Abstract
Denitrification and respiratory ammonification are two competing, energy-conserving NO3−/NO2− reduction pathways that have major biogeochemical consequences for N retention, plant growth and climate. Batch and continuous culture experiments using Shewanella loihica strain PV-4, a bacterium possessing both the denitrification and respiratory ammonification pathways, revealed factors that determine NO3−/NO2− fate. Denitrification dominated at low carbon-to-nitrogen (C/N) ratios (that is, electron donor-limiting growth conditions), whereas ammonium was the predominant product at high C/N ratios (that is, electron acceptor-limiting growth conditions). pH and temperature also affected NO3−/NO2− fate, and incubation above pH 7.0 and temperatures of 30 °C favored ammonium formation. Reverse-transcriptase real-time quantitative PCR analyses correlated the phenotypic observations with nirK and nosZ transcript abundances that decreased up to 1600-fold and 27-fold, respectively, under conditions favoring respiratory ammonification. Of the two nrfA genes encoded on the strain PV-4 genome, nrfA0844 transcription decreased only when the chemostat reactor received medium with the lowest C/N ratio of 1.5, whereas nrfA0505 transcription occurred at low levels (≤3.4 × 10−2 transcripts per cell) under all growth conditions. At intermediate C/N ratios, denitrification and respiratory ammonification occurred concomitantly, and both nrfA0844 (5.5 transcripts per cell) and nirK (0.88 transcripts per cell) were transcribed. Recent findings suggest that organisms with both the denitrification and respiratory ammonification pathways are not uncommon in soil and sediment ecosystems, and strain PV-4 offers a tractable experimental system to explore regulation of dissimilatory NO3−/NO2− reduction pathways.
Introduction
Phylogenetically diverse groups of microorganisms use the nitrogen (N) oxyanions nitrate (NO3−) and nitrite (NO2−) as terminal electron acceptors in anoxic environments (Sørensen, 1978; Tiedje et al., 1982; Zumft, 1997; Burgin and Hamilton, 2007). During denitrification, NO3− is reduced to the gaseous products, nitrous oxide (N2O) and dinitrogen gas (N2), in a step-wise manner via NO2− and nitric oxide (NO) as intermediates (Zumft, 1997; Burgin and Hamilton, 2007). N2O and N2 release to the atmosphere causes N loss from terrestrial and aquatic environments, and N2O is an ozone-depleting greenhouse gas (Lashof and Ahuja, 1990; Ravishankara et al., 2009). Alternatively, many microbes reduce NO3− via respiratory ammonification (also referred to as dissimilatory nitrate reduction to ammonium), a pathway that shares the NO3− to NO2− reaction step with denitrification but reduces NO2− to NH4+ (Tiedje et al., 1982; Silver et al., 2001, 2005; Templer et al., 2008; Simon and Klotz, 2013). In contrast to NO3− and NO2−, positively charged ammonium (NH4+) is retained in soils and sediments and has a higher tendency for incorporation into microbial or plant biomass (Laima et al., 1999; Silver et al., 2001; Fitzhugh et al., 2003). Hence, the relative contributions of denitrification versus respiratory ammonification activities have important consequences for N retention, plant growth and climate.
Because of the biogeochemical consequences of denitrification versus respiratory ammonification, many studies aimed at elucidating the environmental controls of the two competing NO3−/NO2− reduction pathways (Tiedje et al., 1982; Nägele and Conrad, 1990; Ogilvie et al., 1997; Fazzolari et al., 1998; Stevens et al., 1998; Silver et al., 2001; Nizzoli et al., 2010; Dong et al., 2011). Based on empirical observations, Tiedje et al. (1982) suggested that the carbon-to-nitrogen (C/N) ratio regulates NO3− fate. Theoretically, denitrification yields more free energy per electron but respiratory ammonification yields more free energy per molecule of NO3− reduced than complete denitrification to N2 (Strohm et al., 2007). Pure culture growth experiments demonstrated that heterotrophic denitrifier biomass yields were lower than those obtained with organisms performing respiratory ammonification, suggesting more efficient energy conservation in the latter process (Strohm et al., 2007). Therefore, it is sensible that denitrification occurs under electron donor-limiting conditions (that is, low C/N ratios), whereas respiratory ammonification is preferred under N oxyanion-limiting conditions (that is, high C/N ratios). In support of this hypothesis, Fazzolari et al. (1998) demonstrated that high glucose-to-NO3− ratios increased NH4+ and lowered N2O production. Nijburg et al. (1997) observed that increased NO3− loading favored denitrifying bacteria, and Schmidt et al. (2011) observed a strong correlation between C/N ratio and respiratory ammonification activity in arable soils; however, other studies failed to establish a relationship between C/N ratios and NO3− fate (Kelso et al., 1997; Stevens et al., 1998). Furthermore, the effects of varying C/N ratios on the expression of key genes implicated in denitrification and respiratory ammonification are unclear.
The C/N ratio is not the only environmental parameter hypothesized to affect the environmental fate of NO3−/NO2−. Increased temperature has been linked with elevated respiratory ammonification activity (Ogilvie et al., 1997; Silver et al., 2001; Tomaszek and Rokosz, 2007; Nizzoli et al., 2010; Dong et al., 2011). pH also influenced the relative contributions of denitrification versus respiratory ammonification, although with no consistent patterns. Stevens et al. (1998) observed significantly higher respiratory ammonification activity in a surface water gley soil at pH 8.0 than at pH 6.5 and 6.0, whereas Nägele and Conrad (1990) reported elevated NH4+ production under acidic conditions. Many studies have explored how microbial community composition and geochemical parameters affect the fate of NO3−/NO2− via the two competing dissimilatory pathways but these efforts yielded inconsistent results and have led to conflicting conclusions.
Until recently, the general understanding had been that denitrification and respiratory ammonification pathways do not coexist within a single organism. This apparent pathway incompatibility limited experimental designs exploring the environmental factors controlling dissimilatory NO3−/NO2− reduction pathways to mixed cultures, or, at best, co-cultures of microorganisms performing either pathway (Rehr and Klemme, 1989). Because of organism-specific characteristics in terms of growth kinetics and growth yields under different cultivation conditions, experiments aimed at delineating conditions that favor denitrification over respiratory ammonification, or vice versa, yielded inconclusive results. Opitutus terrae, Marivirga tractuosa and Shewanella loihica possess the complete sets of genes encoding both pathways and 16S rRNA gene surveys, as well as metagenomic analyses, suggested that bacteria harboring the pathways for denitrification and respiratory ammonification might not be rare in the environment (Hengstmann et al., 1999; Sanford et al., 2012; Mania et al., 2014). S. loihica strain PV-4 possesses two copies of nrfA, as well as the complete suite of genes encoding denitrification enzymes (nirK, norB and nosZ) (Sanford et al., 2012; Yoon et al., 2013). Inconsistent with the genome information, S. loihica was initially characterized as a non-NO3−/NO2−-reducing bacterium (Gao et al., 2006) but a recent study demonstrated growth via denitrification (Yoon et al., 2013). Here, we demonstrate that strain PV-4 also performs respiratory ammonification and provide evidence that a single organism can reduce NO3−/NO2− to NH4+ and/or N2O/N2. Using batch and continuous (chemostat) cultures of S. loihica strain PV-4, the effects of C/N ratio, pH and temperature on the selection of a dissimilatory NO3−/NO2− reduction pathway were explored.
Materials and methods
Media and culture conditions
For batch experiments with S. loihica strain PV-4, completely synthetic, anoxic, phosphate-buffered basal salt medium was prepared as previously described (Yoon et al., 2013). The medium (100 ml) was distributed to 160 ml serum bottles using the Hungate technique, and the bottles were sealed with black butyl-rubber stoppers (Geo-Microbial Technologies, Inc., Ochelata, OK, USA). After autoclaving, vitamins (Wolin et al., 1964) were added from a degassed and filter-sterilized 200-fold concentrated stock solution. Electron donor (varying amounts of lactate), electron acceptors (varying amounts of NO3−) and NH4+ (0.1 mM) were added from sterilized and degassed stock solutions. Sodium nitrate (⩾99%, Fisher Scientific, Pittsburg, PA, USA) and ammonium chloride (⩾99%, Fisher Scientific) stock solutions were prepared in distilled water at concentrations of 0.1 M. Sodium lactate stock solutions (0.5 M) were prepared from a 60% lactate syrup (Sigma-Aldrich, St Louis, MO, USA). The initial substrate concentrations and the medium pH varied depending on the objectives of each experiment (see below). Early stationary phase S. loihica strain PV-4 cultures grown with 2.0 mM lactate and 1.0 mM NO3− served as inocula (0.5%, vol/vol) after NO3− and NO2− were depleted. The sum of the amounts of lactate and acetate transferred with the inocula to the fresh medium did not exceed 0.01 μmol. To inhibit nitrous oxide reductase (NosZ) activity and measure N2O as a proxy for denitrification activity, 6 ml N2 headspace was replaced with acetylene gas (99.6%, Airgas, Knoxville, TN, USA) (Yoshinari et al., 1977). Unless mentioned otherwise, all batch experiments were performed at room temperature (21 °C). The medium for the chemostat experiment was slightly modified and used higher phosphate (25 mM) and ammonium chloride (0.5 mM) concentrations to increase the buffering capacity and provide sufficient N for assimilation and cell growth. To prevent precipitate formation, trace metals were added to the chemostat from a degassed and autoclaved 200-fold concentrated stock solution.
