Background: CBMs are, generally, functionally and structurally autonomous from their associated catalytic domains.
Results: The structure of a novel cellulase, BhCel5B, reveals that the appended carbohydrate-binding module, CBM46, extends the enzyme catalytic cleft.
Conclusion: CBM46 targets BhCel5B to xyloglucan and is part of the catalytic cleft required for the hydrolysis of β-1,3–1,4-glucans.
Significance: CBM46 has a dual role in the hydrolysis of complex carbohydrates by BhCel5B.
Keywords: Carbohydrate-binding Protein, Cellulase, Cellulose, Plant Cell Wall, Protein Structure
Abstract
Structural carbohydrates comprise an extraordinary source of energy that remains poorly utilized by the biofuel sector as enzymes have restricted access to their substrates within the intricacy of plant cell walls. Carbohydrate active enzymes (CAZYmes) that target recalcitrant polysaccharides are modular enzymes containing noncatalytic carbohydrate-binding modules (CBMs) that direct enzymes to their cognate substrate, thus potentiating catalysis. In general, CBMs are functionally and structurally autonomous from their associated catalytic domains from which they are separated through flexible linker sequences. Here, we show that a C-terminal CBM46 derived from BhCel5B, a Bacillus halodurans endoglucanase, does not interact with β-glucans independently but, uniquely, acts cooperatively with the catalytic domain of the enzyme in substrate recognition. The structure of BhCBM46 revealed a β-sandwich fold that abuts onto the region of the substrate binding cleft upstream of the active site. BhCBM46 as a discrete entity is unable to bind to β-glucans. Removal of BhCBM46 from BhCel5B, however, abrogates binding to β-1,3–1,4-glucans while substantially decreasing the affinity for decorated β-1,4-glucan homopolymers such as xyloglucan. The CBM46 was shown to contribute to xyloglucan hydrolysis only in the context of intact plant cell walls, but it potentiates enzymatic activity against purified β-1,3–1,4-glucans in solution or within the cell wall. This report reveals the mechanism by which a CBM can promote enzyme activity through direct interaction with the substrate or by targeting regions of the plant cell wall where the target glucan is abundant.
Introduction
Deconstruction of plant cell wall carbohydrates is a process of considerable biological importance but is relatively inefficient due to the interlocking organization of polysaccharides within this macromolecular assembly (1, 2). Reflecting the complex organization of plant cell walls, which restricts enzyme access to their target substrates (3), hydrolysis of structural polysaccharides requires the cooperative action of a large consortia of carbohydrate active enzymes (CAZYmes),3 primarily glycoside hydrolases but also polysaccharide lyases, carbohydrate esterases, and polysaccharide oxidases (4–6). These enzymes have recently acquired a significant biotechnological significance in different industries, particularly in the emerging bioenergy and biorefinery sectors (7).
CAZYmes acting on recalcitrant substrates often present a modular architecture comprising a catalytic domain connected through flexible linker sequences to one or more noncatalytic carbohydrate-binding modules (CBMs). CBMs potentiate the activity of their appended catalytic modules by promoting a close interaction between the associated catalytic domains and their target substrates (8–13). Glycoside hydrolases and CBMs are grouped in families based on primary sequence similarities in the continuously updated CAZy database (14–16). Currently (July 2014), there are 133 families of GHs and 69 families of CBMs. GH5, a large glycoside hydrolase family, contains members from a broad spectrum of organisms that display a diversity of specificities against substrates comprising β-linked oligo- and polysaccharides and glycoconjugates (17). GH5 was recently organized into more than 51 evolutionary different subfamilies, which may be mono- or poly-specific. One example of a poly-specific GH5 subfamily is GH5_4 that includes endo-β-1,4-glucanases (EC 3.2.1.4), xyloglucanase (EC 3.2.1.151), β-1,3–1,4-glucanases (EC 3.2.1.73), and xylanases (EC 3.2.1.8). GH5 catalytic domains, including those of subfamily GH5_4, are generally fused to CBMs of different families, which display a ligand specificity that reflects the substrate specificity of the associated catalytic module.
Based on structure/function studies, CBMs were classified into three types. Type A CBMs recognize the surfaces of crystalline polysaccharides such as cellulose and are located in cellulases and also in noncellulosic enzymes (18). Type B CBMs bind internally single carbohydrate chains (endo-type). In contrast, type C CBMs bind the termini of a large variety of polysaccharides (exo-type) (13). Thus, CBMs are generally structurally independent of the associated catalytic domain and may express a variety of ligand specificities supported from three major surface topologies. One notable exception to this general trend is CBM3 subfamily c (CBM3c) (19). Members of subfamilies a and b were shown to bind strongly to crystalline cellulose (20, 21). In contrast, CBM3cs, as discrete entities, do not bind crystalline cellulosic substrates. Instead, CBM3c members are always associated with a subgroup of GH9 catalytic domains and have been shown to alter GH9 function from the standard endo-acting mode to a processive endo-mode of action (22, 23). Structural data revealed that CBM3c extends the catalytic site of the associated GH9 catalytic domains (22). More recent examples of CBMs modulating catalytic specificity is provided by a type C fructan-binding CBM66 that directs the cognate enzyme toward highly branched glucans rather than linear fructose polymers (24) and a CBM48 that contributes to substrate binding at the active site of a glucan phosphatase (25).
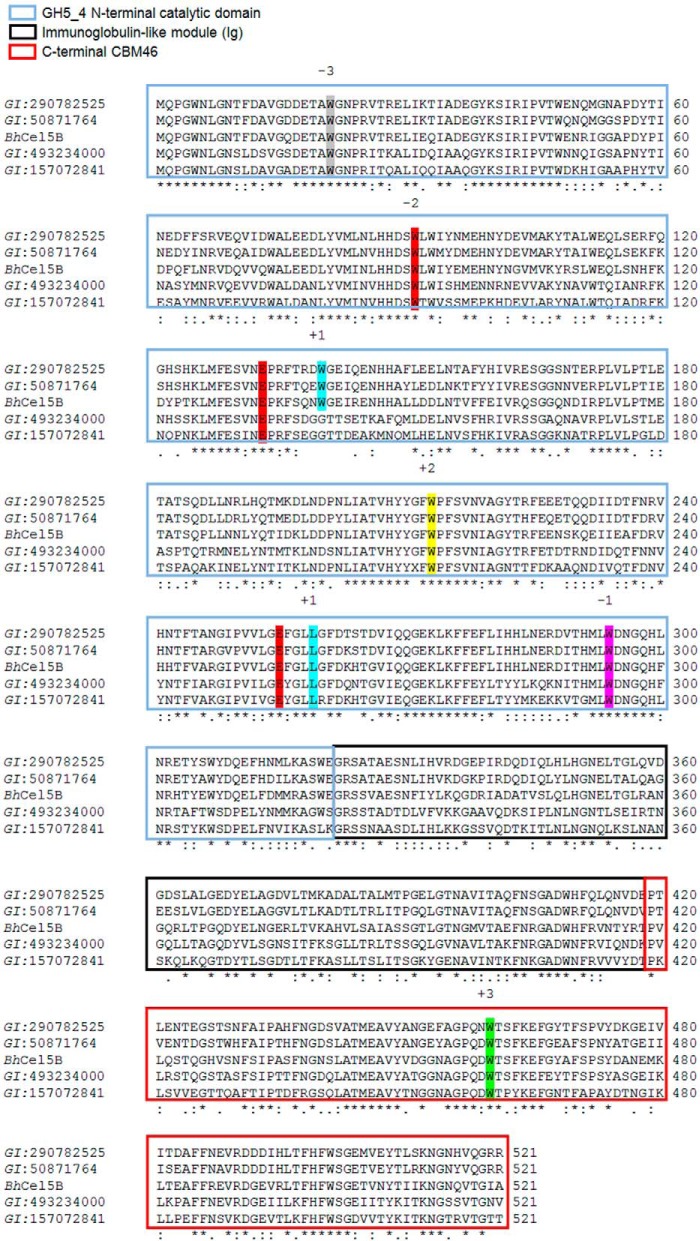
Although in general CBMs function independently from their appended catalytic domains, it is possible that there are other examples, similar to CBM3c, CBM66, and CBM48, in which CBMs are structurally integrated into the associated catalytic domains and make a direct contribution to enzymatic activity and specificity. Inspection of the CAZy database revealed a structurally uncharacterized CBM family, CBM46, where all members are associated with GH5_4 catalytic domains and are part of proteins displaying identical molecular architectures. Thus, all CBM46 members are found at the C terminus of CAZymes containing an N-terminal GH5_4 catalytic domain, followed by an internal immunoglobulin (Ig)-like module. Conservation in the molecular architectures of proteins containing CBM46 suggests a functional relevance for the association of GH5_4 and CBM46. Here, we report the biochemical, structural, and functional characterization of endo-β-1,4-glucanase B (BhCel5B) from B. halodurans (Fig. 1) (26). B. halodurans is a rod-shaped, Gram-positive soil bacterium that in comparison with Bacillus subtilis is known to colonize more alkaline environments (26). This study reveals that BhCBM46 as a discrete entity does not bind soluble or insoluble polysaccharides. The crystal structure of BhCel5B reveals that CBM46 is tightly associated with the catalytic module and, dependent on the glucan presented to the enzyme, can contribute directly to substrate binding or play a targeting role in directing the enzyme to regions of the plant cell wall rich in the polysaccharide hydrolyzed by the enzyme.
