Abstract
Neuron navigator 2 (Nav2) was first identified as an all-trans retinoic acid (atRA)-responsive gene in human neuroblastoma cells (retinoic acid-induced in neuroblastoma 1, RAINB1) that extend neurites after exposure to atRA. It is structurally related to the Caenorhabditis elegans unc-53 gene that is required for cell migration and axonal outgrowth. To gain insight into NAV2 function, the full-length human protein was expressed in C. elegans unc-53 mutants under the control of a mechanosensory neuron promoter. Transgene expression of NAV2 rescued the defects in unc-53 mutant mechanosensory neuron elongation, indicating that Nav2 is an ortholog of unc-53. Using a loss-of-function approach, we also show that Nav2 induction is essential for atRA to induce neurite outgrowth in SH-SY5Y cells. The NAV2 protein is located both in the cell body and along the length of the growing neurites of SH-SY5Y cells in a pattern that closely mimics that of neurofilament and microtubule proteins. Transfection of Nav2 deletion constructs in Cos-1 cells reveals a region of the protein (aa 837-1065) that directs localization with the microtubule cytoskeleton. Collectively, this work supports a role for Nav2 in neurite outgrowth and axonal elongation and suggests this protein may act by facilitating interactions between microtubules and other proteins such as neurofilaments that are key players in the formation and stability of growing neurites.
Keywords: retinoic acid, microtubules, neurite outgrowth, neurofilament, unc-53
Introduction
The vitamin A metabolite, all-trans retinoic acid (atRA) is essential for normal development of the vertebrate nervous system. Roles for atRA in patterning the nervous system during early development and in later neuronal specification are well established (reviewed in: Gavalas and Krumlauf, 2000; Clagett-Dame and DeLuca, 2002; Appel and Eisen, 2003; Maden, 2006). There is also a growing body of literature to support a role for atRA in promoting neurite outgrowth, axonal pathfinding and neuronal regeneration (reviewed in Clagett-Dame et al., 2006; Mey and McCaffery, 2004; Mey, 2006).
Neurite outgrowth requires a partnership between the actin cytoskeleton and the microtubule network. To initiate neurite formation, microtubules align and form a tight bundle while actin filaments reorganize to produce the growth cone, thereby providing the force required for initiating outgrowth. Microtubules then act to stabilize and maintain the neurites resulting in neurite elongation (da Silva and Dotti, 2002). The intermediate filament neurofilament proteins play important structural roles and influence protein trafficking, cellular motility, and intracellular signaling, all of which contribute to neurite outgrowth and cell survival (Helfand et al., 2003).
atRA induces neurite outgrowth of human neuroblastoma SH-SY5Y cells. The atRA-responsive gene, retinoic acid-induced in neuroblastoma 1 (RAINB1, a.k.a. Nav2, Pomfil2 and Helad1), was first identified by screening a subtractive SH-SY5Y library and is rapidly induced (within 4 h) by atRA (Merrill et al., 2002). Additionally, Nav2 mRNA was detected in the developing rat nervous system where its expression is sensitive to both high and low levels of atRA (Merrill et al., 2002). These studies, however, did not address whether Nav2 was obligate for atRA to elicit effects on neurite outgrowth.
Nav2 is one of three members of the neuron navigator family (Maes et al., 2002). The largest Nav2 ORF encodes a full-length protein of 261 kDa with several putative functional domains, including a calponin-homology (CH) domain at the N-terminus, a domain that is often found in cytoskeletal and signal transduction proteins (Gimona and Mital, 1998; Stradal et al., 1998); and several coiled-coil regions, as well as a SH3-binding motif, capable of mediating protein-protein interactions. Additionally, Nav2 contains an ATP/GTP nucleotide-binding site (AAA-domain) at the C-terminus. The AAA-domain is found in a large number of proteins and is associated with a wide variety of cellular activities related to conformational remodeling of substrate proteins leading to protein degradation, DNA replication, membrane fusion and microtubule motor movement (Hanson and Whiteheart, 2005). NAV2 and NAV3 both contain a calponin homology domain whereas NAV1 does not. The physiological function(s) of Nav2 are largely unknown. In addition to its induction by atRA in human neuroblastoma cells, Nav2 (Helad 1) has been reported to have helicase and endonuclease activities in vitro, and has been implicated in colorectal carcinogenesis by the APC signaling pathway (Ishiguro et al., 2002). Nav2 is the closest human homolog of C. elegans unc-53 and there is significant conservation of functional domains between these proteins. The C. elegans unc-53 mutant shows disrupted longitudinal migration of several cell types including neurons, developing sex myoblasts and the excretory cell (Stringham et al., 2002). In the nervous system, there is abnormal sprouting of the mechanosensory neurons including truncation of the posterior lateral microtubule PLM neurons (Hekimi and Kershaw, 1993). Evidence that NAV2 plays a role in nervous system function in vivo was also recently provided by the group led by Moehars, who demonstrated that mice hypomorphic for this gene have impaired sensory function (Peeters et al., 2004).
In order to better understand how Nav2 functions, the ability of the human NAV2 protein to rescue the defect in mechanosensory neuron elongation in the C. elegans unc-53 mutant was studied. A loss-of-function approach was used to evaluate the importance of Nav2 in the atRA induction of neurite outgrowth in SH-SY5Y cells. The cellular location of both the endogenous and ectopically over expressed protein was determined, and transfection of Nav2 deletion constructs in Cos-1 cells was performed to determine regions of the Nav2 sequence involved in specifying the subcellular location of the protein.
Methods
Plasmids
For mammalian expression the Nav2 cDNA (7.29 kb, accession no. AF466144) was cloned into the Not I-EcoR I site of pIRES-hrGFP-1α (Stratagene) yielding the 2429 aa NAV2 protein containing a 3× C-terminal flag tag, construct A (Merrill, 2004). The full length NAV2 ORF was also cloned into the Xba I-EcoR I site of the IRES-EGFP plasmid pMES (kind gift from Dr. Catherine Krull, University of Michigan) driving expression from the chick β Actin promoter and containing a 1× flag tag (Swartz et al., 2001). For expression in C. elegans, the Nav2 ORF (7.29 kb, accession no AF466144) contained within the pIRES-hrGFP-1α expression vector was removed by digestion with Age I and Mfe I and subcloned into the Age I-EcoR I site of the C. elegans expression vector pPD96.41. This resulted in a construct with the human Nav2 containing a single flag tag at the C-terminus expressed under the control of the mec-7 promoter (Pmec-7∷NAV-2∷FLAG). The pPD96.41 plasmid was obtained from the Fire Laboratory vector kit, Carnegie Institute of Washington, Baltimore, MD.
