Abstract
During the past decade, DNA sequencing output has been mostly dominated by the second generation sequencing platforms which are characterized by low cost, high throughput and shorter read lengths for example, Illumina. The emergence and development of so called third generation sequencing platforms such as PacBio has permitted exceptionally long reads (over 20 kb) to be generated. Due to read length increases, algorithm improvements and hybrid assembly approaches, the concept of one chromosome, one contig and automated finishing of microbial genomes is now a realistic and achievable task for many microbial laboratories. In this paper, we describe high quality sequence datasets which span three generations of sequencing technologies, containing six types of data from four NGS platforms and originating from a single microorganism, Clostridium autoethanogenum. The dataset reported here will be useful for the scientific community to evaluate upcoming NGS platforms, enabling comparison of existing and novel bioinformatics approaches and will encourage interest in the development of innovative experimental and computational methods for NGS data.
Subject terms: DNA sequencing, Next-generation sequencing, Bacterial genomics
Background & Summary
It has been a decade since the release of the initial Next Generation Sequencing (NGS) platform by 454 Life Sciences (now Roche)1. During these ten years several NGS platforms including 454, Illumina, SOLiD, Ion Torrent and Pacific Biosciences (PacBio) have been released and improved2. Currently, Illumina offers the highest throughput and the lowest per base cost3, while third generation sequencing technology provider Pacific Biosciences (PacBio) has median read lengths in range of 4–5 kb and reads length >20 kb4. A performance comparison of various NGS platforms and recent advances are summarised2,3,5. In general, the second generation sequencing platforms are characterized by shorter read lengths while third generation platforms generate significantly longer, but fewer and more error prone reads.
The majority of published draft genomes have been sequenced using second generation sequencing technologies (Illumina and 454) and this data is readily available6. Since its introduction, the PacBio sequencing platform has become more widely used due to the utility of its longer read lengths7 and range of applications8. A limitation for earlier versions of PacBio technology for producing accurate genome assemblies was high error rates (> 15%) combined with lower sequence output (100 Mb)9. To address this, efficient algorithms were developed9,10, which require either >100x PacBio sequence coverage or accurate Illumina reads for error correction. Therefore, development of hybrid approaches which utilize previous sequencing data and also provide an option to employ long-read data remains as the major scientific focus area. An evaluation of various hybrid assembly strategies was recently published in mid-2014 (ref. 11) and within a short time frame the field continued to progress with the release of newer hybrid algorithms12–16 and updates to existing ones17,18. This underlines the requirement and utility of hybrid approaches to the scientific community. The long-read PacBio platform was speculated to be increasingly used to produce finished microbial genome assemblies4,6, supported by several recent examples19–23 and the utility of long-read sequencing for microbial genomes has been reviewed recently24. Combined developments of greater sequence output and longer reads, together with the random nature of PacBio errors have facilitated improved de novo assembly outputs. PacBio has the ability to detect DNA base modifications such as 4-methylcytosine (4-mC), 5-methylcytosine (5-mC) or 6-methyladenine (6-mA)25. This methylome information can be useful to understand biological processes such as gene expression and to aid development and optimization of transformation protocols26–28.
Examples of former NGS platforms include Helicos Biosciences29, and upcoming platforms include examples such Qiagen-intelligent Biosystems30, Oxford Nanopore31, and Quantum Biosystems32 platforms. Oxford Nanopore has released its portable sequencer MinION, and a recent publication describes the nature of data produced33. Many of these newer platforms are still in the initial development stages and especially for customized methods for alignment, consensus, variant calling, de novo assembly and scaffolding. During the maturation of these upcoming platforms, evaluations and assessments for sequence data error rates, accuracy, length, output, cost and performance will be critical, as will the development and assessment of bioinformatics tools. Therefore, datasets which contain high-quality data from various generations of sequencing platforms for a single microorganism will be useful for others to test, compare and contrast existing and novel experimental and computational advances and benchmark automated bioinformatics pipelines.
To facilitate further assessments and tool development for current and future NGS technologies, we report and describe in detail the methods, data and quality measurements for five sequencing technologies used to sequence the biofuel producing Clostridium autoethanogenum genome. This dataset represents three generations of sequencing technologies, and contains six types of data from four NGS platforms; 454 GS FLX, Illumina MiSeq, Ion Torrent, and PacBio RS-II; and Sanger sequence data. The PacBio data alone was sufficient to obtain the complete genome assembly of C. autoethanogenum. Several datasets were initially released into the NCBI Sequence Read Archive (SRA) with the finished C. autoethanogenum genome4. At present the NCBI SRA supports deposition of PacBio fastq files, but not the raw files required by certain software. The earlier study showed that assemblies utilizing shorter read DNA technologies were confounded by the nine copies of the 5 kb rRNA gene operons and other repetitive sequences4. Raw Ion Torrent and 454 shotgun sequence data for the draft genome sequence were not been previously released34, nor were C. autoethanogenum DNA methylation data.