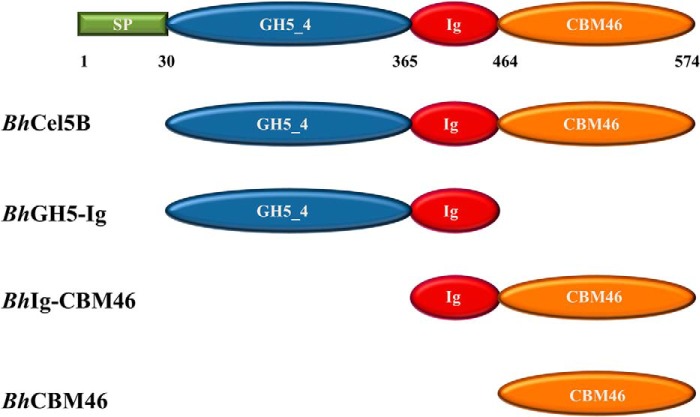
FIGURE 1.
Architectural arrangement of BhCel5B and truncated derivatives produced in this work. Green, signal peptide; blue, N-terminal glycoside hydrolase family 5 catalytic module; red, immunoglobulin-like module; orange, C-terminal family 46 CBM.
EXPERIMENTAL PROCEDURES
Carbohydrates
All carbohydrates were purchased from Megazyme International (Bray, County Wicklow, Ireland), except hydroxyethylcellulose that was obtained from Sigma and Avicel from Merck. Regenerated cellulose was prepared as described by Ref. 27.
Cloning, Expression, and Purification
DNA encoding full-length BhCel5B (residues 30–574; accession no. BA000004) and its truncated derivatives, BhGH5-Ig (residues 30–463), BhCBM46 (residues 464–574), and BhIg-CBM46 (residues 365–574), were amplified by PCR from B. halodurans genomic DNA using the thermostable DNA polymerase NZYProof (NZYTech Ltd., Portugal) and primers described in Table 1 (see Fig. 1 for molecular architecture of the proteins). Primers contained engineered restriction sites for direct cloning into the prokaryotic expression vector. Thus, the amplified genes were digested with NheI and XhoI and cloned into pET28a. The gene encoding BhCel5B_W501A_F504A_F507A_Y509A_R531A_E296A (Table 1) was synthesized in vitro (NZYTech Ltd.) with a codon usage optimized for expression in Escherichia coli. The synthesized gene was cloned into pET28a as described above. All recombinant proteins contained N-terminal His6 tags. Recombinant plasmids encoding BhCel5B derivatives were used to transform E. coli BL21 (DE3) cells. Expression of all proteins was achieved by adding isopropyl β-d-thiogalactopyranoside (1 mm final concentration) to mid-exponential phase (A600 nm = 0.6) grown cells with incubation for a further 16 h at 19 °C. The His6-tagged recombinant proteins, and their respective mutant derivatives, were purified from cell-free extracts by immobilized metal affinity chromatography as described previously (28). For crystallization, proteins were further purified by size exclusion chromatography. Following immobilized metal affinity chromatography, fractions containing the purified proteins were buffer-exchanged, using PD-10 Sephadex G-25 M gel filtration columns (GE Healthcare), into 50 mm Na-HEPES buffer, pH 7.5, containing 200 mm NaCl and 5 mm CaCl2. Recombinant proteins were subjected to gel filtration using a HiLoad 16/60 Superdex 75 column (GE Healthcare) at a flow rate of 1 ml/min. Preparation of E. coli to generate selenomethionylated BhCBM46 (SeMet-BhCBM46) was performed as described previously (29), and the protein was purified using the same procedures as employed for the native BhCBM46. Purified proteins were concentrated using an Amicon 10-kDa molecular mass centrifugal concentrator and washed three times with 5 mm DTT (for the Se-Met proteins) or water (for native BhCBM46 and BhCel5B), containing 1 mm CaCl2. Protein purity was analyzed through SDS-PAGE.
TABLE 1.
Primers used to clone the genes in this study
Mutation points are shown in bold.
| Genes | Residues | Primers | Direction |
|---|---|---|---|
| BhCel5B | 26–574 | GGATCCGTTAGTTCTGCTCATGAGGATGTG | Forward |
| GTCGACATTCGGGTAACACCATAGAAAGC | Reverse | ||
| BhCBM46 | 457–574 | TGGGATCCTATCGTACGCCTGTATTGC | Forward |
| GTCGACGGGTAACACCATAGAAAGCGCTT | Reverse | ||
| BhGH5-Ig | 30–463 | CACACGCTAGCGCTCATGAGGATGTGAAAAC | Forward |
| CACACCTCGAGTTGCAATACAGGCGTACG | Reverse | ||
| BhIg-CBM46 | 365–574 | CACACGCTAGCTCCGTTGCCGAGTCAAAC | Forward |
| CACACCTCGAGTGCGGCCGCAAGCTTGTCG | Reverse | ||
| BhCel5B_E296A | 26–574 | GGAATTCCAGTCGTTCTAGGT[b]GCGTTCGGCTTGCTTGGATTTG | Forward |
| CAAATCCAAGCAAGCCGAA[b]CGCACCTAGAACGACTGGAATTCC | Reverse | ||
| BhGH5-Ig_E296A | 30–463 | CACACGCTAGCGCTCATGAGGATGTGAAAAC | Forward |
| CACACCTCGAGTTGCAATACAGGCGTACG | Reverse | ||
| BhCel5B_W501A_E296A | 26–574 | GGAAATGCTGGCCCGCAAGAC[b]GCTACTTCCTTTAAGGAGTTTGG | Forward |
| CCAAACTCCTTAAAGGAAGT[b]AGCGTCTTGCGGGCCAGCATTTCC | Reverse | ||
| BhCel5B_F504A_E296A | 26–574 | GGCCCGCAAGACTGGACTTCC[b]GCCAAGGAGTTTGGCTATGCC | Forward |
| GGCATAGCCAAACTCCTT[b]GGCGGAAGTCCAGTCTTGCGGGCC | Reverse | ||
| BhCel5B_F507A_E296A | 26–574 | GACTGGACTTCCTTTAAGGAG[b]GCCGGCTATGCCTTCTCTCCTTC | Forward |
| GAAGGAGAGAAGGCATAGCC[b]GGCCTCCTTAAAGGAAGTCCAGTC | Reverse | ||
| BhCel5B_Y509A_E296A | 26–574 | CCTTTAAGGAGTTTGGC[b]GCCGCCTTCTCTCCTTCATATGATGC | Forward |
| GCATCATATGAAGGAGAGAAGGC[b]GGCGCCAAACTCCTTAAAGG | Reverse | ||
| BhCel5B_R531A_E296A | 26–574 | GGCGTTTTTTCGTGAGGTG[b]GCCGATGGTGAAGTTCGGTTAACC | Forward |
| GGTTAACCGAACTTCACCATC[b]GGCCACCTCACGAAAAAACGCC | Reverse | ||
| BhCel5B_W501A_F504A_F507A_Y509A_R531A_E296A | 30–574 | Gene synthesized | |
Site-directed Mutagenesis
Site-directed mutagenesis was carried out using the PCR-based NZYMutagenesis site-directed mutagenesis kit (NZYTech Ltd., Portugal) deploying the primers listed in the Table 1. BhCBM46, BhGH5-Ig, and BhCel5B were used as DNA templates. The generated DNA sequences were sequenced to ensure that only the engineered mutations had been incorporated into the nucleic acid.
Affinity Gel Electrophoresis
The binding to soluble polysaccharides was evaluated by affinity gel electrophoresis following the method described previously (30). Polysaccharide ligands were used at a concentration of 0.3% (w/v), unless otherwise stated. Electrophoresis was carried out at room temperature in native 10% (w/v) polyacrylamide gels. The gels were also loaded with BSA, which acts as a noninteracting negative control. After electrophoresis, proteins were visualized through staining with Coomassie Blue.
Isothermal Titration Calorimetry (ITC)
ITC experiments were carried out essentially as described previously (30), except that proteins were in 50 mm Na-HEPES buffer, pH 7.5, containing 200 mm NaCl at 25 °C. The reaction cell contained protein at 50 μm, and the syringe contained the polysaccharide at 20 mg/ml, unless stated otherwise. For experiments with regenerated cellulose, the ligand was retained in the cell at 12 mg/ml, and the protein (200 μm) was injected. Titrations were carried out at same conditions. Integrated heat effects, after correction for heats of dilution, were analyzed by nonlinear regression using a single site-binding model (Microcal ORIGIN, Version 5.0; Microcal Software). The fitted data yielded the association constant (Ka) and the enthalpy of binding (ΔH). Other thermodynamic parameters were calculated by using the standard thermodynamic equation: −RTlnKa = ΔG = ΔH − TΔS.
