Abstract
Prochlorococcus is the numerically dominant phototroph in the oligotrophic subtropical ocean and carries out a significant fraction of marine primary productivity. Although field studies have provided evidence for nitrate uptake by Prochlorococcus, little is known about this trait because axenic cultures capable of growth on nitrate have not been available. Additionally, all previously sequenced genomes lacked the genes necessary for nitrate assimilation. Here we introduce three Prochlorococcus strains capable of growth on nitrate and analyze their physiology and genome architecture. We show that the growth of high-light (HL) adapted strains on nitrate is ∼17% slower than their growth on ammonium. By analyzing 41 Prochlorococcus genomes, we find that genes for nitrate assimilation have been gained multiple times during the evolution of this group, and can be found in at least three lineages. In low-light adapted strains, nitrate assimilation genes are located in the same genomic context as in marine Synechococcus. These genes are located elsewhere in HL adapted strains and may often exist as a stable genetic acquisition as suggested by the striking degree of similarity in the order, phylogeny and location of these genes in one HL adapted strain and a consensus assembly of environmental Prochlorococcus metagenome sequences. In another HL adapted strain, nitrate utilization genes may have been independently acquired as indicated by adjacent phage mobility elements; these genes are also duplicated with each copy detected in separate genomic islands. These results provide direct evidence for nitrate utilization by Prochlorococcus and illuminate the complex evolutionary history of this trait.
Introduction
The unicellular cyanobacterium Prochlorococcus is the smallest known free-living oxygenic phototroph (Chisholm et al., 1992; Partensky et al., 1999; Coleman and Chisholm, 2007; Partensky and Garczarek, 2010). It is numerically dominant in the tropical and subtropical regions of the world's oceans and responsible for 5–10% of marine primary productivity (Campbell et al., 1994; Partensky et al., 1999; Buitenhuis et al., 2012; Flombaum et al., 2013). Prochlorococcus has undergone a process of genome reduction following divergence from its closest relatives, the marine Synechococcus (Rocap et al., 2002; Kettler et al., 2007). These streamlined genomes are often considered an adaptation to the oligotrophic environments they occupy (Dufresne et al., 2003; Rocap et al., 2003). Even though individual genomes are small, the collective of all Prochlorococcus cells possesses a vast reservoir of genetic and physiological diversity (Kettler et al., 2007). Prochlorococcus is composed of a polyphyletic group of low-light (LL) adapted clades (LLI–LLVI and NC1), and a more recently diverged monophyletic group of high-light (HL) adapted clades (HLI–HLVI) (Moore et al., 1998; Moore and Chisholm, 1999; Rocap et al., 2002; Martiny et al., 2009c; Lavin et al., 2010; Shi et al., 2011; Huang et al., 2012; Malmstrom et al., 2013). Some of these clades are known to be differentially distributed along gradients of light intensity, temperature and nutrient concentrations (Bouman et al., 2006; Johnson et al., 2006; Zinser et al., 2006; Zwirglmaier et al., 2007, 2008; Malmstrom et al., 2010, 2013).
Nitrogen availability often limits primary productivity in marine systems (Tyrrell, 1999), and organisms have evolved diverse mechanisms for uptake of various chemical forms of nitrogen. Nitrate is one of the more abundant sources of inorganic nitrogen available to phytoplankton (Gruber, 2008), and the majority of cyanobacteria possess pathways for the uptake and assimilation of nitrate (Herrero et al., 2001; García-Fernández et al., 2004; Ohashi et al., 2011). Early reports on the vertical distributions of Prochlorococcus noted a subsurface maximum in abundance at the base of the euphotic zone, suggesting that Prochlorococcus was sensitive to nitrogen depletion and might be assimilating nitrate supplied from deep waters (Olson et al., 1990; Vaulot and Partensky, 1992). Therefore, it was surprising that nearly all isolates of Prochlorococcus could not use nitrate and lacked the genes required for this function (Moore et al., 2002; Coleman and Chisholm, 2007; Kettler et al., 2007), even though most isolates of Synechococcus are capable of using nitrate (Fuller et al., 2003; Ahlgren and Rocap, 2006). Only a single Prochlorococcus culture, PAC1 isolated in 1992, was reported to utilize nitrate (Williams et al., 1999), but because of the presence of other bacteria in that culture, direct nitrate uptake by Prochlorococcus could not be conclusively demonstrated.
Several pieces of evidence indicated that nitrate assimilation was a more common trait within Prochlorococcus populations than previously thought. Field experiments demonstrated the uptake of isotopically labeled nitrate by Prochlorococcus cells in the Sargasso Sea (Casey et al., 2007), and nitrate assimilation genes were found to be associated with uncultivated Prochlorococcus genomes from many regions of the subtropical oceans (Martiny et al., 2009b). A scaffold assembled from metagenomic data from the Global Ocean Sampling (GOS) expedition indicated that all the genes required for nitrate assimilation were colocalized in a specific region of the genomes of HL adapted Prochlorococcus. The metagenomic data primarily identified nitrate utilization genes in the HLII clade of Prochlorococcus as sequences from this clade comprised the majority of Prochlorococcus-like sequences in the GOS data set (Rusch et al., 2007).
These past observations raised two important questions about nitrate assimilation in Prochlorococcus. (1) Can axenic strains grow on nitrate as the sole nitrogen source? (2) What is the evolutionary history of nitrate assimilation genes in this group? To address these questions, we isolated and sequenced Prochlorococcus strains capable of nitrate assimilation and examined their growth on different nitrogen sources. We then used comparative genomics to better understand how this trait had evolved in Prochlorococcus.
Materials and methods
Strains and enrichments
Five strains of Prochlorococcus (SB, MIT0604, PAC1, MIT9301 and MED4), one strain of Synechococcus (WH8102) and two Prochlorococcus enrichment cultures (P0902-H212 and P0903-H212) were used in this study. MIT9301, MED4 and WH8102 have previously been rendered axenic (free of heterotrophic contaminants). All axenic cultures were routinely assessed for purity by confirming a lack of turbidity after inoculation into a panel of purity test broths: ProAC (Morris et al., 2008), MPTB (Saito et al., 2002) and ProMM (Pro99 medium (Moore et al., 2007) supplemented with 1 × Va vitamin mix (Waterbury and Willey, 1988) and 0.05% w/v each of pyruvate, acetate, lactate and glycerol). ProMM is a modified version of the PLAG medium (Morris et al., 2008), but uses 100% sea water as the base.
