expressing reduced levels of MCM2-7 have fewer dormant origins, are genomically unstable and are cancer-prone (Alver et al., 2014; Kawabata et al., 2011; Kunnev et al., 2010; Pruitt et al., 2007; Shima et al., 2007). Interestingly, in precancerous and cancer cells, the aberrant expression of oncogenes significantly decreases cellular nucleotide levels (Bester et al., 2011); this nucleotide deficiency leads to reduced replication fork speeds and more frequent fork stalling, placing a higher requirement on dormant origin firing to alleviate replication stress in cancer cells. These studies demonstrate that dormant origin firing is a physiologically important mechanism to maintain normal DNA replication rates in order to prevent genomic instability and tumorigenesis. The signaling network that regulates the firing of dormant origins upon replication stress is currently unknown.
Fanconi anemia (FA) is a human chromosome instability syndrome characterized by progressive bone marrow failure and cancer predisposition (D'Andrea, 2010; Moldovan and D'Andrea, 2009). FA is a genetically heterogeneous disorder, caused by mutations in one of at least 16 genes. The FA gene products all function in a common FA genome stability pathway critical for interstrand crosslink (ICL) repair (Kottemann and Smogorzewska, 2013; Moldovan and D'Andrea, 2009; Wang, 2007). A large set of the FA proteins form a multi-subunit nuclear ubiquitin ligase complex required to monoubiquitinate and activate two downstream FA components, FANCD2 (Garcia-Higuera et al., 2001), and its interacting partner, FANCI (Sims et al., 2007a; Smogorzewska et al., 2007). Monoubiquitination of FANCI-FANCD2 is reversed by the deubiquitinating enzyme (DUB) USP1 (Nijman et al., 2005; Sims et al., 2007a). The role of the FA pathway in DNA repair has been intensely studied and a unifying model has emerged describing how FA proteins coordinate the convergence of multiple DNA repair pathways, including homologous recombination (HR) and translesion synthesis (TLS), for the repair of ICLs (Knipscheer et al., 2009; Kottemann and Smogorzewska, 2013; Räschle et al., 2008). Cells derived from either FA patients or USP1 knockout cells are hypersensitive to the ICL-inducing agent mitomycin C (MMC), but they also exhibit chromosome aberrations, including gaps and breaks, a sign of DNA damage that is reminiscent of incomplete DNA replication (Auerbach and Wolman, 1976; Kim et al., 2009).
The FA pathway is strongly activated by hydroxyurea (HU) (Taniguchi et al., 2002), which unlike ICL-inducing agents (such as MMC or reactive aldehydes) (Garaycoechea et al., 2012), does not elicit DNA lesions that require removal, but induces replication fork slowing or stalling through the depletion of the nucleotide pool (Petermann et al., 2010). Additionally, recent studies found that FANCD2 and FANCI associated with the replisome in response to replication fork arrest and that the FA pathway is critical for protecting stalled replication forks from nuclease-mediated degradation of nascent strands in the presence of high dose (4 mM) HU treatment (Lossaint et al., 2013; Schlacher et al., 2012). Unlike high dose HU, however, low dose (≤200 µM) treatment remains permissive for DNA replication, whereby slower-moving or stalled forks can be compensated for or rescued by the activation of adjacent dormant origins (Ge et al., 2007). Despite our greater understanding of the role of FA in ICL repair, how the FA pathway protects against additional forms of replication stress is not well understood.
Herein, we report that FANCI plays a critical role in the regulation of dormant origin firing upon replication stress and this occurs through an FA pathway-independent mechanism. We found that ATR-mediated phosphorylation of FANCI is a negative regulator of dormant origin firing despite still being able to promote replication restart/DNA repair, which is FA pathway-dependent. Furthermore, cellular resistance to ICLs requires both FA pathway-dependent and – independent functions of FANCI. Finally, we employed super-resolution (SR) microscopy to show that the phosphorylation status of FANCI may be key in localizing FANCI to the proper chromatin-binding complexes for its control of dormant origin firing.
Results
FANCI is enriched at sites of replication origins
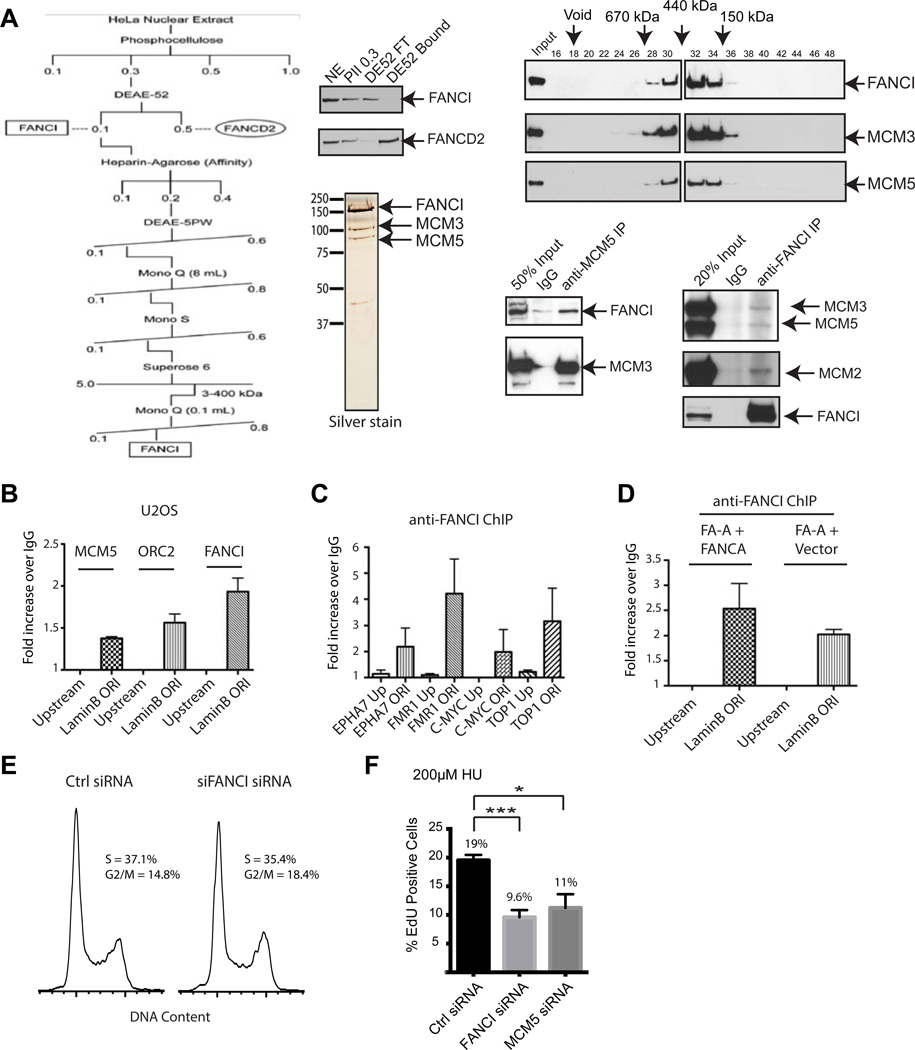
Upon activation of the FA pathway, the level of monoubiquitinated FANCI is low in comparison to FANCD2 (Sims et al., 2007a). To determine whether this unmodified pool of FANCI interacts and functions with a separate protein complex, we purified native FANCI from the soluble nuclear fraction of HeLa extracts using conventional purification techniques as previously described (Sims et al., 2007b; Sims et al., 2011), and analyzed the FANCI-associated proteins by mass spectrometry (MS) (Figure 1A). We found that the soluble, unmodified FANCI associated with MCM3 and MCM5, two subunits of the MCM2-7 replicative helicase complex, and confirmed this interaction by co-IP experiments in U2OS cells (Figure 1A). Indeed, a recent study showed that both FANCI and FANCD2 interact with MCM2-7 at the replisome (Lossaint et al., 2013). In addition to its essential role as a replicative helicase of actively elongating replication forks, MCM2-7 also plays a critical genome-stabilizing role in dormant origin firing under replication stress (Ge et al., 2007; Kawabata et al., 2011). Using chromatin immunoprecipitation (ChIP), we showed that FANCI can be found enriched at sites of well-characterized replication origins (Salsi et al., 2009), but not at DNA regions that are upstream of these origins. FANCI was also enriched at the Lamin B origin in FANCA-deficient cells, suggesting that the complete FA core complex is not required for the targeting of FANCI to replication origins (Figure 1B–D).
Figure 1. FANCI interacts with the MCM replicative helicase complex.
(A) Purification scheme of the FANCI complex from HeLa nuclear extracts. After DEAE-cellulose chromatography, FANCI and FANCD2 are separated. The polypeptides that co-eluted with FANCI at the last chromatographic step were visualized by silver stain and identified by MS. Gel filtration fractions from Superose 6 were probed with antibodies against FANCI, MCM3 and MCM5. Co-IP was done using the indicated antibodies from U2OS nuclear extracts. (B-D) Enrichment of DNA obtained with antibodies against FANCI, MCM5, or ORC2 as determined by ChIP analysis using chromatin prepared from HeLa or FA-A (FANCA-deficient patient) cells corrected with FANCA WT or empty vector. Quantitative PCR analysis was performed using primer sequences for validated origin sites or their upstream counterpart as indicated (Salsi et al., 2009). Results are displayed as fold increase as compared to IgG Ctrl (anti-GFP). In D, differences in fold increase between FA-A + FANCA WT and FA-A + Vector for LaminB Ori is not significant (p>0.05). (E) FACS analysis showing normal cell cycle distribution between Ctrl and FANCI siRNA knockdown U2OS cells. (F) U2OS cells were transfected with the indicated siRNAs and treated with 200 µM HU for 4 h followed by a 30 min EdU pulse. DNA and EdU content were analyzed by FACS and displayed as % EdU-positive cells. For all of the figures, the asterisks represent either *p<0.05, **p<0.01, ***p<0.001, or ****p<0.0001.
