Abstract
Background and Purpose
Colorectal and prostate cancers are two of the most common types and cause of a high rate of deaths worldwide. Therefore, any strategy to stop or at least slacken the development and progression of malignant cells is an important therapeutic choice. The aim of the present work is the identification of novel cancer chemotherapy agents. Nowadays, many different drug discovery approaches are available, but this paper focuses on Molecular Topology, which has already demonstrated its extraordinary efficacy in this field, particularly in the identification of new hit and lead compounds against cancer. This methodology uses the graph theoretical formalism to numerically characterize molecular structures through the so called topological indices. Once obtained a specific framework, it allows the construction of complex mathematical models that can be used to predict physical, chemical or biological properties of compounds. In addition, Molecular Topology is highly efficient in selecting and designing new hit and lead drugs. According to the aforementioned, Molecular Topology has been applied here for the construction of specific Akt/mTOR and β-catenin inhibition mathematical models in order to identify and select novel antitumor agents.
Experimental Approach
Based on the results obtained by the selected mathematical models, six novel potential inhibitors of the Akt/mTOR and β-catenin pathways were identified. These compounds were then tested in vitro to confirm their biological activity.
Conclusion and Implications
Five of the selected compounds, CAS n° 256378-54-8 (Inhibitor n°1), 663203-38-1 (Inhibitor n°2), 247079-73-8 (Inhibitor n°3), 689769-86-6 (Inhibitor n°4) and 431925-096 (Inhibitor n°6) gave positive responses and resulted to be active for Akt/mTOR and/or β-catenin inhibition. This study confirms once again the Molecular Topology’s reliability and efficacy to find out novel drugs in the field of cancer.
Introduction
The US-National Institute of Health estimated the direct cost on oncology care to be $89 billion in 2007 [1]. Surgery, hospitalization, physician visits, imaging, chemotherapy, radiation and biologic therapy are derived costs from oncology care [2]. In 2014, approximately 18% of US gross domestic product has been spent on healthcare and 5% of that keeps being for cancer care [3]. Ten to fifteen percent of total spending on oncology care is linked to cancer drugs [2]. Therefore, developing quality cost-saving strategies for cancer care is an imperative [3].
There are many approaches to fight cancer ranging from chemoprevention (strategy of blocking or slowing the onset of premalignant tumors with relatively nontoxic chemical substances [4]) to chemotherapy, radiotherapy or finally surgical oncology.
Ones of the most common and deadly forms of cancer, colorectal cancer (CRC) and prostate cancer (PtC) were selected as main targets for this study. In order to give an idea of the importance of these forms of cancer, a brief description of their incidence and burden is following.
Worldwide, about 2.1 million people were diagnosed with CRC in 2008, placing it second among the most frequent cancer in women and third in men [5]. Moreover, it is the third most common cause of cancer death worldwide, with more than 600,000 deaths per year [6]. The prevalence of CRC is expected to increase significantly in most developed countries as a result of the growing population belonging to the elderly, since the incidence of CRC increases with age [5].
On the other hand, prostate cancer is the second most common malignancy in men, with more than 900,000 newly diagnosed cancer cases and about 260,000 cancer deaths in 2008 [7]. The incidence of this deadly disease has significantly increased in recent years because of the widespread screening for prostate-specific antigen (PSA), which allows early detection of tumors that otherwise, might remain undetected. In the USA, 90% of patients with prostate cancer who presented localized lesions at the diagnosis, usually have a good prognosis after treatment. However, by 5 years, nearly 30% of treated patients exhibit a rise in PSA levels and evidence of recurrent disease [7].
Considering what has been said up to now, it is not difficult to understand the importance of trying to reduce the incidence and progression of CRC and PtC, so to prevent the inevitable health-spending burden that their treatment and follow-up of patients would imply. Again, the main objective of this work consists in try to discover novel drugs for cancer treatment.
There are many targets involved in CRC and PtC which could be potentially interesting in order to select novel chemotherapeutic compounds. This work focused on two fundamental signaling pathways: PI3K/Akt/mTOR and Wnt/β-catenin (Fig 1).
Fig 1. PI3K/Akt/mTOR pathway related to cancer onset and progression [11].
The PI3K/Akt/mTOR Pathway
Phosphatidylinositol-3-kinase (PI3K) is a key enzyme in the control of cell growth and proliferation. The most common form of this enzyme is activated by the actions of growth factors receptors. By forming triply phosphorylated inositols attracts Akt which becomes phosphorylated by phosphorinositide-dependent kinase (PDKs). Akt then proceeds to phosphorylate a variety of substrates, including the mammalian target of rapamycin (mTOR) regulating cell proliferation, survival and size.
Several studies have shown changes in Akt activity or expression in human precancerous tissues in precancerous prostatic intraepithelial neoplasia and neoplastic colonic epithelium [8]. Moreover, the PI3K/Akt and mTOR signaling pathways are demonstrated to be hyper-activated signaling pathways in CRC and PtC cancers [8–15]. Therefore, these signaling pathways are potent targets for inducing cancer cell death [12].
The Wnt/β-Catenin Pathway
In the absence of Wnt, β-catenin is associated with the multi-protein β-catenin destruction complex that includes constitutively active glycogen synthase kinase 3 (GSK3). GSK3 phosphorylates β-catenin, triggering its degradation through the ubiquitin-proteasome pathway. The presence of Wnt, induces β-catenin dissociation of the degradation complex and its translocation into the nucleus, where it binds to T-cell factor (TCF) transcription factors regulating the expression of many genes including Cyclin D1 (CycD1).
Deregulation of Wnt/β-catenin signaling is a hallmark of the majority of sporadic forms of colorectal cancer and results in increased stability of the protein β-catenin [16]. There are several reasons for targeting the Wnt/β-catenin pathway in CRC and PtC. Approximately 90% of colorectal cancers (CRCs) has mutations on the Wnt/β-catenin (canonical pathway). Those are mainly found in the adenomatous polyposis coli (APC) and β-catenin genes and both lead to pathway activation, although other pathway components can also harbor mutations [16]. Accumulation of β-catenin in the nucleus can be detected in > 80% of CRC tumors [16]. Moreover, high levels of nuclear β-catenin have been correlated with a poor prognosis in CRC patients [16].
In PtC, it has been widely demonstrated how androgen receptor (AR) is the major therapeutic target. However, targeting AR alone can result in drug resistance and disease recurrence. Simultaneous targeting of multiple pathways could in principle be an effective approach to treating prostate cancer [17]. Growing evidence indicates that the canonical Wnt/β-catenin pathway plays an important role in prostate tumor-genesis [18]. Especially because AR binds β-catenin directly to stimulate AR-mediated gene transcription, and importantly, the AR gene itself is a transcriptional target of β-catenin [17]. So, the inhibition of both the AR and β-catenin–signaling pathways that are often unregulated in prostate cancer may represent an effective way for PtC treatment.
Molecular Topology in Cancer
Once it is clear the importance to find new chemotherapy agents against CRC and PtC, the question to be asked is: how could it be done? In the field of Computer Aided Drug Design (CADD) and Quantitative structure–activity relationship (QSAR), the possibilities are many, but the results obtained are seldom satisfactory, especially in the field of cancer. Since, the Drug Design and Molecular Topology Research Unit at the University of Valencia (Spain), has a consolidated background of achievements in cancer drug discovery [19–26], including several patents [27–29], thanks to a computational methodology based on Molecular Topology. In addition to the identification of novel active molecules in PtC [26], our research unit also succeeded in finding new molecules active on the initial stages of colorectal cancer. Moreover, our team has obtained significant results in IBD (inflammatory bowel disease) treatment, by the identification of several compounds inhibiting inflammation mediators like IL-6, NF-ĸβ and COX-2 [30–34].
The use of QSAR with topological indices as regression variables has proved to be an excellent approach for a fast and accurate prediction of many physicochemical and biological properties [35–37]. Molecular Topology (MT) can be defined as a part of mathematical chemistry consisting of the topological description of molecular structures. Such description deals basically with the connectivity of the atoms forming the molecule that must yield numerical values which are invariant under deformation of the structure. Features such as bond lengths, dihedral angles, energies or any other sort of physical or geometrical magnitudes are not being considered in this scenario. Molecular descriptors are ‘numbers that characterize a specific aspect of a molecule’ [38]. That is, they are numbers containing information derived from the structural representation of the molecule.
MT’s singularity lies in the following reasons:
Molecular structure is described within a mathematical framework [39].
It may find out new drugs, either by screening compound’s databases or by designing novel compounds following the reverse process (properties → structure).
It can be easily computerized.
The use of MT as a tool for the design and selection of new drugs has been our team’s task since the early 1980’s [38–42].
Materials and Methods
Molecular Topology Model
Obtaining a MT-QSAR model for the identification of novel cancer chemotherapeutic agents consists of the following steps (Fig 2):
Step I: Creating an in-house database of compounds active on the selected targets (Akt and β-catenin).
Step II: Calculation of topological descriptors using Dragon software [43].
Step III: Splitting data set into two groups: training and test set.
- Step IV: Models were built up following the linear discriminant analysis (LDA) approachment just considering the training set. The model’ labels were:
- Model #1: Akt inhibition model for natural compounds.
- Model #2: Akt inhibition model for comercial compounds.
- Model #3: β-catenin inhibition model for natural compounds.
- Model #4: β-catenin inhibition model for comercial compounds.
Step V: Development of an external model validation using the test set for each discriminant function.
Step VI: Carrying out a virtual screening in SPECS libraries. Search for potential Akt and β-catenin inhibitors as novel chemotherapeutics cancer agents.
Step VII: In vitro testing of the selected compounds in colorectal and prostate cancer cell lines.
Fig 2. Scheme of Akt and β-catenin inhibitors research through Molecular Topology by virtual screening on SPECS databases.
Compound analysis
Model #1: Akt inhibition for natural compounds
The QSAR model was developed from a training set composed by 155 compounds (32 actives and 123 inactives) with heterogeneous molecular structures. This model was then tested against a set of 39 compounds consisting of 8 natural Akt inhibitors and 31 Akt-inactive compounds.
While we are confident that the active group compounds have been tested and exhibit pharmacological activity against Akt, regarding the inactive group compounds we don’t know for sure if they have been tested against such pharmacological activity. However, given that the number of compounds in the inactive set is significantly large, the risk of misclassification is negligible.
All compounds were obtained from different sources [44–80] and from a database named MicroSource Pure Natural Products Collection [81].
Model #2: Akt inhibition for comercial compounds
The QSAR model was developed from a training set composed by 647 compounds (20 actives and 627 inactive) with significant structural heterogeneity. External validation of this model was performed with a set of 205 compounds consisting of 6 active on Akt inhibition and 199 inactive.
As mentioned above, whereas we cannot ensure inactivity for all compounds belonging to the inactive set, the risk of false inactive is very low.
All compounds were obtained from Akt Selleckchem database [82], literature [83–97] and MicroSource US Drugs Collection [98].
Model #3: β-catenin inhibition for natural compounds
The QSAR model was developed from a structurally heterogeneous training set made of 153 compounds (50 actives and 103 inactives). This model was validated against a set of 27 compounds consisting of 10 natural β-catenin inhibitors and 17 17 inactive.
All compounds were obtained from literature [99–159] and from a database named MicroSource Pure Natural Products Collection [81].
Model #4: β-catenin inhibition for comercial compounds
Finally, the last QSAR model developed was formed by a training set composed by 1047 commercial compounds (93 actives and 954 inactive). External validation of the model was made on a test set of 207 compounds consisting of 17 β-catenin inhibitors and 190 inactive compounds.
As in the previous cases, the probability of activity within the inactive set is negligible due to its size.
All compounds were obtained from β-catenin Selleckchem database [143], literature [160–201] and the database MicroSource US Drugs Collection [98].
Molecular descriptors
Topological descriptors codify information about molecular structure in a purely numerical way. This numerical format significantly eases the search of other molecules showing similar properties, thereby making easier the discovery of new drugs.
The 2D structure of each compound was drawn using the ChemDraw Ultra package (version 10.0) [202]. Every compound was characterized by a set of descriptors including constitutional and topological descriptors. Among the last stand the edge adjacency indices, walk and path counts, connectivity indices and topological charge indices [203]. Other graph-theoretical descriptors were also calculated but are not depicted here because their lack of effectiveness. All indices were calculated with Dragon software version 5.4 [43] and their values for every compound included in this study (training, test, and virtual screening set) are shown in the Supplementary Material.
Other complementary models (not shown here) were also applied to find molecules showing drug-like profiles. Among the properties considered in these models stand water solubility, toxicity, oral absorption, etc… (details on these additional models can be disclosed under request). Dragon software version 5.4 and in-house software were used to obtain the models.
Modeling techniques
Linear discriminant analysis (LDA) was used to distinguish between the active and inactive compounds. It is a statistical method to find the best linear combination of variables (TIs in our case) that better distinguish between two or more categories or objects (in our case active or inactive as Akt or β-catenin inhibition). We start from a training set of compounds in which everyone is classified either as active or inactive. LDA was then applied to these two groups to get the discriminant function (DF) that better separate the two categories. The software used was Statistica 9.0 [204].
TIs were used as the independent variables and the two groups were balanced so that both (active and inactive) show the same weight, regardless of the number of compounds in each one.
The discriminant capability was assessed as the percentage of correct classifications in each set of compounds. The classification criterion was based on the minimum Mahalanobis distance (distance of each case to the mean of all the cases in a category) [205] and the quality of the discrimination was evaluated using the Wilks’ parameter, λ [206], which was obtained by multivariate analysis of variance that tests the equality of group means for the variable in the discriminant model. The shorter the Wilks’ parameter value, the smaller the overlap of the active and inactive (λ = 0 would mean a perfect separation between the groups).
The descriptors’ selection was carried out according to the value of the Fisher–Snedecor parameter (F) [207], which establishes the relevance of candidate variables. The descriptors input to compute the linear classification function are chosen in a stepwise-manner: at each step, the variable making the largest contribution to the division of the groups is introduced into the discriminant equation (or the variable that makes the smallest contribution is removed).