PAC1 was enriched from sea water collected from the deep chlorophyll maximum in the North Pacific Ocean at Station ALOHA (22.75°N, 158°W) on Hawai'i Ocean Time-series (HOT) cruise 36. Sea water was passed through a 0.6 μm Nucleopore filter twice, and the filtrate was serially diluted into K/10 medium (Chisholm et al., 1992), but with the following modifications for final nutrient concentrations: 5 μM urea, 5 μM ammonium and 1 μM β-glycerophosphate replacing inorganic phosphate, 0.01 μM Na2MoO4 and 0.05 μM NiCl2. MIT0604 was derived from an enrichment culture initiated with Pro2 nutrient additions (Moore et al., 2007) to sea water obtained at Station ALOHA on HOT cruise 181, but with all nitrogen sources replaced by 0.217 mM sodium nitrate. The P0902-H212 and P0903-H212 enrichments were initiated with Pro2 nutrient additions (Moore et al., 2007) to sea water obtained from Station ALOHA on HOT cruise 212, but with all nitrogen sources replaced by 0.05 mM sodium nitrate.
Purification of Prochlorococcus strains
SB and MIT0604 were rendered axenic in this study using a modified dilution to extinction method. Prochlorococcus from exponential phase cultures were enumerated using an Influx Cell Sorter (BD Biosciences, San Jose CA, USA) or a FACSCalibur flow cytometer (BD Biosciences) as previously described (Olson et al., 1985; Cavender-Bares et al., 1999). Cultures consisting of >80% Prochlorococcus cells were serially diluted into multiple multiwell plates at final concentrations of 1–10 cells per well in at least 200 μl of ProMM medium. Axenic Prochlorococcus do not grow from such low cell densities in Pro99 medium without ‘helper' heterotrophic bacteria (Morris et al., 2008, 2011); however, they do grow when diluted into ProMM. The main ingredient in ProMM that promotes the growth of cells from low densities is pyruvate, and we suspect that in this context pyruvate serves as a potent hydrogen peroxide scavenger (Giandomenico et al., 1997). Wells contaminated with heterotrophic bacteria were identified by the appearance of turbidity. The multiwell plates were monitored by eye and by fluorometry using a Synergy HT Microplate Reader (BioTek, Winooski, VT, USA), and nonturbid wells were monitored by flow cytometry using a FACSCalibur flow cytometer. Wells that appeared green or had Prochlorococcus cells as determined by flow cytometry were immediately transferred to Pro99 medium directly, or into fresh ProMM medium until consistent growth was observed, at which point the cultures were introduced back into Pro99 medium. Cultures were examined for heterotrophic bacteria contaminants by flow cytometry and by inoculation into the panel of purity test broths as described above.
PCR screen for the nitrate reductase gene
Based on an alignment of GOS reads coding for the Prochlorococcus narB sequence (Martiny et al., 2009b), degenerate primers 30narB175f (5′-TGYGTDAAAGGMGCAACAGTNTG-3′) and 30narB574r (5′-GACAYTCWGCBGTATTWGTHCC-3′) were designed to specifically amplify the narB gene from HLII clade Prochlorococcus, and degenerate primers 40narB1447f (5′-TATTGYCCAGCWTTYMGDCCDTG-3′) and 40narB1766r (5′-AKAGGWTGYTTWGTRTARAAYTG-3′) were designed to specifically amplify the narB gene from LLI clade Prochlorococcus. PCR used annealing temperatures of 52.5 °C for the HLII narB sequence and 56 °C for the LLI narB sequence. Reactions contained 1 × PCR buffer, 2.5 mM MgCl2, 0.2 mM each of dATP, dTTP, dCTP and dGTP, 0.2 μM of each primer, 1 unit of Platinum Taq DNA polymerase (Life Technologies, Grand Island, NY, USA) and 1 ng of genomic DNA prepared from Prochlorococcus cultures in the MIT Cyanobacteria Culture Collection (Chisholm Laboratory, MIT, Cambridge, MA, USA). DNA from Synechococcus WH8102, which contains a narB gene, was used as a negative control. Reactions were cycled 30 times at 94 °C for 15 s, the primer-specific annealing temperature for 15 s and 72 °C for 60 s. PCR products with the expected size were sequenced at the Dana-Farber/Harvard Cancer Center DNA Resource Core (Boston, MA, USA) to confirm amplification of the narB gene.
Growth in the presence of alternative nitrogen sources
Axenic Prochlorococcus strains SB, MIT0604, MIT9301 and MED4, and axenic Synechococcus strain WH8102 were acclimated to Pro99 medium (Moore et al., 2007) prepared with sea water from the South Pacific Subtropical Gyre and grown at 24 °C and 50 μmol photons m−2 s−1 continuous illumination for at least 10 generations or until growth rates were similar between successive transfers. Bulk culture fluorescence was measured as a proxy for biomass using a 10-AU fluorometer (Turner Designs, Sunnyvale, CA, USA). Triplicate cultures of each strain were initiated in Pro99 that contained 0.8 mM ammonium chloride. Once cultures had reached mid-exponential phase, they were transferred into Pro99 medium containing 0.8 mM ammonium chloride, 0.8 mM sodium nitrate, 0.8 mM sodium cyanate or no nitrogen additions as a control to monitor utilization of carry-over ammonium. Cultures were successively transferred at mid-exponential phase until growth in the cultures lacking nitrogen additions had arrested because of nitrogen limitation. Specific growth rates were estimated from the log-linear portion of the growth curve for the final transfer. Two tailed homoscedastic t-tests were conducted in Microsoft Excel (Microsoft Corporation, Redmond, WA, USA) in order to evaluate the likelihood of significantly different growth rates in each strain for each pair of nitrogen sources and for strains grown on the same nitrogen source.
Genome data
A total of 41 Prochlorococcus and 15 Synechococcus genomes (Biller et al., 2014) that include the genomes of the nitrate assimilating strains SB, MIT0604 and PAC1 were used in this study. Sequence data were also obtained for the P0902-H212 and P0903-H212 enrichment cultures as described in the Supplementary Methods. These enrichment assemblies had total sequence lengths approximately twice the size of previously sequenced Prochlorococcus genomes, suggesting the presence of at least two unique strains dominating each enrichment. Binning contigs based on average sequencing coverage yielded a subset of highly covered contigs in each assembly with a total sequence length similar to that of previously sequenced Prochlorococcus genomes. In the highly covered subsets for each assembly, the complete set of nitrate assimilation genes were only found on a single contig. For the purpose of this study, only these contigs were relevant and entered into our analysis.