Interaction with Insoluble Polysaccharides
The binding of BhCBM46 to insoluble polysaccharide (Avicel) was carried out as follows. 30 μg of protein in 5 mm Tris-HCl buffer, pH 8.0, containing 0.05% (v/v) Tween 20 and 5 mm CaCl2 (Buffer A) were mixed with 20 mg of Avicel in a final reaction volume of 200 μl. The reaction mixture was incubated for 2 h at 4 °C with gentle shaking, after which time the insoluble ligand was precipitated by centrifugation at 13,000 × g for 5 min. The supernatant was removed, and the pellet was washed three times with 200 μl of Buffer A. Bound and unbound fractions were analyzed by SDS-PAGE using a 14% acrylamide gel. BSA (Sigma) and CBM3a from Clostridium thermocellum (NZYTech Ltd.) were included as negative and positive controls, respectively.
Enzyme Assays
BhCel5B and BhGH5-Ig were assayed for enzyme activity using the 3,5-dinitrosalicylic acid assay, described previously (31), to detect the release of reducing sugar. To explore the pH profile of BhCel5B, 50 mm MES, pH 4.5–7, 50 mm Tris-HCl, pH 7–9.5, 50 mm NaHCO3, pH 9–11, buffers were used in enzyme assays that employed 0.2% barley β-glucan as the substrate. The activity was determined at 55 °C by measuring the amount of reducing sugar released after a 10-min incubation period, using glucose to construct the standard curve. Determination of temperature of maximal enzyme activity for BhCel5B was performed by incubating the enzyme for 10 min at temperatures ranging from 20 to 80 °C and measuring reducing sugar release from barley β-glucan. For thermostability experiments, BhCel5B and BhGH5-Ig were incubated at temperatures ranging from 20 to 70 °C for 20 min, and residual activity was measured as described above at 30 °C. To determine kinetic parameters, assays with BhCel5B and BhGH5-Ig were carried out in 50 mm Tris-HCl buffer, pH 7, at 30 °C. Kinetic parameters were determined by nonlinear regression analysis using the Michaelis-Menten equation in GraphPad Prism 5.
Thin Layer Chromatography (TLC)
The qualitative analysis of BhCel5B and BhGH5-Ig hydrolysis products was performed by TLC on a silica gel-coated aluminum plate for detecting the released sugars. Reactions were performed in 20 mm sodium phosphate, pH 8, 0.1 mg/ml BSA, 0.3% (w/v) of substrate at 37 °C. Enzymes were incubated for 4 h, and at different time points the reactions were stopped by incubation at 100 °C for 10 min. Enzyme-substrate reaction product and standard and negative control were loaded onto the TLC plate.
Preparation of Plant Materials, Enzymatic Treatments, and Polysaccharide Immunodetection Procedures
Excised regions of tobacco and Miscanthus stems were fixed in 50 mm Pipes buffer, pH 6.9, containing 5 mm EGTA, 5 mm MgSO4, and 4% (v/v) paraformaldehyde and were embedded in wax and sectioned as described previously for tobacco stems (8). As access to xyloglucan can be masked by pectic homogalacturonan (32), tobacco stem sections were treated with pectate lyase (CjPel10A; 10 μg/ml) in 50 mm CAPS buffer, pH 10, containing 2 mm CaCl2 for 2 h before the activity of BhCel5B and its derivatives were assessed. Enzyme treatments were carried out as described previously (8) at concentrations ranging from low nanomolar to low micromolar for 30 min in phosphate-buffered saline (PBS) at 30 °C. Sections not treated with the enzymes were incubated for an equivalent time with the corresponding buffers. All sections were subsequently treated for 20 min with 5 μg/ml proteinase K (Sigma) in PBS before substrate detection to remove any enzymes still attached to the cell walls through their CBMs. Xyloglucan was detected with a rat monoclonal antibody LM15 (32), and β-1,3–1,4-glucan was detected with a mouse monoclonal antibody, deploying appropriate FITC-labeled secondary antibodies as described (8).
Immunofluorescence Microscopy and Quantification of Enzyme Impact on Cell Wall Polysaccharides
Immunofluorescence analysis was carried out with an Olympus BX-61 microscope equipped with epifluorescence irradiation, and all micrographs were captured with an ORCA 285 camera (Hamamatsu) using Volocity software (PerkinElmer Life Sciences). The relative capacities of the enzymes to degrade their substrates within cell walls were determined by quantitative assessments of the immunofluorescence intensities, which were captured in equivalent micrographs using a protocol that has been described (8). Briefly, using Volocity quantitation software, the absolute level of fluorescence contained in the micrographs was determined. For cell wall deconstruction, the modulation of signal is the disappearance of epitopes after polysaccharide degradation. Control micrographs obtained without enzymatic treatment were designated as 100% of initial fluorescence. In all cases the fluorescence quantification derives from the analysis of micrographs obtained from a minimum of four assessments for each enzyme concentration.
Crystallization and Data Collection
BhCBM46 and BhCel5B were crystallized by the sitting-drop vapor-phase diffusion method using the Oryx8 robotic nanodrop dispensing system (Douglas Instruments) (49, 50). The crystals of native BhCBM46 (28 to 14 mg/ml with 10 mm 1,4-β-d-cellohexaose (C6)) were obtained in 0.4 m potassium sodium tartrate tetrahydrate with equal volumes (0.7 μl) of protein and reservoir solution. Crystals of SeMet-BhCBM46 were obtained with equal volumes (0.7 μl) of protein (18 mg/ml) and reservoir solution (0.2 m ammonium sulfate, 30% (w/v) polyethylene glycol 4000). Crystals of BhCel5B grew in the following conditions: 0.2 m calcium acetate, 0.1 m cacodylate, pH 6.5, and 8% polyethylene glycol 8000 and 0.2 m CaCl2, 0.1 m HEPES, pH 7.4, and 25% PEG 4K. All crystals were cryo-cooled in liquid nitrogen using 30% (v/v) glycerol as cryoprotectant added to the harvesting solution.
All data sets were collected at the Diamond Light Source (Harwell, UK). Data sets were collected from BhCBM46 crystals on beamline I04-1 and on beamline I04, from BhCel5B crystals on beamlines I04 and 102. Data sets were processed for BhCBM46 using iMosflm (33) and AIMLESS (34) from the CCP4 suite (35). Data sets were processed for BhCel5B using the Fast_dp and xia2 (36) packages, which use the programs XDS (37), POINTLESS (38), and SCALA (38) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994) (35). BhCBM46 crystals belong to the tetragonal space group (I41 2 2), and BhCel5B crystal belongs to the orthorhombic space group P 21 21 2.
Structural Determination and Refinement
Data were collected for the native BhCBM46 to 2.46 Å resolution (Protein Data Bank code 4uzn). The data collected for the SeMet-BhCBM46 were used to solve the BhCBM46 structure. The crystal diffracted to a resolution of 2.3 Å (Protein Data Bank code 4uz8). The SeMet-BhCBM46 structure was determined by a single-wavelength anomalous dispersion experiment using AutoSol (39) from the PHENIX suite (40). The three-dimensional structure for the native BhCBM46 was solved by molecular replacement using Phaser (41) with the SeMet-derivative model as a search model, giving a TFZ of 22.0 and an LLG of 2404. Data collection and refinement statistics are presented in Table 2. The BhCel5B crystals diffracted to a resolution beyond 1.5 and to 2.75 Å. The best crystal was processed to a resolution of 1.64 Å (Protein Data Bank code 4v2x). BALBES was used to carry out molecular replacement (42). The best solution was found using the GH5 catalytic domain of endoglucanase D from Clostridium cellulovorans (PDB code 3ndz, with a sequence identity of 31.1%). An ARP/wARP (43) run after BALBES gave an almost complete model with 521 amino acid residues identified in a single chain, with an estimated correctness of 98%. Structure refinement and analysis are presented in Table 2.
TABLE 2.
