Abstract
Mutualistic symbioses shape the evolution of species and ecosystems and catalyze the emergence of biological complexity, yet how such symbioses first form is unclear. We show that an obligate mutualism between the yeast Saccharomyces cerevisiae and the alga Chlamydomonas reinhardtii—two model eukaryotes with very different life histories—can arise spontaneously in an environment requiring reciprocal carbon and nitrogen exchange. This capacity for mutualism is phylogenetically broad, extending to other Chlamydomonas and fungal species. Furthermore, we witnessed the spontaneous association of Chlamydomonas algal cells physically interacting with filamentous fungi. These observations demonstrate that under specific conditions, environmental change induces free-living species to become obligate mutualists and establishes a set of experimentally tractable, phylogenetically related, synthetic systems for studying the evolution of symbiosis.
Mutualistic symbioses—beneficial associations between different species involving persistent physical contact and physiological coupling—are central to many evolutionary and ecological innovations (1–3). These include the origin of eukaryotic cells, the colonization of land by plants, coral reefs, and the gut microbiota of insects and animals (4, 5). Despite their ubiquity and importance, we understand little about how mutualistic symbioses form between previously free-living organisms (5, 6). Like speciation, the birth of novel symbioses has rarely been witnessed, making it difficult to determine if co-evolution occurs before symbiosis begins or if chance ecological encounters initiate new symbioses (5, 7). Such “ecological fitting” (8, 9) occurs when both a particular environment and previously evolved traits allows a set of species to complement each other, giving rise to novel interactions without the need for prior coevolutionary adaptation.
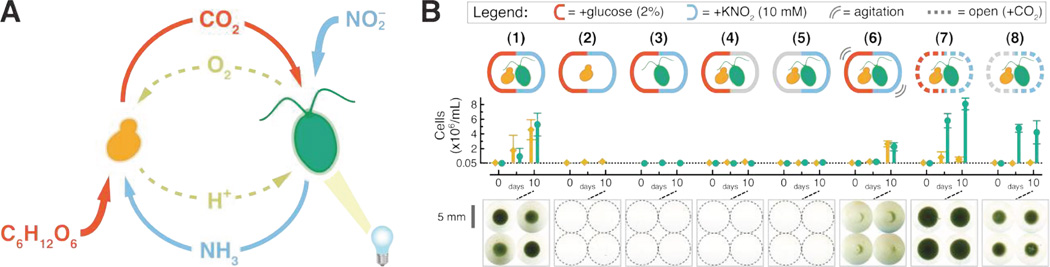
We tested two genetically tractable organisms, the budding yeast Saccharomyces cerevisiae and the green alga Chlamydomonas reinhardtii, to determine if a reciprocal exchange of carbon and nitrogen would lead to obligate mutualism between algae and fungi such as those which occur naturally (10–13). In our scheme (Fig. 1A), S. cerevisiae metabolizes glucose to carbon dioxide (CO2), a carbon source that C. reinhardtii fixes via photosynthesis, and C. reinhardtii reduces nitrite (NO2−) into ammonia (NH3) (14), which yeast can use as a nitrogen source. Co-culturing experiments (15) indicate that by preventing access to atmospheric CO2, S. cerevisiae and C. reinhardtii become obligate mutualists (Fig. 1B). This mutualism depends on the metabolic capabilities of the two organisms: S. cerevisiae cannot use nitrite as a nitrogen source and C. reinhardtii cannot use glucose as a carbon source. Cell proliferation did not require genetic engineering or fine-tuning of nutrient concentrations or starting ratios of the two species (Figs. 1B, S1, S2) and failed when either species (Fig. 1B, conditions 2–3), glucose, or nitrite was omitted from the experiment (Fig. 1B, conditions 4–5). Agitation attenuates this mutualism (Fig. 1B, condition 6), suggesting the importance of cell-cell proximity and spatial structure in establishing successful cooperation (16). Thus, a simple environmental change can induce free-living organisms to be mutualistic without requiring adaptive co-evolution.
Figure 1. A synthetic mutualism between S. cerevisiae and C. reinhardtii.
