Abstract
One factor limiting the success of non-viral gene therapy vectors is the relative inability to target genes specifically to a desired cell type. To address this limitation, we have begun to develop cell-specific vectors whose specificity is at the level of the nuclear import of the plasmid DNA. We have recently shown that nuclear import of plasmid DNA is a sequence-specific event, requiring the SV40 enhancer, a region known to bind to a number of general transcription factors. From these studies we developed a model whereby transcription factor(s) bind to the DNA in the cytoplasm to create a protein–DNA complex that can enter the nucleus using the protein import machinery. Our model predicts that by using DNA elements containing binding sites for transcription fac tors expressed in unique cell types, we should be able to create plasmids that target to the nucleus in a cell-specific manner. Using the promoter from the smooth muscle gamma actin (SMGA) gene whose expression is limited to smooth muscle cells, we have created a series of reporter plasmids that are expressed selectively in smooth muscle cells. Moreover, when injected into the cytoplasm, plas mids containing portions of the SMGA promoter localize to the nucleus of smooth muscle cells, but remain cytoplas mic in fibroblasts and CV1 cells. In contrast, a similar plasmid carrying the SV40 enhancer is transported into the nuclei of all cell types tested. Nuclear import of the SMGA promoter-containing plasmids could be achieved when the smooth muscle specific transcription factor SRF was expressed in stably transfected CV1 cells, supporting our model for the nuclear import of plasmids. Finally, these nuclear targeting sequences were also able to promote increased gene expression in liposome- and polycation-transfected non-dividing cells in a cell-specific manner, similar to their nuclear import activity. These results pro vide proof of principle for the development of cell-specific non-viral vectors for any desired cell type.
Keywords: nuclear import, non-viral vectors, cell specificity, plasmid DNA
Introduction
A variety of techniques and vectors for gene therapy have been developed to target genes to cells, including replication-deficient recombinant retroviruses, adeno-viruses, and adeno-associated viruses, as well as nonviral vectors such as ligand–DNA conjugates or DNA lipoplexes. However, most targeting techniques developed to date have only addressed the ability to internalize the DNA into the cytoplasm of the cell. This is especially true in the case of vectors that are to be targeted to a unique cell or tissue type. A fact that has been largely overlooked until recently is that gene therapy relies on the ability of exogenous genes to enter the nucleus, regardless of how the DNA or RNA is targeted to the cell. Once within the cytoplasm, the gene must become nuclear to be transcribed, replicated and maintained either in an integrated or episomal state, yet there has been little attention directed toward discovering and exploiting the mechanisms used by the cell to direct DNA to the nucleus.
Recent work from our laboratory has begun to address the nuclear targeting and entry of plasmid DNA. Using cultured cells, we have shown that plasmid DNA is able to enter the nuclei of cells in the absence of cell division and its accompanying nuclear envelope breakdown.1 As for all other macromolecular exchange between the cytoplasm and nucleus (for a review, see Ref. 2), DNA nuclear entry appears to be mediated by the nuclear pore complex.1,3–5 Furthermore, we have identified a 366 bp sequence of DNA containing the simian virus 40 (SV40) origin of replication and early promoter that is absolutely necessary for the nuclear entry of plasmid DNA in non-dividing cultured cell lines derived from monkey, rat, mouse, hamster and human origin, as well as non-transformed primary cells from rat, chicken and human tissues.1 Thus, nuclear import of plasmid DNA is signal-dependent and occurs in all eukaryotic cells tested to date. We have further localized this DNA nuclear targeting signal to regions within the 366 bp DNA fragment, and have found that regions of DNA containing multiple binding sites for various general transcription factors mediate the best nuclear import of plasmid DNA. These results demonstrate that transport of DNA into the nucleus is sequence-specific and it is our hypothesis that this import is mediated by sequences containing binding sites for eukaryotic transcription factors.6 Since transcription factors bind to specific DNA sequences and contain nuclear localization signals (NLSs) for their nuclear import, it is likely that these proteins coat the DNA with NLSs, thereby allowing the DNA–protein complex to utilize the NLS-mediated import machinery for nuclear entry.
A corollary to this hypothesis is that expression of the transcription factor is necessary for DNA import. More-over, if a plasmid contains the binding site for a transcription factor that is expressed in a specific cell type such as a smooth muscle cell (SMC), but not in any other cell type, the DNA should be targeted to the nucleus only in SMCs. The ubiquitous transcription factors that may participate in SV40 DNA nuclear import such as AP1, AP2 and NF-kB, would be replaced in the smooth muscle cell example by smooth muscle cell-specific transcription factors whose binding sites are on the plasmid we want to target to the nucleus.
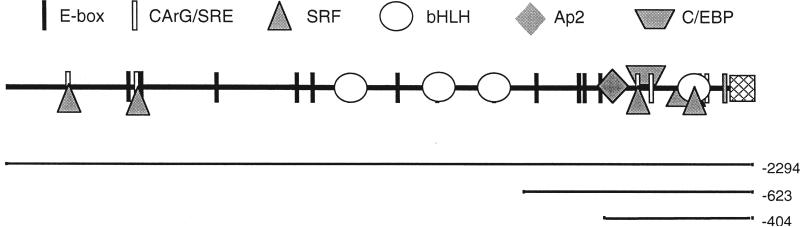
Smooth muscle gamma-actin (SMGA) is an actin iso-form that is expressed only in visceral and vascular SMCs.7,8 This selective expression is transcriptional and depends on the presence of multiple positive and negative DNA elements that interact with combinations of ubiquitous and cell type-specific transcription factors.8 As in the vascular smooth muscle alpha-actin gene and the promoters of all smooth muscle genes examined to date, two cis elements are needed for smooth muscle cell-specific expression, the CArG/SRE motif and the E-Box.9–13 The avian SMGA gene promoter contains six CArG/SRE motifs (Figure 1), four of which are known to be conserved in structure and location with the human and mouse SMGA genes.14,15 Moreover, serum response factor (SRF), a highly conserved protein that is developmentally regulated in smooth muscle myogenesis and which binds to DNA as a heterodimer with multiple distinct partners, binds four of the six CArG/SRE motifs of the SMGA promoter.7,8,16,17 The promoter also contains 13 E-box motifs, three of which bind to a class of transcription factors, the bHLH family, exemplified by the muscle-specific factor MyoD.18 Thus, the SMGA promoter provides us with an excellent candidate with which to test our model. In this report, we have tested our hypothesis using DNA sequence elements from the SMGA promoter. Our studies clearly demonstrate that nuclear import of plasmids containing the SMGA promoter occurs only in smooth muscle cells. Consistent with the main tenet of our hypothesis, specific nuclear import of DNA containing segments of the SMGA promoter can be induced in non-smooth muscle cells via the forced expression of SRF. Thus smooth muscle cell-specific nuclear import of plasmid DNA is achieved, at least in part, by the expression of transcription factors within these cells.