Structure statistics
| Dataset | CBM46-SeMet peak | CBM46 native | Cel5B |
|---|---|---|---|
| Wavelength (Å) | 0.9798 | 0.9200 | 0.9795 |
| Resolution range (Å) | 85.45–2.3 (2.382–2.3) | 85.69–2.46 (2.548–2.46) | 70.97–1.64 (1.699–1.64) |
| Space group | I 41 2 2 | I 41 2 2 | P 21 21 2 |
| Unit cell | 120.85 120.85 76.38 90 90 90 | 121.19 121.19 77.28 90 90 90 | 74.22, 141.84, 50.82 90 90 90 |
| Total reflections | 246,067 (9270) | 87,997 (9188) | 480,050 (33,097) |
| Unique reflections | 12861 (1250) | 10723 (1059) | 66600 (4833) |
| Multiplicity | 19.1 (7.5) | 8.2 (7.7) | 7.2 (6.8) |
| Completeness (%) | 99.96 (99.68) | 99.74 (99.91) | 99.9 (99.8) |
| Mean I/σ(I) | 17.08 (3.83) | 9.66 (1.24) | 19.7 (2.6) |
| Wilson B-factor | 34.00 | 56.48 | 13.99 |
| Rmergea | 12.4 (79.9) | 12 | 7.3 (74.3) |
| Rp.i.m. b | 2.9 (30.4) | 4.6 (61.5) | 4.4 (45.7) |
| CC½c | 0.997 (0.89) | 0.998 (0.613) | 0.998 (0.60) |
| Average mosaicity | 0.23 | 1.53 | 0.77 |
| Reflections used for Rfree | 629 (54) | 519 (51) | 2943 (211) |
| Rwork | 0.2039 (0.2737) | 0.2143 (0.4199) | 0.1543 (0.1809) |
| Rfree | 0.2545 (0.3116) | 0.2400 (0.4382) | 0.2460 (0.256) |
| CC(work) | 0.945 | 0.956 | 0.970 |
| CC(free) | 0.920 | 0.947 | 0.962 |
| No. of non-hydrogen atoms | 1752 | 1669 | 4979 |
| Macromolecules | 1688 | 1646 | 4372 |
| Ligands | 5 | 0 | 68 |
| Water | 59 | 23 | 499 |
| Protein residues | 213 | 210 | 534 |
| r.m.s. (bonds) | 0.016 | 0.012 | 0.050 |
| r.m.s. (angles) | 1.83 | 1.41 | 1.07 |
| Ramachandran favored (%) | 90 | 90 | 97 |
| Ramachandran allowed (%) | 5 | 9 | 3 |
| Ramachandran outliers (%) | 5 | 1 | 1 |
| ClashScore | 15.60 | 2.20 | 5.08 |
| Average B-factor | 56.40 | 70.10 | 19.39 |
| Macromolecules | 56.80 | 70.30 | 19.10 |
| Ligands | 52.70 | 0 | 38.60 |
| Solvent | 44.40 | 55.30 | 31.60 |
| PDB code | 4uz8 | 4uzn | 4v2x |
a Rmerge = ΣhklΣi(Ii(hkl) − 〈Ihkl)〉)/ΣhklΣi(Ii(hkl), where Ii(hkl) is the ith intensity measurement of reflection hkl, including symmetry-related reflections, and 〈(I(hkl)〉 is its average.
b Rp.i.m. = (Σhkl√1/n − 1/Σj = 1n|Ihkl, j − 〈Ihkl〉|)/(ΣhklΣ Ihkl, j, where 〈Ihkl〉 is the average of symmetry-related observations of a unique reflection.
c CC½ is the half-data set correlation coefficient (51).
d Values for the outer shell are given in parentheses. Structure refinement was carried out using REFMAC5 (52), interspersed with manual rebuilding using COOT (53). In the penultimate round, the models were optimized using the PDB_REDO server (54). In the final round, each module (GH5, Ig_Like and CBM46) was treated as a single group for anisotropic TLS refinement.
RESULTS
Ligand Specificity of BhCBM46
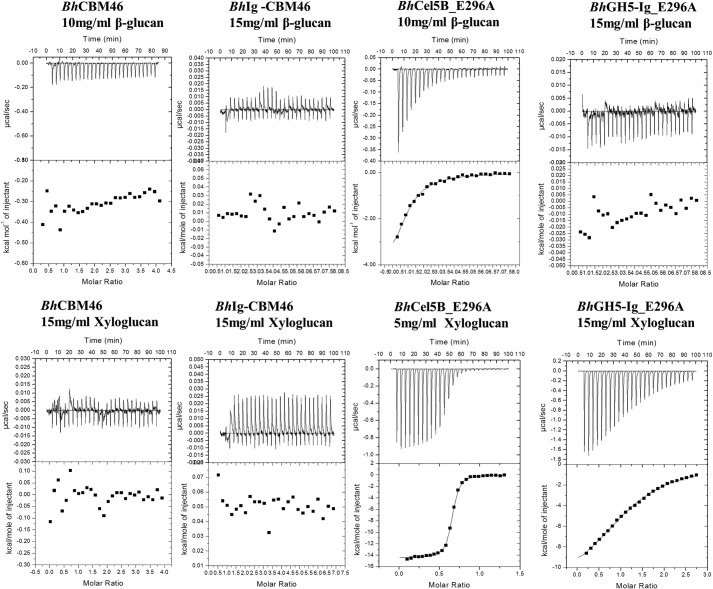
Previous qualitative studies suggested that the CBM46 of the endoglucanase BhCel5B displayed moderate binding to Avicel (44). Here, we have confirmed that the CBM displays some binding to Avicel; however, the affinity was too low to accurately quantify. Affinity gel electrophoresis showed that BhCBM46 fused to the central Ig domain (BhIg-CBM46) did not bind to a range of polysaccharides, including β-1,3–1,4-glucans (mixed linkage glucan), β-1,4-glucans, highly decorated β-1,4-glycans such as xyloglucan, mannan, glucomannan, xylans or to galactans, pectins, or α-glucans (data not shown). ITC confirmed that both BhIg-CBM46 and BhCBM46 did not bind to barley β-glucan (a β-1,3–1,4-glucan with an average sequence of Glc-β1,4-Glc-β1,4-Glc-β1,3-Glc) or to tamarind xyloglucan, which contains a core structure of a β1,4-linked Glc backbone that is decorated at O6 with β-d-Xyl units in the repeating sequence of XXXG (X is a Glc decorated with Xyl and G is an undecorated Glc), see Table 3. Taken together, the data suggest that BhCBM46 is unable to bind significantly to either soluble or insoluble carbohydrates.
TABLE 3.
Affinity and thermodynamic parameters of the binding of BhCBM46, BhCel5B_E296A and its variants to polysaccharide ligands
HEC is hydroxyethylcellulose and RC is regenerated cellulose.
| Ligand | Ka | ΔG | ΔH | TΔS | n | |
|---|---|---|---|---|---|---|
| m−1 | kcal mol−1 | kcal mol−1 | ||||
| BhCBM46 | Xyloglucan | No binding | ||||
| HEC | No binding | |||||
| β-Glucan | No binding | |||||
| RC | No binding | |||||
| BhIg-CBM46 | Xyloglucan | No binding | ||||
| BhCel5B_E296A | Xyloglucan | 1.27 (±0.06) × 107 | −9.7 ± 0.6 | −14.4 | −4.7 ± 0.1 | 0.6 ± 0.2 |
| BhGH5-Ig_E296A | Xyloglucan | 4.89 (±0.20) × 104 | −6.4 ± 0.3 | −12.1 ± 0.2 | −5.7 ± 0.2 | 1.2 ± 0.1 |
| BhCel5B_W501A_E296A | Xyloglucan | 8.91 (±1.41) × 105 | −8.1 ± 0.4 | −10.5 ± 0.3 | −2.4 ± 0.1 | 1.1 |
| BhCel5B_F504A_E296A | Xyloglucan | 2.64 (±0.41) × 106 | −8.8 ± 0.7 | −12.5 ± 0.2 | −3.7 ± 0.1 | 0.9 |
| BhCel5B_F507A_E296A | Xyloglucan | 8.89 (±1.60) × 106 | −9.4 ± 0.6 | −16.9 ± 0.2 | −7.5 ± 0.2 | 0.7 ± 0.1 |
| BhCel5B_Y509A_E296A | Xyloglucan | 5.58 (±1.51) × 106 | −9.1 ± 0.7 | −8.5 ± 0.2 | 0.6 | 0.9 |
| BhCel5B_R531A_E296A | Xyloglucan | 6.40 (±0.60) × 106 | −9.2 ± 0.6 | −9.8 ± 0.1 | −0.6 | 0.9 |
| BhCel5B_W501A_F504A_F507A_ | Xyloglucan | 4.96 (±0.41) × 105 | −7.8 ± 0.6 | −9.4 ± 0.2 | −1.6 ± 0.1 | 1 |
| Y509A_ R531A_E296A | ||||||
| BhIg-CBM46 | β-Glucan | No binding | ||||
| BhCel5B_E296A | β-Glucan | 1.05 (±0.11) × 105 | −6.8 ± 0.5 | −5.5 ± 0.6 | ||
To explore the function of BhCBM46 in the context of the full-length enzyme, two inactive variants of the endoglucanase were generated, BhCel5B-E296A and BhGH5-Ig-E296A, in which the catalytic nucleophile (Glu-296) had been substituted for alanine. BhCel5B-E296A includes the full-length enzyme, whereas BhGH5-Ig-E296A consists of the GH5_4 catalytic module and the central Ig domain but lacks the CBM46 module. Analysis of glycan binding of the two proteins by ITC (full dataset presented in Table 3 with example titrations in Fig. 2) showed that BhCel5B-E296A bound to barley β-glucan and xyloglucan with affinities of ∼105 and ∼107 m−1, respectively. Although BhGH5-Ig-E296A displayed no affinity for the β-1,3–1,4-glucan, the truncated derivative lacking CBM46 bound to xyloglucan with an affinity (∼4 × 104 m−1) 400-fold lower than BhCel5B-E296A. For both glycans, ligand binding was driven primarily by the change in enthalpy, a feature that is common to virtually all proteins that bind to soluble glycans (12). Overall, the data indicate that synergistic interactions between the GH5_4 catalytic domain and CBM46 of BhCel5B play a critical role in glucan binding. The contribution of the enhanced substrate binding afforded by BhCBM46 to catalytic activity is described below.