Various fragments generated by digestion of construct A were subcloned into the pIRES-hrGFP-1α plasmid to generate constructs C (BstZ17 I and Cla l, with T4 polymerase fill in), D (Ale I- Ale I) and E (Stu I- Stu I). Construct F was prepared by removing the Sma I fragment from construct D, followed by the insertion into this site of a Sma I digest of a Ale I- Ale I fragment obtained from construct A, generating Nav2 lacking sequence encoding for amino acids 451-836. Construct G (719-1331 aa) was PCR-amplified from construct A using the following primers: forward 5′ GGA TCC ATG CCT GAG GCT CGG CGG CTG CG and reverse 5′ GTC GAC GGG TGG CAT GAG GAT TGG AGA TGA, followed by subcloning into pIRES-hrGFP-1α.
Construct B (1066-2429) was designed based on a shorter transcript identified by PCR from SH-SY5Y cells, in which an alternate start site/alternate exon 1 was identified that produced 16 unique amino acids and then joined to exon 14 of the full-length Nav2 sequence (accession no. AF466144). This 97 bp sequence along with 215 bp of the rest of the transcript starting at exon 14 was PCR amplified from a reverse transcribed cDNA template of SH-SY5Y cells treated with RA (1 μM) for 24 h. The PCR primers used were: forward 5′ GCG GCC GCA TCC TTC AGC ACT TCA GAG G and reverse 5′ TGA TCA TGG CCA GGC CGG CGG CGG AAC. The 312 bp PCR product was cloned into TOPO-PCRII (Invitrogen), and sequenced (GeneBank accession no. EU625578). This construct was cut with Not I and Bcl I and cloned into Not I-Bcl I digested NAV2 in pMES.
Expression of NAV2 in C. elegans unc-53(e404)
All nematodes were cultured using standard techniques (Brenner, 1974). The C. elegans strain EG1194 (lin-15(n765ts); oxls1[Pmec-7∷GFP;lin-15(+)]), a kind gift from Dr. E. Jorgensen (University of Utah), was designated as “wild-type”. EG1194 was crossed with unc-53(e404) (Brenner, 1974) to generate PT500 (unc-53(e404); oxIs1), allowing direct observation of mechanosensory neurons using fluorescent microscopy. Transgenic worms were created by injecting PT500 hermaphrodites with Pmec-7∷NAV-2:FLAG or pPD96.41 along with a co-transformation marker DNA (Pmyo-2∷GFP) (Okkema PG, 1993; Mellow and Fire, 1995). Transgenic strains that stably expressed the marker plasmid were further characterized. In transgenic worms expressing Pmec-7∷NAV-2:FLAG, the mec-7 promoter drives strong expression of NAV-2 in the mechanosensory neurons and weak expression in the FLPs and ALNs. unc-53(e404) worms are egg-laying deficient, so eggs were isolated from gravid adults using a hypocholorite treatment (Sulston and Hodgkin, 1988). Twenty-four hours after hatching, animals from each transgenic line were mounted on agarose pads formed on microscope slides and photographed with a Spot2 camera (Diagnostic Instruments Inc.) attached to an inverted Nikon Diaphot 200 microscope and analyzed in Metamorph v4.5 (Universal Imaging Corporation). The distance from the center of the PLM cell body to the anterior most end of the axonal process was measured and divided by the distance from the grinder of the pharynx to the anus yielding the relative length of PLM measure.
Cell culture and transfection
The human neuroblastoma cell line SH-SY5Y was maintained as previously described (Clagett-Dame et al., 1993); Cos-1 (ATCC) and HEK-293FT (Invitrogen) were grown in DMEM (4.5 g/L glucose) with 10% FBS. All three cell lines were cultured at 37° C with 5% CO2. Cos-1 and HEK-293FT cells were transfected using Fugene 6 (Roche) as per the manufacturer's guidelines.
Antibody production, immunocytochemistry, and immunoblotting
A polyclonal antibody was generated in rabbit to a 15-mer peptide (anti-NAV2 PGSKWRRNPSDMSDE) conjugated to keyhole limpet hemocyanin. The antibodies were affinity purified (Sigma-Genosys, USA). The antibody detects the 261 kDa human NAV2 protein overexpressed in Cos1 cells as assessed by Western blotting (data not shown).
For immunostaining endogenous NAV2 in SH-SY5Y cells and in Cos-1 cells transfected with Nav2 and other deletion constructs, cells were rinsed once in ice-cold phosphate-buffered saline (PBS) and fixed in 100% methanol for 5 min at -20° C. SH-SY5Y cells used for quantitative analyses of neurite outgrowth were fixed in 4% paraformaldhyde (PFA) in PBS at room temperature for 30 min.
The primary antibodies used were: anti-NAV2 affinity purified rabbit polyclonal antibody (2 μg/ml), monoclonal anti-neurofilament medium chain 160 kDa, RMO 270 (ascites, 5 μg/ml; Zymed, Invitrogen), affinity purified rabbit polyclonal anti α-tubulin to C-terminal amino acids 426-450 (0.2 μg/ml, Abcam), affinity purified monoclonal anti α-tubulin, Clone B-5-1-2 (1.25 μg/ml, Sigma), and affinity purified monoclonal anti-Flag, M2 (0.5 μg/ml, Sigma). The secondary antibodies used were: goat anti-rabbit IgG H+L Alexa 488 (2 μg/ml) or Alexa 594 (2 μg/ml), goat anti-mouse IgG H+L Alexa 350 (40 μg/ml) or Alexa 488 (2 μg/ml) or Alexa 594 (2 μg/ml) (Invitrogen).
All immunocytochemistry was performed at room temperature. Cells fixed in methanol were rehydrated in PBS for 5 min followed by blocking for 1 h in 5% normal goat serum in PBS. Primary antibodies were diluted in 5% goat serum in PBS, incubated with cells for 2 h followed by washing in PBS and secondary antibody incubation for 1 h. Cells were washed three times (5 min each) in PBS and the coverslips were then mounted in gel/mount permanent aqueous mounting media (Biomeda). For counterstaining cell nuclei, 300 nM DAPI in 0.9% saline replaced the third washing step, and cells were then briefly rinsed twice in PBS and mounted. For quantitation of neurite outgrowth, SH-SY5Y cells fixed in 4% PFA were incubated in 5% goat serum in PBS with 0.5% Triton X-100 for 10 min followed by blocking, primary (monoclonal alpha-tubulin) and secondary antibodies as above.
Total cell extracts in Laemmli sample buffer (60 mM Tris HCl, pH 6.8, 5% sodium dodecylsulfate, 5% BME, 10% glycerol and 0.05% bromophenol blue) were resolved by SDS PAGE, and proteins were transferred to a nitrocellulose membrane (Bio-Rad). The membranes were rinsed in PBS and then blocked in 5% skim milk powder in PBS for 1 h at room temperature followed by primary antibodies in blocking buffer for another 1 h. Membranes were washed three times in PBS before incubating with secondary antibodies conjugated to HRP in block buffer for 1 h. Antigen-antibody conjugates were visualized by Supersignal West Pico Chemiluminecent kit (Pierce), and exposed to X-ray film.