Next, we tested whether DNA synthesis levels were affected in FANCI knockdown U2OS cells by measuring nucleotide analog (EdU) incorporation by FACS analysis. In the absence of HU, the cell cycle profile and replication rates of FANCI-depleted cells were similar to Ctrl knockdown cells (Figure 1E and Figure S1A–B). However, after treatment with low dose HU for 4 hours, FANCI knockdown resulted in fewer EdU-positive cells, suggesting that FANCI is required to maintain normal DNA replication during conditions of replication stress (Figure 1F). These results are in agreement with previous studies demonstrating the role of MCM2-7 (MCM5 knockdown in Figure 1F) in promoting dormant origin firing in order for cells to ensure timely completion of genomic DNA replication in the presence of low dose HU (Ge et al., 2007), and allows us to test whether FANCI behaves similarly as MCM2-7 in regulating dormant origin firing.
FANCI is required for dormant origin firing during mild replication stress
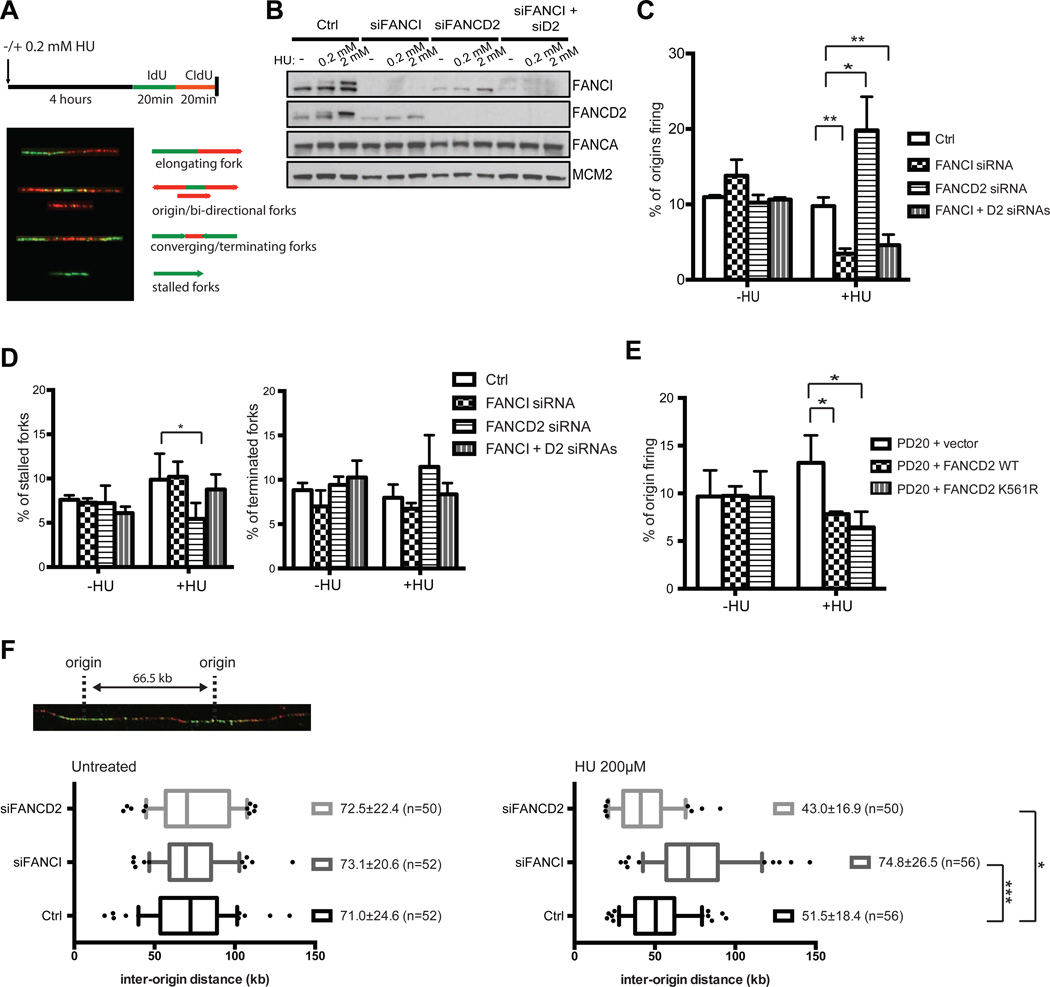
To gain insight into the possible functions of FANCI during replication stress, we employed single-molecule DNA fiber analysis to assess replication fork dynamics in human cells (Bianco et al., 2012; Michalet et al., 1997). We analyzed individual DNA fibers from untreated or low dose HU-treated asynchronous growing human retinal pigment epithelial cells (RPE) after sequential pulse-labeling of cells with IdU and CldU (See schematic, Figure 2A). In the presence of mild replication stress, FANCI knockdown reduced the percentage of active replication origins without affecting other types of replication fork structures, suggesting that FANCI is required for origin firing under mild replication stress (Figure 2B–D). Interestingly, the depletion of FANCD2 enhanced origin firing in a manner that is dependent on FANCI, as co-depletion of both FANCI and FANCD2 reduced origin firing to that of FANCI knockdown alone (Figure 2B–D). The loss of FANCD2 also resulted in reduced stalled forks in the presence of low-dose HU, which is likely due to increased origin firing to help alleviate replication stress (Figure 2D). We also showed that the role of FANCD2 for restricting origin firing in response to low dose HU is independent of its monoubiquitination status, as the K561R monoubiquitination-defective mutant can suppress origin firing at similar levels to its wildtype counterpart (Figure 2E). The depletion of FANCD2, while inhibiting FANCI monoubiquitination, led to an increase in the number of active origins (Figure 2B). This suggests that the FA core complex (ubiquitin ligase complex for FANCI and FANCD2 monoubiquitination) is likely not required for FANCI-mediated origin firing during replication stress, and only a sub-population of FANCI is necessary for origin firing. As predicted, the depletion of FANCA (Figure S1D) or other FA core complex members (data not shown) in either U2OS or RPE cells had no effect on the number of active origins in the presence of low dose HU. Interestingly, only the loss of FANCD2, and not members of the FA core complex, enhanced origin firing in a FANCI-dependent manner, suggesting that FANCD2 binding to FANCI may be inhibitory for origin firing. This supports the notion that an intermediary modification on FANCI preceding monoubiquitination could be the trigger for modulating FANCI function at dormant origins.
Figure 2. FANCI is required for dormant origin firing during conditions of replication stress.
(A) Schematic for DNA fiber analysis. RPE cells were treated with 200 µM HU for 4 h and then pulse-labeled sequentially with IdU (green) and CldU (red) for 20 min each in the presence of HU after separate washes. Images depicting the different classes of replication structures used to determine the frequency of replication origins. DNA fibers were extracted, denatured and stained by antibodies on the slides. (B) RPE cells were transfected with the indicated siRNAs and treated with different HU doses for 4 or 18 h. Samples were then analyzed by Western blot to confirm knockdown efficiency. The monoubiquitinated form of either FANCI or FANCD2 is represented by the slower-migrating band. (C) The percentage of new origin firing (counted as the sum of second label firing and bi-directional forks over the total number of different replication structures) was measured in RPE cells transfected with the indicated siRNAs and left untreated or treated with 200 µM HU. (D) The percentage of stalled and terminated forks were also measured and quantified as in (C). (E) The percentage of new origin firing was measured in FANCD2-deficient patient cells (PD20) complemented with either vector only, FANCD2 WT or FANCD2 K561R mutant and left untreated or treated with HU as in (C). (F) RPE cells were transfected with the indicated siRNAs and left untreated or treated with 200 µM HU and DNA fibers were analyzed for changes in inter-origin distances. Inter-origin distances were measured as the distance between two adjacent initiation sites during IdU pulse, and median values are indicated in kilobases (kb). Over 50 fibers were analyzed in each condition. Box and whiskers in all graphs indicate 25%-75% and 10%-90% percentiles, respectively. The lines represent the median values. For the measurement of inter-origin distances, the p value was calculated by the Mann-Whitney rank sum t-test.
A reduction in the number of active origins in FANCI-depleted cells under mild replication stress suggests a dormant origin firing defect. If this were due to a loss of dormant origin firing, the distance between adjacent replication origins would increase. After assessing the inter-origin distance (IOD) in normal vs FANCI-depleted RPE cells, we found that the median IOD in HU-treated FANCI-depleted cells was significantly increased (74.8 kb) relative to that observed in HU-treated WT cells (51.5 kb), suggesting that FANCI depletion causes a loss in dormant origin firing in response to replication stress (Figure 2F). In contrast, FANCD2 knockdown reduced the IOD, suggesting that FANCD2 is a negative regulator of dormant origin firing. The median IOD in untreated cells was relatively similar in either FANCD2 or FANCI knockdown conditions, as compared to normal RPE cells. Collectively, our data suggests that FANCI functions independently of the canonical FA pathway to promote dormant origin firing under mild replication stress.
