Abstract
The entry of new all-oral direct acting antiviral therapy for hepatitis C provides an opportunity to scale up HCV care in low- and middle-income countries. In HIV, use of dried blood spots (DBS) has facilitated the diagnosis and management of HIV in resource-poor settings. DBS may be used in a similar way to facilitate diagnosis and management of HCV. Here, we present a systematic review of the literature of DBS for HCV RNA detection and genotyping. Using an a priori review protocol, three databases were searched for studies published up to August 2013 that reported the use of dried blood and serum spots in genotyping, detection and measurement of HCV RNA, as well as the rate of degradation of HCV RNA when stored in DBS at room temperature. Nine papers were eligible for inclusion; eight studied DBS and one dried serum. Two studies measured concordance between genotype and subtype determined by DBS and whole plasma and both found 100% concordance. Four studies measured endpoint detection limits of HCV RNA-positive samples by DBS and found positive predictive values of 100% down to 250, 334, 2500 and 24160 IU/mL. Two studies found deterioration of HCV RNA in DBS samples stored at room temperature, while two others failed to detect such deterioration. These results support the potential use of DBS for genotyping and HCV RNA detection. Studies of the use of DBS for HCV RNA viral load measurement and of the rate of degradation of HCV RNA when stored in DBS at ambient temperatures remain inconclusive.
Keywords: dried blood spot, hepatitis C, LMIC, RNA storage, viral load quantification
Globally, there are approximately 130–150 million people living with hepatitis C (HCV), and the majority of these live in low- and middle-income countries (LMICs) [1,2]. Due in part to the complexity and cost of the current algorithm for diagnosis and treatment using pegylated interferon, the majority of LMICs do not support HCV programming and as a result, without accessible and effective treatment, nearly half a million people die of HCV annually [2,3]. However, the more effective and tolerable oral direct acting antivirals (DAAs) offer the opportunity to significantly simplify both the treatment and the diagnostic algorithm, enabling the implementation, decentralization and scale-up of HCV care in LMICs. To further simplify diagnosis and monitoring of HCV, dried blood spots (DBS) may be considered.
DBS has been used to aid in the diagnosis of a wide variety of pathogens, including assessment of antibody to viral or bacterial infections such as HIV, hepatitis B, HPV and measles virus [4,5] and qualitative and quantitative viral load detection in HIV [6]. In particular, use of DBS in detecting and measuring HIV RNA has aided in decentralization for HIV services in low-resource areas and expanded the ability to perform early infant diagnosis for children at risk of vertical transmission [7].
DBS for detection and monitoring of HCV viral load has a number of advantages. Especially salient for resource-poor settings is the possibility of using DBS to store and transport samples to a central laboratory without having to use refrigeration or dry ice, which is necessary for serum/plasma samples (when stored at room temperature, serum/plasma must be processed within 6h of venous blood draw, necessitating transportation within that time) [8–10]. DBS can also be prepared using capillary blood, which obviates the need, seen in venipuncture, for centrifugation to separate blood cells from serum/plasma [11]. In injection drug users (IDUs), venipuncture can be complicated by the difficulty in finding an accessible vein, and thus, capillary blood from fingerprick samples may be easier to obtain [12]. DBS holds advantages over oral fluid sampling, which has also been used to detect HCV, as it has been shown that HCV RNA in saliva is independent of plasma viral load [13], and patients with low serum HCV RNA viral loads are less likely to have detectable HCV in saliva [14]. Other advantages of DBS over venipuncture include lower cost, minimal storage facility and transport requirements, decreased donor discomfort, and decreased risk to health care workers [10]. If DBS could be used for HCV RNA detection and monitoring, it would facilitate the simplification and decentralization of HCV diagnosis and monitoring [9,15].
