Abstract
Purpose
We tested the validity of a freely available segmentation pipeline to measure compartmental brain volumes from 3T MRI in patients with multiple sclerosis (MS). Our primary focus was methodological to explore the effect of segmentation corrections on the clinical relevance of the output metrics.
Methods
Three-dimensional T1-weighted images were acquired to compare 61 MS patients to 30 age- and gender-matched normal controls (NC). We also tested the within patient MRI relationship to disability (eg, expanded disability status scale [EDSS] score) and cognition. Statistical parametric mapping v. 8 (SPM8)-derived gray matter (GMF), white matter (WMF), and total brain parenchyma fractions (BPF) were derived before and after correcting errors from T1 hypointense MS lesions and/or ineffective deep GM contouring.
Results
MS patients had lower GMF and BPF as compared to NC (P<.05). Cognitively impaired patients had lower BPF than cognitively preserved patients (P<.05). BPF was related to EDSS; BPF and GMF were related to disease duration (all P<.05). Errors caused bias in GMFs and WMFs but had no discernable influence on BPFs or any MRI-clinical associations.
Conclusions
We report the validity of a segmentation pipeline for the detection of MS-related brain atrophy with 3T MRI. Longitudinal studies are warranted to extend these results.
Keywords: Brain atrophy, multiple sclerosis, MRI, segmentation, lesions, gray matter
Introduction
Multiple sclerosis (MS) is a chronic progressive disease of the central nervous system (CNS) often associated with brain and spinal cord atrophy.1 Both whole brain and compartment-specific cerebral gray matter (GM) atrophy are common and highly relevant to clinical status.2–4 Most previous studies of brain atrophy, including nearly all therapeutic trials in MS, have employed 1.5T scanners for quantification of brain atrophy.5 However, there is a continuing shift of MS clinical care and research studies to 3T platforms, approved several years ago by the Food and Drug Administration, to take advantage of higher resolution and faster scan times.5–7
The use of high-field 3T MRI has the potential to improve the accuracy and reproducibility of tissue classification due to the increased signal-to-noise ratio and contrast-to-noise ratio compared to lower fields.7,8 This results in improved white matter (WM) to GM contrast over the entire brain,6 particularly when optimized sequences are used.7 Furthermore, the tissue boundaries are better defined as a result of high-resolution images, which have lower partial volume effects.6 However, the segmentation of deep central GM nuclei (DGM) structures is still challenging due to poor contrast versus surrounding WM tissue.9
In MS, the true MRI estimation of brain volume is further confounded by the presence of hypointense lesions on T1-weighted images, which are often misclassified as GM in commonly used T1-based segmentation algorithms.10 Classification errors can also occur in the tissue surrounding T1 hypointensities due to partial volume effects.11 If these MS-related misclassifications are left uncorrected, estimates of GM volumes may be biased proportionally to the amount of lesion volume present, which could ultimately lead to a misinterpretation of the results obtained. Further studies are warranted to determine the necessity (ie, clinical relevance) of such corrections.
Several groups are currently engaged in the development of efficient, reliable, and valid tools for segmentation of brain tissue from 3T MRI to set the stage for efficient patient monitoring of global atrophy.8,9,12–14 In this study, we employed a freely available segmentation tool to test its validity in the measurement of compartment-specific brain segmentation, both with and without manual intracerebral misclassification corrections. For validation, we compared MS patients to healthy controls, and tested the within patient relationship between MRI and clinical status, including physical disability and cognitive function. We defined validity of the candidate MRI measures as the strength of their ability to differentiate patients from controls and the strength of their associations with clinical measures of MS disease severity.
Materials and Methods
Subjects
Demographic and clinical characteristics of the subjects are summarized in Table1. For this study, we identified 61 consecutive patients with MS based on the following criteria: (1) age 18-55; (2) established MS diagnosis of either relapsing-remitting (RRMS), secondary progressive, primary progressive, or clinically isolated syndrome by the International Panel criteria;15 (3) no other major medical, neurologic, or neuropsychiatric disorder; (4) no relapse or corticosteroid use in the 4 weeks prior to MRI to avoid transient confounding effects on MRI; (5) not initiated on disease-modifying therapy in the 6 months preceding MRI, to avoid transient confounding effects on MRI of newly started therapy such as pseudoatrophy;16 and (6) no history of substance abuse or smoking. Forty-nine patients (80.3%) were receiving disease-modifying treatment at the time of MRI. Within 3 months of MRI, each patient underwent a neurologic examination by an MS specialist, including expanded disability status scale (EDSS) and timed 25-foot walk (T25FW) testing, and a formal cognitive evaluation by a PhD in Clinical Psychology and a research fellow under her supervision (B.I.G., A.A.). Thirty normal controls (NC) with a similar distribution of age and gender to the subject group (Table1), with no neurological symptoms or known neurological or major medical disorders, were also included.17 Our institutional review board approved this study and all subjects gave informed consent.
Table 1.
Demographics, Clinical, and Conventional MRI Characteristics
| Multiple Sclerosis | Normal Controls | |
|---|---|---|
| (N = 61) | (N = 30) | |
| Women | N = 42 (69%) | N = 21 (70%) |
| Age (years) | 41.0 ± 8.6 (21-55) | 43.9 ± 6.3 (30-53) |
| Disease duration (years) | 8.3 ± 7.2 (0.2-29.0) | - |
| EDSS score | 1.6 ± 1.7 (0-8.0) | - |
| Timed 25 foot walk | 4.8 ± 4.5 (2.9-38.5) | - |
| T2 (FLAIR) hyperintense lesion volume (ml) | 13.6 ± 11.4 (2.6-49.3) | .44 ± 0.57 (0-2.76) |
| T1 (MDEFT) hypointense cerebral lesion volume (ml) | 6.4 ± 7.4 (0.5-34.0) | .19 ± 0.30 (0-1.28) |
| Disease course—number (%) | ||
| Clinically isolated syndrome | N = 4 (6.6%) | - |
| Relapsing remitting | N = 51 (83.6%) | - |
| Secondary progressive | N = 5 (8.2%) | - |
| Primary progressive | N = 1 (1.6%) | - |
| Receiving disease-modifying therapy | N = 49 (80.3%) | - |
Values in table are: mean ± standard deviation (range); number of subjects (percentage).