MCM4 N-terminal mutant bypasses the requirement of FANCI for dormant origin firing
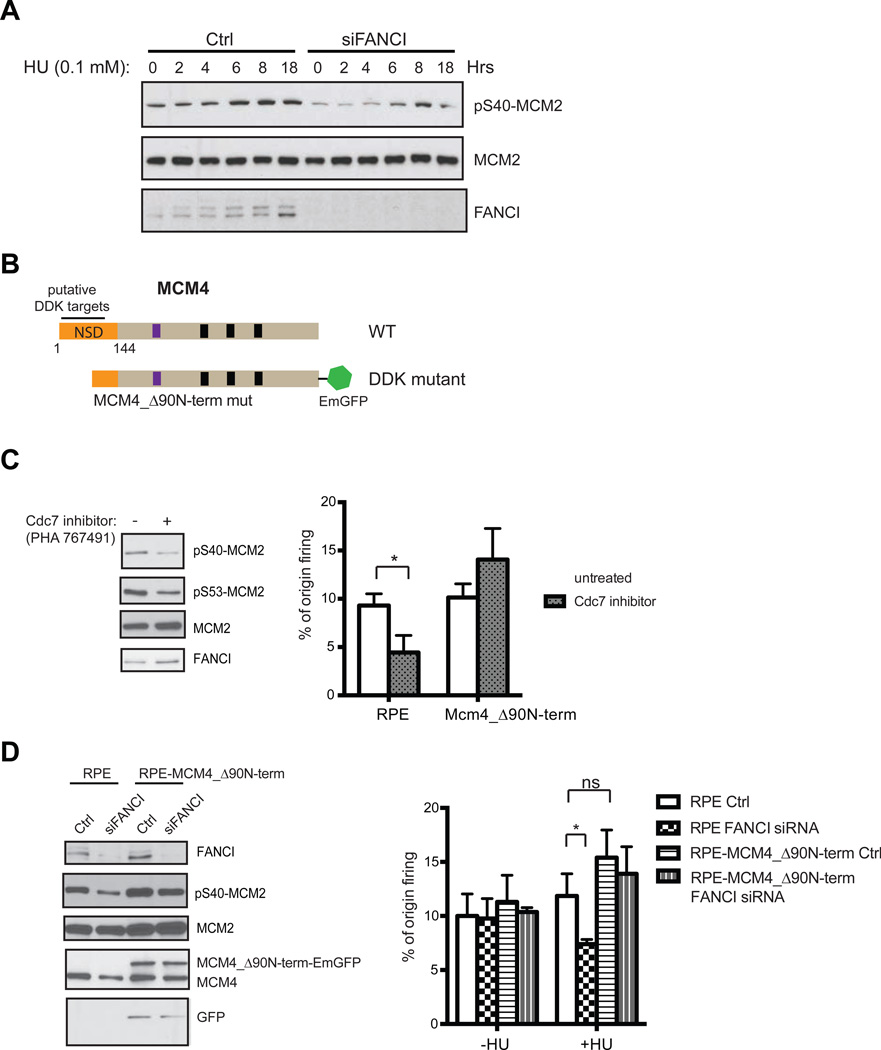
We have shown that FANCI interacts with MCM proteins and localizes to replication origins, however, it is unclear at which step is FANCI needed to facilitate dormant origin firing. The DDK phosphorylation of MCM proteins activates the MCM2-7 replicative helicase by facilitating the loading of additional components, including Cdc45 and the GINS complex (Masai et al., 2006; Owens et al., 1997; Zou and Stillman, 2000). DDK phosphorylation sites have been mapped to MCM2 Ser 40 and additional residues on MCM2 and MCM4 (Cho et al., 2006; Francis et al., 2009; Lei et al., 1997; Montagnoli et al., 2006). To assess whether FANCI plays a role in modulating DDK activity, we examined the levels of MCM2 Ser 40 phosphorylation in FANCI-depleted cells. Interestingly, the levels of MCM2 phosphorylation were significantly reduced in FANCI-depleted untreated cells or cells treated with low dose HU (Figure 3A and 3D). Genetic models in lower eukaryotes have shown that DDK activity is required to initiate DNA replication through the phosphorylation of the N-terminal serine/threonine-rich domain (NSD) of MCM4, which is essential to relieve the inhibitory function of the NSD to initiate DNA unwinding (Sheu et al., 2014; Sheu and Stillman, 2006, 2010). In yeast, mutations in the NSD of MCM4 could partially bypass the requirement of DDK for the initiation of DNA replication. To determine whether the suppression of origin activation observed when FANCI is depleted are the result of reduced DDK activity (Figure 3A), we tested whether generating an RPE cell line over-expressing an N-terminal deletion mutant in mouse MCM4 could compensate and restore origin activation in FANCI-depleted cells treated with low dose HU. The N-terminus of MCM4 is highly divergent making the NSD of budding yeast difficult to accurately define within the mouse MCM4 sequence. Despite these differences, it is clear that the N-terminal region of mouse MCM4 does contain many S/T residues and can be defined as residues 1–144 prior to the highly conserved AAA+ ATPase family and zinc finger motifs involved in double-hexamer formation (Figure 3B and data not shown) (Sheu and Stillman, 2010). To define the inhibitory domain within the N-terminus of mammalian MCM4 capable of bypassing the requirement of DDK, we generated two MCM4 N-terminal deletion mutants (Δ90N-term and Δ144N-term) (Figure 3B). These mutants were expressed with a C-terminal emerald GFP tag (EmGFP) (Kuipers et al., 2011). After checking the localization of both mutants, we found that the Δ90N-term mutant was expressed exclusively in the nucleus, similar to WT, while the Δ144N-term was distributed between both the nucleus and the cytoplasm (data not shown). Therefore, we decided to focus on the functional assessment of the Δ90N-term mutant. The stable expression of the MCM4 Δ90N-term mutant in RPE cells did not alter its cell cycle distribution (Figure S1E) or affect protein expression of critical replication factors in comparison to normal RPE cells (data not shown). To functionally validate that the MCM4 Δ90N-term mutant can indeed bypass the requirement of DDK activity to activate origin firing, we used the Cdc7 inhibitor PHA 767491 to assess the levels of MCM2 phosphorylation and origin firing with DNA fiber analysis in comparison of normal RPE with the MCM4 Δ90N-term mutant-expressing cells (Montagnoli et al., 2008) (Figure 3C). We showed that normal RPE cells displayed a reduction in MCM2 phosphorylation and the number of active replication origins when treated briefly (4 hours) with the Cdc7 inhibitor (Figure 3C). In contrast, MCM4 Δ90N-term mutant-expressing RPE cells could maintain stable levels of origin firing when treated with the Cdc7 inhibitor (Figure 3C). Importantly, we then showed that the decrease in the levels of origin firing caused by FANCI knockdown during mild replication stress could be rescued in RPE cells expressing the MCM4 Δ90N-term mutant (Figure 3D). This demonstrates that FANCI is likely to function as a regulator of DDK activity to help activate the MCM2-7 helicase.
Figure 3. MCM4 N-terminal mutant bypasses the requirement of FANCI for dormant origin firing.
(A) RPE cells transfected with either Ctrl or FANCI siRNAs were treated with 100 µM HU for the indicated times (h). Western blot analysis was performed and probed with the indicated antibodies. (B) Schematic of MCM4 N-terminal deletion mutant (MCM4_Δ90N-term). (C) RPE cells or RPE cells stably expressing MCM4_Δ90N-term were treated with or without the Cdc7 inhibitor (PHA 767491) at 5 uM for 4 h and analyzed by Western blot (RPE cells only) and by DNA fiber analysis for % origin firing. (D) Western blot analysis of normal RPE cells or RPE cells stably expressing MCM4_Δ90N-term after transfection with either Ctrl or FANCI siRNAs (untreated) were probed with the indicated antibodies. Comparison of % origin firing in RPE cells versus RPE cells stably expressing MCM4_Δ90N-term were transfected with Ctrl or FANCI siRNA and treated with or without 200 µM HU.