Figure 1.
Organization of cis-acting elements within the SMGA promoter. Cartoon depicting the smooth muscle specific transcription factor binding sites identified by sequence homology and experimentally within the SMGA promoter14–16,18 and constructs used in these experiments.
Results
Smooth muscle-specific gene expression of the smooth muscle gamma-actin gene
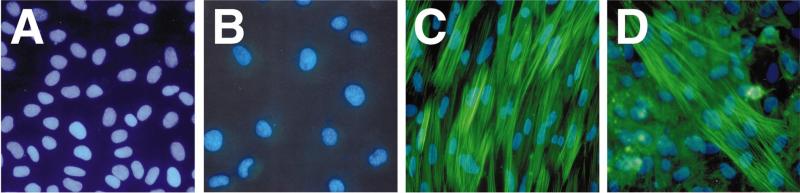
To test our hypothesis that DNA nuclear import could be made cell specific, we first established that the SMGA gene was indeed expressed in a smooth muscle-specific manner in our cell systems. Cells were grown as described and the presence of SMGA was determined by immunofluorescence (Figure 2). SMGA was not detected in CV1 cells (Figure 2A) or chicken embryo fibroblasts (Figure 2B). Significant expression was detected in differentiated cultures of chicken gizzard smooth muscle cells (Figure 2C) and in human pulmonary artery smooth muscle cells (Figure 2D). Both intimal and medial smooth muscle cells from the human pulmonary artery displayed similar staining patterns with the anti-SMGA antibody (not shown). Thus, the endogenous SMGA gene is expressed in an appropriate manner in our cell systems and supports previous findings with chicken culture systems.7,8,16
Figure 2.
Expression of SMGA in smooth muscle and non-smooth muscle cells. The endogenous expression of SMGA was characterized in the cell types used in this study. (A) CV1 cells, (B) chicken embryo fibroblasts, (C) differentiated chicken gizzard SMCs and (D) human pulmonary artery intimal SMCs were fixed in 3% paraformaldehyde, permeabilized with methanol:acetone, and stained for the presence of SMGA by immunofluorescence. FITC-labeled secondary antibodies were used to detect SMGA and the nuclei were stained with DAPI.
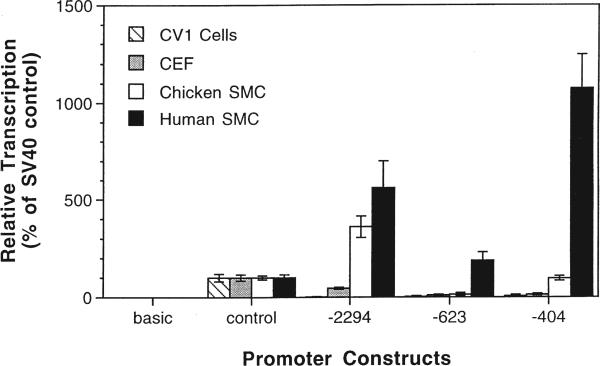
We next created a series of promoter constructs containing various lengths of the SMGA promoter upstream of either a CAT or luciferase reporter gene. By transfecting cells with CAT and luciferase reporter constructs, we analyzed the transcriptional activity of the SMGA promoter. For this promoter to function as a model in our system, its activity must be cell-specific when present on a multicopy plasmid. Three SMGA constructs were chosen for study: (1) the full-length SMGA promoter (2294 nucleotides); (2) a construct containing 623 nucleotides of the SMGA promoter that contains portions of the SMGA negative regulatory region; and (3) the first 404 nucleotides of the SMGA promoter that contains the SMGA specifier domain.7,8,16,18 All three constructs contain binding sites for SMC-specific and general transcription factors. When the full-length or truncated SMGA promoter was used, maximal expression of the reporter was observed in differentiated chicken SMCs, whereas less than 10% of these levels of expression was observed in either fibroblasts or CV1 epithelial cells (Figure 3). The full-length promoter gave maximal expression and the −404 construct displayed approximately 50% of this activity in chicken visceral SMCs. Interestingly, these activities were reversed in human vascular SMCs, with the shorter construct giving maximal activity around 10-fold higher than the SV40 promoter driven control and the full-length promoter giving about 50% of this, or five times greater expression than the control. pCAT-623 showed very little expression, supporting our previous findings that the region from −950 to −404 contains inhibitory sequences that modulate SMGA expression.7,8,16,18 Expression of the SV40 promoter/enhancer constructs was roughly equivalent in all cell types tested. Plasmids containing no promoter or enhancer elements (pCAT-basic and pGL2-basic) gave no activity. Since the SMGA promoter contains a number of conserved putative regulatory elements including several CArG/SRE and E-box motifs (Figure 1), we expect that these sequences mediate transcription through trans-factors.18 Based on these results, the SMGA promoter and the SV40 enhancer were prime candidates to test our model for plasmid DNA nuclear import: one should be cell-specific and the other should be promiscuous in their import activity.
Figure 3.
The SMGA promoter is expressed preferentially in differentiated SMCs. Cells were transfected by DEAE dextran with reporter plasmids containing no promoter (basic), the SV40 promoter and enhancer (control), or various lengths of the SMGA promoter. Forty-eight hours after transfection, cell lysates were prepared and CAT activity was measured as described in Materials and methods. The values are the means of two to four independent experiments performed in triplicate.
Plasmid DNA localizes to the nuclei of smooth muscle cells in a sequence-specific manner
We have previously shown that the nuclear import of plasmid DNA is sequence specific: plasmids carrying portions of the SV40 promoter/enhancer region are transported into the nucleus while similar plasmids lacking this sequence remain cytoplasmic.1 All of these experiments were performed with transformed cell lines from a variety of organisms and tissues. Thus, we wanted to demonstrate that primary cultures of non-transformed smooth muscle cells from chicken viscera and human vasculature were capable of similar sequence-specific nuclear import of plasmid DNA.
