Abstract
We highlight the various domains of the flavivirus virulence factor NS1 and speculate on potential implications of the NS1 3D structure in understanding its role in flavivirus pathogenesis. Flavivirus non-structural protein 1 (NS1) is a virulence factor with dual functions in genome replication and immune evasion. Crystal structures of NS1, combined with reconstructions from electron microscopy (EM), provide insight into the architecture of dimeric NS1 on cell membranes and the assembly of a secreted hexameric NS1-lipid complex found in patient sera. Three structural domains of NS1 likely have distinct roles in membrane association, replication complex assembly, and immune system avoidance. A conserved hydrophobic inner face is sequestered either on the membrane or in the interior of the secreted hexamer and contains regions implicated in viral replication. The exposed variable outer face is presented to cellular and secreted components of the immune system in infected patients and contains candidate regions for immune system modulation. We anticipate that knowledge of the distinct NS1 domains and assembly will lead to advances in elucidating virus-host interactions mediated through NS1 and in dissecting the role of NS1 in viral genome replication.
Introduction
Flaviviruses, which are transmitted to humans by mosquito and tick vectors, are responsible for a large number of diseases, including dengue, West Nile, and yellow fever. As the range of their insect vectors has increased, the range of many flavivirus-caused diseases has likewise increased. Flaviviruses are positive-strand RNA viruses whose genomes encode a large polyprotein that is processed into three structural (E, M and C) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins [1-3]. Of the non-structural proteins, NS3 and NS5 have well defined catalytic roles in genome duplication and protein processing. NS2A, NS2B, NS4A and NS4B are short transmembrane proteins thought to anchor the catalytic proteins to a replication complex (RC) within special vesicle packets located on the cytoplasmic side of the ER membrane [4]. NS1 is essential to genome replication and can be complemented in trans [5], although it has no known catalytic function. The location of NS1 on the lumenal face of the ER membrane suggests an organizational role in the formation of the RC. Interactions identified through genetic methods between NS1 and both NS4A and NS4B [6,7], and confirmed physically for NS4B [6], suggest that the organizational role is mediated through these components.
NS1 is found both as a membrane-associated dimer and as a secreted, lipid-associated, hexamer. Dimeric NS1 is found on membranes both in cellular compartments and on the cell surface. Secreted NS1 (sNS1) is present in patient serum, often at very high levels, where, in the case of dengue viruses, the level of NS1 correlates with the onset of hemorrhagic fever [8]. In addition to the role in genome replication, NS1 has effects on the innate immune response. NS1 has been observed in multiple interactions with the complement system through factor H [9], C1s and C4 [10], and C4 binding protein [11]. Flavivirus propagation is modulated by dsRNA-sensing pathways through the antiviral RIG-I and MDA5 pattern-recognition receptors [12-14], and by the TLR3 dsRNA-response system [15,16]. While direct effects of NS1 on the RIG-I/MDA5 pathway have not been reported, NS1 has been directly implicated in the TLR3 response [17-19], although this latter effect is controversial [20].
Several recent reviews have covered biological and low-resolution structural information available for NS1 in some detail [21-23]. Here we incorporate the structural information gleaned from the recently published atomic-resolution crystal structures for West Nile virus (WNV) and dengue virus type 2 (DENV2) NS1 [24,25] with the pertinent biological knowledge. Distinct domains of the NS1 structure appear suited to its cellular compartments and functions. These observations lead to testable hypotheses to further dissect the role of NS1 in virus replication and immune evasion.