Analytical procedures
The Dionex ICS-2100 system (Sunnyvale, CA, USA) was used to measure NO3− and NO2− and the Dionex ICS-1100 system was used to measure NH4+ concentrations (Yoon et al., 2013). Lactate and acetate were quantified using an Agilent 1200 Series high-performance liquid chromatography system (Palo Alto, CA, USA). For N2O measurements, 1 ml of headspace gas was collected for analysis with an Agilent 3000A MicroGC. Aqueous concentrations of N2O were calculated using a dimensionless Henry's constant for a temperature of 21 °C that was corrected for the medium's ionic strength to 1.751 (Schumpe et al., 1982; Schumpe, 1993; Sander, 1999). N2O measurements for the culture bottles incubated at 30 °C and 37 °C were made after the bottles had been equilibrated to room temperature (21 °C). Biomass estimates assumed a mass of 2.77 × 10−13 g for a single strain PV-4 cell (Yoon et al., 2013). Cell numbers were calculated from quantitative real-time PCR (qPCR) enumeration of 16S rRNA genes corrected for the presence of nine 16S rRNA gene operons on the S. loihica strain PV-4 genome (NCBI Reference Sequence: NC_009092). The C and N content calculations used the empirical formula C5H7O2N for biomass.
Batch experiments
The effects of the C/N ratio (the ratio of C atoms in the electron donor to N atoms in electron acceptor), pH and temperature were explored in batch systems. To examine the effect of C/N ratios, the 160 ml culture vessels were amended with 0.2 mM sodium nitrate and varying concentrations of sodium lactate (0.1, 0.2, 0.5, 1.0, 2.0 and 10.0 mM). The vessels were incubated at room temperature (21 °C) without shaking. Liquid (1.5 ml) and headspace samples (1 ml) were withdrawn immediately after inoculation and after 5 days when NO3− reduction was complete.
For the pH experiments, 5.0 mM potassium phosphate or 20 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris) buffers were used. The ratios of monobasic potassium phosphate (KH2PO4) (Fisher Scientific, St Louis, MO, USA) and dibasic potassium phosphate (K2HPO4) (JT Baker, Phillipsburg, NJ, USA) were varied to achieve pH values of 6.0, 6.5, 7.0, 7.5 and 8.0. If necessary, 10.0 M HCl or 5.0 M NaOH solutions were used to adjust the medium pH to the desirable values. The Tris buffer was adjusted to pH values of 8.0, 8.5 and 9.0 with 10.0 M HCl. The degassed medium was dispensed under a stream of nitrogen gas and autoclaved. The pH measurements at the conclusions of the experiments verified that the pH values remained unchanged during the incubation period. Temperature effects were examined by incubating culture vessels amended with 5.0 mM sodium lactate and 1.0 mM sodium nitrate at 21 °C, 30 °C and 37 °C. All batch experiments were performed in triplicate and repeated in at least one independent experiment to verify reproducibility.
Chemostat experiments
Continuous culture experiments were performed in an anoxic chemostat reactor (DS0200TBSC, DASGIP, Jülich, Germany). The total capacity of the reactor was 475 ml and the volume of the aqueous phase was maintained at 200 ml. A syringe pump (PHD 2000, Harvard Apparatus, Holliston, MA, USA) was employed to simultaneously deliver fresh medium and remove bioreactor waste at a constant rate of 20 ml h−1 that equals a dilution rate of 0.1 h−1. The headspace of the bioreactor and the influent medium was constantly flushed with N2 to maintain anoxic conditions. The medium was prepared as described above in 2 l glass bottles that were subsequently connected to the chemostat influent (Supplementary Figure S1). C/N ratios of 1.5, 3.0, 4.5, 6.0 and 7.5 were established by amending the medium with 1, 2, 3, 4 or 5 mM lactate and 2 mM NO3− as the electron acceptor. To initiate each chemostat experiment, 2 mM lactate, 1 mM NO3−, the vitamin solution and trace metal solution were added to the autoclaved reactor containing 200 ml of medium. The reactor vessel was flushed with N2 for 1 h before inoculation with S. loihica strain PV-4. Following a 2-day incubation period, the syringe pump was turned on, and the bioreactor was continuously stirred to evenly distribute substrates and cells. Constant concentrations of NO3−, NO2−, lactate and acetate over a 6-h time interval indicated steady-state conditions that were reached in 72 h for all experimental conditions tested. Under steady-state reactor operating conditions, the cell density and the concentrations of substrates and products remained constant over time. Lactate, acetate, NO3− NO2− and NH4+ concentrations were measured in 1.5 ml samples collected from the bioreactor. After each sampling event, the valve controlling the outflow was closed for 4.5 min to adjust the aqueous volume to 200 ml. To quantify denitrification activity, the valves controlling the flow of N2 gas were closed and 10% of the headspace gas was replaced with acetylene. Headspace N2O concentrations were measured every 30–40 min to determine N2O production rates. The loss of dissolved N2O from the reactor vessel via the effluent was calculated by integrating the N2O effluent rate (aqueous N2O concentration multiplied by the flow rate) over time. The N2O production rate was corrected by this N2O loss rate to calculate the actual N2O production rate. After sampling, the reactor operation continued overnight before a 15-ml sample was collected for gene expression analyses. After sampling, the effluent valve was closed for 45 min to adjust the reactor volume to 200 ml. The measurements of steady-state lactate, acetate, NO3−, NO2−, and NH4+ concentrations, as well as N2O production rates, were repeated 24 h following the initial sampling event.
RNA extraction, purification and reverse transcription
Biomass for RNA extraction was collected from steady-state chemostat reactors operated under different feeding regimes (that is, C/N ratios of 1.5, 3.0, 4.5, 6.0 and 7.5; fumarate as electron acceptor). Sample aliquots (0.5 ml) were immediately mixed with 1.0 ml of RNA Protect Bacteria Reagent (Qiagen, Germantown, MD, USA), centrifuged for 10 min at 5000 × g and stored at −80 °C after the supernatant had been removed. Total RNA was extracted from the frozen samples within 1 week of sampling using an established protocol (Amos et al., 2008) with the following modifications. Luciferase control mRNA (Promega, Madison, WI, USA) was diluted to 1010 copies per ml and 1 μl of the diluted control mRNA was added as an internal standard to account for RNA loss during the extraction and purification process (Amos et al., 2008; Ritalahti et al., 2010). Cell pellets were then suspended in 350 μl of buffer RLT provided with RNeasy Mini Kit (Qiagen). The cell suspensions were transferred to 2 ml safe-lock tubes containing 50 mg of 200 μm zirconium beads (OPS Diagnostics, Lebanon, NJ, USA) and disrupted with the Omni Bead Ruptor 24 Homogenizer (Omni, Kennesaw, GA, USA) at 5.65 m s−1 for 5 min. After a brief 10-s centrifugation step (16 000 × g), the supernatants of each tube were transferred to new 1.5 ml tubes. The RNeasy Mini Kit (Qiagen) was then used following the manufacturer's recommendations to obtain the RNA in a final volume of 60 μl RNase-free water. The RNase-free DNase Set Kit (Qiagen) was used to remove any residual DNA. To 60 μl of the RNA solution, 10 μl of buffer RDD, 26.5 μl of RNase-free water and 3.5 μl of DNase I stock solution (all supplied with the Set Kit) were added. The reaction was incubated at room temperature for 15 min. After digestion, RNA was purified using the RNA MinElute Kit (Qiagen) according to the Qiagen protocol. The final step used 20 μl of nuclease-free water to elute the RNA from the column. Reverse transcription was performed with Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). One microliter of 10 mM dNTP mix (Invitrogen) and 1 μl of random hexamers (Invitrogen) diluted to 50 ng μl−1 were added to 11 μl of the purified RNA solution. After incubation at 65 °C for 5 min, the reaction vials were immediately placed on ice for 1 min. Then, 4 μl of fivefold concentrated first-strand buffer, 1 μl of 0.1 M dithiothreitol and 1 μl of RNaseOUT (40 U μl−1; Invitrogen) were added. After incubation at room temperature for 2 min, 1 μl of Superscript III Reverse Transcriptase (200 U μl−1) was added. The mixture was incubated at 25 °C for 10 min, at 42 °C for 3 h and at 72 °C for 15 min (Ritalahti et al., 2010). Then, 1 μl of RNase H (2 U μl−1; Invitrogen) was added to remove remaining RNA during a 20-min incubation period at 37 °C.