All sequence data were annotated using the RAST server (Aziz et al., 2008) with FIGfam release 49 in order to facilitate comparison between genomes by ensuring a uniform methodology for gene calling and functional annotation. Clusters of orthologous groups of proteins (COGs) were identified as previously described (Kelly et al., 2012). These clusters are included in the ‘V4' CyCOGs on the ProPortal website (http://proportal.mit.edu) (Kelly et al., 2012; Biller et al., 2014).
Genome phylogeny
We translated 537 single-copy core genes to amino acid sequences, aligned each gene individually in protein space using ClustalW (Larkin et al., 2007), and then back-translated the sequences using TranslatorX (Abascal et al., 2010). Using the principle previously described (Kettler et al., 2007), we randomly concatenated 100 of these aligned genes and built maximum likelihood and neighbor-joining phylogenies using PHYLIP v3.69 (Felsenstein, 2005). We repeated the random concatenation and tree generation 100 times.
Estimation of gene gain and loss
Using a maximum parsimony approach (Mirkin et al., 2003), the patterns of gene gain and loss were mapped onto the topology of the maximum likelihood nucleotide tree using WH5701 as an outgroup. Utilizing 13 590 non-core single-copy COGs, we reconstructed ancestral character states of gene absence and presence on our guide tree and minimized the cost of gains and losses given a gene gain equal to twice a gene loss. We used the program DendroPy to implement the tree traversal portion of the algorithm (Sukumaran and Holder, 2010).
Phylogenies of genes involved in the transport and reduction of nitrate and nitrite
COGs corresponding to the nirA, narB, focA and napA genes were aligned in protein space using ClustalW. Phylogenetic trees were estimated with PHYLIP v3.69 (Felsenstein, 2005) using the programs SEQBOOT, PROTDIST with the Jones–Taylor–Thornton matrix and a constant rate of variability among sites and NEIGHBOR on the aligned amino acid sequences with Synechococcus WH5701 used as an outgroup for nirA and narB and Synechococcus CB0101 used as an outgroup for focA and napA. We included GOS consensus sequences: GOS nirA, GOS narB and GOS napA (Martiny et al., 2009b).
Results and discussion
Isolates of Prochlorococcus are capable of nitrate assimilation
To identify possible cultures capable of nitrate assimilation, we screened existing Prochlorococcus cultures for the assimilatory nitrate reductase gene, narB, using PCR. We found that the LL adapted PAC1 strain (Penno et al., 2000) and the HL adapted SB strain (Shimada et al., 1995) each contained the gene. In search of additional strains capable of utilizing nitrate, we performed selective enrichments from sea water obtained from the subtropical North Pacific Ocean using nitrate as the sole added nitrogen source. This yielded one HL adapted strain (Prochlorococcus MIT0604) and two mixed Prochlorococcus cultures (P0902-H212 and P0903-H212) with the narB gene (Table 1).
Table 1. Prochlorococcus strains and enrichments capable of growth in the presence of nitrate as the sole nitrogen source.
| Name | Clade | Axenic | Isolation depth (m) | Isolation coordinates | Region | Isolation date | Assembly size (bp) | Contigs | % GC | Genbank accession | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unialgal cultures (complete genome sequences) | |||||||||||
| SB | HL II | Yes | 40 | 35°N, 138.3°E | Suruga Bay, Japan | October 1992 | 1 668 514 | 3 | 31.5 | JNAS00000000 | Shimada et al. (1995); Biller et al. (2014) |
| MIT0604 | HL II | Yes | 175 | 22.75°N, 158°W | North Pacific | May 2006 | 1 780 061 | 1 | 31.2 | CP007753 | This study |
| PAC1 | LL I | No | 100 | 22.75°N, 158°W | North Pacific | April 1992 | 1 825 493 | 15 | 35.1 | JNAX00000000 | Penno et al. (2000); Biller et al. (2014) |
| Mixed enrichments (partial genome assemblies) | |||||||||||
| P0902-H212 | LL I | No | 175 | 22.75°N, 158°W | North Pacific | July 2009 | 501 825 | 1 | 35.4 | KJ947870 | This study |
| P0903-H212 | LL I | No | 200 | 22.75°N, 158°W | North Pacific | July 2009 | 291 739 | 1 | 35.2 | KJ947871 | This study |
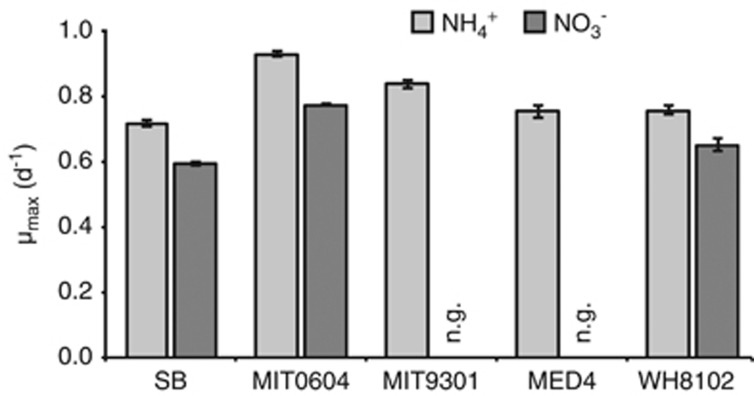
We then rendered SB and MIT0604 axenic and examined their growth in the presence of nitrate or ammonium. As hypothesized, both SB and MIT0604 can grow on nitrate as the sole source of nitrogen, but with a significant reduction in growth rate (18% and 17%, respectively), compared with growth on ammonium (Figure 1 and Supplementary Figure S1). Although the slower growth on nitrate could be explained by the greater amount of reducing power required to assimilate more oxidized N sources (García-Fernández et al., 2004), we assume that these cultures were growing at saturating light intensities based on previous measurements of light saturating irradiances for the growth of Prochlorococcus (Moore and Chisholm, 1999); thus energy supply and reducing power were likely not limiting. Furthermore, recent work has shown that the growth rates and chemical composition of some marine cyanobacteria are not directly related to the oxidation state of the cells' N source (Collier et al., 2012). Under light-limiting conditions, for example, the growth rate and chemical composition of Synechococcus grown on ammonium was the same as that on nitrate; however, under light-saturating conditions, cells grown on nitrate had a higher carbon-to-nitrogen ratio (Collier et al., 2012). This perhaps suggests a bottleneck in the uptake and conversion of nitrate compared with ammonium when energy is sufficient (Collier et al., 2012), and may explain the slower growth of Prochlorococcus on nitrate compared with ammonium.