NS1 has three distinct domains and forms “inner” and “outer” faces
The NS1 structure comprises three domains (Figure 1A): the hydrophobic “β-roll” (amino acids 1 – 29, Figure 1B: left), followed by an α/β “wing” domain (38 – 151, Figure 1B: right) with its RIG-I-like fold, and finally the central β-ladder containing an extended β-sheet on one face and a “spaghetti loop”, which although ordered lacks defined secondary structure elements, on the opposite face (181 – 352, Figure 1B: center) [25]. The intervening segments (30 – 37 and 152 – 180) form a 3-stranded β-sheet “connector” subdomain connecting the wing to the central β-roll and β-ladder domains. NS1 is stabilized by twelve conserved cysteines, which form six disulfide bonds [26,27], and it is post-translationally modified at two or three conserved N-linked glycosylation sites per monomer [28]. The crystal structures confirm four of the six predicted disulfides, with the observed linkage for the remaining two (C291-C312 and C313-C316) differing by a switch of adjacent cysteines from the predicted linkage (C291-C313 and C312-C316). Upon translocation into the ER, NS1 rapidly dimerizes and is glycosylated [29,30]. When assembled into a dimer, NS1 has a distinct cross shape, with the two RIG-I-like wing domains extending from the central β-ladder. Two distinct faces are formed. The “inner face” is hydrophobic and displays the β-roll domain and the exposed β-sheet from the central β-ladder. The “outer face” is hydrophilic and displays the glycosylation sites and the spaghetti loop. A number of sulfate binding sites identified in the crystal structures also localize to the hydrophilic outer face [24,25].
Figure 1.
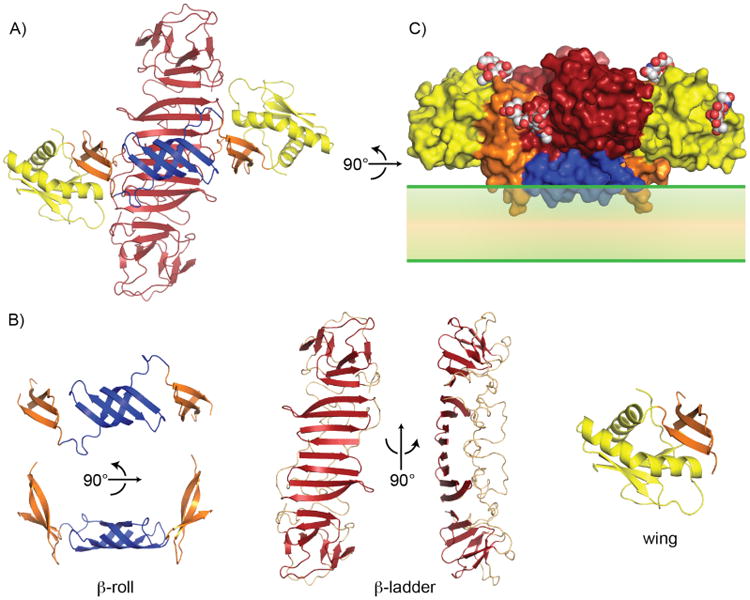
NS1 dimer structure. (A) NS1 dimer with “inner face” towards viewer. Blue: β-roll, yellow: RIG-I like wing domain, and red: β-ladder. (B) Dissection of NS1 structure highlighting three domains. (left) β-roll and connector subdomain (orange), (center) β-ladder with “spaghetti” loop (pale salmon), and (right) wing domain with connector subdomain. (C) NS1 dimer surface (colored as in A) with inner face placed on membrane (green). Hydrophobic protrusion, including residues that affect virus RNA replication (10 – 11 and 159 – 162), interacts with the membrane surface. Sugar residues decorate the outer surface (white, red and blue spheres).
As both the isolated β-ladder domain and intact NS1 form a stable dimer, any monomeric form of NS1 is likely transient and of minimal biological relevance. We suggest that the inner face of the NS1 dimer associates with the membrane surface with a “hydrophobic protrusion” (formed by the β-roll and residues 159 – 162 of the connector subdomain) in contact with the membrane (Figure 1C). This arrangement orients the outer hydrophilic face of the dimer, including the spaghetti loop, glycosylation sites and sulfate binding sites, towards the lumen, and subsequently the extracellular space. Additionally, the most variable regions of the NS1 surface map to the hydrophilic outer face which would be exposed both when NS1 is associated with membranes, and on the secreted NS1 hexamer (see below). Membrane association is an inherent property of NS1, as recombinant wild-type NS1 is found in the membrane fraction of insect cells, separated from membranes by mild detergent treatment and, when purified, coats and remodels lipid vesicles [25]. A proposed GPI anchor [31,32] is not required for membrane association, as it was not present in the recombinant protein. Residues at the β-roll (10 – 11 on a β-turn) and in the connector subdomain (159 – 162 on a β-turn, Figure 1B) are important for genome replication [6,25], and the same β-roll residues are also implicated in a direct interaction with NS4B [6]. These results imply that the hydrophobic protrusion faces the membrane and interacts with other proteins of the replication complex. Interestingly, NS1 variants with substitutions at 159 – 162 from hydrophobic to charged character, although capable of binding to and remodeling liposomes, are deficient in supporting viral replication, and when expressed recombinantly these NS1 variants are secreted as dimers into the media of insect cell cultures, in contrast to wild-type NS1, which is consistently membrane associated. [25]. The increased production of secreted dimers from these constructs suggests both that the less hydrophobic forms of NS1 are poorly retained in the ER lumen – perhaps accounting for the decreased viral production – and that it is not necessary for NS1 to form an intracellular hexamer prior to secretion.