Microtubule depolymerization-repolymerization
Cos-1 cells were transfected with full-length Nav2 (construct A), and after 24 h spent media was replaced with fresh 4° C media. The culture plates were kept on an ice-cold metal block for 30 min. After 30 min cells were fed with warm media (37° C) and returned to the incubator. Cells were removed and washed for 15 sec in microtubule extraction buffer (80 mM PIPES pH 7.4, 5 mM EGTA, 1 mM MgCl2, 0.5% Triton X-100) before being fixed in methanol at -20° C for 5 min at time points 0, 15, and 30 min.
RNA interference studies
Three oligonucleotides (19-21 bp in length) were cloned downstream of the H1 promoter in a tetracycline inducible lentiviral plasmid pLVTHM (Wiznerowicz and Trono, 2003). shNAV2 #1 (119-138 nt of hNav2 coding sequence) GTT GGG AAG CAA GGT GGA G, shNAV2 #2 (753-772 nt) GCT GAA ATG CAG TCC AGA C, and shNAV2 #3 (4398-4419 nt) AAGTTCTGCAGAAGTACTCTG. The efficacy of these short hairpin oligos in knocking down NAV2 protein was assessed by co-transfection of the lentiviral plasmid containing the shRNA and the full-length Nav2 (construct A) in Cos-1 cells followed by immunoblotting. DNA (20 μg total containing: pLVTHM-shRNA or pLV-tTRKRAB-Red-10 μg, packaging plasmid psPAX2-7.5 μg and VSVG envelope plasmid pMD2G-2.5 μg) per 100 mm dish was transfected into HEK-293FT cells for the production of lentivirus. Transfection media was discarded 16 h later, cells were briefly rinsed once in PBS, and fresh media was added. The virus-laden media was collected 24 h later and a second collection was made 24 h post media replenishment. The viral media from the two collection points was pooled and centrifuged at 1000 × g for 20 min to pellet cells and cellular debris, and the supernatant was filtered through a 0.45 μm syringe filter. Cells to be transduced were plated in 25 cm2 tissue culture flask (1-5 × 105). The cells were infected twice in 48 h by replacing the media with virus-laden cleared media every 24 h.
Quantitation of neurite outgrowth and cell aggregation
SH-SY5Y cells were first transduced as described above with the lentiviral-tTRKrab-Red expressing the tetracycline repressor protein fused to Krab, a transcriptional repression module found in many zinc finger containing proteins (Wiznerowicz and Trono, 2003). These cells were grown for 3 d, then split, and again transduced with one of the three shNAV2 expressing viruses, lentiviral-shNAV2 #1, #2, and #3. The cells were then dosed with 0 or 5 μg/ml of doxycycline for 4 d followed by vehicle (ethanol) or atRA (1 μM) for 5 d in the presence or absence of doxycycline. Cells were fixed in 4% paraformaldehyde in PBS and immunostained with anti α-tubulin antibodies. Nuclei were counterstained with DAPI. A Nikon (TE2000) epifluorescence microscope coupled to either Spot 2 (Diagnostic Instruments Inc.) or CoolSNP EZ (Roper Scientific Inc.) was used to acquire 12-bit calibrated images. The neurite outgrowth module in Metamorph v6.3 was used to measure the average neurite length (μm per cell). In each experiment three separate coverslips were prepared for every treatment group. For each group at least 2,000 cells were measured representing a minimum of 10 non-overlapping images.
To assess aggregation in the doxycycline-treated groups, the total number of cells in 4 quadrants of 5 images/treatment group were counted, and the coefficient of variation was calculated.
Results
NAV2 is an ortholog of C. elegans unc-53
To identify the functional role of human Nav2 in neurite outgrowth, we utilized the C. elegans model system. Nav2 shows significant sequence similarity to C. elegans unc-53 including conservation of several putative functional domains (Merrill et al., 2002). The morphology of the PLM axons in unc-53(e404) mutants are abnormal; axons are shorter than in wild-type and are often misguided (Fig. 1B; Hekimi and Kershaw, 1993). When human Nav2 was expressed in the mechanosensory PLM neurons of unc-53 mutants using a neuron-specific promoter (Pmec-7), axonal elongation and branching was restored to wild-type levels. In wild-type worms, the PLM neuronal cell bodies differentiate near the tail, and their axons extend anteriorly towards and just slightly beyond the ALM cell body during larval development (Fig. 1A). The majority of wild-type worms showed a relative PLM axonal length between 0.6-0.7, whereas the extension of this axon in the unc-53 mutants was shortened to 20 to 50% that of wild-type (relative PLM length of 0.2 and 0.3; Fig. 1B). The transgenic worms expressing full-length NAV2 showed a partial rescue with the PLM length approaching that of the wild-type worm (Fig. 1A). The distribution of relative length of the PLM axon shows that the best rescue in the unc-53-NAV2-expressing group was equivalent to the length observed most often in the wild-type worms expressing the native UNC-53 protein (0.6 to <0.7; Fig. 1 B). This work supports the conclusion that NAV2 and UNC-53 are orthologs, and provides strong evidence that NAV2 plays a role in axonal elongation.
Fig. 1. Expression of NAV2 protein in C. elegans rescues the PLM elongation defect in the mechanosensory neurons of unc-53 mutant worms.
(A) Top panel: a diagram of the mechanosensory neurons in wild-type C. elegans (adapted from Savage et al., 1989) with the PLM neuron highlighted in green; bottom panels: representative fluorescent images showing the extension of the PLM neuron in wild-type (EG1194), unc-53(e404) mutant, and unc-53(e404) worms expressing the full length human NAV2 protein under the regulation of the mec7 promoter. Expression of NAV2 in unc-53 mutants rescues the mechanosensory neuron defects as indicated by an increase in the relative length of the PLM. The PLM and ALM cell bodies are labeled (white line) and the most anterior axonal projection of the PLM (arrow) is shown in the panels. Scale bar is 50 μm. (B) The distribution of the relative PLM axon length is shown for wild-type (empty bars), unc-53 (filled bars) and unc-53 rescued with the Pmec7∷NAV2∷FLAG extra-chromosomal array (red); data compiled from two transgenic lines.