Today, a significant literature supports the use of DBS as an alternative to serum/plasma obtained via venipuncture for the detection of HCV antibody [16–18], yet the use of DBS in the detection and monitoring of HCV RNA and genotyping HCV, especially following storage at room temperature, has not been systematically reviewed [19]. To address the question of whether DBS can be used in the diagnosis of HCV in resource limited settings, we undertook a systematic review of use of DBS for HCV RNA detection and genotyping, examining a range of storage conditions.
Materials and Methods
Search strategy
Using a sensitive search strategy, as part of a predefined protocol (Appendix 1), we searched MEDLINE, CAB Abstracts and Web of Science (ISI Citation Index) published up to August 2013 for studies meeting our prespecified inclusion criteria. No date or geographical exclusions were applied; only English-language publications were included. Following an initial screen of abstracts by two separate reviewers, full text copies of potential eligible articles were reviewed independently by two reviewers. A title search of references was performed on articles meeting the inclusion criteria to determine potential articles for inclusion not identified during the initial database search. After all articles meeting the inclusion criteria were identified, data were abstracted by two reviewers according to prespecified categories.
Inclusion/exclusion criteria
We sought studies that reported on the use of DBS as a tool for monitoring and genotyping HCV RNA that included at least one of the following: the sensitivity and/or specificity of DBS in HCV RNA viral load quantification; the accuracy of DBS for genotyping HCV RNA; and the rate of degradation of HCV RNA during transport and storage at room temperature. Nine of the 73 articles met these criteria.
Data analysis
For studies measuring HCV RNA presence and quantification, studies were analysed for viral load endpoint detection, sensitivity, specificity, PPV and NPV of DBS compared to HCV RNA in whole plasma. For the question of HCV RNA deterioration at room temperature in DBS, reviewers extracted reports of the rate of deterioration and the definition of ‘room temperature’. Studies determining HCV genotype were analysed for measures of genotype concordance between whole plasma and DBS, proportion of HCV RNA-positive DBS samples successfully genotyped, and genotypes observed.
Results
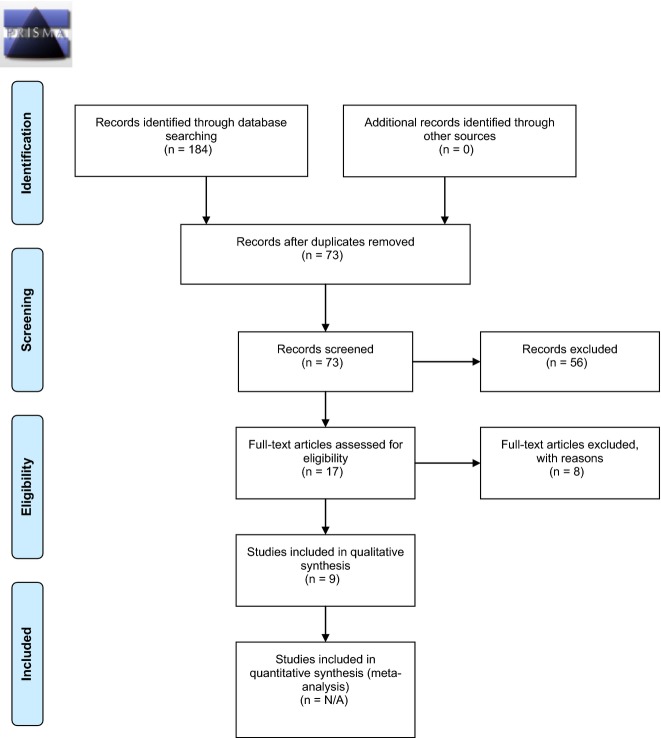
Seventy-three titles were identified using the initial database search and taken for abstract review. Seventeen were selected for full text review. Overall, 9 met eligibility criteria for the review (See flow table in Fig.1 [20]). Characteristics and findings of the studies included are summarized in Table1 (for additional information, see a more complete table in the online appendices).
Figure 1.
PRISMA 2009 Flow Diagram for systematic literature review. From reference [20]. For more information, visit http://www.prisma-statement.org.
Table 1.