The NS1 hexamer exposes an outer variable face
Two EM reconstructions have been reported for secreted hexameric NS1 [33,34]. Although they differ in several details, both structures suggest similarly sized barrel shaped hexamers with a hollow interior. A 30-Å cryo-EM reconstruction [34] was generated with the assumption that secreted NS1 exhibits perfect hexameric (D3) symmetry while a 23-Å negative-stain EM reconstruction [33] assumed only 3-fold symmetry. Thus the differences in the resultant maps may be due to the combination of sample preparation (cryo vs. negative stain) and reconstruction protocol (D3 vs. 3-fold symmetry). The major difference between the two reconstructions is a barrel with one closed end in the negative-stain model and a hollow pore extending through the barrel center in the cryo-EM model. A cargo of lipids in the interior channel of the NS1 hexamer has been characterized using chromatographic techniques, and is similar in composition to that found in high-density lipoproteins [34].
Another atomic model for the secreted NS1 hexamer comes from a second crystal form of WNV NS1, as well as a structure of the complete DENV2 NS1. These two independent crystal structures yielded almost identical hexamers. In conjunction with the 2-fold symmetry inherent to NS1, the resultant assembly has hexameric D3 symmetry (Figure 2A,B). In this hexamer, no obvious protein-protein contacts form inter-dimer associations, and the hexamers are likely held together by a heterogeneous mixture of bound detergent from the protein purification and molecules of the polyethylene glycol (PEG) used in crystallization. Both hexamers have similar overall dimensions, and the pitch of the β-ladder is the same in both forms. Two-dimensional projections from negative-stain EM for the lipid-free recombinant hexamer are consistent with the crystallographic hexamer [25], and more importantly, the crystallographic hexamer is consistent with the 30-Å cryo-EM reconstruction of secreted hexamers (Figure 2A,B) [34], including the pitch of the β-ladder motif. This is remarkable given that the crystallized hexamer lacks lipid and the secreted hexamer in the cryo-EM reconstruction is lipid replete. Despite the uniform appearance of secreted NS1 lipoprotein hexamers that were visualized in the cryo-EM reconstruction, they are apparently heterogeneous, as they have thus far eluded all attempts at crystallization. This is consistent with the loose association of three recombinant, lipid-free NS1 dimers in the crystal structures.
Figure 2.

NS1 hexamer. (A,B) Orthogonal views of crystallographic NS1 hexamer (cartoon – dimers in blue, yellow and green) superposed on cryo-EM map from [34]. Wing domains (in red circles) are not accounted for in cryo-EM map. (C) Outer surface of NS1 hexamer decorated with sulfate-binding, antibody-binding and glycosylation sites. Blue dimer from A and B is shown in grey. Glycosylation (black, red spheres) and sulfate sites (yellow circles) are indicated. Epitope for the neutralizing Fab 22NS1 is shown in orange.