(A) A metabolic circuit for mutualism based on carbon and nitrogen exchange. S. cerevisiae (orange, left) metabolizes glucose (C6H12O6) and releases carbon dioxide (CO2), which is assimilated photosynthetically by C. reinhardtii (green, right) to release oxygen (O2); C. reinhardtii metabolizes nitrite (NO2−) and releases ammonia (NH3) as a nitrogen source for S. cerevisiae. An intrinsic, near-neutral pH balance between 6.8–7.4 is maintained by a metabolic exchange of protons between yeast and alga (15). (B) Proliferation of S. cerevisiae and C. reinhardtii under different co-culture conditions demonstrates that obligate mutualism can arise without any genetic engineering of metabolic pathways. Top: cartoons of the different conditions tested; middle: cell density of yeast and alga over the course of the experiment (mean ± 95% confidence interval; N=4); bottom: images of the cell populations from four representative examples of each culture condition (after 10 days). The dark green hue of the pellets is due to C. reinhardtii cells; S. cerevisiae cells are off-white and are interspersed throughout the pellet. See (15) for further details.
In our scheme, mutualism can be obligate or facultative depending on the environment. Access to atmospheric CO2 makes C. reinhardtii a facultative mutualist by removing its dependence on S. cerevisiae for carbon (Fig. 1B, condition 7), but the yeast remains dependent on the alga for nitrogen. In this environment, algal proliferation is improved by the presence of glucose-metabolizing, CO2-generating budding yeast while yeast proliferation is reduced, although not extinguished (Fig. 1B, conditions 7 vs. 8). Conversely, adding ammonia (as ammonium chloride) to airtight co-cultures allows budding yeast to proliferate independently of the alga while the alga remains dependent on the yeast for carbon. Under these conditions, S. cerevisiae (~4 hrs doubling time in our conditions) out-proliferates C. reinhardtii (≥12 hrs doubling time) and drives the alga to near extinction (Fig. S1, condition 15). These results suggest that stable metabolic mutualisms require that the faster growing species be obligately dependent on nutrients produced by its slower growing partner.
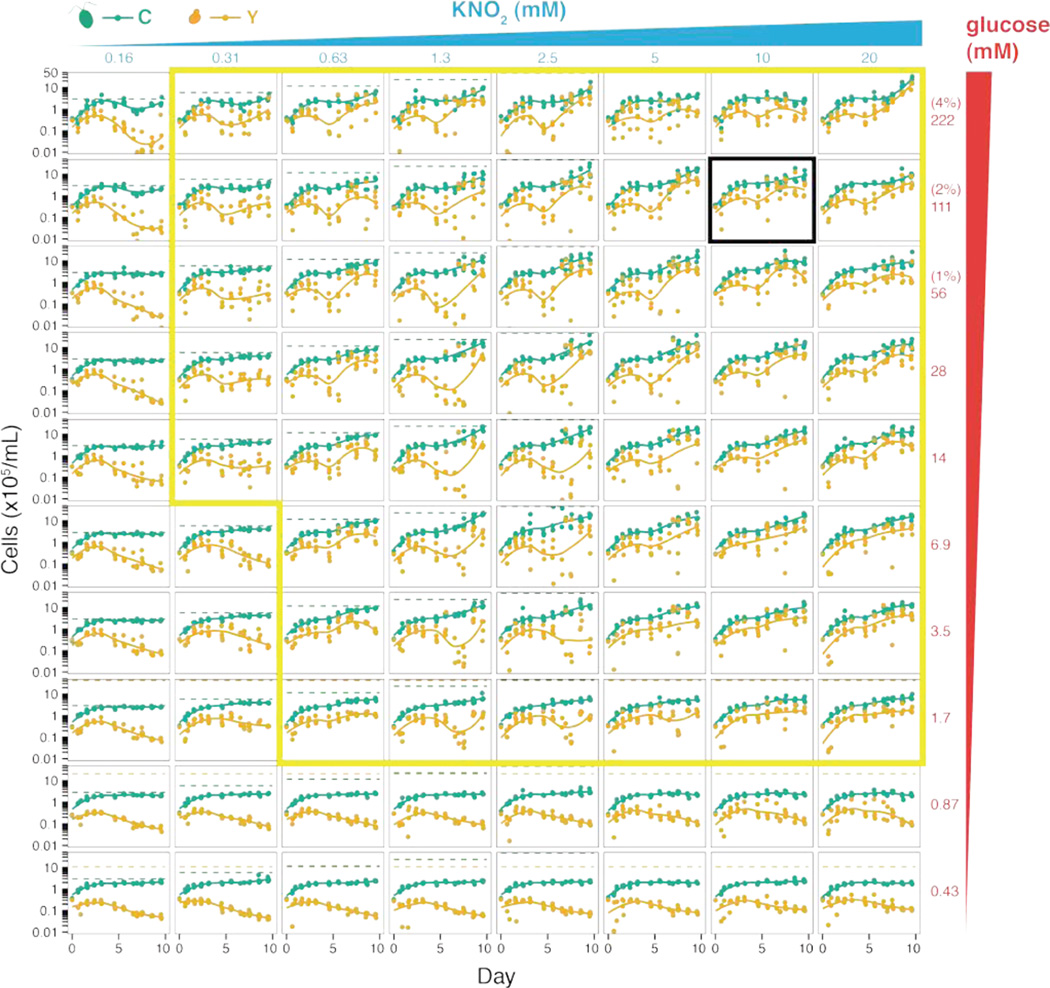