Genomic DNA extraction
For DNA extraction, the biomass from 1.5 ml aliquots was collected by centrifugation at 16 000 × g for 5 min at room temperature, and the cell pellets were immediately stored at −80 °C. Genomic DNA was extracted from S. loihica strain PV-4 cell pellets using the DNeasy Blood and Tissue Kit (Qiagen) as previously described (Yoon et al., 2013) and quantified spectrophotometrically with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Quantitative real-time PCR
Primer sets targeting the S. loihica strain PV-4 16S rRNA, nirK, nosZ and nrfA genes were designed using Primer3 software (Rozen and Skaletsky, 2000) (Table 1). In silico PCR (Bikandi et al., 2004) and Primer-BLAST (Ye et al., 2012) were used to assess primer specificity. S. loihica strain PV-4 harbors two nonidentical nrfA gene copies, nrfA0505 (Shew_0505) and nrfA0844 (Shew_0844). The translated proteins share 44% amino acid identity, and both genes/transcripts were targeted in this study. These targets were amplified using complementary DNA (cDNA) and genomic DNA as templates and quantified using Power SYBR Green detection chemistry (Life Technologies, Carlsbad, CA, USA) and the ABI ViiA7 real-time PCR system (Life Technologies) using default parameters (Yoon et al., 2013). PCR amplicons obtained with these primers were inserted into the PCR2.1 vector using the TOPO TA Cloning Kit (Invitrogen). The plasmids were extracted with the QIAprep Spin Miniprep Kit (Qiagen) and used to construct qPCR calibration curves (Table 1). Triplicate serial 10-fold dilutions yielded 108 to 102 target genes per μl and were used as template DNAs in qPCR assays. Using the default instrument parameters and automated Cq determination, standards resulted in linear calibration curves and amplification efficiencies ranging from 92.1% to 99.6% (Hatt and Löffler, 2012). The standard deviation of Cq values for 102 copies (Cq∼29.0) were lower than 0.16 for all calibration qPCR runs, indicating the limit of detection was below 102 target sequences. All qPCR assays were performed in triplicate for each cDNA and genomic DNA sample. Luciferase cDNA recovery was used to correct for mRNA losses during extraction, purification and the reverse transcription step (Johnson et al., 2005). Consistent melting temperatures suggested qPCR assay specificity and no amplification occurred in no-template controls. The calibration curve for luciferase DNA was constructed by reverse transcription of the luciferase control mRNA and insertion of the PCR product (amplified with primers lucrefF and lucrefR) into the PCR2.1 vector (Amos et al., 2008). Based on the amount of luciferase mRNA recovered, the RNA recovery ranged from 11.3% to 20.7%. The qPCR assay results were analyzed with ViiA7 Software (Life Technologies) provided with the system. The two-tailed Student's t-tests were performed using the SPSS 21.0.0 software package (IBM Corp, Armonk, NY, USA) to verify statistical significance of the qPCR results.
Table 1. Primers used for RT-qPCR analyses and qPCR calibration curve parameters.
| Primer set | Target gene (locus tag) | Amplicon length (bp) | Slope | y-Intercept | Amplification efficiency | R2 | Reference |
|---|---|---|---|---|---|---|---|
| SlonirK853f: 5′-AAGGTGGGTGAGTCTGTGCT-3′SlonirK1040r: 5′-GGCTGGCGGAAGGTGTAT-3′ | nirK (Shew_3335) | 188 | −3.526 | 35.565 | 92.1 | 0.999 | This study |
| SlonrfA1083f: 5′-GGATATCCGTCACGCTCAAT-3′SlonrfA1308r: 5′-GTCCATACCCAATGCAGCTT-3′ | nrfA (Shew_0844) | 226 | −3.384 | 34.555 | 97.5 | 0.998 | Yoon et al. (2013) |
| SlonrfA818f: 5′-AGGGCAAGGCCTATACCAAC-3′SlonrfA950r: 5′-TTCTCGGCTATCTGCGACTT-3′ | nrfA (Shew_0505) | 149 | −3.332 | 35.975 | 99.6 | 0.996 | This study |
| SlonosZ599f: 5′-ATGGTAAGGAGACGCTGGAA-3′SlonosZ758r: 5′-TTGTAGCAGGTAGAGGCGAAG-3′ | nosZ (Shew_3400) | 160 | −3.515 | 34.451 | 92.3 | 0.996 | This study |
| Slo16Sf: 5′-CACACTGGGACTGAGACACG-3′Slo16Sr: 5′-TGCTTCTTCTGCGAGTAACG-3′ | 16S rRNA genesa | 191 | −3.380 | 34.240 | 97.6 | 0.991 | This study |
| lucrefA: 5′-TACAACACCCCAACATCTTCGA-3′lucrefB: 5′-GGAAGTTCACCGGCGTCAT-3′ | luciferase control mRNA | 67 | −3.380 | 36.817 | 97.6 | 0.999 | Johnson et al. (2005) |
Abbreviation: RT-qPCR, reverse-transcriptase real-time quantitative PCR.
Shewanella loihica strain PV-4 possesses nine 16S rRNA gene copies that all amplified with the Slo16Sf and Slo16Sr primer sets, as suggested by in silico PCR (Bikandi et al., 2004).
Results
Effects of C/N ratios on NO3−/NO2− reduction pathways
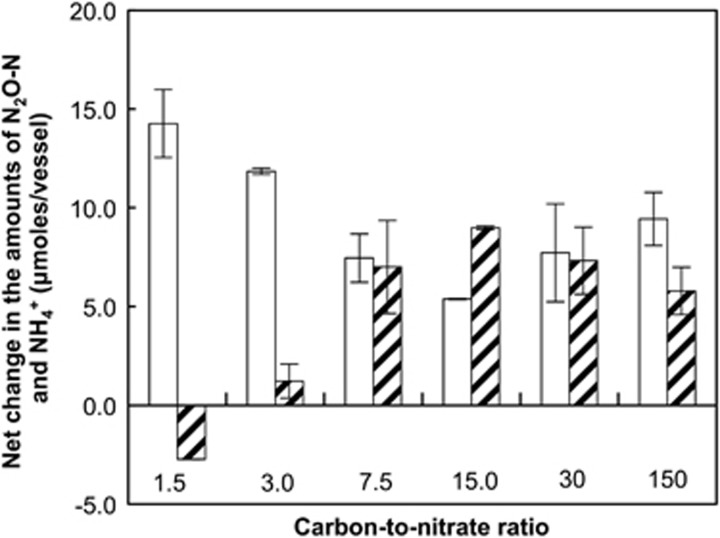
S. loihica strain PV-4 batch culture experiments with 0.1–10 mM lactate and 0.2 mM NO3− suggested that denitrification predominated at low C/N ratios (Figure 1). At an initial C/N ratio of 1.5, the product of dissimilatory NO3− reduction was exclusively N2O, with 14.3±1.7 μmol N2O-N recovered from 19.5±0.1 μmol of NO3−. The amount of NH4+ in the medium decreased during cultivation, presumably because of NH4+ incorporation into biomass. In batch incubations with initial C/N ratios higher than 1.5, both N2O and NH4+ were produced concomitantly. Significantly more N2O-N and less NH4+ were produced at an initial C/N ratio of 3.0 than at a C/N ratio of 7.5 (P<0.01); however, increasing the C/N ratio beyond 7.5 did not affect the denitrification/respiratory ammonification ratio significantly (one-way analysis of variance, P>0.05). When the NO3− concentration was increased to 1 mM, N2O was the predominant product regardless of the C/N ratio. These observations suggested that the batch systems cannot resolve C/N ratio effects on pathway selection presumably because of changing C/N ratios during growth.
Figure 1.
Net production of N2O (white bars) and NH4+ (hatched bars) over a 5-day incubation period at 21 °C of S. loihica strain PV-4 batch cultures with 0.2 mM NO3− and 0.1, 0.2, 0.5, 1.0, 2.0 and 10.0 mM lactate. Each bar represents the average of triplicate samples, with error bars representing the s.d.
To explore NO3− reduction pathways under controlled conditions with consistent limitation of either electron donor or electron acceptor (that is, a constant C/N ratio in the feed medium), S. loihica strain PV-4 was grown in a chemostat vessel. Supplying feed solutions with C/N ratios ⩾6.0 resulted in steady-state conditions with no measurable NO3−/NO2− in the reactor, indicating the electron acceptor is limiting (Table 2). Chemostat vessel operation at C/N ratios between 3.0 and 4.5 resulted in steady-state conditions with no measurable lactate or NO3−/NO2− concentrations in the reactor. At a C/N ratio of 1.5, the steady-state nitrate concentration was 0.18±0.14 mM, indicating electron donor-limiting conditions. Acetate concentrations ranged from 1.47±0.06 to 3.98±0.09 mM in the reactor at C/N ratios of ⩾3.0 but dropped to below the detection limit of ∼10 μM at a C/N ratio of 1.5.
