Abstract
We report observations on the dynamics of bacterial communities in response to methane stimulus in laboratory microcosm incubations prepared with lake sediment samples. We first measured taxonomic compositions of long-term enrichment cultures and determined that, although dominated by Methylococcaceae types, these cultures also contained accompanying types belonging to a limited number of bacterial taxa, methylotrophs and non-methylotrophs. We then followed the short-term community dynamics, in two oxygen tension regimens (150 μM and 15 μM), observing rapid loss of species diversity. In all microcosms, a single type of Methylobacter represented the major methane-oxidizing partner. The accompanying members of the communities revealed different trajectories in response to different oxygen tensions, with Methylotenera species being the early responders to methane stimulus under both conditions. The communities in both conditions were convergent in terms of their assemblage, suggesting selection for specific taxa. Our results support prior observations from metagenomics on distribution of carbon from methane among diverse bacterial populations and further suggest that communities are likely responsible for methane cycling, rather than a single type of microbe.
Introduction
Methane is recognized as one of the major contributors to the greenhouse effect, with its atmospheric concentration steadily increasing (Singh et al., 2010; Nisbet et al., 2014). Freshwater lakes are environments characterized by dynamic cycling of methane (Borrel et al., 2011). The main functional guild of microbes involved in methane consumption in these environments is the aerobic methanotrophic bacteria. Classically, these are divided into two major groups, Type I (Gammaproteobacteria) and Type II (Alphaproteobacteria) (Trotsenko and Murrell, 2008; Chistoserdova and Lidstrom, 2013). Recently, methanotrophic bacteria have been described belonging to the phylum Verrucomicrobia (Op den Camp et al., 2009) and candidate phylum NC10 (Ettwig et al., 2010). Methane can also be oxidized by specific lineages of archaea (Haroon et al., 2013; Offre et al., 2013).
In Lake Washington sediment, oxidation of methane takes place within the top 0.7 cm–0.8 cm layer characterized by steep counter-gradients of methane and oxygen (Lidstrom and Somers, 1984; Kuivila et al., 1988; Auman et al., 2000). Methanotrophs previously cultivated from Lake Washington sediment represented species belonging to the genera of Methylomonas and Methylosarcina (Type I) and Methylocystis and Methylosinus (Type II; Auman et al., 2000). However, metagenomic sequencing targeting active methanotrophs uncovered a dominant response by uncultivated Methylobacter species (Kalyuzhnaya et al., 2008; Chistoserdova, 2011a; Beck et al., 2013). Another intriguing discovery from functional metagenomics was the positive correlation between the populations of Methylobacter and Methylotenera in methane-fed microcosms (Beck et al., 2013). Methylotenera species are unable of methane oxidation (Chistoserdova and Lidstrom, 2013). Thus, the presence of their signatures in the labeled DNA suggested cooperative behavior through which Methylotenera can obtain methane-derived carbon (Chistoserdova et al., 2013). Under microoxic conditions, label from methane was distributed among a broad range of bacterial taxa (Beck et al., 2013). A hypothesis potentially explaining this pattern has recently been proposed, suggesting an alternative mode of methanotrophy that involves fermentation (Kalyuzhnaya et al., 2013). Under this scenario, a small population of methanotrophic bacteria can provide carbon for a variety of microbes.
In this study we focused on the temporal variability of co-occurrence patterns for methanotrophic and non-methanotropic bacteria, in order to test a hypothesis of interspecies cooperation in methane oxidation.
Materials and methods
Sample collection and experimental setup
Samples of Lake Washington sediment were collected on 27 July 2011, 15 August 2011 and 15 July 2013, as previously described (Kalyuzhnaya et al., 2004). For long-term enrichment incubations, samples collected in 2011 were used as follows. 250 ml vials were filled with either 15 ml or 25 ml of filtered lake water and inoculated with either 15 ml or 75 ml of sediment sludge, respectively. These liquid/headspace ratios resulted in oxygen concentrations of 330 μM and 225 μM, respectively. Samples were supplemented with either 200 μM or 400 μM of CH4 and incubated at 10 °C with shaking (220 r.p.m.). Additional enrichment cultures were supplemented with nitrate minimal salts medium (5% v/v; Ojala et al., 2011). Methane consumption was monitored as described below. Enrichments with active methane consumption were serially transferred into nitrate minimal salts medium (0.3 × strength) diluting 1:50, 20 ml of methane added to the headspace weekly.
For short-term community dynamics experiments, seven independent top layer sediment samples collected in 2013 were mixed and the mixture used to set up replicate microcosms. Each microcosm contained 25 ml of the sediment and 25 ml of Lake Washington water and supplemented with 0.5 mM NaNO3. Samples were placed into 250 ml vials sealed with rubber stoppers. Six microcosms were incubated under ‘high' O2 condition (HO; dissolved oxygen concentration of 150 μM). An additional six microcosms were incubated ‘low' O2 condition (LO; dissolved oxygen concentration of 15 μM). To create HO and LO conditions, the vials were flushed with N2 for 2 min, after which N2 pressure was allowed to equalize. For HO, 150 ml of headspace were pulled out from the vial using a syringe and 50 ml of CH4 and 100 ml of air were injected into the headspace. For LO, 60 ml of headspace were pulled out and 50 ml of CH4 and 10 ml of air were injected. Vials were incubated with shaking (250 r.p.m.) at 15 °C. Cultures were transferred weekly, diluting 10-fold, into nitrate minimal salts medium (0.5 × strength) modified by lowering its buffering strength to 20%. HO and LO conditions were recreated, respectively, weekly and daily. After 1 week of incubation, optical density of cultures (OD600) was typically 0.4 in HO microcosms and 0.3 in LO microcosms, with a few exceptions (see Results). These microcosms were incubated for additional 2 weeks. Biomass was collected by centrifugation at 5000 r.p.m. for 12 min. Pellets were used immediately or stored at −20 °C.