The validation of the selected function was done using an external test set. Approximately 20% of the data set was randomly selected as test set and therefore they were not used to create the model.
Pharmacological activity distribution diagrams
A pharmacological distribution diagram (PDD) is a graphical representation that provides a direct way of visualizing the zones of minimum overlap between active and inactive compounds, as well as the region in which the probability of finding active compounds is maximum [208].
From a different perspective, a PDD is a frequency distribution diagram of dependent variables in which the ordinate represents the expectancy (probability of activity) and the abscissa the DF values in the range. For an arbitrary range of values of a given function, the expectancy of activity can be defined as Ea = a/(i +1), where a is the number of active compounds in the range divided by the total number of active compounds and i is the number of inactive compounds in the interval divided by the total number of inactive compounds. The expectancy of inactivity is defined likewise as Ei = i/(a+1). By means of these diagrams, it is easy to visualize the intervals in which there is a maximum probability to find new active compounds as well as the minimum probability to find inactive compounds.
Topological virtual screening
Once the models are set up, it is possible to carry out a virtual screening in databases to select novel Akt and β-catenin inhibitors. Two commercial SPECS databases were screened, the first was made of some one thousand natural compounds and the second containing about 200 000 drug-like small molecules [209].
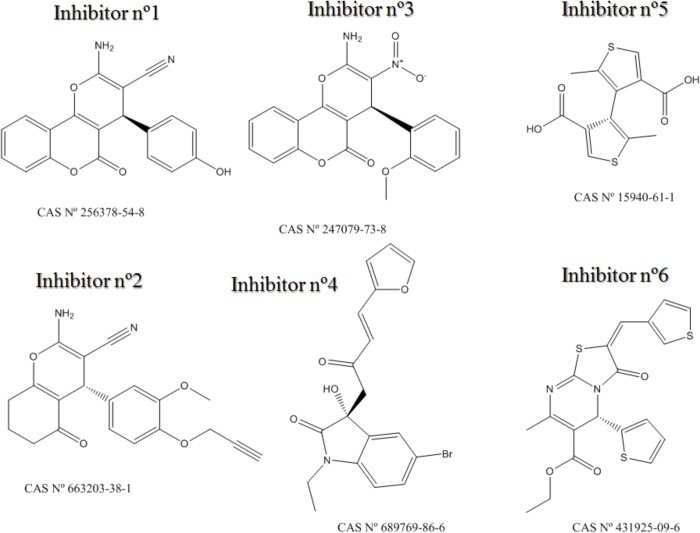
This screening led to the identification of 6 compounds showing potential chemotherapeutic activity as Akt and β-catenin inhibitors. Later on, the selected molecules underwent in vitro tests to confirm the expected activity.
In Vitro Assays
Reagents and antibodies
Chemicals compounds with CAS n° 256378-54-8 (Inhibitor n°1), 663203-38-1 (Inhibitor n°2), 247079-73-8 (Inhibitor n°3), 689769-86-6 (Inhibitor n°4), 15940-61-1 (Inhibitor n°5) and 431925–096 (Inhibitor n°6) were obtained from Specs (Zoetermeeer, The Netherlands) and dissolved in DMSO. LY294002 and AT7519 were supplied by Deltaclon (Madrid, Spain) a Selleckchem supplier. Akt inhibitor IV and FH535 were obtained from Sigma-Aldrich (Madrid, Spain). Recombinant Human Wnt-3a (R&D Systems, Minneapolis, USA) was reconstituted at 200 μg/mL in sterile PBS containing 0.1% bovine serum albumin. The polyclonal antibodies against phospho-mTOR, mTOR, phospho-Akt-ser473, Akt and anti-rabbit IgG were from Cell Signaling Technology (Danvers, MA, USA) and anti-β-catenin was obtained from Santa Cruz Biotechnology (Bergheimer, Heidelberg, Germany). Monoclonal antibody against cyclin D1 was from EMD Millipore (Darmstadt, Germany) and anti-β-tubulin and anti-mouse IgG were from Sigma (Madrid, Spain). Anti-rabbit IgG conjugated to Alexa488 was from Invitrogen (Life Technologies, Alcobendas, Spain).
Cell cultures
PC3 cell line derived fron human prostate adenocarcinoma was obtained from ATCC (CRL-1435, Rockville, MD, USA) and maintained in RPMI medium containing 10% foetal bovine serum and 1% penicillin/streptomycin. Cells were used between passages 10 and 20 and seeded at a density of 20,000 cells/cm2. Sixteen hours post-seeding, medium was changed to serum free medium and treatments were performed 24 hours later. HT-29 cell line derived from a human colon adenocarcinoma (ATCC, Rockville, MD), was used between passages 15 and 25 and cultured in DMEM glucose concentration 4.5 g/L supplemented with 20% fetal bovine serum, penicillin (100 U/mL) and streptomycin sulfate (100 μg/mL) in a humidified 5% CO2 atmosphere at 37 °C. According to the solubility, the compounds assayed were dissolved in a maximum DMSO concentration of 0.5%.
Cytotoxicity assay
The effect of compounds on cell viability was evaluated with the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide (MTT) assay [210]. After 24 h seeding of HT-29 cells, the medium was replaced with fresh DMEM + 0.5% FBS. Two hours later, cells were exposed to compounds at the concentrations (1, 10, or 100 μM) in a 96-well microplate at 37 °C for 24 h. Same protocol was applied to PC-3 cells but cells were seeded in 12-well plates and mantained 24 hours in a serum-free medium prior the treatments (20, 50, 100, 150 and 200 μM).
The medium was then removed and 100 μL per well of a 0.5 mg/mL solution of MTT was added. The resulting solution was incubated at 37 °C until blue deposits were visible and then, this colored metabolite was dissolved in dimethyl sulfoxide (DMSO). Absorbance was measured at 490 nm with a Labsystems Multiskan EX plate reader (Helsinki, Finland). The results were expressed in absolute absorbance readings; a decrease in absorbance indicated a reduction in cell viability. The percentage of cell viability was determined as follows:
The experiments were performed a minimum of three times with three replicates per concentration.
Western blot
After 1, 4 or 48 hours of treatments according to the experiments, cells were lysed in ice-cold lysis buffer (50 mM Tris pH 7.4, 0.8 M NaCl, 5 mM MgCl2, 0.1% Triton X-100, 1 mM PMSF, 10 μg/mL soybean trypsin inhibitor, 1 μg/mL aprotinin, and 5 μg/mL leupeptin), and cleared by microcentrifugation. Protein concentration was determined by the Bradford method and equivalent protein amounts of each sample were loaded onto SDS-PAGE gels and transferred to PVDF membrane. Immunoblot analysis was performed followed by enhanced chemoluminescence detection as previously described [211–212].
Confocal microscopy
Cells seeded onto 1.5 mm glass coverslips were treated for 4 hours with 50 μM of the some of the selected potential chemotherapy agents: Inhibitors n°1-6. Then cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and incubated with β-catenin antibody overnight at 4˚C. The cells were washed and incubated first with secondary antibody conjugated to Alexa488 and then with 4′-6-Diamidino-2-phenylindole (DAPI) as a nuclear counterstain. Cells were visualized in a Leica TCS SP5 laser-scanning confocal microscope with LAS-AF imaging software, using a 63X oil objective.
Statistical analysis
Statistical analysis was performed with a one-way analysis of variance (ANOVA) and Dunnett’s t test. The results are presented as the mean ± SEM. GraphPad Prism 4.0 software (GraphPad Software Inc., San Diego, CA, USA) was used for all calculations. Data related with PC3 cell lines are presented as the mean ± S.D. of the number of experiments indicated.
Results and Discussion
Topological Models
Model #1: Akt inhibition for natural compounds
The discriminant function (DF1) includes three variables, as shown below:
The parameters accounting for the significance of this equation were:
where N is the number of data compounds; F, Fisher–Snedecor parameter; λ, Wilks’ lambda; p, statistical significance; nR09, number of 9-membered rings; Wap, all path Wiener index; EEig11r, eigenvalue 11 from edge adj. matrix weighted by resonance integrals.
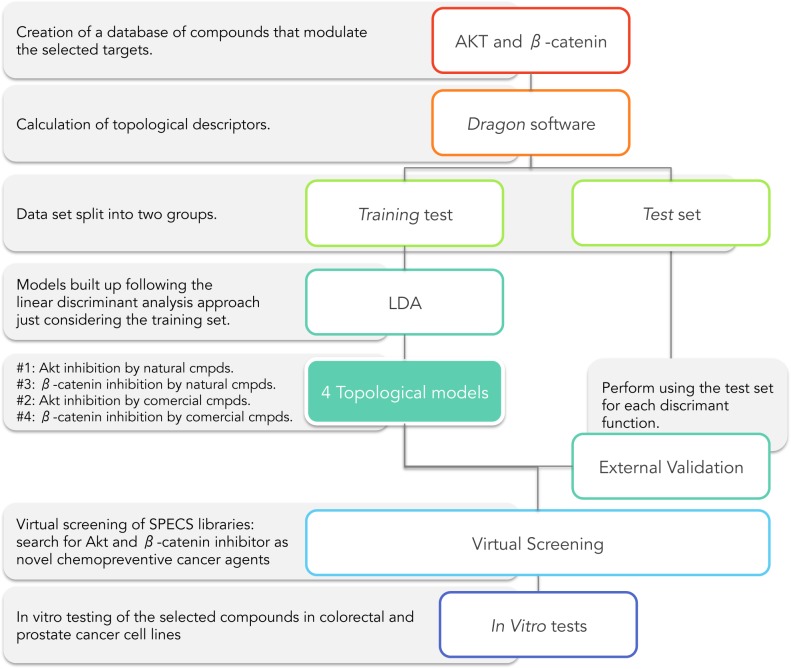
According to DF1, a compound is classified as Akt natural inhibitor if DF1 takes values from 0 to 4; on the other hand, a compound will be labelled as inactive when DF1 takes values between -0.5 and -6 (see Fig 3). Finally, molecules scoring DF1 > 4 or DF1 < -6 will be non-classified by this model. By applying this criterion to the training set composed by 153 compounds, 26 out of 30 natural Akt inhibitors were correctly classified (81% accuracy) while 101 out of 123 inactive compounds were also matched (82% accuracy), as can be seen in Table 1.
Fig 3. Pharmacological distribution diagram for natural Akt inhibitors obtained using the DF1 (the black colour represents Akt inhibitors and the white colour, the compounds without Akt inhibition activity).
Table 1. Classification matrix obtained through the selected DF1 for training and test set.
| PERCENT-CORRECT CLASSIFICATION (%) | COMPOUNDS | |||
|---|---|---|---|---|
| Classified as active | Classified as inactive | Non-classified | ||
| TRAINING SET | ||||
| Active group | 81 | 26 | 4 | _ |
| Inactive group | 82 | 22 | 101 | _ |
| TEST SET | ||||
| Active group | 63 | 5 | 3 | _ |
| Inactive group | 87 | 3 | 27 | 1 |
The best way to evaluate the quality of any DF is to apply it to an external validation group (test set). In our case, this group was made up of 39 compounds (8 active and 31 inactive as Akt inhibitors) which were not included in the training set. Test set was randomly selected with a percentage of 20 of all data set. The model enabled a correct classification of 63% of active group (5 out of 8 compounds) and 87% of the inactive set (27 out of 31 compounds), as outlined in Table 1.
Furthermore, although the strict DF application leads to the loss of some of the active compounds (37%), the important point is that 87% of the inactive compounds were correctly classified, so that the risk of including “false active” is minimized, which means that our model has a high specificity in discovering new Akt natural inhibitors.
Model #2: Akt inhibition for comercial compounds
A new discriminant function (DF2) was used for Akt inhibition by commercial compounds. This function includes five variables as follows:
Statistical data and parameters accounting for the significance of this equation were:
where N is the number of data compounds; F, Fisher–Snedecor parameter; λ, Wilks’ lambda; p, statistical significance; T(O…Br),topological distance between oxygen and bromide; SRW08, self-returning walk count of order 08; MPC04, molecular path count of order 04; piPC02, molecular multiple path count of order 02; piPC05, molecular multiple path count of order 05.
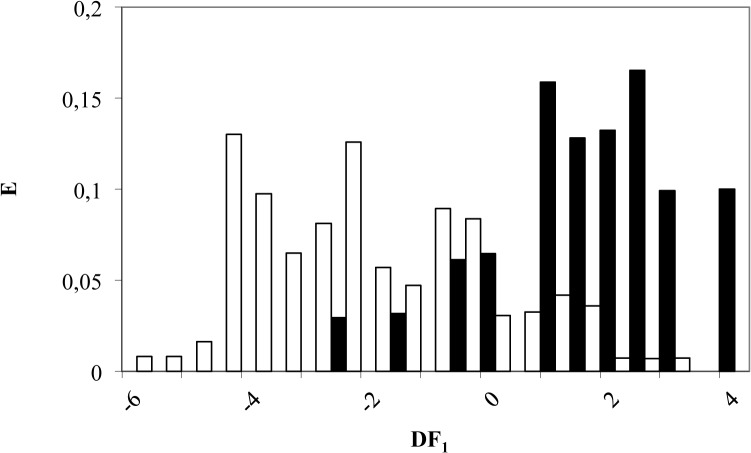
According to DF2, a compound is classified as Akt inhibitor if its DF values range from 1.5 to 7.5. On the other hand, a compound will be labeled as inactive if DF2 stands between 1.5 and -6 (see Fig 4). Finally, molecules scoring DF2> 7.5 or DF2 < -6 are considered as non-classified by this model. By applying the model criterion to the training set composed by 647 compounds, 16 out of 20 (80%) were correctly classified as Akt inhibitors, while 452 out of 627 inactive compounds were also well classified (72% accuracy), as can be seen in Table 2.
Fig 4. Pharmacological distribution diagram for Akt inhibitors obtained using the DF2 (the black colour represents Akt inhibitors and the white colour, the compounds without Akt inhibition activity).