Figure 1.
Maximum specific growth rates (μ max) of Prochlorococcus strains SB, MIT0604, MIT9301, MED4 and Synechococcus WH8102 in the presence of ammonium or nitrate. Values represent the mean and s.d. of three biological replicates. Growth rate differences for each strain grown on ammonium compared with nitrate as well as growth rate differences between strains on the same nitrogen source were significant (P<0.05) in a two-tailed homoscedastic t-test; n.g., no growth.
In the early days of research on Prochlorococcus, the absence of cultures known to utilize nitrate resulted in a distorted view of the role of Prochlorococcus in marine ecosystems; ecosystem models and ecophysiological interpretations were guided by the assumption that most, if not all, Prochlorococcus were incapable of nitrate assimilation (García-Fernández et al., 2004; Fuller et al., 2005; Follows et al., 2007). Why have nitrate-utilizing Prochlorococcus appeared so infrequently in culture collections in the past? Is it because we were selecting against them in isolations using media containing ammonium but not nitrate (Moore et al., 2007)? We think not because SB and MIT0604—both narB-containing strains—grow at equal or better rates on ammonium compared with other HL adapted Prochlorococcus strains (Figure 1 and Supplementary Figure S1). An alternative explanation is that most of the early cultures of Prochlorococcus were isolated from environments that are relatively nitrogen replete—that is, thought to be more limited by phosphorus or iron availability (for example, the Sargasso Sea, Mediterranean Sea and the Equatorial Pacific) (Vaulot et al., 1996; Mann and Chisholm, 2000; Wu et al., 2000; Marty et al., 2002; Kettler et al., 2007; Rusch et al., 2010). We now know that Prochlorococcus cells capable of nitrate assimilation are more likely to be found in ocean regions with lower average nitrate concentrations, such as the Caribbean Sea and Indian Ocean (Martiny et al., 2009b). Indeed, PAC1 and SB (both narB-containing strains that were isolated on medium containing ammonium but lacking nitrate) were isolated from N-poor regions (Shimada et al., 1995; Penno et al., 2000; Wu et al., 2000; Iwata et al., 2005). Thus, we believe that the probability of obtaining a narB-containing strain using medium containing ammonium is in large part a function of the particular water sample used to start enrichment cultures.
Nitrate assimilation is found in diverse lineages of Prochlorococcus
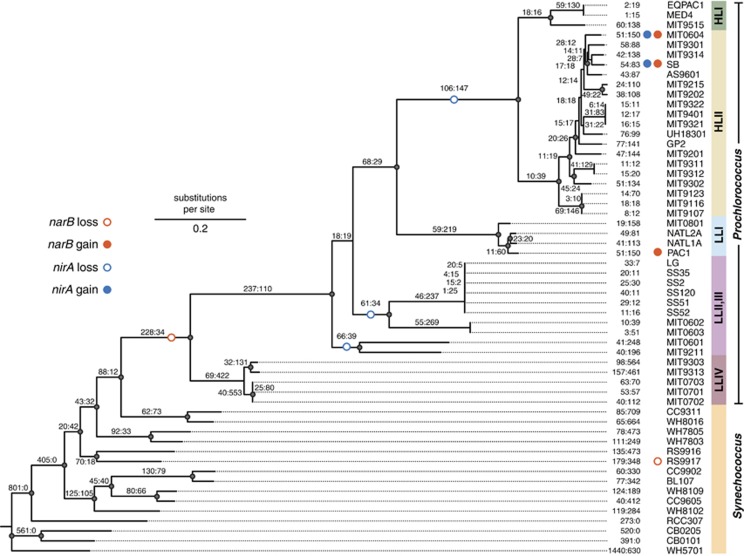
What can the features of the nitrate assimilation genes in Prochlorococcus tell us about how they have been gained or lost during the evolution of this group? The genomes of PAC1, SB and MIT0604, along with contigs containing nitrate assimilation genes from the P0902-H212 and P0903-H212 enrichment cultures, were informative in this regard. These Prochlorococcus belong to both the LL adapted LLI clade (PAC1, P0902-H212 and P0903-H212) and the HL adapted HLII clade (SB and MIT0604) (Figure 2 and Supplementary Figures S2 and S3), demonstrating that nitrate utilization is found in multiple and diverse lineages of Prochlorococcus and suggesting a complex evolutionary history. The presence of nitrite and nitrate metabolism in Prochlorococcus follows that of Synechococcus in that some strains are able to reduce nitrite and some are able to reduce both nitrite and nitrate. Because these traits are not monophyletic, a model of gene gain and loss events provides evidence for three gains and two losses for the narB nitrate reductase gene and two gains and three losses for the nirA nitrite reductase gene (Figure 2). With the limited number of genomes available, it appears that there is evidence for multiple gains and losses of nitrogen assimilation traits through the evolution of Prochlorococcus and Synechococcus, with narB found in at least three distinct Prochlorococcus lineages.
Figure 2.
Maximum likelihood phylogeny of Prochlorococcus and Synechococcus based on the similarity of 100 randomly concatenated single-copy core genes. Nodes are marked by closed circles to indicate that the associated taxa clustered together in at least 75% of 100 replicate trees. Genes lost and gained in the evolution of Prochlorococcus and Synechococcus are indicated at each node by values representing losses followed by gains. Predicted losses (open circles) or gains (closed circles) of nirA (blue) or narB (orange) are labeled on their respective branches.
The genomic context of the nitrate assimilation gene cluster suggests a complex evolutionary history
To look for features that might help us interpret the gains and losses of nitrate and nitrite assimilation genes in Prochlorococcus, we examined the local genomic context of these genes. Although the full complement of nitrate assimilation genes was predicted to be localized in a single region of the highly syntenic HLII clade genomes from metagenomic assemblies (Martiny et al., 2009b), it was unclear whether this context would be found in any individual cell. Furthermore, given that these genes were found in a different region in Prochlorococcus compared with marine Synechococcus, we were curious as to whether we might find evidence for rearrangements or lateral gene transfer.