Table 2. Concentrations of carbon (C) and nitrogen (N) compounds in the influent medium and the steady-state reactor in experiments examining the effect of C/N ratios on respiratory ammonification and denitrification pathways in Shewanella loihica strain PV-4.
| C/N ratio |
Measured influent concentrations (mM) |
Measured steady-state concentrations (mM) |
||||||
|---|---|---|---|---|---|---|---|---|
| Lactate | NO3− | NH4+ | NO3− | NO2− | NH4+ | Lactate | Acetate | |
| 7.5 | 5.04 (0.03) | 1.96 (0.10) | 0.46 (0.03) | 0 | 0 | 1.79 (0.01) | 0.51 (0.01) | 3.98 (0.09) |
| 6.0 | 3.91 (0.18) | 2.03 (0.01) | 0.47 (0.01) | 0 | 0 | 1.48 (0.01) | 0.05 (0.01) | 2.93 (0.05) |
| 4.5 | 2.82 (0.07) | 2.03 (0.02) | 0.44 (0.02) | 0 | 0 | 0.88 (0.01) | 0 | 2.33 (0.10) |
| 3.0 | 2.15 (0.00) | 1.89 (0.00) | 0.50 (0.02) | 0 | 0 | 0.29 (0.05) | 0 | 1.47 (0.06) |
| 1.5 | 1.04 (0.03) | 1.90 (0.03) | 0.48 (0.03) | 0.18 (0.15) | 0.14 (0.13) | 0.20 (0.02) | 0 | 0 |
The averages of duplicate measurements are presented with the s.d. indicated in parentheses.
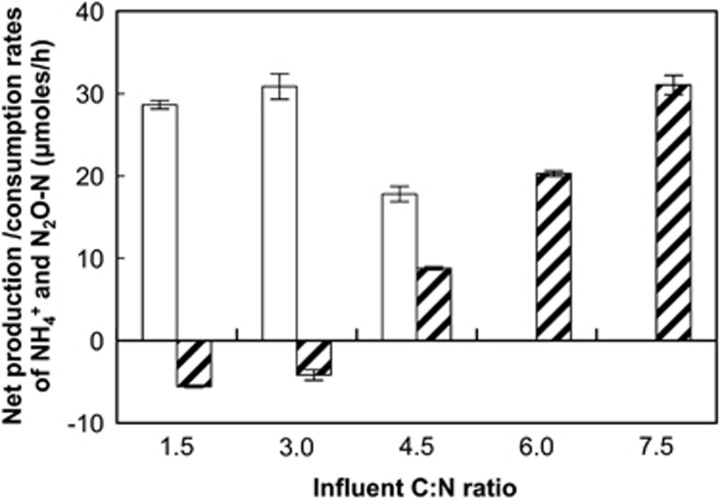
The net NH4+ and N2O production rates served as measures of respiratory ammonification and denitrification activities, respectively. At C/N ratios of ≤3.0 in the chemostat feed, production of N2O, but not of NH4+, was observed (Figure 2). At a C/N ratio of 1.5, denitrification was the predominant pathway with 76±10% of the consumed NO3− recovered as N2O. Assimilation of NH4+ exceeded production, resulting in a net NH4+ consumption (Figure 2). Previous growth experiments with S. loihica strain PV-4 suggested that approximately half of the biomass N originated from NH4+ and the other half from NO3− when grown under denitrifying conditions with acetate as electron donor (Yoon et al., 2013). Assuming an even split of assimilated N compounds with lactate as electron donor, NH4+ production from respiratory ammonification was negligible at a C/N ratio of 1.5. Similar observations were made at a C/N ratio of 3.0, with 82% of NO3− reduced to N2O and a net NH4+ consumption indicating the absence of significant respiratory ammonification activity. Higher C/N ratios in the chemostat feed led to increased NH4+ formation and correspondingly lower N2O production rates. Simultaneous production of NH4+ and N2O was observed at a C/N ratio of 4.5. At C/N ratios of 6.0 and 7.5, only NH4+ and no N2O was produced (Figure 2). Similar results were obtained in an independent replicate chemostat experiment (Supplementary Figure S2). Under the experimental conditions promoting denitrification activity (that is, C/N ratio ≤3.0), biomass yields were lower than under conditions favoring respiratory ammonification (that is, C/N ratio ⩾6.0; Table 3 and Supplementary Figure S3), indicating that more energy was conserved during NO3− to NH4+ reduction than during NO3− to N2O reduction. N mass balances for the different C/N ratios ranged from 98.5% to 106.8% in terms of NO3−-N added to the bioreactor and recovered as NO2−, NH4+, N2O and biomass N (Table 3). The DNA yields mirrored the biomass production rate estimates (Table 3).
Figure 2.
Net production of N2O (white bars) and NH4+ (shaded bars) in S. loihica strain PV-4 chemostat cultures with C/N ratios varying from 1.5 to 7.5 in the feed solution. The feed solution contained 2 mM NO3− and was delivered at a rate of 20 ml h−1. Rates of respiratory ammonification were calculated from the input and steady-state concentrations of NH4+, whereas denitrification rates were calculated by measuring N2O production rates following the addition of 10% (vol/vol) acetylene gas to the reactor vessel headspace. At C/N ratios of 1.5 and 3.0, net consumption of NH4+ occurred. The bars represent the averages of two separate measurements taken on different days after steady-state conditions had been reached. The error bars show the s.d. of two measurements taken with a 24-h interval.
Table 3. Substrate consumption and metabolite production rates in the steady-state reactor receiving feed with different carbon-to-nitrogen (C/N) ratios.
| C/N ratio |
Rates of production/consumptiona
in the steady-state reactor (μmol h−1) |
DNA (ng μl−1) b | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Lactate | Acetate | NO3− | NO2− | NH4+ | N2O-N | Biomass N | Biomass C | ||
| 7.5 | −90.6 (0.4) | 79.6 (1.9) | −39.2 (2.0) | 0 (0) | 26.5 (0.2) | 0 | 14.5 (1.2) | 72.5 (6.0) | 10.2 (1.2) |
| 6.0 | −77.2 (2.3) | 58.56 (1.1) | −40.6 (0.1) | 0 (0) | 20.2 (0.3) | 0 | 19.8 (0.8) | 98.8 (4.1) | 14.5 (1.1) |
| 4.5 | −56.4 (1.5) | 46.6 (2.0) | −40.6 (0.1) | 0 (0) | 8.8 (0.2) | 17.8 (0.9) | 14.1 (0.6) | 70.5 (3.0) | 9.8 (0.5) |
| 3.0 | −43.0 (0.1) | 29.4 (1.1) | −37.8 (0.1) | 0 (0) | −4.2 (0.6) | 30.9 (1.6) | 11.1 (0.5) | 55.7 (2.4) | 9.0 (0.2) |
| 1.5 | −20.8 (0.6) | 0 (0) | −34.4 (2.4) | 2.8 (2.7) | −5.6 (0.1) | 28.6 (0.5) | 11.3 (0.1) | 56.7 (0.6) | 8.4 (0.4) |
The production rates of dissolved compounds and biomass N and C were calculated from the concentrations in the influent medium and steady-state aqueous phase concentrations. The reported N2O production rates account for N2O loss because of the flux of the dissolved N2O with the effluent (see Materials and methods section for details). Except for biomass C and N, the values are the average of duplicate measurements and the s.d. is indicated in parentheses. Biomass C and N data were calculated from the cell quantification data presented in Supplementary Figure S2.
A minus sign denotes consumption.
Data shown are average and s.d. of triplicate DNA extracts; 260/280 ratios were between 1.78 and 1.87.
At an influent C/N ratio of 7.5, a steady-state acetate concentration of 3.98±0.09 mM was observed that was significantly higher (P<0.05) than the steady-state lactate concentration of 0.51±0.01 mM (Table 3). S. loihica strain PV-4 can use acetate for denitrification (Yoon et al., 2013); however, the presence of acetate did not lead to N2O production, indicating that acetate did not affect dissimilatory pathway selection (that is, denitrification vs respiratory ammonification) under the chemostat cultivation conditions. When the feed contained both 1.5 mM NO2− and 0.5 mM NO3−, only NH4+ or N2O were formed at C/N ratios above or below 4.5, respectively, and both products were observed at a C/N ratio of 4.5 (data not shown). These findings were consistent with the observations made in the NO3−-fed reactor, indicating that the electron acceptor (that is, NO3− vs NO2−) did not alter reduced product(s) formation.