Description of key variables in the included studies
| References | Diagnostic question | Study site and population | DBS specimen collection method | Assay | HCV RNA+ samples sucessfully genotyped (types) | DBS VL endpoint detection limit (IU/mL) | DBS Sens/Spec (samples) | DBS PPV/NPV (samples) | Stability during storage at room temp |
|---|---|---|---|---|---|---|---|---|---|
| Hope et al. [24] |
Genotype | UK 299 IDUs > 15 years 177/299 anti-HCV+ 117/288 RNA+ |
Capillary blood on Whatman 903 card |
Dye Terminator Cycle Sequencing with Quick Start Kit (Beckman Coulter); CEQ 8000 genetic analysis system (Beckman Coulter); SEQMAN (DNAS-TAR) | 114/117 (1, 2, 3, 4) | – | – | – | – |
| Bennett et al. [21] |
VL, stability | UK 80 anti-HCV+ |
50 μL whole blood on Whatman 903 cards |
ABI 7500 (Applied Biosystems); ABI 9700 (Applied Biosystems) | – | ≥150 | 100% (57/57)/ 95.7% (22/23) |
98.3% (57/58)/ 100% (22/22) |
no significant variation in cycle threshold over 1 year (2 DBS from 1 patient) |
| Santos et al. [11] |
VL | Brazil 100 on Tx 68 and 24 weeks on Tx |
Four drops capillary blood on SS903 card |
pCR-II-TOPO plasmid (Invitrogen) | – | – | 98.0% (99/101)/ 94.0% (63/67) |
96.1% (99/103)/ 96.9% (63/65) |
– |
| Mahfoud et al. [25] |
Genotype | Lebanon 56 IDUs, anti-HCV+ 28 RNA+ |
Capillary blood on SS903 card |
LINEAR ARRAY HCV Genotyping Test (Roche); COBAS AMPLICOR Analyzer (Roche) |
28/28 (1, 3, 4) |
– | – | – | – |
| De Crignis et al. [9] |
VL | Italy 13 HIV+/HCV+ adults 4 HIV+ adults 3 HCV+ adults 5 HIV+/HCV− blood donors |
50 μL whole blood on Whatman no. 3 card |
Versant HCV RNA 3.0 b-DNA Assay (Siemens); QIAamp DNA Mini kit (Qiagen); Quantitect SYBR Green PCR Master Mix (Qiagen); Taq polymerase (Invitrogen) |
– | 2500 | 93.8% (15/16)/ 100% (9/9) |
100% (15/15)/ 90% (9/10) |
– |
| Tuaillon et al. [15] |
Genotype, VL, stability |
France 100 anti-HCV+ 100 ant-HCV− |
3 drops (50 μL) capillary blood on Whatman 903 card |
Cobas Ampliprep Total Nucleic Acid Isolation 100 kit (Roche); Cobas TaqMan HCV test and real-time PCR COBAS TaqMan 48 instrument plus COBAS Ampliprep analyzer (Roche); One Step RT-PCR kit from Qiagen (Qiagen) |
14/14 (1, 2, 3, 4) |
≥178 | 96.8%(60/62)/– | – | 3-fold decrease in RNA in 6 days (no. of samples not reported) |
| Plamondon et al. [23] |
Genotype | Guinea-Bissau >50 years |
Capillary blood on Whatman no. 3 card |
One-Step RT-PCR System (Roche) |
57/65 (1, 2) |
– | – | – | – |
| Solmone et al. [8] |
Genotype, VL, stability |
Italy 39 anti-HCV+ 34/39 RNA+ 16 anti-HCV− |
50 μL EDTA whole blood on SS903 card |
In-house RT-PCR | 8/8 (1, 2, 3, 4) |
≥960 | 100% (124/124)/ 100% (24/24) |
100% (124/124)/ 100% (24/24) |
No loss of RNA positivity after 11 months (16 paired samples) |
| Abe & Konomi [22] |
Stability | Japan 8 anti-HCV+ 4 anti-HCV− |
50 μL serum on filter paper |
ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer) |
– | – | – | – | 10-fold reduction in virus yield in 4 weeks (in 6 of 8 samples, no samples lost HCV RNA positivity) |
Study characteristics
The selected studies were published between 1998 and 2012; all but two were published between 2007 and 2012. Two of the studies were located in the UK, two in Italy and one each in France, Lebanon, Brazil, Guinea-Bissau and Japan. Four studies reported outcomes in terms of endpoint sensitivity of HCV RNA detection using DBS when compared to plasma [8,9,15,21]. Four studies provided data sufficient to calculate the sensitivity, specificity, positive predictive value and negative predictive value of DBS in HCV RNA detection against the standard of paired plasma samples [8,9,11,21]; one study provided only enough information to measure sensitivity [15]. Four studies measured the stability of HCV RNA in DBS at room temperature [8,15,21,22]. Two studies reported outcomes in terms of concordance between DBS and whole plasma in genotyping [8,15]. Five studies reported the proportion of HCV RNA-positive samples that could be genotyped [8,15,23–25].