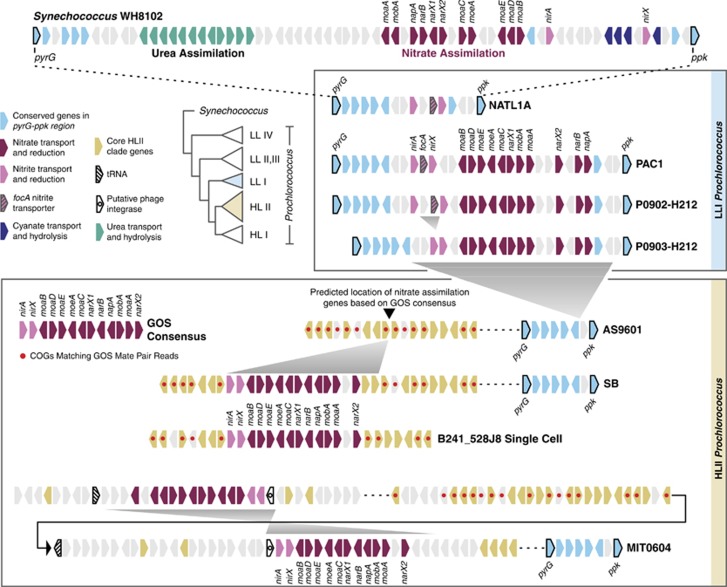
The nitrate assimilation genes in PAC1 and the P0902-H212 and P0903-H212 contigs are syntenic and also found in the same genomic region as the nitrite assimilation genes in NATL1A and the nitrate assimilation genes in Synechococcus WH8102 (Figure 3). This region is bounded by a pyrimidine biosynthesis gene (pyrG) and a polyphosphate kinase gene (ppk) between which many nitrogen assimilation genes are located in marine Synechococcus. Although gene gains and losses have been observed in this region (Scanlan et al., 2009), our data indicate that the genomic location of the nitrate and nitrite assimilation genes is reasonably well fixed in LLI Prochlorococcus and closely related Synechococcus. Although our model of gene gain and loss events suggests the loss of nitrate assimilation genes early in the evolution of Prochlorococcus (Figure 2), the local genomic features of these genes are consistent with the interpretation that some lineages may have retained these genes following the divergence of Prochlorococcus from Synechococcus.
Figure 3.
Architecture of the nitrite and nitrate assimilation genes in LL adapted (LLI clade) and HL adapted (HLII clade) Prochlorococcus relative to Synechococcus WH8102. Similar to Synechococcus, the nitrite and nitrate assimilation genes in the LLI clade of Prochlorococcus are found within the region between the pyrG (pyrimidine biosynthesis) and ppk (polyphosphate kinase) genes. Most LLI clade Prochlorococcus, with the exception of the P0903-H212 contig, possess a focA nitrite transporter in this region (possibly acquired from proteobacteria (Rocap et al., 2003)). Metagenome data (Martiny et al., 2009b), a partial genome from a single cell (B241-528J8) (Kashtan et al., 2014) and a culture genome (Prochlorococcus SB) indicate that the nitrate assimilation genes within HLII clade Prochlorococcus are commonly found in a syntenic region adjacent to genomic island ISL3 (see Figure 4). Prochlorococcus MIT0604 is an exception in that it possesses duplicate nitrate assimilation gene clusters located within genomic islands ISL3 and ISL4 (see Figure 4), with phage integrase genes immediately adjacent to each copy of the nirA (nitrite reductase) gene.
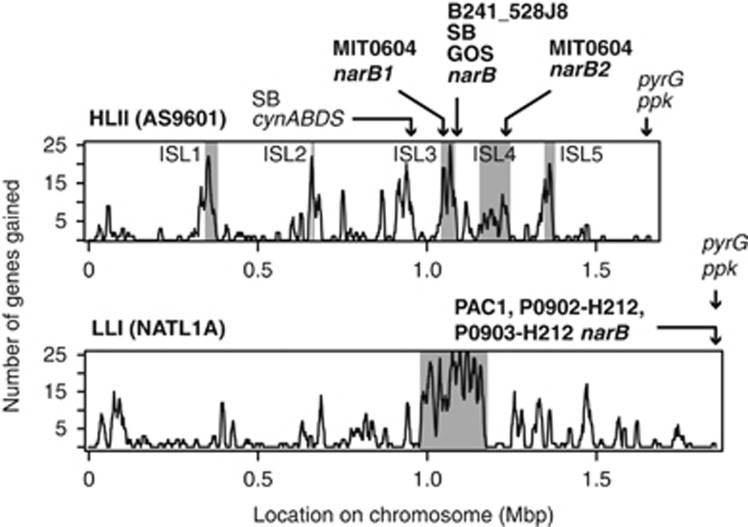
Analysis of metagenomic data from GOS (Martiny et al., 2009b) suggested that the nitrate utilization genes in HLII Prochlorococcus should be located in a different genomic region compared with LLI genomes, indicating an alternative evolutionary origin. Based on a scaffold of mate-paired metagenomic reads, it was inferred that this cluster should be located ∼500 kb downstream of the pyrG-ppk region containing the nitrate assimilation genes in WH8102 and the nitrite assimilation genes in NATL1A (Martiny et al., 2009b). We found a high degree of similarity between the nitrate assimilation gene cluster in SB and the scaffold derived from GOS metagenome sequences obtained from multiple individual cells from multiple sampling stations. This similarity manifested itself not only in the gene order and chromosomal location, but also the phylogeny of the nitrate assimilation genes (Figures 3, 4, 5), placing the nitrate assimilation gene cluster in a genomic region that is syntenic with other HLII genomes and adjacent to a known genomic island (ISL3) in this clade (Figure 4). Furthermore, a partial genome from a Prochlorococcus single cell belonging to the HLII clade (B241-528J8; Genbank JFLE01000089.1) (Kashtan et al., 2014) also possesses a nitrate assimilation gene cluster in the same location and in the same order. The striking similarity between the nitrate assimilation gene clusters of these individual Prochlorococcus and the GOS consensus indicates that the order and location of nitrate assimilation genes are stable within HLII genomes.
Figure 4.
Locations of nitrate and cyanate assimilation genes in strains of Prochlorococcus capable of nitrate assimilation relative to the known genomic islands (shaded regions) observed in the HLII and LLI clades of Prochlorococcus; plots modified from Kettler et al. (2007). Prochlorococcus genomes are highly syntenic and genomic islands have been identified in HL adapted genomes (for example, AS9601) by conserved breaks in gene synteny among strains (Coleman et al., 2006; Kettler et al., 2007). Genomic islands have also been identified (for example, the large region within LLI clade genomes such as NATL1A) by predicted gene gain events along the chromosome (Kettler et al., 2007).