Methods
Microorganism and genomic DNA preparation
Clostridium autoethanogenum strain JA1-1 (DSMZ 10061) was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ).
In order to prepare genomic DNA for 454 paired-end (PE), Illumina PE and PacBio sequencing the strain was cultured in PETC medium as described35. A single JA1-1 colony was purified and its 16S rDNA sequence confirmed before genomic DNA was prepared for Illumina and PacBio sequencing35. Genomic DNA for 454 paired-end, Illumina PE and PacBio sequencing was prepared as described previously4. Genomic DNA for 454 shotgun and Ion Torrent shotgun sequencing was prepared using the UltraClean Microbial DNA Isolation kit (catalog# 12224-250) from MoBio Laboratories, Inc (Carlsbad, CA). Prior to library preparation DNA quality was assessed by Nanodrop analysis (Thermo Scientific) and visualization on an agarose gel. Quality samples have an A260/280 ratio above 1.8, and appear on a gel as a single high molecular weight band. The quantity was determined by Qubit broad range double stranded DNA assay (Life Technologies, Grand Island, NY).
Illumina TruSeq library preparation and sequencing
Illumina TruSeq libraries were prepared as described in the manufacturer’s protocols (Part #15005180 RevA) following the low throughput protocol. In short, 3 μg of DNA was sheared to a size between approximately 200 and 1,000 bp by nebulization (using nitrogen as the carrier gas) for 1 min at 30 PSI. Sheared DNA was purified on a QIAquick Spin column (Qiagen). The quantity of sheared material was accessed with a broad range double stranded DNA assay from Qubit (Life Technologies) and visualized on an Agilent Bioanalyzer DNA 7500 chip (Agilent). One microgram of sheared DNA was used in the end repair reaction, and subsequently cleaned up by Agencourt AMPure XP bead purification (Beckman Coulter). The ends of the DNA were modified by adenylation of the 3′ ends and Illumina adapters were then ligated to the DNA. The DNA was cleaned up using Agencourt AMPure XP beads, and samples were then run for 2 h at 120 Volts on a 2% agarose gel containing SYBR Gold (Life Technologies). Ligation products were then purified from the sample by excising a band from the gel from approximately 350–450 bp. The DNA from the gel slice was then purified using a MinElute Gel Extraction kit (Qiagen) for each library/band. The DNA fragments were enriched by performing 10 cycles of amplification [98 °C–30 s, 10 cycles of: 98 °C for 10 s, 60 °C for 30 s, 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. Amplified products were then cleaned up using Agencourt AMPure XP beads. Final libraries were validated by Qubit (Life Technologies) and visualized by Agilent Bioanalyzer for appearance and size determination. Samples were normalized using the Illumina’s Library dilution calculator to a 10 nM stock, and subsequently run on an Illumina MiSeq Instrument (M02014R).
454 shotgun library preparation and sequencing
The 454 shotgun library was prepared using Roche's GS FLX Titanium Rapid Library Preparation Kit and was run on the Titanium platform according to manufacturer's specifications. Briefly, DNA was fragmented under gas pressure and the ends repaired. Adapters were ligated onto the fragments and then small fragments were selected out of the library. The library was then assessed for quality and concentration (including size length assessment and contaminating fragments of inappropriate size) using an Agilent Bioanalyzer 2100 prior to running on the 454 instrument.
454 3 kb library preparation and sequencing
A 454 3 kb paired end library was prepared following the manufacturer’s instructions (Roche- Paired End Library Preparation Method Manual—3 kb Span GS FLX Titanium Series- Oct 2009)36. Five micrograms of high quality, high molecular weight DNA was sheared to an average fragment size of 3 kb using a HydroShear apparatus (Genomic Solutions). The sheared material was then purified using Angencourt AMPure XP magnetic beads (Beckman Coulter). A portion of the sheared DNA was run on an Agilent Bioanalyzer 2100 to verify the size of the fragments. The fragment ends were polished and purified. The circularization adapters were appended and the product was again purified. Size selection of the material was completed followed by a fill in reaction and circularization. The sample was sheared by nebulization, purified, and checked for size on an Agilent Bioanalyzer 2100. The fragment ends were again polished and purified. The library was immobilized on Dynal M270 Streptavidin beads (Life Technologies) and the library adapters were ligated and gaps were filled. The library was amplified and a final purification step yielded a single stranded paired end library. The final library was amplified using emulsion PCR (emPCR); the products were purified, and then sequenced on a Roche 454 GS FLX system using Titanium chemistry according to the manufacturer’s instructions (Roche).