Effects of C/N ratios on the expression of genes encoding denitrification and respiratory ammonification pathways
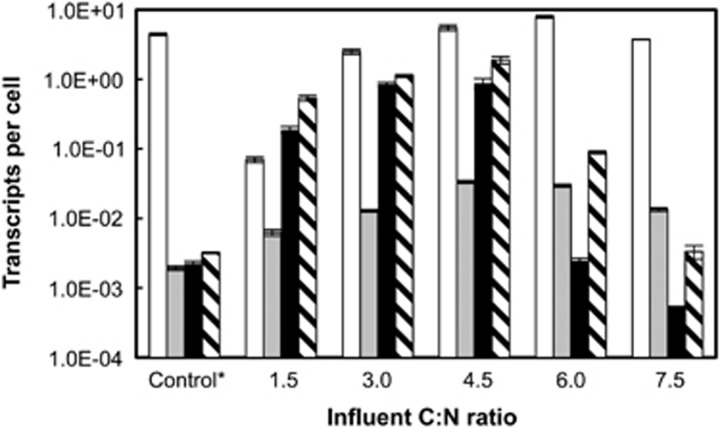
Reverse-transcriptase qPCR (RT-qPCR) was used to enumerate nirK, nosZ and two nrfA transcripts in cDNA derived from RNA of steady-state chemostat cultures grown under different C/N ratios. The mRNA abundances in cells incubated with fumarate as the electron acceptor benchmarked the expression of genes involved in denitrification and/or respiratory ammonification (Supplementary Table S1). No more than 0.01 nrfA0505, nirK (Shew_3335) and nosZ (Shew_3400) transcripts per strain PV-4 cell were measured in fumarate-grown control cultures, whereas the abundance of nrfA0844 transcripts was significantly higher (4.44 transcripts per cell) (Supplementary Table S1). In lactate/NO3−-fed chemostat cultures with a C/N ratio of 1.5, the abundance of nrfA0844 mRNA was two orders of magnitude lower than in fumarate-grown cells, suggesting an effect of the C/N ratio on nrfA0844 gene expression (Figure 3). At a C/N ratio of 3.0, no NH4+ was produced but nrfA0844 was expressed and 2.5±0.2 transcripts per cell were measured. nrfA0844 transcripts at higher C/N ratios were in the same order of magnitude and the maximum abundance of nrfA0844 transcripts was observed at a C/N ratio of 6.0 with 8.0±0.35 transcripts per cell. Interestingly, the abundance of nrfA0844 at a C/N ratio of 7.5 (3.8±0.04 transcripts per cell) was only 1.5-fold higher (P<0.05) than at the C/N ratio of 3.0 despite the significant difference observed in N2O versus NH4+ production (Figure 2). nrfA0505 was expressed at orders of magnitude lower levels than nrfA0844 under all growth conditions tested.
Figure 3.
Reverse-transcriptase (RT)-qPCR analyses of nrfA0844 (white bars), nrfA0505 (gray bars; respiratory ammonification), nirK (black bars) and nosZ (hashed bars; denitrification) transcripts in S. loihica strain PV-4 under varying C/N ratios. The error bars represent the s.d. of triplicate per-cell transcript copy numbers calculated from the s.d. of qPCR measurements for cDNA and genomic DNA using the error propagation method. The results of an independent replicate experiment are shown in Supplementary Figure S3. *The control is the transcript copy number of each gene in the sample extracted from the steady-state reactor incubated with fumarate as the electron acceptor.
In contrast to nrfA0505 and nrfA0844 expression, pronounced nirK transcriptional changes were observed in response to changing the C/N ratios (Figure 3). At C/N ratios of 6.0 and 7.5, the nirK mRNA abundances exceeded those measured at lower C/N ratios by up to three orders of magnitude (that is, ∼1600-fold). The nosZ expression followed a profile similar to that of nirK, and at C/N ratios of 6.0 and 7.5, nosZ transcription was up to 27-fold lower compared with the transcript levels observed at lower C/N ratios. The observation of active nirK transcription (that is, ⩾0.19 transcript per cell) coincided with N2O production, whereas cells with ≤2.5 × 10−3 nirK transcripts per cell produced no N2O. An independent chemostat experiment demonstrated the reproducibility of the reverse-transcriptase qPCR analysis. The nrfA844 mRNA abundances at C/N ratios of 3.0 and 6.0 were <3% different from the nrfA844 expression data of the first experiment and a 2.5-fold difference was observed at a C/N ratio of 1.5 (Supplementary Figure S4). The nirK transcript abundance data exhibited <25% deviation from the nirK expression data presented in Figure 3 for all three C/N ratios tested.
Effects of pH and temperature on NO3−/NO2− reduction pathways
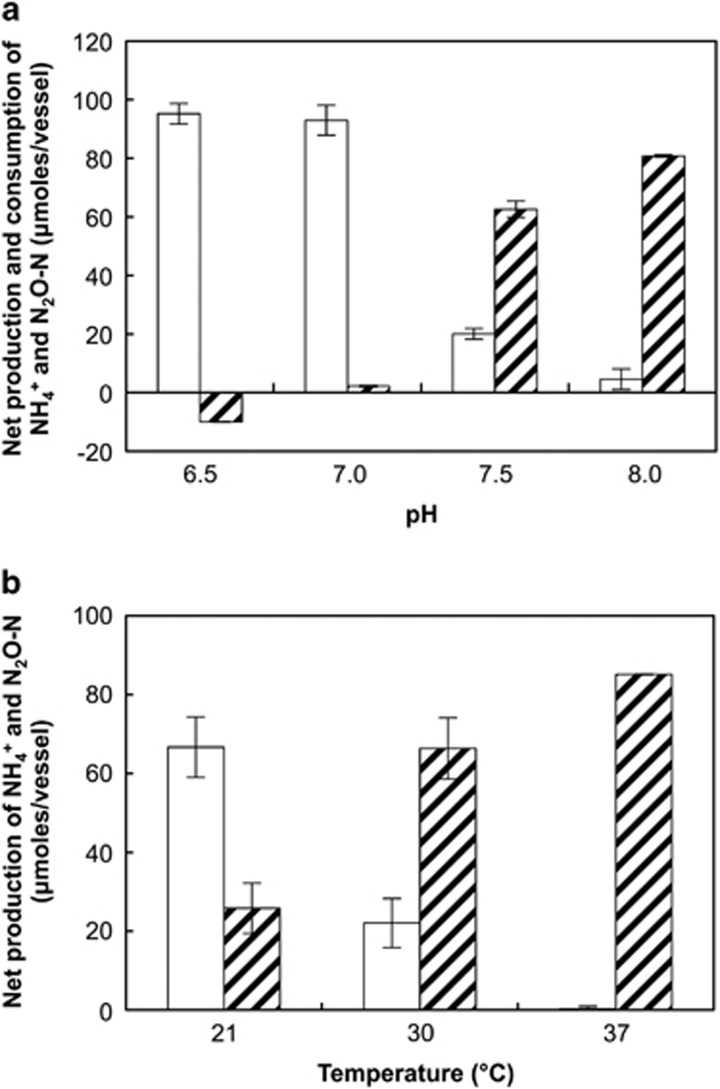
For assessing the effects of pH and temperature on dissimilatory NO3−/NO2− reduction pathways, the medium received 5 mM lactate and 1 mM NO3− (C/N=15) to avoid electron donor limitations. A pH increase from 6.5 to 8.0 shifted the reduced product distribution from entirely N2O to predominantly NH4+. At pH 6.5, 92.1±3.4 μmol of the 105.9 μmol of NO3− were reduced to N2O (Figure 4a). In these cultures, NH4+ decreased by 10.0±0.03 μmol, presumably because of assimilation into biomass. The proportion of NH4+ increased as the pH was raised in 0.5 increments, and at pH 8.0, NH4+ was the predominant product of NO3− reduction with 80.8±0.36 μmol formed, whereas N2O accounted for only 4.4±3.4 μmol (Figure 4a). In Tris-buffered medium with pH ⩾8.0, exclusively NH4+ was produced (data not shown). Consistent results obtained with different buffer systems in overlapping pH ranges indicated that the buffer type (that is, phosphate versus Tris buffer) did not affect NO3−/NO2− reduction pathways in S. loihica strain PV-4. These findings demonstrate that pH has a strong effect on the selection of dissimilatory NO3− reduction pathways in strain PV-4, and N2O is the major product under acidic conditions whereas NH4+ predominated at alkaline pH.
Figure 4.
Net change of N2O (white bars) and NH4+ (hatched bars) in S. loihica strain PV-4 batch cultures amended with 5.0 mM lactate and 1.0 mM NO3− (C/N=15:1) under varying (a) pH and (b) temperature conditions. The bars represent the average of triplicate samples, with error bars representing the s.d.