Table 2. Classification matrix obtained through the selected DF2 for training and test set.
| PERCENT-CORRECT CLASSIFICATION (%) | COMPOUNDS | |||
|---|---|---|---|---|
| Classified as active | Classified as inactive | Non-classified | ||
| TRAINING SET | ||||
| Active group | 80 | 16 | 4 | _ |
| Inactive group | 72 | 173 | 452 | 2 |
| TEST SET | ||||
| Active group | 83 | 5 | 1 | _ |
| Inactive group | 81 | 38 | 161 | 1 |
An external validation of DF2 was performed over 206 compounds (6 active and 200 inactive molecules as Akt inhibitors) not included in the training set. Twenty-percent of all data were used for test set. DF2 was able to classify correctly 83% of the active group (5 out of 6 compounds) and 81% of inactive (161 out of 200 compounds) as illustrated in Table 2.
Therefore, application of DF leads to discard just 17% of the active compounds and 19% of inactive compounds. In other words, this model has a high specificity and sensitivity.
Model #3: β-catenin inhibition for natural compounds
To select β-catenin natural inhibitors another discriminant function was developed (DF3). This function includes five variables, as shown below:
Statistical data and parameters accounting for the significance of this equation were as follows:
where N is the number of data compounds; F, Fisher–Snedecor parameter; λ, Wilks’ lambda; p, statistical significance; Dz, Pogliani index; S2K, 2-path Kier alpha-modified shape index; PCR, ratio of multiple path count over path count; X2sol, solvation connectivity index chi-2; JGI4, mean topological charge index of order 4.
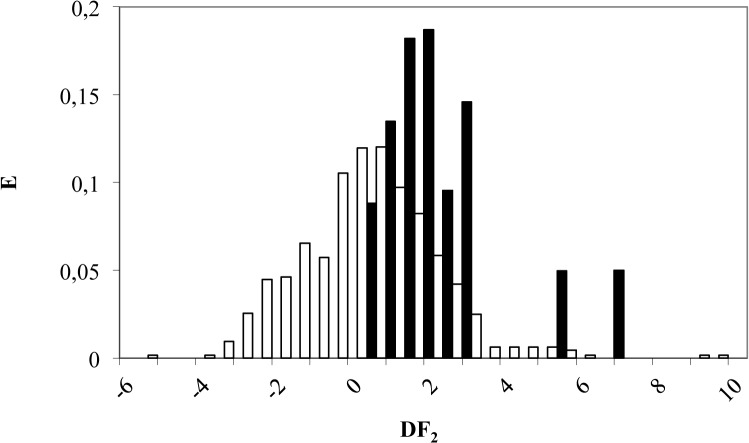
According to DF3, a compound is classified as β-catenin natural inhibitor if it takes values between 0 and 5. On the other hand, a compound will be labeled as inactive when DF3 adopt values from 0 to -5. Finally, molecules scoring DF3 > 5 or DF3< -5 will be non-classified by this model (see Fig 5). Applying the above criteria to a training set composed by 111 compounds, 31 out of 40 natural β-catenin inhibitors were correctly classified for the model (78% correctly classified) while 52 out of 71 inactive compounds were also well classified (73% accuracy), as can be seen in Table 3.
Fig 5. Pharmacological distribution diagram for natural β-catenin inhibitors obtained using the DF3 (the black color represents natural β-catenin inhibitors and the white color, the compounds without β-catenin inhibition activity).
Table 3. Classification matrix obtained through the selected DF3 for training and test set.
| PERCENT-CORRECT CLASSIFICATION (%) | COMPOUNDS | |||
|---|---|---|---|---|
| Classified as active | Classified as inactive | Non-classified | ||
| TRAINING SET | ||||
| Active group | 78 | 31 | 9 | _ |
| Inactive group | 73 | 19 | 52 | _ |
| TEST SET | ||||
| Active group | 67 | 16 | 6 | 2 |
| Inactive group | 88 | 2 | 15 | _ |
The validation of the DF was performed on an external group (test set). The group was made up of 41 compounds (24 active and 17 inactive as β-catenin inhibitors) not previously included in the training set. The test set was randomly selected as 20% of all data set. In this case, the model was able to correctly classify 67% of active group (16 out of 24 compounds) and 88% of the inactive one (15 out of 17 compounds) as can be seen in Table 3.
As in previous cases, the strict DF application results in the loss of 33% of the active compounds but 88% of the inactive natural compounds were correctly classified, thereby minimizing the risk of including “false active” compounds. That confirms our model’s high specificity to predict β-catenin natural inhibitors.
Model #4: β-catenin inhibition by comercial compounds
A discriminant function modeling β-catenin inhibition by commercial molecules was finally calculated (DF4). This equation includes four variables, as shown below:
Statistical data and parameters accounting for the significance of this equation were as follows:
where N is the number of data compounds; F, Fisher–Snedecor parameter; λ, Wilks’ lambda; p, statistical significance; SCBO, sum of conventional bond orders (H-depleted); nN, number of Nitrogen atoms; ZM1, First Zagreb index M1; GGI4, topological charge index of order 4.
A compound is classified as β-catenin inhibitor if DF4 takes values from 0 to 4.5, whereas a compound will be labeled as inactive if DF4 adopt values between 0 and -4. Finally, molecules scoring DF4 > 4.5 or DF4 < -4 will be non-classified (see Fig 6). For the entire set of 840 compounds, 48 out of 73 β-catenin inhibitors (66%) were correctly classified by the model, whilst 570 out of 764 inactive compounds were also well classified (75% accuracy), as illustrated in Table 4.
Fig 6. Pharmacological distribution diagram for β-catenin inhibitors obtained using the DF4 (the black color represents β-catenin inhibitors and the white color, the compounds without β-catenin inhibition activity).
Table 4. Classification matrix obtained through the selected DF4 for training and test set.
| PERCENT-CORRECT CLASSIFICATION (%) | COMPOUNDS | |||
|---|---|---|---|---|
| Classified as active | Classified as inactive | Non-classified | ||
| TRAINING SET | ||||
| Active group | 66 | 48 | 25 | _ |
| Inactive group | 75 | 192 | 570 | 2 |
| TEST SET | ||||
| Active group | 88 | 14 | 2 | _ |
| Inactive group | 74 | 49 | 141 | _ |
In this case, the external validation set included 206 compounds (16 active and 190 inactive). Test set was a random selection of 20% of all data set. The model was able to correctly classify 88% of the active group (14 out of 16 compounds) and 74% of the inactive one (141 out of 190 compounds) as shown in Table 4.
According to this results, we can conclude that this model presents a higher sensitivity (88%) but a less satisfactory specificity since only 74% of the inactive compounds were recorded, missing the remaining 26%. This could lead to select “false active” compounds, but it can be avoided by applying the four models stepwise.
Topological virtual screening
Based on the four discriminant models described (DF1-4), a virtual screening was carried out on two commercial chemical libraries from SPECS. Table 5 shows the compounds selected (naturals and synthetics). DF1 and DF3 were applied only to the natural compounds.
Table 5. Values of DF and probability of activity as Akt and β-catenin inhibitors of the selected anti-cancer agents after carried on a virtual screening of SPECS natural and screening compounds databases.
| COMPOUNDS | CAS registry n° | Akt natural inh model | Akt inh. model | β-catenin natural inh. model | β-catenin inh. model | ||||
|---|---|---|---|---|---|---|---|---|---|
| DF1 | P. (Activ.) | DF2 | P. (Activ.) | DF3 | P. (Activ.) | DF4 | P. (Activ.) | ||
| Inhibitor n°1* | 256378-54-8 | 1.68 | 0.843 | 1.67 | 0.699 | 1.75 | 0.851 | 1.55 | 0.82 |
| Inhibitor n°2 | 663203-38-1 | 3.97 | 0.67 | 1.31 | 0.781 | ||||
| Inhibitor n°3 | 247079-73-8 | 2.52 | 0.769 | 1.24 | 0.769 | ||||
| Inhibitor n°4 | 689769-86-6 | 3.02 | 0.921 | 0.36 | 0.582 | ||||
| Inhibitor n°5 | 15940-61-1 | 2.24 | 0.62 | 0.77 | 0.679 | ||||
| Inhibitor n°6 | 431925-09-6 | 4.28 | 0.914 | 0.19 | 0.539 | ||||
DF: discriminant function; P.(Activ.): probability of being classified as active by the model.
*Natural compound.
In vitro assays
Cytotoxicity assay: HT-29 and PC3 cell line
HT-29 cells were exposed to the selected Akt and β-catenin potential inhibitors selected by the DF1-4 models, in order to determine cell viability. As shown in Table 6, several among the selected compounds were better or similar to the reference β-catenin inhibitor (FH535), Akt inhibitor (LY294002) and multi-CDK inhibitor (AT7519). For instance, Inhibitor n°4 at 100 μM is able to kill 48% of HT-29 cells. This compared with FH535, LY294002 and AT7519 at 100 μM inhibition rates represent 38%, 51% and 36%, respectively. It’s quite a promising result in colorectal cancer cells inhibition.
Table 6. Effect of selected anti-cancer agents on the HT-29 cell viability.
| Group | Concentration (μM) | A ± SEM | Cell viability (%) |
|---|---|---|---|
| Blank | 0 | 0.88 ± 0.02 | - |
| Vehicule (DMSO) | 1 | 0.86 ± 0.01 | 97 |
| 10 | 0.86 ± 0.07 | 98 | |
| 100 | 0.80 ± 0.10 | 91 | |
| Inhibitor N°1 | 1 | 0.84 ± 0.10 | 95 |
| 10 | 0.77 ± 0.07 | 87 | |
| 100 | 0.86 ± 0.06 | 98 | |
| 1 | 0.87 ± 0.13 | 99 | |
| Inhibitor N°2 | 10 | 0.79 ± 0.11 | 89 |
| 100 | 0.82 ± 0.11 | 93 | |
| Inhibitor N°3 | 1 | 0.84 ± 0.10 | 95 |
| 10 | 0.78 ± 0.05 | 88 | |
| 100 | 0.63 ±0.08 | 72 | |
| Inhibitor N°4 | 1 | 0.77 ± 0.05 | 87 |
| 10 | 0.75 ± 0.08 | 85 | |
| 100 | 0.46 ± 0.06** | 52** | |
| Inhibitor N°5 | 1 | 0.85 ± 0.12 | 96 |
| 10 | 0.78 ± 0.09 | 89 | |
| 100 | 0.71 ± 0.07 | 80 | |
| FH535 | 1 | 0.80 ±0.02 | 91 |
| 10 | 0.72 ± 0.06 | 81 | |
| 100 | 0.55 ± 0.08* | 62* | |
| LY294002 | 1 | 0.90 ± 0.03 | 102 |
| 10 | 0.71 ± 0.04 | 81 | |
| 100 | 0.43 ± 0.05*** | 49*** | |
| AT7519 | 1 | 0.59 ± 0.05 | 67 |
| 10 | 0.59 ± 0.02 | 67 | |
| 100 | 0.56 ± 0.01 | 64 |
Values were expressed in function of the blank (untreated cells) and represent mean absorbance ± SEM,
and are representative of at least three independent experiments per group. Differentiation with the blank
group (untreated cells) were determined by mean of a one-way analysis of variance (ANOVA) followed by
Dunnett's multiple comparisons test Dunnett’s t test.
No significant difference with the blank group (P>0.05);
*Significant difference with the blank group (P values from 0.01 to 0.05);
**Very significant (P values from 0.001 to 0.01);
***Extremely significant (P values from 0.0001 to 0.001) n = 3; Dunnett’s t test).
It was investigated whether the selected cancer chemotherapeutics agents had any effect on the proliferation of human prostate cancer cells. PC-3 cells were treated with different doses of the Inhibitors n°1-6 for 48 h and cell viability was monitored by MTT. Results show that Inhibitor n°4 totally blocked the viability of prostate cancer cells (Table 7). Inhibitors n°1 and n°6 also decreased cell viability although in a lesser extent; Inhibitor n°3 exerted a slight decrease whereas compounds Inhibitor n°2 and n°5 didn’t have any effect (Table 7).
Table 7. Effect of selected anti-cancer agents on the PC-3 cell viability.
| Group | Concentration (μM) | A ± SEM | Cell viability (%) |
|---|---|---|---|
| Blank | 0 | 0.4 ± 0.02 | |
| Vehicule (DMSO) | 10 | 0.4 ± 0.03 | 100 |
| Inhibitor N°1 | 50 | 0.35 ± 0.04 | 88 |
| 100 | 0.24± 0.03 | 61** | |
| 200 | 0.16 ± 0.03 | 42*** | |
| Inhibitor N°2 | 50 | 0.37 ± 0.02 | 101 |
| 100 | 0.37± 0.02 | 101 | |
| 200 | 0.34± 0.02 | 92 | |
| Inhibitor N°3 | 50 | 0.32± 0.02 | 83** |
| 100 | 0.32± 0.03 | 83* | |
| 200 | 0.28± 0.02 | 72** | |
| Inhibitor N°4 | 50 | 0.22± 0.01 | 60*** |
| 100 | 0.16± 0.01 | 44** | |
| 200 | 0.01± 0.001 | 3*** | |
| Inhibitor N°5 | 50 | 0.34± 0.001 | 101 |
| 100 | 0.36± 0.001 | 106 | |
| 200 | 0.36± 0.005 | 107 | |
| Inhibitor N°6 | 50 | 0.34± 0.06 | 88 |
| 100 | 0.32± 0.05 | 83* | |
| 200 | 0.27± 0.06 | 69* |
Values were expressed in function of the vehicle-treated cells and represent mean absorbance ± SD, and are representative of at least two experiments performed in duplicate. Differentiation with the blank group (untreated cells) were determined by mean of a Student ‘s T test. No significant difference with the blank group (P>0.05)
*Significant difference with the blank group (P values from 0.01 to 0.05)
**Very significant (P values from 0.001 to 0.01)
***Extremely significant (P values from 0.0001 to 0.001) n = 4)
Akt inhibition
Akt activation involves the phosphorylation of two residues: threonine 308 (Thr308) in the activation loop and serine 473 (Ser473) in the C-terminal hydrophobic motif. The phosphorylation of both of these regulatory sites leads to complete activation of the enzyme. To evaluate the inhibitory ability of the selected compounds against Akt we measured the phosphorylation of the enzyme in serine 347. As shown in Fig 7, Inhibitor n°4 at 1 hour of treatment, induced a decrease in Akt phosphorylation at Ser473. This effect may be compared with the well-known Akt inhibitor IV, a cell-permeable and reversible benzimidazole compound that inhibits Akt phosphorylation/activation. When cells were incubated with the compounds for 48 hours, this effect was even greater and was also observed for Inhibitors n°3 and n°6 (Fig 7).