The engineered obligate mutualism between S. cerevisiae and C. reinhardtii is not limited to our initial choice of input nutrient concentrations. Successful mutualisms were established over nearly two orders of magnitude in glucose and nitrite concentrations (Fig. 2). However, this resulted in complex population dynamics. We observed undulations and variations in stability across time similar to density-dependent population cycles predicted for mutualistic systems (17). Other carbon (e.g., galactose) or nitrogen (e.g., nitrate) sources, although less effective, also sustain mutualism between S. cerevisiae and C. reinhardtii (Fig. S3).
Figure 2. Landscape of mutualistic productivity.
Cell densities over time for each species grown in co-culture (C. reinhardtii in green; S. cerevisiae in orange) grown from an initial inoculum of ~0.3×105 cells/mL for each species; irradiance=110 µmol/m2/s. Each of 4 replicate point pairs (green and orange) are plotted. Local polynomial regression fits (by robust linear regression in R with y~x) for both cell types are plotted as a visual guide of cell proliferation. Co-culture conditions are denoted on the left in black and show increasing, respectively, left to right KNO3 and bottom to top glucose concentrations. Dashed lines indicate the maximum predicted cell densities expected for C. reinhardtii (green) and S. cerevisiae (orange) for each co-culture (15). Net positive proliferation of both yeast and algae is supported within the region bounded by the yellow outline. The limited proliferation of C. reinhardtii under conditions outside this region (in days 1–2) indicates residual atmospheric CO2 in the wells of the sealed microtiter plate.
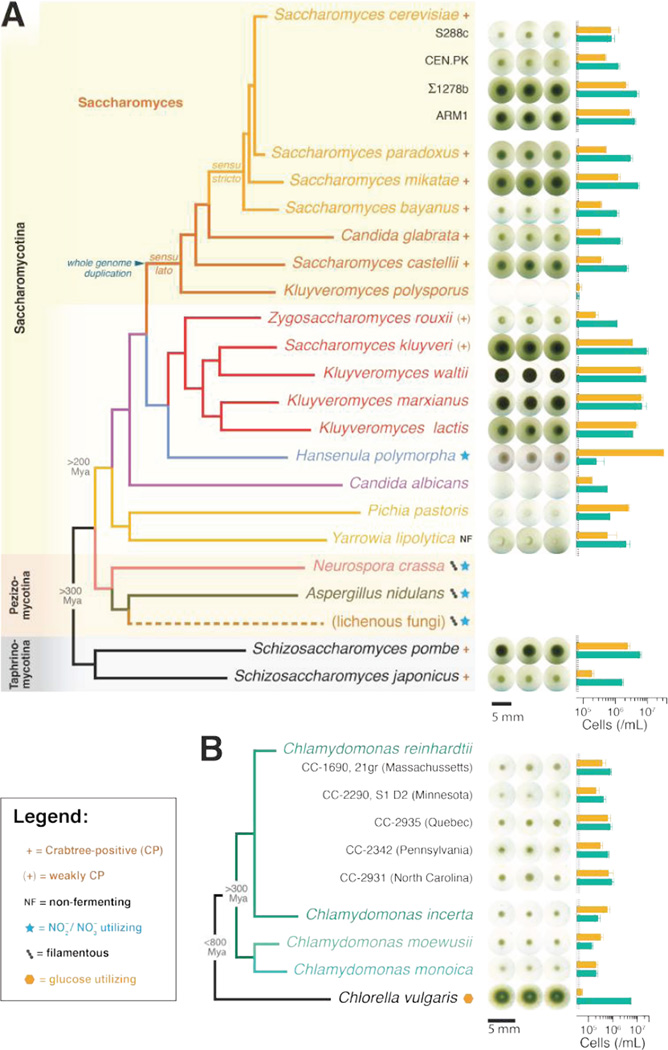
We also demonstrate that many different ascomycetous yeast and four Chlamydomonas species, spanning over 300 million years of evolutionary divergence in each clade, can form mutualisms (Fig. 3). Nearly all yeast species we examined form synthetic obligate mutualisms with C. reinhardtii, although with different degrees of productivity (Fig. 3A). Mutualistic productivity, as assessed by total cell counts, did not correlate with a yeast’s preference for a fermentative or respiratory lifestyle (Fig. 3A), whether a yeast strain was isolated from soil (a potential habitat shared with C. reinhardtii), had an intrinsic growth rate, or nitrite-mediated inhibition of growth (Fig. S4, Table S1). Thus, we observe that mutualisms can be phylogenetically broad, but that the degree of success depends on species-specific traits.
Figure 3. The capacity for mutualism is phylogenetically broad.
