Background: Lignocellulose-degrading microorganisms utilize binding modules associated with glycosidic enzymes to attach to polysaccharides.
Results: Structurally unique, discrete proteins (tāpirins) bind to cellulose with a high affinity.
Conclusion: Tāpirins represent a new class of proteins used by Caldicellulosiruptor species to attach to cellulose.
Significance: The tāpirins establish a new paradigm for how cellulolytic bacteria adhere to cellulose.
Keywords: Adhesin, Bacteria, Biodegradation, Cellulose, Extreme Thermophile, Protein Structure, Caldicellulosiruptor, Cellulose-binding Protein
Abstract
A variety of catalytic and noncatalytic protein domains are deployed by select microorganisms to deconstruct lignocellulose. These extracellular proteins are used to attach to, modify, and hydrolyze the complex polysaccharides present in plant cell walls. Cellulolytic enzymes, often containing carbohydrate-binding modules, are key to this process; however, these enzymes are not solely responsible for attachment. Few mechanisms of attachment have been discovered among bacteria that do not form large polypeptide structures, called cellulosomes, to deconstruct biomass. In this study, bioinformatics and proteomics analyses identified unique, discrete, hypothetical proteins (“tāpirins,” origin from Māori: to join), not directly associated with cellulases, that mediate attachment to cellulose by species in the noncellulosomal, extremely thermophilic bacterial genus Caldicellulosiruptor. Two tāpirin genes are located directly downstream of a type IV pilus operon in strongly cellulolytic members of the genus, whereas homologs are absent from the weakly cellulolytic Caldicellulosiruptor species. Based on their amino acid sequence, tāpirins are specific to these extreme thermophiles. Tāpirins are also unusual in that they share no detectable protein domain signatures with known polysaccharide-binding proteins. Adsorption isotherm and trans vivo analyses demonstrated the carbohydrate-binding module-like affinity of the tāpirins for cellulose. Crystallization of a cellulose-binding truncation from one tāpirin indicated that these proteins form a long β-helix core with a shielded hydrophobic face. Furthermore, they are structurally unique and define a new class of polysaccharide adhesins. Strongly cellulolytic Caldicellulosiruptor species employ tāpirins to complement substrate-binding proteins from the ATP-binding cassette transporters and multidomain extracellular and S-layer-associated glycoside hydrolases to process the carbohydrate content of lignocellulose.
Introduction
Interest in producing biofuels from lignocellulosic substrates has intensified focus on the mechanisms by which microorganisms degrade and utilize plant biomass. To date, most attention has been focused on cellulolytic enzymes implicated in this process, but it has been established for some time that noncatalytic, biomolecular contributions are critical to the degradation of crystalline cellulose (1, 2). In addition to catalytic domains, glycoside hydrolases (GHs)3 capable of this difficult biotransformation typically contain carbohydrate-binding modules (CBMs) that are currently classified into at least 71 families, based on amino acid sequence homology (3, 4). CBMs play a role in maintaining proximity between the active site and substrate surface, as well as in modifying the electronic structure of cellulose to promote hydrolysis (3). Metagenomic screening of microbial communities growing on cellulosic materials has typically expanded known inventories of GHs and CBMs (5–7) and has also facilitated the identification of novel CBM families (8) and GH families (9).
Species-level analysis of plant biomass deconstruction has revealed synergism between cellular and enzymatic processes in degrading lignocellulose. Indeed, within the expanding genome sequence databases, novel, often cell membrane-bound, cellulose-degrading systems are being discovered. For example, in cellulolytic members of the Fibrobacteres-Chlorobi-Bacteroidetes phyla, the ruminal bacterium Fibrobacter succinogenes (10) lacks identifiable cellobiohydrolases but does possess a novel, outer membrane-bound, modular endo-glucanase (CBM30-CBM11-CBM11-GH51) (11). Additionally, a large (180 kDa) glycosylated protein from F. succinogenes binds cellulose (12) through a Fibrobacter-unique “fibro-slime domain” (11). Another ruminal bacterium, Ruminococcus albus, uses a unique CBM family (CBM37) to facilitate anchoring of enzymes to the cell surface (13, 14). Other glycosylated proteins from R. albus also play a role in cellulose binding, including PilA1 homologs (15) from strains 8 (16) and 20 (17). Perhaps not surprisingly, type IV pili have also been demonstrated to be involved in adhesion of cells to cellulose in F. succinogenes (11) and Ruminococcus flavefaciens (18).
The genus Caldicellulosiruptor employs a variety of multifunctional enzymes, both cell-anchored and “free,” to deconstruct plant biomass at high temperatures (65–80 °C, see Refs. 19–21). In fact, one of the notable characteristics of the genus is their large modular enzymes, some of which are the largest single polypeptide glycoside hydrolases known (22, 23). Twelve genome sequences are now available for members of the genus, including eight genomes from species ranging from weakly to strongly cellulolytic (24). Previous comparative genomics analyses confirmed the enzymatic determinants for a strongly cellulolytic phenotype, namely the presence of modular enzymes that include a GH family 48 catalytic domain and CBM family 3 (24, 25). When one of these prominent modular cellulase genes (celA) was deleted from the genome, the ability of Caldicellulosiruptor bescii to grow on cellulose was significantly impacted (26). Remarkably, recent biochemical characterization of CelA indicates that the modular nature of this enzyme is beneficial such that it outperforms commercial cellulases (21).
Adherence to lignocellulosic substrates has been observed for the cellulolytic members of the genus Caldicellulosiruptor, including C. saccharolyticus (27), C. bescii (28), and C. obsidiansis (29). Although the mechanism by which this occurs has not been established, insights along these lines have been described. Some of the modular enzymes from the genus Caldicellulosiruptor are cell-anchored using S-layer homology domains (30) and are hypothesized to mediate cell-substrate proximity. In addition, substrate-binding proteins (in some cases, components of ATP-binding cassette transporters (24, 31)) may also play a role in orienting Caldicellulosiruptor species toward carbohydrate moieties in plant biomass, possibly through positively charged amino acid residues interacting with negatively charged areas (32). However, comparative proteogenomics analysis of whole cell, extracellular, and cellulose-bound fractions of Caldicellulosiruptor species revealed that previously annotated hypothetical proteins (referred to here as “tāpirins” taken from the Māori verb “to join”) bound tightly to cellulose, thereby implicating yet another element in the mechanism by which these bacteria attach to plant biomass. Recombinant versions of both classes of tāpirins confirmed their strong and specific affinity for cellulose. Moreover, three-dimensional structural information for one of the tāpirins demonstrated that these proteins are unique, not only from the perspective of amino acid sequence, but structurally as well.