FIGURE 2.
Representative ITC data of BhCBM46, BhCel5B_E296A, and variants binding to soluble ligands. Titrations were conducted in 50 mm Na-HEPES buffer, pH 7.5, containing 200 mm NaCl at 25 °C. Top, ligand barley β-glucan in the syringe was titrated into cell contained protein (50 μm). Bottom, ligand xyloglucan in the syringe was titrated into cell contained protein (50 μm).
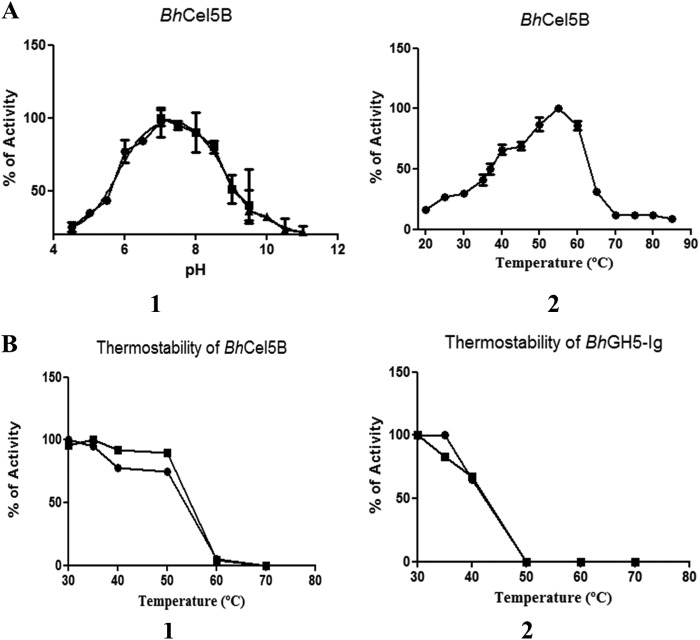
Effect of BhCBM46 on Enzyme Activity in Solution
The activities of recombinant forms of BhGH5-Ig and BhCel5B were evaluated. Consistent with the previous data of Wamalwa et al. (44), only the full-length enzyme was functional when assayed at 55 °C. A temperature stability profile of the endo-glucanase, however, showed that the CBM mediated a substantial increase in thermostability, and it was this stabilization effect that had a profound influence on enzyme activity (Fig. 3). We therefore explored the influence of BhCBM46 on the activity of the endoglucanase at a permissive temperature (30 °C), where the truncated enzyme, BhGH5-Ig, did not undergo thermal inactivation (Fig. 3). The data, presented in Table 4, showed that BhCel5B was ∼5-fold more active against the barley β-glucan than xyloglucan. TLC analysis of the products generated by BhCel5B against the two glucans revealed a range of differently sized oligosaccharides, indicative of an endo-acting mode of action (data not shown). The removal of the CBM46 had little impact on the activity of the enzyme against xyloglucan; however, BhGH5-Ig was ∼60-fold less active against barley β-glucan than the full-length enzyme (Table 4). These data show that in solution the CBM46 contributes to the degradation of β-1,3–1,4-glucans, but the module contributes little to xyloglucan hydrolysis.
FIGURE 3.
pH and temperature profile of BhCel5B (A) and thermostability of BhCel5B and BhGH5-Ig (B). A, panel 1, BhCel5B was incubated with 0.2% barley β-glucan at standard conditions in MES (●), Tris (■), and NaHCO3 (▴) buffers, and the activity was determined at 55 °C. A, panel 2, BhCel5B activity was determined with 0.2% barley β-glucan at different temperatures (●). B, panel 1, for thermostability, BhCel5B was incubated with 0.3% xyloglucan (●) or 0.3% barley β-glucan (■) for 30 min at different temperatures, and residual activity was determined at 30 °C. B, panel 2, BhGH5-Ig was incubated with 0.2% xyloglucan (●) or 0.7% barley β-glucan (■) for 30 min at different temperatures, and residual activity was determined at 30 °C.
TABLE 4.
Enzyme kinetics of BhCel5B and BhGH5-Ig against xyloglucan and barley β-glucan substrates
Errors reported are standard errors generated from triplicate results. Data were generated by nonlinear regression using the Michaelis-Menten equation in GraphPad Prism 5.
| Substrate |
BhCel5B |
BhGH5-Ig |
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | μm | |||
| Barley β-Glucan | 0.2578 ± 0.05 | 2.0152 ± 0.20 | 0.1819 ± 0.07 | 0.0224 ± 0.00 |
| Xyloglucan | 0.2740 ± 0.09 | 0.4043 ± 0.070 | 0.6416 ± 0.22 | 0.4547 ± 0.09 |
Effect of BhCBM46 on Enzyme Activity against Plant Cell Walls
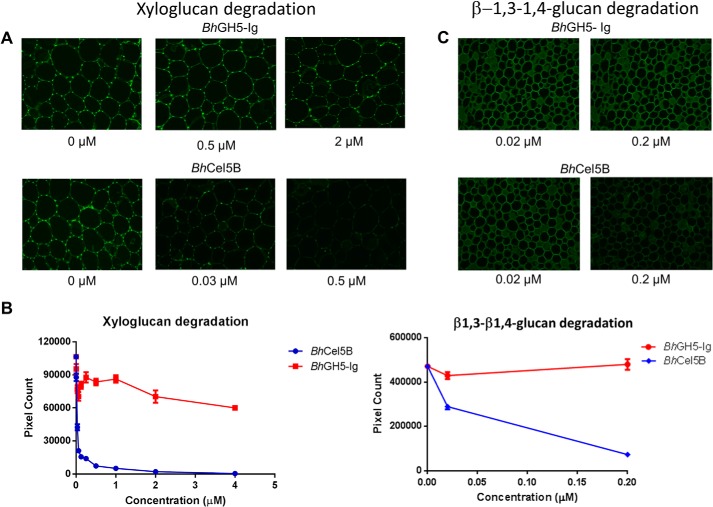
To assess the activity of BhGH5-Ig and BhCel5B against plant cell walls, the two enzymes at a range of concentrations were incubated with sections of tobacco (pretreated with pectate lyase to expose the xyloglucan (32)) and miscanthus stems. After 1 h, the reactions were stopped, and the amounts of xyloglucan and β-1,3–1,4-glucan remaining in the tobacco and miscanthus sections, respectively, were determined by immunohistochemistry using antibodies specific for the two polysaccharides. The data (Fig. 4) showed that BhCel5B at 30 nm removed 50% of the xyloglucan from tobacco cells, although even at 4 μm BhGH5-Ig was only able to remove ∼40% of the polysaccharide. With respect to miscanthus, BhCel5B at 200 nm removed ∼75% of the mixed linked, although at the same concentration BhGH5-Ig did not appear to release any of the polysaccharide. These data show that BhCBM46 enhances the enzymatic degradation of β-1,3–1,4-glucans. In contrast to the in vitro solution experiments described above, in the context of plant cell walls the CBM46 mediated a substantial increase in the xyloglucanase activity of the enzyme.
FIGURE 4.
Activity of BhCel5B and BhGH5-Ig against plant cell walls. The experiments were carried out as described under “Experimental Procedures.” A and C show representative micrographs of stem sections probed with antibodies (A shows tobacco stem sections probed with rat monoclonal antibody LM15 directed to xyloglucan, and C shows miscanthus stem sections probed with a mouse monoclonal antibody directed to β-1,3–1,4-glucan). B shows the quantified fluorescence intensities of the antibodies binding to equivalent stem sections after enzymatic treatments.
Crystal Structure of BhCBM46
To explore the structural basis for the different roles of BhCBM46 against xyloglucan and the β-1,3–1,4-glucan, the crystal structure of BhCBM46 and the full-length enzyme BhCel5B were determined. The crystal structure of BhCBM46 yielded an initial trace of the polypeptide backbone, which was further refined in this report yielding a structure determined to 2.2 Å. The CBM displays a classic β-sandwich jelly roll fold. The two β-sheets contain four anti-parallel β-strands. The order of the β-strands in β-sheet 1 and β-sheet 2 are β1, β2, β5, β4 and β3, β6, β7, β8, respectively. The β-strands are connected primarily by loops, although there is a small helix extending from residues Glu-524 to Val-530 (Fig. 5). Inspection of the β-sheet presenting a slight concave surface revealed an absence of aromatic residues, which generally play a central role in ligand recognition by CBMs (12). In contrast, the loop connecting β-strand 3 and 4 contains four aromatic residues comprising Trp-501, Phe-504, Phe-507, and Tyr-509. This loop may participate in carbohydrate recognition; however, such interactions are likely mediated only by Trp-501 and Tyr-509, as Phe-504 and Phe-507 are predominantly buried at the interface with the catalytic domain (see below). Three-dimensional structural comparison using the SSM site revealed that the closest structural homologue of BhCBM46 is the filamin immunoglobulin-like repeat from Homo sapiens (PDB code 2rgh), with a Z score of 4.9, r.m.s.d. of 2.85 Å over 97 aligned residues. Several other immunoglobulin-like modules with a β-sandwich fold showed similar levels of structural identity with BhCBM46. BhCBM46 presents a lower degree of homology with functionally relevant CBMs presenting a β-sandwich fold.