(A) Annotated phylogenetic tree of select ascomycetous fungal species (adapted and modified from published work (15)) paired with C. reinhardtii (CC-1690, 21gr), in three representative 9 day-old cocultures (indicated by dashed lines in the histogram of cell densities) grown under 110 µmol/m2/s of light. Measured cell counts (mean ± 95% confidence interval; N=4) for yeast (orange) and alga (green) are shown to the right of cell culture images. Subphyla of Ascomycota are indicated on the far left. Crabtree-positive yeasts (exhibiting a preference for fermentation over respiration even under aerobic conditions) are indicated with a “+”, and weakly Crabtree-positive yeasts with a “(+)”. Y. lipolytica is a non-fermenting (NF) yeast. Nitrate/nitrite utilizing fungi are indicated by a light blue star and filamentous fungi by three connected dots (see Fig. S5). (B) Annotated phylogenetic tree (adapted and modified from published work (15)) of select algal species and Chlamydomonas cultivars (green bars at right) paired with S. cerevisiae (S288C) (orange bars, right) in representative 7 day-old co-cultures grown under 110 µmol/m2/s of light (mean ± 95% confidence interval; N=12). Chlorella vulgaris, is an asexual alga, distantly related to C. reinhardtii, able to use glucose as a carbon source (orange hexagon). Descriptions of strains are provided in Table S1.
Two yeast species and the alga Chlorella vulgaris did not form obligate mutualisms (Fig. 3). C. vulgaris, which can use glucose as a carbon source, out-proliferated S. cerevisiae, whereas Hansenula polymorpha, a yeast that can use nitrite as a sole nitrogen source, out-proliferated C. reinhardtii. The yeast Kluveromyces polysporus failed to form an obligate mutualism with C. reinhardtii. This yeast can grow in an ammonium-supplemented co-culture medium, suggesting that it fails to cooperate with C. reinhardtii likely because it either cannot grow at the low ammonia levels produced by C. reinhardtii or is more sensitive to nitrite inhibition at such low ammonia levels (Fig. S4). Neurospora crassa and Aspergillus nidulans are genetically tractable filamentous fungi that can use nitrite as a nitrogen source (18). The ability of these fungi to reduce nitrite keeps wild type strains from forming obligate mutualisms with C. reinhardtii. However, mutants that cannot reduce nitrite did form obligate mutualisms (Fig. S5), suggesting that a loss of gene function in one species could be complemented through mutualism (11, 19).