EXPERIMENTAL PROCEDURES
Microorganisms, Plasmids, and Reagents Used
Axenic strains of C. bescii, Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor lactoaceticus, Caldicellulosiruptor owensensis, and C. saccharolyticus were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. Cloning (NovaBlue and DH10-β) and protein expression (Rosetta2[DE3] B834[DE3]) strains were purchased from EMD Millipore (Merck). The pRARE2 plasmid from Escherichia coli Rosetta2[DE3] was transferred to E. coli B834[DE3] for selenomethionine-labeled protein production. Genes of interest were cloned into pET46 Ek/LIC for protein production (EMD Millipore). Saccharomyces cerevisiae strain EBY100 and vector pCTCON were used for surface display of proteins. Yeast strains and vectors were a kind gift from Dr. Balaji Rao (North Carolina State University, Raleigh, NC). Carbohydrates and biomass used in this study include the following: Avicel PH-101 (50 μm) and PH-102 (100 μm, FMC Biopolymer, Philadelphia, PA); birchwood xylan (X0502, Sigma); ground Populus trichocarpa × Populus deltoides (80 mesh), dilute acid-pretreated (DAP) P. trichocarpa × P. deltoides and DAP switchgrass (Panicum virgatum) (National Renewable Energy Laboratory, Golden, CO).
Cultivation of Caldicellulosiruptor sp. for Microscopy and Proteomics Screening
Modified DSMZ medium 640 and culturing conditions used in this study are described elsewhere (25). Proteomics screening was conducted, as described previously (24). Cells were fixed for electron microscopy using a 4:1 (v/v) mixture of formaldehyde and glutaraldehyde, respectively. Scanning and transmission electron micrographs were captured at the Laboratory for Advanced Electron and Light Optical Methods (College of Veterinary Science, North Carolina State University). Epifluorescent microscopy was conducted as described previously (25) using Sytox Green (Invitrogen) to stain the cells.
Bioinformatic Analysis
Additional tāpirins were identified in related Caldicellulosiruptor species from the GenBankTM database nr and IMG database using BLASTP analysis (33) with known protein sequences as the query. Alignment of protein sequences used the Muscle algorithm (34). Neighbor-Joining phylogenetic trees were estimated and drawn using the MEGA version 6.0 software package, with 500 bootstraps used (35). Signal peptide leader sequences were predicted using SignalP 4.1 trained for Gram-positive bacteria (36). Transmembrane domains were predicted using the TMHMM Server, version 2.0, in conjunction with signal peptide data predicted by SignalP (37). InterPro (38) was used to scan for functional protein domain signatures.
Cloning, Production, and Purification of Recombinant Tāpirins
Calkro_0844 (GenBankTM accession number YP_004023543) and Calkro_0845 (GenBankTM accession number YP_004023544) were cloned without their respective signal peptides or transmembrane domains into the expression vector pET46 Ek/LIC (EMD Millipore). Oligonucleotide primer sequences used for cloning, including for Csac_1073 (GenBankTM accession number YP_001179878), are listed in Table 1. All tāpirin proteins were produced with N-terminal His6 tags for purification via immobilized nickel affinity columns (5 ml of HisTrap, GE Healthcare), according to manufacturer's protocols. Autoinduction medium (39) was used for induction of protein production, including higher concentrations of kanamycin (100 μg/ml) as recommended for higher phosphate media. Concentration of purified protein was determined using the bicinchoninic acid assay (Thermo Scientific Pierce) using bovine serum albumin for the standard curve.
TABLE 1.
Primers used in this study
Underlined sequences are either vector-specific or restriction enzyme sites.
| Primer name | Sequence | Cloning vector |
|---|---|---|
| Csac_1073 F | GACGACGACAAGATGTCTGCTGTGTTGGCATCACTG | pET46 Ek/LIC |
| Csac_1073 R | GAGGAGAAGCCCGGTTACTTTATAACAATGTTTCGTC | pET46 Ek/LIC |
| Calkro_0844 F | GACGACGACAAGATGTCGGCTGTGTTAGCATCGCTG | pET46 Ek/LIC |
| Calkro_0844 R | GAGGAGAAGCCCGGTTACCTTGTAACCATGTTTCGTC | pET46 Ek/LIC |
| Calkro_0845F | GACGACGACAAGATGTCGGCTGTGTTAGCATCGCTG | pET46 Ek/LIC |
| Calkro_0845R | GAGGAGAAGCCCGGTTATTTAACAACAATACTTCTTC | pET46 Ek/LIC |
| Calkro_0844 F GA | GGTGGTTCTGCTAGCGCATCGCTGAACCAGAGCACATCTATA | pCTCON |
| Calkro_0844 R GA | GCTTTTGTTCGGATCCCCTTGTAACCATGTTTCGTCTTAA | pCTCON |
| pCTCON F-BamHI | GACGAAACATGGTTACAAGGGGATCCGAACAAAAGCTTATTTCTGAAGAG | pCTCON |
| pCTCON R-NheI | GCTCTGGTTCAGCGATGCGCTAGCAGAACCACCACCACCAGA | pCTCON |
| YSD-Calkro_0845-F | GTCAGCTAGCATGTCGGCTGTGTTAGCATCGCTGAA | pCTCON |
| YSD-Calkro_0845-R | GTCACTCGAGCTATTATTTAACAACAATACTTCTTCTTAC | pCTCON |
Binding of Tāpirins to Substrates
Binding of tāpirins to insoluble substrates included the following: 40 μg of purified protein (Csac_1073, Calkro_0844, or Calkro_0845) and 9 mg of substrate in a total volume of 100 μl. For binding experiments, all of the substrates were washed overnight (10 g/liter substrate) with binding buffer (50 mm MES, 3.9 mm NaCl, pH 7.2) at 70 °C and dried at 70 °C in an oven prior to weighing them out for the binding assays. Binding was allowed to proceed at 70 °C and 750 rpm in a Thermo-mixer (Eppendorf). After 1 h of incubation, the bound and unbound portions of protein were separated via centrifugation, and the bound fraction was washed three times with binding buffer, centrifuging again after each wash. One volume of 2× Laemmli sample buffer was added to the unbound sample, and an equal volume of 1× Laemmli sample buffer diluted in the binding buffer was added to the bound sample. Samples were boiled for 30 min and then loaded onto an SDS-polyacrylamide gel for separation. SDS-polyacrylamide gels were stained with Gel Code Blue (Pierce) for visualization, with a protein ladder for reference (Benchmark, Life Technologies). Images are representative of three replicates.