FANCD2 restricts replication fork speed during replication stress
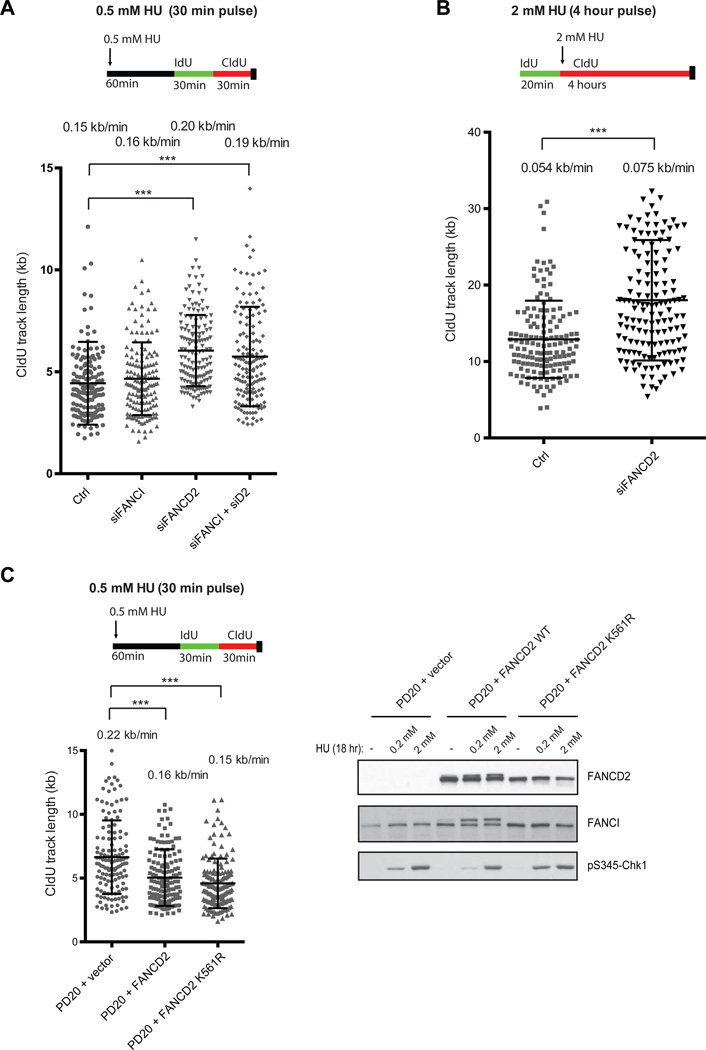
Although we showed that FANCD2 plays a critical role in suppressing dormant origin firing in response to mild replication stress (Figure 1), it was unclear whether FANCD2 also functions to control replication fork speed. We monitored DNA chain elongation from replication tracks labeled with consecutive pulses of IdU and CldU using increasing doses of HU (Figure S1C and Figure 4). The depletion of FANCD2 in RPE cells resulted in longer track lengths and faster replication fork speeds at higher doses of HU (Figure S1C and Figure 4A–B). Surprisingly, FANCD2 monoubiquitination is not required for this function, as both FANCD2 WT and K561R mutant are capable of restricting replication fork speed in PD20 cells (Figure 4C). In contrast, FANCI does not share this trait with FANCD2 at the replisome (Figure 4A).
Figure 4. FANCD2 restricts replication fork speed after HU treatment in a monoubiquitination-independent manner.
(A-C) Schematics of treatment with HU and pulse labeling with IdU and CldU are shown. Replication fork lengths (Jackson and Pombo, 1998) were obtained by converting the CldU track size in uM to kb and analyzed in the siRNA-transfected RPE cells or PD20 cells treated with the indicated dose of HU. Average fork speeds are calculated by dividing the track lengths by the labeling time and depicted above the track lengths. Western blot analysis of PD20 cells is shown and probed with the indicated antibodies. For the measurement of track length distributions for replication fork speeds, the p value was calculated by the Mann-Whitney rank sum t-test.
ATR-mediated FANCI phosphorylation inhibits dormant origin firing
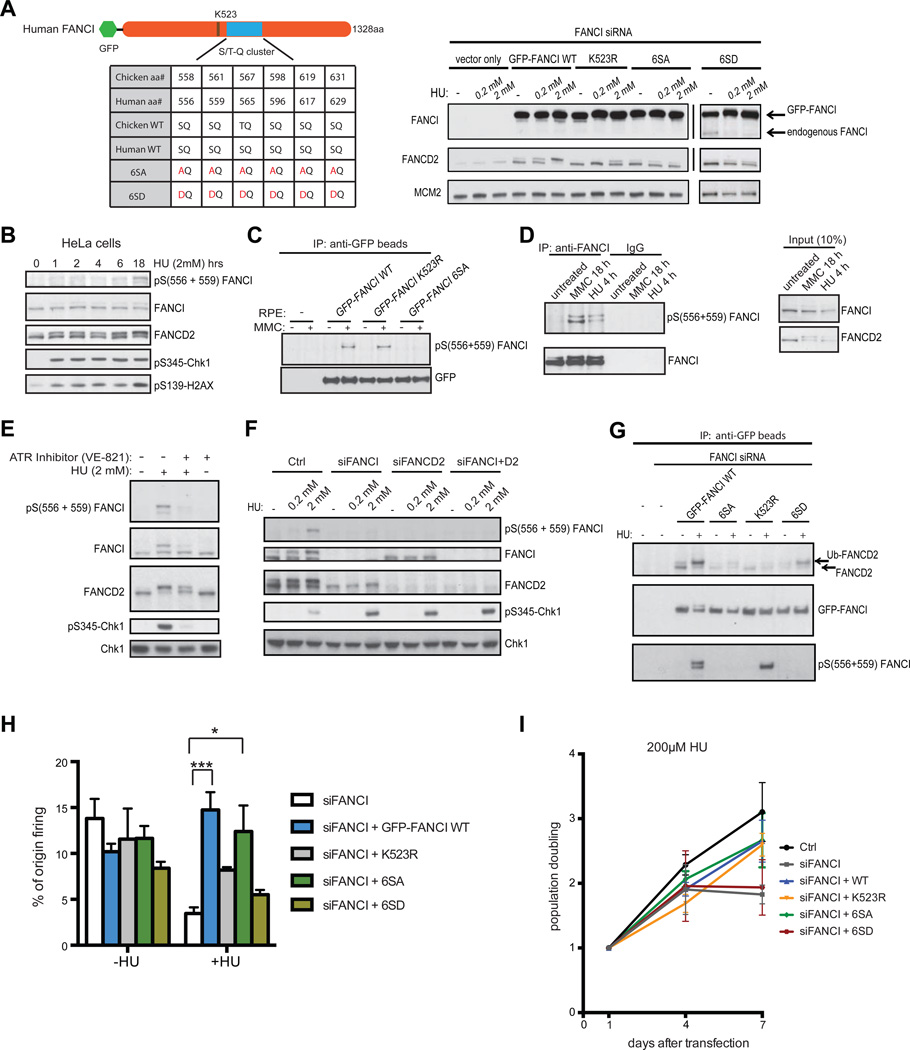
We showed that FANCA (member of the FA core complex) and FANCD2 are not required for dormant origin firing in the presence of low dose HU (Figure 2C and Figure S1D). This suggests that the FANCI monoubiquitination-defective mutant should behave similarly to FANCI WT in its ability to promote dormant origin firing. To functionally analyze different FANCI mutants, we used GFP-FANCI expression constructs containing silent mutations that render them resistant to the FANCI siRNA sequence (Figure 5A) (Colnaghi et al., 2011). Unexpectedly, the K523R monoubiquitination-defective mutant was only able to partially rescue dormant origin firing in both FANCI-deficient RPE and FA-I (F010191) patient-derived cells (Figure 5H and Figure S1F). It was previously shown that the corresponding K525R mutant in chicken DT40 cells, while incapable of being monoubiquitinated, was still proficiently phosphorylated by the Ataxia Telangiectasia and Rad3-related (ATR) kinase in MMC-treated cells, as analyzed by Phos-tag gels (Ishiai et al., 2008). In chicken DT40 cells, ATR phosphorylates FANCI on a cluster of at least six Ser/Thr-Gln (S/T-Q) phospho-consensus sites adjacent to the K525 monoubiquitination site to induce efficient chicken FANCI and FANCD2 monoubiquitination, nuclear foci formation and ICL repair (see schematic, Figure 5A) (Ishiai et al., 2008). However, the role of this phosphorylation event has not been properly demonstrated in human cells. In addition, ATR plays a critical inhibitory role in the regulation of dormant origin firing in response to mild replication stress (Ge and Blow, 2010; Schwab et al., 2010), but the downstream phosphorylation target(s) of ATR have not be fully elucidated.
Figure 5. ATR-mediated Phosphorylation of FANCI inhibits dormant origin firing.
(A) Schematic of the conserved ATR phospho-consensus S/T-Q sites on human FANCI. Rescue of FANCI-depleted RPE cells with stably expressed siRNA-resistant GFP-FANCI WT, K523R (monoubiquitination deficient), 6SA and 6SD mutants; probed with the indicated antibodies for Western blot analysis. (B) Time-course experiment of HU-treated (2mM) HeLa cells was analyzed by Western blot and probed with the pS(556 + 559) FANCI phospho-specific antibody and other antibodies as indicated. Slower-migrating phosphorylated FANCI band corresponds to the monoubiquitinated FANCI band. (C) RPE cells transfected with FANCI siRNA and complemented with either GFP-FANCI WT, K523R, or 6SA were untreated or treated with MMC (1 µM). Extracts were subjected to co-IP with anti-GFP beads and analyzed by Western blot with the indicated antibodies. (D) RPE cells treated with either HU (2 mM) or MMC (1 µM) for the indicated times were subjected to co-IP with anti-FANCI antibody or IgG control and analyzed by Western blot with the indicated antibodies. (E) RPE cells were treated with HU (2 mM) for 18 h with or without the ATR inhibitor (VE-821) at 10 uM for 4 h as indicated and analyzed by Western blot. (F) RPE cells transfected with the indicated siRNAs and treated with the indicated dose of HU for 18 h and analyzed by Western blot. (G) RPE cells transfected with Ctrl or FANCI siRNA and complemented with either GFP-FANCI WT, 6SA, K523R, or 6SD mutants were treated with HU (2 mM) and subjected to co-IP with anti-GFP beads and analyzed by Western blot with the indicated antibodies. (H) Measurement of % origin firing in FANCI-depleted RPE cells complemented with different GFP-FANCI expression constructs and treated with 200 µM HU. (I) Measurement of cell proliferation in HU-treated RPE cells as indicated. P values for Ctrl vs siFANCI or siFANCI vs siFANCI + WT, + 6SA, or + K523R are < 0.05 for the 7 day time point. P value for siFANCI vs siFANCI + 6SD is > 0.05.