S. loihica strain PV-4 was reported to grow over a temperature range of 0 °C to 42 °C (Gao et al., 2006), and the effects of temperature on the two NO3− reduction pathways were explored. Batch cultures incubated at 21 °C transformed 98.7±0.8 μmol of NO3− to 33.2±3.1 μmol N2O and 22.1±7.8 μmol NH4+ (Figure 4b). Higher incubation temperatures shifted the reduced product distribution toward NH4+ and 65.6±6.1 μmol NH4+ and 10.9±3.9 μmol N2O were produced at 30 °C. At 37 °C, the highest temperature tested, NH4+ (84.3±0.9 μmol) was the predominant product and <0.3 μmol of N2O was detected at the end of the incubation. Despite numerous attempts, cultivation of S. loihica strain PV-4 with NO3− as electron acceptor was not successful at temperatures below 20 °C.
Discussion
The batch culture experiments with S. loihica strain PV-4 had shortcomings in unraveling C/N ratio effects on the selection of denitrification versus respiratory ammonification pathways. At C/N ratios of ⩾7.5, N2O and NH4+ were produced in similar amounts (one-way analysis of variance, P>0.05). Respiratory ammonification yields more biomass per molecule of NO3−/NO2− reduced than denitrification and thus should be a competitive process under NO3−/NO2−-limiting conditions (Tiedje et al., 1982; Strohm et al., 2007). Hence, the prediction is that organisms select the pathway that maximizes energy conservation, in particular under conditions of substrate limitation. In batch cultures with 0.2 mM NO3− and low initial cell numbers (<106 cells per ml), NO3−/NO2− was, at least initially, not limiting and therefore should not have affected pathway selection. At C/N ratios of ≤3.0, the formation and consumption of acetate complicated data interpretation. Shewanella spp. couple denitrification to acetate oxidation but acetate does not support NO2− reduction to NH4+ (Yoon et al., 2013). As lactate oxidation to acetate was insufficient to deplete the NO3−/NO2− pool at these C/N ratios, the ensuing utilization of acetate yielded exclusively N2O. A similar bias because of selective acetate oxidation was previously observed in a mixed culture experiment where denitrification coupled to acetate oxidation obscured C/N ratio effects (Rehr and Klemme, 1989). Therefore, the batch culture results could not resolve the effects of different C/N ratios on pathway selection. The chemostat experiments in conjunction with gene-targeted reverse-transcriptase qPCR expression analyses resolved these complications and demonstrated the C/N ratio effects on the selection of dissimilatory NO3−/NO2− reduction pathways in S. loihica strain PV-4. Chemostats can achieve steady-state substrate concentrations near zero while maintaining a constant substrate supply, and cellular responses to substrate limitations can be observed (Friedrich, 1982; Durner et al., 2000). In the experiments with S. loihica strain PV-4, high C/N ratios in the influent resulted in NO3− limitations that reduced nirK expression levels and led to the predominance of respiratory ammonification. Thus, the chemostat cultures demonstrated the effects of high C/N ratios on pathway selection, and the findings are consistent with previous observations that respiratory ammonification predominates in carbon-rich environments.
Although S. loihica strain PV-4 can couple acetate oxidation to denitrification but not respiratory ammonification, the chemostat supplied with a C/N ratio of 7.5 demonstrated that the presence of acetate (up to 3.92 mM) did not result in increased nirK/nosZ expression levels or denitrification activity (Figures 2 and 3). Thus, acetate oxidation occurred only under conditions that favored denitrification, suggesting that acetate did not affect the outcome and interpretation of the chemostat experiments. Although the steady-state lactate concentrations in the reactor were below the method detection limit of ∼10 μM at C/N ratios of ≤3.0 in the feed solution, lactate was constantly introduced into the reactor at rates of 20–40 μmol min−1, and lactate oxidation sustained denitrification activity but not respiratory ammonification. Consistent with these observations, the reverse-transcriptase qPCR results demonstrated elevated nirK and reduced nrfA expression levels under lactate-limiting conditions (that is, at low C/N ratios).
The transcription levels of nrfA and nirK did not decrease at intermediate C/N ratios of 3.0 and 4.5, at which lactate and nitrate concentrations in steady-state chemostats were both below the detection limits (Table 3). Nevertheless, the influent C/N ratio caused a subtle (for example, twofold and statistically distinguishable) shift in nrfA or nirK transcription levels (Figure 3 and Supplementary Table S2), resulting in significantly different outcomes at these two C/N ratios (that is, exclusively N2O production at a C/N ratio of 3.0 versus production of both N2O and NH4+ at a C/N ratio of 4.5; Figure 2). Taken together, the phenotypic observations and the nrfA0844, nirK and nosZ transcription levels (Figure 3) indicated that both pathways were active at a C/N ratio of 4.5. This ambivalent regulation of the two pathways at intermediate C/N ratios is a likely cause for the inconsistent batch experiment results, where neither electron donor nor electron acceptor was limiting until the end of the experiment.
S. loihica strain PV-4 possesses two nonidentical nrfA genes, nrfA0505 and nrfA0844, and the expression levels of nrfA0844 exceeded those of nrfA0505 by at least one order of magnitude. NrfA0844 shares 78% amino acid identity to the NrfA of S. oneidensis strain MR-1, whose function as an ammonia-forming nitrite reductase has been confirmed (Cruz-Garcia et al., 2007). On the other hand, NrfA0505 has not been functionally characterized but shares up to 50% similarity with functionally verified NrfA proteins from S. oneidensis strain MR-1 and Escherichia coli. The low expression level of nrfA0505 suggests that NrfA0844 is responsible for the observed respiratory ammonification activity; however, a detailed biochemical characterization of NrfA505-type proteins to elucidate their functional roles, if any, in respiratory ammonification has yet to be accomplished. A microarray-based transcriptome analysis of S. oneidensis strain MR-1 (Beliaev et al., 2005) found the nrfA0844-like gene highly expressed when grown with NO3−; however, our study found that nrfA0844 was also expressed in cultures grown with fumarate in the absence of NO3−. The chemostat experiments demonstrated downregulation of nrfA0844 under electron donor-limiting conditions (that is, at a C/N ratio of 1.5), suggesting that carbon availability may be a controlling factor in nrfA0844 gene transcription. At a C/N ratio of 4.5 with simultaneous production of NH4+ and N2O, nrfA0844 transcription was higher than at a C/N ratio of 7.5 (P<0.05), where the product was predominantly NH4+. These observations indicate that the measurement of nrfA transcripts as a proxy for respiratory ammonification activity in an environmental sample must be interpreted cautiously, and should be at least accompanied by additional measurements such as nirK and nirS mRNA abundance.
Although pure cultures studies have limitations to predict environmental processes, the observations made with S. loihica strain PV-4 can explain at least some previous field observations. Soils and sediments receiving high NO3− loadings showed increased denitrification rates, whereas elevated respiratory ammonification rates were observed in soils and sediments receiving high carbon loadings (Koop-Jakobsen and Giblin, 2010; Nizzoli et al., 2010; Schmidt et al., 2011; Fernandes et al., 2012; Dunn et al., 2013). In carbon-rich environments, the N oxyanion availability in anoxic zones depends on NO3− or NO2− fluxes from the oxic zone (that is, through organic N compound degradation and nitrification). Hence, carbon-rich environments are generally NO3− and NO2− limited, conditions that were mimicked in chemostats operated under high C/N ratios, under which respiratory ammonification activity predominated. In events of heavy N input such as fertilizer application, bioavailable electron donor and carbon sources in anoxic zones may become limiting (DeSimone and Howes, 1996; Inwood et al., 2007; Koop-Jakobsen and Giblin, 2010). Such conditions were simulated in chemostat experiments receiving feed with low C/N ratios that increased NH4+ production rates but decreased N2O production rates and nirK transcription levels. A recent study using a chemostat inoculated with a tidal flat sediment community also reported that the C/N ratio was a major factor determining the end product of bacterial NO3− reduction (Kraft et al., 2014). Nevertheless, a number of studies found no consistent correlation between C/N ratios and dissimilatory NO3− reduction pathways (Nizzoli et al., 2006; Schmidt et al., 2011). This inconsistency may be attributed to balanced input of C and N, as observed in the chemostats receiving intermediate C/N ratios, the degradability of the bioavailable C substrate or the heterogeneity of the soil and sediment environment studied. Another possibility is that respiratory ammonification activity is only upregulated when labile C sources, such as root exudates, are available (Schmidt et al., 2011). To further explore the effects of C or N limitation on NO3−/NO2− fate via dissimilatory pathways, controlled mesocosm-scale experiments should be performed to carefully determine whether the strain PV-4 pure culture results have bearing for predicting NO3−/NO2− fate in complex environmental systems. Other NO3−/NO2− reduction pathways may also play a role, including the recently discovered reverse-HURM (hydroxylamine:ubiquinone reductase module) pathway in Nautilia and Campylobacter spp. (Hanson et al., 2013). This NO3−/NO2− ammonification pathway provides the organism with ammonium for biomass synthesis; however, its significance for dissimilatory NO3−/NO2− reduction remains to be determined. Nevertheless, the possibility that dissimilatory processes in addition to denitrification and respiratory ammonification contribute to NO3−/NO2− fate should be considered.