Protein Adsorption to Cellulose
Recombinant tāpirin proteins (Calkro_0844 or Calkro_0845) were buffer-exchanged into binding buffer using 10-kDa molecular mass cutoff polyethersulfone ultrafiltration membranes (Millipore). Triplicate samples were established over a range of protein concentrations in microcentrifuge tubes with 3 mg each of substrate for 1 h at 70 °C and 700 rpm in a Thermo-mixer (Eppendorf). As a control, protein, lacking substrate, was also incubated in a microcentrifuge tube. Protein concentrations of each data point, for unbound or free protein, were calculated by averaging technical triplicates using the bicinchoninic acid assay. Calculated unbound protein concentrations were then corrected for any protein adsorbing to the microcentrifuge tube. Triplicate data points were fit to a Langmuir isotherm, using Equation 1,
 |
where Eb is the concentration of bound protein (μmol·g cellulose−1); Ef is the concentration of unbound protein (μm); Ka is the association constant (μm−1); and Bmax is the maximum amount of protein bound by cellulose (μmol·g cellulose−1). Parameters of association (Ka) and maximal binding capacity (Bmax) were estimated using JMP (version 9, SAS, Cary, NC).
Yeast Surface Display of Tāpirin Proteins
Tāpirins (Calkro_0844, Calkro_0845) were cloned into the vector pCTCON (40), using conventional ligation (Calkro_0845) or Gibson Assembly® master mix (Calkro_0844) (New England Biolabs), according to the manufacturer's directions. The cloning strategy excluded signal peptides and/or transmembrane domains; oligonucleotide primers used for cloning are listed in Table 1. Resulting clones were transformed into competent S. cerevisiae strain EBY100, using the Frozen-EZ Yeast Transformation II kit (Zymo Research). Transformed yeast cells were directly plated on selective SDCAA medium (per 1 liter of medium: 5 g of casamino acids, 6.7 g of yeast nitrogen base (Difco), 20 g of d-glucose, 7.45 g of monobasic sodium phosphate, 5.4 g dibasic sodium phosphate, 15 g of agar, 182 g of sorbitol). Fusion-protein expression and yeast binding to Avicel was conducted as described by Nam et al. (41). Briefly, for induction of recombinant protein, yeast cultures were initially subcultured into liquid SDCAA (as above, including 1:100 penicillin/streptomycin (Invitrogen) without sorbitol and agar) and allowed to grow overnight at 30 °C and 250 rpm in a shaking incubator. Cultures were harvested using centrifugation (2,500 × g for 5 min, 4 °C) and resuspended to an absorbance of 1 (A600) in liquid SGCAA medium (as above, substituting 5 g/liter d-galactose for d-glucose). Protein expression continued at 20 °C and 250 rpm for 20 h; afterward, the cells were pelleted by centrifugation as above and washed three times with phosphate-buffered saline (PBS) buffer, as described elsewhere (41). Cells were resuspended to an A600 of 3 in PBS with 10 mg/ml Avicel. Attachment to Avicel proceeded at 4 °C for 18 h with end-over-end rotation.
Immunofluorescence Microscopy with Yeast
Polyclonal antibodies raised against recombinant Calkro_0844 and Calkro_0845 were used for immunofluorescence of yeast cells expressing either tāpirin (GeneTel Laboratories, Madison, WI). For blocking, the yeast cell/Avicel slurry was resuspended in a blocking solution of 0.1% (w/v) bovine serum albumin in PBS and incubated on ice for 45 min. The cell slurry was then resuspended in primary antibody diluted 100× in blocking solution and incubated with end-over-end rotation at room temperature for 1 h. The cell slurry was then pelleted and washed with blocking solution three times, after which the slurry was incubated with goat anti-rabbit DyLight488 conjugate (ImmunoReagents) diluted 100× in blocking solution for 30 min at room temperature with end-over-end rotation. Cells were washed one time in PBS and mounted in SlowFade Gold (Invitrogen) prior to epifluorescence microscopy using ×40 magnification.
Truncated C-terminal Tāpirin Purification
Using the pET46 Ek/LIC-derived expression vector described above, selenomethionine-labeled (Acros Organics) Calkro_0844 was produced using protein production strain E. coli B834[DE3], pRARE2. A defined autoinduction medium (42) was used to incorporate selenomethionine into recombinant Calkro_0844. Purified Calkro_0844 was weakly digested with thermolysin (Promega) in the following reaction buffer: 50 mm Tris-Cl, pH 8.0, 0.15 m NaCl, and 0.5 mm CaCl2. Thermolysin was also resuspended at 1 mg/ml in the same reaction buffer. Protein and enzyme were mixed in a mass ratio of 1:500 and incubated for 1 h at 70 °C in a thermocycler. After 1 h, the reaction mixture was chilled on ice and immediately loaded on a Sephacryl HR size exclusion column (S-100, GE Healthcare), connected to a BioLogic LP System (Bio-Rad) to purify a roughly 45-kDa fragment. Purity of the fragment was confirmed using SDS-PAGE.
Crystallization
Calkro_0844_C crystals were obtained by sitting drop vapor diffusion using a 96-well plate with PEG ion HT screen from Hampton Research (Aliso Viejo, CA). Fifty μl of well solution was added to the reservoirs, and drops were made with 0.2 μl of well solution and 0.2 μl of protein solution using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, CA). The crystals were grown at 20 °C with 0.07 m citric acid, 0.03 m Bistris propane, pH 3.4, and 16% (w/v) polyethylene glycol (PEG) 3350 as the well solution. The protein solution contained 5.5 mg/ml protein in 50 mm Tris, pH 8, and 150 mm NaCl.
Data Collection and Processing
The Calkro_0844_C crystal was flash-cooled in a nitrogen gas stream at 100 K before data collection. The crystallization solution with the PEG 3350 concentration increased to 30% (w/v) was used for freezing the crystal. Data were collected using in-house Bruker X8 MicroStar X-Ray generator with Helios mirrors and Bruker Platinum 135 CCD detector. Data were then indexed and processed with the Bruker Suite of programs version 2013.8-1 (Bruker AXS, Madison, WI).
Structure Solution and Refinement
Intensities were converted into structure factors, and 5% of the reflections were flagged for Rfree calculations using programs SCALEPACK2MTZ, CTRUNCATE, MTZDUMP, Unique, CAD, FREERFLAG, and MTZUTILS from the CCP4 package of programs (43) version 6.4.0. PHASER EP from the CCP4 interface with HySS (44, 45) Phaser (46) was employed for finding the selenium sites using single wavelength anomalous dispersion. Phaser EP failed to build the model using the resulting phases, but using the selenium sites found by Phaser EP Crank2 (47), it successfully produced a model with a figure of merit of 0.9235. The structure was refined and manually rebuilt using REFMAC5 (48) version 5.8.0073 and Coot (49) version 0.7.2. The MOLPROBITY method (50) was used to analyze the Ramachandran plot, and root mean square deviations of bond lengths and angles were calculated from ideal values of Engh and Huber stereochemical parameters (51). Wilson B-factor was calculated using ctruncate version 1.15.5. Average B-factors were calculated using program ICM version 3.8 (Molsoft LLC, La Jolla, CA). The data collection and refinement statistics are shown in Table 2. Programs Coot, PyMOL, and ICM were used for comparing and analyzing structures. This structure has been deposited to the Protein Data Bank with entry code 4WA0.