SMRTbell library preparation and PacBio sequencing
Ten micrograms of DNA were sheared using G-tubes (Covaris, Inc., Woburn, MA, USA), targeting 20 kb fragments. SMRTbell libraries were prepared with the DNA Template Kit 1.0 (Pacific Biosciences, Menlo Park, CA, USA) and library fragments above 4 kb were isolated using the BluePippin system (Sage Science, Inc., Beverly, MA, USA). The average SMRTbell library insert size (including adapters) was approximately 19 kb. Sequencing primers were annealed to the SMRTbell template and samples were sequenced on a PacBio RS II system (2013) using Magbead loading, C2 chemistry, Polymerase version P4, and SMRT analysis software version 2.2. DNA base modifications analysis was performed by ‘RS Modification and Motif Analysis’ workflow with default settings. Detailed information about detection of DNA base modifications workflow is available as online documentation37.
Ion torrent library preparation and sequencing
Genomic libraries were prepared separately for each genomic sample from 100 ng of DNA. DNA was fragmented with Ion Shear Plus Reagents, Ion Torrent specific adapters Ion Xpress P1 (5′—CCTCTCTATGGGCAGTCGGTGAT-3′) and Ion Xpress Barcode X Adapters (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′) were ligated to DNA using DNA ligase (Life Technologies, Grand Island, NY). The Ion Xpress Barcode X Adapters contain a 10 bp sequence, Ion Xpress Barcode (Life Technologies) unique to each of the samples. Ligated DNA was nick repaired using Nick Repair Polymerase (Life Technologies) and purified with Agencourt AMPure XP Reagent (Beckman Coulter, Indianapolis, IN). The ligated and nick repaired DNA was size-selected individually with the E-GelR SizeSelect Agarose Gel (Life Technologies). The size selected libraries were amplified using PlatinumR PCR SuperMix High Fidelity and Library Amplification Primer Mix (Life Technologies). The thermal profile for the amplification of each sample had an initial denaturing step at 94 °C for 5 min, followed by a cycling of denaturing of 95 °C for 15 s, annealing at 58 °C for 15 s and a 1 min extension at 70 °C (5 cycles) and a final hold at 4 °C. Each sample was again purified individually using Agencourt AMPure XP Reagent (Beckman Coulter, Indianapolis, IN) and standardized prior to pooling. Template-Positive Ion OneTouch 200 Ion Sphere Particles were prepared from the library pool using the Ion OneTouch DL system (Life Technologies, Invitrogen division). Prepared template was sequenced on an Ion Torrent PGM instrument (Microbiome Core Facility, Chapel Hill NC) using the Ion PGM 300 Sequencing reagents and protocols (Life Technologies). Initial data analysis, base pair calling and trimming of each sequence was performed on an Ion Torrent browser to yield high quality reads.
Sanger sequencing
Prior to PacBio sequencing, limited manual finishing of C. autoethanogenum was performed using PCR and Sanger sequencing. PCR reactions were performed using Phusion High-Fidelity PCR Kit (New England Biolabs, Ipswich, MA) following the standard protocol. Sanger sequencing was performed at Molecular Biology Research Facility, University of Tennessee, Knoxville using ABI 3730 Genetic Analyzer Instrument (Life Technologies).
Data Records
Raw data from each sequencing platform was submitted to the Sequence Read Archive (SRA) at NCBI under Project ID SRP030033 (Data Citation 1). Raw data deposited at the SRA and Dryad repository is organized by the type of sequencing platforms and corresponding accessions and file sizes are provided in Table 1. Data deposited in Dryad (Data Citation 2) are available under a project deposition and details for different datasets are summarized (Table 1).