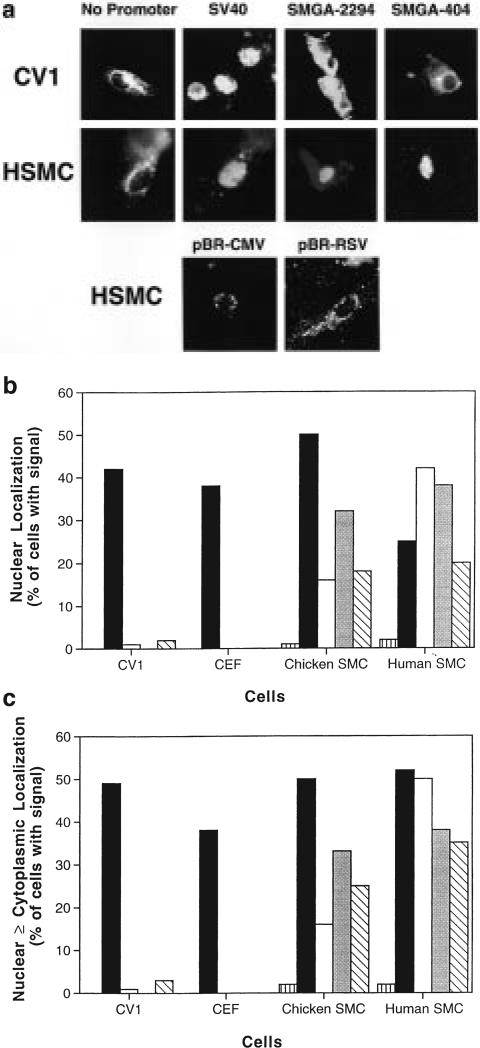
When DNAs were micro-injected into the cytoplasm, both visceral and vascular smooth muscle cells were able to transport SV40 DNA and a pCAT plasmid containing the SV40 promoter/enhancer into their nuclei, while a plasmid lacking the SV40 sequences (pCAT-basic, denoted ‘no promoter’) was unable to localize to the nucleus (Figure 4). No differences were detected between replicating chicken SMCs and post-mitotic, differentiated SMCs; SV40 DNA and pCAT-SV40 localized to the nuclei of both cultures and pCAT-basic remained completely cytoplasmic. Virtually identical results were seen in chicken embryo fibroblasts and CV1 cells (Figure 4), confirming our previous results. In addition, several other strong viral promoters that have been shown not to support plasmid nuclear import in CV1 cells6 were tested for their ability to mediate nuclear import in SMCs. As expected, both the CMV immediate–early promoter/enhancer and the Rous sarcoma virus LTR remained cytoplasmic at 8 h in micro-injected SMCs (Figure 4a). Since localization is assayed by in situ hybridization and does not rely on gene expression, the presence or absence of promoter sequences will not affect our findings.
Figure 4.
Sequence- and cell-specific nuclear import of plasmid DNAs. At least 100 cells of each indicated cell type were cytoplasmically injected with pCAT reporter plasmids containing no promoter (pCAT-basic  ), the SV40 promoter and enhancer (pCAT-control ■), or various lengths of the SMGA promoter (pCAT-2294 □, pCAT-623
), the SV40 promoter and enhancer (pCAT-control ■), or various lengths of the SMGA promoter (pCAT-2294 □, pCAT-623  , and pCAT-404
, and pCAT-404  ). Eight hours later, the subcellular localization of the plasmid DNA was detected by in situ hybridization. (a) Representative cells showing nuclear or cytoplasmic localization of the plasmids. (b) The percentage of cells displaying only nuclear staining of the plasmids were determined from three to five independent experiments for each cell type. (c) Cells showing nuclear only (as in b) in addition to those with both nuclear and cytoplasmic staining were determined as in (b).
). Eight hours later, the subcellular localization of the plasmid DNA was detected by in situ hybridization. (a) Representative cells showing nuclear or cytoplasmic localization of the plasmids. (b) The percentage of cells displaying only nuclear staining of the plasmids were determined from three to five independent experiments for each cell type. (c) Cells showing nuclear only (as in b) in addition to those with both nuclear and cytoplasmic staining were determined as in (b).
Cell-specific DNA nuclear import in smooth muscle cells
To test our hypothesis that DNA nuclear import can be made cell-specific, we next micro-injected the SMGA promoter constructs into the cytoplasm of CV1, chicken embryo fibroblasts and differentiated chicken and human smooth muscle cells, and measured the nuclear localization of the plasmids by in situ hybridization 8 h after injection (Figure 4). Unlike plasmids containing portions of the SV40 promoter/enhancer, essentially none of the plasmids containing portions of the SMGA promoter were imported into the nuclei of CV1 cells or chicken embryo fibroblasts; between 98 and 100% of the injected DNA remained in the cytoplasm. The sole exception was pCAT-404, which showed some nuclear and cytoplasmic localization in fewer than 5% of the injected CV1 cells. In chicken embryo fibroblasts, no cells were detected with this plasmid in the nucleus.