To determine whether FANCI was indeed phosphorylated in the K523R monoubiquitination-defective mutant, we generated a phospho-specific antibody targeting the highly conserved phosphorylated Ser 556 and 559 residues on FANCI (pS556+559 FANCI antibody). We showed that in both whole cell and FANCI-enriched cell lysates, the phospho-specific antibody was able to recognize the endogenous and ectopically expressed FANCI phosphorylated species in MMC- or HU-treated samples, but not in untreated cells (Figure 5B–D). Importantly, phosphorylation on exogenous FANCI was detectable in GFP-FANCI K523R-expressing cells treated with either MMC or HU (Figure 5C and G). Curiously, FANCI phosphorylation is not sufficient for enhanced FANCD2 binding, as the K523R mutant does not bind to FANCD2 as robustly as wildtype FANCI (Figure 5G).
A possible explanation as to why the K523R mutant is only partially functional for dormant origin firing is that phosphorylation of FANCI can still occur in the absence of monoubiquitination, and that phosphorylation is the more relevant trigger to inhibit dormant origin firing. To address this, all six of the S/T-Q ATR phospho-consensus sites on human FANCI were changed from Ser to Ala (6SA, phospho-dead mutant) or to Asp (6SD, phospho-mimic mutant) and tested for their ability to prevent FANCI phosphorylation and/or rescue dormant origin firing in RPE cells depleted of endogenous FANCI (Figure 5A and H). The 6SA mutant was indeed unable to be phosphorylated using the phospho-specific FANCI antibody in the presence of MMC or HU (Figure 5C and G). FANCI phosphorylation on Ser residues 556 and 559 was dependent on ATR kinase activity since treatment with an ATR inhibitor (VE-821) (Reaper et al., 2011) potently blocks FANCI phosphorylation (Figure 5E). Importantly, ATR-mediated FANCI phosphorylation requires the presence of FANCD2, as FANCD2 knockdown abrogates FANCI phosphorylation (Figure 5F). While expression of the GFP-FANCI 6SA mutant could not fully complement HU-induced FANCD2 monoubiquitination when compared to WT, the 6SA mutant was able to rescue dormant origin firing and cell growth defects in cells treated with low dose HU (Figure 5H–I and Figure S2A). Complementation of FA-I cells with WT or 6SA also gave similar results as in the RPE cells for the rescue of dormant origin firing (Figure S1F). In contrast, expression of the 6SD mutant could not rescue dormant origin firing and cell growth defects under conditions of replication stress (Figure 5H–I). These data demonstrate that ATR-mediated FANCI phosphorylation is indeed inhibitory for dormant origin firing and normal cell proliferation, and suggest that the unmodified FANCI serves a critical role in activating dormant origins in response to mild replication stress.
ATR-mediated FANCI phosphorylation is required for replication restart in response to replication stress
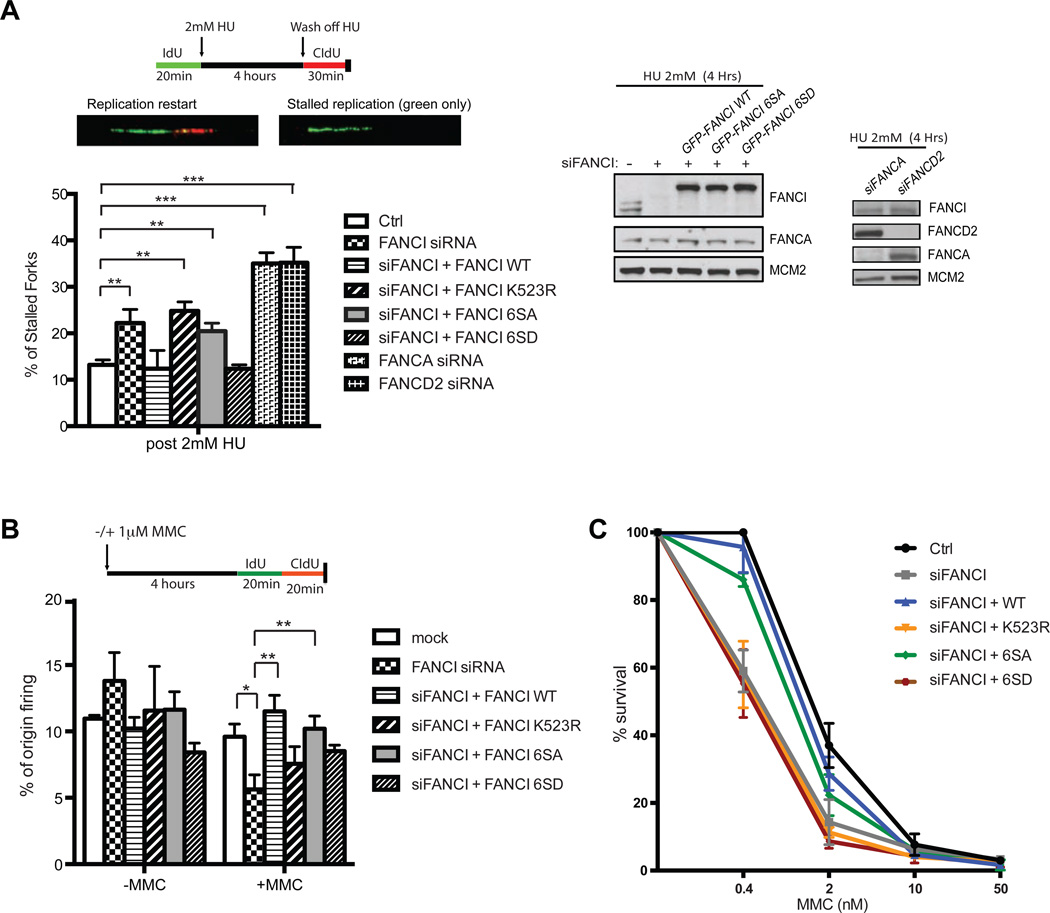
Next, we determined the functional consequences of the phosphorylation site mutants in promoting replication fork stability/restart during severe replication stress conditions. Previous studies have shown that the canonical FA pathway (FANCD2 monoubiquitination) is critical for many aspects of replication fork stability under conditions of severe replication stress (³2 mM HU) (Chaudhury et al., 2013; Lossaint et al., 2013; Schlacher et al., 2012). However, it is unclear whether phosphorylation of FANCI is required for these FA pathway-related functions. Unlike low dose HU treatment, which allows DNA replication to continue through the firing of dormant origins, long-term (18 h) treatment of cells with high dose HU will cause the majority of forks to irreversibly stall or collapse (Petermann et al., 2010). Exposing cells to high dose HU for only a short period of time (4 h) will enable most of the stalled forks to properly restart or recover (Petermann et al., 2010). Treatment of HeLa cells with high dose HU stimulates FANCI phosphorylation on the S/T-Q sites (pS556 + 559 FANCI) and also for other ATR substrates, including Chk1 (pS-345) and H2AX (pS139) (Figure 5B). To analyze the functional consequence of FANCI phosphorylation during conditions of transient high dose HU exposure, DNA fiber analysis were implemented to measure replication fork restart/recovery in RPE cells. To measure replication fork restart, the double nucleotide analog labeling protocol was modified to allow for detection of fork recovery after the removal of HU (Figure 6A). Unlike the FANCI 6SD mutant, the 6SA and K523R mutant-expressing RPE cells displayed reduced levels of fork restart, as reflected by an increase in the percentage of stalled forks (Figure 6A). We also show that depletion of different upstream FA pathway components, including FANCD2 or FANCA, also resulted in elevated levels of stalled forks (Figure 6A). These findings are consistent with the FA pathway performing important genome protective roles at stalled replication forks (Chaudhury et al., 2013; Schlacher et al., 2012). Collectively, these data suggest that depending on the magnitude of replication stress, phosphorylated and monoubiquitinated FANCI may function concomitantly as both a positive regulator of replication fork restart at stalled forks and as an inhibitor of dormant origin firing in the vicinity of stalled forks.
Figure 6. Phosphorylation of FANCI is required for replication fork restart during replication stress.