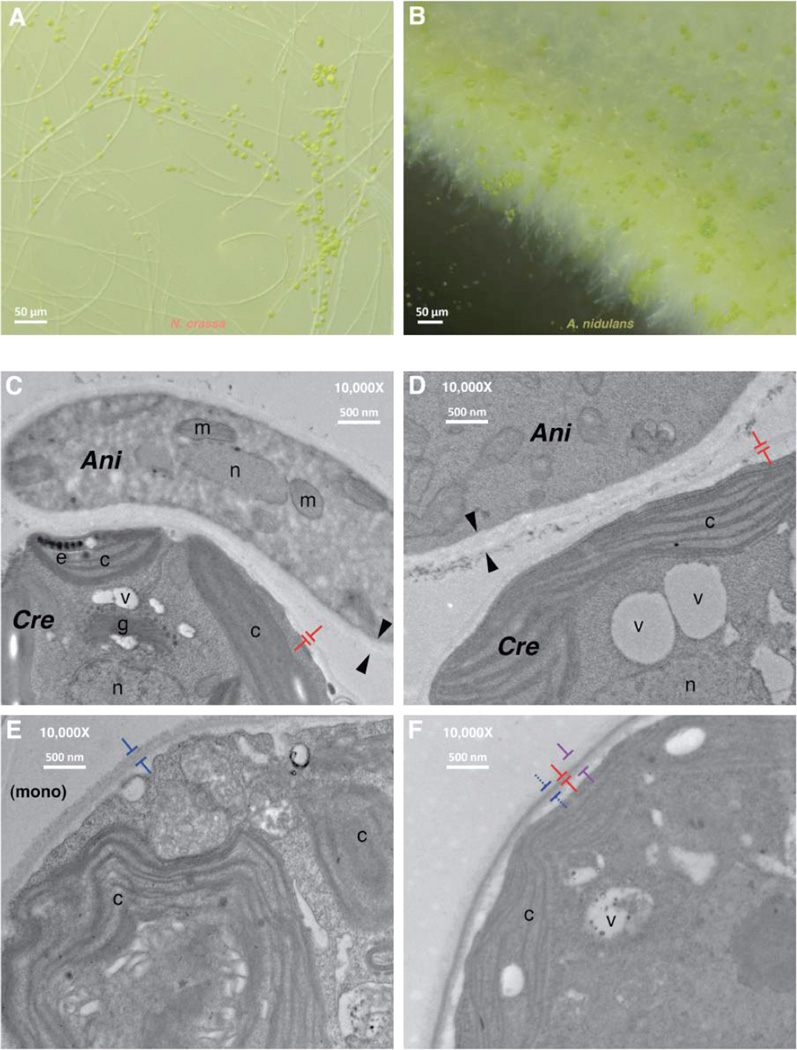
We observed that the filamentous fungi formed macroscopic structures such that the fungal hyphae were decorated with C. reinhardtii cells (Figs. S5, 4A–B, Movies S1–S6). However, physical associations between fungus and alga form even in the absence of any metabolic dependency (Figs. S6, S7, Movies S7–S16). Electron microscopy of interactions between C. reinhardtii and A. nidulans, which shares a most recent common ancestor with lichenous fungi within the class Eurotiomycetes (10), revealed a tight fungal-algal contact interface (Fig. 4C–D) reminiscent of wall-to-wall interfaces between fungal and algal cells in extant lichens (11). The walls of C. reinhardtii cells in contact with A. nidulans hyphae are less heavily stained and appear thinner than C. reinhardtii cells cultured separately (Fig. 4E) possibly due to locally secreted A. nidulans cell wall remodeling enzymes. We saw no evidence of any morphologically complex tissue structures, such as those seen in many lichens, nor of fungal hyphae penetrating algal cells (11, 20). Thus, these synthetic mutualisms may result in physical complexes but they do not form elaborate morphological structures at the cellular or organismal level.
Figure 4. C. reinhardtii physically associates with N. crassa and A. nidulans.