TABLE 2.
X-ray data collection and refinement statistics
| Data collection | |
| Space group | P212121 |
| Unit cell, Å, ° | a = 61.51, b = 75.29, c = 77.45 α = β = γ = 90.00 |
| Wavelength, Å | 1.54178 |
| Temperature (K) | 100 |
| Resolution, Å | 25-1.7 (1.8-1.7)a |
| Unique reflections | 40244 (6215) |
| Rintb | 0.071 (0.539) |
| Average redundancy | 8.2 (5.7) |
| 〈I〉/〈σ(I)〉 | 17.5 (2.4) |
| Completeness, % | 99.8 (98.8) |
| Structure refinement | |
| Resolution, Å | 25-1.7 (1.74-1.70) |
| R/Rfree | 0.150 (0.257)/ 0.194 (0.345) |
| Protein atoms | 2904 |
| Water molecules | 583 |
| Other atoms | 1 |
| r.m.s.d.c from ideal bond length, Å (51) | 0.019 |
| r.m.s.d. from ideal bond angles, ° (51) | 1.969 |
| Wilson B-factor | 14.3 |
| Average B-factor for protein atoms, Å2 | 19.2 |
| Average B-factor for water molecules, Å2 | 32.5 |
| Ramachandran plot statistics, % (50) | |
| Allowed | 99.8 |
| Favored | 96.5 |
| Outliers | 1 (Asn-562) |
a Statistics for the highest resolution bin are in parentheses.
b Rint = Σ| I − 〈I〉|/Σ|I|, where I is the intensity of an individual reflection and 〈I〉 is the mean intensity of a group of equivalents, and the sums are calculated over all reflections with more than one equivalent measured.
c r.m.s.d. means root mean square deviation.
RESULTS
Proteomic Analysis of Caldicellulosiruptor Species Growing on Avicel Reveals Novel Cellulose-binding Proteins (Tāpirins)
Fig. 1 shows the attachment of a highly cellulolytic Caldicellulosiruptor species, C. kronotskyensis, to Avicel and dilute acid-pretreated biomass (DAP P. trichocarpa × P. deltoides or switchgrass) particles during growth. When observed under transmission electron microscopy, the outer cell surface appears rough, with structures protruding outside of the peptidoglycan layer (see Fig. 1, A and B). Additionally, C. kronotskyensis cells anchored in a web-like matrix were observed using scanning electron microscopy (Fig. 1C). Using epifluorescence microscopy (Fig. 1, D and E), C. kronotskyensis cells can be observed adhering to both Avicel and DAP biomass, implicating these ultrastructural features in attachment to lignocellulose. This observation is also representative of other Caldicellulosiruptor species growing on plant biomass-based substrates (28, 52). Previous studies showed that these bacteria are capable of forming biofilms on cellulosic substrates (29), but the intrinsic basis for their direct attachment to solid surfaces is unknown.
FIGURE 1.
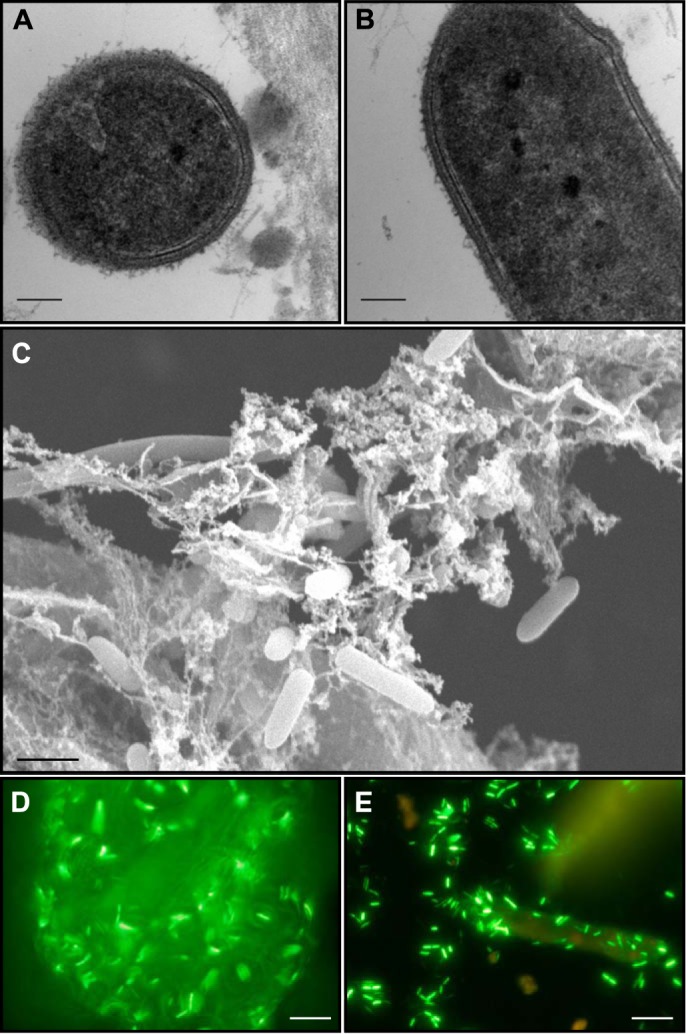
Micrographs of C. kronotskyensis grown on plant biomass or cellulose. C. kronotskyensis grown on dilute acid-pretreated P. trichocarpa × P. deltoides viewed as transmission electron micrographs from transverse (A) and longitudinal (B) sections and with scanning electron microscopy (C). With transmission electron microscopy, a fuzzy outer coat is observed through both sections. Using scanning electron microscopy, the cells appear embedded in a fibrous matrix. Epifluorescence microscopy of C. kronotskyensis grown on Avicel (D) and DAP switchgrass (E). In both cases, cells are adhering to cellulosic substrates. Scale bars, 100 nm (A and B), 1 μm (C), and 100 μm (D and E).
To further explore mechanisms underlying cell-surface attachment, a proteomics screen was conducted for several weakly to strongly cellulolytic Caldicellulosiruptor species growing on Avicel (see Fig. 2). From this screen, several Avicel-bound proteins could be identified (tāpirins), albeit annotated as hypothetical proteins in Caldicellulosiruptor genomes (24). The species previously determined to be moderately to strongly cellulolytic produce these proteins, which were highly enriched in the Avicel-bound (SB) fraction, although only the most cellulolytic species produced a second paralogous protein that was also enriched in the SB fraction (Fig. 2). In addition to normalized spectral counts, peptide coverage over the two tāpirin classes indicated that the first protein class was more abundant (46–73%) than the second one (9–43%). Two weakly cellulolytic Caldicellulosiruptor species (see Fig. 2) produced related proteins that were either enriched in the supernatant fraction (e.g. C. owensensis) or otherwise poorly expressed (e.g. C. hydrothermalis).