Methane and oxygen detection
Methane and oxygen concentrations were measured for selected samples using a gas chromatograph GC2014 (Shimadzu Instruments, Portland, OR, USA) using the FID and the TCD detectors, with detection limits of 0.1 p.p.m. and 100 p.p.m., respectively. In this setup, samples pass through a 3 m hayesepQ column and through a 3 m molecular sieve 5A column carried by argon, both columns maintained at 90 °C. Authentic Shimadzu syringes were used for injection, with sample volumes (10 ml) exceeding the volume of the column by fivefold, to eliminate ambient air contamination, as instructed by the manufacturer.
16S rRNA gene amplicon sequencing
DNA was isolated using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) and submitted to MR DNA service facility (http://www.mrdnalab.com/). 16S rRNA gene amplicon pyrosequencing was carried out as previously described (Dowd et al., 2008). Briefly, 16S rRNA gene universal primers 27F and 519r were utilized in a single-step PCR using HotStarTaq Plus Master Mix Kit (Qiagen, Valencia, CA, USA). The reaction conditions were: 3 min at 94 °C, followed by 28 cycles of 94 °C for 30 s, 53 °C for 40 s and 72 °C for 60 s, followed by the final step of 5 min at 72 °C. The amplicon products from different samples were mixed in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, Beverly, MA, USA). The sequencing was carried out on a Roche (San Francisco, CA, USA) 454 FLX Titanium Instrument using the manufacturer's reagents and following the manufacturer's instructions. Sanger sequencing of 16S rRNA gene PCR fragment libraries was carried out as previously described (Kalyuzhnaya et al., 2009). The data have been archived with the NCBI (Bioproject 253595, http://www.ncbi.nlm.nih.gov/bioproject/253595).
Bioinformatics
The UPARSE method was used for sequence processing and operational taxonomic unit (OTU) clustering with USEARCH version 7.0.1001 (Edgar, 2013). Clustering was performed at 95% and chimeras were identified against the ChimeraSlayer reference database in the Broad Microbiome Utilities version r20110519 obtained from the UCHIME distribution (Edgar et al., 2011). Taxonomic assignments were made using the RDP Classifier from the Ribosomal Database Project downloaded on 22 October 2013 (Wang et al., 2007). The samples were scaled so that the number of reads in each sample were equal. Hierarchical clustering of samples and OTUs was performed using the percentage of reads per OTU for the most abundant taxa, that is, >2.5% population in at least one sample. Bray–Curtis distances, Shannon indices, ordination plots and multivariate analyses were calculated using the vegan library version 2.0-10 (Oksanen et al., 2013) in R version 3.0.2 (http://www.R-project.org/).
Illumina MiSeq sequencing
DNA libraries were prepared from 1 ng of purified DNA using Nextera XT DNA Sample Prep Kit with Nextera XT Index Kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer's protocol (version October 2012). DNA libraries were normalized to 2 nM, pooled in equal volumes, denatured with 0.2 N NaOH and then diluted with Hybridization buffer according to the Nextera XT protocol (version March 2013; Illumina, Inc.). Libraries at 6 pM final concentration were sequenced using MiSeq Reagent Kit v3 (150 cycles) on MiSeq Sequencer (Illumina Inc.) running MiSeq Control Software version 2.3.0.3. Nonindexed PhiX library (PhiX Control v3, Illumina) was used at 5% for performance control. Illumina SAV (Sequencing Analysis Viewer) showed cluster density of 751 K mm−2 with 94.5% cluster passing filter, 95.3% ⩾Q30 at read 4, 3.8 Gb total yield. The data have been archived with the NCBI (Bioproject 253699; http://www.ncbi.nlm.nih.gov/bioproject/253699).
Alignment of MiSeq reads to genomic scaffolds
The paired end MiSeq reads were aligned to the genomes of Methylobacter tundripaludum 31/32, Methylophilus methylotrophus 1, Methylotenera mobilis 13 and Methylotenera versatilis 301 obtained from the IMG database (Markowitz et al., 2014). Alignments were computed with BWA version 0.7.5a-r405 (Li and Durbin, 2010) allowing for up to eight mismatches in each 75-bp read with seeding disabled (options -n 8 –l 100000). The results were postprocessed, including single nucleotide polymorphism calling, with SAMtools version 0.1.19-44428cd (Li et al., 2009). Visualization of the alignments for manual inspection was performed with Integrative Genomics Viewer version 2.3.34 (Thorvaldsdóttir et al., 2013).