FIGURE 5.
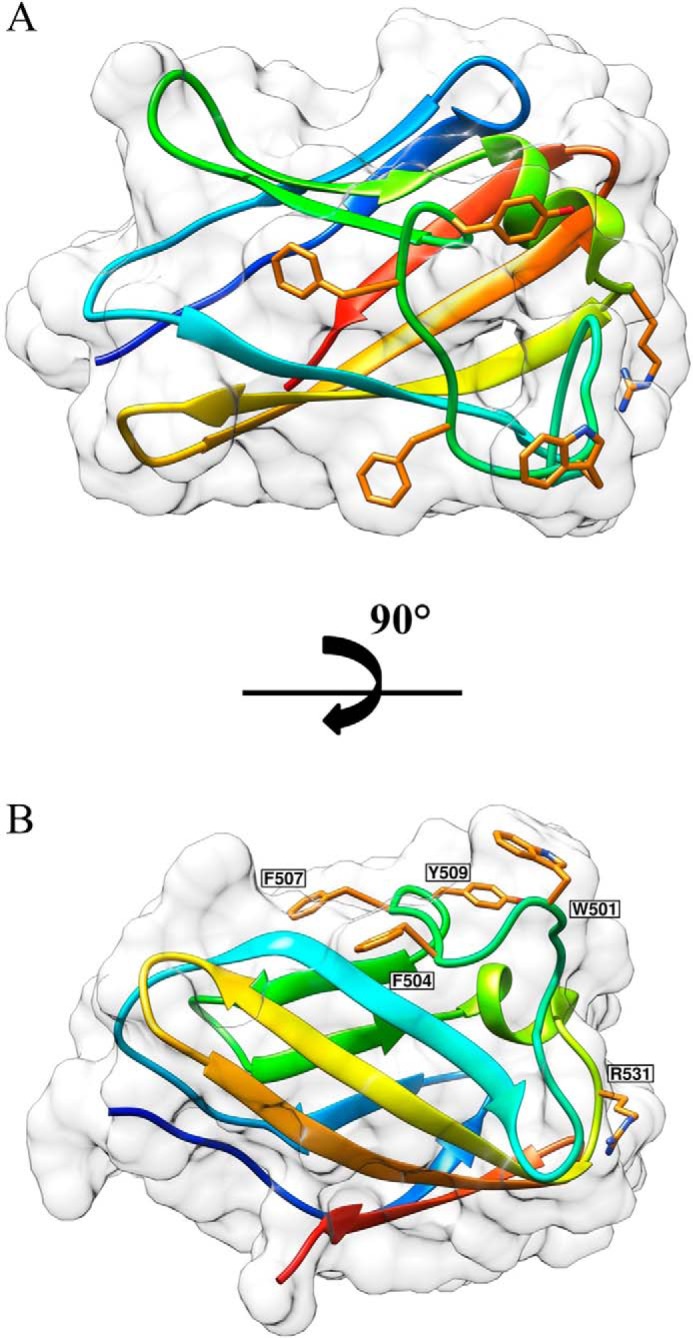
Structure of BhCBM46. A, BhCBM46 shows an open cleft. B, mutations are drawn as sticks. The pictures were prepared using Chimera (48).
The crystal structure of BhCBM46 suggests that residues Trp-501 and Tyr-509 may constitute the CBM46 ligand-binding site. Arg-531 is located at the surface of the protein in the vicinity of the aromatic residues and thus may also play a role in ligand recognition. Substituting these three residues with alanine individually in BhCel5B-E296A revealed that only the W501A mutation resulted in a substantial reduction in affinity for both xyloglucan and barley β-glucan, suggesting that the tryptophan makes an important contribution to ligand recognition. In conclusion, it would appear that evaluating the binding of mutants of CBM46 to glucans has little relevance to the biological role of these amino acids in a catalytically competent enzyme.
Crystal Structure of BhCel5B
To visualize the contribution of BhCBM46 to substrate recognition in the full-length enzyme, the structure of the trimodular β-1,4-glucanase BhCel5B (GH5-Ig-CBM46) was solved to a resolution of 1.64 Å by molecular replacement (Fig. 6). The structure of BhCBM46 reported above was used as the search model. The polypeptide chain is visible from Lys-31 to the C-terminal residue Gln-564.
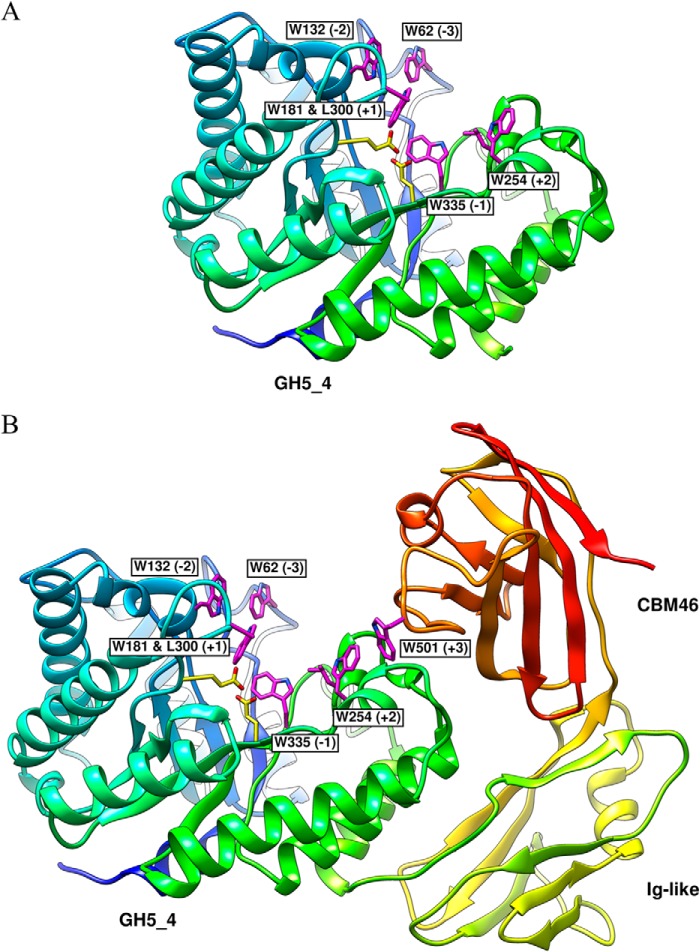
FIGURE 6.
Structure of BhCel5B. A, all important residues required for substrate recognition and catalysis presented on GH5_4 catalytic domain are drawn as sticks. B, BhCel5B is a tri-modular protein, composed of an N-terminal glycoside hydrolase family 5 catalytic module (GH5_4) followed by an immunoglobulin-like module (Ig) and a C-terminal family 46 CBM. Catalytic residues on GH5_4, Glu-170, and Glu-296, are drawn as sticks.