EDSS = expanded disability status scale; FLAIR = fluid-attenuated inversion-recovery; MDEFFT = modified driven-equilibrium Fourier transform.
Neuropsychological Evaluation
Cognitive performance was assessed using the minimal assessment of cognitive function in MS (MACFIMS), according to consensus panel recommendations.18 Patients were also evaluated for depressive symptoms using the Center for Epidemiologic Studies Depression (CES-D) scale. We decided a priori to control for depressive symptoms in the analysis of MRI-cognition relationships due to the fact that depression may affect cognition (see Statistical Analysis section). Subjects had not been previously exposed to any of the tests in the MACFIMS battery. Given the small sample size of the NC group, demographically adjusted T-scores were calculated for MS patients using the regression-based norms as previously derived from a single sample of 100 healthy controls, demographically matched to a sample of 395 representative MS clinical patients.19 Impairment on a single neuropsychological measure was defined as a T-score ≤ 35.19 A cut-point of T = 35 is roughly equivalent to a z-score of -1.5 or a percentile rank of 5th percentile. Cognitive impairment was defined as impairment on 2 or more cognitive measures. The use of defective performance on 2 or more measures to define cognitive impairment derives from the standard approach in field where impairment on roughly 1/5 of the battery tests would define the subject as impaired. This led to the classification of our MS patients as cognitively impaired (n = 23) or cognitively preserved (n = 38).
MRI Acquisition
All participants underwent MRI on the same scanner (3T Signa, General Electric Healthcare, Milwaukee, WI, USA) using a receive-only phase array head coil with the same MRI protocol. Contiguous slices covering the whole brain were acquired as follows:
Coronal 3-dimensional modified driven-equilibrium Fourier transform (MDEFT):20 TR = 7.9 ms, TE = 3.14 ms, flip angle = 150, slice thickness = 1.6 mm (124 slices—no gap), matrix size = 256 × 256, pixel size =.938 mm ×.938 mm, acquisition time = 7.5 minutes, number of signal averages = 1. We used this sequence for its utility in morphometric studies.8,10
Axial T2-weighted fast fluid-attenuated inversion-recovery (FLAIR): TR = 9,000 ms, TE = 151 ms, TI = 2,250 ms, slice thickness = 2 mm (70 slices—no gap), matrix size = 256 × 256, pixel size =.976 mm ×.976 mm, acquisition time = 9 minutes, number of signal averages = 1.
Image Analysis
All MDEFT and FLAIR images were inspected for major artifacts (eg, motion and ghosting) and to establish quality of the images. No major artifacts were found. FLAIR images of a subset of the NC group showed focal incidental WM hyperintensities17 that sometimes corresponded to hypointense areas on MDEFT images (Table1). Brain MRI analysis was performed using Jim (v. 5, Xinapse Systems, Northants, UK, http://www.xinapse.com) and statistical parametric mapping (SPM; v. 8, Wellcome Department of Cognitive Neurology, London, UK, http://www.fil.ion.ucl.ac.uk/spm). The latter employed the unified segmentation algorithm21 as improved in SPM8, to obtain volume measurements for GM, WM, and cerebrospinal fluid (CSF) tissue compartments. Figure1 shows the flow chart and operator time required for each key step of the image processing steps applied to obtain compartment-specific tissue-segmented images. Figure2 shows sample outputs from a patient with MS.
Figure 1.
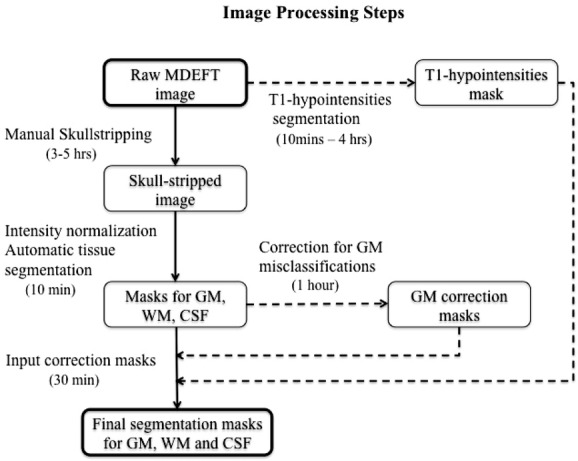
This flow chart represents the image processing steps that were followed to obtain final segmentation for masks gray matter (GM), white matter (WM), and CSF. Raw MDEFT images were manually deskulled, intensity normalized, and automatically segmented into GM, WM, and CSF. The obtained segmentation masks were corrected for misclassifications of deep GM and T1 hypointensities. The generated GM correction mask and T1 hypointensities mask were used to correct the original tissue masks, thus obtaining final corrected segmentations for GM, WM, and CSF.
Figure 2.
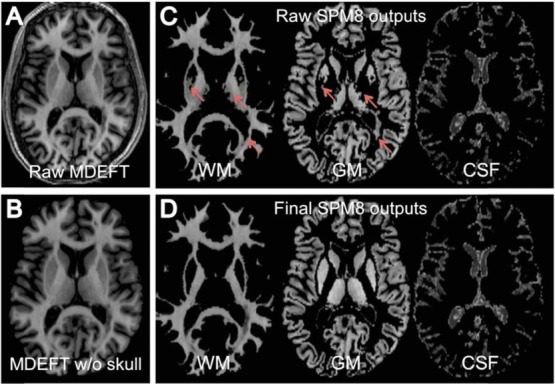
Panels A and B show a representative slice of a raw axial MDEFT image before and after manual skull stripping in a patient with MS. Panel C shows the corresponding uncorrected SPM8-derived segmented images obtained for white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) tissue compartments; note the images from left to right show: underestimation of the volume of the putamen and thalamus (anterior arrows) and misclassification of T1 hypointensities in WM as GM (posterior arrows). Panel D shows the corresponding segmentations obtained for the same tissue dually corrected compartments (ie, after manually correcting both types of errors).