Analysis of NAV2 ShRNA on RA-induced neurite outgrowth
Nav2 (RAINB1) was originally reported as an mRNA that is rapidly induced by atRA in the human neuroblastoma cell line, SH-SY5Y (Merrill et al., 2002). This cell line differentiates in response to atRA by increasing the length of neurite extensions. In order to determine whether NAV2 plays an essential role downstream of atRA to produce this change in cell morphology, shRNAs to NAV2 were tested for knockdown and then used in a lentivirus-based system enabling the regulated expression of shNAV2 under the control of doxycycline in SH-SY5Y cells. It was necessary to use inducible shRNAs because constitutive expression of the shNAV2 was lethal in neuroblastoma cells (data not shown). Three short hairpin oligos (shNAV2 #1, 2, 3) were cloned into tetracycline inducible lentiviral plasmids behind the Pol III H1 promoter. When co-transfected with the NAV2 expression plasmid in Cos-1 cells, the oligos produced either complete (shNAV2 #2 and #3) or partial (shNAV2 #1) knockdown of the NAV2-3x flag-tagged protein (Fig. 2A). The ability of doxycycline to induce knockdown of NAV2 was then shown in HEK293FT cells that had been transduced by the shNAV2 #2 containing lentivirus and transfected with the NAV2-pIRES-hrGFP-1α plasmid. After exposure of cells to doxycycline for 4 d, the expression of NAV2-3x flag protein was virtually eliminated when compared to the cells that had not been exposed to the drug (Fig. 2B). Finally, prior to testing the effect of the ShNav2 knockdown on atRA-induced neurite outgrowth, we verified that the shRNAs were effective in knocking down Nav2 mRNA in SH-SY5Y cells in a doxycycline-dependent manner. In the control cells (SH-SY5Y transduced with tTR-Krab and the non-specific shRNA), the induction of Nav2 mRNA by atRA was not altered in the presence or absence of doxycycline (Fig. 2C). However, in SH-SY5Y cells transduced with shNAV2 #2, induction of NAV2 mRNA by atRA was decreased from approximately 3.8 to 1.8 fold when doxycycline was present, indicating that knockdown had occurred. Taken together (Fig. 2B and 2C), these control experiments demonstrate that the lentiviral constructs are capable of knocking down NAV2 mRNA and protein in a doxycycline-dependent manner.
Fig.2. NAV2 knockdown is achieved through drug inducible lentivirus vector mediated RNA interference.
(A) Immunoblot analysis of the full-length human NAV2 protein containing a C-terminal 3× flag tag. Cos-1 cells were cotransfected with NAV2-pIRES-hrGFP-1α and a shNAV2 construct (pLV-shNAV #1, #2, or #3) or empty vector pLVTHM. Cells were lysed 24 h later. NAV2 protein was detected using a flag antibody. shNAV2 constructs #2 and #3 are completely effective at knocking down overexpressed NAV2 protein, whereas shNav2 #1 is partially effective. (B) Knockdown of over-expressed NAV2 protein in HEK293FT cells is inducible with the addition of doxycycline. (C) The atRA-mediated induction of Nav2 mRNA is reduced in SH-SY5Y cells co-transduced with LV-tTRKrab-Red and LV-ShNAV2 #2 viruses and exposed to doxycycline. SH-SY5Y cells were dosed with 0 or 0.5 μg/ml of doxycycline for 4 d followed by either vehicle (ethanol) or atRA (1 μM) for another 48 h, followed by the isolation of mRNA and analysis of Nav2 mRNA by Q-PCR. (D) Doxycycline-induced knockdown of Nav2 results in reduced neurite outgrowth in atRA-treated SH-SY5Y cells. Four SH-SY5Y cell lines (tTRKrab, and tTRKrab plus either ShNAV2 #1, #2, or #3) were dosed with 0 or 5 μg/ml of doxycycline for 4 d followed by atRA (1 μM) for 5 d. Results in panel D are presented as mean ± standard error. The assay was performed in triplicate, and at least 2,000 cells were measured per treatment group. The results shown are representative of three independent experiments. Asterisks indicate a statistically significant difference (p-value<0.0001) compared to empty vector control.
The effect of the shNav2 lentivirus on neurite outgrowth was then examined. In control atRA-treated SH-SY5Y cells (tTR-Krab), the mean neurite length per cell was unaffected by the presence or absence of doxycycline (Fig. 2D). However, atRA-treated SH-SY5Y cells transduced with any one of the three shNAV2-expressing viruses showed a significant reduction in neurite length in the presence of doxycycline (p-value<0.001; Fig. 2D). Thus, three independent shRNAs to human NAV2 reduced overall neurite length in atRA-exposed cells.
Because SH-SY5Y cells contain small neurites even in the absence of retinoid treatment (Fig. 3B panel a), the next experiment was directed at determining how effective the shNAV2 shRNAs were in knocking down the neurite outgrowth produced specifically in response to atRA in SH-SY5Y cells. Mean neurite length was increased in control cells (tTR-Krab) after atRA treatment compared to vehicle-treated cells, and the response to atRA was similar in the absence and presence of doxycycline (Fig. 3A). The cell lines transduced with either shNAV2 #2 or #3 that were not treated with doxycycline (i.e. the shNAV2 is not expressed) showed the same atRA-mediated increase in neurite outgrowth as that observed in the control tTR-Krab retinoid-treated cells. However in SH-SY5Y cells transduced with either shNAV2 #2 or #3 and exposed to doxycycline to induce expression of the shRNA, neurite outgrowth was reduced to the level of the vehicle-treated cells (Fig. 3A). Thus, the expression of shNAV2 shRNAs to two different regions of the NAV2 sequence in SH-SY5Y cells completely eliminated the ability of atRA to increase neurite outgrowth. It is interesting that although NAV2 knockdown did not alter the mean neurite length of pre-existing neurites in vehicle-treated cells, it did appear to alter the adhesive properties of SH-SY5Y cells, both in the vehicle and atRA-treated groups as evidenced by an increase in cell aggregation when either shNAV2 was present (compare Fig. 3B panel a to c & e and panel b to d & f). In summary, NAV2 appears to play a role in cell adhesion as well as in retinoid-mediated neurite outgrowth, and knocking down NAV2 significantly eliminates the ability of atRA to increase neurite length in SH-SY5Ycells.
Fig. 3. Knocking down NAV2 eliminates RA-induced neurite outgrowth in SH-SY5Y cells.
Knockdown of NAV2 prevents neurite outgrowth from occurring in response to atRA, and also produces changes in cell adhesion. (A) The addition of doxycycline for 4 d does not alter the ability of atRA to increase mean neurite length in the control tTR-Krab cells. atRA is also fully effective in inducing neurite outgrowth when cells are transduced with either shNAV2-containing virus but not exposed to doxycycline. In contrast, when either shNAV2 #2 or #3 is expressed, the ability of atRA to induce neurite outgrowth is completely inhibited. Neurite outgrowth was evaluated as described in Fig. 2, and the results shown are representative of two independent experiments. Asterisks indicate a statistically significant (p-value < 0.001) difference compared to the no doxycycline atRA treated control. Scale bar is 100 μm. (B) The effect of knocking down NAV2 by two separate shNav2-containing viruses on SH-SY5Y cell morphology in cells treated with or without atRA for 5 d is shown. The knockdown of NAV2 not only reduces atRA induced neurite outgrowth, but also increases cell aggregation (with and without atRA). To quantify the dispersion of cells, the coefficient of variation of the number of cells in each quadrant of a 116mm × 139mm image was calculated. Five images, each divided into quadrants, were used for each treatment group. The tTR-Krab control cell line has a coefficient of variation of 41% in vehicle treated cells indicating cells are aggregated in certain quadrants. atRA treatment resulted in the disaggregation of the control cell line and that is reflected in the decrease of the coefficient of variation (29%). This is consistent with the fact that atRA is known to cause SH-SY5Y cells to disaggregate. In both NAV2 knockdown cell lines there is increased cell aggregation with and without atRA as indicated by a coefficient of variation of 67% and 76% for vehicle treated and 57% and 65% for atRA treated groups in the shNAV2#2 and ShNAV2 #3 cell lines, respectively.