Representative light micrographs of the periphery of algal-fungal associations formed in obligate mutualistic co-culture. C. reinhardtii cells (green) stick to hyphae (white filaments) of (A) N. crassa (FGSC 11007 Δnit-4) or (B) A. nidulans (TS003 crnA- crnB-). (C–F) Representative transmission electron micrographs reveal a simple wall-to-wall interface between C. reinhardtii (Cre) cells and A. nidulans (Ani) hyphae. Opposed arrows indicate the thickness of fungal cell walls and opposed colored T-bars indicate those of algal cells ((C): 51 ± 10 nm; (D): 60 ± 7 nm; mean ± SD). (E) C. reinhardtii grown in mono-culture (160 ± 20 nm; blue T-bars) or (F) unattached C. reinhardtii isolated from the supernatant of the same co-culture [T demarcations: reference mono-culture cell wall thickness (red dashed; see E); (2) core (heavy) cell wall staining (blue): 50 ± 4 nm; (3) diffuse cell wall staining (purple): 260 ± 30 nm)] (15). Labeled intracellular components: m, mitochondria; c, chloroplast; e, eyespot; g, Golgi; n, nucleus; and v, vacuole.
The ease with which fungal-algal mutualisms were created suggests that ecological interactions may be relatively easy to establish (21). Furthermore, they do not require a prior facultative, commensal, or parasitic stage, or co-evolutionary adaptation (5–7, 22, 23). Our understanding of how “ecologically framed” pairs of species can be created due to environments that force them to depend on each other will be useful in the emerging field of synthetic ecology (24, 25) as well as for understanding the assembly of microbial communities in cases of disturbed or invaded habitats.
Supplementary Material
Acknowledgments
We thank Q. Justman, B. Stern, A. Pringle, S. Sasso, M. Dayel, N. Collins, J. Hess, M. Mueller, G. Frenkel, S. Kryazhimskiy, M. McDonald, D. Van Dyken, E. Wallace, K. Zimmerman, P. Boynton, J. Calarco, D. Chiang, Y. Eun, K. Foster, R. Losick, W. Tong, Y. Katz, and members of the Murray and Nelson labs for helpful feedback. We thank D. Thompson, M. Dunham, F. Winston, and N. Rhind for yeast strains; T. Schinko and J. Strauss for A. nidulans strains; and the FGSC (Kansas City, MO) for fungal strains. We thank P. Rogers, M. Tam, and B. Tilton (FAS Center for Systems Biology FACS Core); B. Goetze, C. Kraft, and D. Richardson (Harvard Center for Biological Imaging); M. Yankova and S. King (Central Electron Microscopy Facility, University of Connecticut Health Center) for their resources and assistance; and U. Goodenough for her help in interpreting EM micrographs. Supported in part by a Jane Coffin Childs postdoctoral fellowship to E. H. and by the NIGMS Center for Modular Biology (NIH grant P50-GM068763). Additional data described in this work can be found in the online supplementary material. E. H. conceived the project, performed the experiments, and analyzed the data. E. H. and A. M. devised the research and wrote the manuscript. A.M. supported and provided input throughout all stages of this work.
References and Notes
- 1.Thompson JN. Science. 1999;284:2116–2118. doi: 10.1126/science.284.5423.2116. [DOI] [PubMed] [Google Scholar]
- 2.Bronstein JL. In: The Princeton Guide to Ecology. Levin SA, editor. Princeton N.J.: Princeton University Press; 2009. pp. 233–238. [Google Scholar]
- 3.Brucker RM, Bordenstein SR. Trends Ecol Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Paracer S, Ahmadjian V. Symbiosis: An Introduction to Biological Associations. USA: Oxford University Press; 2000. [Google Scholar]
- 5.Douglas AE. The Symbiotic Habit. Princeton N.J.: Princeton University Press; 2010. [Google Scholar]
- 6.Sachs JL, Skophammer RG, Regus JU. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris JJ, Lenski RE, Zinser ER. mBio. 2012;3:e00036–e00012. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janzen DH. Oikos. 1985;45:308–310. [Google Scholar]
- 9.Agosta SJ, Klemens JA. Ecol Lett. 2008;11:1123–1134. doi: 10.1111/j.1461-0248.2008.01237.x. [DOI] [PubMed] [Google Scholar]
- 10.Lutzoni F, Pagel M, Reeb V. Nature. 2001;411:937–940. doi: 10.1038/35082053. [DOI] [PubMed] [Google Scholar]
- 11.Honegger R. In: Fungal Associations. Hock B, editor. Berlin: Springer; 2012. pp. 287–339. [Google Scholar]
- 12.Hawksworth DL. Botan J Linn Soc. 1988;96:3–20. [Google Scholar]
- 13.Kohlmeyer J, Kohlmeyer EJA. Marine Mycolog y The Higher Fungi. New York: Academic Press; 1979. pp. 70–78. [Google Scholar]
- 14.Azuara MP, Aparicio PJ. Plant Physiol. 1983;71:286–290. doi: 10.1104/pp.71.2.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods are available as supplementary material on Science Online.