Results
Long-term microcosm enrichments with methane select for Methylobacter and for specific accompanying bacterial types
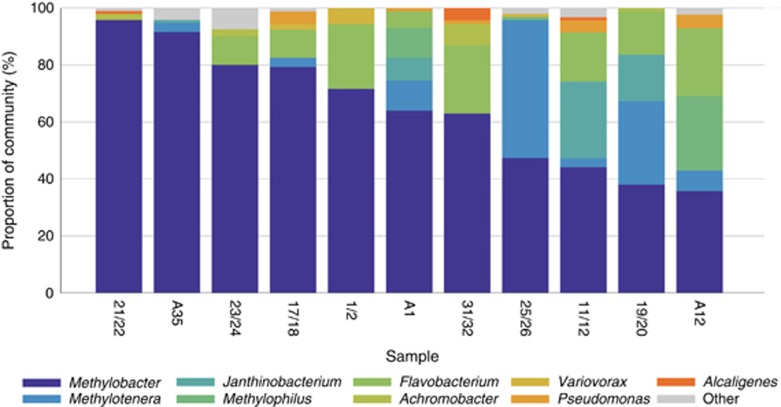
We established a series of microcosms at 10 °C, as described in Materials and methods, and followed methane consumption over time. Of the microcosms that showed active methane consumption, 11 were randomly chosen for 16S rRNA gene profiling. This analysis indicated low species diversity in all communities. Each was dominated by a single Methylococcaceae species, closely related to Methylobacter tundripaludum (99.7% sequence identity; Svenning et al., 2011). Other species differed somewhat from microcosm to microcosm. These belonged to a total of 18 taxa, at the genus level (Figure 1 and Supplementary Table 1). Some taxa were more persistent among the microcosms analyzed, and these were also present at higher relative abundances. Seven microcosms contained Methylotenera species, constituting up to 48% of total sequences; eight microcosms contained Flavobacterium (up to 24% of total sequences); five contained Pseudomonas, albeit at low relative abundance. Four microcosms contained Janthinobacterium, and in two of them they were present at high relative abundances (up to 27% of all sequences);four contained Achromobacter (up to 7.6% of all sequences). Methylophilus sequences were detected in only three microcosms, but in two of them they were the most prominent accompanying species (10% and 26% of all sequences, respectively). The remaining satellite types were found at lesser relative abundances. Overall, the narrow and overlapping range of the satellite types suggested that these types were nonrandom, and likely they were selected under specific environmental pressures.
Figure 1.
Community composition of long-term methane-fed microcosms. The samples were ordered from the highest to the lowest content of Methylobacter OTUs. Sample named starting with ‘A' originated from August 2011 enrichments, and the rest of the samples originated from June 2011 enrichments. Other, taxa found in fewer than three samples (Beggiatoa, Methylovorus, Acidovorax, Caulobacter, Polaromonas, Comamonadaceae, Allicyclobacillus, Bosea, Denitratisoma and Rhodobacter). See Supplementary Table 1 for details.
Short-term community dynamics support nonrandom occurrence of the methanotroph-accompanying types
Short-term community dynamics were followed under two oxygen tension regimens, HO and LO. Under HO, microcosms were exposed to relatively high initial oxygen tension (150 μM, corresponding to concentration at ∼1 mm depth in native sediment; Auman et al., 2000), decreasing weekly to under detection limit. Under LO, microcosms were microoxic at all times, with the initial oxygen tension of 15 μM (corresponding to concentration at ∼5 mm of native sediment), typically decreasing to below detection within 24 h (Supplementary Figure 1). These conditions were chosen to mimic the situation in situ, where methane-oxidizing microbes are limited by oxygen (Kuivila et al., 1988).
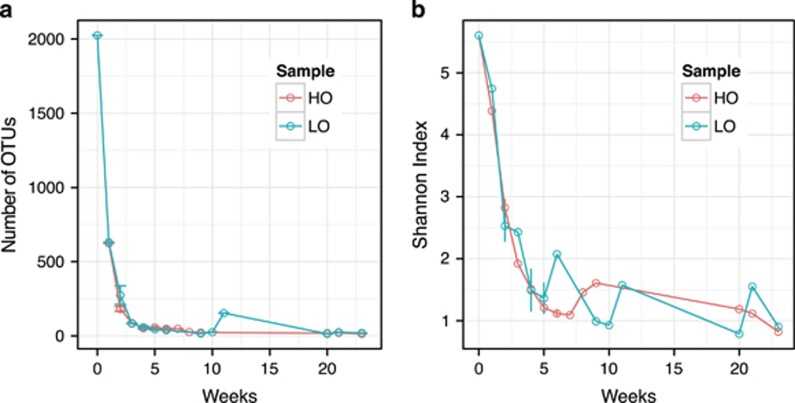
The composition of the communities was monitored by pyrosequencing, for a total of 23 weeks. The community originating directly from the sediment was used as a zero point benchmark. Sample key, total DNA concentrations and sequencing statistics are shown in Supplementary Table 2. As expected (Kalyuzhnaya et al., 2008; Beck et al., 2013), high phylogenetic complexity was observed for zero point samples, of up to 2025 operational taxonomic units (OTU; 95% DNA sequence identity cutoff; Supplementary Table 3). In the microcosms, the community complexity rapidly decreased with time. After 1 week, the number of OTUs reduced by ∼threefold under both HO and LO, and by week 2 the number of OTUs further reduced by another fivefold, followed by further reductions (Figure 2).
Figure 2.
Community complexity as a function of time. A total number of OTUs at 95% cutoff (a) and a diversity index (Shannon; b) are shown for each sample. Errors are shown for replicate samples (weeks 2, 4 and 6 for HO and weeks 2, 4 and 5 for LO).