In contrast to the lack of import seen in CV1 cells and chicken embryo fibroblasts, nuclear import of the full-length SMGA promoter construct was detected in human SMCs. Moreover, the level of nuclear import observed in human SMCs with pCAT-2294 was almost equal to that obtained with the SV40 promoter/enhancer, with import occurring in 43% of the cells. Interestingly, when the same construct was injected into the cytoplasm of chicken SMCs, it only localized to the nucleus in 16% of the cells with detectable signal. When decreasing lengths of the SMGA promoter were tested for their import capacity in human SMCs, they displayed somewhat decreased nuclear import compared with the full-length promoter. Even with the smallest promoter fragment tested, however, nuclear import was seen at the same level as the SV40 promoter in these cells. The percentages of chicken SMCs displaying nuclear localization of pCAT-623 and pCAT-404 were almost indistinguishable from those of the same plasmids in the human SMCs. Thus, in chicken SMCs, the smaller promoter fragments were imported into the nuclei more efficiently than the full-length promoter. Essentially similar patterns of nuclear import activity were seen when the numbers of cells displaying both nuclear and cytoplasmic expression in the same cell were scored (Figure 4c). In this case, only minimal numbers of cell showed plasmid in both compartments; the vast majority of cells displayed either all nuclear or all cytoplasmic DNA localization, a pattern similar to what we have reported in TC7, HeLa, CHO and Vero cells.1
SRF plays a role in the nuclear localization of SMGA promoter containing plasmids
The fact that all three SMGA promoter constructs were able to mediate nuclear import uniquely in SMCs supported our model for plasmid nuclear targeting. Thus, it is likely that a unique transcription factor, or set of factors, expressed in SMCs but not other cell types may be responsible for transporting the DNA–protein complex into the nucleus. Since all three lengths of the SMGA promoter supported nuclear import, the factor(s) must bind to the smallest fragment common to all constructs, namely the proximal 404 bp. Further, the ability of the longer fragments to drive greater nuclear localization suggests that additional binding sites for these factor(s) or additional factors are present in the distal regions of the promoter. One candidate that fulfills these criteria is SRF which binds to CArG/SRE sites within the 404 bp fragment and throughout the SMGA promoter.16,18 Thus, we expressed the SRF gene from the CMV promoter in stably transfected CV1 cells. SRF was expressed in nine clones as detected by Western blot and no SRF was detected in untransfected CV1 cells (not shown). We have previously demonstrated that when SRF is expressed in transfected CV1 cells, expression of a full length SMGA promoter reporter construct is stimulated in a dose-dependent manner.16
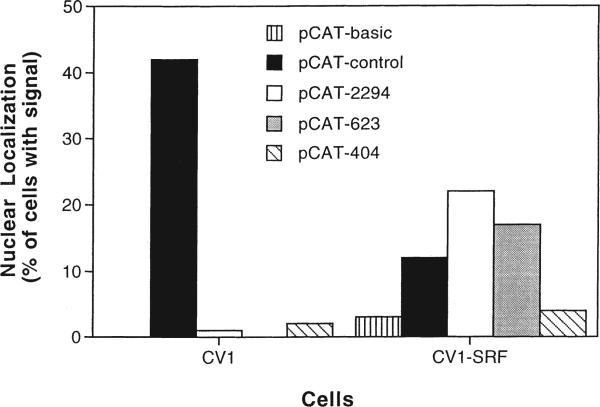
When pCAT-SV40 was micro-injected into the cytoplasm of the SRF-expressing CV1 cells, it localized to the nucleus in 10 to 15% of the injected cells, a level two- to four-fold lower than detected in CV1 cells, fibroblasts, or SMCs, while the plasmid with no promoter remained almost exclusively in the cytoplasm (Figure 5). The full-length SMGA promoter construct localized to the nuclei of 22% of the cells, compared with only 1% in non-transfected CV1 cells. Plasmids containing the two smaller fragments of the SMGA promoter showed lower capacity for nuclear targeting. Although all the SMGA reporter plasmids were imported into the nuclei of the SRF-expressing CV1 cells, the level of import was reduced by a factor of two for all plasmids except the smallest construct in which case import was reduced five-fold compared with human and chicken SMCs. These results indicate that SRF does indeed play a role in SMC-specific nuclear import of SMGA promoter-containing plasmids.
Figure 5.
SRF can mediate the nuclear localization of SMGA promoter-containing plasmids. SRF-expressing stable transfectants of CV1 cells (CV1-SRF) were isolated and used for micro-injection experiments. The various pCAT plasmids were injected into the cytoplasm of CV1-SRF cells and their subcellular localization was determined 8 h later by in situ hybridization. The percentage of cells showing nuclear localization of the plasmids were based on three independent experiments. Plasmid localization in non-transfected CV1 cells (Figure 4) are shown for comparison.
Nuclear targeting sequences stimulate gene expression in transfected non-dividing cells
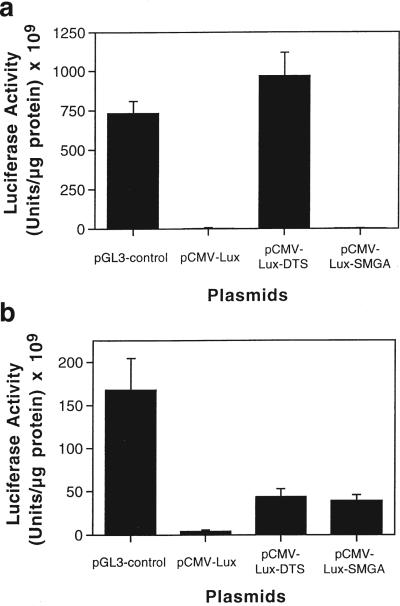
Since the ultimate goal is to exploit these nuclear targeting signals to increase gene transfer to cells, we tested whether these DNA sequences also functioned to stimulate gene expression in transfected cells. Constructs were made taking advantage of the finding that the CMV promoter does not support nuclear import in CV1 cells or SMCs. Thus, it was used to drive expression of the firefly luciferase gene. Either a 240 bp fragment of the SV40 promoter and enhancer region (DNA nuclear targeting sequence, DTS) or the full-length 2294 bp SMGA promoter was cloned downstream of the luciferase gene, so as not to play a direct role in transcription. However, since both DNA sequences have classic enhancer activity, any increase in gene expression could be due to a combination of increased nuclear localization and/or transcription. Since the greatest effect of the nuclear targeting sequences is expected in non-dividing cells, CV1 cells were arrested with aphidicolin before and during the transfections. When CV1 cells were transfected using the cationic lipid lipofectin, significant activity was seen with pGL3-control, a luciferase vector essentially identical to the pCAT-control plasmid (Figure 6a). When the CMV-driven vector lacking any nuclear targeting sequence was transfected into cells, very little activity was detected, but when the SV40 promoter/enhancer region was included, gene expression increased by over 500-fold. Similar results were obtained with arrested HeLa cells and HUVECs (not shown). In contrast, the SMGA-containing construct had no effect on CMV-driven luciferase expression, as expected, presumably because it does not cause nuclear import in these cells.
Figure 6.