In addition to C/N ratios, other environmental factors, including pH and temperature, can influence pathway selection. Previous reports about pH effects on NO3− fate were largely contradictory (Nägele and Conrad, 1990; Stevens et al., 1998). The pure culture experiments with S. loihica strain PV-4 revealed that respiratory ammonification was favored at elevated pH and denitrification was favored under lower pH conditions. The catalytic subunit (NirK) of the copper-dependent nitrite reductase (CuNIR) and the ammonia-forming nitrite reductase, NrfA, are both located in the periplasm. The periplasmic pH of E. coli, a neutrophilic Gammaproteobacterium like S. loihica, is influenced by the extracellular pH (Wilks and Slonczewski, 2007), suggesting that the in vivo activities of these enzymes could be affected by the pH of the surrounding matrix. Previous enzyme characterization studies demonstrated that CuNIR and NrfA proteins have distinct pH optima and ranges for activity. For example, the pH optima of characterized CuNIR enzyme systems are below 7 (Iwasaki and Matsubara, 1972; Abraham et al., 1997; Jacobson et al., 2007). The CuNIR isolated from the Betaproteobacterium Alcaligenes xylosoxidans showed optimal catalytic performance at pH 5.2 that diminished to negligible levels above pH 7.5 (Abraham et al., 1997). Interestingly, the other group of NO-forming nitrite reductases, the periplasmic cytochrome cd1 nitrite reductases (catalytic subunit encoded by nirS) (Yamazaki et al., 1995), also showed maximum activity under acidic pH conditions (Lam and Nicholas, 1969; Singh, 1974; Richter et al., 2002), suggesting that NO2− reduction to NO is favored at pH<7. In contrast, the E. coli strain K-12 NrfA protein exhibited optimal activity around pH 7.5, with negligible activity below pH 7.0, and the Desulfovibrio desulfuricans NrfA protein demonstrated maximum activity between pH 8.0 and 9.5 (Liu and Peck, 1981; Kajie and Anraku, 1986). The S. loihica strain PV-4 NirK shares 76% amino acid sequence similarity with the NirK of Alcaligenes xylosoxidans, whereas NrfA0844 is 76% similar to the E. coli NrfA. Thus, it is possible that the physicochemical properties of NirK and NrfA contributed to the observed pH effects on denitrification and respiratory ammonification activity in strain PV-4. Although temperature effects on dissimilatory NO3−/NO2− reduction pathways were observed with S. loihica strain PV-4, the mechanistic underpinning is unclear. A possible explanation for the observed temperature effects may be the direct responses of the NrfA and NirK enzyme systems. The E. coli NrfA showed maximum activity at 57 °C (Kajie and Anraku, 1986); however, no information regarding temperature effects on NirK activity is available. Although the mechanism for temperature regulation is unknown, the strain PV-4 experimental findings are in agreement with previous field observations, as respiratory ammonification activity had a stronger influence on NO3− fate in tropical climates and during the warmer summer season (Ogilvie et al., 1997; Nizzoli et al., 2006; Dong et al., 2011; Dunn et al., 2013).
In summary, the experiments with S. loihica strain PV-4 capable of NO3−/NO2− reduction to N2 or NH4+ revealed the specific conditions that favor either denitrification or respiratory ammonification. NO3−/NO2− limitations at high C/N ratios decreased transcription levels of denitrification genes (nirK and nosZ) and led to the predominance of the respiratory ammonification pathway, whereas electron acceptor limitations at low C/N ratios increased nirK and nosZ transcription, leading to the predominance of denitrification. At elevated temperatures and alkaline pH conditions, the respiratory ammonification pathway predominated over the denitrification pathway. The S. loihica strain PV-4 experiments suggest that pure culture studies can contribute to a better understanding of the environmental controls governing the fate of NO3−/NO2− in soils and sediments.
Acknowledgments
This research was supported by the US Department of Energy, Office of Biological and Environmental Research, Genomic Science Program, Award DE-SC0006662.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abraham ZH, Smith BE, Howes BD, Lowe DJ, Eady RR. pH-dependence for binding a single nitrite ion to each type-2 copper centre in the copper-containing nitrite reductase of Alcaligenes xylosoxidans. Biochem J. 1997;324:511–516. doi: 10.1042/bj3240511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos BK, Ritalahti KM, Cruz-Garcia C, Padilla-Crespo E, Löffler FE. Oxygen effect on Dehalococcoides viability and biomarker quantification. Environ Sci Technol. 2008;42:5718–5726. doi: 10.1021/es703227g. [DOI] [PubMed] [Google Scholar]
- Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF, Tiedje JM, et al. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J Bacteriol. 2005;187:7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikandi J, Millan RS, Rementeria A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics. 2004;20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- Burgin AJ, Hamilton SK. Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Front Ecol Environ. 2007;5:89–96. [Google Scholar]
- Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol. 2007;189:656–662. doi: 10.1128/JB.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone LA, Howes BL. Denitrification and nitrogen transport in a coastal aquifer receiving wastewater discharge. Environ Sci Technol. 1996;30:1152–1162. [Google Scholar]
- Dong LF, Sobey MN, Smith CJ, Rusmana I, Phillips W, Stott A, et al. Dissimilatory reduction of nitrate to ammonium, not denitrification or anammox, dominates benthic nitrate reduction in tropical estuaries. Limnol Oceanogr. 2011;56:279–291. [Google Scholar]
- Dunn RJK, Robertson D, Teasdale PR, Waltham NJ, Welsh DT. Benthic metabolism and nitrogen dynamics in an urbanised tidal creek: domination of DNRA over denitrification as a nitrate reduction pathway. Est Coast Shelf Sci. 2013;131:271–281. [Google Scholar]
- Durner R, Witholt B, Egli T. Accumulation of Poly[(R)-3-hydroxyalkanoates] in Pseudomonas oleovorans during growth with octanoate in continuous culture at different dilution rates. Appl Environ Microbiol. 2000;66:3408–3414. doi: 10.1128/aem.66.8.3408-3414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzolari É, Nicolardot B, Germon JC. Simultaneous effects of increasing levels of glucose and oxygen partial pressures on denitrification and dissimilatory nitrate reduction to ammonium in repacked soil cores. Eur J Soil Biol. 1998;34:47–52. [Google Scholar]
- Fernandes SO, Bonin PC, Michotey VD, Garcia N, LokaBharathi PA. Nitrogen-limited mangrove ecosystems conserve N through dissimilatory nitrate reduction to ammonium. Sci Rep. 2012;2:419. doi: 10.1038/srep00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzhugh RD, Lovett GM, Venterea RT. Biotic and abiotic immobilization of ammonium, nitrite, and nitrate in soils developed under different tree species in the Catskill Mountains, New York, USA. Glob Change Biol. 2003;9:1591–1601. [Google Scholar]
- Friedrich CG. Depression of hydrogenase during limitation of electron donors and derepression of ribulosebisphosphate carboxylase during carbon limitation of Alcaligenes eutrophus. J Bacteriol. 1982;149:203–210. doi: 10.1128/jb.149.1.203-210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Obraztova A, Stewart N, Popa R, Fredrickson JK, Tiedje JM, et al. Shewanella loihica sp. nov., isolated from iron-rich microbial mats in the Pacific Ocean. Int J Syst Evol Microbiol. 2006;56:1911–1916. doi: 10.1099/ijs.0.64354-0. [DOI] [PubMed] [Google Scholar]
- Hanson TE, Campbell BJ, Kalis KM, Campbell MA, Klotz MG. Nitrate ammonification by Nautilia profundicola AmH: experimental evidence consistent with a free hydroxylamine intermediate. Front Microbiol. 2013;4:180. doi: 10.3389/fmicb.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatt JK, Löffler FE. Quantitative real-time PCR (qPCR) detection chemistries affect enumeration of the Dehalococcoides 16S rRNA gene in groundwater. J Microbiol Methods. 2012;88:263–270. doi: 10.1016/j.mimet.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Hengstmann U, Chin KJ, Janssen PH, Liesack W. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl Environ Microbiol. 1999;65:5050–5058. doi: 10.1128/aem.65.11.5050-5058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inwood SE, Tank JL, Bernot MJ. Factors controlling sediment denitrification in midwestern streams of varying land use. Microb Ecol. 2007;53:247–258. doi: 10.1007/s00248-006-9104-2. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Matsubara T. A nitrite reductase from Achromobacter cycloclastes. J Biochem. 1972;71:645–652. [PubMed] [Google Scholar]