The nitrate assimilation genes in strain MIT0604 had a different local genome structure compared with strain SB and the partial single-cell genome, B241-528J8. MIT0604 has duplicate clusters of these genes that are inversely oriented and located upstream and downstream of the GOS-predicted location (Figure 3 and 4). A Southern blot confirmed that MIT0604 does indeed contain two copies of narB whereas SB contains only one (Supplementary Figure S4), and they are located within genomic islands ISL3 and ISL4 of HLII clade Prochlorococcus (Figure 4). Genomic islands are common features of Prochlorococcus genomes, particularly within the HL adapted clades (Coleman et al., 2006; Kettler et al., 2007). They harbor much of the variability in gene content between members of the same clade and are hot spots for lateral gene transfer. Phage integrase genes are located proximal to both nitrate assimilation gene clusters in MIT0604, and a transfer RNA gene is adjacent to one of these clusters (Figure 3). The transfer RNA genes are known to serve as sites for insertion of phage DNA in bacteria (Williams, 2002), and thus the location of these phage integrase and transfer RNA genes suggests transduction as a possible mechanism by which MIT0604 has acquired the nitrate assimilation gene cluster. Notably, duplication of such a large region of the chromosome has not been observed previously in Prochlorococcus, and, thus far, MIT0604 is the only Prochlorococcus or Synechococcus strain possessing two complete copies of the genes required for nitrate assimilation.
The phylogenies of nitrate assimilation genes are similar to the phylogeny of genomes
Given the evidence for both a stable arrangement of the nitrate assimilation genes in some Prochlorococcus and possible gene transfer leading to acquisition of the nitrate assimilation trait in MIT0604, we were curious to know whether the phylogenies of these genes were congruent with whole genome phylogenies (Figure 2 and Supplementary Figure S2), as well as the phylogeny of GyrB (Supplementary Figure S3) that has been identified as a useful phylogenetic marker for Prochlorococcus (Mühling, 2012). Thus, we reconstructed the amino acid phylogenies of the NirA and NarB reductases, the FocA nitrite transporter and the NapA nitrite/nitrate transporter (Figure 5). The NirA phylogeny is largely consistent with our observations based on the GOS metagenome data (Martiny et al., 2009b), such that the NirA proteins from genomes in the LLIV clade are more closely associated with marine Synechococcus than with other Prochlorococcus sequences. In all phylogenetic trees, the PAC1, P0902-H212 and P0903-H212 sequences are in a separate clade distinct from that of the SB and MIT0604 sequences, reinforcing the HL versus LL differentiation (Figure 5). The NirA and NarB sequences from SB are consistently more closely affiliated with the GOS consensus sequence (Martiny et al., 2009b) than with the MIT0604 sequences. NapA sequences from SB and MIT0604 are also both closely related to the GOS NapA consensus sequence (Figure 5). Similar to the GyrB phylogeny (Supplementary Figure S3), the P0903-H212 sequences fall outside the clade containing the other LLI sequences. With the exception of the LLIV NirA sequences, the phylogenies of these nitrite and nitrate assimilation proteins (Figure 5) are congruent with whole genome and GyrB phylogenies (Figure 2 and Supplementary Figures S2 and S3) at a resolution defining the major Prochlorococcus clades.
Figure 5.
Neighbor-joining phylogeny of four proteins involved in the transport and reduction of nitrate and nitrite in marine cyanobacteria: (a) NirA; nitrite reductase, (b) NarB; nitrate reductase, (c) FocA; nitrite transporter and (d) NapA; nitrite/nitrate transporter. The percentage of 100 replicate trees in which the associated taxa clustered together is indicated at nodes by closed circles (>75%) or open circles (>50%). Scale bars represent substitutions per site.
Nitrate assimilating Prochlorococcus possess a diverse set of nitrogen acquisition pathways
Gene content in Prochlorococcus has been shown, for several traits, to reflect the selective pressures in the specific environments from which they (or their genes) were captured (Martiny et al., 2006; Rusch et al., 2007; Coleman and Chisholm, 2010; Feingersch et al., 2012; Malmstrom et al., 2013). Thus, we wondered whether other nitrogen assimilation traits might co-occur with nitrate assimilation in Prochlorococcus, and examined the potential for PAC1, SB and MIT0604 to access alternative sources of nitrogen based on their gene content (Supplementary Table S1 and Supplementary Figure S5).
Like other members of the LLI clade, PAC1 possesses genes for the assimilation of ammonium and urea, but lacks cyanate transporter genes. In addition to the napA nitrite/nitrate transporter, the focA nitrite transporter is found in both PAC1 and the contig from P0902-H212. However, the focA gene is absent from HL adapted strains SB and MIT0604, and most surface water metagenomic samples (Martiny et al., 2009b). Some Synechococcus strains (for example, WH8102) (Supplementary Figure S5) also lack focA; thus, this gene is clearly subject to gain and loss. Although focA is also similar to formate transporters, evidence implicates its role in nitrite uptake in Prochlorococcus; for example, the gene is located near other nitrite assimilation genes (Figure 3), it is upregulated under nitrogen stress (Tolonen et al., 2006) and it is absent from Prochlorococcus that cannot grow on nitrite (Moore et al., 2002; Coleman and Chisholm, 2007; Kettler et al., 2007) (Supplementary Figure S5). As PAC1 possesses both a nitrite transporter (focA) and the dual-function nitrate/nitrite transporter (napA), it is possible that focA provides some advantage to LL adapted cells that are often maximally abundant near the nitrite maxima in the oceans (Scanlan and West, 2002; Lomas and Lipschultz, 2006). LL adapted cells that possess the dual-function nitrite/nitrate transporter may benefit from having an additional transporter for nitrite. Given that HL adapted Prochlorococcus strains capable of nitrate utilization lack the focA gene, these cells may be less reliant on nitrite as a nitrogen source.
SB and MIT0604 possess urea assimilation genes and can utilize urea as a sole nitrogen source (Supplementary Figure S6). Furthermore, SB possesses cyanate transporter genes that are rare in both Prochlorococcus and Synechococcus strains (Kamennaya et al., 2008), and it can indeed grow utilizing cyanate (Supplementary Figure S1) as the sole source of nitrogen. Although very little is known about cyanate concentrations in marine systems, cynA genes (encoding the periplasmic component of the cyanate ABC-type transporter system) were relatively abundant in the seasonally stratified and nitrogen depleted waters of the northern Red Sea (Kamennaya et al., 2008). The cynA gene of SB clusters with clones obtained from the Red Sea (Supplementary Figure S7), supporting their origin in HLII clade genomes as hypothesized by Kamennaya et al. (2008).