The crystallized hexamers of both DENV2 NS1 and WNV NS1 direct the hydrophobic protrusion to the hexamer interior, where presumably the lipid cargo would be sequestered. The sugar residues, spaghetti loops and sulfate binding sites are thus directed to the exterior. Additionally, the structure of the DENV2 β-ladder in complex with the Fab of a neutralizing antibody (22NS1) [24] is consistent with this hexamer arrangement (Figure 2C), as the 22NS1 epitope is on the hexamer exterior. The 22NS1 epitope overlaps with epitopes mapped for human patient antibodies showing that the hexamer exterior is recognized by secondarily infected dengue patients [35]. Additionally, regions of NS1, which are most varied among the four major dengue serotypes, map almost exclusively to the outer face (Figure 3). Despite these similarities, the cryo-EM reconstruction appears to lack the overall cross shape of the NS1 hexamer (Figure 2 – red circles). This may be accounted for by the differences in sample preparation as the NS1 crystal hexamers have been stripped of lipids and reconstituted with detergent and PEG while the EM reconstructions are from secreted NS1 with presumably a more native lipid cargo. Alternatively, heterogeneity in the secreted NS1 lipoprotein may have obscured structural details in the EM reconstruction. With the recent insights into NS1 structure and solution behavior and improvements in electron microscopy instrumentation and methodology, it may be feasible to generate EM reconstructions of native secreted NS1 particles in which particle heterogeneity is the sole resolution-limiting factor.
Figure 3.
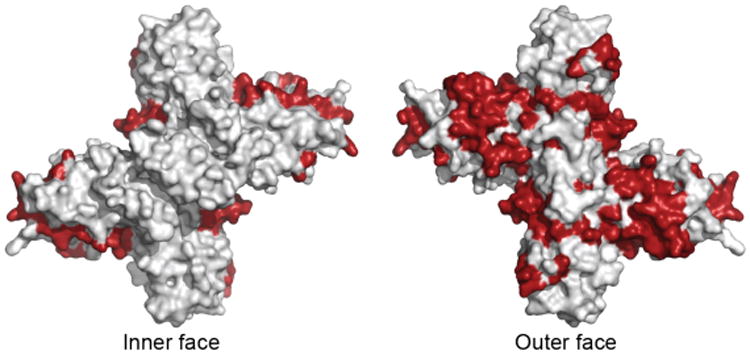
Dengue virus NS1 variable regions (red), identified from an alignment of consensus sequences from the four major dengue serotypes, mapped onto dimer surface.
Membrane-associated NS1 is both enriched at lipid rafts [32] and found on the luminal side of invaginated vesicle packets where replication occurs [4]. It is not clear whether NS1 localization is a physical property of NS1 itself, a property of one of the transmembrane non-structural proteins (i.e. NS4B [36]), or of some other, possibly host-derived, targeting mechanism. The observed enrichment in cholesterol of the lipid cargo [34] may be a result of NS1 association with lipid rafts [32] prior to hexamer formation. The transition from membrane-associated dimeric NS1 to secreted, hexameric NS1 could occur when three dimers associate on a membrane surface, pass through a tripod intermediate, and pinch off the lipid cargo as they associate into the barrel shaped particle [34].
Wing and β-ladder domains - players in immune system interaction?
Secreted NS1 interacts with the cell surface through glycosaminoglycans (GAG) in a sulfation-dependent manner [37,38]. Multiple sulfate binding sites identified in the NS1 crystal structures [24,25] are on the exterior of the proposed hexamer where sulfate recognition would be unimpeded (Figure 2C). Thus a rather simple system for cell surface interactions could be achieved through NS1 association with sulfated GAG. Whether the hexamer remained intact on the cell surface or dissociated into membrane-associated dimers, interactions with cell surface receptors and complement components could occur while NS1 is associated with the cell surface.
An important role for NS1 in complement mediation has been observed through interactions with factor H, C1s, C4 and C4 binding protein [9-11]. A common element of these proteins is repeats of the complement control protein (CCP) domain, a short (∼60 residue) disulfide-bonded motif. Several structures of pathogen proteins in complex with CCP motifs (also called sushi domains) have been reported (PDB: 2W80, 3O8E, 4J38 and related entries) [39-41]. Interestingly, these examples have in common an anti-parallel β-sheet motif through which the pathogenic protein interacts with the CCP domain. The β-roll and β-ladder domains of NS1, both rich in anti-parallel β-strands, are thus likely candidates for complement protein binding. These interactions would occur on the cell surface where they could be facilitated by the membrane association of NS1.