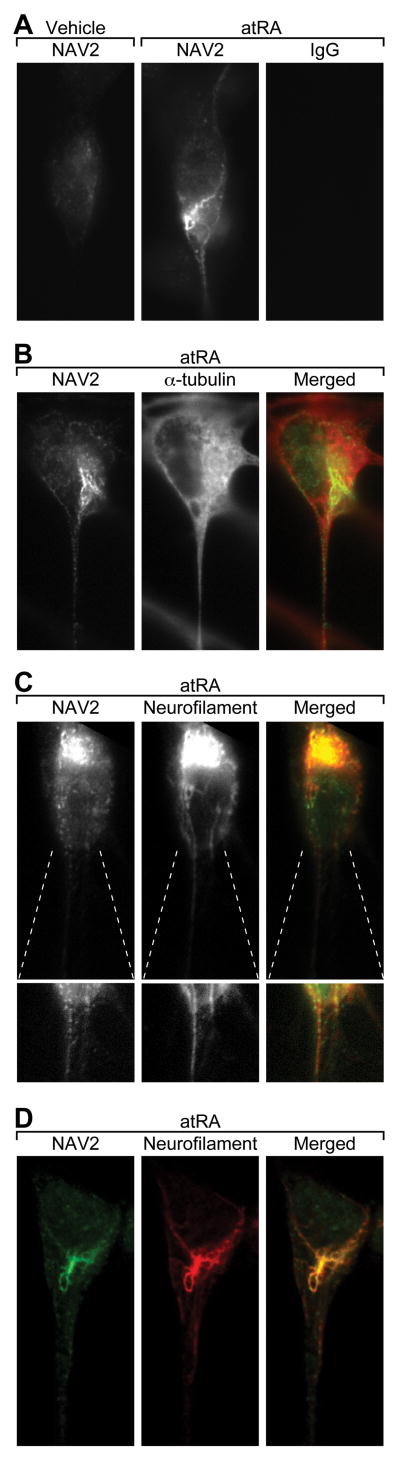
Endogenous NAV2 protein in SH-SY5Y cells is distributed similarly to that of neurofilament proteins
Immunocytochemical studies using a polyclonal anti-NAV2 peptide antibody were conducted in SH-SY5Y cells to probe the subcellular localization of NAV2, and to determine whether exposure to atRA would alter its distribution. Epifluorescent images of vehicle-treated SH-SY5Y cells show NAV2 distributed in a punctate fashion both in the cytoplasm and in the few neurites present (Fig. 4A, left). After exposure to atRA for 72 h, NAV2 antibody staining adjacent to the nucleus appeared brighter and more highly condensed, and staining within the neurite was also enhanced. The NAV2 staining in atRA-treated SH-SY5Y cells extended throughout the length of the neurite, and was often most prominent in the longest neurite extending from the cell (Fig. 4A center).
Fig. 4. NAV2 protein is localized in the cell body and along the length of the growing neurites in RA-treated SH-SY5Y cells.
(A) Epifluorescence images of SH-SY5Y cells exposed to vehicle or atRA for 72 h, and stained with affinity-purified NAV2 antibody or rabbit IgG. (B) Epifluorescent images of atRA treated SH-SY5Y cells (72 h) stained with NAV2 affinity-purified antibody (green in merged panel) along with anti α-tubulin (red in merged panel). (C) Epifluorescent images of atRA-treated (72 h) SH-SY5Y cells stained with affinity purified NAV2 antibody (green in merged panel) and an antibody to phosphorylated medium chain neurofilament (RMO270, red in merged panel). A region where the neurite is emerging from the cell is shown below each panel in which the light intensity levels are optimized to show staining in the neurite. (D) Confocal analysis of NAV2 (green) and neurofilament (red) protein show punctate NAV2 protein distribution in cell body and along the length of the neurite co-localizing with neurofilament protein which appears as smooth fibers. The merged image shows colocalization of NAV2 immunostaining with neurofilament in SH-SY5Y cells (yellow).
SH-SY5Y cells were then co-stained with the NAV2 antibody and antibodies to neurofilament and α-tubulin (Fig. 4B-D). Staining for NAV2 overlapped in some regions of the cell with staining for the microtubule protein, α-tubulin (Fig. 4B). However, NAV2 immunostaining most closely mirrored the cellular distribution of the phosphorylated intermediate chain neurofilament protein as assessed using the RMO-270 antibody (Fig. 4C & 4D). This neurofilament antibody showed continuous staining along the filament whereas NAV2 staining was more punctate. Confocal analysis of atRA treated SH-SY5Y cells labeled with anti-NAV2 and anti-RMO 270 showed intense staining in a region closely associated with the nucleus as well as in fibers extending into the neurite process (Fig. 4D). Similar results were shown using an antibody to the neurofilament associated protein 3A10 (data not shown). Additional confocal studies showed that markers/antibodies for the ER and Golgi did not colocalize with NAV2 antibody. Staining with mitotracker showed some similarity to NAV2 staining, but only in a subset of cells (data not shown).
Because microtubules are discontinuous structures, SH-SY5Y cells were stained with an antibody to EB1, a protein that associates with the plus-ends of microtubules, and no significant overlap with the expression of NAV2 was observed (data not shown). In summary, NAV2 expression in SH-SY5Y cells is most similar to that of neurofilament protein, which is also found in neural cells in close association with microtubules.
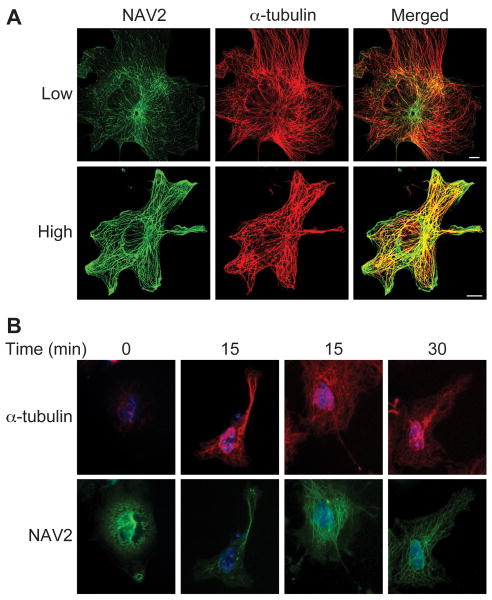
Over expressed NAV2 protein has a subcellular distribution similar to microtubules in Cos-1 cells
When NAV2 was over expressed in Cos-1, monkey kidney cells, NAV2 staining appeared in regions of the cell where α-tubulin was found (Fig. 5A). In Cos-1 cells showing low NAV2 expression levels, NAV2 staining was distributed in a punctate fashion along the length of the microtubules (Fig. 5A). As the expression of NAV2 increased, staining was distributed along the length of microtubules in a more continuous fashion, and the microtubules appeared thickened when compared to cells expressing lower levels of NAV2 (Fig. 5A). Similar results were observed in NTera2 cells, a human embryonal carcinoma cell line, transfected with the full-length NAV2 protein (data not shown).