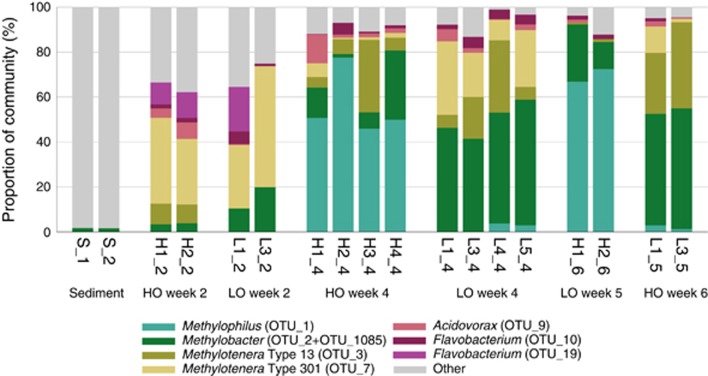
Weekly analysis of community composition was carried out for one HO and one LO microcosm over the period of complexity reduction (9 weeks for HO and 11 weeks for LO, with a 2-week delay between two of the samples in the latter case due to low optical density). Two replicates from each condition were sampled at three time points, and four replicates were sampled at one point, in order to estimate the degree of variance in OTU composition/relative abundance (Figure 3). From these analyses, it was apparent that organisms persisting in the microcosms were not random, the community composition being skewed toward specific bacterial types. The major persisting type was Methylobacter (OTU_2; Supplementary Table 3, Supplementary Figures 2, 3) previously observed in the long-term enrichments (99.7% 16S rRNA gene identity), and this type increased in relative abundance with time. Under HO, its relative abundance increased to reach up to 30% of total sequences in 4 weeks, and under the LO, its relative abundance increased to reach up to 55% of the total sequences (Figure 3; Supplementary Table 3). Another OTU was present across samples classified as Methylobacter (OTU_1085). Alignment of this sequence with the sequence of OTU_2 revealed two insertions of six nucleotides in OTU_1085 (Supplementary Figure 4). Comparisons with the public databases revealed that sequences aligning with OTU_1085 without gaps were all sequences amplified from environmental samples, none belonged to cultivated microbes. From this we concluded that OTU_1085 must be a product of PCR amplification error. Thus, in our analyses we bundled OTU_2 and OTU_1805 together. OTUs representing other bona fide methanotrophs (Methylosarcina, Methylomonas, Methylosoma and Methylocystis) were found at low relative abundances, and these OTUs did not reveal dynamic response to methane (Supplementary Table 3).
Figure 3.
Community composition of replicate samples. Major (most abundant and most persistent) OTUs were considered in this analysis. ‘Other' indicates a sum of minor (less abundant/less persistent) OTUs. For details, see Supplementary Table 3.
Two other OTUs persistently present in the microcosms, OTU_7 and OTU_3, represented Methylotenera, members of Methylophilaceae, non-methane-utilizing methylotrophs. After 1 week, the population of Methylotenera increased ∼200-fold in both conditions compared with the original community (0.1% of total sequences), demonstrating a more dramatic response to methane compared with Methylobacter. The proportion of the Methylotenera sequences continued to increase, and by week 2 these constituted up to 50% of total reads in some of the microcosms (Figure 3 and Supplementary Table 3). By week 4, however, the population of Methylotenera under HO decreased, being replaced by Methylophilus (OTU_1; up to 77.7% of total sequences), whereas under LO the Methylotenera types persisted (Figures 4b and c; Supplementary Table 3). The trends for appearance and for persistence of these types under HO and in LO were well replicated (Figure 4a). However, their specific ratios fluctuated, reflecting individual community trajectories (Figure 4a; Supplementary Table 3). To further test reproducibility of occurrence of specific major taxa, we set up an independent LO experiment, starting with a frozen sediment sample, diluting the sample ∼10-fold compared with the samples of the fresh sediment. This microcosm was probed at 3, 7 and 9 weeks, revealing the presence of two major OTUs, OTU_2 (Methylobacter) and OTU_7 (Methylotenera), the latter gradually replaced by OTU_3 (Methylotenera), that is, following the trajectory observed with LO communities inoculated by the fresh sediment. (Supplementary Figure 5, Supplementary Tables 2, 3).
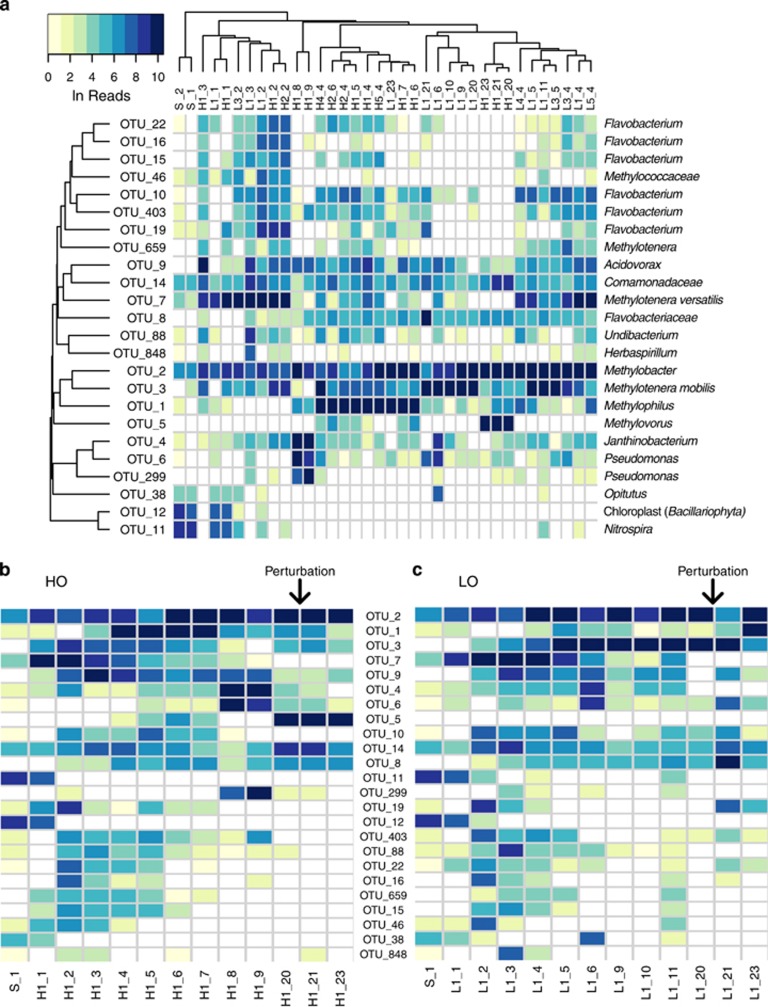
Figure 4.