NS1 may interact with two dsRNA recognition systems of the innate immune response. Among eukaryotic proteins, the NS1 wing domain is most similar to the double-stranded RNA (dsRNA) sensor (DExD/H box helicase domain) from the RIG-I and MDA5 dsRNA response elements of the immune system. RIG-I and MDA5, which are implicated in attenuating flavivirus replication [12,13], are located exclusively in the cytoplasm and thus are not likely candidates for direct NS1 mimicry. However, the similarity of the NS1 wing domain with an RNA sensor may provide clues to the role of NS1 in attenuation of dsRNA sensing. When the NS1 wing domain is aligned with RIG-I/MDA5, the resultant β-ladder domain placement is equivalent to that for dsRNA in RIG-I-like receptor (RLR) - dsRNA complexes. This and the negatively charged surface of the wing domain suggest that the NS1 wing does not bind dsRNA but rather the wing domain in conjunction with the β-ladder may interfere with RLR-dsRNA signaling in some as yet undetermined fashion. At present it is not possible to propose a role for the NS1 wing domain in the RLR-mediated innate immune response, but the structural similarity may prove to be informative as our understanding of the complex RLR system improves.
In contrast with the RLR receptors, a direct role for NS1 interference with dsRNA sensing through the extracellular TLR3 sensor has been reported [13,26,37]. Amino acid positions found to affect antiviral signaling were mapped to the C-terminal region of the β-ladder. Mutations at the conserved positions Pro320 and Met333 were found to have the largest effect on antiviral signaling while still allowing viral replication. In contrast to the RLR pathways, the TLR3 dsRNA-sensing domain is extracellular and no topological constraints exist. Thus we can speculate that the β-ladder may interfere with dsRNA sensing through TLR3, but a direct physical interaction between NS1 and TLR3 has not been reported, and the NS1 connection to the TLR3 system has been disputed [5].
Conclusions and outlook
The NS1 crystal structures reveal a remarkable segregation into domains and surfaces that appear to be related to specific biological functions. These lead to directly testable hypotheses about the still-enigmatic functions of NS1. The identification and characterization of the three-domain architecture of NS1 will facilitate mutational analysis that can help to associate the multiple functions of NS1 with specific regions of the protein. We anticipate a number of informative experiments directed at dissecting interactions of NS1 with other viral factors in viral replication and with host factors in immune system modulation. For example, the β-ladder domain may be a useful probe for additional interaction studies, as it is stable and well behaved in isolation [24]. The role of NS1 in dsRNA sensing pathways needs to be further elucidated. A direct role in the TLR3 pathway is controversial, but the tools are available to test for direct interactions with NS1 and components of the TLR3 pathway. Identification of additional dsRNA-sensing elements and their association, if any, with NS1 would be very exciting. The established associations of NS1, such as with the complement proteins, could be dissected. Additionally, advances in NS1 preparation and in EM instrumentation may allow for higher-resolution visualization of native secreted NS1. Eventually NS1 or its constituent domains may be useful for development of flavivirus vaccines and antiviral drugs.
Acknowledgments
Supported by a grant from the National Institutes of Health (P01AI055672) to RJK and JLS, by the Martha L. Ludwig Professorship of Protein Structure and Function to JLS.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bollati M, Alvarez K, Assenberg R, Baronti C, et al. Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Antiviral Res. 2010;87:125–48. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinton MA. Replication cycle and molecular biology of the West Nile virus. Viruses. 2014;6:13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–40. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–17. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youn S, Li T, McCune BT, Edeling MA, et al. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol. 2012;86:7360–71. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–21. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libraty DH, Young PR, Pickering D, Endy TP, et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis. 2002;186:1165–8. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- 9.Chung KM, Liszewski MK, Nybakken G, Davis AE, et al. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A. 2006;103:19111–6. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010;207:793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avirutnan P, Hauhart RE, Somnuke P, Blom AM, et al. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol. 2011;187:424–33. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errett JS, Suthar MS, McMillan A, Diamond MS, et al. The essential, non-redundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013 doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazear HM, Pinto AK, Ramos HJ, Vick SC, et al. The pattern recognition receptor MDA5 modulates CD8+ T cell-dependent clearance of West Nile virus from the central nervous system. J Virol. 