Lesion Segmentation
Brain FLAIR hyperintense and MDEFT hypointense lesions were segmented using a semiautomated edge-finding tool based on local thresholding with expert manual corrections applied as needed in Jim5, as previously described.10,22 The presence of T1 hypointense lesions in WM on MDEFT images (Fig 2) was determined by consensus of 2 trained observers. T2 (FLAIR) hyperintense and T1 hypointense lesion volumes (T2LV, T1LV) were obtained as described previously.10
Intracranial Volume
Raw MDEFT images were skull-stripped using Jim5 (Fig 2) and the intracranial volume (ICV) was calculated from the sum of GM, WM, and CSF, after creating masks of the intracranial cavity. All of the native MDEFT images acquired from the whole head were applied to skull stripping, which was expert-reviewed and manually corrected. ICV was used to normalize each subject's brain tissue compartment and mask the raw output of SPM8 segmentation.
Tissue Segmentation
Manually deskulled MDEFT images were brought into approximate alignment with the International Consortium for Brain Mapping template21 in SPM8 running in MATLAB (v. 2009a, The MathWorks Inc., Natick, MA, USA). To account for higher image intensity nonuniformity at 3T, images were corrected using N3 (nonparametric nonuniform intensity normalization),23 with a smoothing distance of 50 mm. All scans were automatically segmented into GM, WM, and CSF probability maps using the unified segmentation model implemented in the segmentation routine of SPM8 with the bias field correction tool disabled. Mutually exclusive segmentation masks for GM, WM, and CSF were initially obtained (Fig 2) from the SPM8-derived tissue probability maps using maximum likelihood criteria (ie, pixels were assigned to the tissue compartments that had the greatest probability) as described previously.24 Two expert readers, that compared the segmentation label maps obtained for each tissue and the underlying grayscale MDEFT image, assessed the quality of the derived segmentation.
Mutually exclusive uncorrected raw segmentation GM masks (uGMM), WM masks (uWMM), and CSF masks (uCSFM) and related volumes (uGMV, uWMV, uCSFV) were automatically obtained in MATLAB. The uWMV, uGMV, and uCSFV were used to compute uncorrected WM fraction (uWMF) (uWMF = uWMV/ICV), GM fraction (uGMF) (uGMF = uGMV/ICV), and brain parenchymal fraction (uBPF) [uBPF = (uWMV + uGMV)/ICV].
Correction for Segmentation Misclassifications
After visual inspection of SPM8-derived tissue segmentations masks, we detected areas of segmentation error related to underestimation of the DGM contour and misclassification in some areas of T1 hypointense MS lesions (Fig 2). We therefore focused on testing 4 candidate outputs to measure compartment-specific brain volumes:
Raw (uncorrected—the only manual step is skull stripping): uBPF, uGMF, uWMF.
Only DGM-corrected: c1BPF, c1GMF, c1WMF.
Only T1 lesions-corrected: c2BPF, c2GMF, c2WMF.
Both DGM and T1 lesion-corrected (dually corrected): cBPF, cGMF, cWMF.
Underestimation of DGM structures was corrected by an expert trained observer on uGMM using a drawing tool in 3-dimensional Slicer (v. 3.4, http://www.slicer.org). Using MATLAB, we obtained the GM-change mask from the subtraction of manually corrected GM mask (c1GMM) and uGMM for each subject. The GM-change mask included pixels of DGM misclassified as WM as well as pixels of DGM misclassified as CSF. The GM-change mask was used to obtain corrected masks for WM and CSF (c1WMM, c1CSFM) from uWMM and uCSFM by setting pixels in the GM-change mask to background values in c1WMM and c1CSFM. Global brain volumes (c1GMV, c1WMV, c1BPV) and the corresponding corrected brain fractions (c1GMF, c1WMF, c1BPF) were recalculated.
T1 hypointensities were corrected using their mask within the WM compartment in uGMM, uWMM, and uCSFM by reassigning misclassified pixels to the proper tissue compartment in corrected masks (c2GMM, c2WMM, c2CSFM). Global brain volumes (c2GMV, c2WMV, c2BPV) and the corresponding corrected brain fractions (c2GMF, c2WMF, c2BPF) were recalculated.
Combined corrected masks (cGMM, cWMM, cCSFM) were derived from uGMM, uWMM, and uCSFM by correcting for underestimation both of DGM and T1 hypointensities (dually corrected). Final corrected global brain volumes (cGMV, cWMV, cBPV) and their corresponding dually corrected “gold standard” brain fractions (cGMF, cWMF, cBPF) were recalculated.
Assessment of Segmentation
In each subject or for each type of correction applied, we assessed the volume of tissue that was misclassified and to what extent each compartment (GM, WM, or CSF) was affected by the misclassification. We also assessed how these misclassifications affected the differences between the final compartment-specific fractional data as follows: (1) the uncorrected maps versus the other 3 maps and (2) the dually corrected maps versus the other 3 methods.