Heat map of major OTU relative abundances across samples. Abundances were measured as Ln of reads. Names of samples indicate the condition (S, untreated sediment; H, ‘high' oxygen; L, ‘low' oxygen; the first numeral indicates the number of the replicate, and the second indicates the number of weeks of incubation. Sample details are shown in Supplementary Table 2. Note that samples taken on weeks 21 and 23, while named based on the original condition, are postperturbation samples, thus representing an alternative incubation condition. (a) Hierarchical clustering (average linkage) of samples and OTUs based on Bray–Curtis dissimilarity index of relative abundance profiles. (b) ‘High' oxygen (HO) condition samples arranged chronologically, with OTUs sorted by decreasing sum of read counts across all samples. (c) ‘Low' oxygen (LO) condition samples arranged chronologically, with OTUs sorted as in (b). For (b) and (c), the microcosm perturbation (oxygen condition switch) performed at week 20 is denoted by the arrow.
Community trajectories and stability appear to be dependent on environmental factors
Overall, 52 OTUs were found most persistent across the microcosms, accounting for OTUs present at over 1% in more than one sample. The number of persistent OTUs was further reduced to 24 by considering only those constituting over 2.5% of total sequences in at least one sample. This demonstrates that communities under HO and LO conditions were convergent in terms of species composition (Figure 4a; Supplementary Figures 2, 3). Among these 24 OTUs, 3 (OTU_11, OTU_12 and OTU_38) were only relatively abundant in nontreated and early treatment samples. The remaining 21 OTUs were classed among a very limited number of taxa. Two represented Methylococcaceae, five represented Methylophilaceae, seven represented Flavobacteriaceae, five represented Burkholderiales and two represented Pseudomonas.
Although only a few OTUs represented the highly relatively abundant species forming the dynamically changing microbial consortia, we noted differential trajectories for the communities placed under two different conditions. Under HO, while Methylobacter remained the only methane-oxidizing organism, we observed successions of accompanying taxa, early in the enrichment represented by two different ecotypes of Methylotenera (OTU_7 and OTU_3), followed by Acidovorax (OTU_9), succeeded by Methylophilus (OTU_1), followed by Janthinobacterium and Pseudomonas (OTU_4, OTU_6 and OTU_299) and finally by Methylovorus (OTU_5; Figure 4). At week 20, we perturbed the community by switching it to an alternative oxygen regimen (HO to LO) and measured community composition at 1 and 3 weeks after the disturbance, noting no significant changes in the community (Figure 4b, Supplementary Table 3).
The LO community trajectory was notably different (P-value of 0.01 by analysis of variance). Although the same Methylobacter (OTU_2) represented the only methanotroph type, the dynamics of accompanying types mostly involved Methylotenera, with OTU_7 being gradually replaced by OTU_3. The community was perturbed at week 20, switching from LO to HO. The community composition measured 1 week later showed a significant change compared to predisturbance, with the Methylobacter (OTU_2) relative abundance dropping and with the population of Flavobacterium (OTU_8, OTU_19) increasing (Figure 4c). By week 23, the community was transformed to resemble the original HO community during weeks 4–7 stage, with the dominant presence of Methylophilus (OTU_1; Figure 4a). This result suggests that a shift in a single environmental parameter has the potential to significantly change the assemblage of microbes involved in methane metabolism, further suggesting that the rare biosphere that is part of this community has the potential to play a role in these dynamic changes. The trends in community dynamics in response to the switch in oxygen tension were highly reproducible (Supplementary Figure 6). Analysis of variance using the canonical correspondence analysis model (oxygen+week+replicate) yielded a P-value of 0.77 for the replicate term, whereas oxygen and week were significant at 0.01. Partialling out the oxygen and week terms strengthened the significance (P-value⩾0.99).
Metagenomic analysis identifies cultivated species proxies and provides insights into metabolic capabilities
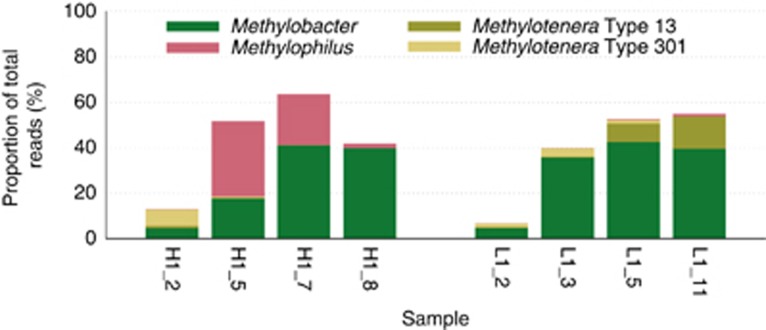
In order to obtain a better understanding of the physiology of the major types involved in community dynamics, we analyzed DNA samples from eight microcosms (four HO and four LO) by Illumina MiSeq-based metagenomic sequencing. The reads were aligned along the genomes of Methylobacter, various ecotypes of Methylophilaceae and the genomes of the closest relatives of the non-methylotrophic heterotrophs (Supplementary Table 4). The following genomes were identified that matched significantly with the metagenomic sequences: Methylobacter tundripaludum (100% 16S rRNA gene identity), Methylophilus methylotrophus strain 1 (100% 16S rRNA gene identity), Methylotenera versatilis strain 301 (type 301; 97% 16S rRNA gene identity) and Methylotenera mobilis strain 13 (type 13; 99% 16S rRNA gene identity; Figure 5). On the basis of read abundance and the pyrosequencing data, these proxy genomes represented, respectively, OTU_2, OTU_1, OTU_7 and OTU_3.