- Jacobson F, Pistorius A, Farkas D, De Grip W, Hansson Ö, Sjölin L, et al. pH dependence of copper geometry, reduction potential, and nitrite affinity in nitrite reductase. J Biol Chem. 2007;282:6347–6355. doi: 10.1074/jbc.M605746200. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajie S, Anraku Y. Purification of a hexaheme cytochrome c552 from Escherichia coli K 12 and its properties as a nitrite reductase. Eur J Biochem. 1986;154:457–463. doi: 10.1111/j.1432-1033.1986.tb09419.x. [DOI] [PubMed] [Google Scholar]
- Kelso B, Smith RV, Laughlin RJ, Lennox SD. Dissimilatory nitrate reduction in anaerobic sediments leading to river nitrite accumulation. Appl Environ Microbiol. 1997;63:4679–4685. doi: 10.1128/aem.63.12.4679-4685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop-Jakobsen K, Giblin AE. The effect of increased nitrate loading on nitrate reduction via denitrification and DNRA in salt marsh sediments. Limnol Oceanogr. 2010;55:789–802. [Google Scholar]
- Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, et al. Nitrogen cycling. The environmental controls that govern the end product of bacterial nitrate respiration. Science. 2014;345:676–679. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- Laima MJC, Girard MF, Vouve F, et al. Distribution of adsorbed ammonium pools in two intertidal sedimentary structures, Marennes-Oléron Bay, France. Mar Ecol Prog Ser. 1999;182:29–35. [Google Scholar]
- Lam Y, Nicholas DJD. A nitrite reductase with cytochrome oxidase activity from Micrococcus denitrificans. Biochim Biophys Acta. 1969;180:459–472. doi: 10.1016/0005-2728(69)90025-5. [DOI] [PubMed] [Google Scholar]
- Lashof DA, Ahuja DR. Relative contributions of greenhouse gas emissions to global warming. Nature. 1990;344:529–531. [Google Scholar]
- Liu MC, Peck HD. The isolation of a hexaheme cytochrome from Desulfovibrio desulfuricans and its identification as a new type of nitrite reductase. J Biol Chem. 1981;256:13159–13164. [PubMed] [Google Scholar]
- Mania D, Heylen K, van Spanning RJM, Frostegård Å. The nitrate-ammonifying and nosZ carrying bacterium Bacillus vireti is a potent source and sink for nitric and nitrous oxides under high nitrate conditions. Environ Microbiol. 2014;16:3196–3210. doi: 10.1111/1462-2920.12478. [DOI] [PubMed] [Google Scholar]
- Nägele W, Conrad R. Influence of soil pH on the nitrate-reducing microbial populations and their potential to reduce nitrate to NO and N2O. FEMS Microbiol Ecol. 1990;7:49–57. [Google Scholar]
- Nijburg JW, Coolen M, Gerards S, Gunnewiek P, Laanbroek HJ. Effects of nitrate availability and the presence of Glyceria maxima on the composition and activity of the dissimilatory nitrate-reducing bacterial community. Appl Environ Microbiol. 1997;63:931–937. doi: 10.1128/aem.63.3.931-937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzoli D, Carraro E, Nigro V, Viaroli P. Effect of organic enrichment and thermal regime on denitrification and dissimilatory nitrate reduction to ammonium (DNRA) in hypolimnetic sediments of two lowland lakes. Water Res. 2010;44:2715–2724. doi: 10.1016/j.watres.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Nizzoli D, Welsh DT, Fano EA, Viaroli P. Impact of clam and mussel farming on benthic metabolism and nitrogen cycling, with emphasis on nitrate reduction pathways. Mar Ecol Prog Ser. 2006;315:151–165. [Google Scholar]
- Ogilvie BG, Rutter M, Nedwell DB. Selection by temperature of nitrate-reducing bacteria from estuarine sediments: species composition and competition for nitrate. FEMS Microbiol Ecol. 1997;23:11–22. [Google Scholar]
- Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326:123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- Rehr B, Klemme J-H. Competition for nitrate between denitrifying Pseudomonas stutzeri and nitrate ammonifying enterobacteria. FEMS Microbiol Lett. 1989;62:51–57. [Google Scholar]
- Richter CD, Allen JWA, Higham CW, Koppenhöfer A, Zajicek RS, Watmough NJ, et al. Cytochrome cd1, reductive activation and kinetic analysis of a multifunctional respiratory enzyme. J Biol Chem. 2002;277:3093–3100. doi: 10.1074/jbc.M108944200. [DOI] [PubMed] [Google Scholar]
- Ritalahti KM, Cruz-Garcia C, Padilla-Crespo E, Hatt JK, Löffler FE.2010RNA extraction and cDNA analysis for quantitative assessment of biomarker transcripts in groundwaterIn: Timmis KN, McGenity T, Meer JR, Lorenzo V, (eds)Handbook of Hydrocarbon and Lipid Microbiology Springer: Berlin; 3671–3685. [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sander R.1999. Compilation of Henry's law constants for inorganic and organic species of potential importance in environmental chemistry. www.henrys-law.org/henry.pdf .
- Sanford RA, Wagner DD, Wu Q, Chee-Sanford JC, Thomas SH, Cruz-García C, et al. Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proc Natl Acad Sci USA. 2012;109:19709–19714. doi: 10.1073/pnas.1211238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CS, Richardson DJ, Baggs EM. Constraining the conditions conducive to dissimilatory nitrate reduction to ammonium in temperate arable soils. Soil Biol Biochem. 2011;43:1607–1611. [Google Scholar]
- Schumpe A. The estimation of gas solubilities in salt solutions. Chem Eng Sci. 1993;48:153–158. [Google Scholar]
- Schumpe A, Quicker G, Deckwer W-D. Gas solubilities in microbial culture media. Adv Biochem Eng. 1982;24:1–38. [Google Scholar]
- Silver WL, Herman DJ, Firestone MK. Dissimilatory nitrate reduction to ammonium in upland tropical forest soils. Ecology. 2001;82:2410–2416. [Google Scholar]
- Silver WL, Thompson AW, Reich A, Ewel JJ, Firestone MK. Nitrogen cycling in tropical plantation forests: potential controls on nitrogen retention. Ecol Appl. 2005;15:1604–1614. [Google Scholar]
- Simon J, Klotz MG. Diversity and evolution of bioenergetic systems involved in microbial nitrogen compound transformations. Biochim Biophys Acta. 2013;1827:114–135. doi: 10.1016/j.bbabio.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Singh J. Cytochrome oxidase from Pseudomonas aeruginosa. III. Reduction of hydroxylamine. Biochim Biophys Acta. 1974;333:28–36. doi: 10.1016/0005-2728(74)90159-5. [DOI] [PubMed] [Google Scholar]
- Sørensen J. Capacity for denitrification and reduction of nitrate to ammonia in a coastal marine sediment. Appl Environ Microbiol. 1978;35:301–305. doi: 10.1128/aem.35.2.301-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens RJ, Laughlin RJ, Malone JP. Soil pH affects the processes reducing nitrate to nitrous oxide and di-nitrogen. Soil Biol Biochem. 1998;30:1119–1126. [Google Scholar]
- Strohm TO, Griffin B, Zumft WG, Schink B. Growth yields in bacterial denitrification and nitrate ammonification. Appl Environ Microbiol. 2007;73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templer PH, Silver WL, Pett-Ridge J, DeAngelis KM, Firestone MK. Plant and microbial controls on nitrogen retention and loss in a humid tropical forest. Ecology. 2008;89:3030–3040. doi: 10.1890/07-1631.1. [DOI] [PubMed] [Google Scholar]
- Tiedje JM, Sexstone AJ, Myrold DD, Robinson JA. Denitrification: ecological niches, competition and survival. Antonie van Leeuwenhoek. 1982;48:569–583. doi: 10.1007/BF00399542. [DOI] [PubMed] [Google Scholar]
- Tomaszek JA, Rokosz GÄR. Rates of dissimilatory nitrate reduction to ammonium in two Polish reservoirs: impacts of temperature, organic matter content, and nitrate concentration. Environ Technol. 2007;28:771–778. doi: 10.1080/09593332808618834. [DOI] [PubMed] [Google Scholar]
- Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin EA, Wolfe RS, Wolin MJ. Viologen dye inhibition of methane formation by Methanobacillus omelianskii. J Bacteriol. 1964;87:993–998. doi: 10.1128/jb.87.5.993-998.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Oyanagi H, Fujiwara T, Fukumori Y. Nitrite reductase from the magnetotactic bacterium Magnetospirillum magnetotacticum: a novel cytochrome cd1 with Fe(II):nitrite oxidoreductase activity. Eur J Biochem. 1995;233:665–671. doi: 10.1111/j.1432-1033.1995.665_2.x. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Sanford RA, Löffler FE. Shewanella spp. use acetate as an electron donor for denitrification but not ferric iron or fumarate reduction. Appl Environ Microbiol. 2013;79:2818–2822. doi: 10.1128/AEM.03872-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari T, Hynes R, Knowles R. Acetylene inhibition of nitrous oxide reduction and measurement of denitrification and nitrogen fixation in soil. Soil Biol Biochem. 1977;9:177–183. [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.