Table 1. Summary of datasets.
| Sequencing Platform | Data type | SRA Accession/ Dryad doi | Size |
|---|---|---|---|
| Datasets described in this manuscript, which can be accessed using the accession numbers provided |
|||
| Accession linking all SRA data for this project |
SRP030033 | — | |
| Roche 454 shotgun | Raw data in SFF format | SRR1748017 | 1.5 Gb |
| Roche 454 3 kb | Raw data in SFF format | SRR989497 | 1.4 Gb |
| Illumina | Raw data in fastq format | SRR989790 | (669x2) Mb* |
| Ion Torrent | Raw data in SFF format | SRR1748018 | 858 Mb |
| PacBio RS II | Filtered subreads in fastq format | SRR1740585 | 1.2 Gb |
| Dryad doi linking all depositions for this project |
doi: 10.5061/dryad.6fm1p | — | |
| PacBio RS II | Raw PacBio data in tar.gz format | doi:10.5061/dryad.6fm1p/4 | 8.5 Gb |
| PacBio RS II | DNA methylation motifs in gff format | doi:10.5061/dryad.6fm1p/2 | 1.99 Mb |
| Sanger Sequencing | Chromatogram files in ABI format | doi:10.5061/dryad.6fm1p/1 | 4.39 Mb |
*There are two files for Illumina data corresponding read_1 and read_2. Detailed instructions for downloading data from SRA are provided in the Supplementary Information.
Illumina sequencing instruments generate raw image files which are automatically processed through instrument control software to output sequence data in fastq format. More details about different types of data files generated by the instrument and fastq conversion steps are described in online documentation38. The 150 bp paired-end (PE) Illumina reads in fastq format were deposited to SRA with run ID SRR989790. The fastq is standard file format which can be directly used to perform several downstream applications such as de novo assembly or mapping to a reference genome. The 454 Pyrosequencing and Ion Torrent instrument generates the sequencing data in Standard Flowgram Format (SFF). The SRA deposition for 454 shotgun, 454 3 kb PE and Ion Torrent data was made in SFF format under run ID SRR1748017, SRR989497 and SRR1748018, respectively. For validation purposes, quality statistics were determined for each short-read dataset using CLC Genomics Workbench (CLC) software version 7.5.1 and a complete report is provided as Supplementary Information.
The PacBio sequencing was performed using two SMRT cells. Each SMRT cell generates a metadata.xml file which contains information about run conditions and barcodes. Three bax.h5 files containing base calls and quality information of actual sequencing data and one bas.h5 file that acts as a pointer to consolidate three bax.h5 files8. A typical raw read from PacBio sequencing is composed of DNA insert with both ends flanked by the adapter sequences8. During downstream processing through SMRT Analysis software, the adapter sequences are removed and subreads are created which contains only the DNA sequence of interest. The PacBio filtered subreads were deposited at SRA in fastq format under run ID SRR1740585. Additionally, all the primary analysis data in the original formats as provided by the PacBio RS-II instrument are now made available on an external server (Table 1). Methylation in bacteria generally occurs at specific sequence motifs that are recognized by methyltransferases. Genome wide analysis of DNA base modifications was performed and a high level summary of the motifs discovered is provided in Table 2. Additionally, a ‘motifs.gff’ file is provided (Table 1), which shows all of the sites in the genome that are methylated, all the sites with one of the discovered motifs and the overlap between the methylation and the motifs as detected by SMRT analysis software version 2.2. Prior to PacBio sequencing, a manual finishing strategy for C. autoethanogenum generated high-quality Sanger sequence data and it is available to download (Table 1).
Table 2. Summary of DNA methylation motif patterns discovered across the C. autoethanogenum genome.
| Motif | Modified Position | Modification Type | % Motifs Detected | No. of Motifs Detected | No. of Motifs In Genome | Mean Modification QV | Mean Motif Coverage |
|---|---|---|---|---|---|---|---|
| Modified base within each motif is shown in bold. | |||||||
| CAAAAAR | 6 | m6A | 95.44 | 4,190 | 4,390 | 68.4 | 56.8 |
| GWTAAT | 5 | m6A | 93.87 | 7,975 | 8,496 | 78.5 | 58.1 |
| SNNGCAAT | 7 | m6A | 85.27 | 3,242 | 3,802 | 75.9 | 57.8 |
Raw reads represent the actual output from sequencing instruments. However, quality based trimming of Illumina and 454 data is recommended and often yields better results with downstream applications such as de novo assembly11,39. On the other hand, PacBio raw read filtering to generate subreads is a necessary step to remove adapter sequences8. Quality based trimming of Illumina and 454 data was performed using CLC software while PacBio filtering and mapping was performed using SMRT analysis version 2.2. The post-filter summary statistics for Illumina, 454 and Ion Torrent datasets are listed in Table 3 and for PacBio dataset in Table 4. The Illumina and PacBio datasets were sequenced to higher coverages (>100x), while 454 and Ion Torrent datasets had lower coverages (<50x). See the Technical Validation section for details on quality statistics and filtering parameters used.