Fig 7. Effect of of selected anti-cancer agents on Akt/mTOR signaling pathway.
PC-3 cells were treated with vehicle (C) or 50 μM of the selected compounds and proteins were detected by Western blot. Upper panel, a representative image of three different experiments. Lower panel, densitometric values represented as the mean ± S.D. of the three experiments.
To assess the impact of the compounds on downstream molecules of the Akt signaling pathway, we used Western blot analysis to observe phosphorylation status and total protein expression of the mammalian target of rapamycin (mTOR), a key effector downstream of Akt. As observed in Fig 7, all the agents tested except Inhibitor n°5, decreased mTOR phosphorylation, being the Inhibitors n°2, n°4 and n°6 the most effective at 1 hour and Inhibitors n°1, n°2 and n°4 at 48 hours.
These results indicate that all the compounds selected, except Inhibitor n°5, were able to inhibit Akt signaling pathway although with different efficacy.
β-Catenin inhibition
To investigate whether the compounds also inhibited β-catenin pathway we measured β-catenin levels as well as its downstream well-characterized target gene cyclin D1.
In the basal situation, much of the cellular β-catenin is bound to E-cadherin on the cell membrane. Cytosolic β-catenin maintained in an inactive state through its interaction with a large protein complex with several proteins including the GSK-3β. In this situation β-catenin is phosphorylated mainly by GSK-3β and labelled for polyubiquitination and degradation into proteasome. Under these conditions, β-catenin targeted genes are repressed. When the pathway is activated, GSK-3β is inhibited leading to the stabilization and accumulation of cytoplasmic β-catenin, which then enters the nucleus, and induces the expression of its target genes. One of the better characterized β-catenin-regulated gene is Cyclin D1 which promote cell cycle progression. To test the effect of selected compounds on the β-catenin pathway levels of β-catenin and Cyclin D1 were determined by Western blot. Fig 8 shows that the Inhibitors n°4 and n°6 were the most efficacious in inhibiting the expression of Cyclin D1, although Inhibitors n°1, n°2 and n°3 also exhibited a weak repression.
Fig 8. Effect of selected anti-cancer agents on β-Catenin/CyclinD1 signaling pathway.
PC-3 cells were treated with vehicle (C) or 50 μM of the selected compounds and proteins were detected by Western blot. Upper panel, a representative image of three different experiments. Lower panel, densitometric values represented as the mean ± S.D. of the three experiments.
To further investigate the inhibitor role of the selected compounds, PC-3 cells were stimulated with the well-stablished β-catenin/Cyclin D1 pathway activator Wnt3a (Wnt). When cells were stimulated with Wnt, β-catenin was recruited by membrane adherens junctions and accumulated in the nucleus (Fig 9). When cells were pre-treated with the selected compounds β-catenin redistribution was inhibited. This effect was well apreciated in Inhibitors n°2, n°4 and n°6 (Fig 9). Moreover, levels of β-catenin in Inhibitor n°6-treated cells almost desappeared (Fig 9).
Fig 9. Effect of selected anti-cancer agents on β-catenin cellular distribution in PC-3 cells.

PC-3 cells were seeded on glass coverslips and pre-treated for 30 min with 50 μM of the inhibitors and then co-treated with 40 ng/ml Wnt3 (Wnt) for 4 h. Cells were stained with polyclonal antibody anti-beta-catenin followed by Alexa-Fluor488-conjugated anti rabbit IgG as described in methods. Confocal image shown is representative of two experiments.
As seen in the Figs 8 and 9, the selected cancer chemotherapeutic agents were able to impact both the subcellular redistribution and the activity of β-catenin as they were able to inhibit its nuclear-cytoplasmic shuttling and its recruitment at plasma membrane as well as the expression of its target gene CyclinD1. The Inhibitors n°2, n°4 and n°6 were the most effective.
Notably, the compounds that inhibited both Akt and β-catenin had the greatest impact on cell viability supporting the idea that dual inhibitor of the Akt/mTOR pathway and β-catenin may result in a potent anti-proliferative effect against human prostate cancer PC-3 cells, although the putative impact of the compounds in other signaling pathways is not ruled out.
Therefore, after applying several models developed by Molecular Topology approach, six compounds has been selected as potential cancer chemotherapeutic agents. After in vitro study whether they inhibit Akt, mTOR, β-catenin and/or Cyclin D1 at different times of incubation; we can affirm that five out of these six compounds (see Fig 10) inhibit at least one of the studied targets. We can also state that these five compounds affect PC3 cell viability with different potency. And, we can remark also that Inhibitor n°4 also reduce cell viability on HT-29 cell line.
Fig 10. Cancer chemotherapeutic agents selected by Molecular Topology.
Finally, we can confirm that this five selected compounds could be used as cancer agents interacting with Akt/mTOR and Wnt/β-catenin pathways in colorectal and prostate human cancer cell lines.
Although it is evident that complex properties, such as inhibition of Akt or β-catenin, cannot be discussed in such simple “structural” terms, thanks to the QSAR study results, some interesting consequences can be pointed out.
Thus, in equations DF1 to DF4, there are indices more or less explicitly related to the existence of high conjugation (as for instance EEig11r, piPC02 or SCBO). Other descriptors, such as nR09, evaluate the presence of condensed rings. It is noteworthy that most selected compounds show condensed ring with large conjugation due to aromatic rings such as benzene, furan or thiophene. In this regard, it is significant that the only molecule not showing condensed rings (Inhibitor n°5) is inactive despite having two highly aromatic thiophene rings.
Furthermore, the two molecules having six-five member condensed rings are by far the most potent ones. Indeed Inhibitor n°4 exhibits the dihydro-indolone moiety and Inhibitor n°6 the thiazolopirimidinone ring. The next in the ranking of potency is Inhibitor n°2, which has two condensed six-member rings. Interestingly, the two less active compounds (Inhibitors n°1 and n°3) contain three condensed rings, namely the amino-pyrano-chromenone moiety, what points toward a maximum of two condensed rings for optimal activity.
It is also interesting to note that all selected molecules include one chiral carbon, which suggests that the activity of the enantiomers might not be as significant as predicted by the topological models, since these models do not take into account the chirality.
Conclusions
Molecular Topology has been used to select novel potential anti-cancer compounds with prevention or therapeutic activity. The in vitro evaluation of the selected compounds, CAS n° 256378-54-8 (Inhibitor n°1), 663203-38-1 (Inhibitor n°2), 247079-73-8 (Inhibitor n°3), 689769-86-6 (Inhibitor n°4), 15940-61-1 (Inhibitor n°5) and 431925–096 (Inhibitor n°6), demonstrated significant activity as Akt and β-catenin inhibitors for all compounds except for Inhibitor n°5. However, particularly Inhibitors n°4 and n°6, can be considered as novel chemotherapy hits by dually inhibit Akt and β-catenin. Moreover, they also have demonstrated activity over prostate cancer cell line (Inhibitors n°4 and n°6) and on a human colorectal cancer cell line (Inhibitor n°4).
Topological Virtual Screening upon four discriminant models (DF1-4) resulted in an overall accuracy of five out of six compounds which showed the predicted activity. Accordingly, it can be concluded that Molecular Topology is a reliable and useful tool in the search and discovery of novel chemotherapeutic agents acting on Akt and beta-catenin pathways. The presented exhaustive performance opens new horizons for MT as a time-cost effective resource. Finally, it must be point out that the next step could be perform rational drug design based on the structures and then, carry on in vivo test to confirm their effectiveness as potential cancer chemotherapeutic agents.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
IDL is supported by grants from Spanish Minneco (grant BFU2012-31444), Comunidad de Madrid (grant S2010/BMD-2308) Junta de Castilla-La Mancha (grant POII-2014-011-P), Fundación Tatiana Pérez de Guzmán (grant 2013-001). MCR is supported by Valencia University (Especials Accions Grant UV-INV-AE13-139455). MGL is supported by University of Valencia (Atraction of Talent Grant UV-INV-PDOC11-39853). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Meropol NJ, Schrag D, Smith TJ, Mulvey TM, Langdon RM Jr, Blum D, et al. American Society of Clinical Oncology guidance statement: the cost of cancer care. J Clin Oncol 2009;27(23):3868–3874. 10.1200/JCO.2009.23.1183 [DOI] [PubMed] [Google Scholar]
- 2. Eagle D. The cost of cancer care: part I. Oncology-NY 2012;26(10):918 [PubMed] [Google Scholar]
- 3. Scalo JF, Rascati KL. Trends and issues in oncology costs. Expert Rev Pharmacoecon Outcomes Res 2014;14(1):35–44. 10.1586/14737167.2014.864561 [DOI] [PubMed] [Google Scholar]
- 4. Surh Y. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer 2003;3(10):768–780. [DOI] [PubMed] [Google Scholar]
- 5. Yabroff KR, Borowski L, Lipscomb J. Economic studies in colorectal cancer: challenges in measuring and comparing costs. J Natl Cancer Inst Monogr 2013;2013(46):62–78. 10.1093/jncimonographs/lgt001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haug U, Engel S, Verheyen F, Linder R. Estimating Colorectal Cancer Treatment Costs: A Pragmatic Approach Exemplified by Health Insurance Data from Germany. PLoS one 2014; 9(2): e88407 10.1371/journal.pone.0088407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edlind MP, Hsieh AC. PI3K-AKT-mTOR signaling in prostate cancer progression and androgen deprivation therapy resistance. Asian J Androl 2014;16(3):378–386. 10.4103/1008-682X.122876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crowell JA, Steele VE, Fay JR. Targeting the AKT protein kinase for cancer chemoprevention. Mol Cancer Ther 2007;6(8):2139–2148. [DOI] [PubMed] [Google Scholar]
- 9. Seo BR, Min K, Cho IJ, Kim SC, Kwon TK. Curcumin Significantly Enhances Dual PI3K/Akt and mTOR Inhibitor NVP-BEZ235-Induced Apoptosis in Human Renal Carcinoma Caki Cells through Down-Regulation of p53-Dependent Bcl-2 Expression and Inhibition of Mcl-1 Protein Stability. PloS one 2014;9(4):e95588 10.1371/journal.pone.0095588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang H, Duan L, Zou Z, Li H, Yuan S, Chen X, et al. Activation of the PI3K/Akt/mTOR/p70S6K Pathway is Involved in S100A4-induced Viability and Migration in Colorectal Cancer Cells. Int J Med Sci 2014;11(8):841 10.7150/ijms.8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson SM, Gulhati P, Rampy BA, Han Y, Rychahou PG, Doan HQ, et al. Novel expression patterns of PI3K/Akt/mTOR signaling pathway components in colorectal cancer. J Am Coll Surg 2010;210(5):767–776. 10.1016/j.jamcollsurg.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baba Y, Nosho K, Shima K, Hayashi M, Meyerhardt JA, Chan AT, et al. Phosphorylated AKT expression is associated with PIK3CA mutation, low stage, and favorable outcome in 717 colorectal cancers. Cancer 2011;117(7):1399–1408. 10.1002/cncr.25630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu Q, Wang L, Zhong M. Clinical significance of mTOR and p-mTOR protein expression in human colorectal carcinomas. Asian Pac J Cancer Prev 2011;12:2581–2584. [PubMed] [Google Scholar]
- 14. Nozawa H, Watanabe T, Nagawa H. Phosphorylation of ribosomal p70 S6 kinase and rapamycin sensitivity in human colorectal cancer. Cancer Lett 2007;251(1):105–113. [DOI] [PubMed] [Google Scholar]
- 15. Gao N, Zhang Z, Jiang B, Shi X. Role of PI3K/AKT/mTOR signaling in the cell cycle progression of human prostate cancer. Biochem Biophys Res Commun 2003;310(4):1124–1132. [DOI] [PubMed] [Google Scholar]
- 16. Yang Y, Yang J, Tao H, Jin W. New perspectives on β-catenin control of cell fate and proliferation in colon cancer. Food Chem Toxicol 2014;74:14–19. 10.1016/j.fct.2014.08.013 [DOI] [PubMed] [Google Scholar]
- 17. Lee E, Madar A, David G, Garabedian MJ, Dasgupta R, Logan SK. Inhibition of androgen receptor and beta-catenin activity in prostate cancer. Proc Natl Acad Sci U S A 2013;110(39):15710–15715. 10.1073/pnas.1218168110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yardy G, Brewster S. Wnt signalling and prostate cancer. Prostate Cancer Prostatic Dis 2005;8(2):119–126. [DOI] [PubMed] [Google Scholar]
- 19. Gálvez J, Gomez-Lechón M, García-Domenech R, Castell J. New cytostatic agents obtained by molecular topology. Bioorg Med Chem Lett 1996;6(19):2301–2306. [Google Scholar]
- 20. Gálvez J, Garcıa-Domenech R, Gómez-Lechón M, Castell J. Use of molecular topology in the selection of new cytostatic drugs. Journal of Molecular Structure: THEOCHEM 2000;504(1):241–248. [Google Scholar]
- 21. Villagra S, Jáuregui E, Gálvez J. New Anti-Neoplastics Obtained by a Molecular Connectivity Method. Molecules 2000;5(3):330–331. [Google Scholar]
- 22. Llacer MT, Gálvez J, Garcia-Domenech R, Gómez-Lechón M, Más-Arcas C, de Julian-Ortiz J. Topological virtual screening and pharmacological test of novel cytostatic drugs. Internet Electron J Mol Des 2006;5:306–319. 16249936 [Google Scholar]
- 23. Jasinski P, Welsh B, Galvez J, Land D, Zwolak P, Ghandi L, et al. A novel quinoline, MT477: suppresses cell signaling through Ras molecular pathway, inhibits PKC activity, and demonstrates in vivo anti-tumor activity against human carcinoma cell lines. Invest New Drugs 2008;26(3):223–232. [DOI] [PubMed] [Google Scholar]
- 24. Jasinski P, Zwolak P, Vogel RI, Bodempudi V, Terai K, Galvez J, et al. MT103 inhibits tumor growth with minimal toxicity in murine model of lung carcinoma via induction of apoptosis. Invest New Drugs 2011;29(5):846–852. 10.1007/s10637-010-9432-4 [DOI] [PubMed] [Google Scholar]
- 25. López ME, Pitarresi VLF, Valero SS, Gálvez J, García-Domenech R. Application of Molecular Topology to the Prediction of the Reaction Yield and Anticancer Activity of Imidazole and Guanidine Derivatives. IJCCE 2013;3(2):64–74. [Google Scholar]
- 26. Caboni L, Galvez-Llompart M, Galvez J, Blanco F, Rubio-Martinez J, Fayne D, et al. Molecular topology applied to the discovery of 1-benzyl-2-(3-fluorophenyl)-4-hydroxy-3-(3-phenylpropanoyl)-2H-pyrrol-5-one as a non-ligand binding pocket anti-androgen. J Chem Inf Model 2014; 54(10):2953–2966. 10.1021/ci500324f [DOI] [PubMed] [Google Scholar]
- 27.Galvez J, Llompart J, Pal K, inventors. Spain, assignee. N,N-dicyclohexyl-(1S)-isoborneol-10-sulfonamide (MT103) and related compounds for the treatment of cancer. Patent Application Country: Application: US; US; Priority Application Country: US patent US20040266732. 2004 1230; Patent Application Date: 20040430.; Priority Application Date: 20020920.