Fig. 5. NAV2 protein overexpressed in Cos-1 cells distributes similarly to that of endogenous α-tubulin.
Construct NAV2-pIRES-hrGFP-1α (C-terminal 3× flag tag) was transfected into Cos-1 cells, and 24 h later cells were fixed and stained with mouse monoclonal anti-flag and rabbit polyclonal anti α-tubulin antibodies. (A) Confocal images of cells expressing different amounts of NAV2 protein defined as low or high. At low levels, NAV2 is distributed in a punctate fashion along the length of the microtubules; at high levels the protein appears distributed along the length of the microtubules and causes thickening or bundling. (B) Cos-1 cells were transfected with full-length Nav2 pMES construct, and 24 h later microtubules were depolymerized and repolymerized as described in Methods. NAV2 protein lags in its apparent re-association with the polymerizing microtubules. At 0 min, NAV2 protein is diffusely distributed throughout the cell (panel B, column 1). At 15 min some cells show a focused distribution in association with reforming microtubules whereas other NAV2 staining remains diffuse (panel B, columns 2 and 3). By 30 min NAV2 protein shows an expression pattern similar to α-tubulin (5B, column 4). Scale bar is 10 μm.
In order to determine if NAV2 was found in association with microtubules as they formed, a depolymerization-repolymerization experiment was conducted. Microtubules are formed by polymerization of α and β tubulin monomers, and this process is thermo sensitive. At 4° C the microtubules depolymerize releasing the two tubulin monomers. Raising the temperature back to 37°C enables the α and β tubulins to repolymerize, thereby reconstructing the microtubules. At time 0 in NAV2 transfected Cos-1 cells exposed to low temperature, the microtubules were nearly completely depolymerized, and little α-tubulin monomer remained in cells after detergent extraction (Fig. 5B). At time 0, exogenous NAV2 protein was distributed in a diffuse manner throughout the cytoplasm (Fig. 5B, column 1). Fifteen minutes after the initiation of repolymerization, two types of cells were observed. A growing network of microtubules could be seen in some cells while the NAV2 protein remained largely dispersed in the cytoplasm, with a small amount of protein associated with the polymerizing microtubules (Fig. 5B, column 2). However, at 15 min other cells showed a significant amount of NAV2 localizing with the microtubules indicating that this was a time point at which NAV2 was beginning to associate with microtubules (Fig. 5B, column 3). By 30 min, the microtubules were very distinct, and a significant amount of the NAV2 protein was found associated with the forming tubules (Fig. 5B, column 4). After 2 h results were similar to those at 30 min (data not shown). The finding that microtubules reformed with only a very small subset of tubules showing NAV2 co-staining (Fig. 5B, column 2) suggests that NAV2 does not play a role in microtubule formation. It is possible that another factor or factors may need to be present at the microtubule before NAV2 can associate with the complex.
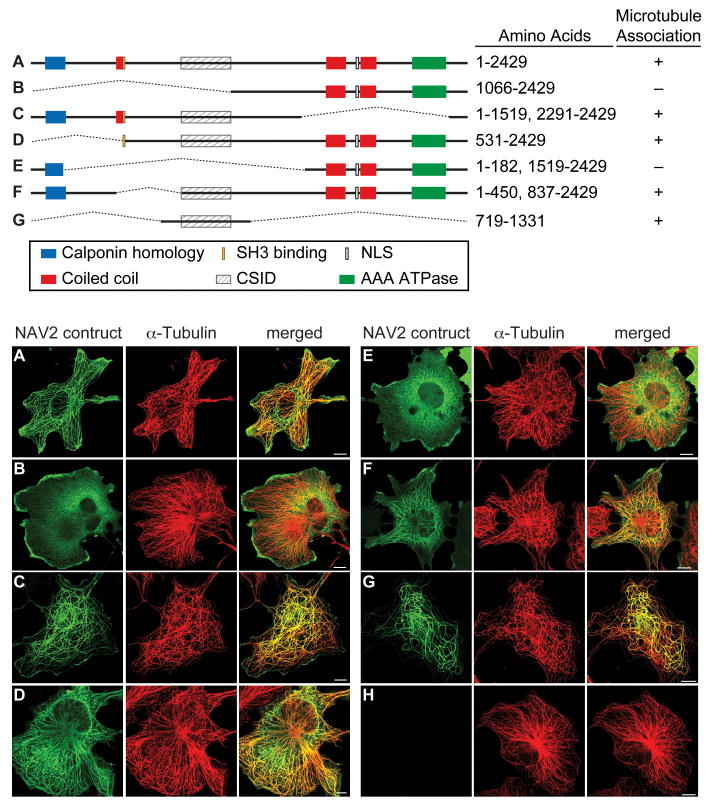
Identification of a NAV2 protein domain that is required for microtubule localization
In order to determine which regions of the NAV2 sequence are needed to direct it to the microtubule, deletion mutants were constructed and transfected into Cos-1 cells. The effect on protein distribution relative to that of α-tubulin was examined. The proteins resulting from these NAV2 deletion constructs were expressed and examined by immunoblotting, and all were found to be of the expected sizes based on sequencing (data not shown). The full-length NAV2 protein was distributed in the cell in similar fashion to that of α-tubulin (Fig. 6 construct A). Deleting N-terminal amino acids 1-1065 led to a random distribution of NAV2 throughout the cytoplasm (Fig. 6B), whereas deletion of much of the C-terminal region containing several putative coil-coil domains (red boxes) and all of the AAA-ATPase domain of the protein did not alter subcellular distribution from that of the full-length protein (Fig. 6C). A more detailed study of the N-terminus of the NAV2 protein showed that deletion of only the first 530 amino acids did not alter the subcellular distribution of the truncated protein relative to that of the full-length protein (Fig. 6D). Deletion of the interior of the protein sequence from amino acids 183-1518, but not of the calponin-homology domain in the N-terminus, disrupted the microtubule-like expression (Fig. 6E). Constructs with smaller deletions, lacking amino acids 451-836 (Fig. 6F), showed staining that overlapped that of α-tubulin. Interestingly, the expression of amino acids 719-1331 (Fig. 6G) was sufficient to direct NAV2 to the microtubule resulting in a NAV2 staining pattern that overlapped that of α-tubulin. Thus, the results of deletion analysis studies identify a region in the NAV2 protein (aa 837-1066) that confers microtubule localization (Fig. 6; cytoskeletal interacting domain or CSID).
Fig. 6. NAV2 protein contains a putative cytoskeletal-interacting domain.