Table 3. Summary of quality trimming statistics for Illumina, 454 and Ion Torrent data.
| Sequencing Platform | Type | No. of reads | Average length | No. of reads after Trim | Average length after Trim | Total Trimmed bases | Fold Coverage |
|---|---|---|---|---|---|---|---|
| Roche 454 | Singletons* Paired end readsShotgun Data | 128,856764,756462,052 | 275151289 | 128,806764,744458,340 | 261144249 | 33,631,416110,124,864114,126,660 | 46x 26x |
| Ion Torrent | Single end reads | 453,686 | 215 | 419,010 | 188 | 78,773,880 | 18x |
| Illumina | Paired end reads | 3,689,644 | 150 | 3,682,655 | 149 | 549,756,956 | 126x |
*The singleton sequences are generated from 454 3 kb sequencing run.
Table 4. Post-filter quality statistics for PacBio data.
| Sequencing Platform | Type | No. of filtered subreads | N50 filtered subread length | Maximum filtered subread length | Total filtered bases | Fold Coverage |
|---|---|---|---|---|---|---|
| PacBio RSII | Single end reads | 94,408 | 9,196 | 26,777 | 631,598,400 | 145x |
Technical Validation
DNA and sample preparation
All samples were required to pass a quantity and quality assessment using a Qubit (Life Technologies), Nanodrop (ThermoFisher) and gel electrophoresis. Samples were required to have readings indicative of pure DNA and of sufficient quantity to move forward with library preparations. DNA was visualized by gel electrophoresis and was required to be high molecular weight DNA without shearing or RNA contamination.
Each sequencing library preparation method includes specific technical validation to determine quality and quantity of the final libraries to ensure high quality output from the various sequencing platforms. This technical validation typically involves assessment of the final libraries with a Qubit assay (Life Technologies) to determine quantity and visualization of the final libraries on an Agilent Bioanalyzer chip to determine quality.
Quality determination and analysis
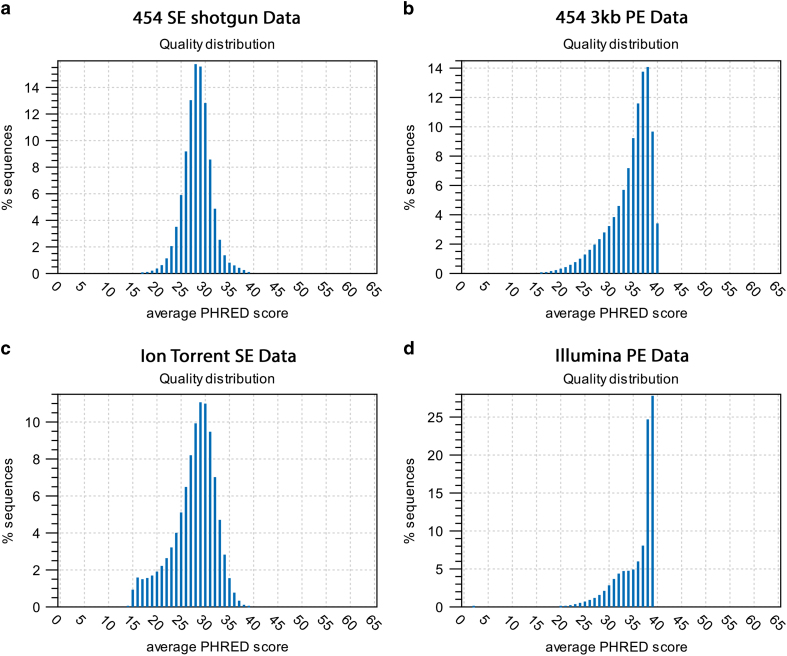
To assess the quality of the libraries sequenced, we determined basic quality statistics for Illumina, 454 and Ion Torrent datasets using CLC software. This includes the calculation of sequence lengths distribution, GC-content, Ambiguous base-content, PHRED quality score distribution, nucleotide contributions, kmer distribution analysis and sequence duplication levels. The quality statistics are calculated for every read, averaged for each dataset and provided in a complete quality report (Supplementary Information). More than 95% of the Illumina, 454 and Ion Torrent reads have PHRED scores above 20 (Fig. 1) with a very low percentage of ambiguous bases and sequence duplication levels detected (See section 2.3 and 4.2 for each dataset—Supplementary Information). Quality based trimming of these short-read datasets was performed at a stringent cut-off value of 0.02. More details about the trimming algorithm used by CLC and an example can be found in online documentation40. After quality trimming, only a few reads were discarded and minor changes in average read lengths were observed (Table 3). The PacBio data was processed through SMRT analysis software version 2.2. Filtering conditions applied were read quality score>0.8, read length >500 bp, subread length >500 bp. In addition, adapter sequences were removed and ends of the reads were removed when found outside of the high-quality region8,41. PacBio data retained 72% of the bases after filtering. The PacBio data by itself was sufficient to generate finished genome sequence. The complete genome sequence of C. autoethanogenum strain DSM10061 and de novo and hybrid assembly comparison using QUAST, REAPR, CGAL and Mauve tools have been described previously4. The Sanger sequencing data were found to be in agreement with the finished genome sequence of strain DSM10061 and provide additional validation for the high quality of PacBio dataset4.