Statistical Analysis
Linear regression modeling adjusted for age and gender compared MS and NC in terms of uncorrected and corrected whole brain fractions for WMF, GMF, and BPF. In addition, MS patients and NCs were compared in terms of T1LV and T2LV using linear regression after log transformation of each of the lesion volumes. Among MS patients, the cognitively impaired and cognitively preserved groups were compared using linear regression to adjust for age-, gender-, and CES-D-defined depressive symptoms. Finally, associations between MRI-derived measures and measures of clinical status (EDSS score, disease duration, T25FW) were assessed using the Spearman's correlation coefficient in patients with MS. A P-value less than.05 was considered significant and less than.10 a trend to significance in this exploratory study. All statistical analyses were performed in R (R Development Core Team RR Foundation for Statistical Computing, Vienna, Austria, http://www.R-project.org). Effect size (d) was also calculated for group comparisons.25
Results
Errors due to Segmentation Misclassification
Table2 shows the relevant comparisons of differences in GMF, WMF, and BPF associated with the type of segmentation method in the MS group. For GMF, the total absolute error was (mean) 1.04%, and, for WMF, the total absolute error was 1.67%. Both of these effects were driven primarily by DGM misclassification. For BPF, the total error was only.026%, and was driven primarily by lesion misclassification. Regarding the tissue-specific effect of the 2 types of errors in the MS group that underlie the differences in compartmental fractions: (1) DGM was predominantly misclassified as WM (mean ± SD: 9.59 ± 1.93 ml displaced from GM to WM), and much less commonly as CSF (.01 ±.02 ml displaced from GM to CSF); (2) T1 WM hypointensities were predominantly misclassified as GM (1.99 ± 1.84 ml displaced from WM to GM), and much less commonly as CSF (.29 ±.42 displaced from WM to CSF). Because these effects were in opposite directions, they had a cancelling effect (DGM error reduced but T1 lesion error increased GMV [and vice versa with regard to WMV]); this limited the magnitude of the overall effect on final outputs as reflected in the small rates of total absolute errors. The relatively small effect of misclassification on BPF was due to the fact that errors typically did not involve misclassification of brain tissue as CSF. DGM misclassification errors were similar in magnitude between MS and NS; also, as expected, because T1 hypointense lesions were much more common in the MS group, the absolute value of the combined effects of errors was greater in the MS group (NC error data not shown).
Table 2.
Errors in Global Normalized Brain Volumes per Subject due to Segmentation Misclassifications in the MS Group
| Segmentation | % Difference | % Difference |
|---|---|---|
| Outputs Compared | (Mean) | (SD) |
| cGMF-uGMF | 1.04 | .33 |
| cGMF-c1GMF | −.28 | .26 |
| cGMF-c2GMF | 1.31 | .27 |
| uGMF-c1GMF | −1.33 | .27 |
| uGMF-c2GMF | .28 | .26 |
| cWMF-uWMF | −1.67 | .58 |
| cWMF-c1WMF | 0.53 | .52 |
| cWMF-c2WMF | −2.20 | .42 |
| uWMF-c1WMF | 2.16 | .41 |
| uWMF-c2WMF | −0.53 | .52 |
| cBPF-uBPF | .026 | .04 |
| cBPF-c1BPF | .025 | .04 |
| cBPF-c2BPF | .0006 | .002 |
| uBPF-c1BPF | −.0007 | .002 |
| uBPF-c2BPF | −.025 | .04 |
Examples of differences between segmentation output types; uncorrected (u, raw); correction of misclassifications related to either deep gray matter contouring (c1), T1 hypointensities (c2), or both (c, dually corrected).
GMF = gray matter fraction; WMF = white matter fraction; BPF = brain parenchymal fraction.
Percent differences were calculated using the intrasubject differences between the 2 values divided by that subject's first value listed in each row (eg, cGMF - uGMF = [(cGMF - uGMF)/cGMF)]*100%).
Group Differences in MRI Data
Table3 shows the group differences for compartment-specific volumes, before and after corrections for errors, in patients with MS and NC. GMF and BPF derived using all 4 methods were significantly lower in patients with MS compared to NC; while none of the WMFs showed any significant group differences. Regarding the effect sizes of these differences, the BPF changes were identical regardless of segmentation method. The dually corrected GM fraction (cGMF) demonstrated a slightly larger effect size than the other 3 methods; however, all 4 showed significance in detecting GM atrophy in the MS group. As expected, no significant group differences were found with regard to ICV (MS 1,412.2 ± 134.2, NC 1,386 ± 118.3 ml; age- and gender-adjusted P =.36).
Table 3.
Differences in Global Normalized Brain Volume Measures between Patients and Controls
| Multiple Sclerosis | Normal Controls | P-Value | Effect Size | |
|---|---|---|---|---|
| Compartmental Volume | (n = 61) | (n = 30) | (d) | |
| uGMF | .514 ±.028 | .522 ±.021 | .019* | −.323 |
| c1GMF | .521 ±.028 | .529 ±.021 | .014* | −.323 |
| c2GMF | .513 ±.028 | .522 ±.021 | .011* | −.364 |
| cGMF | .520 ±.028 | .529 ±.021 | .008* | −.364 |
| uWMF | .315 ±.019 | .324 ±.020 | .108 | −.461 |
| c1WMF | .308 ±.019 | .316 ±.020 | .131 | −.410 |
| c2WMF | .317 ±.018 | .324 ±.020 | .200 | −.368 |
| cWMF | .310 ±.018 | .316 ±.020 | .238 | −.315 |
| uBPF | .830 ±.031 | .846 ±.017 | .002* | −.640 |
| c1BPF | .830 ±.031 | .846 ±.017 | .002* | −.640 |
| c2BPF | .830 ±.030 | .846 ±.017 | .002* | −.656 |
| cBPF | .830 ±.030 | .846 ±.017 | .002* | −.656 |
Values in table are mean ± standard deviation regarding the uncorrected (u, raw) versus corrected segmentations after correcting misclassifications related to either deep gray matter contour (c1), T1 hypointensities (c2), or both (c, dually corrected).
GMF = gray matter fraction; WMF = white matter fraction; BPF = brain parenchymal fraction.
P-values are group differences based on linear regression modeling controlling for age and gender
P <.05.