FIGURE 2.
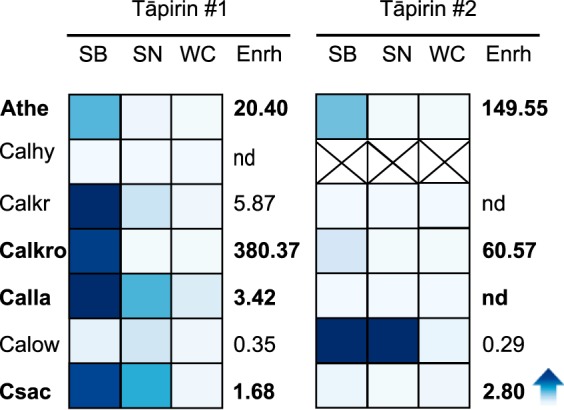
Heat plot of normalized spectral counts for Caldicellulosiruptor tāpirin proteins from selected species grown on Avicel. Darker shading of blue indicates a higher normalized spectral count. Species abbreviations are as follows: Athe, C. bescii; Calhy, C. hydrothermalis; Calkr, C. kristjanssonii; Calkro, C. kronotskyensis; Calla, C. lactoaceticus; Calow, C. owensensis; Csac, C. saccharolyticus. Column headings are abbreviated as follows: WC, whole cell; SN, supernatant; SB, substrate-bound; Enrh, Avicel enrichment score: (SB/(SN + WC)). For C. hydrothermalis, the × symbol indicates that there is no second class of tāpirin present. nd, not detected.
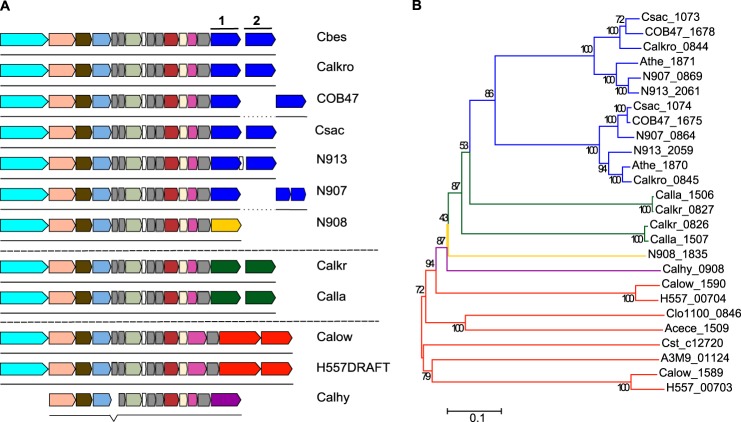
Bioinformatics analysis indicated that the tāpirins mapped to a locus in Caldicellulosiruptor genomes downstream of a locus predicted to encode a type IV pilus (Fig. 3A). Additionally, the predicted proteins are larger (ranging from 69 to 100 kDa) than typical type IV pilus-associated proteins. The genomes of strongly cellulolytic species (C. bescii, C. kronotskyensis, C. saccharolyticus, and C. obsidiansis) (24) all contained two paralogous classes of tāpirins, delineated by a phylogenetic tree built from alignments of their amino acid sequences (Fig. 3B). Interestingly, no other homologous genes or proteins to these two classes of tāpirins can be found in GenBankTM, outside of the genus Caldicellulosiruptor, confirming that these hypothetical proteins are unique to the genus. Furthermore, no protein domain signatures were detected from any homologs of the highly cellulolytic tāpirin proteins. Within each class of tāpirins, the orthologous proteins shared an amino acid sequence identity of greater than 80% identity over 95% or more of the protein (supplemental Table 1). The genomes of C. kristjanssonii and C. lactoaceticus also contained two genes encoding for proteins that were no more than 41% identical to the tāpirins from the four most cellulolytic Caldicellulosiruptor species (supplemental Table 1). These two tāpirin-like proteins can also be separated into two classes, because they shared even less amino acid homology (∼27%) to each other (supplemental Table 1 and Fig. 3).
FIGURE 3.
Genomic locus containing type IV pilus-related genes and Caldicellulosiruptor tāpirin. A, layout of genes theorized to encode type IV pili and tāpirins include from left to right: response regulator (aqua); PulE (peach); twitching mobility protein (dark olive); PulF (light blue); hypothetical proteins with prepilin-type N-terminal cleavage domains (gray); prepilin peptidase (mint); ComFB (white); pilus assembly protein PilM (dark pink); fimbrial assembly protein (cream); and pilus assembly protein PilO (magenta). Caldicellulosiruptor tāpirins are characterized as two classes of proteins by amino acid identity. Colors of the tāpirin genes indicate whether the families are from strongly cellulolytic (blue and yellow), moderately to weakly cellulolytic (green), or weakly cellulolytic (orange and purple) Caldicellulosiruptor species. Members of each color-differentiated tāpirin class (1 or 2) are categorized by 80% or more amino acid sequence identity over more than 60% of the query protein (see supplemental Table 1). B, phylogenetic tree was built using amino acid sequences from all sequences identified in Caldicellulosiruptor genomes through homology or position in relation to the type IV pilus operon. MEGA (version 6.0) was used to align amino acid sequences and build a neighbor-joining phylogenetic tree. Branches are colored to correspond with the groups of tāpirin genes noted in A. Species abbreviations follow gene locus tags when possible: Acece, Acetivibrio cellulolyticus CD2; A3M9, Caloramator sp. ALD01; H557, C. acetigenus; Athe, C. bescii; Calhy, C. hydrothermalis; Calkr, C. kristjanssonii; Calkro, C. kronotskyensis; Calla, C. lactoaceticus; COB47, C. obsidiansis; Calow, C. owensensis; Csac, C. saccharolyticus; N908, Caldicellulosiruptor sp. Rt8.B8; N913, Caldicellulosiruptor sp. Wai35.B1; Cst_c, Clostridium stercorarium subsp. stercorarium; Clo1100, Clostridium sp. BNL1100; N907, Thermoanaerobacter cellulolyticus.
Both C. owensensis and C. acetigenus, neither of which degrades cellulose to any extent (25, 53), each contained two genes downstream of the type IV pilus locus which had highly divergent amino acid sequences from the other tāpirins and from each other. Finally, C. hydrothermalis, also not capable of significant cellulose degradation (24, 25), contained just one protein, which shared little homology to either class of tāpirins from the strongly cellulolytic Caldicellulosiruptor species. However, this protein does share weak amino acid homology, 24%, with a tāpirin from a newly sequenced species, Caldicellulosiruptor sp. strain Rt8.B8 (supplemental Table 1). Another Caldicellulosiruptor species that is not fully sequenced, Caldicellulosiruptor sp. strain Tok7.B1, also encodes for a protein that has homology with part of this type of tāpirin (GenBankTM accession number AAD30365), sharing 91% homology over 29% of the protein from Caldicellulosiruptor sp. Rt8.B8. This indicates that there is further diversity among the tāpirins from Caldicellulosiruptor species in nature.