GH5_4
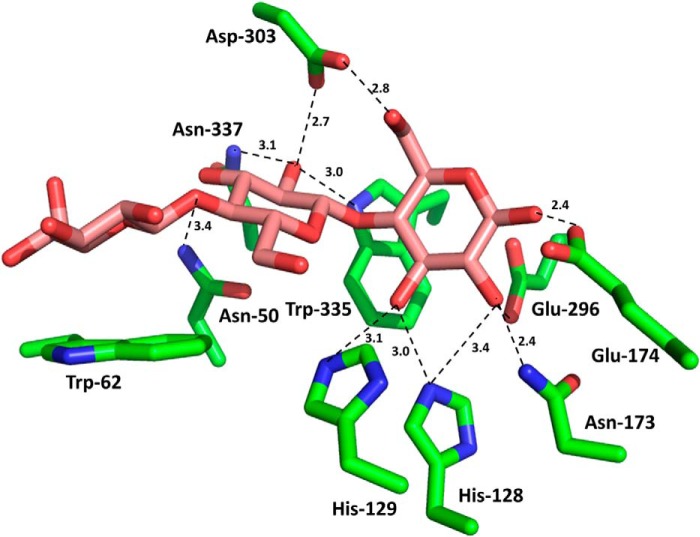
As expected, the N-terminal GH5_4 module displayed a (β/α)8 barrel architecture. GH5 enzymes are members of clan GH-A, in which the two catalytic residues are invariant glutamates presented at the end of β-strands 4 and 7 (45). From the structure of BhCel5B, the catalytic acid base is likely to be Glu-174 (end of β-strand 4) and the catalytic nucleophile Glu-274 (end of β-strand 7). The catalytic role of Glu-296 was confirmed by the observation that the mutant E296A is inactive (see above). A narrow and deep V-shaped cleft, ∼30 Å in length, extends along the entire length of the GH5_4 module and sits over the top of the β-barrel. Cleft dimensions and the position of the catalytic apparatus suggest that the protein contains ∼5 subsites extending from −3 to +2, although the presence of CBM46 might contribute to an additional subsite extending the number of positive subsites to three (see under “Discussion”). An analysis of structural homologues of GH5_4 using the SSM site identified a large number of GH5 and clan GH-A enzymes that displayed significant structural similarity to GH5_4. The C. cellulovorans endoglucanase D (PDB code 3ndz), with r.m.s.d. of 1.46 Å over 345 Cα atoms and a Z-score of 15.2, and the fungal GH5 endoglucanase of Piromyces rhizinflata (PDB code 3ays), with r.m.s.d. of 1.59 Å over 367 Cα atoms and a Z-score of 13.7, are the closest structural homologues. The structures of the two GH5_4 homologues were previously solved in complex with a cellotriose molecule bound to the −3, −2, and −1 subsites. An overlay of the three structures revealed that the −1 subsite (active site), where the transition state is formed, is similar in the three enzymes. Glu-174 makes a hydrogen bond with His-249, which may be important to both the position and ionization state of the catalytic acid base. The interactions of GH5_4 of BhCel5B with substrate can be predicted from an overlay of cellotriose (derived from PDB code 3ndz, where cellotriose is bound to the negative subsites of C. cellulovorans endoglucanase D) with the GH5–4 domain of BhCel5B. The overlay (Fig. 7) predicts that Asn-173, which is highly conserved in clan GH-A, hydrogen bonds with the O2 of the sugar at the −1 subsite. It is believed that this interaction plays an important role in transition state stabilization (46). The position of the GH5_4 catalytic nucleophile, Glu-296, is stabilized through hydrogen bonds with Tyr-251 and Arg-84, althoughTrp-335 is likely to form the sugar-binding hydrophobic platform at the −1 subsite. In addition to the amino acids coordinating substrate recognition and cleavage at the catalytic center, GH5_4 contains several residues in subsites distal to the active site that likely participate in substrate recognition and are conserved in other GH5s. Thus, at the −3 subsite, Trp-62 is predicted to make hydrophobic interactions with glucose and Asn-50 could make polar contacts with the sugar. At subsite −2, Asn-50 and Asp-303 are within hydrogen bonding distance of the sugar moiety. Significantly, subsite −2 of GH5_4 contains a tryptophan (Trp-132) that is not present in the other cellulases. The aromatic residue is in close proximity with the O6 of the sugar at the −2 subsite, suggesting that the Trp-132 side chain could make hydrophobic interactions with sugar decorations of the glucose backbone exemplified by xyloglucan. Indeed, the −3 subsite is particularly solvent-exposed and could accommodate decorations appended to O2, O3, or O6 of the bound glucose. Several aromatic residues may contribute to substrate recognition at the positive subsites of the substrate binding cleft of BhCel5B, notably Trp-254, Leu-300, and Trp-181. Thus, although Leu-300 and Trp-181 could form a pair of hydrophobic residues that could bind the α- and β-face of the sugar at subsite +1, Trp-254 could play a major role in carbohydrate recognition at the +2 subsite.
FIGURE 7.
Interaction of the negative subsites of the substrate binding cleft of BhCel5B with cellotriose. The amino acids of BhCel5B that are predicted to interact with cellotriose are shown in stick format with the carbons colored green. The cellotriose was derived from an overlay of overlay of BhCel5B with C. cellulovorans endoglucanase D in complex with the trisaccharide (PDB code 3ndZ; the r.m.s.d. for the overlay was 1.46 Å over 345 Cα atoms). The distance of the polar interactions are shown in Å.
Ig-like and CBM46 Modules within BhCel5B
The Ig-like module of BhCel5B consists of two β-sheets arranged around a hydrophobic core in a typical β-sandwich fold (Fig. 6). The structure is highly homologous to other immunoglobulin domains of prokaryotic or eukaryotic origin. The twisted pair of β-sheets contain β-strands β1, β7, β6, and β3 (β-sheet 1) and β-strands β2, β5, and β7 (β-sheet 2), respectively. The β-strands are connected primarily by loops, although there is a small helix extending from Ala-442 to Gly-432 and connecting β5 and β6. β1 and β7 of β-sheet 1 form a planar surface that establishes an extensive network of polar and apolar contacts with GH5–4 α-helices 7 and 8. The role of Ig-like domains in CAZYme function is not completely elucidated, although in the context of BhCel5B it may fulfill a structural anchor by providing the correct spatial organization of the associated protein modules in the context of the full-length enzyme. A small linker sequence (Thr-456 to Thr-459) connects the immunoglobulin like domain and CBM46. The structure of the CBM46 either when expressed individually (described above) or within BhCel5B was essentially identical (r.m.s.d. ∼1.2 Å). Thus, CBM46 does not undergo significant conformational changes when folded in the context of the entire protein. CBM46 β4 (β-sheet 1), β3 (β-sheet 2), and the loop connecting these two β-strands make a large number of contacts with GH5_4 loops connecting α7 and β7 and α6 and β6. In particular, CBM46 Phe-504, Phe-507, and Trp-542 dominate the hydrophobic contacts with the GH5_4 surface.
DISCUSSION
The data presented in this study show that BhCBM46 is not a BhCel5B-stabilizing domain but, in synergy with the catalytic module, binds to glucans and plays an active role in catalysis. The intriguing feature of the role of the CBM is that it appears to be variably dependent on the substrate. Against β-1,3–1,4-glucans the CBM participates in productive substrate binding and thus plays a direct role in the hydrolytic activity of the enzyme. In some ways the CBM46 resembles CBM3c modules that abut onto the substrate binding cleft of GH9 cellulases and are described in detail above. By contrast BhCBM46 was shown not to contribute to the hydrolysis of xyloglucan chains in solution. This suggests that the CBM is unable to bind to xyloglucan molecules that occupy the substrate binding cleft. BhCBM46, however, mediated a substantial increase in activity against xyloglucan contained in plant cell walls. This indicates that the CBM46 potentiates activity by directing the enzyme to regions of the cell wall rich in xyloglucan.
The crystal structure of BhCel5B provides insights into the possible mechanisms by which BhCBM46 makes different contributions to the activity of the enzyme. With respect to β-1,3–1,4-glucans, a β-1,3-linkage between Glc at the +1 and +2 subsites may enable the downstream glucan chain to make productive interactions with BhCBM46, explaining why this module contributes to the activity of BhCel5B against β-1,3–1,4-glucans. In contrast, the linear trajectory of β-1,4-glucan chains, such as xyloglucan, would result in these polymers occupying only the substrate binding cleft. These homopolymers would lack the kink in the backbone, afforded by a β-1,3-linkage, which is required for a glucan chain that occupies the active site and proximal subsites to access the CBM46. Inspection of the crystal structure, however, showed that a xyloglucan chain bound to the aromatic residues presented by the putative ligand-binding site of CBM46 would be too distant from the substrate binding cleft to prevent access to the active site. Thus, the CBM46 could contribute to substrate targeting in the context of the plant cell wall, without sterically hindering access to the active site.
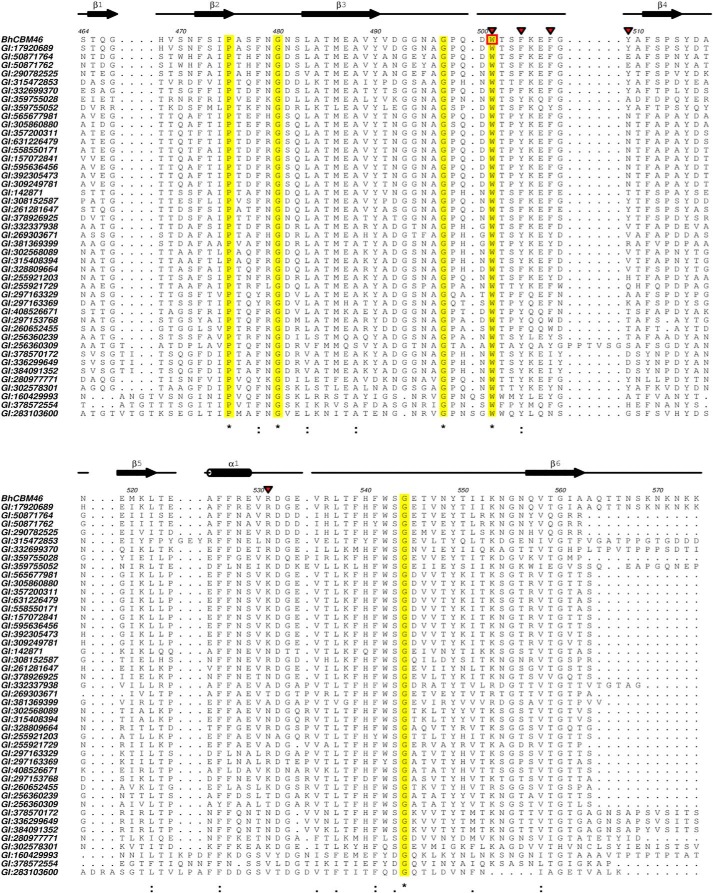
BhCel5B is a tri-modular protein composed of an N-terminal glycoside hydrolase family 5 catalytic module (GH5_4) followed by an immunoglobulin (Ig)-like module and a C-terminal family 46 CBM. Inspection of the CBM46 family revealed that all of its 45 members are located at the C terminus of CAZymes containing an N-terminal GH5_4 catalytic domain and an internal immunoglobulin-like module. This is unusual as generally CBMs of the same family are associated with catalytic modules of different families that display diverse substrate specificities (12), particularly in the context of plant cell wall-degrading enzymes (47). Alignment of CBM46 representatives revealed that Trp-501 is invariant in the family (Fig. 8). Thus, CBM46 may display a conserved function in polysaccharide recognition. Similarly, all the residues that are predicted to play an important role in catalysis or substrate binding in BhCel5B are conserved in five enzymes displaying a GH5-Ig-CBM46 molecular architecture with closest sequence similarity with BhCel5B (Fig. 9), suggesting that these proteins are optimized to mediate xyloglucan and β-1,3–1,4-glucan hydrolysis within plant cell walls. Significantly, the CBM46 contributes to the depolymerization of these glucans through distinct mechanisms.