- 16.Müller MJI, Neugeboren BI, Nelson DR, Murray AW. Proc Natl Acad Sci USA. 2014;111:1037–1042. doi: 10.1073/pnas.1313285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland JN, DeAngelis DL. Ecology. 2010;91:1286–1295. doi: 10.1890/09-1163.1. [DOI] [PubMed] [Google Scholar]
- 18.Slot JC, Hibbett DS. PLoS One. 2007;2:e1097. doi: 10.1371/journal.pone.0001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wade MJ. Nat Rev Genet. 2007;8:185–195. doi: 10.1038/nrg2031. [DOI] [PubMed] [Google Scholar]
- 20.Honegger R. New Phytol. 1986;103:785–795. [Google Scholar]
- 21.Gómez JM, Verdú M, Perfectti F. Nature. 2010;465:918–921. doi: 10.1038/nature09113. [DOI] [PubMed] [Google Scholar]
- 22.Harcombe W. Evolution. 2010;64:2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 23.Hillesland KL, Stahl DA. Proc Natl Acad Sci USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klitgord N, Segrè D. PLoS Comput Biol. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Momeni B, Chen CC, Hillesland KL, Waite A, Shou W. Cell Mol Life Sci. 2011;68:1353–1368. doi: 10.1007/s00018-011-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai HW, Henderson JN, Remington SJ, Campbell RE. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzur AAM, Jorgensen JKA, Shapiro PA, Kirschner HMA, Marc M. PLoS ONE. 2011;6:e16053. doi: 10.1371/journal.pone.0016053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DY, Fiehn O, Lee DY, Fiehn O. Plant Methods. 2008;4:7. doi: 10.1186/1746-4811-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyle NR, Morgan JA. BMC Syst Biol. 2009;3:4. doi: 10.1186/1752-0509-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraenkel DG. Yeast Intermediary Metabolism. Cold Spring Harbor N.Y.: Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- 31.Dykstra MJ. Biological Electron Microscopy: Theory, Techniques, and Troubleshooting. New York: Springer; 1992. pp. 5–78. [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, et al. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dujon B. Nat Rev Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 34.Lutzoni F, Kauff F, Cox CJ, McLaughlin D, et al. Am J Bot. 2004;91:1446–1480. doi: 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- 35.Pröschold T, Marin B, Schlösser UG, Melkonian M. Protist. 2001;152:265–300. doi: 10.1078/1434-4610-00068. [DOI] [PubMed] [Google Scholar]
- 36.Berbee ML, Taylor JW. Fungal Biol Rev. 2010;24:1–16. [Google Scholar]
- 37.Sipiczki M. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-2-reviews1011. reviews1011.1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redecker D, Kodner R, Graham LE. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 39.VanWinkle-Swift K, Baron K, McNamara A, Minke P, et al. Genetics. 1998;148:131–137. doi: 10.1093/genetics/148.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nedelcu AM, Lee RW. In: Advances in Photosynthesis and Respiration: The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Rochaix J, Goldschmidt-Clermont M, Merchant S, editors. Netherlands: Springer; 2004. pp. 63–91. [Google Scholar]
- 41.Herron MD, Hackett JD, Aylward FO, Michod RE. Proc Natl Acad Sci USA. 2009;106:3254–3258. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syrett PJ. Can Bull Fish Aquat Sci. 1981;210:182–210. [Google Scholar]
- 43.Hammer T, Bode R, Schmidt H, Birnbaum D. J Basic Microbiol. 1991;31:43–49. doi: 10.1002/jobm.3620300109. [DOI] [PubMed] [Google Scholar]
- 44.Muñoz-Blanco J, Hidalgo-Martinez J, Cárdenas J. Planta. 1990;182:194–198. doi: 10.1007/BF00197110. [DOI] [PubMed] [Google Scholar]