Confirmation of Cellulose-binding Capacity for C. kronotskyensis Tāpirins
To establish that the tāpirins encoded by Calkro_0844 and Calkro_0845 could bind to cellulose, several approaches were used. First, we sought to determine whether the proteins were capable of binding to cellulose in the absence of any hypothesized interactions with the type IV pilus. Selected representative genes from each class of cellulolytic tāpirin (i.e. Calkro_0844 and Calkro_0845) were expressed as chimeric proteins fused to the C terminus of the yeast α-agglutinin protein to facilitate yeast surface display (40, 54). Previously, yeast surface display systems in S. cerevisiae have successfully expressed and fused cellulose-specific CBM modules (41) and cellulases (55) from cellulolytic fungi to the yeast cell wall. Other protein complexes from bacterial systems can also successfully display on the yeast surface, including assembly of mini-cellulosomes using components from mesophilic (56) and thermophilic Clostridia (57–59).
After protein induction in the yeast host, each tāpirin could mediate cell attachment to Avicel (Fig. 4, C and D), whereas yeast cells not expressing either of the proteins were unable to attach (Fig. 4, A and B). Using polyclonal antibodies raised against one or the other class of tāpirins (rabbit anti-Calkro_0844 and rabbit anti-Calkro_0845), immunofluorescent microscopy demonstrated that the recombinant proteins were produced and linked to the yeast cell surface. Fluorescent signals were observed after binding to a DyLight488-conjugated secondary antibody (goat anti-rabbit) primarily located at the interface between the Avicel particles and yeast cells (Fig. 4, G and H). Therefore, it appears that the inherent binding ability of the tāpirins remains intact. This was the case even when expressed in a eukaryotic yeast surface display system at temperatures approaching 50 °C below the optimal growth temperature of the parent organism and in the absence of type IV pili.
FIGURE 4.
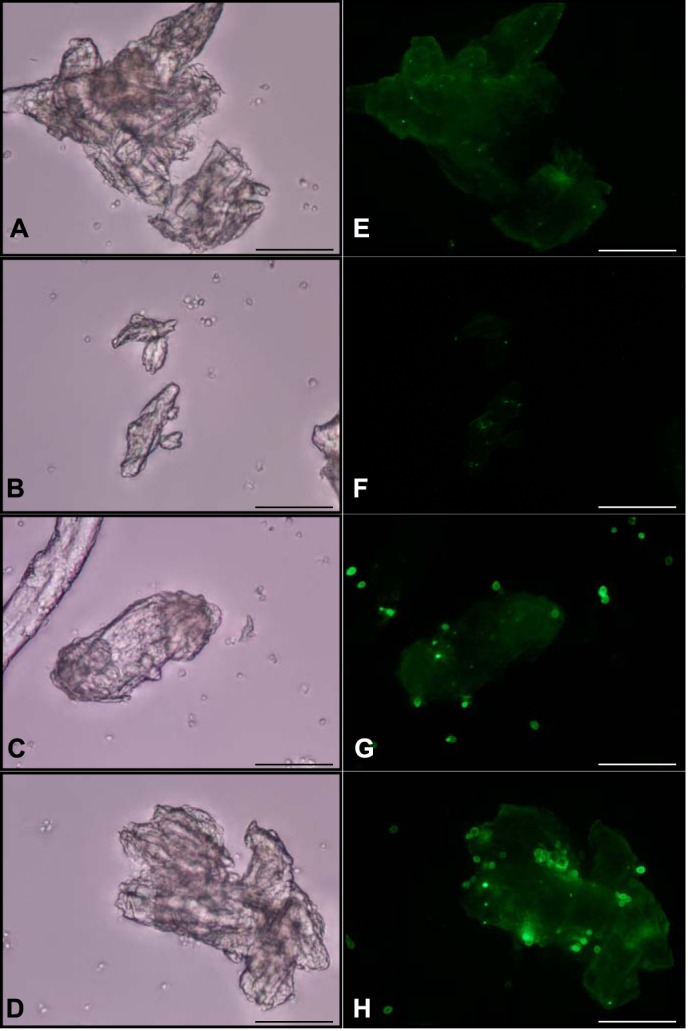
Use of immunofluorescence microscopy to detect tāpirin proteins displayed on the cell wall of yeast. White light and epifluorescent images for S. cerevisiae EBY100 treated with anti-Calkro_0844 (A and E) or anti-Calkro_0845 (B and F) antibodies. S. cerevisiae EBY100 expressing Calkro_0844 was observed under white light (C) and epifluorescence (G) after incubation with anti-Calkro_0844 antibodies. S. cerevisiae EBY100 expressing Calkro_0845 was observed under white light (D) and epifluorescence (H) after incubation with anti-Calkro_0845 antibodies. Goat anti-rabbit conjugated with DyLight488 was used as a secondary antibody. All images were captured at ×40; scale bar in each image is 50 μm.
A second in vitro approach sought to confirm the ability of recombinant versions from both classes of tāpirins to bind to a variety of carbohydrate and plant biomass substrates. Both classes of tāpirin proteins demonstrated some binding affinity to a variety of cellulosic substrates, including Avicel, filter paper, and dilute acid-pretreated plant biomass (Fig. 5). Multiple recombinant members of tāpirin class 1 (Csac_1073 and Calkro_0844, see Fig. 3) were used to confirm that similar binding profiles would occur between orthologs (Fig. 5, A and B). Importantly, the binding to cellulose appears to be specific, as neither of the tāpirins tested bound to xylan (Fig. 5, A–C). Presumably, the weak binding to unpretreated biomass is in part due to xylan masking the majority of available cellulose-binding sites. Because the binding assays were conducted at temperatures characteristic of the environment from which the proteins came, a type IV pilus is not required for the tāpirins to adsorb to cellulose.
FIGURE 5.
SDS-PAGE analysis of tāpirin binding to various plant cell wall components and plant biomass. Tāpirins tested include Csac_1073 (class 1) (A), Calkro_0844 (class 1) (B), and Calkro_0845 (class 2) (C), and thermolysin-digested Calkro_0844 (Calkro_0844_C) (D). Abbreviations for plant biomass substrates are as follows: aSWG, dilute acid-pretreated switchgrass; aPTD, dilute acid-pretreated P. deltoides × P. trichocarpa; PTD, P. deltoides × P. trichocarpa. B, bound protein liberated from the substrate after boiling in 1× Laemmli buffer; U, free protein. 40 μg of protein was used in all conditions tested; image is representative of three replicates.