Figure 1. PHRED quality score distribution.
The distribution of average PHRED quality score is plotted on X-axes and percentage of sequences on Y-axes for (a) 454 single end shotgun data (b) 454 3 kb paired end data (c) Ion Torrent single end data and (d) Illumina paired end data. Quality distribution shows that more than 95% reads from each dataset have average PHRED scores above 20.
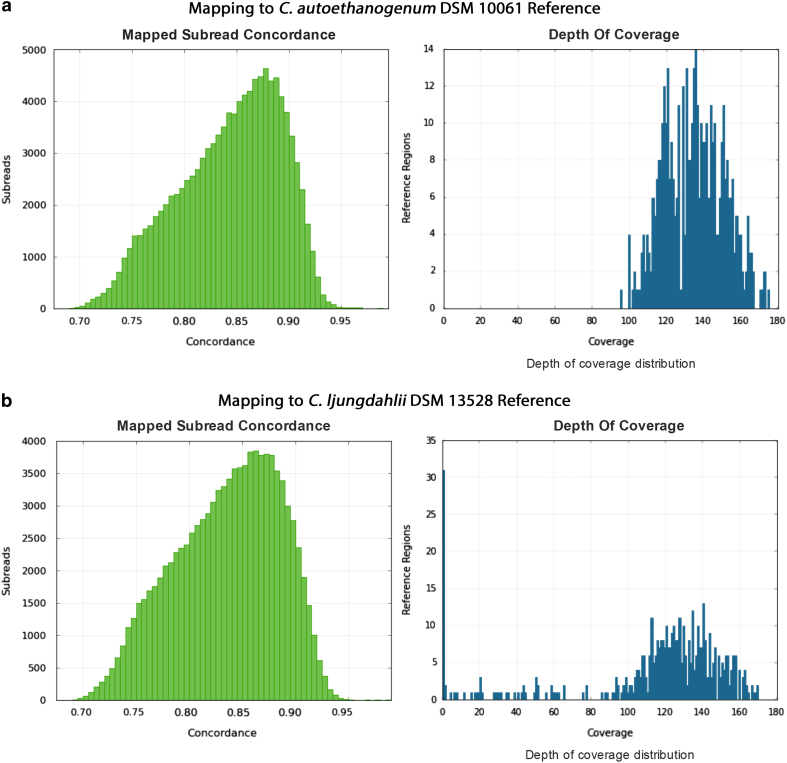
To further ensure that the sequences matched with the model organism of interest, we mapped the post-filtering reads from each dataset to the model organism of interest. We used C. autoethanogenum DSM 10061 genome from NC_022592.1 (Data Citation 3) and C. ljungdahlii DSM 13528 from NC_014328.1 (Data Citation 4) at the NCBI GenBank as reference sequences. Since a finished genome sequence for C. autoethanogenum was obtained using the PacBio reads from the current dataset, we used another independent reference C. ljungdahlii DSM 13528 to avoid any bias. These two genomes have an average nucleotide identity score over 99%. Illumina and 454 reads were mapped to reference using the bowtie2 algorithm42 while PacBio reads were mapped using the BLASR algorithm43 from the SMRT Analysis software. The Illumina and 454 datasets have mapping rates above 90% with C. ljungdahlii and above 97% with the finished genome of C. autoethanogenum. Ion Torrent data have a comparatively lower mapping rate, 86% with C. ljungdahlii and 91% with C. autoethanogenum. For the PacBio dataset, plots showing the distributions of mapped subread concordances and coverage are shown in Fig. 2 and provide an estimate of read agreement with reference genomes. Therefore, the data quality statistics, trimming reports and mapping results articulate the high quality of the datasets described in this manuscript.
Figure 2. Mapped subread concordance and coverage.