The presence of nuclear targeting sequences increases gene expression in transfected cells in a cell-specific manner. (a) CV1 cells were arrested in G1 by 24 h pretreatment with 50 μm aphidicolin and transfected with lipofectin. Wells of a 12-well dish received 3 μl of lipofectin complexed with 1 μg of plasmid. Aphidicolin remained present throughout the experiment. Luciferase activities from quadruplicate wells were measured in lysates prepared at 48 h after transfection and normalized to total protein. (b) Differentiated human SMCs were transfected with DEAE-dextran as described in Materials and methods. Luciferase activities from quadruplicate wells were measured as in (a). These results are representative of at least three independent experiments.
When differentiated human SMCs were transfected using DEAE-dextran, high levels of expression were detected with the SV40 promoter/enhancer driven pGL3-control plasmid (Figure 6b). As in arrested CV1 cells, the pCMV-lux plasmid showed very little activity in SMCs, but the SV40 DTS-containing vector gave a 10-fold stimulation over the parent pCMV-lux. In contrast to the results obtained with CV1 cells, pCMV-lux-SMGA gave levels of gene expression in SMCs similar to those of the SV40-containing pCMV-lux-DTS plasmid. These results indicate that the incorporation of these DNA nuclear targeting sequences into plasmids enhances gene expression in liposome- or polycation-transfected cells according to their cell specificity. Namely, while the SV40 sequence which is active for nuclear import in all cells tested stimulated expression in both non-smooth muscle cells and SMCs, the SMGA sequence acted only in SMCs.
Discussion
The majority of techniques and vectors developed to date to target genes to specific cell types have focused on specific interactions at the cell surface and at the transcriptional level. We have taken a different approach and instead have begun to characterize and exploit the mechanisms of nuclear import of plasmid DNA. In a previous study, we demonstrated that plasmid DNA can gain access to the intact nucleus by being transported through the nuclear pore complex in a DNA sequence-specific manner.1 Based on the ability of the SV40 promoter/enhancer to mediate plasmid DNA nuclear import, we hypothesized that other promoters may have the same activity, and moreover, that this could be a novel approach with which to design cell-specific nuclear targeting DNA vectors. In the present report, we have shown that plasmid DNA containing the chicken smooth muscle gamma-actin promoter is selectively transported into the nuclei of differentiated smooth muscle cells; transport was not observed in any other cell type tested. Our results indicate that this is due at least in part to the transcription factor SRF, since when it is expressed in non-smooth muscle cells, the SMGA promoter-containing plasmids are able to target to the nucleus. Furthermore, the ability of these nuclear targeting sequences to act in liposome- and polycation-transfected cells suggests that their incorporation into non-viral vector systems will enhance gene transfer. These results are the first demonstration of cell-specific nuclear import of plasmid DNA and potentially will lead to the creation of improved vascular gene therapy vectors that are both cell-specific and capable of greater gene transfer efficiencies.
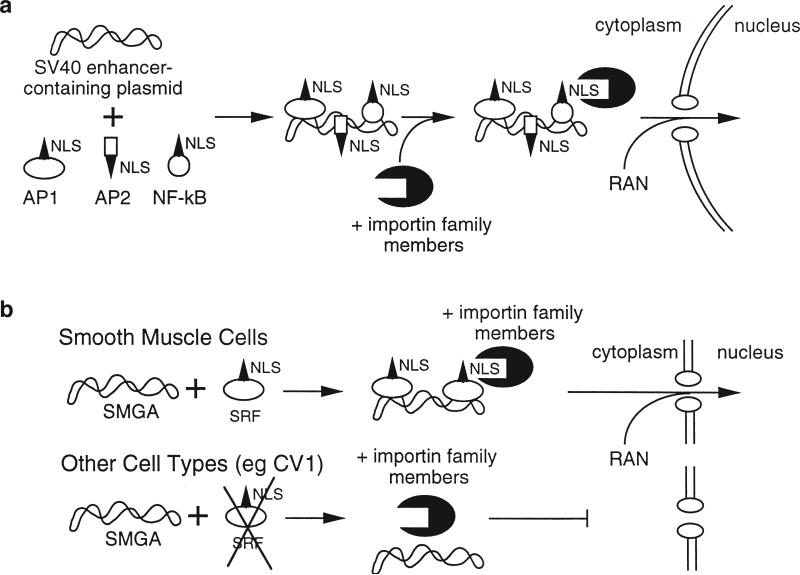
Based on the DNA sequences that appear to be required for the nuclear import of plasmid DNA, we have proposed a model for the import reaction (Figure 7). In the case of general nuclear import, the DNA fragment that gave the best transport activity contained the 72 bp enhancer repeat of the SV40 early promoter,6 a region rich in consensus binding sites for numerous transcription factors. These include AP1, AP2, AP3, AP4, NF-kB, Oct-1 and SP1, among others. Since transcription factors, like all proteins, are translated in the cytoplasm, they must target to the nucleus either after synthesis or upon proper stimulation. To enter the nucleus they must either contain NLSs or form oligomers with other proteins that contain an NLS. Since transcription factors bind to specific DNA sequences, it is possible that if DNA containing the appropriate sequences is present in the cytoplasm, it can be complexed by these proteins, thus coating the DNA with protein NLSs. The NLSs present in this nucleoprotein complex can then interact with the appropriate importin NLS receptor and enter the nucleus using the normal nuclear protein import machinery.2 Whether only one specific transcription factor or multiple factors are capable of mediating import is unknown. We are currently pursuing experiments with a reconstituted permeabilized cell system to identify the protein(s).
Figure 7.
Model for plasmid DNA nuclear import. (a) Sequence-specific nuclear import of SV40 promoter/enhancer containing plasmid DNA occurs in all cell types tested. The SV40 enhancer contains binding sites for ubiquitously expressed NLS-containing transcription factors. Since these proteins are synthesized in the cytoplasm, once plasmid DNA has entered the cytoplasm by transfection, injection, or infection, the newly synthesized proteins can bind to the plasmid DNA to form a DNA–protein complex. Thus, the DNA is coated with NLSs, allowing the NLS-mediated import machinery to recognize DNA as a substrate and target it to the nucleus. (b) Smooth muscle cell-specific nuclear import is achieved by incorporating DNA sequence elements that contain binding sites for transcription factors that are expressed uniquely in SMCs, such as SRF or certain bHLH proteins that bind to E-boxes. These factors act analogously to the ubiquitous transcription factors to couple the DNA to the NLS-mediated nuclear import machinery.