- 45.Vallon O, Bulté L, Kuras R, Olive J, Wollman FA. Eur J Biochem. 1993;215:351–360. doi: 10.1111/j.1432-1033.1993.tb18041.x. [DOI] [PubMed] [Google Scholar]
- 46.Murthy SN, Janardanasarma MK. Mol Cell Biochem. 1999;197:13–23. doi: 10.1023/a:1006906505745. [DOI] [PubMed] [Google Scholar]
- 47.Giordano M, Chen Y-B, Koblizek M, Falkowski PG. Eur J Phycol. 2005;40:345–352. [Google Scholar]
- 48.Fernández E, Llamas A, Galván A. In: The Chlamydomonas Sourcebook, Volume 2: Organellar and Metabolic Processes. Stern D, editor. San Diego, CA: Academic Press; 2009. pp. 69–114. [Google Scholar]
- 49.Azuara MP, Aparicio PJ. Plant Physiol. 1983;71:286–290. doi: 10.1104/pp.71.2.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald TR, Dietrich FS, Lutzoni F. Mol Biol Evol. 2012;29:51–60. doi: 10.1093/molbev/msr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald TR, Mueller O, Dietrich FS, Lutzoni F. BMC Genomics. 2013;14:225. doi: 10.1186/1471-2164-14-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Müller B, Russo VEA. Fungal Gen Newsl. 1989;36:58–60. [Google Scholar]
- 53.Adams TH, Wieser JK, Yu J-H. Microbio Mol Biol Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veiga A, Arrabaça JD, Loureiro-Dias MC. FEMS Microbiol Lett. 2000;190:93–97. doi: 10.1111/j.1574-6968.2000.tb09268.x. [DOI] [PubMed] [Google Scholar]
- 55.Dijken JPV, Bauer J, Brambilla L, Duboc P, et al. Enzyme Microb Technol. 2000;26:706–714. doi: 10.1016/s0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- 56.Liti G, Barton DB, Louis EJ. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merico A, Sulo P, Piskur J, Compagno C. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- 58.Van Urk H, Voll WS, Scheffers WA, Van Dijken JP. Appl Environ Microbiol. 1990;56:281–287. doi: 10.1128/aem.56.1.281-287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Møller K, Christensen B, Förster J, Piskur J, et al. Biotechnol Bioeng. 2002;77:186–193. doi: 10.1002/bit.10122. [DOI] [PubMed] [Google Scholar]
- 60.Gillum AM, Tsay EY, Kirsch DR. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 61.Baumann K, Dato L, Graf AB, Frascotti G, et al. BMC Genomics. 2011;12:218. doi: 10.1186/1471-2164-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colvin HJ, Sauer BL, Munkres KD. J Bacteriol. 1973;116:1322–1328. doi: 10.1128/jb.116.3.1322-1328.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bradshaw RE, Bird DM, Brown S, Gardiner RE, Hirst P. Mol Genet Genomics. 2001;266:48–55. doi: 10.1007/s004380100517. [DOI] [PubMed] [Google Scholar]
- 64.Schinko T, Berger H, Lee W, Gallmetzer A, et al. Mol Microbiol. 2010;78:720–738. doi: 10.1111/j.1365-2958.2010.07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sager R. Genetics. 1955;40:476–489. doi: 10.1093/genetics/40.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gross CH, Ranum LP, Lefebvre PA. Curr Genet. 1988;13:503–508. doi: 10.1007/BF02427756. [DOI] [PubMed] [Google Scholar]
- 67.Spanier JG, Graham JE, Jarvik JW. J Phycol. 1992;28:822–828. [Google Scholar]
- 68.Sack L, Zeyl C, Bell G, Sharbel T, Reboudet X. J Phycol. 1994;30:770–773. [Google Scholar]
- 69.Harris EH. The Chlamydomonas Sourcebook, Volume 1: Introduction to Chlamydomonas and Its Laboratory Use, Second Edition. San Diego, CA: Academic Press; 2009. [Google Scholar]
- 70.Wiese L, Wiese W. Am Nat. 1977;111:733–742. [Google Scholar]
- 71.Deason TR, Ratnasabapathy M. J Phycol. 1976;12:82–85. [Google Scholar]
- 72.Ahmadjian V. Am J Botany. 1962;49:277–283. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.