MRI-Cognition Relationships
As shown in Table4, regarding all brain MRI measures of atrophy and lesions available, only BPF and T1LV were significantly worse in cognitively impaired versus cognitively preserved patients (P <.05). GMF and T2LV showed a trend to significance. No significant differences between the 2 cognitive groups were noted in WMF (P >.05). The magnitude of difference between the 2 cognitive groups was generally similar when using the uncorrected or corrected BPFs and WMFs. The cGMF demonstrated a slightly larger effect size than the uncorrected method; however, no relevant benefit could be realized (ie, neither showed a significance in the group comparison).
Table 4.
MRI Differences Based on Cognitive Status in the Multiple Sclerosis Group (n = 61)
| Cognitively Impaired | Cognitively Preserved | P-Value | Effect size | |
|---|---|---|---|---|
| Brain MRI Metric | (n = 23) | (n = 38) | (d) | |
| uGMF | .505 ±.030 | .520 ±.026 | .121 | −.534 |
| c1GMF | .512 ±.029 | .527 ±.026 | .116 | −.545 |
| c2GMF | .504 ±.030 | .519 ±.026 | .101 | −.534 |
| cGMF | .510 ±.030 | .526 ±.026 | .096 | −.570 |
| uWMF | .312 ±.015 | .317 ±.021 | .208 | −.274 |
| c1WMF | .305 ±.015 | .310 ±.020 | .210 | −.283 |
| c2WMF | .314 ±.014 | .318 ±.020 | .268 | −.232 |
| cWMF | .307 ±.015 | .312 ±.020 | .270 | −.283 |
| uBPF | .817 ±.034 | .837 ±.026 | .033* | −.661 |
| c1BPF | .817 ±.034 | .837 ±.026 | .033* | −.661 |
| c2BPF | .818 ±.034 | .837 ±.026 | .034* | −.628 |
| cBPF | .818 ±.034 | .837 ±.026 | .034* | −.628 |
| T1LV (ml) | 1.70 ± 1.10 | 1.09 ±.983 | .030* | .585 |
| T2LV (ml) | 2.53 ±.811 | 2.16 ±.730 | .078 | .480 |
Values in table are mean ± standard deviation; uncorrected (u, raw) versus corrected segmentations after correcting misclassifications related to either deep gray matter contour (c1), T1 hypointensities (c2), or both (c, dually corrected).
GMF = gray matter fraction; WMF = white matter fraction; BPF = brain parenchymal fraction; T1LV = total brain T1 hypointense lesion volume; T2LV = total brain T2 hyperintense lesion volume.
The 2 cognition groups were defined and analyzed as described in the Methods section.
P-values are comparisons between the 2 groups based on linear regression modeling controlling for age, gender, and depressive symptoms
P <.05.
MRI-Clinical Correlations in the MS Group
As shown in Table5, regarding all brain MRI measures of atrophy and lesions available, we tested their relationship to 3 measures of clinical status—physical disability (EDSS score), ambulatory function (T25FW), and disease duration (time since first MS symptoms). Among all MRI measures, only BPF showed a significant (albeit weak) correlation with EDSS score (r range −.289−.293, P ≤.05); GMF showed a trend to significant correlation with EDSS but was generally weak regardless of using uncorrected or corrected GMFs (r ∼−.224−.231, P <.10). None of the MRI measures showed a significant correlation with T25FW; GMF was the only measure that showed a trend to significant correlation with T25FW, but was generally weak regardless of using uncorrected or corrected GMFs (r ∼−.220−.234, P <.10). Only BPF (r ∼−.302−.303, P <.05) and GMF (r ∼−.341−.354, P <.05) correlated significantly with disease duration. Overall, corrections of MRI segmentation did not impact on the strength of these correlations.
Table 5.
Correlations between MRI and Clinical Status in the Multiple Sclerosis Group (n = 61)
| Expanded Disability | ||||||
|---|---|---|---|---|---|---|
| Status Scale Score |
Timed 25 Foot Walk |
Disease Duration |
||||
| Brain MRI Metric | r | P-Value | r | P-Value | r | P-Value |
| uGMF | −.230 | .075 | −.233 | .071 | −.343 | .007* |
| c1GMF | −.224 | .083 | −.220 | .089 | −.352 | .006* |
| c2GMF | −.231 | .073 | −.234 | .070 | −.341 | .007* |
| cGMF | −.223 | .084 | −.222 | .085 | −.354 | .005* |
| uWMF | −.152 | .242 | −.069 | .596 | −.053 | .688 |
| c1WMF | −.170 | .190 | −.083 | .523 | −.056 | .663 |
| c2WMF | −.141 | .278 | −.050 | .702 | −.036 | .784 |
| cWMF | −.163 | .208 | −.071 | .586 | −.029 | .824 |
| uBPF | −.289 | .024* | −.171 | .187 | −.303 | .018* |
| c1BPF | −.289 | .024* | −.171 | .187 | −.303 | .018* |
| c2BPF | −.293 | .022* | −.171 | .188 | −.302 | .018* |
| cBPF | −.293 | .022* | −.171 | .188 | −.302 | .018* |
| T1LV | .181 | .163 | −.095 | .465 | −.062 | .636 |
| T2LV | .100 | .442 | −.1621 | .212 | −.117 | .368 |
Uncorrected (u, raw) versus corrected segmentations after correcting misclassifications related to either deep gray matter contour (c1), T1 hypointensities (c2), or both (c, dually corrected).
GMF = gray matter fraction; WMF = white matter fraction; BPF = brain parenchymal fraction; T1LV = log of the total brain T1 hypointense lesion volume; T2LV = log of the total brain T2 hyperintense lesion volume.
Spearman rank correlation r and P-values are shown
P <.05.