- 28.Llompart J, Galvez J, inventors. Medisyn Technologies I, Spain, assignee. N,N-dicyclohexyl-(1S)-isoborneol-10-sulfonamide MT103 family members as antitumor and other therapeutic agents and corresponding treatments and compositions. Patent Application Country: Application: US; US; Priority Application Country: US patent US20040059000. 2004 0325; Patent Application Date: 20020920.; Priority Application Date: 20020920.
- 29.Llompart J, Galvez J, Pal K, inventors. Medisyn Technologies I, USA, assignee. Treatment of cancer with MT477 derivatives. Patent Application Country: Application: US; US; Priority Application Country: US patent US20060014770. 2006 0119; Patent Application Date: 20050711.; Priority Application Date: 20040709.
- 30. Galvez-Llompart M, M Giner R, C Recio M, Candeletti S, Garcia-Domenech R. Application of molecular topology to the search of novel NSAIDs: Experimental validation of activity. Lett Drug Des Discov 2010;7(6):438–445. [Google Scholar]
- 31. Gálvez-Llompart M, Recio MC, García-Domenech R. Topological virtual screening: a way to find new compounds active in ulcerative colitis by inhibiting NF-κB. Mol Divers 2011;15(4):917–926. 10.1007/s11030-011-9323-4 [DOI] [PubMed] [Google Scholar]
- 32. Gálvez-Llompart M, Zanni R, Romualdi P, García-Domenech R. Selection of nutraceutical compounds as COX inhibitors by molecular topology. Med Chem Res 2013;22(7):3466–3477. [Google Scholar]
- 33. Galvez-Llompart M, María del Carmen Recio Iglesias, Gálvez J, García-Domenech R. Novel potential agents for ulcerative colitis by molecular topology: suppression of IL-6 production in Caco-2 and RAW 264.7 cell lines. Mol Divers 2013;17(3):573–593. 10.1007/s11030-013-9458-6 [DOI] [PubMed] [Google Scholar]
- 34. García-Domenech R, Gálvez-Llompart M, Zanni R, Recio MC, Gálvez J. QSAR methods for the discovery of new inflammatory bowel disease drugs. Expert Opin Drug Discov 2013;8(8):933–949. 10.1517/17460441.2013.800043 [DOI] [PubMed] [Google Scholar]
- 35. Duart M, Antón-Fos G, de Julian-Ortiz J, Gozalbes R, Gálvez J, Garcıa-Domenech R. Use of molecular topology for the prediction of physico-chemical, pharmacokinetic and toxicological properties of a group of antihistaminic drugs. Int J Pharm 2002;246(1):111–119. [DOI] [PubMed] [Google Scholar]
- 36. Galvez J, Garcia-Domenech R. On the contribution of molecular topology to drug design and discovery. Curr Comput Aided Drug Des 2010;6(4):252–268. [DOI] [PubMed] [Google Scholar]
- 37.Galvez J, Garcia-Domenech R, Castro E. Molecular topology in QSAR and drug design studies. QSPR-QSAR Studies on Desired Properties for Drug Design. Research Signpost 2010:63–94.
- 38. García-Domenech R, Gálvez J, de Julián-Ortiz JV, Pogliani L. Some new trends in chemical graph theory. Chem Rev 2008;108(3):1127–1169. 10.1021/cr0780006 [DOI] [PubMed] [Google Scholar]
- 39. Gálvez J, Gálvez-Llompart M, García-Domenech R. Molecular topology as a novel approach for drug discovery. Expert Opin Drug Discov 2012;7(2):133–153. 10.1517/17460441.2012.652083 [DOI] [PubMed] [Google Scholar]
- 40. Galvez J, Villar V, Galvez-Llompart M, Amigo J. Chemistry explained by topology: An alternative approach. Comb Chem High Throughput Screen 2011;14(4):279–283. [DOI] [PubMed] [Google Scholar]
- 41. Galvez J, Galvez-Llompart M, Garcia-Domenech R. Introduction to molecular topology: basic concepts and application to drug design. Curr Comput Aided Drug Des 2012;8(3):196–223. [DOI] [PubMed] [Google Scholar]
- 42. Galvez J, Galvez-Llompart M, Zanni R, Garcia-Domenech R. Molecular topology–dissimilar similarities. Drug Discov Today Technol 2013;10(4):e475–e481. 10.1016/j.ddtec.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 43.Todeschini R, Consonni V, Mauri A, Pavan M. Dragon for windows (Software for Molecular Descriptor Calculations), version 5.4. Talete srl: Milan, Italy 2006.
- 44. Merla R, Ye Y, Lin Y, Manickavasagam S, Huang MH, Perez-Polo RJ, et al. The central role of adenosine in statin-induced ERK1/2, Akt, and eNOS phosphorylation. Am J Physiol Heart Circ Physiol 2007;293(3):H1918–28. [DOI] [PubMed] [Google Scholar]
- 45. Wang S, Liu Q, Zhang Y, Liu K, Yu P, Liu K, et al. Suppression of growth, migration and invasion of highly-metastatic human breast cancer cells by berbamine and its molecular mechanisms of action. Mol Cancer 2009;8(1):81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lambert JR, Young CD, Persons KS, Ray R. Mechanistic and pharmacodynamic studies of a 25-hydroxyvitamin D 3 derivative in prostate cancer cells. Biochem Biophys Res Commun 2007;361(1):189–195. [DOI] [PubMed] [Google Scholar]
- 47.Xiaonan W, Qing W, Xu Y, Liansheng Z, Yiping W, Yanwen S. Study of Celastrol on Akt Signaling Pathway and Its Roles in the Apoptosis of K562 Cells. J Cancer Ther 2011;2011.
- 48. Xie RH, Yin M, Yin CC, Cheng XG, Xu ZW, Cao XQ, et al. Mechanism of chlorogenic acid on apoptosis of rat nucleus pulposus cells induced by oxidative stress. Zhong Yao Cai 2014;37(3):465–469. [PubMed] [Google Scholar]
- 49. Jayasooriya Rajapaksha Gedara Prasad Tharanga, Dilshara MG, Park SR, Choi YH, Hyun J, Chang W, et al. 18β-Glycyrrhetinic acid suppresses TNF-α induced matrix metalloproteinase-9 and vascular endothelial growth factor by suppressing the Akt-dependent NF-κB pathway. Toxicol in Vitro 2014;28(5):751–758. 10.1016/j.tiv.2014.02.015 [DOI] [PubMed] [Google Scholar]
- 50. Pandey MK, Kale VP, Song C, Sung S, Sharma AK, Talamo G, et al. Gambogic acid inhibits multiple myeloma mediated osteoclastogenesis through suppression of chemokine receptor CXCR4 signaling pathways. Exp Hematol 2014; 42(10):883–896. 10.1016/j.exphem.2014.07.261 [DOI] [PubMed] [Google Scholar]
- 51. Woo KJ, Jeong Y, Park J, Kwon TK. Chrysin-induced apoptosis is mediated through caspase activation and Akt inactivation in U937 leukemia cells. Biochem Biophys Res Commun 2004;325(4):1215–1222. [DOI] [PubMed] [Google Scholar]
- 52. Liu X, Zhao G, Yan Y, Bao L, Chen B, Qi R. Ginkgolide B reduces atherogenesis and vascular inflammation in ApoE−/− mice. PloS one 2012;7(5):e36237 10.1371/journal.pone.0036237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hwang T, Wang C, Kuo Y, Huang H, Wu Y, Kuo L, et al. The hederagenin saponin SMG-1 is a natural FMLP receptor inhibitor that suppresses human neutrophil activation. Biochem Pharmacol 2010;80(8):1190–1200. 10.1016/j.bcp.2010.06.028 [DOI] [PubMed] [Google Scholar]
- 54. Saiprasad G, Chitra P, Manikandan R, Sudhandiran G. Hesperidin induces apoptosis and triggers autophagic markers through inhibition of Aurora-A mediated phosphoinositide-3-kinase/Akt/mammalian target of rapamycin and glycogen synthase kinase-3 beta signalling cascades in experimental colon carcinogenesis. Eur J Cancer 2014;50(14):2489–2507. 10.1016/j.ejca.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 55. Merhi F, Tang R, Piedfer M, Mathieu J, Bombarda I, Zaher M, et al. Hyperforin inhibits Akt1 kinase activity and promotes caspase-mediated apoptosis involving Bad and Noxa activation in human myeloid tumor cells. PloS one 2011;6(10):e25963 10.1371/journal.pone.0025963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yoon SO, Shin S, Lee HJ, Chun HK, Chung AS. Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression. Mol Cancer Ther 2006;5(11):2666–2675. [DOI] [PubMed] [Google Scholar]
- 57. Dung TTM, Kim SC, Yoo BC, Sung G, Yang WS, Kim HG, et al. (5-Hydroxy-4-oxo-4H-pyran-2-yl) methyl 6-hydroxynaphthalene-2-carboxylate, a kojic acid derivative, inhibits inflammatory mediator production via the suppression of Syk/Src and NF-κB activation. Int Immunopharmacol 2014;20(1):37–45. 10.1016/j.intimp.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 58. Shanmugam MK, Nguyen AH, Kumar AP, Tan BK, Sethi G. Targeted inhibition of tumor proliferation, survival, and metastasis by pentacyclic triterpenoids: potential role in prevention and therapy of cancer. Cancer Lett 2012;320(2):158–170. 10.1016/j.canlet.2012.02.037 [DOI] [PubMed] [Google Scholar]
- 59. Chen W, Huang Y, Yang M, Lee C, Chen C, Yeh C, et al. Protective effect of rutin on LPS-induced acute lung injury via down-regulation of MIP-2 expression and MMP-9 activation through inhibition of Akt phosphorylation. Int Immunopharmacol 2014;22(2):409–413. 10.1016/j.intimp.2014.07.026 [DOI] [PubMed] [Google Scholar]
- 60. Morikawa K, Nonaka M, Mochizuki H, Handa K, Hanada H, Hirota K. Naringenin and hesperetin induce growth arrest, apoptosis, and cytoplasmic fat deposit in human preadipocytes. J Agric Food Chem 2008;56(22):11030–11037. 10.1021/jf801965n [DOI] [PubMed] [Google Scholar]
- 61. Bajer MM, Kunze MM, Blees JS, Bokesch HR, Chen H, Brauß TF, et al. Characterization of pomiferin triacetate as a novel mTOR and translation inhibitor. Biochem Pharmacol 2014;88(3):313–321. 10.1016/j.bcp.2014.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolle D, Lee SJ, Li Z, Litan A, Barwe SP, Langhans SA. Inhibition of epidermal growth factor signaling by the cardiac glycoside ouabain in medulloblastoma. Cancer medicine 2014;3(5):1146–1158. 10.1002/cam4.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kwak DH, Lee J, Kim D, Kim T, Lee KJ, Ma JY. Inhibitory effects of Hwangryunhaedok-Tang in 3T3-L1 adipogenesis by regulation of Raf/MEK1/ERK1/2 pathway and PDK1/Akt Phosphorylation. Evid-Based Compl Alt 2013; 2013 (Article ID 413906). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu H, Song J, Gao X, Xu Z, Xu X, Xia Y, et al. Paeoniflorin attenuates lipopolysaccharide-induced permeability of endothelial cells: involvements of F-actin expression and phosphorylations of PI3K/Akt and PKC. Inflammation 2013;36(1):216–225. 10.1007/s10753-012-9537-3 [DOI] [PubMed] [Google Scholar]
- 65. Do MT, Kim HG, Choi JH, Khanal T, Park BH, Tran TP, et al. Antitumor efficacy of piperine in the treatment of human HER2-overexpressing breast cancer cells. Food Chem 2013;141(3):2591–2599. 10.1016/j.foodchem.2013.04.125 [DOI] [PubMed] [Google Scholar]
- 66. Deeb D, Gao X, Liu YB, Pindolia K, Gautam SC. Pristimerin, a quinonemethide triterpenoid, induces apoptosis in pancreatic cancer cells through the inhibition of pro-survival Akt/NF-κB/mTOR signaling proteins and anti-apoptotic Bcl-2. Int J Oncol 2014;44(5):1707–1715. 10.3892/ijo.2014.2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Song NR, Chung M, Kang NJ, Seo SG, Jang TS, Lee HJ, et al. Quercetin suppresses invasion and migration of H-Ras-transformed MCF10A human epithelial cells by inhibiting phosphatidylinositol 3-kinase. Food Chem 2014;142:66–71. 10.1016/j.foodchem.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 68. ZHAO S, DUAN H, XU Y, LU Y, JIANG Z. Effects of rhaponticin on proliferation of SK-BR-3 breast cancer cells. Bulletin of the Academy of Military Medical Sciences 2010;5:017. [Google Scholar]
- 69. Raina K, Agarwal C, Wadhwa R, Serkova NJ, Agarwal R. Energy deprivation by silibinin in colorectal cancer cells: a double-edged sword targeting both apoptotic and autophagic machineries. Autophagy 2013;9(5):697–713. 10.4161/auto.23960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Esmaeili MA, Farimani MM. Inactivation of PI3K/Akt pathway and upregulation of gene are involved in daucosterol, isolated from salvia sahendica, induced apoptosis in human breast adenocarcinoma cells. S Afr J Bot 2014; 93:37–47. [Google Scholar]
- 71. Ou X, Liu M, Luo H, Dong LQ, Liu F. Ursolic Acid Inhibits Leucine-Stimulated mTORC1 Signaling by Suppressing mTOR Localization to Lysosome. PloS one 2014;9(4):e95393 10.1371/journal.pone.0095393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hou L, Sun Y, Zhang X, Zhao D, Wu K. Vitamin E succinate induced protective autophagy in human gastric cancer cells SGC-7901 via the Akt/mTOR signaling pathway. Aibian, Jibian, Tubian 2013;25(5):343–347, 351. [Google Scholar]
- 73. Jin K, Oh YN, Hyun SK, Kwon HJ, Kim BW. Betulinic acid isolated from vitis amurensis root inhibits 3-isobutyl-1-methylxanthine induced melanogenesis via the regulation of MEK/ERK and PI3K/Akt pathways in B16F10 cells. Food Chem Toxicol 2014; 68:38–43. 10.1016/j.fct.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 74. Buitrago CG, Arango NS, Boland RL. 1α, 25 (OH) 2D3-dependent modulation of Akt in proliferating and differentiating C2C12 skeletal muscle cells. J Cell Biochem 2012;113(4):1170–1181. 10.1002/jcb.23444 [DOI] [PubMed] [Google Scholar]