Sample characteristics
The patient characteristics varied across studies. Two studies drew from a population of solely IDUs recruited by respondent-driven sampling [24,25], another from a cross-section of individuals over 50 years of age [23]. One study used DBS to measure the viral load response to treatment [11], while the others did not specify the treatment experience.
Across the four studies assessing the stability of HCV RNA at room temperature, storage time varied between 2 days and twelve months from collection. The definition of ‘room temperature’ across four studies measuring stability of HCV RNA was given in two studies [21,22], although not in two others [8,15].
Finally, the preparation of DBS varied among the samples. Eight of the nine studies used DBS (only one used dried serum). Eight studies used capillary blood, while one used venous blood [8]. The studies used a variety of RNA extraction and elution methods and nucleic acid amplification assays.
DBS and HCV viral load quantification
HCV RNA was detected by DBS in 100% of HCV-positive samples with serum viral loads as low as 150–250 IU/mL [21], 334 IU/mL [15], 2500 IU/mL [9] and 4830–24 160 IU/mL [8]. Studies with paired serum and DBS samples found sensitivities to HCV RNA ranging from 93.8–100% and specificities ranging from 94.0–100% [8,9,11,15,21]. In one study of patients on treatment (100 patients after 4 weeks, and 68 patients after 24 weeks of pegylated interferon alfa-2b + ribavirin), sensitivity was 98%, specificity was 94.0%, PPV was 96.1% and NPV was 96.9% [11]. The one study that compared DBS HCV RNA concentration with paired serum HCV RNA found a strong correlation (r2 = 0.94; P < 0.001) [15].
One study found deterioration of HCV RNA in DBS samples stored at room temperature, while two other studies evidenced far more stability. Tuaillon et al. found a threefold decrease in HCV RNA levels at 6 days compared to those stored at −20°C (although the definition of ‘room temperature’ and number of samples tested were not specified) [15]. One study saw no loss of HCV RNA positivity in 16 DBS samples after 11 months, although titres were not quantified to determine whether there was a decrease in RNA levels (again the definition of ‘room temperature’ was not specified) [8]. Another study using 2 DBS samples tested in duplicate saw no variation in the stability of HCV in DBS at 21°C over a 1-year period using an endpoint limit of detection of 250 IU/mL [21]. The one study of dried serum spots demonstrated a 10-fold reduction in virus yield in six of eight tested samples stored at 20–25°C after 4 weeks as compared with frozen samples [22].
DBS and HCV genotyping
The two studies measuring concordance between genotype and subtype determined by DBS and plasma both found 100% concordance; one study had 8 paired samples [8], and another had 14 paired samples [15]. Four studies were able to successfully genotype most (114/117[24] and 57/65[23]) or all (28/28 [25] and 14/14 [15]) HCV RNA-positive DBS samples.