Discussion
In this study, we sought to assess the performance of a segmentation pipeline in patients with MS using the freely available SPM8 package (requiring skull stripping as the only manual step) from 3T MRI scans. We compared this pipeline to further manual manipulations to correct errors in: (1) the contouring of DGM and (2) the misclassification of T1 hypointense lesions. The major findings of our study were that, whether either or both of these corrections were applied, there was generally no gain in the validity of the results. The ability to detect brain atrophy in MS versus healthy individuals and the ability to detect selective atrophy affecting cognitively impaired versus cognitively preserved MS patients was unaffected by application of these corrections. Furthermore, the ability to correlate brain atrophy with the level of physical disability and disease duration was also similar with the raw versus corrected outputs. Taken together, these results show the utility of this segmentation pipeline in a cross-sectional analysis. With our continuing adoption of newer, efficient skull stripping tools, it is likely that the level of automation will increase and the operator time will decrease in future studies.26,27
It should be noted that our findings might not be generalizable to a more disabled MS population. The vast majority of our patients were receiving treatment with disease-modifying therapy, were largely affected by relapsing (rather than progressive) forms of the disease, and, on average, had generally mild physical disability. Given that both DGM atrophy and T1 hypointense lesion load advance with increasing years of illness and disease severity,28,29 it remains unknown whether corrections for these errors would become relevant at those later stages of the disease. In addition, ours was a cross-sectional study; it is yet to be determined what effect these errors would have on the detection of the intrasubject rate of change of brain volume in longitudinal monitoring.
Segmentation of the DGM contour is marred by the indistinct outer boundaries of these structures in relation to the adjacent WM on MRI scans.30,31 Even though the DGM are typically the site of a large, disproportionate rate of atrophy as compared to other parts of the cerebral GM and the whole brain,32–34 our study showed that correction of the underestimation of DGM did not impact on validity of global brain volumes. This could be related to the small volumetric effect that DGM segmentation had on the magnitude of overall brain volume. Or, perhaps, the areas most vulnerable to atrophy in DGM were the least affected by segmentation error—a substructure effect we did not explore in this study.
Several studies have shown the tissue segmentation bias introduced by the presence of MS T1 hypointense lesions on GM/WM volume measures utilizing postsegmentation lesion corrections or presegmentation lesion filling.2,35,36 These studies uniformly show that GM volume is upwardly biased, at the expense of WM volume, due to these segmentation errors. These findings are in agreement with our study. Also, similar to this study, 1 previous investigation testing the effect of this error on validity of results, showed that both raw and lesion-filled images generated segmentations showing significantly lower global GM and DGM in MS patients versus healthy individuals.36 However, we would agree with the remarks of those investigators—that lesion corrections would be important as lesion load increases (in patients with advanced disability and progressive forms of MS). Such populations have yet to be fully assessed regarding these methodologic issues. In this study, we employed an MDEFT sequence for the depiction of GM/WM tissue differences. This sequence has been proposed as superior to standard 3-dimensional T1-weighted images for voxel-based morphometry.8,10 However, this sequence is not widely used, and may have a different effect on GM and lesion appearance compared to standard 3-dimensional T1-weighted images. In future studies, it would also be important to test the use of a priori “WM-filled” lesions and “GM-filled” misclassified GM as input maps to fully understand the type of corrections and type of algorithm desired for optimization.
In this study, we detected whole brain atrophy in MS patients versus healthy subjects, a finding that is almost universal in other MS studies,1 including our own previous work at 1.5T2 and 3T.37 Furthermore, our data showed that the atrophy was dominated by GM rather than WM change. Nearly all studies examining the compartment-specific effects of GM versus WM global atrophy in MS have similarly found the former to be selectively affected.1 This most likely relates to various differential influences of the MS disease process and other physiologic changes on GM versus WM tissue volume.16
When considering cognitively impaired versus cognitively preserved patients with MS, we found that whole brain volume and cerebral T1 hypointense lesion volume differed significantly between groups but GM volume did not. Several studies have showed a relationship between whole brain atrophy and cognitive impairment in MS.38–42 Our study was not powered to identify the precise anatomic substrate of cognitive impairment, but, rather to compare 4 methods of global brain segmentation. Thus, we did not investigate regional GM atrophy or specific cognitive domains, each of which may have allowed the detection of GM-behavior relationships.33,40–44
With regard to the relationship between MRI and disability, 2 key findings emerged. First, all 4 methods of measuring GMV showed a trend to a significant (P <.10) inverse correlation with ambulatory function, while none of the other MRI measures showed a significant correlation. Nonetheless, these findings are in keeping with previous studies highlighting the contribution of cerebral GM atrophy to MS-related gait impairment to a greater extent than can be explained by whole brain atrophy or cerebral lesion volume.2,45,46 A second key finding was in the evaluation of the relationship between MRI and overall neurologic disability (EDSS score), for which whole brain volume was significantly inversely correlated and GMV showed a trend to correlation. In contrast, WMV and lesion measures showed no correlation. These findings are in keeping with numerous studies showing that brain atrophy is more closely associated with physical disability than are lesion measures.1 However, it is likely that the ability to find any correlation between MRI and either ambulatory function or overall neurologic impairment was limited by the restricted range in this mildly disabled cohort, as was seen in a previous study.47
In conclusion, we report the validity of an SPM-based segmentation pipeline for the detection of MS-related brain atrophy in patients with MS from 3T MRI scans. We identified segmentation errors related to misclassification of MS lesions and ineffective deep GM contouring. However, while these errors caused bias in the segmentation of GM and WM compartments, there was no discernable influence on the measurement of normalized whole brain volume. Furthermore, the corrections did not influence any MRI-clinical associations with physical disability and cognitive impairment. Therefore, in cross-sectional studies, this pipeline has utility and efficiency. However, longitudinal studies are warranted to determine the role of such errors on disease monitoring.