- 75. Choo EJ, Rhee Y, Jeong S, Lee H, Kim HS, Ko HS, et al. Anethole exerts antimetatstaic activity via inhibition of matrix metalloproteinase 2/9 and AKT/mitogen-activated kinase/nuclear factor kappa B signaling pathways. Biological and Pharmaceutical Bulletin 2011;34(1):41–46. [DOI] [PubMed] [Google Scholar]
- 76. McGuire TF, Trump DL, Johnson CS. Vitamin D(3)-induced apoptosis of murine squamous cell carcinoma cells. Selective induction of caspase-dependent MEK cleavage and up-regulation of MEKK-1. J Biol Chem 2001;276(28):26365–26373. [DOI] [PubMed] [Google Scholar]
- 77. Park H, Park K, Lee D, Kang S, Nagappan A, Kim J, et al. Polyphenolic extract isolated from korean lonicera japonica thunb. induce G2/M cell cycle arrest and apoptosis in HepG2 cells: Involvements of PI3K/Akt and MAPKs. Food Chem Toxicol 2012; 50(7):2407–2416. 10.1016/j.fct.2012.04.034 [DOI] [PubMed] [Google Scholar]
- 78. Huang M, Tang S, Upadhyay G, Marsh JL, Jackman CP, Shankar S, et al. Embelin Suppresses Growth of Human Pancreatic Cancer Xenografts, and Pancreatic Cancer Cells Isolated from KrasG12D Mice by Inhibiting Akt and Sonic Hedgehog Pathways. PloS one 2014;9(4):e92161 10.1371/journal.pone.0092161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Oh JH, Kwon TK. Withaferin A inhibits tumor necrosis factor-α-induced expression of cell adhesion molecules by inactivation of Akt and NF-κB in human pulmonary epithelial cells. Int Immunopharmacol 2009;9(5):614–619. 10.1016/j.intimp.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 80. Syed DN, Chamcheu J, Khan MI, Sechi M, Lall RK, Adhami VM, et al. Fisetin inhibits human melanoma cell growth through direct binding to p70S6K and mTOR: Findings from 3-D melanoma skin equivalents and computational modeling. Biochem Pharmacol 2014;89(3):349–360. 10.1016/j.bcp.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MicroSource website (Pure natural products). Available: http://www.msdiscovery.com/natprod.html Accessed 30 March 2015.
- 82.Selleckchem website (Akt inhibitors). Available: http://www.selleckchem.com/pathways_Akt.html?gclid=CLXsnIPglLMCFYXLtAodSWUAIw Accessed 30 March 2015.
- 83. Hu C, Solomon VR, Ulibarri G, Lee H. The efficacy and selectivity of tumor cell killing by Akt inhibitors are substantially increased by chloroquine. Bioorg Med Chem 2008;16(17):7888–7893. 10.1016/j.bmc.2008.07.076 [DOI] [PubMed] [Google Scholar]
- 84. Prinz H, Chamasmani B, Vogel K, Böhm KJ, Aicher B, Gerlach M, et al. N-benzoylated phenoxazines and phenothiazines: synthesis, antiproliferative activity, and inhibition of tubulin polymerization. J Med Chem 2011;54(12):4247–4263. 10.1021/jm200436t [DOI] [PubMed] [Google Scholar]
- 85. Scartabelli T, Gerace E, Landucci E, Moroni F, Pellegrini-Giampietro DE. Neuroprotection by group I mGlu receptors in a rat hippocampal slice model of cerebral ischemia is associated with the PI3K–Akt signaling pathway: A novel postconditioning strategy? Neuropharmacology 2008;55(4):509–516. 10.1016/j.neuropharm.2008.06.019 [DOI] [PubMed] [Google Scholar]
- 86. Garcia-Echeverria C, Sellers W. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene 2008;27(41):5511–5526. 10.1038/onc.2008.246 [DOI] [PubMed] [Google Scholar]
- 87. Toulany M, Minjgee M, Saki M, Holler M, Meier F, Eicheler W, et al. ERK2-dependent reactivation of Akt mediates the limited response of tumor cells with constitutive K-RAS activity to PI3K inhibition. Cancer biology & therapy 2013;15(3):0–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ma L, Zhang G, Miao X, Deng X, Wu Y, Liu Y, et al. Cancer stem-like cell properties are regulated by EGFR/AKT/β–catenin signaling and preferentially inhibited by gefitinib in nasopharyngeal carcinoma. FEBS Journal 2013;280(9):2027–2041. 10.1111/febs.12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Böckmann S, Nebe B. The in vitro effects of H-89, a specific inhibitor of protein kinase A, in the human colonic carcinoma cell line Caco-2. Eur J Cancer Prev 2003;12(6):469–478. [DOI] [PubMed] [Google Scholar]
- 90. Krech T, Thiede M, Hilgenberg E, Schäfer R, Jürchott K. Characterization of AKT independent effects of the synthetic AKT inhibitors SH-5 and SH-6 using an integrated approach combining transcriptomic profiling and signaling pathway perturbations. BMC Cancer 2010;10(1):287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang XD, Gillespie SK, Hersey P. Staurosporine induces apoptosis of melanoma by both caspase-dependent and-independent apoptotic pathways. Mol Cancer Ther 2004;3(2):187–197. [PubMed] [Google Scholar]
- 92. Jeon SJ, Seo JE, Yang S, Choi JW, Wells D, Shin CY, et al. Cellular stress-induced up-regulation of FMRP promotes cell survival by modulating PI3K-Akt phosphorylation cascades. J Biomed Sci 2011;18:17 10.1186/1423-0127-18-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li J, Davies BR, Han S, Zhou M, Bai Y, Zhang J, et al. The AKT inhibitor AZD5363 is selectively active in PI3KCA mutant gastric cancer, and sensitizes a patient-derived gastric cancer xenograft model with PTEN loss to Taxotere. J Transl Med 2013;11:241-5876-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tabernero J, Cervantes A, Saura C, Roda D, Yan Y, Lin K, et al. Targeting the PI3K-Akt-mTOR pathway with GDC-0068, a novel selective ATP competitive Akt inhibitor. Ann Oncol 2011;22. [Google Scholar]
- 95. Jin X, Gossett D, Wang S, Yang D, Cao Y, Chen J, et al. Inhibition of AKT survival pathway by a small molecule inhibitor in human endometrial cancer cells. Br J Cancer 2004;91(10):1808–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Meuillet EJ, Zuohe S, Lemos R, Ihle N, Kingston J, Watkins R, et al. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol Cancer Ther 2010; 9(3):706–717. 10.1158/1535-7163.MCT-09-0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Makhov PB, Golovine K, Kutikov A, Teper E, Canter DJ, Simhan J, et al. Modulation of Akt/mTOR signaling overcomes sunitinib resistance in renal and prostate cancer cells. Mol Cancer Ther 2012;11(7):1510–1517. 10.1158/1535-7163.MCT-11-0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.MicroSource website (US Drugs Collection). Available: http://www.msdiscovery.com/usdrugs.html Accessed 30 March 2015.
- 99. Saifo MS, Rempinski DR Jr, Rustum YM, Azrak RG. Targeting the oncogenic protein beta-catenin to enhance chemotherapy outcome against solid human cancers. Mol Cancer 2010;9:310 10.1186/1476-4598-9-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yoshimitsu T, Ino T, Futamura N, Kamon T, Tanaka T. Total synthesis of the β-catenin inhibitor,(−)-agelastatin A: A second-generation approach based on radical aminobromination. Org Lett 2009;11(15):3402–3405. 10.1021/ol9012684 [DOI] [PubMed] [Google Scholar]
- 101. Liu Y, Wang W, Xu J, Li L, Dong Q, Shi Q, et al. Dihydroartemisinin inhibits tumor growth of human osteosarcoma cells by suppressing Wnt/β-catenin signaling. Oncol Rep 2013;30(4):1723–1730. 10.3892/or.2013.2658 [DOI] [PubMed] [Google Scholar]
- 102. Kawabata K, Murakami A, Ohigashi H. Citrus auraptene targets translation of MMP-7 (matrilysin) via ERK1/2-dependent and mTOR-independent mechanism. FEBS Lett 2006;580(22):5288–5294. [DOI] [PubMed] [Google Scholar]
- 103. Lin B, Wan S, Liu F, Cui Z, Zhang X, Li X. Effect of bergapten on cells cycle in nasopharyngeal carcinoma. Zhongguo Yaoxue Zazhi (Beijing, China) 2014;49(10):837–842. [Google Scholar]
- 104. Chen RH, Ding WV, McCormick F. Wnt signaling to beta-catenin involves two interactive components. Glycogen synthase kinase-3beta inhibition and activation of protein kinase C. J Biol Chem 2000; 275(23):17894–17899. [DOI] [PubMed] [Google Scholar]
- 105. Jia D, Yang W, Li L, Liu H, Tan Y, Ooi S, et al. β-Catenin and NF-κB co-activation triggered by TLR3 stimulation facilitates stem cell-like phenotypes in breast cancer. Cell Death Differ 2015; 22(2):298–310. 10.1038/cdd.2014.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Park S, Gwak J, Han SJ, Oh S. Cardamonin suppresses the proliferation of colon cancer cells by promoting β-catenin degradation. Biol Pharm Bull 2013;36(6):1040–1044. [DOI] [PubMed] [Google Scholar]
- 107. Cui L, Jia X, Zhou Q, Zhai X, Zhou Y, Zhu H. Curcumin affects β‐catenin pathway in hepatic stellate cell in vitro and in vivo. J Pharm Pharmacol 2014; 66(11):1615–1622. 10.1111/jphp.12283 [DOI] [PubMed] [Google Scholar]
- 108. Thamilselvan V, Menon M, Thamilselvan S. Anticancer efficacy of deguelin in human prostate cancer cells targeting glycogen synthase kinase-3 β/β-catenin pathway. Int J Cancer 2011;129(12):2916–2927. 10.1002/ijc.25949 [DOI] [PubMed] [Google Scholar]
- 109. Xiao D, Li M, Herman-Antosiewicz A, Antosiewicz J, Xiao H, Lew KL, et al. Diallyl trisulfide inhibits angiogenic features of human umbilical vein endothelial cells by causing Akt inactivation and down-regulation of VEGF and VEGF-R2. Nutr Cancer 2006;55(1):94–107. [DOI] [PubMed] [Google Scholar]
- 110. Chen B, Zhang Z, Liu Z, Zhao Q, Jiang K, Yang K, et al. EGCG inhibited the proliferation of NP69-LMP1 cells through Wnt1-β-catenin pathway. Xiandai Shengwuyixue Jinzhan 2012;12(36):7034–7039. [Google Scholar]
- 111. Vanella L, Di Giacomo C, Acquaviva R, Barbagallo I, Cardile V, Kim DH, et al. Apoptotic markers in a prostate cancer cell line: Effect of ellagic acid. Oncol Rep 2013;30(6):2804–2810. 10.3892/or.2013.2757 [DOI] [PubMed] [Google Scholar]
- 112. Way T, Huang J, Chou C, Huang C, Yang M, Ho C. Emodin represses TWIST1-induced epithelial–mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the β-catenin and Akt pathways. Eur J Cancer 2014;50(2):366–378. 10.1016/j.ejca.2013.09.025 [DOI] [PubMed] [Google Scholar]
- 113. Lee SY, Lim TG, Chen H, Jung SK, Lee HJ, Lee MH, et al. Esculetin suppresses proliferation of human colon cancer cells by directly targeting beta-catenin. Cancer Prev Res (Phila) 2013;6(12):1356–1364. 10.1158/1940-6207.CAPR-13-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Su T, Lin J, Tsai C, Huang T, Yang Z, Wu M, et al. Inhibition of melanogenesis by gallic acid: Possible involvement of the PI3K/Akt, MEK/ERK and Wnt/β-catenin signaling pathways in B16F10 cells. International journal of molecular sciences 2013;14(10):20443–20458. 10.3390/ijms141020443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pang X, Wu Y, Wu Y, Lu B, Chen J, Wang J, et al. (-)-Gossypol suppresses the growth of human prostate cancer xenografts via modulating VEGF signaling-mediated angiogenesis. Mol Cancer Ther 2011;10(5):795–805. 10.1158/1535-7163.MCT-10-0936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Liagre B, Vergne-Salle P, Leger DY, Beneytout J. Inhibition of human rheumatoid arthritis synovial cell survival by hecogenin and tigogenin is associated with increased apoptosis, p38 mitogen-activated protein kinase activity and upregulation of cyclooxygenase-2. Int J Mol Med 2007; 20:451–460. [PubMed] [Google Scholar]
- 117. Joo YN, Eun SY, Park SW, Lee JH, Chang KC, Kim HJ. Honokiol inhibits U87MG human glioblastoma cell invasion through endothelial cells by regulating membrane permeability and the epithelial-mesenchymal transition. Int J Oncol 2014;44(1):187–194. 10.3892/ijo.2013.2178 [DOI] [PubMed] [Google Scholar]
- 118. Chinni SR, Li Y, Upadhyay S, Koppolu PK, Sarkar FH. Indole-3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene 2001;20(23):2927–2936. [DOI] [PubMed] [Google Scholar]
- 119. Wang G, Fang Y, Lu Z, Pan Z, Wan D. Effects of indomethacin on the proliferation and invasion of colon cancer cell line hct116 and its mechanisms. Shiyong Yixue Zazhi 2012;28(11):1756–1758. [Google Scholar]
- 120. Su Y, Simmen RC. Soy isoflavone genistein upregulates epithelial adhesion molecule E-cadherin expression and attenuates beta-catenin signaling in mammary epithelial cells. Carcinogenesis 2009;30(2):331–339. 10.1093/carcin/bgn279 [DOI] [PubMed] [Google Scholar]