Several recent reports have suggested similar models for the NLS-dependent nuclear import of DNA. In experiments using EBNA-expressing Epstein–Barr virus (EBV) transformed cells, it was demonstrated that plasmids containing the EBV oriP site were up to 100-fold more efficient at gene expression when micro-injected into the cytoplasm or transfected into cells as compared with similar plasmids lacking the oriP site.19 Since the EBNA-1 protein localizes to the nucleus and binds the oriP site, it was hypothesized that this protein mediates the increased gene expression by stimulating nuclear localization of the oriP-containing plasmids. Similar NLS-mediated nuclear import models have been suggested for several viral genomes including HIV, hepadnavirus, adenovirus and SV40 where the NLS-containing proteins include matrix proteins, polymerases, and capsid proteins carried in the incoming virus particle.20–25 Several reports have also shown that synthetic NLS peptides can act to increase nuclear localization of plasmids when either coupled to DNA covalently or simply mixed with the DNA.5,26,27 However, in all of these cases, it is viral or other exogenous proteins or peptides that are needed for the nuclear targeting of the DNA, as opposed to the exploitation of the existing proteins present in the cell, as we report here.
In the case of the cell-specific nuclear import, we have begun a detailed analysis of the chicken SMGA promoter using transfection and gel shift assays to determine which elements are utilized in the cell-specific regulation of this gene. Experiments from our laboratories have indicated that motifs within the proximal promoter of the SMGA gene do play a role in the cell-specific regulation of the gene.16 To date, we have shown that while the four CArG/SRE boxes located within the proximal approximately 400 bp of the SMGA promoter bind nuclear factors, only two of them bind to SRF in vitro.18 That SRF, when expressed in CV1 cells, can facilitate the nuclear localization of plasmids containing portions of the SMGA promoter strongly supports our model for plasmid nuclear import. Since the NLS and the DNA-binding domain or SRF are spatially distinct, SRF should act well as an adapter between the plasmid DNA and the importin nuclear import machinery.28,29 In experiments using SRF-expressing CV1 transfectants, we routinely observed decreased nuclear localization of micro-injected plasmids, including the SV40 promoter/enhancer containing pCAT-control. While it is possible that the aberrant expression of SRF caused a down-regulation of general plasmid nuclear import, perhaps by transcriptionally regulating the expression of one or more transcription factors needed for SV40 DNA nuclear import, it is more likely that the decreased nuclear import of the pCAT-control plasmid was due to experimental variation. Regardless of this, however, the general trend in import seen with the different plasmids is similar in SRF-expressing CV1 cells and SMCs. In all cases, the least capacity for import is detected with the smallest promoter fragment. This suggests that additional transcription factors can bind to the larger fragments increasing their ability to enter the nucleus. In fact, binding of proteins in addition to SRF have been mapped upon the SMGA promoter (Figure 1) and ongoing analyses will determine how these different binding sites regulate expression and nuclear import.8,16,18 Candidates for these factors include Nkx family and other proteins that bind to the E-boxes present in the modifier domain of the promoter.16,18 Similarly, we have previously shown that if SRF is transiently expressed in CV1 cells, transcription of the SGMA promoter is stimulated, although not to the levels seen in SMCs.16 This suggests that while SRF is capable of acting alone to increase transcription (and import), additional proteins, including SRF dimerization partners or other SMC-specific transcription factors, are needed for maximal activity.
Based on these findings, it may be possible to assemble gene delivery vectors that can be made to localize to the nuclei of any desired cell type by incorporating sequences that contain binding sites for transcription factors that are expressed uniquely in the desired cells (Figure 7). Examples could include incorporating a promoter or portions thereof that contain consensus binding sites for Pax-6, a homeodomain transcription factor whose expression is limited to the developing brain and the eye, or AP-2β, a transcription factor expressed preferentially in the adult eye, kidney and skin into a vector to cause its specific nuclear import in cells of the eye.30–32 Alternatively, sequences from the skeletal muscle alpha-actin promoter, which expresses only in skeletal muscle cells, could be incorporated into a plasmid to create a vector that only targets to the nuclei of skeletal muscle cells.9 For specific nuclear targeting in hematopoietic stem cells, portions of the promoter immediately upstream of the β-globin gene which contain binding sites for GATA-1 and other transcription factors that are only found in hematopoietic cells could be incorporated into the new vector.33 Moreover, as illustrated by our study, these binding elements do not necessarily need to participate in transcription to facilitate plasmid DNA nuclear import. Thus, any of a number of combinations can be envisioned that can (1) target to the nuclei of cells in the absence of cell division, and (2) do so in a cell-specific manner. An added advantage of this approach is that the use of vectors containing these type of cell-specific DNA targeting sequences will ensure safety since nuclear import and resulting gene expression will occur only in target cells. Whether all of these promoters will work to mediate nuclear import is unknown. Indeed, two very strong viral promoters, the human cytomegalovirus immediate–early promoter/enhancer and the Rous sarcoma virus LTR have no nuclear import activity.6 However, the ability of the SV40 enhancer and the SMGA promoter to direct nuclear import strongly suggests that other sequences will function as well.
Materials and methods
Plasmids
The plasmid pDD180 was created by digesting pBR322 with EcoRI and HindIII and inserting a multiple cloning site (HindIII–XbaI–NcoI–SmaI–KpnI–EcoRI) to facilitate cloning. The 870 bp immediate–early promoter/enhancer from the human cytomegalovirus (hCMViep) was removed from pBK-CMV (Stratagene, San Diego, CA, USA) by digestion with AflIII and EcoRI and cloned into pDD180 to create pBR-CMV. The 600 bp promoter contained within the Rous sarcoma virus long terminal repeat (RSV LTR) was excised from pRc/RSV (Invitrogen, San Diego, CA, USA) by digestion with BglII and HindIII and cloned into pDD180 to create pBR-RSV.