The top panel shows a schematic of the Nav2 deletion constructs (A, full length; B-G, deletion constructs). The bottom panels (A-G) show the correspondingly transfected Cos-1 cells co-stained for NAV2 (green) and α-tubulin (red) and the merged images (yellow). All deletion constructs with the exception of B and E show staining that overlaps that of α-tubulin. Deletion construct G confers microtubule localization and is proposed to contain a cytoskeletal-interacting domain (CSID; hatched rectangle). The full-length NAV2 (A) and all deletion constructs were stained with mouse anti-flag and rabbit polyclonal α-tubulin antibodies. NAV2 protein is not detected in Cos-1 cells transfected with empty vector (H). Scale bar is 10 μm.
Discussion
Two major lines of evidence including overexpression and loss-of-function studies support the conclusion that human Nav2, a member of the neuron navigator family, functions in neurite outgrowth. First, when human Nav2 is expressed under the control of a C. elegans mechanosensory neuron-specific promoter, the defect in PLM neuron elongation in unc-53 mutants is rescued. unc-53 is known to play an important role in cell migration and axonal extension and guidance (Hedgecock et al., 1987; Hekimi and Kershaw, 1993). Our data is the first to directly demonstrate that human Nav2 is not only a homolog with significant conservation of putative domains, but also an ortholog to unc-53, showing a conserved function in axon elongation. The second line of evidence supporting a role for Nav2 in neurite outgrowth comes from knockdown studies of the endogenous protein in SH-SY5Y cells. Nav2 (RAINB1) was identified by Merrill et al. (2002) as an atRA regulated gene, both in a human neuroblastoma cell line (SH-SY5Y) as well as in the developing rat embryo. It is well established that exposure of SH-SY5Y cells to atRA leads to a robust increase in the length of neurites (Clagett Dame et al., 2006). In the present study, we show that abrogation of NAV2 induction in SH-SY5Y cells eliminates the RA-mediated increase in neurite outgrowth. Multiple Nav2 splice variants have been reported. The full length (∼10.5 kb) transcript reported by Merrill et al. (2002) is most abundant in SH-SY5Y cells, and is also highly expressed in the nervous system both in embryonic and adult tissues (Coy et al., 2002; Ishiguro et al., 2002; Merrill et al., 2002; Peeters et al., 2004). This is the same transcript that is missing in the hypomorphic mouse model resulting in impaired perception in several sensory systems. A shorter transcript starting upstream of exon 13 is still expressed in this mouse, but is largely found in non-neuronal sites (Peeters et al., 2004). In the present study, an shRNA designed to knock down all transcripts (shNAV2 #3) and one specifically directed to the full-length transcript (shNAV2 #2) were both equally effective in eliminating all atRA-induced neurite outgrowth in SH-SY5Y cells, indicating that it is the full-length protein that is responsible for mediating responsiveness to atRA. In summary, full-length NAV2 protein plays an important role in atRA mediated neurite outgrowth in SH-SY5Y cells, axonal elongation in C. elegans, and sensory function in the mouse model.
The manner in which NAV2 functions may involve its interaction, either directly or indirectly, with cytoskeletal proteins, specifically microtubules and neurofilament proteins. Endogenous NAV2 protein is found both in the cell body and neurites of SH-SY5Y cells, and treatment of cells with atRA leads to an increase in the intensity of endogenous NAV2 staining at both sites. NAV2 staining is often punctate in nature in untreated SH-SY5Y cells, and upon exposure of cells to atRA the staining becomes more intensely expressed in the region of the cell body adjacent to the largest neurite into which the staining extends in a more continuous fashion. In the cell body as well as along the neurite of SH-SY5Y cells, NAV2 shows the most similarity in distribution to that of neurofilament proteins. When over expressed in Cos-1, a cell line that does not express neurofilament proteins, NAV2 protein is also found in association with the cytoskeleton but is distributed in a fashion similar to that of microtubules. When expressed at a low level, NAV2 appears punctate in nature, whereas at higher levels of expression it appears to cause bundling of the microtubules. NAV2, however, does not appear to initiate microtubule assembly, as the kinetics of microtubule re-formation after the exposure of cells to cold is faster than the re-association of NAV2 staining with the newly formed microtubule. Interestingly, Nav1, a navigator gene family member, has been shown to associate at the plus- ends of microtubules when expressed ectopically in Cos-1 cells at a low level, throughout the microtubules at intermediate levels, and to induce bundling at high levels of expression (Martinez-Lopez et al., 2005).
The results of deletion studies have defined a sequence that mediates NAV2-microtubule association. The minimal element conferring cytoskeletal localization in the cell includes an internal region containing aa 837-1065 (CSID), whereas the C-terminus of the NAV2 protein (1066-2429) was ineffective in promoting interaction. A domain has recently been defined in the NAV1 protein that mediates association with the plus ends of microtubules. (Martnez-Lopez et al., 2005). The cytoskeletal interacting domain in NAV2 is similarly located to the microtubule-binding domain (MTBD) in the NAV1 protein, and shares 33% identity and 47% similarity (ClustalW alignment) indicating this region may be functionally conserved within the family. The CH domain of UNC-53 has been proposed to interact with actin (Stringham et al., 2002), but this region is not required to promote microtubule association with NAV2, and is absent in NAV1.
In neurons, it is well known that there is a close association between intermediate filament proteins and microtubules. When neurite outgrowth occurs, there is a dramatic influx of microtubules into the growing process. Neurofilaments extend along the length of the growing neurite and are believed to play an important role in organelle and vesicle movement. Additionally, they provide structural stability of the growing process, and ultimately determine the final “girth” of the axon (Chang & Goldman, 2004). Microtubule regulation is a key process in neurite outgrowth, axonal guidance and neuronal migration (Gleeson et al., 1999). The cellular distribution and localization of NAV2 suggests that this protein could play a role in neuronal elongation by facilitating crosstalk between the microtubule and neurofilament components of the cytoskeleton. In the future it will be important to determine if the localization of NAV2 in cells of neural origin is due to a direct interaction with microtubules and/or intermediate filament proteins, or whether intermediary proteins are involved.
In summary, we have provided the first direct evidence that Nav2 functions in axonal elongation acting as an ortholog to C. elegans unc-53. We have shown that Nav2 knockdown eliminates RA induced neurite outgrowth in SH-SY5Y cells. NAV2 is found in association with cytoskeletal elements integral in providing support for neurite extension and outgrowth, and by deletion analysis we have identified a region in the amino terminus of the protein that is involved in microtubule localization.