(A)A schematic for measuring replication fork restart using DNA fiber technique. Cells are labeled first with IdU for 20 min, then treated with 2 mM HU for 4 h to block DNA replication. Following a wash step to remove the HU, cells are then pulsed with CldU for 30 min. Regions on the DNA fibers with both colors labeled demonstrate fork restart, whereas fibers with only the green track represent forks that could not restart. The frequency of “green only” tracks over total forks is displayed as % of stalled forks. FANCI-depleted RPE cells complemented with different FANCI WT and mutant expression constructs or RPE cells depleted of FANCA or FANCD2 by siRNAs were treated with 2 mM HU for 4 h to measure stalled forks and samples were analyzed by Western blot with the indicated antibodies. (B) Schematic of MMC-treated RPE cells that are analyzed for % origin firing. RPE cells depleted of FANCI and complemented with different GFP-FANCI expression constructs as indicated were processed for % origin firing measurements. (C) MMC sensitivity growth assay displayed as % cell survival using Syto60 staining with the indicated MMC dose in RPE cells. P values for Ctrl vs siFANCI or siFANCI vs siFANCI + WT or + 6SA are < 0.05 for the 0.4 nM MMC dose. P values for siFANCI vs siFANCI + 6SD or + K523R are > 0.05 at the same dose.
FANCI-dependent dormant origin firing is required for cellular ICL resistance
Since FANCI functions as an integral part of the FA pathway to confer cellular resistance to ICLs, we asked whether this newly discovered role of FANCI in dormant origin firing alleviates MMC-induced DNA damage. Confirming previous studies (Ishiai et al., 2008), we showed that FANCI is inducibly phosphorylated upon MMC treatment using the phospho-specific FANCI antibody (Figure 5C), suggesting that dynamic regulation of phosphorylated FANCI may be important in mediating both DNA repair and/or dormant origin firing. RPE cells treated with MMC were assessed for changes in replication fork dynamics after FANCI knockdown or rescue with WT, 6SA, or 6SD mutants. The dose of MMC used (1 µM) did not slow the replication fork speed or increase fork stalling dramatically in a global manner (Figure S2B–C). The replication fork speed in MMC-treated cells was also relatively similar between Ctrl and FANCI knockdown cells, suggesting that the loss of FANCI did not lead to acute stalling of replication forks (Figure S2B). Importantly, we found that FANCI, but not FANCD2 or FANCA, is required to maintain the level of origin firing after transient MMC treatment (Figure 6B and Figure S2D). Similar to low dose HU treatment, the expression of either WT or the 6SA mutant in FANCI-depleted cells was capable of rescuing dormant origin firing, but not K523R or the 6SD mutant (Figure 6B). Monoubiquitinated FANCI (and FANCD2) has an established role at the replication fork to initiate ICL repair (Knipscheer et al., 2009). Consistent with its canonical function in ICL repair, the 6SA mutant could only partially rescue MMC-induced DNA damage, as measured by the MMC-induced cytotoxicity and G2/M accumulation assay (indicative of unrepaired ICLs activating the G2/M checkpoint) (Figure 6C and Figure S2E). Surprisingly, the 6SD mutant displayed a more severe phenotype for MMC-induced cytotoxicity and G2/M accumulation as compared to the 6SA mutant (Figure 6C and Figure S2E). Collectively, our data demonstrates the relevance of origin firing as a cellular response to alleviate ICL damage. Whether dormant origin firing adjacent to ICL-induced fork stalling events contribute directly to the repair process remains to be determined.
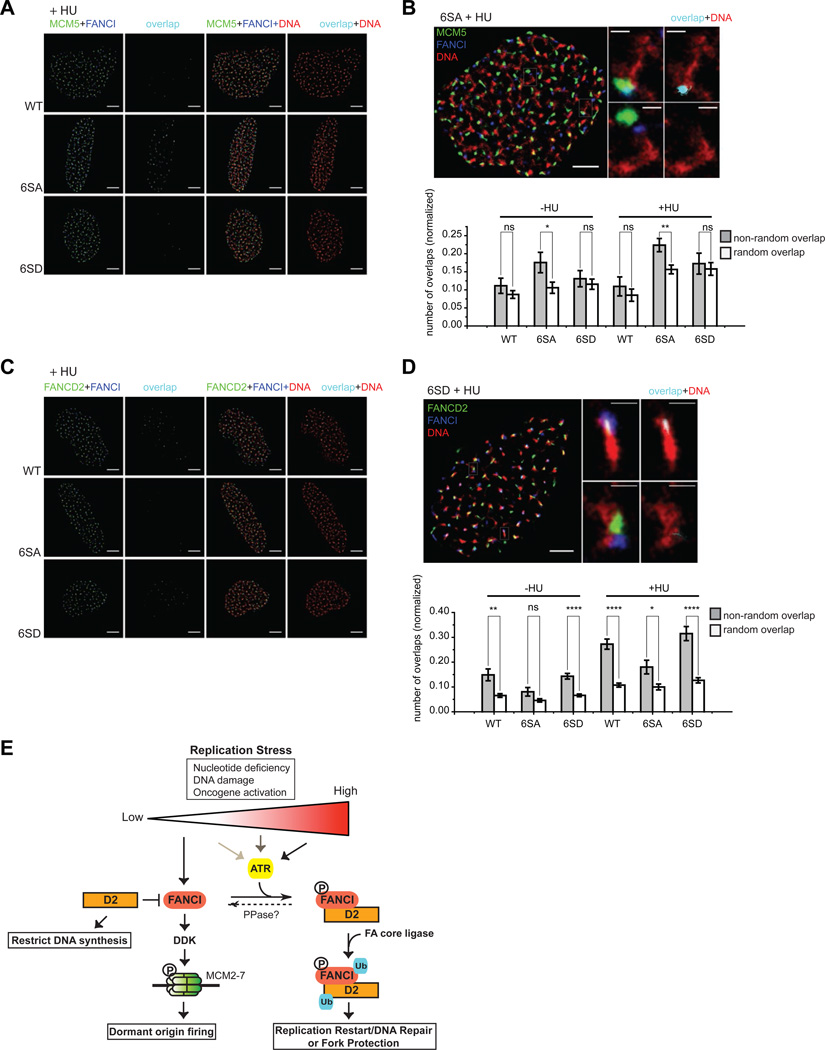
Super-resolution imaging detects FANCI co-localization with the MCM complex
Despite being able to show a functional separation between the different phosphorylation mutants, it remains unclear why the phosphorylated form of FANCI is defective for dormant origin firing in response to replication stress. It is possible that the phosphorylation status of FANCI is a critical modulator of its localization to different functional complexes on chromatin. To accurately map the nuclear organization of FANCI, FANCD2 and the MCM5 proteins in relation to nascent DNA, we employed single-molecule localization based super-resolution (SR) microscopy (Heilemann et al., 2008; Rust et al., 2006) to obtain typically an order of magnitude higher resolution images than conventional microscopy (Agullo-Pascual et al., 2013; Pavlides et al., 2013; van de Linde et al., 2011) (see Figure S3A). To ensure that the observed co-localization/signal overlaps between MCM5 and FANCI or FANCD2 and FANCI do not stem from random events, we carried cross-correlation analysis of intra- and inter-overlap events (Coltharp et al., 2014), which showed that localization of complexes is indeed unique (Figures S3 and S4). Similar S-phase nuclei were picked for analysis based on the EdU pulse-labeled staining pattern. Using this analysis, we found a higher level of co-localization/overlap between the FANCI 6SA mutant and MCM5 proteins in comparison to WT or the 6SD mutant in response to mild replication stress (Figure 7A–B). In contrast, the FANCI 6SA mutant co-localized less often with FANCD2, in comparison to WT or the 6SD mutant (Figure 7C–D). This is in agreement with our co-IP data, showing that the 6SD mutant associates much better with FANCD2 than the 6SA mutant (Figure 5G). Collectively, our data suggests that the phosphorylated form of FANCI is likely excluded from MCM-containing complexes on chromatin (dormant origins) when bound to FANCD2; FANCI in this phosphorylated state is refractory to dormant origin activation. In contrast, unphosphorylated FANCI may remain bound to MCM2-7 but excluded from FANCD2-associated replisomes; FANCI in this unphosphorylated state is still permissive for dormant origin firing. Thus, by using SR microscopy, we were able to resolve co-localization differences between the phosphorylated and nonphosphorylated forms of FANCI and how they correlate spatially with the MCM or FANCD2 proteins.
Figure 7. Super-resolution imaging detects FANCI co-localization with the MCM complex.
(A and C) SR images of the nuclei of U2OS cells stably expressing either FLAP-tagged FANCI WT (top row), 6SA (middle), or 6SD (bottom) that is treated with 200 µM (+HU) and pulsed labeled with EdU for 1 h. Localization of MCM5, FANCD2, FANCI, and newly synthesized DNA were visualized using antibodies against MCM5, FANCD2 or GFP (FANCI) and direct labeling of EdU with Click-iT chemistry (Scale bar, 5 µm). (B and D) Co-localization (overlap) measurements of FANCI with either MCM5 or FANCD2 as revealed by SR imaging. The magnified images depict an SR image of either a FANCI 6SA or 6SD mutant-expressing U2OS nucleus treated with HU (MCM5, green; FANCD2, green; FANCI, blue; newly synthesized DNA, red; scale bar, 3 µm). The magnification of the marked region (i) shows an example of a localization overlap event between MCM5 (or FANCD2) and FANCI in the way of either two-color co-localization, i(a), or two-color overlap, i(b); while the magnification of region (ii) shows an example of a lack of a localization overlap event between the two proteins as in (i) with the two-color co-localization, ii(a), or two-color overlap, ii(b) (scale bar, 500 nm). The graphs in B and D depict the average number of overlap events normalized between MCM5 (or FANCD2) and FANCI from the nuclei of either WT, 6SA or 6SD-expressing U2OS cells. The grey bar shows the average number of overlaps between MCM5 (or FANCD2) and FANCI from the same nuclei (intra-overlap); these events are related and therefore represent a non-random occurrence. In contrast, the white bar shows the average number of overlaps detected between different nuclei (inter-overlap); these events are unrelated and represent a random occurrence. (E) Cartoon model depicting how a phosphorylation switch mediated by the ATR kinase (and an unknown protein phosphatase) can exert control over FANCI to either activate or inhibit dormant origin firing in response to different levels or types of replication stress. Cells exposed to minimal levels of replication stress will result in more dormant origin firing to help rescue slowed or stalled forks. During low levels of replication stress, dormant origin firing is likely operational as a default setting due to the absence of ATR signaling. In contrast, higher exposure to DNA damage or replication stress will result in robust and sustained ATR activation, leading to FANCI and Chk1 phosphorylation and reduced dormant origin firing. This may give cells more time to activate DNA repair and restart replication forks; both of these events could be mediated, in part, through FANCI phosphorylation and monoubiquitination. FANCD2 has an unanticipated role in inhibiting FANCI-mediated dormant origin firing, independent of its monoubiquitination status. FANCI can activate dormant origins by promoting DDK-dependent phosphorylation of MCM complexes to relieve MCM proteins from its inhibitory state. We predict that phosphorylation of FANCI will shift the pool of FANCI from MCM-bound dormant origins to stalled replication forks.