- 121. Wang N, Wang Z, Peng C, You J, Shen J, Han S, et al. Dietary compound isoliquiritigenin targets GRP78 to chemosensitize breast cancer stem cells via beta-catenin/ABCG2 signaling. Carcinogenesis 2014;35(11):2544–2554. 10.1093/carcin/bgu187 [DOI] [PubMed] [Google Scholar]
- 122. Lee Y, Shin S, Jue S, Kwon I, Cho E, Cho E, et al. The Role of PIN1 on Odontogenic and Adipogenic Differentiation in Human Dental Pulp Stem Cells. Stem Cells Dev 2013;23(6):618–630. 10.1089/scd.2013.0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kim M, Song Y, Kim C, Hwang J. Kirenol inhibits adipogenesis through activation of the Wnt/β-catenin signaling pathway in 3T3-L1 adipocytes. Biochem Biophys Res Commun 2014;445(2):433–438. 10.1016/j.bbrc.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 124. Choi H, Gwak J, Cho M, Ryu M, Lee J, Kim SK, et al. Murrayafoline A attenuates the Wnt/β-catenin pathway by promoting the degradation of intracellular β-catenin proteins. Biochem Biophys Res Commun 2010;391(1):915–920. 10.1016/j.bbrc.2009.11.164 [DOI] [PubMed] [Google Scholar]
- 125. Li H, Yang B, Huang J, Xiang T, Yin X, Wan J, et al. Naringin inhibits growth potential of human triple-negative breast cancer cells by targeting β-catenin signaling pathway. Toxicol Lett 2013;220(3):219–228. 10.1016/j.toxlet.2013.05.006 [DOI] [PubMed] [Google Scholar]
- 126. Wei W, Chua M, Grepper S, So S. Small molecule antagonists of Tcf4/β-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int J Cancer 2010;126(10):2426–2436. 10.1002/ijc.24810 [DOI] [PubMed] [Google Scholar]
- 127. Choi J, Jiang X, Jeong JB, Lee S. Anticancer Activity of Protocatechualdehyde in Human Breast Cancer Cells. J Med Food 2014; 17(8):842–848. 10.1089/jmf.2013.0159 [DOI] [PubMed] [Google Scholar]
- 128. Mojsin M, Vicentic JM, Schwirtlich M, Topalovic V, Stevanovic M. Quercetin reduces pluripotency, migration and adhesion of human teratocarcinoma cell line NT2/D1 by inhibiting Wnt/β-catenin signaling. Food Funct 2014;5(10):2564–2573. 10.1039/c4fo00484a [DOI] [PubMed] [Google Scholar]
- 129. Fu Y, Chang H, Peng X, Bai Q, Yi L, Zhou Y, et al. Resveratrol Inhibits Breast Cancer Stem-Like Cells and Induces Autophagy via Suppressing Wnt/β-Catenin Signaling Pathway. PloS one 2014;9(7):e102535 10.1371/journal.pone.0102535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lu W, Lin C, King TD, Chen H, Reynolds RC, Li Y. Silibinin inhibits Wnt/β-catenin signaling by suppressing Wnt co-receptor LRP6 expression in human prostate and breast cancer cells. Cell Signal 2012;24(12):2291–2296. 10.1016/j.cellsig.2012.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Baskar AA, Ignacimuthu S, Paulraj GM, Al Numair KS. Chemopreventive potential of β-sitosterol in experimental colon cancer model-an in vitro and in vivo study. BMC Complement Altern Med 2010;10(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res 2010;16(9):2580–2590. 10.1158/1078-0432.CCR-09-2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wang Y, Liu X, Zhou L, Zhu H, Fan Z, Li Q. Effect of tanshinone IIA on angiogenesis in nude mice with colorectal cancer. Zhongguo Shiyan Fangjixue Zazhi 2013;19(3):167–171. [Google Scholar]
- 134. Lee I, Kamba A, Low D, Mizoguchi E. Novel methylxanthine derivative-mediated anti-inflammatory effects in inflammatory bowel disease. WJG 2014;20(5):1127 10.3748/wjg.v20.i5.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Antony L, van der Schoor F, Dalrymple SL, Isaacs JT. Androgen receptor (AR) suppresses normal human prostate epithelial cell proliferation via AR/β-catenin/TCF-4 complex inhibition of c-MYC transcription. Prostate 2014;74(11):1118–1131. 10.1002/pros.22828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kim J, Kim YH, Song G, Kim D, Jeong Y, Liu K, et al. Ursolic acid and its natural derivative corosolic acid suppress the proliferation of APC-mutated colon cancer cells through promotion of β-catenin degradation. Food Chem Toxicol 2014;67:87–95. 10.1016/j.fct.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 137. Deng J, Miller SA, Wang H, Xia W, Wen Y, Zhou BP, et al. β-catenin interacts with and inhibits NF-κB in human colon and breast cancer. Cancer cell 2002;2(4):323–334. [DOI] [PubMed] [Google Scholar]
- 138. Li Q, Hannah SS. Wnt/β-catenin signaling is downregulated but restored by nutrition interventions in the aged heart in mice. Arch Gerontol Geriatr 2012;55(3):749–754. 10.1016/j.archger.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 139. Dihlmann S, Klein S, Doeberitz Mv M. Reduction of beta-catenin/T-cell transcription factor signaling by aspirin and indomethacin is caused by an increased stabilization of phosphorylated beta-catenin. Mol Cancer Ther 2003;2(6):509–516. [PubMed] [Google Scholar]
- 140. YuJun H, BaoHua L, DeBing X, JuXin Z. Effects of caffeic acid phenethyl ester on the colorectal cancer via the β-catenin pathway. Chinese Journal of Digestive Surgery 2009;8(4):294–297. [Google Scholar]
- 141. Lu W, Jia G, Meng X, Zhao C, Zhang L, Ren Y, et al. Beta-catenin mediates the apoptosis induction effect of celastrol in HT29 cells. Life Sci 2012;91(7):279–283. [DOI] [PubMed] [Google Scholar]
- 142. Xia JJ, Pei LB, Zhuang JP, Ji Y, Xu GP, Zhang ZP, et al. Celecoxib inhibits beta-catenin-dependent survival of the human osteosarcoma MG-63 cell line. J Int Med Res 2010;38(4):1294–1304. [DOI] [PubMed] [Google Scholar]
- 143.Selleckchem website (β-catenin inhibitors). Available: http://www.selleckchem.com/Wnt.html Accessed 30 March 2015.
- 144. Raju J, Bird RP. Diosgenin, a naturally occurring furostanol saponin suppresses 3-hydroxy-3-methylglutaryl CoA reductase expression and induces apoptosis in HCT-116 human colon carcinoma cells. Cancer Lett 2007;255(2):194–204. [DOI] [PubMed] [Google Scholar]
- 145. Liu J, Li G, Liu D, Liu J. FH535 inhibits the proliferation of HepG2 cells via downregulation of the Wnt/β-catenin signaling pathway. Mol Med Rep 2014; 9: 1289–1292. 10.3892/mmr.2014.1928 [DOI] [PubMed] [Google Scholar]
- 146. Syed DN, Afaq F, Maddodi N, Johnson JJ, Sarfaraz S, Ahmad A, et al. Inhibition of human melanoma cell growth by the dietary flavonoid fisetin is associated with disruption of Wnt/β-catenin signaling and decreased Mitf levels. J Invest Dermatol 2011;131(6):1291–1299. 10.1038/jid.2011.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Lee M, Kim WK, Park HJ, Kang SS, Lee SK. Anti-proliferative activity of hydnocarpin, a natural lignan, is associated with the suppression of Wnt/β-catenin signaling pathway in colon cancer cells. Bioorg Med Chem Lett 2013;23(20):5511–5514. 10.1016/j.bmcl.2013.08.065 [DOI] [PubMed] [Google Scholar]
- 148.Liu Y, Zhou L, inventors. Nanfang Hospital, Southern Medical University, Peop. Rep. China, assignee. Application of compound ICG-001 in preparation of drugs for treating chronic kidney disease. Patent Application Country: Application: CN; CN; Priority Application Country: CN patent CN104055779. 2014 0924; Patent Application Date: 20140613.; Priority Application Date: 20140613.
- 149. Bilir B, Kucuk O, Moreno CS. Wnt signaling blockage inhibits cell proliferation and migration, and induces apoptosis in triple-negative breast cancer cells. J Transl Med 2013; 11:280-5876-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Arensman M, Kovochich A, Kulikauskas R, Lay A, Yang P, Li X, et al. WNT7B mediates autocrine Wnt/β-catenin signaling and anchorage-independent growth in pancreatic adenocarcinoma. Oncogene 2013;33(7):899–908. 10.1038/onc.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Mao J, Hu X, Xiao Y, Yang C, Ding Y, Hou N, et al. Overnutrition stimulates intestinal epithelium proliferation through beta-catenin signaling in obese mice. Diabetes 2013; 62(11):3736–3746. 10.2337/db13-0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wang J, Zhou D, He X, Wang Y, Hu W, Jiang L, et al. Effect of downregulated beta-catenin on cell proliferative activity, the sensitivity to chemotherapy drug and tumorigenicity of ovarian cancer cells. Cell Mol Biol (Noisy-le-grand) 2011; 57 Suppl:OL1606–13. [PubMed] [Google Scholar]
- 153. Leow P, Tian Q, Ong Z, Yang Z, Ee PR. Antitumor activity of natural compounds, curcumin and PKF118-310, as Wnt/β-catenin antagonists against human osteosarcoma cells. Invest New Drugs 2010;28(6):766–782. 10.1007/s10637-009-9311-z [DOI] [PubMed] [Google Scholar]
- 154. Matsuzaki S, Darcha C. Involvement of the wnt/β-catenin signaling pathway in the cellula103r and molecular mechanisms of fibrosis in endometriosis. PloS one 2013; 8: e76808 10.1371/journal.pone.0076808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pradhan A, Olsson PE. Juvenile ovary to testis transition in zebrafish involves inhibition of ptges. Biol Reprod 2014;91(2):33 10.1095/biolreprod.114.119016 [DOI] [PubMed] [Google Scholar]
- 156. Lee H, Bae S, Kim K, Kim W, Chung S, Yang Y, et al. Shikonin inhibits adipogenesis by modulation of the WNT/β-catenin pathway. Life Sci 2011;88(7):294–301. 10.1016/j.lfs.2010.12.004 [DOI] [PubMed] [Google Scholar]
- 157. Han A, Song Z, Tong C, Hu D, Bi X, Augenlicht LH, et al. Sulindac suppresses β-catenin expression in human cancer cells. Eur J Pharmacol 2008;583(1):26–31. 10.1016/j.ejphar.2007.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Tenbaum SP, Ordóñez-Morán P, Puig I, Chicote I, Arqués O, Landolfi S, et al. [beta]-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med 2012;18(6):892–901. 10.1038/nm.2772 [DOI] [PubMed] [Google Scholar]
- 159. Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res 2007;13(14):4042–4045. [DOI] [PubMed] [Google Scholar]
- 160. Dehnhardt CM1, Venkatesan AM, Chen Z, Ayral-Kaloustian S, Dos Santos O, Delos Santos E et al. Design and synthesis of novel diaminoquinazolines with in vivo efficacy for beta-catenin/T-cell transcriptional factor 4 pathway inhibition. J Med Chem 2010; 53(2):897–910. 10.1021/jm901370m [DOI] [PubMed] [Google Scholar]
- 161. Hara A, Sakata K, Yamada Y, Kuno T, Kitaori N, Oyama T, et al. Suppression of β-catenin mutation by dietary exposure of auraptene, a citrus antioxidant, in N, N-diethylnitrosamine-induced hepatocellular carcinomas in rats. Oncol Rep 2005;14(2):345–351. [PubMed] [Google Scholar]
- 162. Olivier-Van Stichelen S, Guinez C, Mir AM, Perez-Cervera Y, Liu C, Michalski JC, et al. The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation. Am J Physiol Endocrinol Metab 2012;302(4):E417–24. 10.1152/ajpendo.00390.2011 [DOI] [PubMed] [Google Scholar]
- 163. Panno ML, Giordano F, Mastroianni F, Palma MG, Bartella V, Carpino A, et al. Breast cancer cell survival signal is affected by bergapten combined with an ultraviolet irradiation. FEBS Lett 2010;584(11):2321–2326. 10.1016/j.febslet.2010.04.001 [DOI] [PubMed] [Google Scholar]
- 164. Zeller J, Turbiak AJ, Powelson IA, Lee S, Sun D, Showalter HD et al. Investigation of 3-aryl-pyrimido 5, 4- [e] [1, 2, 4] triazine-5, 7-diones as small molecule antagonists of β-catenin/TCF transcription. Bioorg Med Chem Lett 2013; 23:5814–5820. 10.1016/j.bmcl.2013.08.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Sundram V, Chauhan SC, Ebeling M, Jaggi M. Curcumin attenuates β-catenin signaling in prostate cancer cells through activation of protein kinase D1. PLoS one 2012;7(4):e35368 10.1371/journal.pone.0035368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Jiang S, Liang Z, Wei G, Liu Y. Wnt/β-catenin pathway regulating glucocorticoids-mediated Alzheimer's disease-like pathological changes. Zhonghua Shenjingke Zazhi 2012;45(7):500–504. [Google Scholar]
- 167. Anitha P, Priyadarsini RV, Kavitha K, Thiyagarajan P, Nagini S. Ellagic acid coordinately attenuates Wnt/β-catenin and NF-κB signaling pathways to induce intrinsic apoptosis in an animal model of oral oncogenesis. Eur J Nutr 2013;52(1):75–84. 10.1007/s00394-011-0288-y [DOI] [PubMed] [Google Scholar]
- 168. Lu D, Cottam HB, Corr M, Carson DA. Repression of beta-catenin function in malignant cells by nonsteroidal antiinflammatory drugs. Proc Natl Acad Sci U S A 2005;102(51):18567–18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kaza N. Mechanisms of AT101 [(-)-gossypol] induced cytotoxicity in malignant peripheral nerve sheath tumors. ProQuest Dissertations and Theses.2014.