2013 doi: 10.1128/JVI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suthar MS, Ma DY, Thomas S, Lund JM, et al. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog. 2010;6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daffis S, Samuel MA, Suthar MS, Gale M, Jr, et al. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82:10349–58. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholle F, Mason PW. West Nile virus replication interferes with both poly (I:C)-induced interferon gene transcription and response to interferon treatment. Virology. 2005;342:77–87. doi: 10.1016/j.virol.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Crook KR, Miller-Kittrell M, Morrison CR, Scholle F. Modulation of innate immune signaling by the secreted form of the West Nile virus NS1 glycoprotein. Virology. 2014;458-459:172–82. doi: 10.1016/j.virol.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison CR, Scholle F. Abrogation of TLR3 inhibition by discrete amino acid changes in the C-terminal half of the West Nile virus NS1 protein. Virology. 2014;456-457:96–107. doi: 10.1016/j.virol.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82:8262–71. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baronti C, Sire J, de Lamballerie X, Querat G. Nonstructural NS1 proteins of several mosquito-borne Flavivirus do not inhibit TLR3 signaling. Virology. 2010;404:319–30. doi: 10.1016/j.virol.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Chuang YC, Wang SY, Lin YS, Chen HR, et al. Re-evaluation of the pathogenic roles of nonstructural protein 1 and its antibodies during dengue virus infection. J Biomed Sci. 2013;20:42. doi: 10.1186/1423-0127-20-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller DA, Young PR. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013;98:192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11:115–28. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 24.Edeling MA, Diamond MS, Fremont DH. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc Natl Acad Sci U S A. 2014;111:4285–90. doi: 10.1073/pnas.1322036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akey DL, Brown WC, Dutta S, Konwerski J, et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science. 2014;343:881–5. doi: 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blitvich BJ, Scanlon D, Shiell BJ, Mackenzie JS, et al. Determination of the intramolecular disulfide bond arrangement and biochemical identification of the glycosylation sites of the nonstructural protein NS1 of Murray Valley encephalitis virus. J Gen Virol. 2001;82:2251–6. doi: 10.1099/0022-1317-82-9-2251. [DOI] [PubMed] [Google Scholar]
- 27.Wallis TP, Huang CY, Nimkar SB, Young PR, et al. Determination of the disulfide bond arrangement of dengue virus NS1 protein. J Biol Chem. 2004;279:20729–41. doi: 10.1074/jbc.M312907200. [DOI] [PubMed] [Google Scholar]
- 28.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–88. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 29.Winkler G, Maxwell SE, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171:302–5. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- 30.Winkler G, Randolph VB, Cleaves GR, Ryan TE, et al. Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology. 1988;162:187–96. doi: 10.1016/0042-6822(88)90408-4. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, et al. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. Faseb J. 2000;14:1603–10. doi: 10.1096/fj.14.11.1603. [DOI] [PubMed] [Google Scholar]
- 32.Noisakran S, Dechtawewat T, Avirutnan P, Kinoshita T, et al. Association of dengue virus NS1 protein with lipid rafts. J Gen Virol. 2008;89:2492–500. doi: 10.1099/vir.0.83620-0. [DOI] [PubMed] [Google Scholar]
- 33.Muller DA, Landsberg MJ, Bletchly C, Rothnagel R, et al. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J Gen Virol. 2012;93:771–9. doi: 10.1099/vir.0.039321-0. [DOI] [PubMed] [Google Scholar]
- 34.Gutsche I, Coulibaly F, Voss JE, Salmon J, et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc Natl Acad Sci U S A. 2011;108:8003–8. doi: 10.1073/pnas.1017338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omokoko MD, Pambudi S, Phanthanawiboon S, Masrinoul P, et al. A highly conserved region between amino acids 221 and 266 of dengue virus non-structural protein 1 is a major epitope region in infected patients. The American journal of tropical medicine and hygiene. 2014;91:146–55. doi: 10.4269/ajtmh.13-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufusi PH, Kelley JF, Yanagihara R, Nerurkar VR. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One. 2014;9:e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, et al. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007;3:e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youn S, Cho H, Fremont DH, Diamond MS. A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J Virol. 2010;84:9516–32. doi: 10.1128/JVI.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Persson BD, Reiter DM, Marttila M, Mei YF, et al. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14:164–6. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- 40.Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–3. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharjee A, Oeemig JS, Kolodziejczyk R, Meri T, et al. Structural basis for complement evasion by Lyme disease pathogen Borrelia burgdorferi. J Biol Chem. 2013;288:18685–95. doi: 10.1074/jbc.M113.459040. [DOI] [PMC free article] [PubMed] [Google Scholar]