The promoter for the smooth muscle gamma actin (SMGA) gene was obtained from a genomic clone of the chicken SMGA gene.7 Briefly, a chicken genomic library was constructed in EMBL-3 phage and was screened for SMGA clones using the 32P-dCTP labeled full length cDNA, SMGA15–1, as a probe.34 A unique XbaI restriction site was introduced at position +25 in exon 1 of the SMGA gene by PCR to facilitate cloning. The full length promoter was subcloned into the pCAT-basic vector (Promega, Madison, WI, USA) using the EcoRI and XbaI sites (Figure 1). This plasmid was named pCAT-2294. Using existing ApaI and SmaI restriction sites within the promoter, truncated SMGA promoters containing only the 623 bp or 404 bp immediately upstream of the transcriptional start were also cloned into the pCAT-basic vector to create pCAT-623 and pCAT-404. A similar series of transcription reporter plasmids (pGL-2294 and pGL-404) were created in pGL3-basic (Promega) that express luciferase. pCAT-control contains the SV40 early promoter (nts 5171–130) and enhancer (nts 130–270) and was purchased from Promega.
The human SRF gene was removed from pCGNSRF16,35,36 and inserted into the multiple cloning site of pBK-CMV (Stratagene) to create pBK-SRF.
Plasmid pCMV-lux was made by exchanging the NheI–BamHI fragment of pRL-CMV (Promega) for that of pGL3-basic (Promega) to create a plasmid expressing the firefly luciferase gene from the CMV promoter. pCMV-lux-DTS was constructed similarly, but contained the NheI–BamHI fragment from pGL3-enhancer (Promega) and contains the SV40 enhancer and GC boxes (SV40 nts 44 to 268) downstream of the luciferase gene. pCMV-lux-SMGA was constructed by removing the 2.3 kb SMGA promoter from pCAT-2294 by digestion with XbaI and HindIII followed by treatment with Klenow and insertion into the HpaI site of pCMV-lux, downstream of the luciferase gene.
Plasmid DNA was purified by either alkaline lysis and subsequent CsCl gradient centrifugation or Promega maxiprep columns. DNA purified in either manner displayed the same transfection efficiency and intracellular distribution after cytoplasmic micro-injection.
Cell culture and micro-injection
CV1 cells, a subline of African Green monkey kidney epithelium were grown on coverslips in DMEM containing 10% fetal bovine serum and cytoplasmically micro-injected as described.37 Human pulmonary artery intimal and medial smooth muscle cells were a generous gift from Dr Paul Babal (Department of Pathology, University of South Alabama) and were grown in DMEM containing 10% fetal bovine serum. Primary chicken gizzard smooth muscle cell cultures were established based on the method of Cambell et al with modifications as described.7,38 Cells were grown on collagen type IV-coated, etched coverslips in medium 199 supplemented with 20 % fetal bovine serum, 1 μm insulin, 2 mM l-gluta-mine and 100 units/ml of penicillin/streptomycin. At confluency, cells were differentiated by switching the culture medium to DMEM/F12 supplemented with 2 mm l-glutamine, 1 μm insulin, 100 units/ml of penicillin/streptomycin, 5 units/ml apo-transferrin and 0.2 mm ascorbic acid. Cells were maintained in this medium for 48 h before micro-injection. Chicken embryo fibroblasts were obtained from the American Type Culture Collection and grown as described.39
Purified protein-free DNA was suspended in phosphate-buffered saline and injected at a concentration of 0.25 mg/ml. By micro-injecting radionuclides of known specific activity into a number of cells, we have determined that 3 × 10−10 ml is delivered by micro-injection. This corresponds to approximately 10 000 molecules of plasmid injected per cell.
Transfections
Cells were grown in 12-well tissue culture dishes to 60–70% confluency (CV1, fibroblasts, and replicating SMCs) or were differentiated upon reaching confluency and grown an additional 3 days (differentiated SMCs). Transfections were performed with DEAE-dextran as described previously.16 Cells lysates were prepared 48 h later and CAT activity was measured using a Quanticat kit (Amersham, Arlington Heights, IL, USA). To select stable transfectants, CV1 cells were transfected with pBKSRF using lipofectin (Life Technologies, Gaithersburg, MD, USA) and grown in DMEM containing 10% FBS for 48 h. The cells were trypsinized and replated into G418-containing medium and grown until colonies were detected. G418-resistant clones were isolated with cloning rings and subcultured in the presence of drug. Production of SRF in nine independent clones was confirmed by Western blots using an anti-SRF monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
For transfections of growth-arrested CV1 cells, the cells were grown to 70% confluency and treated for 24 h with 50 μM aphidicolin before lipofectin-mediated transfections. DNA was added to the cells in serum-free DMEM for 4 h and aphidicolin-containing growth medium was added back to the cells. All transfections were performed in triplicate or quadruplicate and normalized to the amount of protein per well. Luciferase activity was measured using the Luciferase Assay System (Promega).
In situ hybridization and indirect immunofluorescence
In situ hybridizations were performed as described.1 After microinjection and incubation for the appropriate time, the cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline at 23°C for 1 min, fixed in acetone:methanol (1:1) at −20°C for 5 min and incubated in 70% formamide in 2 × SSC at 70°C for 2 min to denature the DNA. The cells were then hybridized overnight at 37°C with a fluorescently labeled probe. All samples were treated with RNaseH (8 U/ml) after hybridization and the subsequent washing steps, and the cells were mounted with DAPI and the anti-bleaching reagent DABCO. Fluorescently labeled probes were prepared by nick translation of pBR322, pCAT-basic and SV40 DNA as described except that fluorescein-12-dUTP or Texas Red 5-dUTP (Molecular Probes, Eugene, OR, USA) were incorporated directly into the DNA.40 All photographs were taken with an Olympus BMAX50 epifluorescence microscope equipped with a PM20 photodocumentation system (Olympus, Tokyo, Japan) on 400 ASA Kodak Ektachrome or TriX-PAN film (Eastman Kodak, Rochester, NY, USA). Confocal microscopy was performed on an ACAS 570 laser-scanning confocal microscope.
Acknowledgements
We thank Curtis Browning and Ileana Aragon for establishing and culturing the chicken embryo fibroblasts and smooth muscle cell cultures, Dr Adrienne Kovacs for the SMGA promoter/CAT constructs, and Dr Paul Babal for providing us with cultures of human smooth muscle cells and for helpful discussions. Supported in part by grants ALG 960006 (DAD) from the Alabama Affiliate of the American Heart Association, 9404341 (WEZ) from the USDA, and R01 HL59956–01 (DAD) and 5P60 HL38639 (DAD and WEZ) from the NIHLB.