Acknowledgments
Presented in preliminary form at the 2011 combined meeting of the European and Americas Committees on Treatment and Research in Multiple Sclerosis (ECTRIMS/ACTRIMS), Amsterdam, and the 2012 meeting of the American Academy of Neurology, New Orleans. We thank Mariann Polgar-Turcsanyi, MS, for database management and Jonathan S. Jackson, PhD, for valuable consultation.
References
- 1.Bermel RA, Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol. 2006;5:158–170. doi: 10.1016/S1474-4422(06)70349-0. [DOI] [PubMed] [Google Scholar]
- 2.Sanfilipo MP, Benedict RH, Sharma J. The relationship between whole brain volume and disability in multiple sclerosis: a comparison of normalized gray vs. white matter with misclassification correction. NeuroImage. 2005;26:1068–1077. doi: 10.1016/j.neuroimage.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Dalton CM, Chard DT, Davies GR. Early development of multiple sclerosis is associated with progressive grey matter atrophy in patients presenting with clinically isolated syndromes. Brain. 2004;127:1101–1107. doi: 10.1093/brain/awh126. [DOI] [PubMed] [Google Scholar]
- 4.Pirko I, Lucchinetti CF, Sriram S. Gray matter involvement in multiple sclerosis. Neurology. 2007;68:634–642. doi: 10.1212/01.wnl.0000250267.85698.7a. [DOI] [PubMed] [Google Scholar]
- 5.Bakshi R, Thompson AJ, Rocca MA. MRI in multiple sclerosis: current status and future prospects. Lancet Neurol. 2008;7:615–625. doi: 10.1016/S1474-4422(08)70137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kangarlu A. High-field magnetic resonance imaging. Neuroimag Clin N Am. 2009;19:113–128. doi: 10.1016/j.nic.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Tardif CL, Collins DL, Pike GB. Regional impact of field strength on voxel-based morphometry results. Hum Brain Mapp. 2010;31:943–957. doi: 10.1002/hbm.20908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tardif CL, Collins DL, Pike GB. Sensitivity of voxel-based morphometry analysis to choice of imaging protocol at 3T. NeuroImage. 2009;44:827–838. doi: 10.1016/j.neuroimage.2008.09.053. [DOI] [PubMed] [Google Scholar]
- 9.Wonderlick JS, Ziegler DA, Hosseini-Varnamkhasti P. Reliability of MRI-derived cortical and subcortical morphometric measures: effects of pulse sequence, voxel geometry, and parallel imaging. NeuroImage. 2009;44:1324–1333. doi: 10.1016/j.neuroimage.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceccarelli A, Jackson JS, Tauhid S. The impact of lesion in-painting and registration methods on voxel-based morphometry in detecting regional cerebral gray matter atrophy in multiple sclerosis. AJNR Am J Neuroradiol. 2012;33:1579–1585. doi: 10.3174/ajnr.A3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura K, Fisher E. Segmentation of brain magnetic resonance images for measurement of gray matter atrophy in multiple sclerosis patients. NeuroImage. 2009;44:769–776. doi: 10.1016/j.neuroimage.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neema M, Goldberg-Zimring D, Guss ZD. 3T MRI relaxometry detects T2 prolongation in the cerebral normal-appearing white matter in multiple sclerosis. Neuroimage. 2009;46:633–641. doi: 10.1016/j.neuroimage.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasan KM, Walimuni IS, Abid H. Multi-modal quantitative MRI investigation of brain tissue neurodegeneration in multiple sclerosis. J Magn Reson Imaging. 2012;35:1300–1311. doi: 10.1002/jmri.23539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones BC, Nair G, Shea CD. Quantification of multiple-sclerosis-related brain atrophy in two heterogeneous MRI datasets using mixed-effects modeling. NeuroImage Clin. 2013;13:171–179. doi: 10.1016/j.nicl.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polman CH, Reingold SC, Edan G. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria. Ann Neurol. 2005;58:840–846. doi: 10.1002/ana.20703. et al. ”. [DOI] [PubMed] [Google Scholar]
- 16.Khoury S, Bakshi R. Cerebral pseudoatrophy or real atrophy after therapy in multiple sclerosis. Ann Neurol. 2010;68:778–779. doi: 10.1002/ana.22254. [DOI] [PubMed] [Google Scholar]
- 17.Neema M, Guss ZD, Stankiewicz JM. Normal findings on brain fluid-attenuated inversion recovery MR images at 3T. AJNR Am J Neuroradiol. 2009;30:911–916. doi: 10.3174/ajnr.A1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benedict RH, Cookfair D, Gavett R. Validity of the minimal assessment of cognitive function in multiple sclerosis (MACFIMS) J Int Neuropsychol Soc. 2006;12:549–558. doi: 10.1017/s1355617706060723. [DOI] [PubMed] [Google Scholar]
- 19.Parmenter BA, Testa SM, Schretlen DJ. The utility of regression-based norms in interpreting the minimal assessment of cognitive function in multiple sclerosis (MACFIMS) J Int Neuropsychol Soc. 2010;16:6–16. doi: 10.1017/S1355617709990750. [DOI] [PubMed] [Google Scholar]
- 20.Deichmann R, Schwarzbauer C, Turner R. Optimisation of the 3D MDEFT sequence for anatomical brain imaging: technical implications at 1.5 and 3 T. NeuroImage. 2004;21:757–767. doi: 10.1016/j.neuroimage.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 21.Ashburner J, Friston KJ. Unified segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Stankiewicz JM, Glanz BI, Healy BC. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging. 2011;21:e50–e56. doi: 10.1111/j.1552-6569.2009.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE T Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 24.Chard DT, Griffin CM, Parker GJ. Brain atrophy in clinically early relapsing-remitting multiple sclerosis. Brain. 2002;125:327–337. doi: 10.1093/brain/awf025. [DOI] [PubMed] [Google Scholar]