Figure 5.
Proportion of metagenomic reads matched to four proxy genomes, Methylobacter tundripaludum, Methylophilus methylotrophus, Methylotenera mobilis (Type 13) and Methylotenera versatilis (Type 301).
Aligning the metagenomic sequence reads along the proxy genomes allowed us to observe the differences between each genome pair and to evaluate conservation of the important metabolic modules. Alignments with the M. tundripaludum genomes were possible for each of the eight samples, with the highest mean coverage for samples H1_7 and L1_5 (over 40% of the total reads mapped at 90% identity). The alignments produced multiple gaps. The gaps contained a variety of apparently nonessential functions such as DNA integration and recombination. All of the important functions, including functions essential in methanotrophy previously reconstructed for the Lake Washington Methylobacter population (Kalyuzhnaya et al., 2008) were identified. However, comparisons of sample-specific data sets in terms of alignment to the M. tundripaludum genome as well as in terms of single nucleotide polymorphisms uncovered the presence of distinct populations of Methylobacter. Between the eight samples, up to six distinct populations of Methylobacter were identifiable (Supplementary Figure 7 and Supplementary Table 5). Notably, one of the populations was either devoid of the pxmAB genes that are homologs of pmoAB genes (Tavormina et al., 2011) or possessed pxmAB diverging more than 10% (Supplementary Figure 7a). Of the pmo genes, pmoAB were highly conserved among the populations, while pmoC exhibited a number of single nucleotide polymorphisms allowing for distinguishing among different populations, in combination with other select genomic regions (Supplementary Figure 7b; Supplementary Table 5).
The genome of OTU_1 aligned well with the genome of M. methylotrophus strain 1, representing one of the previously defined ecotypes of M. methylotrophus (Beck et al., 2014). As expected, it was present at high coverage in samples H1_5 and H1_7, at low coverage in samples H1_8 and L1_11 and practically absent from the remaining samples. Alignments showed a number of gaps that mostly encoded apparently nonessential functions (DNA recombination, transport and unknown). For example, a gene for the single (and likely not functional) subunit of nitric oxide reductase previously identified in the genome of M. methylotrophus (Beck et al., 2014) was absent from the OTU_1 genome.
The genome of M. versatilis strain 301 (type 301; Beck et al., 2014) was the least similar proxy genome, and likely many reads were not aligned at the stringency utilized. However, alignments with samples H1_2 and L1_3 produced sufficient coverage to evaluate the contents of the OTU_7 genome. Remarkably, one gap in the alignment contained a pair of genes considered key in assimilatory metabolism of Methylophylaceae, edd and eda, encoding, respectively, 6-phosphogluconate dehydratase and 2-keto 3-deoxy 6-phosphogluconate aldolase and enabling the EDD/EDA variant of the ribulose monophosphate pathway (Anthony, 1982; Chistoserdova, 2011b). However, this genome appeared to encode a pair of enzymes that enable an alternative variant of the ribulose monophosphate pathway, phosphofructokinase and fructose bisphosphate aldolase (Anthony, 1982). Thus, OTU_7 represents the first example of a Methylophilaceae microbe utilizing this metabolic arrangement. Other omissions from the OTU_7 genome were predicted to be nonessential.
Alignments with the genome of M. mobilis strain 13 (type 13; Beck et al., 2014), most highly covered in sample L1_11, revealed similar trends of showing gaps encoding apparently nonessential functions, whereas all the essential functions were conserved. Interestingly, one function that distinguished the genome of M. mobilis 13 from all other characterized Methylophilaceae genomes, respiratory denitrification complete with the nitrous oxide reduction step, was missing from the closely-related genome of OTU_3, as no sequence reads matching the respective genomic region were detected in sample L1_11 highly enriched in this type. However, the respective genomic locus was covered in samples H1_2, L1_2, L1_3 and L1_5. One explanation for this phenomenon is that the island missing from the type 13-like genome is present in the type 301-like genome, OTU_7. The island in question has been previously identified as a result of lateral transfer (Beck et al., 2014). From these observations, the two Methylotenera types that appear in succession in the evolving microcosms are physiologically different. The type 301-like organism can potentially denitrify, likely does not possess an MxaFI methanol dehydrogenase (Beck et al., 2014) and utilizes a more efficient variant of the ribulose monophosphate pathway, whereas the type 13-like organism is not capable of respiratory denitrification and possesses MxaFI type methanol dehydrogenase. In terms of methylotrophy and nitrate metabolism potentials, the phenotype of the Methylophilus OTU_1 is predicted to resemble the phenotype of the type 13-like Methylotenera rather than that of the type 301-like Methylotenera (Table 1). It is interesting to note that during microcosm progression, OTU_1 under HO acted as a counterpart of OTU_3 under LO. No good proxies were found among the publically available genomes for Flavobacterium, Pseudomonas, Acidovorax or Burkholderiales.
Table 1. Major physiological traits of the Methylophilaceae OTUs as predicted from metagenomic sequencing.
| Function/OTU | MxaFI-MDH | XoxF-MDH | Respiratory denitrification | Assimilatory denitrification | EDD/EDA RuMP | PFK/FBA RuMP |
|---|---|---|---|---|---|---|
| OTU_1 | + | + | − | + | + | − |
| OTU_3 | + | + | − | + | + | − |
| OTU_7 | − | + | + | + | − | + |
Abbreviations: EDD/EDA, Entner–Doudoroff; MDH, methanol dehydrogenase; RuMP, ribulose monophosphate; PFK/FBA, glycolytic variants of ribulose monophosphate cycle.