SB contains the most extensive suite of nitrogen acquisition pathways of any cultured Prochlorococcus strain examined to date. Why might this be? A useful analogy can be drawn from our understanding of selection pressures that have shaped Prochlorococcus genomes with respect to adaptations involved in phosphorus assimilation. Individual cells and populations from phosphorus-limited environments possess accessory phosphorus acquisition genes, such as alkaline phosphatase (phoA) and phosphonate utilization (phnYZ) genes, at a higher frequency than Prochlorococcus from phosphorus-replete environments (Martiny et al., 2006, 2009a; Coleman and Chisholm, 2010; Feingersch et al., 2012). Thus, we hypothesize that the nitrogen assimilation traits present in Prochlorococcus SB were likely shaped by frequent nitrogen limitation in its original habitat (Iwata et al., 2005); that is, cells capable of accessing a wide pool of nitrogen compounds may be at a selective advantage in nitrogen-limited environments.
Conclusions
Given the large standing stock of Prochlorococcus in the subtropical oceans and the extent to which nitrogen limits primary production in these regions (Tyrrell, 1999; Moore et al., 2013), the absence of nitrate assimilation capabilities in cultured strains of Prochlorococcus has long puzzled biological oceanographers. This motivated field studies (Casey et al., 2007; Martiny et al., 2009b) and the use of models to help us understand the selection pressures driving the loss of nitrate assimilation genes in Prochlorococcus relative to Synechococcus (Bragg et al., 2010). In this study we show unequivocally that some strains of Prochlorococcus are indeed capable of growth using nitrate as the sole nitrogen source. Future studies of these strains will help elucidate the physiological tradeoffs of carrying these genes and help refine the nitrogen inventory in biogeochemical models of the global ocean (Follows et al., 2007). Correlations between environmental nitrate concentrations and ribotype phylogeny (Martiny et al., 2009c) and the striking similarity between Prochlorococcus SB and the GOS consensus sequence both suggest that the trait for nitrate assimilation could be tied to distinct ribotype lineages. Still, evolution has many ways of introducing genomic complexity: the MIT0604 genome suggests that these genes are also subject to horizontal gene transfer, allowing further diversification of this trait in other lineages. This is reminiscent of the phylogenetic characteristics of phosphorus acquisition traits that are nearly independent of ribotype phylogeny (Martiny et al., 2009c)—with extensive diversity in the ‘leaves of the tree'. As we learn more about these layers of diversity, it will inform parameterizations of the relationship between light, temperature and nutrient acquisition traits for ocean simulation modeling.
Acknowledgments
We thank the captain and crew of the R/V Kilo Moana and members of the Hawai'i Ocean Time-series program (HOT181 and HOT212) for technical support with field operations. We also thank Robert D Harper and Hassan Shaleh (University of Southern Maine, Portland, ME, USA) for culturing assistance as well as Libusha Kelly (Albert Einstein College of Medicine, Bronx, NY, USA) for advice on bioinformatics analyses. This work was funded in part by the Gordon and Betty Moore Foundation through Grant GBMF495 to SWC and by the National Science Foundation (Grants OCE-1153588 and DBI-0424599 to SWC, OCE-0928544 to ACM, OCE-0851288 to LRM and OCE-9417071 to LC). AGK was supported by the NSF Graduate Research Fellowship Program (DGE-1321846). This article is a contribution from the NSF Center for Microbial Oceanography: Research and Education (C-MORE).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren NA, Rocap G. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl Environ Microbiol. 2006;72:7193–7204. doi: 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biller SJ, Berube PM, Berta-Thompson JW, Kelly L, Roggensack SE, Awad L, et al. Genomes of diverse isolates of the marine cyanobacterium Prochlorococcus. Scientific Data. 2014;1:140034. doi: 10.1038/sdata.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WK, Platt T, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- Bragg JG, Dutkiewicz S, Jahn O, Follows MJ, Chisholm SW. Modeling selective pressures on phytoplankton in the global ocean. PLoS One. 2010;5:e9569. doi: 10.1371/journal.pone.0009569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitenhuis ET, Li WKW, Vaulot D, Lomas MW, Landry MR, Partensky F, et al. Picophytoplankton biomass distribution in the global ocean. Earth Syst Sci Data. 2012;4:37–46. [Google Scholar]
- Campbell L, Nolla HA, Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol Oceanogr. 1994;39:954–961. [Google Scholar]
- Casey JR, Lomas MW, Mandecki J, Walker DE. Prochlorococcus contributes to new production in the Sargasso Sea deep chlorophyll maximum. Geophys Res Lett. 2007;34:L10604. [Google Scholar]
- Cavender-Bares KK, Mann EL, Chisholm SW, Ondrusek ME, Bidigare RR. Differential response of equatorial Pacific phytoplankton to iron fertilization. Limnol Oceanogr. 1999;44:237–246. [Google Scholar]
- Chisholm SW, Frankel SL, Goericke R, Olson RJ, Palenik B, Waterbury JB, et al. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch Microbiol. 1992;157:297–300. [Google Scholar]
- Coleman ML, Chisholm SW. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15:398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc Natl Acad Sci USA. 2010;107:18634–18639. doi: 10.1073/pnas.1009480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, DeLong EF, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- Collier JL, Lovindeer R, Xi Y, Radway JC, Armstrong RA. Differences in growth and physiology of marine Synechococcus (Cyanobacteria) on nitrate versus ammonium are not determined solely by nitrogen source redox state. J Phycol. 2012;48:106–116. doi: 10.1111/j.1529-8817.2011.01100.x. [DOI] [PubMed] [Google Scholar]
- Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingersch R, Philosof A, Mejuch T, Glaser F, Alalouf O, Shoham Y, et al. Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 2012;6:827–834. doi: 10.1038/ismej.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the Author. Department of Genome Sciences, University of Washington: Seattle; 2005. [Google Scholar]
- Flombaum P, Gallegos JL, Gordillo RA, Rincón J, Zabala LL, Jiao N, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follows MJ, Dutkiewicz S, Grant S, Chisholm SW. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315:1843–1846. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]
- Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol. 2003;69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller NJ, West NJ, Marie D, Yallop M, Rivlin T, Post AF, et al. Dynamics of community structure and phosphate status of picocyanobacterial populations in the Gulf of Aqaba, Red Sea. Limnol Oceanogr. 2005;50:363–375. [Google Scholar]