Supplementary Material
Immunoblot detection of recombinant full-length NAV2 flag-tagged protein using polyclonal antibody-containing NAV2 serum or monoclonal antibody to the Flag sequence (A) or affinity-purified NAV2 antibody (B). Both the serum and purified NAV2 antibodies, and Flag antibody detect a protein with the predicted molecular weight for full-length NAV2 protein (261 kDa) in the lane loaded with NAV2 containing extract, but not in the lane containing the pIRES control. NAV2-containing extracts were prepared from Cos-1 cells (A) and HEK-293FT (B) cells transfected with the Nav2 cDNA (7.29 kb) containing a 3× C-terminal flag tag (construct A). Control extract was generated from cells transfected with the parent pIRES-hrGFP-1α vector. Extracts were loaded on 6% (A) or 10% (B) SDS polyacrylamide gels and transferred to nitrocellulose followed by immunoblotting with either rabbit NAV2 antibody-containing serum (1:500) or purified NAV2 antibody (0.5 μg/ml), or the Flag antibody; followed by incubation with goat anti-rabbit IgG-HRP or goat anti-mouse IgG-HRP (Southern Biotech).
Acknowledgments
We thank L. Vanderploeg in the Biochemistry Media Lab for the artwork. E.M.McNeill was supported in part by a fellowship from NIH T32 DK07665.
References
- Appel B, Eisen JS. Retinoids run rampant: multiple roles during spinal cord and motor neuron development. Neuron. 2003;40:461–464. doi: 10.1016/s0896-6273(03)00688-3. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, DeLuca HF. The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, McNeill EM, Muley PD. Role of all-trans retinoic acid in neurite outgrowth and axonal elongation. J Neurobiol. 2006;66:739–756. doi: 10.1002/neu.20241. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, Verhalen TJ, Biedler JL, Repa JJ. Identification and characterization of all-trans-retinoic acid receptor transcripts and receptor protein in human neuroblastoma cells. Arch Biochem Biophys. 1993;300:684–693. doi: 10.1006/abbi.1993.1095. [DOI] [PubMed] [Google Scholar]
- Coy JF, Wiemann S, Bechmann I, Bachner D, Nitsch R, Kretz O, Christiansen H, Poustka A. Pore membrane and/or filament interacting like protein 1 (POMFIL1) is predominantly expressed in the nervous system and encodes different protein isoforms. Gene. 2002;290:73–94. doi: 10.1016/s0378-1119(02)00567-x. [DOI] [PubMed] [Google Scholar]
- da Silva JS, Dotti CG. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Krumlauf R. Retinoid signalling and hindbrain patterning. Curr Opin Genet Dev. 2000;10:380–386. doi: 10.1016/s0959-437x(00)00100-3. [DOI] [PubMed] [Google Scholar]
- Gimona M, Mital R. The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J Cell Sci. 1998;111(Pt 13):1813–1821. doi: 10.1242/jcs.111.13.1813. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Hekimi S, Kershaw D. Axonal guidance defects in a Caenorhabditis elegans mutant reveal cell-extrinsic determinants of neuronal morphology. J Neurosci. 1993;13:4254–4271. doi: 10.1523/JNEUROSCI.13-10-04254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Mendez MG, Pugh J, Delsert C, Goldman RD. A role for intermediate filaments in determining and maintaining the shape of nerve cells. Mol Biol Cell. 2003;14:5069–5081. doi: 10.1091/mbc.E03-06-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Shimokawa T, Tsunoda T, Tanaka T, Fujii Y, Nakamura Y, Furukawa Y. Isolation of HELAD1, a novel human helicase gene up-regulated in colorectal carcinomas. Oncogene. 2002;21:6387–6394. doi: 10.1038/sj.onc.1205751. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoids and spinal cord development. J Neurobiol. 2006;66:726–738. doi: 10.1002/neu.20248. [DOI] [PubMed] [Google Scholar]
- Maes T, Barcelo A, Buesa C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 2002;80:21–30. doi: 10.1006/geno.2002.6799. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez MJ, Alcantara S, Mascaro C, Perez-Branguli F, Ruiz-Lozano P, Maes T, Soriano E, Buesa C. Mouse neuron navigator 1, a novel microtubule-associated protein involved in neuronal migration. Mol Cell Neurosci. 2005;28:599–612. doi: 10.1016/j.mcn.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Merrill RA. Biochemistry. Madison University of Wisconsin; 2004. Retinoic Acid Regulated Genes in Neuronal Development; p. 277. [Google Scholar]
- Merrill RA, Plum LA, Kaiser ME, Clagett-Dame M. A mammalian homolog of unc-53 is regulated by all-trans retinoic acid in neuroblastoma cells and embryos. Proc Natl Acad Sci USA. 2002;99:3422–3427. doi: 10.1073/pnas.052017399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey J. New therapeutic target for CNS injury? The role of retinoic acid signaling after nerve lesions. J Neurobiol. 2006;66:757–779. doi: 10.1002/neu.20238. [DOI] [PubMed] [Google Scholar]
- Mey J, McCaffery P. Retinoic acid signaling in the nervous system of adult vertebrates. Neuroscientist. 2004;10:409–421. doi: 10.1177/1073858404263520. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters PJ, Baker A, Goris I, Daneels G, Verhasselt P, Luyten WH, Geysen JJ, Kass SU, Moechars DW. Sensory deficits in mice hypomorphic for a mammalian homologue of unc-53. Brain Res Dev Brain Res. 2004;150:89–101. doi: 10.1016/j.devbrainres.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Savage C, Hamelin M, Culotti JG, Coulson A, Albertson DG, Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3:870–881. doi: 10.1101/gad.3.6.870. [DOI] [PubMed] [Google Scholar]
- Stradal T, Kranewitter W, Winder SJ, Gimona M. CH domains revisited. FEBS Lett. 1998;431:134–137. doi: 10.1016/s0014-5793(98)00751-0. [DOI] [PubMed] [Google Scholar]
- Stringham E, Pujol N, Vandekerckhove J, Bogaert T. unc-53 controls longitudinal migration in C. elegans. Development. 2002;129:3367–3379. doi: 10.1242/dev.129.14.3367. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhoabditis elegans. Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Swartz ME, Eberhart J, Pasquale EB, Krull CE. EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development. 2001;128:4669–4680. doi: 10.1242/dev.128.23.4669. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblot detection of recombinant full-length NAV2 flag-tagged protein using polyclonal antibody-containing NAV2 serum or monoclonal antibody to the Flag sequence (A) or affinity-purified NAV2 antibody (B). Both the serum and purified NAV2 antibodies, and Flag antibody detect a protein with the predicted molecular weight for full-length NAV2 protein (261 kDa) in the lane loaded with NAV2 containing extract, but not in the lane containing the pIRES control. NAV2-containing extracts were prepared from Cos-1 cells (A) and HEK-293FT (B) cells transfected with the Nav2 cDNA (7.29 kb) containing a 3× C-terminal flag tag (construct A). Control extract was generated from cells transfected with the parent pIRES-hrGFP-1α vector. Extracts were loaded on 6% (A) or 10% (B) SDS polyacrylamide gels and transferred to nitrocellulose followed by immunoblotting with either rabbit NAV2 antibody-containing serum (1:500) or purified NAV2 antibody (0.5 μg/ml), or the Flag antibody; followed by incubation with goat anti-rabbit IgG-HRP or goat anti-mouse IgG-HRP (Southern Biotech).