To further investigate the specificity of adsorption for both classes of tāpirins to cellulose, binding affinities of both C. kronotskyensis proteins were modeled using Langmuir isotherms (Fig. 6). Affinity data for both classes of tāpirins revealed that each protein bound to filter paper with an association constant (Ka) of ∼0.7 μm−1, with more total protein binding to filter paper from tāpirin class 2. Filter paper appears to be the better binding substrate when both measures of affinity (Figs. 5, B and C, and 6) are considered. In contrast, the tāpirin from Calkro_0845 exhibited a higher affinity for Avicel (Ka of 0.94 μm−1) than that of Calkro_0844 (Ka of 0.05 μm−1), although more total protein from Calkro_0844 bound to Avicel. These association constants are within the range (μm−1) previously reported for cellulose-binding proteins associated with Avicel or filter paper, including Trichoderma reesei CbhI (0.15 μm−1, Ref. 60), cellulose-binding CBM families 2, 3, and 17 (0.3–3.33 μm−1, Refs. 60–63), fungal swollenin (1.14 μm−1, Ref. 64), and bacterial expansin (0.45 μm−1, Ref. 61). Because there is no sequence similarity between these tāpirins and other cellulose-binding proteins previously reported, it appears that the genus Caldicellulosiruptor has developed a unique mechanism through which cells may attach to cellulose.
FIGURE 6.
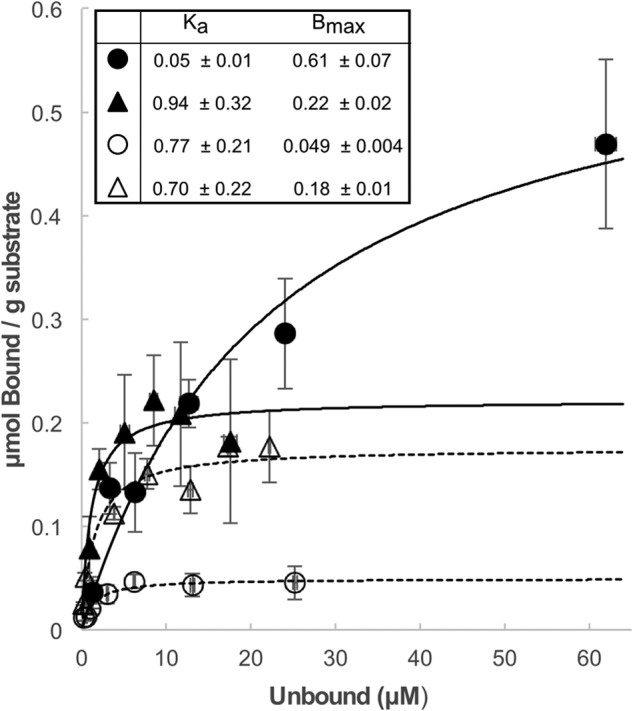
Binding isotherm of selected tāpirin to filter paper and Avicel. MES (50 mm), pH 7.2, NaCl (3.9 mm) as used as the binding buffer. Equilibrium association constant (Ka, μm−1) and binding capacity (Bmax, μmol g−1) for each protein and substrate combination are indicated in the inset table. Binding isotherms are represented by the following: solid circles for Calkro_0844 and Avicel; empty circles for Calkro_0844 and filter paper; solid triangles for Calkro_0845 and Avicel; empty triangles for Calkro_0845 and filter paper.
Three-dimensional Crystal Structure of Calkro_0844
To determine how the structure of the tāpirins influenced their interaction with cellulose, a crystal structure was solved for a truncated version of the protein encoded by Calkro_0844. Previous attempts to crystallize the full-length tāpirin without the transmembrane domains were unsuccessful because of protein lysis resulting in unpredictable, nonreproducible, and very slow (10 months and longer) crystal growth. Upon analysis of preliminary x-ray diffraction data from these crystals, it was determined that the asymmetric unit of the crystal cell was too small for the full-length protein. This finding indicated that only a protein fragment could have been crystallized, so a limited proteolysis approach was used. This strategy for obtaining crystals for recalcitrant proteins using proteolysis has been previously described (65, 66). In this case, in situ proteolysis proved unsuccessful. However, digestion of the recombinant Calkro_0844 consistently yielded a fragment with the same molecular mass. Therefore, selenomethionine-labeled Calkro_0844 was digested, thus creating discrete domains that would crystallize. Specifically, treatment with low concentrations of various proteases, and specifically thermolysin, resulted in a protein fragment with an estimated molecular mass of 45 kDa that crystallized successfully after purification.
The structure of digested Calkro_0844 C-terminal domain (Calkro_0844_C) was refined to a resolution of 1.7 Å with an R and Rfree of 0.150 and 0.194, respectively (Table 1 and Fig. 7). There is one molecule in the asymmetric unit (Fig. 7A), and it contains one magnesium ion coordinated by main chain carbonyls of Asp-473, Val-475, Ser-477, and Leu-480, as well as two side-chain oxygen atoms of Asp-473 and Glu-509. This is in contrast to some cellulose-binding CBMs (3) or polysaccharide lyases (67) that complex calcium ions. The core of Calkro_0844_C is a β-helix comprised of 11 complete turns in total, plus a few extra β-strands. The longest β-helix contains 14 strands. Furthermore, the ends of the β-helix are capped with α-helices (α1 and α3; Fig. 7A), and the turns of the β-helix are not consecutive. It appears that turns from 3 to 11 are formed at the N terminus, and turns 1 and 2 are formed closer to the C terminus of the construct (Fig. 7A).
FIGURE 7.
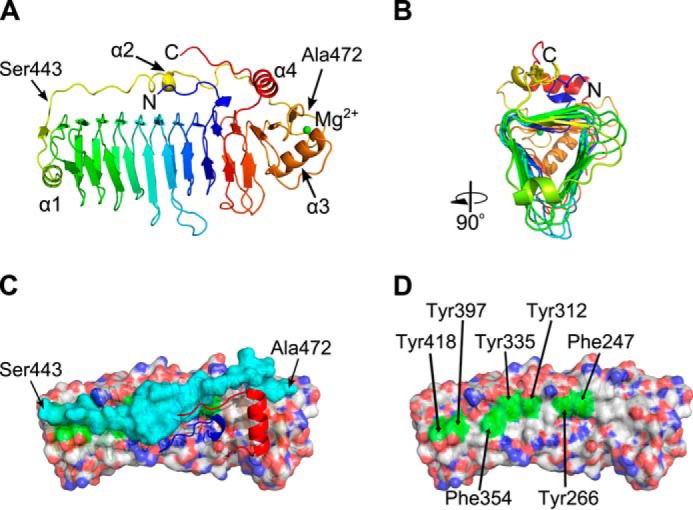
Crystal structure of thermolysin-digested Calkro_0844_C. A, schematic representation in spectrum colors from blue on the N terminus to red on the C terminus. A single magnesium ion is depicted as a green sphere. Four α-helices are marked as well as first and last residues of the protective loop. B, cartoon representation rotated 90° to illustrate the triangular shape of the β-helix core as well as two exposed and one protected surfaces. C, view from the top onto hydrophobic surface of the β-helix core (semi-transparent surface representation, CPK colors), protective loop (semi-transparent surface, cyan), and N and C termini (cartoon, blue and red, respectively). The first and last residues of the protective loop are marked. D, view from the top onto hydrophobic surface of the β-helix core with protective loop, N and C termini removed. Exposed aromatic residues are highlighted in green and are labeled.