- 170. Singh T, Katiyar SK. Honokiol inhibits non-small cell lung cancer cell migration by targeting PGE2-mediated activation of β-catenin signaling. PloS one 2013;8(4):e60749 10.1371/journal.pone.0060749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Greenspan EJ, Madigan JP, Boardman LA, Rosenberg DW. Ibuprofen inhibits activation of nuclear {beta}-catenin in human colon adenomas and induces the phosphorylation of GSK-3{beta}. Cancer Prev Res (Phila) 2011;4(1):161–171. 10.1158/1940-6207.CAPR-10-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Blum CA, Xu M, Orner GA, Fong AT, Bailey GS, Stoner GD, et al. beta-Catenin mutation in rat colon tumors initiated by 1,2-dimethylhydrazine and 2-amino-3-methylimidazo[4,5-f]quinoline, and the effect of post-initiation treatment with chlorophyllin and indole-3-carbinol. Carcinogenesis 2001;22(2):315–320. [DOI] [PubMed] [Google Scholar]
- 173. Fila C, Metz C, van der Sluijs P. Juglone inactivates cysteine-rich proteins required for progression through mitosis. J Biol Chem 2008;283(31):21714–21724. 10.1074/jbc.M710264200 [DOI] [PubMed] [Google Scholar]
- 174. Canter RJ, Kesmodel SB, Heitjan DF, Veeramachaneni NK, Mokadam NA, Drebin JA, et al. Suppression of β-Catenin by Antisense Oligomers Augments Tumor Response to Isolated Limb Perfusion in a Rodent Model of Adenomatous Polyposis Coli–Mutant Colon Cancer. Annals of surgical oncology 2005;12(9):733–742. [DOI] [PubMed] [Google Scholar]
- 175. Brown JB, Lee G, Managlia E, Grimm GR, Dirisina R, Goretsky T, et al. Mesalamine inhibits epithelial β-catenin activation in chronic ulcerative colitis. Gastroenterology 2010;138(2):595–605. e3. 10.1053/j.gastro.2009.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wang Y, Zhang X, Wang H. Involvement of BMPR2 in the protective effect of fluoxetine against monocrotaline-induced endothelial apoptosis in rats. Can J Physiol Pharmacol 2011;89(5):345–354. 10.1139/y11-024 [DOI] [PubMed] [Google Scholar]
- 177. Watson S. Oncogenic targets of β-catenin-mediated transcription in molecular pathogenesis of intestinal polyposis. The Lancet 2001;357(9256):572–573. [DOI] [PubMed] [Google Scholar]
- 178. Roy HK, Karolski WJ, Wali RK, Ratashak A, Hart J, Smyrk TC. The nonsteroidal anti-inflammatory drug, nabumetone, differentially inhibits β-catenin signaling in the MIN mouse and azoxymethane-treated rat models of colon carcinogenesis. Cancer Lett 2005;217(2):161–169. [DOI] [PubMed] [Google Scholar]
- 179. Suh N, Reddy BS, DeCastro A, Paul S, Lee HJ, Smolarek AK, et al. Combination of atorvastatin with sulindac or naproxen profoundly inhibits colonic adenocarcinomas by suppressing the p65/beta-catenin/cyclin D1 signaling pathway in rats. Cancer Prev Res (Phila) 2011;4(11):1895–1902. 10.1158/1940-6207.CAPR-11-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Li J, Chen X, Ding X, Cheng Y, Zhao B, Lai Z, et al. LATS2 Suppresses Oncogenic Wnt Signaling by Disrupting β-Catenin/BCL9 Interaction. Cell reports 2013;5(6):1650–1663. 10.1016/j.celrep.2013.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang C, Dai J, Sun Z, Shi C, Cao H, Chen X, et al. Targeted inhibition of dishevelled PDZ domain via NSC668036 Depresses fibrotic process. Exp Cell Res 2015; 331(1):115–22. 10.1016/j.yexcr.2014.10.023 [DOI] [PubMed] [Google Scholar]
- 182. Piazza GA, Keeton AB, Tinsley HN, Whitt JD, Gary BD, Mathew B, et al. NSAIDs: old drugs reveal new anticancer targets. Pharmaceuticals 2010;3(5):1652–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Saini MK, Sanyal SN. Piroxicam and c-phycocyanin prevent colon carcinogenesis by inhibition of membrane fluidity and canonical Wnt/β-catenin signaling while up-regulating ligand dependent transcription factor PPARγ. Biomed Pharmacother 2014; 68 (5): 537–550. 10.1016/j.biopha.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 184. Mologni L, Brussolo S, Ceccon M, Gambacorti-Passerini C. Synergistic effects of combined Wnt/KRAS inhibition in colorectal cancer cells. PloS one 2012;7(12):e51449 10.1371/journal.pone.0051449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Shan B, Wang M, Li R. Quercetin inhibit human SW480 colon cancer growth in association with inhibition of cyclin D1 and survivin expression through Wnt/β-catenin signaling pathway. Cancer Invest 2009;27(6):604–612. 10.1080/07357900802337191 [DOI] [PubMed] [Google Scholar]
- 186. Xiong N, Cao X, Zhang Z, Huang J, Chen C, Zhang Z, et al. Long-term efficacy and safety of human umbilical cord mesenchymal stromal cells in rotenone-induced hemiparkinsonian rats. Biology of Blood and Marrow Transplantation 2010;16(11):1519–1529. 10.1016/j.bbmt.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 187. Muche S, Kirschnick M, Schwarz M, Braeuning A. Synergistic effects of beta-catenin inhibitors and sorafenib in hepatoma cells. Anticancer Res 2014;34(9):4677–4683. [PubMed] [Google Scholar]
- 188. Bao-ying H, Luo-yuan C, Xian-guo F. Effects of tanshinone IIA on wnt/β-catenin signaling pathway of high glucose induced renal tubular epithelial cell transdifferentiation. Chinese Journal of Integrated Traditional and Western Medicine 2012; 32(7):965–969. [PubMed] [Google Scholar]
- 189. Stein U, Fleuter C, Siegel F, Smith J, Kopacek A, Scudiero D, et al. Impact of mutant β-catenin on ABCB1 expression and therapy response in colon cancer cells. Br J Cancer 2012;106(8):1395–1405. 10.1038/bjc.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Mao Y, Tian W, Huang Z, An J. Convenient Synthesis of Toxoflavin that Targets β-Catenin/Tcf4 Signaling Activities. J Heterocycl Chem 2014;51(3):594–597. [Google Scholar]
- 191. Yadav VR, Prasad S, Aggarwal BB. Cardamonin sensitizes tumour cells to TRAIL through ROS-and CHOP-mediated up-regulation of death receptors and down-regulation of survival proteins. Br J Pharmacol 2012;165(3):741–753. 10.1111/j.1476-5381.2011.01603.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Li L, Wang Z, Wang Z. Combination of IL-24 and Cisplatin Inhibits Cervical Cancer Growth in a Xenograft Nude Mice Model. Asian Pac J Cancer Prev 2011;12:3293–3298. [PubMed] [Google Scholar]
- 193. Suzuki M, Nakagawa-Goto K, Nakamura S, Tokuda H, Morris-Natschke SL, Kozuka M, et al. Cancer preventive agents. Part 5. anti-tumor-promoting effects of coumarins and related compounds on Epstein-Barr virus activation and two-stage mouse skin carcinogenesis. Pharm Biol 2006;44(3):178–182. [Google Scholar]
- 194. Kaur J, Sanyal S. Oxidative stress and stress-signaling in chemoprevention of early colon cancer by diclofenac. Am J Biomed Sci 2010;2(1):63–78. [Google Scholar]
- 195. Way T, Huang J, Chou C, Huang C, Yang M, Ho CT. Emodin represses TWIST1-induced epithelial–mesenchymal transitions in head and neck squamous cell carcinoma cells by inhibiting the β-catenin and akt pathways. Eur J Cancer 2014; 50(2): 366–378. 10.1016/j.ejca.2013.09.025 [DOI] [PubMed] [Google Scholar]
- 196. Behari J, Zeng G, Otruba W, Thompson MD, Muller P, Micsenyi A, et al. R-Etodolac decreases β-catenin levels along with survival and proliferation of hepatoma cells. J Hepatol 2007;46(5):849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Tinsley HN, Grizzle WE, Abadi A, Keeton A, Zhu B, Xi Y, et al. New NSAID targets and derivatives for colorectal cancer chemoprevention Prospects for chemoprevention of colorectal neoplasia: Springer; 2013. p.105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Haegele L, Ingold B, Naumann H, Tabatabai G, Ledermann B, Brandner S. Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol Cell Neurosci 2003;24(3):696–708. [DOI] [PubMed] [Google Scholar]
- 199. Gardner H, Hawcroft G, Hull MA. Effect of nonsteroidal anti-inflammatory drugs on beta-catenin protein levels and catenin-related transcription in human colorectal cancer cells. Br J Cancer. 2004; 91(1): 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Hallett RM, Kondratyev MK, Giacomelli AO, Nixon AM, Girgis-Gabardo A, Ilieva D, et al. Small molecule antagonists of the Wnt/beta-catenin signaling pathway target breast tumor-initiating cells in a Her2/Neu mouse model of breast cancer. PloS one 2012;7(3):e33976 10.1371/journal.pone.0033976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Maier TJ, Janssen A, Schmidt R, Geisslinger G, Grosch S. Targeting the beta-catenin/APC pathway: a novel mechanism to explain the cyclooxygenase-2-independent anticarcinogenic effects of celecoxib in human colon carcinoma cells. FASEB J 2005;19(10):1353–1355. [DOI] [PubMed] [Google Scholar]
- 202.ChemDraw Ultra package (version 10.0). 2009. CambridgeSoft.
- 203. Todeschini R, Consonni V. Handbook of molecular descriptors2008. Wiley-VCH Verlag GmbH, Weinheim: 10.1002/9783527613106.oth2 [DOI] [Google Scholar]
- 204.StatSoft I. Statistica (data analysis software system), version 9.2009. Tulsa, USA.
- 205. De Maesschalck R, Jouan-Rimbaud D, Massart D. The mahalanobis distance. Chemom Intell Lab Syst 2000; 50(1):1–18. [Google Scholar]
- 206. Klecka WR. Discriminant analysis. Sage University Paper Series on Quantitative Applications in the Social Sciences,1980. Beverly Hills, CA: Sage Publications. [Google Scholar]
- 207. Furnival GM. All possible regressions with less computation.Technometrics 1971; 13:403–408. [Google Scholar]
- 208. Gálvez J, García-Domenech R, de Gregorio Alapont C, de Julián-Ortiz J, Popa L. Pharmacological distribution diagrams: a tool for de novo drug design. J Mol Graph 1996; 14:272–276. [DOI] [PubMed] [Google Scholar]
- 209.SPECS Database; SPECS NV: Delft, The Netherlands.
- 210. Mosmann T. Rapid colorimetric assay for celular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63. [DOI] [PubMed] [Google Scholar]
- 211. Vara D, Salazar M, Olea-Herrero N, Guzmán M, Velasco G, Díaz-Laviada I. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: role of AMPK-dependent activation of autophagy. Cell Death & Differentiation 2011;18(7):1099–1111. 10.1038/cdd.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Vara D, Morell C, Rodríguez-Henche N, Diaz-Laviada I. Involvement of PPARγ in the antitumoral action of cannabinoids on hepatocellular carcinoma. Cell death & disease 2013;4(5):e618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.