Discussion
In this work, we followed up on our prior observations from metagenomics, regarding the strong correlation between the populations of bona fide methane-oxidizing bacteria with those of non-methanotrophic bacteria, primarily representatives of Methylophilaceae (Kalyuzhnaya et al., 2008; Beck et al., 2013). The broader goal of this study was to test for other associations that methanotrophs might form. A top–down approach was implemented, in which a complex natural community was manipulated under selective pressure of methane as the only source of carbon and energy, with a single variable being amount of oxygen provided. Oxygen concentrations are referred to as ‘high' and ‘low' for convenience. Oxygen was a limiting factor in both conditions, at least at times. Whereas the LO communities were always limited by oxygen, the HO communities were exposed to oxygen limitation only temporarily.
Both conditions selected for a single methanotroph type, a close relative of M. tundripaludum, whereas other known methanotrophs became rapidly diluted. Remarkably, the most significant initial response to methane was revealed by the Methylotenera species, with type 301-like organism being the early responder and type 13-like organism being a later responder. After ∼4 weeks, the community trajectories diverged between the two conditions. Although the HO communities progressed through a series of population sweeps along with the oscillating population of Methylobacter, the LO communities remained dominated, in addition to Methylobacter, by the two different ecotypes of Methylotenera, in succession. However, sweeps in the populations of Methylobacter also occurred, based on metagenomic sequencing of select samples. Although the condition perturbation as applied to the simplified communities (switch in oxygen tension) had no effect on the original HO communities, it caused a notable effect on the original LO communities.
Overall, although the two conditions employed caused different community trajectories and affected community stability in a different fashion, in both conditions, we observed remarkably rapid simplification of the original community in response to methane, with only a handful of major OTUs being involved in the dynamic response. These results suggest that whereas methane oxidation may rely on more than one type of microbe, it must be contained to a closely-knit, simple community of cooperating functional types. This study supports prior observations from metagenomics suggesting that Methylococcaceae and Methylophilaceae may participate in cooperative behavior that involves carbon (and potentially other metabolite) sharing (Beck et al., 2013). Other taxa prominently present in the communities were Flavobacteriaceae, Burkholderiales and Pseudomonas. These types were also found in long-term enrichments, suggesting that associations of methanotrophs with non-methylotrophs are stable over time.
Methanotrophs have been previously implicated as key players in driving microbial food webs in environments rich in methane (Hutchens et al., 2004; Murase and Frenzel, 2007; Jensen et al., 2008), and correlation between the populations of methanotrophs and non-methanotrophs has been noted in both natural and manipulated environments (Jensen et al., 2008; Redmond et al., 2010; He et al., 2012; Beck et al., 2013; Dubinsky et al., 2013; Rivers et al., 2013; van der Ha et al., 2013). Moreover, direct carbon transfer among organisms was determined via stable isotope probing (Hutchens et al., 2004; Redmond et al., 2010; He et al., 2012; Beck et al., 2013). In freshwater environments the major associated groups are Methylococcaceae, Methylophilaceae and Flavobacteriaceae, whereas in marine environments these are Methylococcaceae, Priscirickettsiaceae (Methylophaga), Methylophilaceae and Flavobacteriaceae (references above).
As oxygen tension was the only variable between the two types of microcosm incubations we implemented, likely, this variable was responsible for the alternative choices in community memberships and community trajectories. It appears that Methylophilus and Methylovorus are more competitive under high oxygen, in which they outcompete Methylotenera, whereas Methylotenera species are more competitive under low oxygen. The concept of micro-niche adaptation (Reim et al., 2012) would provide a good explanation for how efficient methane oxidation could take place over steep counter-gradients of methane and oxygen characterizing lake sediments.
Although carbon sharing between methanotrophs and non-methanotrophs has been mostly interpreted as cross-feeding, the partnerships may be more intimate. The cross-feeding scenario involving either methanol/formaldehyde or organic acids would not explain selection for the specific groups of methylotrophic and non-methylotrophic heterotrophs, respectively. For example, a variety of methylotroph taxa are present in Lake Washington sediment (Chistoserdova et al., 2013), but only Methylophilaceae respond strongly to the methane stimulus. Likewise, a wide variety of non-methylotrophic heterotrophs are present (Beck et al., 2013), but only a handful of OTUs, especially Flavobacterium, are detected in the microcosms described here. Importantly, the results of the experiments described here pinpoint the prominent models and form a foundation for the future bottom-up approaches in which pure cultures may be used to establish fully controlled synthetic communities.
We suggest that it is necessary to start addressing the factors involved in the observed co-occurrence patterns, in order to understand the mechanisms selecting for specific taxa, as well as the very nature of these relationships. Hence, the dynamics among these partnering groups, rather than the size of methanotroph populations, may be more informative in evaluating and predicting response to local increase in methane concentration. This knowledge will be important for better understanding the specific roles of microbes in methane emission mitigation, via a better detection of the methane-oxidizing potential and better informed modeling.