Discussion
Overall, this review shows that DBS has potential for use in HCV RNA detection, quantification and genotyping. DBS is likely sufficient to detect and quantify HCV RNA for the purpose of HCV diagnosis. Viral load endpoint detection limits varied widely, yet the highest-powered study designed specifically to measure endpoint sensitivity found the lowest threshold of 150–250 IU/mL [21]. This remains higher than the threshold of 50 IU/mL for plasma samples, a difference multiple authors attribute to the limited sample collected by DBS using a standard 1-cm-diameter DBS punch size holding 50 μL and less efficient nucleic acid extraction[11,21,26]. The strong correlation between DBS HCV RNA concentration and paired serum HCV RNA provides additional evidence that the comparatively lower sensitivity of DBS to HCV RNA detection is attributable to this difference in sample amount [9,15]. Given that most untreated patients with HCV have viral loads >1000 IU/mL[15], the difference in sensitivity between DBS and serum/plasma may be more important for monitoring of patients during treatment than in initial diagnosis.
While one study found a strong correlation between HCV RNA concentration on DBS and serum viral load [15], every other author using DBS for viral load assumed the correlation was imprecise and relied instead on DBS for a qualitative determination of the presence or absence of HCV RNA in blood. This qualitative use does not allow for the precise evaluation of virologic response that does not reach undetectable levels [8,9,11,21]. However, given that DAA-based treatment will rely less on response-guided treatment, HCV monitoring guidelines may change and decrease the importance of quantitative monitoring. In the near future, only qualitative virologic testing may be necessary to confirm infection, undetectable viral load at end of treatment, and sustained virologic response (SVR). Thus, the increasing use of DAAs may enable use of DBS for monitoring and confirmation of SVR.
False-positive findings for HCV RNA with DBS are not yet entirely explicable, although Santos [11] conjectures that the four such ‘false-positive’ DBS samples found in patients early in their treatment course (pegylated interferon alfa-2b + ribavirin) might be attributable to the presence of HCV in B lymphocytes found in the DBS samples. This would account for the reproducibly negative results for the plasma samples of these patients even while their DBS specimens contained HCV RNA. Indeed, Stapleton et al. found that whole-blood-based HCV RNA detection was more sensitive than plasma-based methods and hypothesized that the difference was due to the presence of cell-associated HCV RNA in whole blood [27]. As cell-associated HCV RNA has been shown to persist long after spontaneous clearance or successful treatment, this may prove to be a problem for determining SVR by DBS [28]. However, techniques to reduce the measurement of cell-associated virus are being developed for measurement of HIV RNA using DBS, and these same techniques may be applied to HCV DBS [29].
The most persistent unanswered question in the literature is the stability of HCV RNA in DBS when stored at room temperature. Since the first study of this question was published fifteen years ago, only three additional studies have appeared in the peer-reviewed literature. One study found a threefold decrease in RNA within 6 days but did not report the number of samples tested [15]; another reported a 10-fold decrease in 6 of 8 samples within 4 weeks [22]; yet another found no significant quantitative degradation in two samples over the course of a year [21]; and the fourth reported 16 samples with no loss of qualitative RNA positivity after 11 months. Only two of the four studies actually specify the definition of ‘room temperature,’ and none specified humidity levels. The small aggregate number of samples and the wide discrepancy in results begs additional study of this important question. Even the most rapid of the reported rates of degradation might allow for the use of DBS for HCV RNA detection in the diagnosis (if not the management) of patients without refrigeration in settings removed from central laboratories, provided transport is available within days. Still, the rate of HCV RNA degradation will determine the need to store and transport DBS cards at −20°C soon after collection in many settings where immediate transport is not possible [19].
Use of DBS for HCV genotyping is extremely promising. Most studies demonstrated that DBS provided sufficient sample and good correlation for genotyping. The only study where PCR amplification was insufficient to genotype at least 97% of HCV RNA-positive DBS samples used a single, short, non-coding region and drew samples from a geographically broad section of the general population of Guinea-Bissau.