The distribution of mapped subread concordances and mapped subread coverages are plotted with (a) C. autoethanogenum DSM 10061 finished genome and (b) C. ljungdahlii DSM 13528 as reference. These graphs suggest good agreement between reads and reference genomes.
Usage Notes
The five NGS datasets described can be downloaded from the SRA with accession numbers provided in Table 1. Detailed instructions for downloading each dataset from NCBI SRA and md5 checksum values are provided in the Supplementary Information. The fastq/SFF formatted files from second generation sequencing data are sufficient to use for any downstream analysis using most third-party tools. On the other hand, original data formats are necessary for analysing the PacBio data through SMRT analysis software or other algorithms and these are provided (Table 1). Currently the SRA allows depositions of fastq formatted PacBio reads only. Therefore, all the primary analysis data in original formats as generated by the PacBio RS II instrument (*.metadata.xml, *.bas.h5, *.bax.h5 files), as well as DNA methylation motifs detected by PacBio sequencing are available in Dryad (Data Citation 2). The sequence IDs provided in primary analysis files are different than those available through SRA because SRA uses an internal naming convention which changes existing sequence IDs. The sequence IDs in original format contain information about run and the naming convention is described in detail here8. Sanger data are posted in Dryad (Data Citation 2).
Some of the datasets described here were initially released with the manuscripts describing the draft34 and finished genome of C. autoethanogenum 4, with primary focus on genomic features and characteristics of this microorganism. Previous manuscripts did not include Ion torrent/454 shotgun data release and detailed quality evaluation and usage instructions were not provided. In addition, DNA modification data for C. autoethanogenum from the PacBio is provided, identifying three m6A adenosine methylation patterns (Table 2). The ‘motifs.gff’ file is a text file which can be opened in most of the graphical sequence viewer software. This data descriptor in Scientific Data provides an opportunity to present the collection of these five different datasets which are originated from a single microorganism and spans three generations of sequencing technologies. Here we provide the detailed characteristics for each dataset and appropriate instructions to download and use the data. Since sequencing technologies are rapidly evolving, this legacy dataset can be used as a benchmark to compare the data from newer NGS technologies and will encourage the development of new and existing hybrid algorithms.
Additional information
How to cite this article: Utturkar, S. M. et al. Sequence data for Clostridium autoethanogenum using three generations of sequencing technologies. Sci. Data. 2:150014 doi: 10.1038/sdata.2015.14 (2015).
Supplementary Material
Acknowledgments
LanzaTech supported the generation of the 454 PE, Illumina and PacBio data. Ion Torrent and 454 shotgun data generation was supported by the North Carolina State University Department of Microbiology, Department of Biological and Agricultural Engineering, and in part by the North Carolina Biotechnology Center. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research (BER) in the DOE Office of Science. S.M.U. is supported by the BER Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov) and part of his stipend was also supported by LanzaTech. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the DOE under Contract DE-AC05-00OR22725. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Footnotes
LanzaTech has a commercial interest in Clostridium autoethanogenum.
Data Citations
- 2014. NCBI Sequence Read Archive. SRP030033
- Utturkar S. M. 2015. Dryad. http://dx.doi.org/10.5061/dryad.6fm1p
- Brown S. D. 2014. GenBank. NC_022592.1
- Koepke M. 2010. GenBank. NC_014328.1
References
- Margulies M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Auger H., Jaszczyszyn Y. & Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 30, 418–426 (2014). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. 2012, 251364 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. et al. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant Clostridia. Biotechnol. Biofuels 7, 40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M. A. et al. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 14, R101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Carneiro M. O. & Schatz M. C. The advantages of SMRT sequencing. Genome Biol. 14, 405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. E. et al. Long-read, whole-genome shotgun sequence data for five model organisms. Sci. Data 1, 140045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 30, 693–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- Utturkar S. M. et al. Evaluation and validation of de novo and hybrid assembly techniques to derive high quality genome sequences. Bioinformatics 30, 2709–2716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmela L. & Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics 30, 3506–3514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. et al. Error correction and assembly complexity of single molecule sequencing reads. Preprint at BioRxiv http://dx.doi.org/10.1101/006395 (2014). [Google Scholar]