Acknowledgments
This work was funded by the grants from the National Science foundation (MCB-0950183) and from the Department of Energy (DE-SC0005154 and DE-SC0010556) and facilitated through the use of advanced computational storage and networking infrastructure provided by the Hyak supercomputer system supported in part by the University of Washington eScience Institute. Alexey Gilman is acknowledged for help with gas chromatograph operation, and Dr. Sascha Krause is acknowledged for critical reading of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anthony C. The Biochemistry of Methylotrophs. Academic Press: London; 1982. [Google Scholar]
- Auman AJ, Stolyar S, Costello AM, Lidstrom ME. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl Environ Microbiol. 2000;66:5259–5266. doi: 10.1128/aem.66.12.5259-5266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, Kalyuzhnaya MG, Malfatti S, Tringe SG, Glavina del Rio T, Ivanova N, et al. A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ. 2013;1:e23. doi: 10.7717/peerj.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DAC, McTaggart TL, Setboonsarng U, Vorobev A, Kalyuzhnaya MG, Ivanova N, et al. The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLoS One. 2014;9:e102458. doi: 10.1371/journal.pone.0102458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Jézéquel D, Biderre-Petit C, Morel-Desrosiers N, Morel JP, Peyret P, et al. Production and consumption of methane in freshwater lake ecosystems. Res Microbiol. 2011;162:832–847. doi: 10.1016/j.resmic.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L. Methylotrophy in a lake: from metagenomics to single organism physiology. Appl Environ Microbiol. 2011;77:4705–4711. doi: 10.1128/AEM.00314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L, Lidstrom ME.2013Aerobic methylotrophic prokaryotesIn: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E, (eds), The Prokaryotes4th ednSpringer: Berlin, Hiedelberg; 267–285. [Google Scholar]
- Chistoserdova L, Kalyuzhnaya MG, Lidstrom M. Cycling single-carbon compounds: from omics to novel concepts. Microbe. 2013;8:395–400. [Google Scholar]
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubinsky EA, Conrad ME, Chakraborty R, Bill M, Borglin SE, Hollibaugh JT, et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the gulf of Mexico. Environ Sci Technol. 2013;47:10860–10867. doi: 10.1021/es401676y. [DOI] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, et al. 2013Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage Nature 500567–570.Erratum in: Nature 2013 501: 578. [DOI] [PubMed] [Google Scholar]
- He R, Wooller MJ, Pohlman JW, Catranis C, Quensen J, Tiedje JM, et al. Identification of functionally active aerobic methanotrophs in sediments from an arctic lake using stable isotope probing. Environ Microbiol. 2012;14:1403–1419. doi: 10.1111/j.1462-2920.2012.02725.x. [DOI] [PubMed] [Google Scholar]
- Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol. 2004;6:111–120. doi: 10.1046/j.1462-2920.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- Jensen S, Neufeld JD, Birkeland NK, Hovland M, Murrell JC. High diversity of microplancton surrounds deep-water coral reef sediment off the coast of Norway. FEMS Microbiol Ecol. 2008;66:320–330. doi: 10.1111/j.1574-6941.2008.00575.x. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lidstrom ME, Chistoserdova L. Utility of environmental probes targeting ancient enzymes: methylotroph detection in Lake Washington. Microb Ecol. 2004;48:436–472. doi: 10.1007/s00248-004-0212-6. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, et al. High resolution metagenomics targets major functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Martens-Habbena W, Wang T, Hackett M, Stolyar SM, Stahl DA, et al. Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ Microbiol Rep. 2009;1:385–392. doi: 10.1111/j.1758-2229.2009.00046.x. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Yang S, Rozova ON, Smalley NE, Clubb J, Lamb A, et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun. 2013;4:2785. doi: 10.1038/ncomms3785. [DOI] [PubMed] [Google Scholar]
- Kuivila KM, Murray JW, Devol AH, Lidstrom ME, Reimers CE. Methane cycling in the sediments of Lake Washington. Limnol Oceanogr. 1988;33:571–581. [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools.1000 genome project data processing subgroup. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom ME, Somers L. Seasonal study of methane consumption in Lake Washington. Appl Environ Microbiol. 1984;47:1255–1260. doi: 10.1128/aem.47.6.1255-1260.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase J, Frenzel P. A methane-driven microbial food web in a wetland rice soil. Environ Microbiol. 2007;9:3025–3034. doi: 10.1111/j.1462-2920.2007.01414.x. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet EG, Dlugokencky EJ, Bousquet P. Atmospheric science. Methane on the rise–again. Science. 2014;343:493–495. doi: 10.1126/science.1247828. [DOI] [PubMed] [Google Scholar]
- Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Annu Rev Microbiol. 2013;67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- Ojala DS, Beck DA, Kalyuzhnaya MG. Genetic systems for moderately halo(alkali)philic bacteria of the genus Methylomicrobium. Meth Enzymol. 2011;495:99–118. doi: 10.1016/B978-0-12-386905-0.00007-3. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2013. Vegan: community ecology package.
- Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, et al. Environmental, genomic and taxonomic perspectives on methanotrophic verrucomicrobia. Environ Microbiol Rep. 2009;1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL, Sessions AL. Identification of novel methane-, ethane-, and propane-oxidizing bacteria at marine hydrocarbon seeps by stable isotope probing. Appl Environ Microbiol. 2010;76:6412–6422. doi: 10.1128/AEM.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim A, Lüke C, Krause S, Pratscher J, Frenzel P. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 2012;6:2128–2139. doi: 10.1038/ismej.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7:2315–2329. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–790. doi: 10.1038/nrmicro2439. [DOI] [PubMed] [Google Scholar]
- Svenning M.M, Hestnes A.G, Wartiainen I, Stein L.Y, Klotz M.G, Kalyuzhnaya M.G, et al. Genome sequence of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193:6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Orphan VJ, Kalyuzhnaya MG, Jetten MS, Klotz MG. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. doi: 10.1111/j.1758-2229.2010.00192.x. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotsenko YA, Murrell JC. Metabolic aspects of aerobic obligate methanotrophy. Adv Appl Microbiol. 2008;6:183–229. doi: 10.1016/S0065-2164(07)00005-6. [DOI] [PubMed] [Google Scholar]
- van der Ha D, Vanwonterghem I, Hoefman S, De Vos P, Boon N. Selection of associated heterotrophs by methane-oxidizing bacteria at different copper concentrations. Antonie Van Leeuwenhoek. 2013;103:527–537. doi: 10.1007/s10482-012-9835-7. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.