- 25.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale, NJ: Lawrence Erlbaum Associates; 1988. [Google Scholar]
- 26.Galdames FJ, Jaillet F, Perez CA. An accurate skull stripping method based on simplex meshes and histogram analysis for magnetic resonance images. J Neurosci Methods. 2012;206:103–119. doi: 10.1016/j.jneumeth.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 27.Arora A, Moscufo N, Neema M. Increasing efficiency of intracranial cavity determination from high-resolution 3 tesla(T) MRI scans of the brain. Neurology. 2013;79:106–121. [Google Scholar]
- 28.Fisher E, Lee JC, Nakamura K. Gray matter atrophy in multiple sclerosis: a longitudinal study. Ann Neurol. 2008;64:255–265. doi: 10.1002/ana.21436. [DOI] [PubMed] [Google Scholar]
- 29.Ceccarelli A, Rocca MA, Pagani E. A voxel-based morphometry study of grey matter loss in MS patients with different clinical phenotypes. NeuroImage. 2008;42:315–322. doi: 10.1016/j.neuroimage.2008.04.173. [DOI] [PubMed] [Google Scholar]
- 30.Helms G, Draganski B, Frackowiak R. Improved segmentation of deep brain grey matter structures using magnetization transfer (MT) parameter maps. NeuroImage. 2009;47:194–198. doi: 10.1016/j.neuroimage.2009.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derakhshan M, Caramanos Z, Giacomini PS. Evaluation of automated techniques for the quantification of grey matter atrophy in patients with multiple sclerosis. NeuroImage. 2010;52:1261–1267. doi: 10.1016/j.neuroimage.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 32.Bermel RA, Innus MD, Tjoa CW. Selective caudate atrophy in multiple sclerosis: a 3D MRI parcellation study. NeuroReport. 2003;14:335–339. doi: 10.1097/00001756-200303030-00008. [DOI] [PubMed] [Google Scholar]
- 33.Houtchens MK, Benedict RHB, Killiany R. Thalamic atrophy and cognition in multiple sclerosis. Neurology. 2007;69:1213–1223. doi: 10.1212/01.wnl.0000276992.17011.b5. [DOI] [PubMed] [Google Scholar]
- 34.Bergsland N, Horakova D, Dwyer MG. Subcortical and cortical gray matter atrophy in a large sample of patients with clinically isolated syndrome and early relapsing-remitting multiple sclerosis. AJNR Am J Neuroradiol. 2012;33:1573–1578. doi: 10.3174/ajnr.A3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chard DT, Jackson JS, Miller DH. Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. J Magn Reson Imaging. 2010;32:223–228. doi: 10.1002/jmri.22214. [DOI] [PubMed] [Google Scholar]
- 36.Gelineau-Morel R, Tomassini V, Jenkinson M. The effect of hypointense white matter lesions on automated gray matter segmentation in multiple sclerosis. Hum Brain Mapp. 2012;33:2802–2814. doi: 10.1002/hbm.21402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klawiter EC, Ceccarelli A, Arora A. Corpus callosum atrophy correlates with gray matter atrophy in patients with multiple sclerosis. J Neuroimaging. 2014 doi: 10.1111/jon.12124. et al. [Epub ahead of print May 9,; DOI: 10.1111/jon.12124 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benedict RH, Carone DA, Bakshi R. Correlating brain atrophy with cognitive dysfunction, mood disturbances, and personality disorder in multiple sclerosis. J Neuroimaging. 2004;14:36S–45S. doi: 10.1177/1051228404266267. [DOI] [PubMed] [Google Scholar]
- 39.Amato MP, Zipoli V, Portaccio E. Multiple sclerosis-related cognitive changes: a review of cross-sectional and longitudinal studies. J Neurol Sci. 2006;245:41–46. doi: 10.1016/j.jns.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 40.Sanfilipo MP, Benedict RH, Weinstock-Guttman B. Gray and white matter brain atrophy and neuropsychological impairment in multiple sclerosis. Neurology. 2006;66:685–692. doi: 10.1212/01.wnl.0000201238.93586.d9. [DOI] [PubMed] [Google Scholar]
- 41.Benedict RH, Bruce JM, Dwyer MG. Neocortical atrophy, third ventricular width, and cognitive dysfunction in multiple sclerosis. Arch Neurol. 2006;63:1301–1306. doi: 10.1001/archneur.63.9.1301. [DOI] [PubMed] [Google Scholar]
- 42.Calabrese M, Agosta F, Rinaldi F. Cortical lesions and atrophy associated with cognitive impairment in relapsing-remitting multiple sclerosis. Arch Neurol. 2009;66:1144–1150. doi: 10.1001/archneurol.2009.174. [DOI] [PubMed] [Google Scholar]
- 43.Morgen K, Sammer G, Courtney SM. Evidence for a direct association between cortical atrophy and cognitive impairment in relapsing-remitting MS. NeuroImage. 2006;30:891–898. doi: 10.1016/j.neuroimage.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 44.Sicotte NL, Kern KC, Giesser BS. Regional hippocampal atrophy in multiple sclerosis. Brain. 2008;131:1134–1141. doi: 10.1093/brain/awn030. [DOI] [PubMed] [Google Scholar]
- 45.Tjoa CW, Benedict RHB, Weinstock-Guttman B. MRI T2 hypointensity of the dentate nucleus is related to ambulatory impairment in multiple sclerosis. J Neurol Sci. 2005;234:17–24. doi: 10.1016/j.jns.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Shiee N, Bazin PL, Zackowski KM. Revisiting brain atrophy and its relationship to disability in multiple sclerosis. PLoS One. 2012;7:e37049. doi: 10.1371/journal.pone.0037049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen AB, Neema M, Arora A. The relationships among MRI-defined spinal cord involvement, brain involvement, and disability in multiple sclerosis. J Neuroimaging. 2012;22:122–128. doi: 10.1111/j.1552-6569.2011.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