FIGURE 8.
Alignments of CBM46 with all 45 representatives members. The alignment was made using Aline 011208. Residues that are invariant within the family are shaded in yellow and indicated by an asterisk. Mutations are indicated by inverted red triangle. The most important residue in carbohydrate recognition is reported with a red box.
FIGURE 9.
Alignments of BhCel5B with four proteins. The alignment was made using ClustalW2. Residues required for substrate recognition and catalysis are conserved in the five proteins. The residues occupying the subsites are indicated.
Acknowledgments
We acknowledge Diamond Light Source, Harwell, UK for provision of synchrotron-radiation facilities (beamlines I04, I04-1, and I02).
Note Added in Proof
Table 2 was incomplete, and Refs. 49 through 54 were inadvertently excluded in the version of this article that was published as a Paper in Press on February 23, 2015. Table 2 and the reference list have been corrected.
This work was supported by the Fundação para a Ciência e a Tecnologia, Portugal through Project PTDC/BIA-PRO/103980/2008. This work was also supported by European Union Seventh Framework Programme FP7 2007-2013 under the WallTraC Project Grant 263916 and BioStruct-X Grant Agreement 283570, Biostruct- X_4399.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 4uzn, 4uz8, and 4v2x) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CAZYmes
- carbohydrate active enzymes
- r.m.s.d.
- root mean square deviation
- CBM
- carbohydrate-binding module
- PDB
- Protein Data Bank
- Se-Met
- selenomethionylated
- ITC
- isothermal titration calorimetry
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
REFERENCES
- 1. Himmel M. E., Bayer E. A. (2009) Lignocellulose conversion to biofuels: current challenges, global perspectives. Curr. Opin Biotechnol. 20, 316–317 [DOI] [PubMed] [Google Scholar]
- 2. Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 [DOI] [PubMed] [Google Scholar]
- 3. Mohnen D. (2008) Pectin structure and biosynthesis. Curr. Opin Plant Biol. 11, 266–277 [DOI] [PubMed] [Google Scholar]
- 4. Gilbert H. J. (2010) The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Minic Z., Jouanin L. (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 44, 435–449 [DOI] [PubMed] [Google Scholar]
- 6. Gowen C. M., Fong S. S. (2010) Exploring biodiversity for cellulosic biofuel production. Chem. Biodivers 7, 1086–1097 [DOI] [PubMed] [Google Scholar]
- 7. Horn S. J., Vaaje-Kolstad G., Westereng B., Eijsink V. G. (2012) Novel enzymes for the degradation of cellulose. Biotechnol Biofuels 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boraston A. B., Kwan E., Chiu P., Warren R. A., Kilburn D. G. (2003) Recognition and hydrolysis of noncrystalline cellulose. J. Biol. Chem. 278, 6120–6127 [DOI] [PubMed] [Google Scholar]
- 10. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henrissat B., Davies G. J. (2000) Glycoside hydrolases and glycosyltransferases. families, modules, and implications for genomics. Plant Physiol. 124, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilbert H. J., Knox J. P., Boraston A. B. (2013) Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr. Opin. Struct. Biol. 23, 669–677 [DOI] [PubMed] [Google Scholar]
- 14. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 16. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aspeborg H., Coutinho P. M., Wang Y., Brumer H., 3rd, Henrissat B. (2012) Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 12, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Creagh A. L., Ong E., Jervis E., Kilburn D. G., Haynes C. A. (1996) Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U.S.A. 93, 12229–12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bayer E. A., Chanzy H., Lamed R., Shoham Y. (1998) Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 8, 548–557 [DOI] [PubMed] [Google Scholar]
- 20. Gilad R., Rabinovich L., Yaron S., Bayer E. A., Lamed R., Gilbert H. J., Shoham Y. (2003) CelI, a noncellulosomal family 9 enzyme from Clostridium thermocellum, is a processive endoglucanase that degrades crystalline cellulose. J. Bacteriol. 185, 391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., Steitz T. A. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 15, 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 22. Sakon J., Irwin D., Wilson D. B., Karplus P. A. (1997) Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 4, 810–818 [DOI] [PubMed] [Google Scholar]
- 23. Irwin D., Shin D. H., Zhang S., Barr B. K., Sakon J., Karplus P. A., Wilson D. B. (1998) Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J. Bacteriol. 180, 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cuskin F., Flint J. E., Gloster T. M., Morland C., Baslé A., Henrissat B., Coutinho P. M., Strazzulli A., Solovyova A. S., Davies G. J., Gilbert H. J. (2012) How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meekins D. A., Raththagala M., Husodo S., White C. J., Guo H. F., Kötting O., Vander Kooi C. W., Gentry M. S. (2014) Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc. Natl. Acad. Sci. U.S.A. 111, 7272–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takami H., Nakasone K., Takaki Y., Maeno G., Sasaki R., Masui N., Fuji F., Hirama C., Nakamura Y., Ogasawara N., Kuhara S., Horikoshi K. (2000) Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28, 4317–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boraston A. B. (2005) The interaction of carbohydrate-binding modules with insoluble non-crystalline cellulose is enthalpically driven. Biochem. J. 385, 479–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Najmudin S., Guerreiro C. I., Carvalho A. L., Prates J. A., Correia M. A., Alves V. D., Ferreira L. M., Romão M. J., Gilbert H. J., Bolam D. N., Fontes C. M. (2006) Xyloglucan is recognized by carbohydrate-binding modules that interact with β-glucan chains. J. Biol. Chem. 281, 8815–8828 [DOI] [PubMed] [Google Scholar]
- 29. Carvalho A. L., Goyal A., Prates J. A., Bolam D. N., Gilbert H. J., Pires V. M., Ferreira L. M., Planas A., Romão M. J., Fontes C. M. (2004) The family 11 carbohydrate-binding module of Clostridium thermocellum Lic26A-Cel5E accommodates β-1,4- and β-1,3–1,4-mixed linked glucans at a single binding site. J. Biol. Chem. 279, 34785–34793 [DOI] [PubMed] [Google Scholar]
- 30. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) The family 6 carbohydrate binding module CmCBM6-2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 31. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 [Google Scholar]
- 32. Marcus S. E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J. J., Farkas V., Pedersen H. L., Willats W. G., Knox J. P. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P. R. (2011) An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 67, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winter G., Lobley C. M., Prince S. M. (2013) Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69, 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 39. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wamalwa B. M., Sakka M., Shiundu P. M., Ohmiya K., Kimura T., Sakka K. (2006) Essentiality of a newly identified carbohydrate-binding module for the function of CelB (BH0603) from the alkaliphilic bacterium Bacillus halodurans. Appl. Environ. Microbiol. 72, 6851–6853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. U.S.A. 92, 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams S. J., Notenboom V., Wicki J., Rose D. R., Withers S. G. (2000) A new, simple, high-affinity glycosidase inhibitor: analysis of binding through x-ray crystallography, mutagenesis, and kinetic analysis. J. Am. Chem. Soc. 122, 4229–4230 [Google Scholar]
- 47. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., et al. (2009) Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 49. Venditto I., Santos H., Ferreira L. M. A., Sakka K., Fontes C. M. G. A., Najmudin S. (2014) Overproduction, purification, crystallization and preliminary x-ray characterization of the family 46 carbohydrate-binding module (CBM46) of endo-β-1,4-glucanase B (CelB) from Bacillus halodurans. Acta Crystallogr. F Struct. Biol. Commun. 70, 754–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Venditto I., Santos H., Ferreira L. M. A., Sakka K., Fontes C. M. G. A., Najmudin S. (2014) Overproduction, purification, crystallization and preliminary X-ray characterization of the tri-modular endo-β-1,4-glucanase B (Cel5B) from Bacillus halodurans. Acta Crystallogr. F Biol. Crystallogr. 70, 1628–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diederichs K., Karplus P. A. (2013) Better models by discarding data? Acta Crystallogr. D Biol. Crystallogr. 69, 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Joosten R. P., Long F., Murshudov G. N., Perrakis A. (2014) The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 1, 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]