DISCUSSION
In this study, we provide evidence supporting a new, unanticipated role for FANCI in regulating dormant origin firing in response to replication stress (Figure 7E). By using either 6SA or 6SD ATR phospho-site mutants of FANCI in a cell-based reconstitution assays and a phospho-specific FANCI antibody, we were able to demonstrate that phosphorylation at the S/T-Q cluster of FANCI inhibits the ability of FANCI to promote dormant origin firing in response to mild replication stress. Unmodified FANCI is likely proficient for dormant origin firing, but is insufficient for promoting replication fork restart under severe replication stress or DNA repair after MMC treatment. Regions of the DNA with slower moving forks may require unphosphorylated FANCI to promote dormant origin firing, enabling the cell to complete replication in a timely manner. Larger numbers of stalled forks will trigger ATR activation, which stabilizes and repairs replication forks through unknown mechanisms (Byun et al., 2005). Here, we demonstrate that FANCI phosphorylation by ATR promotes replication fork restart/recovery. Severe replication stress may cause cells to preferentially suppress dormant origin firing to preserve licensed origins during cell cycle checkpoint activation and allow for the stalled replication forks more time to restart or recover (Petermann and Helleday, 2010). It could be too dangerous for cells to initiate more replication forks, as once these backup origins are done firing, no new origins can be licensed until after the cells undergo mitosis. On the other hand, mild replication stress can be easily overcome through dormant origin firing in the absence of checkpoint activation, and likely does not require more time-consuming and potentially error-prone HR-dependent DNA transactions involved during replication fork restart (Carr and Lambert, 2013; Guirouilh-Barbat et al., 2014). Thus, dormant origin firing can only be effective as a quasi-repair strategy under mild replication stress conditions where the newly established replication forks have a low probability of stalling.
Studies on the DNA repair-independent role of the FA pathway have only recently been appreciated. Work was largely confined to the cellular response necessary to counteract high levels of replication stress, where stalled forks can be substrates for nuclease attack or DSB formation, which would then require mechanisms for fork stabilization/protection or replication restart (Lossaint et al., 2013; Schlacher et al., 2012; Yeo et al., 2014). Prior to this study, very little was known about whether members of the FA pathway provide additional function(s) to alleviate mild replication stress that persists in the absence of checkpoint activation. Importantly, conditions that mimic mild replication stress is recapitulated during cancer initiation and progression model systems whereby oncogenes induce replication stress via perturbation of the available nucleotide pools in these cells to promote uncontrolled DNA replication, leading to genomic instability and malignancy (Bester et al., 2011). Understanding the molecular pathways that help alleviate mild replication stress will inevitably provide key insights into a physiological process designed to counteract replication stress in normal and malignant cells. Based on our findings, we showed that FANCI has at least two non-overlapping roles in counteracting replication stress (see model, Figure 7E): First, in cells encountering severe replication stress (high dose HU), ATR-mediated FANCI phosphorylation and subsequent FA core complex and FANCD2-dependent monoubiquitination of FANCI signifies the critical molecular event that promotes replication fork protection and/or restart of stalled forks. We speculate that this event could also be linked to DNA repair, as ICL repair requires many of the same components of the FA pathway as for replication fork restart, although supporting evidence for this remains to be seen. Second, in cells exposed to mild replication stress (low dose HU), the unphosphorylated form of FANCI participates in dormant origin firing likely through the activation of the DDK, and this occurs independently of the FA core complex and FANCD2 (Figure 7E). Interestingly, we found that loss of FANCD2 elevates dormant origin firing, but not in the absence of FANCI, suggesting that FANCD2 acts as an inhibitor of dormant origin firing through FANCI. This is consistent with previous studies showing that the loss of FANCD2 stimulates new origin firing in cells treated with HU or APH in other human cell lines (Chaudhury et al., 2013; Panneerselvam et al., 2014). It remains to be determined whether FANCD2 is able to inhibit this function of FANCI through direct binding to the FANCI or the MCM complex, or whether other intermediary proteins are involved. We favor the former prediction as an ubiquitin-binding domain of FANCD2 is suggested to bind to monoubiquitinated FANCI (Rego et al., 2012). It is unknown whether other FA proteins regulate dormant origin firing differentially through FANCI or whether FANCI is truly unique amongst the FA members to promote dormant origin firing in response to replication stress (Tumini et al., 2011).
We found that dormant origin activation plays an unanticipated role in alleviating replication stress caused by MMC-induced DNA damage. According to the current model of replication-dependent ICL repair, a second adjacent fork converging onto the ICL may participate in the ICL repair process (Huang et al., 2013; Jones and Huang, 2012; Knipscheer et al., 2009; Kottemann and Smogorzewska, 2013; Zhang and Walter, 2014). This critical second fork may arise from the firing of additional adjacent origins. If this is indeed the case, it will be important to understand mechanistically how FANCI and/or other FA proteins are able to couple DNA repair and adjacent origin firing together in the vicinity of the ICL damaged regions of the genome. Mutations in the factor(s) that regulate this coupling event could contribute to the FA disease and/or cancer susceptibility. Furthermore, whether dormant origin activation plays an important part in the repair and cellular tolerance to other types of DNA damaging agents, including UV damage, reactive aldehydes or double-strand break inducers, is unknown.
It remains unclear how FANCI promotes dormant origin firing in response to replication stress. When local replication forks are inhibited or slowed, MCM2-7 complexes are activated by the DDK (Diffley, 2010), leading to the firing of dormant origins to ensure that genomic replication is completed in a timely manner. It has been suggested that ATR signaling intersects with DDK-mediated activation of the MCM2-7 helicase complex (Yamada et al., 2013), but the molecular details have remained vague. In our study, we showed that FANCI likely works at or upstream of DDK-mediated activation of the MCM complex, as the loss of the inhibitory domain of MCM4 can rescue FANCI-depleted cells for dormant origin activation. Interestingly, we observed a reduction in MCM4 protein levels in FANCI-depleted cells. We speculate that FANCI may directly associate with MCM4 to prevent its turnover in the pre-RC complex. How N-terminal phosphorylation of MCM4 by different upstream kinases regulates FANCI or DDK recruitment to the MCM2-7 complex remains to be determined.
By focusing on the mechanistic role of FANCI during replication stress, we have been able to demonstrate a new FA pathway-independent function for FANCI in the regulation of dormant origin firing, which is a key component needed to manage the deleterious effects of replication stress. Importantly, it was recently highlighted that replication stress is a potent driver of functional decline in aging hematopoietic stem cells (HSCs) (Flach et al., 2014). As HSCs are particularly vulnerable to replication stress, these are the same cells that are defective in the FA disease (D'Andrea, 2010; Garaycoechea et al., 2012; Kottemann and Smogorzewska, 2013). In the future, understanding precisely how the FA pathway and FANCI responds to and protects against replication stress will be of high clinical relevance, especially for the development of improved therapeutic interventions for FA patients and others requiring bone marrow transplantations.
EXPERIMENTAL PROCEDURES
Cell culture, Antibodies, Purifications and Reagents
U2OS, HeLa, and Phoenix-GP cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) with 10% fetal bovine serum (FBS), 1% Pen-Strep, 1% Glutamine at 5% CO2 in 37°C incubator. FA patient-derived FANCI-deficient (F010191), FANCD2-deficient (PD20) and FANCA-deficient (GM6914) fibroblasts were grown in 15% FBS. hTERT immortalized RPE-1 cells (ATCC) were grown in DMEM/F-12 medium (Life Technologies) supplemented with 10% FBS, 3% sodium bicarbonate and 1% Pen-Strep. The Cdc7 inhibitor, PHA 767491, was purchased from Santa Cruz Biotech and the ATR inhibitor, VE-821, from EMD Millipore. For conventional purification of FANCI complexes, standard chromatographic methods were used (Sims et al., 2007b; Sims et al., 2011). Transfections, western blotting, and co-immunoprecipitation methods and sources of antibodies, reagents, and plasmids used in this study are presented in the Supplemental Experimental Procedures.