Conclusion
Use of DBS may help to enable diagnosis and monitoring of HCV in resource-limited settings and facilitate decentralization of care within these settings. DBS for measurement of HCV RNA appears to be a reliable tool in qualitative determination of the presence or absence of HCV RNA in the blood, although DBS is less specific than serum or plasma due to a higher endpoint detection limit. With new oral medicines reaching the market, there may be less of a need for stringent quantification of viral load, so DBS may be useful in monitoring and confirmation of sustained virologic response. Because degradation of HCV RNA could cause positive samples to fall below limits of detection, the rate of degradation of HCV RNA when stored in DBS at ambient temperatures requires systematic study. The current literature supports the use of DBS in HCV genotyping. Further study and field validation are needed to help the scale-up of HCV care in LMICs.
Funding
The authors have no personal or funding interests to declare.
Glossary
- DBS
dried blood spots
- DAAs
direct acting antivirals
- DBS
dried blood spots
- LMICs
low- and middle-income countries
- SVR
sustained virologic response
Appendix
Appendix 1
References
- 1.Hanafiah KM, Groeger J, Flaxman AD, et al. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 2014. Hepatitis C Factsheet. April. Available at: http://www.who.int/mediacentre/factsheets/fs164/en/ (Accessed 16 August, 2014)
- 3.WHO. 2013. Global policy report on the prevention and control of viral hepatitis. Available at: http://www.who.int/csr/disease/hepatitis/global_report/en/ (accessed 17 August, 2014)
- 4.Parker SP, Cubitt WD. The use of the dried blood spot sample in epidemiological studies. J Clin Pathol. 1999;52:633–639. doi: 10.1136/jcp.52.9.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waterboer T, Dondog B, Michael KM, et al. Dried blood spot samples for seroepidemiology of infections with human papillomaviruses, Helicobacter pylori, and hepatitis C virus. Cancer Epidemiol Biomark Prev. 2012;21:287–293. doi: 10.1158/1055-9965.EPI-11-1001. [DOI] [PubMed] [Google Scholar]
- 6.Johannessen A, Garrido C, Zahonero N, et al. Dried blood spots perform well in viral load monitoring of patients who receive antiretroviral treatment in rural tanzania. Clin Infect Dis. 2009;49(6):976–981. doi: 10.1086/605502. [DOI] [PubMed] [Google Scholar]
- 7.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS, et al. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. JAIDS. 2005;38(5):615–617. doi: 10.1097/01.qai.0000143604.71857.5d. [DOI] [PubMed] [Google Scholar]
- 8.Solmone M, Girardi E, Costa F, Pucillo L, Ippolito G, Capobianchi MR. Simple and reliable method for detection and genotyping of Hepatitis C virus RNA in dried blood spots stored at room temperature. J Clin Microbiol. 2002;40(9):3512. doi: 10.1128/JCM.40.9.3512-3514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. HIV-1 and HCV detection in dried blood spots by SYBR Green multiples real-time PCR. J Virol Methods. 2010;165:51–56. doi: 10.1016/j.jviromet.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 10.McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44:899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- 11.Santos C, Reis A, dos Santos CV, et al. The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. J Virol Methods. 2012;179:17–20. doi: 10.1016/j.jviromet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Hickman M, McDonald T, Judd A, Nichols T, Hope V, Skidmore S, Parry JV. Increasing the uptake of hepatitis C virus testing among injecting drug users in specialist drug treatment and prison settings using dried blood spots for diagnostic testing: a cluster randomized controlled trial. J Viral Hepat. 2008;15:250–254. doi: 10.1111/j.1365-2893.2007.00937.x. [DOI] [PubMed] [Google Scholar]