References
- 1.Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 3.Dowty ME, et al. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc Natl Acad Sci USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagstrom JE, et al. Nuclear import of DNA in digitonin-permeabilized cells. J Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 5.Sebestyén MG, et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nature Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 6.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid DNA nuclear entry. doi: 10.1006/excr.1999.4716. (Submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovacs AM, Zimmer WE. Cell specific transcription of the smooth muscle γ-actin gene requires both positive and negative acting cis-elements. Gene Exp. 1998;7:115–129. [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmer WE, Browning C, Kovacs AM. Cell-specific regulation of the SMGA gene. Dev Biol. 1996;175:399. Abstr. [Google Scholar]
- 9.Shimizu RT, et al. The smooth muscle α-actin gene promoter is differentially regulated in smooth muscle versus non-smooth muscle cells. J Biol Chem. 1995;270:7631–7643. doi: 10.1074/jbc.270.13.7631. [DOI] [PubMed] [Google Scholar]
- 10.Kallmeier R, Somasundaram C, Babji P. A novel smooth muscle enhancer regulates transcription of the smooth muscle myosin heavy chain gene in vascular smooth muscle cells. J Biol Chem. 1995;270:30949–30957. doi: 10.1074/jbc.270.52.30949. [DOI] [PubMed] [Google Scholar]
- 11.Herring B, Smith A. Telokin expression is mediated by a smooth muscle specific promoter. Am J Physiol. 1996;270:C1656–C1665. doi: 10.1152/ajpcell.1996.270.6.C1656. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Miano J, Mercer B, Olson E. Expression of the SM22 α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samaha F, et al. Developmental pattern of expression and genomic organization of the calponin-hi gene. J Biol Chem. 1996;271:395–403. doi: 10.1074/jbc.271.1.395. [DOI] [PubMed] [Google Scholar]
- 14.Miwa T, et al. Structure, chromosome location, and expression of the human smooth muscle (enteric type) gamma-actin gene: evolution of six human actin genes. Mol Cell Biol. 1991;11:3296–3306. doi: 10.1128/mcb.11.6.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szucsick J, Lessard J. Cloning and sequence analysis of the mouse smooth muscle γ-enteric actin gene. Genomics. 1995;28:154–162. doi: 10.1006/geno.1995.1126. [DOI] [PubMed] [Google Scholar]
- 16.Browning CL, et al. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- 17.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 18.Browning C, Zimmer WE. Multiple trans-acting factors bind with promoter elements of the smooth muscle γ-actin gene to regulate cell specific transcription. (Submitted for publication) [Google Scholar]
- 19.Langle-Rouault F, et al. Up to 100-fold increase of apparent gene expression in the presence of Epstein–Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Schwedler U, Kornbluth RS, Trono D. The nuclear localization signal of the matrix protein of human immunodeficiency virus type 1 allows the establishment of infection in macrophages and quiescent T lymphocytes. Proc Natl Acad Sci USA. 1994;91:6992–6996. doi: 10.1073/pnas.91.15.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukrinsky MI, et al. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bukrinsky MI, et al. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kann M, Bischof A, Gerlich WH. In vitro model for the nuclear transport of the hepadnavirus genome. J Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greber UF, Kasamatsu H. Nuclear targeting of SV40 and aden-ovirus. Trends Cell Biol. 1996;6:189–195. doi: 10.1016/0962-8924(96)10016-7. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi A, et al. Association with capsid proteins promotes nuclear targeting of simian virus 40. Proc Natl Acad Sci USA. 1996;93:96–100. doi: 10.1073/pnas.93.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collas P, Husebye H, Alestrom P. The nuclear localization sequence of the SV40 T antigen promotes transgene uptake and expression in zebrafish embryo nuclei. Transgenic Res. 1996;5:451–458. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- 27.Collas P, Alestrom P. Rapid targeting of plasmid DNA to zebrafish embryo nuclei by the nuclear localization signal of SV40 T antigen. Mol Mar Biol Biotechnol. 1997;6:48–58. [PubMed] [Google Scholar]
- 28.Pelligrini L, Tan S, Richmond TJ. Structure of serum response factor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 29.Gauthier-Rouvière C, et al. The serum response factor nuclear localization signal: general implications for cyclic AMP-dependent protein kinase activity in control of nuclear localization. Mol Cell Biol. 1995;15:433–444. doi: 10.1128/mcb.15.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 31.Mansouri A, Stoykova A, Gruss P. Pax genes in development. J Cell Sci Suppl. 1994;18(Suppl.):35–42. doi: 10.1242/jcs.1994.supplement_18.5. [DOI] [PubMed] [Google Scholar]
- 32.Moser M, et al. Cloning and characterization of a second AP-2 transcription factor: AP-2β. Development. 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 33.Orlic D, Anderson S, Nishikawa S-I, Nienhuis AW. Gene expression in purified murine hematopoietic stem cells. Blood. 1992;80:245a. [Google Scholar]
- 34.Kovacs AM, Zimmer WE. Molecular cloning and expression of the chicken smooth muscle γ-actin mRNA. Cell Mot Cytoskel. 1993;24:67–81. doi: 10.1002/cm.970240108. [DOI] [PubMed] [Google Scholar]
- 35.Chen CY, et al. Activation of the cardiac a-actin promoter depends upon serum response factor, tinman homologue, NKx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Croissant JD, et al. Avian serum response factor expression restricted primarily to muscle specific lineages is required for α-actin gene transcription. Dev Biol. 1996;177:250–264. doi: 10.1006/dbio.1996.0160. [DOI] [PubMed] [Google Scholar]
- 37.Dean DA, Li PL, Lee LM, Kasamatsu H. Essential role of the Vp2 and Vp3 DNA-binding domain in SV40 morphogenesis. J Virol. 1995;69:1115–1121. doi: 10.1128/jvi.69.2.1115-1121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cambell GR, Chamley JH, Burnstock G. Development of smooth muscle cells in tissue culture. J Anat. 1974;117:295–312. [PMC free article] [PubMed] [Google Scholar]
- 39.Hayward LJ, Schwartz RJ. Sequential expression of chicken actin genes during myogenesis. J Cell Biol. 1986;102:1485–1493. doi: 10.1083/jcb.102.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: implications for genome organization. Meth Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]