- Hackl T., Hedrich R., Schultz J. & Forster F. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C., Hill C., Koren S., Ruan J., Zhanshan M., Yorke J. A. & Zimin A. DBG2OLC: Efficient assembly of large genomes using the compressed overlap graph. Preprint at arXiv http://arxiv.org/abs/1410.2801 (2014). [Google Scholar]
- Prjibelski A. D. et al. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics 30, i293–i301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. C., Salerno W. J. & Reid J. G. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15, 180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou K. et al. Complete genome sequences of eight Helicobacter pylori strains with different virulence factor genotypes and methylation profiles, isolated from patients with diverse gastrointestinal diseases on Okinawa Island, Japan, determined using PacBio Single-Molecule Real-Time Technology. Genome Announc. 2, 2 e00286–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnaz S., Bauer J. S. & Gross H. Complete genome sequence of the sugar cane endophyte Pseudomonas aurantiaca PB-St2, a disease-suppressive bacterium with antifungal activity toward the plant pathogen Colletotrichum falcatum. Genome Announc. 2, 1 e01108–e01113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhay G. P. et al. Complete closed genome sequences of three Bibersteinia trehalosi nasopharyngeal isolates from cattle with shipping fever. Genome Announc. 2, 1 e00084–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckweiler D., Bunk B., Sproer C., Overmann J. & Haussler S. Complete genome sequence of highly adherent Pseudomonas aeruginosa small-colony variant SCV20265. Genome Announc. 2, 1 e01232–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. D. et al. Complete genome sequence of Pelosinus sp. strain UFO1 assembled using Single-Molecule Real-Time DNA sequencing technology. Genome Announc. 2, 5 e00881–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. & Phillippy A. M. ONE chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr. Opin. Microbiol. 23C, 110–120 (2014). [DOI] [PubMed] [Google Scholar]
- Davis B. M., Chao M. C. & Waldor M. K. Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr. Opin. Microbiol. 16, 192–198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak J. M., Liebl W. & Ehrenreich A. Development of an in vivo methylation system for the solventogen Clostridium saccharobutylicum NCP 262 and analysis of two endonuclease mutants. J. Biotechnol. 188C, 97–99 (2014). [DOI] [PubMed] [Google Scholar]
- Mermelstein L. D. & Papoutsakis E. T. In vivo methylation in Escherichia coli by the Bacillus subtilis phage phi 3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 59, 1077–1081 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne M. E., Moo-Young M., Chung D. A. & Chou C. P. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol. Biofuels 6, 50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkarev D., Neff N. F. & Quake S. R. Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 27, 847–850 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J. et al. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc. Natl Acad. Sci. USA 103, 19635–19640 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009). [DOI] [PubMed] [Google Scholar]
- BusinessWire. Quantum Biosystems Demonstrates First Reads Using Quantum Single Molecule Sequencing. http://www.businesswire.com/news/home/20140127005012/en/Quantum-Biosystems-Demonstrates-Reads-Quantum-Single-Molecule#.VIH5dDHF8fU (2014).
- Quick J., Quinlan A. R. & Loman N. J. A reference bacterial genome dataset generated on the MinION portable single-molecule nanopore sequencer. Gigascience 3, 22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno-Barcena J. M., Chinn M. S. & Grunden A. M. Genome sequence of the autotrophic acetogen Clostridium autoethanogenum JA1-1 strain DSM 10061, a producer of ethanol from carbon monoxide. Genome Announc. 1, 4 e00628–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopke M. et al. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl Acad. Sci. USA 107, 13087–13092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Klingeman D. M. & Brown S. D. Microbial Metabolic Engineering: Methods and Protocols Vol. 834, 111–136 (Springer, 2012). [DOI] [PubMed] [Google Scholar]
- Pacific-BioSciences. Detecting DNA Base Modifications. http://www.pacb.com/pdf/TN_Detecting_DNA_Base_Modifications.pdf (2012).
- Illumina-Inc. CASAVA v1.8.2 User Guide http://support.illumina.com/content/dam/illumina-support/documents/myillumina/a557afc4-bf0e-4dad-9e59-9c740dd1e751/casava_userguide_15011196d.pdf (2011).
- Salzberg S. L. et al. GAGE: A critical evaluation of genome assemblies and assembly algorithms. Genome Res. 22, 557–567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLC, CLC Genomics Workbenach Manual—Trimming using the Trim tool. http://www.clcsupport.com/clcgenomicsworkbench/800/index.php?manual=Trimming_using_Trim_tool.html (2015).
- Pacific-Biosciences, Statistics Output Guide. http://files.pacb.com/software/instrument/1.3.1/Statistics%20Output%20Guide.pdf (2014).
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J. & Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2014. NCBI Sequence Read Archive. SRP030033
- Utturkar S. M. 2015. Dryad. http://dx.doi.org/10.5061/dryad.6fm1p
- Brown S. D. 2014. GenBank. NC_022592.1
- Koepke M. 2010. GenBank. NC_014328.1