- 13.L Lins, H Almeida, L Vitvisk, T Carmo, R Paraná, MG Reis. Detection of hepatitis C virus RNA in saliva is not related to oral health status or viral load. J Med Virol. 2005;77(2):216–220. doi: 10.1002/jmv.20438. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Omata K, Satoh T, et al. Quantitative detection of hepatitis C virus (HCV) RNA in saliva and gingival crevicular fluid of HCV-infected patients. J Clin Microbiol. 2005;43(9):4413–4417. doi: 10.1128/JCM.43.9.4413-4417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuaillon E, Mondain A, Meroueh F, et al. Dried blood spot for Hepatitis C virus serology and molecular testing. Hepatology. 2010;51(3):752–758. doi: 10.1002/hep.23407. [DOI] [PubMed] [Google Scholar]
- 16.Parker SP, Cubitt WD, Ades AE. A method for the detection and confirmation of antibodies to hepatitis C in dried blood spots. J Virol Methods. 1997;68:199–205. doi: 10.1016/s0166-0934(97)00127-4. [DOI] [PubMed] [Google Scholar]
- 17.Judd A, Parry J, Hickman M, et al. Evaluation of a modified commercial assay in detecting antibody to hepatitis C virus in oral fluids and dried blood spots. J Med Virol. 2003;71:49–55. doi: 10.1002/jmv.10463. [DOI] [PubMed] [Google Scholar]
- 18.Parker SP, Khan HI, Cubitt WD. Detection of antibodies to hepatitis C virus in dried blood spot samples from mothers and their offspring in Lahore, Pakistan. Journal of Clinical Microbiology. 1999;37:2061–2063. doi: 10.1128/jcm.37.6.2061-2063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snijdewind IJ, van Kampen JJ, Fraaij PL, van der Ende ME, Osterhaus AD, Gruters RA. Current and future applications of dried blood spots in viral disease management. Antiviral Res. 2012;93:309–321. doi: 10.1016/j.antiviral.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG PRISMA Group. Preferred reporting items for systematic meta-analyses: the PRISMA statement. PLoS Med. 6(6):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett S, Gunson RN, McAllister GE, Hutchinson SJ, Goldberg DJ, Cameron SO, Carman WF. Detection of hepatitis C virus RNA in dried blood spots. J Clin Virol. 2012;54:106–109. doi: 10.1016/j.jcv.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Abe K, Konomi N. Hepatitis C virus RNA in dried serum spotted onto filter paper is stable at room temperature. J Clin Microbiol. 1998;36(10):3070. doi: 10.1128/jcm.36.10.3070-3072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plamondon M, Labbé AC, Frost E, Deslandes S, Alves AC, Bastien N, Pepin J. Hepatitis C virus infection in Guinea-Bissau: a sexually transmitted genotype 2 with parenteral amplification? PLoS ONE. 2007;2(4):e372. doi: 10.1371/journal.pone.0000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hope VD, Hickman M, Ngui SL, et al. Measuring the incidence, prevalence and genetic relatedness of hepatitis C infections among a community recruited sample of injecting drug users, using dried blood spots. J Viral Hepatitis. 2011;18:262–270. doi: 10.1111/j.1365-2893.2010.01297.x. [DOI] [PubMed] [Google Scholar]
- 25.Mahfoud Z, Kassak K, Kreidieh K, Shamra S, Ramia S. Distribution of hepatitis C virus genotypes among injecting drug users in Lebanon. Virol J. 2010;7:96. doi: 10.1186/1743-422X-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevaliez S, Rodriguez C, Pawlotsky JM. New virologic tools for management of chronic hepatitis B and C. Gastroenterology. 2012;142:1303–1313. doi: 10.1053/j.gastro.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stapleton JT, Klinzman D, Schmidt WN, et al. Prospective comparison of whole-blood- and plasma-based hepatitis C virus RNA detection systems: improved detection using whole blood as the source of viral RNA. J Clin Microbiol. 1999;37:484–489. doi: 10.1128/jcm.37.3.484-489.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham T, et al. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78(11):5867–5874. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Baum P, Crask M, Do T, Honisch C, Will S. 2014. A Simple Method to Elute Cell-Free HIV from Dried Blood Spots Improves the Ascertainment of Virologic Suppression. International AIDS Conference. June 25–27,, Melbourne, Australia. [DOI] [PubMed]