- García-Fernández JM, de Marsac NT, Diez J. Streamlined regulation and gene loss as adaptive mechanisms in Prochlorococcus for optimized nitrogen utilization in oligotrophic environments. Microbiol Mol Biol Rev. 2004;68:630–638. doi: 10.1128/MMBR.68.4.630-638.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico AR, Cerniglia GE, Biaglow JE, Stevens CW, Koch CJ. The importance of sodium pyruvate in assessing damage produced by hydrogen peroxide. Free Radical Bio Med. 1997;23:426–434. doi: 10.1016/s0891-5849(97)00113-5. [DOI] [PubMed] [Google Scholar]
- Gruber N.2008The marine nitrogen cycle: overview and challengesIn: Capone DG, Bronk DA, Mulholland MR, Carpenter EJ, (eds)Nitrogen in the Marine Environment Academic Press: Burlington, MA; 1–50. [Google Scholar]
- Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in Cyanobacteria. Biochim Biophys Acta. 2001;183:411–425. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Wilhelm SW, Harvey HR, Taylor K, Jiao N, Chen F. Novel lineages of Prochlorococcus and Synechococcus in the global oceans. ISME J. 2012;6:285–297. doi: 10.1038/ismej.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Shinomura Y, Natori Y, Igarashi Y, Sohrin R, Suzuki Y. Relationship between salinity and nutrients in the subsurface layer in the Suruga Bay. J Oceanogr. 2005;61:721–732. [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Kamennaya NA, Chernihovsky M, Post AF. The cyanate utilization capacity of marine unicellular Cyanobacteria. Limnol Oceanogr. 2008;53:2485–2494. [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- Kelly L, Huang KH, Ding H, Chisholm SW. ProPortal: a resource for integrated systems biology of Prochlorococcus and its phage. Nucleic Acids Res. 2012;40:D632–D640. doi: 10.1093/nar/gkr1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lavin P, González B, Santibáñez JF, Scanlan DJ, Ulloa O. Novel lineages of Prochlorococcus thrive within the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol Rep. 2010;2:728–738. doi: 10.1111/j.1758-2229.2010.00167.x. [DOI] [PubMed] [Google Scholar]
- Lomas MW, Lipschultz F. Forming the primary nitrite maximum: nitrifiers or phytoplankton. Limnol Oceanogr. 2006;51:2453–2467. [Google Scholar]
- Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER, et al. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 2010;4:1252–1264. doi: 10.1038/ismej.2010.60. [DOI] [PubMed] [Google Scholar]
- Malmstrom RR, Rodrigue S, Huang KH, Kelly L, Kern SE, Thompson A, et al. Ecology of uncultured Prochlorococcus clades revealed through single-cell genomics and biogeographic analysis. ISME J. 2013;7:184–198. doi: 10.1038/ismej.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann EL, Chisholm SW. Iron limits the cell division rate of Prochlorococcus in the eastern equatorial Pacific. Limnol Oceanogr. 2000;45:1067–1076. [Google Scholar]
- Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11:1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009;106:10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Tai AP, Veneziano D, Primeau F, Chisholm SW. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- Marty J-C, Chiavérini J, Pizay M-D, Avril B. Seasonal and interannual dynamics of nutrients and phytoplankton pigments in the western Mediterranean Sea at the DYFAMED time-series station (1991-1999) Deep Sea Res Part II. 2002;49:1965–1985. [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, Lindell D, et al. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr Meth. 2007;5:353–362. [Google Scholar]
- Moore LR, Post AF, Rocap G, Chisholm SW. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol Oceanogr. 2002;47:989–996. [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–710. [Google Scholar]
- Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One. 2011;6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühling M. On the culture-independent assessment of the diversity and distribution of Prochlorococcus. Environ Microbiol. 2012;14:567–579. doi: 10.1111/j.1462-2920.2011.02589.x. [DOI] [PubMed] [Google Scholar]
- Ohashi Y, Shi W, Takatani N, Aichi M, Maeda SI, Watanabe S, et al. Regulation of nitrate assimilation in cyanobacteria. J Exp Bot. 2011;62:1411–1424. doi: 10.1093/jxb/erq427. [DOI] [PubMed] [Google Scholar]
- Olson RJ, Chisholm SW, Zettler ER, Altabet MA, Dusenberry JA. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep Sea Res Part I. 1990;37:1033–1051. [Google Scholar]
- Olson RJ, Vaulot D, Chisholm SW. Marine phytoplankton distributions measured using shipboard flow cytometry. Deep Sea Res Part I. 1985;32:1273–1280. [Google Scholar]
- Partensky F, Blanchot J, Vaulot D.1999Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a reviewIn: Charpy L, Larkum AWD (eds)Marine CyanobacteriaBulletin de l Institut oceanographique de Monaco, No. spécial 19Musée océanographique: Monaco; 457–475. [Google Scholar]
- Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar Sci. 2010;2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- Penno S, Campbell L, Hess WR. Presence of phycoerythrin in two strains of Prochlorococcus (Cyanobacteria) isolated from the subtropical north Pacific Ocean. J Phycol. 2000;36:723–729. doi: 10.1046/j.1529-8817.2000.99203.x. [DOI] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito MA, Moffett JW, Chisholm SW, Waterbury JB. Cobalt limitation and uptake in Prochlorococcus. Limnol Oceanogr. 2002;47:1629–1636. [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan DJ, West NJ. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol. 2002;40:1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- Shimada A, Nishijima M, Maruyama T. Seasonal appearance of Prochlorococcus in Suruga Bay, Japan. J Oceanogr. 1995;51:289–300. [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, Delong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- Tolonen AC, Aach J, Lindell D, Johnson ZI, Rector T, Steen R, et al. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Syst Biol. 2006;2:53. doi: 10.1038/msb4100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature. 1999;400:525–531. [Google Scholar]
- Vaulot D, Lebot N, Marie D, Fukai E. Effect of phosphorus on the Synechococcus cell cycle in surface Mediterranean waters during summer. Appl Environ Microbiol. 1996;62:2527–2533. doi: 10.1128/aem.62.7.2527-2533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, Partensky F. Cell cycle distributions of prochlorophytes in the north western Mediterranean Sea. Deep Sea Res Part I. 1992;39:727–742. [Google Scholar]
- Waterbury JB, Willey JM. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 1988;167:100–105. [Google Scholar]
- Williams EZ, Campbell L, DiTullio G.1999. The nitrogen specific uptake of three strains of Prochlorococcus. Presented at the American Society of Limnology and Oceanography Aquatic Sciences Meeting; 4 February 1999, Santa Fe, NM.
- Williams KP. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sunda W, Boyle EA, Karl DM. Phosphate depletion in the western North Atlantic Ocean. Science. 2000;289:759–762. doi: 10.1126/science.289.5480.759. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EM, Zubkov MV, Scanlan DJ. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.