DNA Fiber Analysis
DNA fibers were prepared as described previously (Terret et al., 2009). In brief, cells were labeled sequentially with IdU and CldU. Cells were trypsinized/resuspended and spotted onto a glass slide and lysed. Slides were then tilted at an angle to allow the fibers to spread by gravity flow. Slides were then labeled with fluorescent antibodies and imaged on a Deltavision microscope. To obtain extended fiber tracks for measuring inter-origin distances, agarose plugs containing DNA were prepared as described previously (Aris and Pommier, 2012),(Conti et al., 2001). The DNA was then stretched onto silane-treated slides (Genomic Vision) by Molecular Combing System (Genomic Vision). Images were captured and analyzed using SoftWoRX (Applied Precision, General Electric). Results were based on three independent experiments. At least 150 pulse-labeled forks were scored in each experiment unless otherwise indicated. Fork speed was calculated by dividing the track length in kilobase (kb) by the labeling time. Details can be found in the Supplemental Experimental Procedures.
Cell Growth, Cytotoxicity, and FACS Assays
For cell growth assay, cells were transfected with siRNAs and then replated in the presence or absence of HU and then counted subsequently , treated with or without HU, and then counted manually with a haemocytometer on different days. For MMC cytotoxicity assay, cells were fixed with 4% paraformaldehyde and stained with SYTO60 fluorescence probe (Life Technologies) and quantified with a Li-Cor Odyssey. Results were based on three independent experiments. Detailed conditions for Cell Growth, Cytotoxicity and FACS methods are described in the Supplemental Experimental Procedures.
Super-Resolution Microscopy
Details on microscopy setup for super-resolution imaging and experimental details for image capture and analyses are described in the Supplemental Experimental Procedures.
Statistical Analysis
Error bars represent standard deviation of at least three independent experiments. P values were calculated using a two-tailed Student’s t test, if not specified.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. D’Andrea and T. Taniguchi for the FA patient-derived PD20 and GM6914 cells, B. Stillman for the anti-MCM3 antibody that was useful during our early studies, K. Burns-Huang for critical reading of the manuscript, and M. Bekes and A. Sims for technical assistance. We are grateful to the NYU proteomics core (T. Neubert) for mass spectrometry analysis. We are also grateful to members of the P. Jallepalli, E. Rothenberg, M. Pagano, D. Bar-Sagi, D. Reinberg and T. Huang labs for reagents, equipment and helpful discussions. Work in E.R. laboratory is supported by grants from the NIH (GM057691 and GM110385), the Arnold and Mabel Beckman Foundation and the Shifrin-Meyer Breast Cancer Discovery award. Work in T.H. laboratory is supported by grants from the NIH (GM084244) and from ACS (RSG-12-158-DMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information contains Supplemental Experimental Procedures and four supplemental figures.
AUTHOR CONTRIBUTIONS
Y.C., assisted by M.J., performed all of the DNA fiber analysis and cell growth/cytotoxicity assays. M.J. generated all the FANCI and MCM4 mutant cell lines and contributed to the early stages of this project. Y.Y. performed super resolution microscopy analysis under the guidance of E.R. S.C. tested and performed experiments with the phospho-specific FANCI antibody. L.C. performed ChIP experiments and R.S. purified the mammalian FANCI complex. Y.C., M.J., Y.Y., E.R., P.J. and T.H. worked on data analysis, formulated the hypothesis and supervised experiments. Y.C., M.J., and T.H. wrote the paper.
REFERENCES
- Agullo-Pascual E, Reid DA, Keegan S, Sidhu M, Fenyö D, Rothenberg E, Delmar M. Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin-2 inside the connexin43 plaque. Cardiovasc Res. 2013;100:231–240. doi: 10.1093/cvr/cvt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alver RC, Chadha GS, Blow JJ. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA Repair (Amst) 2014;19:182–189. doi: 10.1016/j.dnarep.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris SM, Pommier Y. Potentiation of the novel topoisomerase I inhibitor indenoisoquinoline LMP-400 by the cell checkpoint and Chk1-Chk2 inhibitor AZD7762. Cancer Res. 2012;72:979–989. doi: 10.1158/0008-5472.CAN-11-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- Bell SD, Botchan MR. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol. 2013;5:a012807. doi: 10.1101/cshperspect.a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco JN, Poli J, Saksouk J, Bacal J, Silva MJ, Yoshida K, Lin YL, Tourrière H, Lengronne A, Pasero P. Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods. 2012;57:149–157. doi: 10.1016/j.ymeth.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733–4744. doi: 10.1016/j.jmb.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Chaudhury I, Sareen A, Raghunandan M, Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 2013;41:6444–6459. doi: 10.1093/nar/gkt348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WH, Lee YJ, Kong SI, Hurwitz J, Lee JK. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proc Natl Acad Sci U S A. 2006;103:11521–11526. doi: 10.1073/pnas.0604990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnaghi L, Jones MJ, Cotto-Rios XM, Schindler D, Hanenberg H, Huang TT. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood. 2011;117:2247–2256. doi: 10.1182/blood-2010-07-295758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltharp C, Yang X, Xiao J. Quantitative analysis of single-molecule superresolution images. Curr Opin Struct Biol. 2014;28C:112–121. doi: 10.1016/j.sbi.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C, Caburet S, Schurra C, Bensimon A. Molecular combing. Curr Protoc Cytom. 2001 doi: 10.1002/0471142956.cy0810s16. Chapter 8, Unit 8.10. [DOI] [PubMed] [Google Scholar]
- D'Andrea AD. Susceptibility pathways in Fanconi's anemia and breast cancer. N Engl J Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Diffley JF. The many faces of redundancy in DNA replication control. Cold Spring Harb Symp Quant Biol. 2010;75:135–142. doi: 10.1101/sqb.2010.75.062. [DOI] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014 doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Ge XQ, Blow JJ. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol. 2010;191:1285–1297. doi: 10.1083/jcb.201007074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS. Is homologous recombination really an error-free process? Front Genet. 2014;5:175. doi: 10.3389/fgene.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, Tinnefeld P, Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Schwob E, Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MJ, Huang TT. The Fanconi anemia pathway in replication stress and DNA crosslink repair. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D’Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers MA, Stasevich TJ, Sasaki T, Wilson KA, Hazelwood KL, McNally JG, Davidson MW, Gilbert DM. Highly stable loading of Mcm proteins onto chromatin in living cells requires replication to unload. J Cell Biol. 2011;192:29–41. doi: 10.1083/jcb.201007111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnev D, Rusiniak ME, Kudla A, Freeland A, Cady GK, Pruitt SC. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010;29:3630–3638. doi: 10.1038/onc.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Décaillet C, Gari K, Constantinou A. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–690. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J Biol Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- McIntosh D, Blow JJ. Dormant origins, the licensing checkpoint, and the response to replicative stresses. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J Biol Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V, Tibolla M, Tenca P, Brotherton D, Albanese C, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4:357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JC, Detweiler CS, Li JJ. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci U S A. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J, Pickering A, Han B, Li L, Zheng J, Zhang J, Zhang Y, Fei P. Basal level of FANCD2 monoubiquitination is required for the maintenance of a sufficient number of licensed-replication origins to fire at a normal rate. Oncotarget. 2014;5:1326–1337. doi: 10.18632/oncotarget.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlides SC, Huang KT, Reid DA, Wu L, Blank SV, Mittal K, Guo L, Rothenberg E, Rueda B, Cardozo T, et al. Inhibitors of SCF-Skp2/Cks1 E3 ligase block estrogen-induced growth stimulation and degradation of nuclear p27kip1: therapeutic potential for endometrial cancer. Endocrinology. 2013;154:4030–4045. doi: 10.1210/en.2013-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- Rego MA, Kolling FW, Vuono EA, Mauro M, Howlett NG. Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood. 2012;120:2109–2117. doi: 10.1182/blood-2012-02-410472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Räschle M, Knipscheer P, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi V, Ferrari S, Ferraresi R, Cossarizza A, Grande A, Zappavigna V. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol Cell Biol. 2009;29:5775–5788. doi: 10.1128/MCB.00509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Kinney JB, Lengronne A, Pasero P, Stillman B. Domain within the helicase subunit Mcm4 integrates multiple kinase signals to control DNA replication initiation and fork progression. Proc Natl Acad Sci U S A. 2014;111:E1899–E1908. doi: 10.1073/pnas.1404063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007a;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007b;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV. Cohesin acetylation speeds the replication fork. Nature. 2009;462:231–234. doi: 10.1038/nature08550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumini E, Plevani P, Muzi-Falconi M, Marini F. Physical and functional crosstalk between Fanconi anemia core components and the GINS replication complex. DNA Repair (Amst) 2011;10:149–158. doi: 10.1016/j.dnarep.2010.10.006. [DOI] [PubMed] [Google Scholar]
- van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Yamada M, Watanabe K, Mistrik M, Vesela E, Protivankova I, Mailand N, Lee M, Masai H, Lukas J, Bartek J. ATR-Chk1-APC/CCdh1-dependent stabilization of Cdc7-ASK (Dbf4) kinase is required for DNA lesion bypass under replication stress. Genes Dev. 2013;27:2459–2472. doi: 10.1101/gad.224568.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo JE, Lee EH, Hendrickson E, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.