Structural Comparison
Calkro_0844_C is a truly unique structure. A sequence search for known structures from the PDB found no similar entries. Pairwise secondary-structure matching of structures with at least 70% secondary structure similarity by PDBfold (68) found only partial matches to structures with similar folds from the PDB, such as pectate lyases. However, these matches were similar to only a portion of the β-helix core of Calkro_0844_C. Recently, a smaller (∼30 kDa) β-helix protein from Clostridium thermocellum was identified as possessing polysaccharide binding abilities; however, it also shares no appreciable similarity to the structure of Calkro_0844_C (69).
Calkro_0844_C has a long loop (residues Ser-443 to Ala-472) connecting the ends of the β-helix between strands β25 and β26 (Fig. 7, A and C). This is a remarkable feature of the protein structure that might play a role in cellulose binding. First, this rigid loop may hold the uniquely folded β-helix domains together, increasing protein stability in the harsh environment. Another hypothesis would have the loop contributing to the cellulose-binding properties of the protein, possibly directly interacting with cellulose. Co-crystallization of Calkro_0844_C with a mixture of cellobiose, cellotetraose, and cellohexaose, as well as extensive soaking of the existing crystals in the same oligosaccharides mixture, did not reveal soluble oligosaccharides binding in the crystal structure. Binding of the C-terminal fragment to insoluble cellulose and biomass was tested (Fig. 5D), and Calkro_0844_C was found to bind to both cellulose and DAP biomass. This suggests that binding of the Calkro_0844_C is specific for longer insoluble chains of cello-oligosaccharides.
The cross-section of the right-handed β-helix core is triangular in shape (Fig. 7B) with three relatively flat faces. Two of the faces are exposed to the solvent and are almost exclusively dominated by hydrophilic residues. The third face is mainly protected by the long loop and partially by the N and C termini. The interface between the long loop and the rest of the protein is lined up with hydrophobic residues on both sides. Curiously, there are only five hydrogen bonds between the 30-residue loop and the surface of the β-helix. Multiple alternative conformations of the residues in that range, including main chain atoms, further emphasize the flexible nature of this peptide loop. Movements of this peptide loop would expose the hydrophobic surface of the β-helix to bind to cellulose (Fig. 7, C and D).
Along this region, there is a line of seven aromatic side chains (tyrosines and phenylalanines, Fig. 7D) over nine β-strands flat on the surface and spaced ∼5–10 Å apart. That arrangement, possibly together with the hydrophobic residues of the loop Ser-443–Ala-472, could act as an effective binding platform for cellodextrins similar to the flat binding faces of some carbohydrate-binding modules (3, 61). When all tāpirin amino acid sequences were aligned against each other, aromatic residues at these seven positions are conserved across both classes of tāpirins from strongly cellulolytic members of the genus Caldicellulosiruptor. The only exception is Tyr-418, which is conserved only among class 1 tāpirins. In class 2 tāpirins, that tyrosine residue is shifted seven amino acids toward the C terminus (data not shown). Moreover, it is possible that these aromatic residues interact with cellulose, after the protective loop 443–472 shifts away and exposes the predicted cellulose-binding platform. Such a structural mechanism may be needed for solubility or to prevent nonspecific binding. Although binding assays clearly show that Calkro_0844_C is able to bind insoluble cellulose (Fig. 5D), it should be noted that the N-terminal domain is not present in the crystal structure. It would be in close proximity to the loop area and may have an effect on the loop conformations. The tāpirins described here thus represent a new class of cellulose-binding proteins, and further efforts are warranted to fully understand their unique binding mechanisms at the molecular level.
DISCUSSION
Previous studies from various cellulolytic bacteria have identified proteins that are theorized to aid in attachment to cellulose (11, 12, 15), indicating that there exist different mechanisms that bacteria may use to maintain proximity to their substrate. Cellulolytic Clostridia have been described as using large polypeptide structures, coined cellulosomes, to facilitate attachment and hydrolysis of plant biomass (22, 70). Highly cellulolytic members from the genus Caldicellulosiruptor have previously been demonstrated to adhere to plant biomass (28–30, 52), now including C. kronotskyensis (Fig. 1). The genus Caldicellulosiruptor, while related to cellulosomal bacteria, does not encode for cellulosomes, indicating that these species must use an alternative mechanism to attach to biomass. Extracellular S-layer-associated GHs and substrate-binding proteins from ATP-binding cassette transporters have previously been theorized to participate in mediating cellular adherence to plant biomass (30–32); however, many of these proteins are also present in the Caldicellulosiruptor core genome and are not exclusively used by highly cellulolytic members of the genus (24). Here, a combination of comparative genomics, proteomics, functional characterization, and x-ray crystallography was employed to further understand attachment mechanisms used by highly cellulolytic Caldicellulosiruptor species.
Comparative genomics analysis of the genus Caldicellulosiruptor (24) had previously highlighted two hypothetical proteins downstream of a well conserved type IV pilus locus (Fig. 3A). The possibility of type IV pili being involved in attachment to cellulose is plausible, because pilin proteins from both R. albus and R. flavefaciens were demonstrated to bind to cellulose (16, 71). However, the Caldicellulosiruptor tāpirins (roughly 70 kDa) are roughly three times as large as the individual pilin subunits from Ruminococcus species (roughly 20–25 kDa). Furthermore, no protein domain signatures were detected, including pilin signatures, making it unlikely that these proteins are pilin subunits. In the absence of typical pilin processing signals, it is also unlikely that these proteins are incorporated into type IV pili. In addition, these genes (tāpirins) encode for proteins that are not homologous to any other protein outside of the genus Caldicellulosiruptor. As such, the tāpirins cannot be classified with other known cellulose-binding proteins, such as CBMs or substrate-binding proteins from ATP-binding cassette transporters.
Using proteomics screening, these proteins were found to be highly enriched on the Avicel-bound protein fraction and are hypothesized to be involved in maintaining cell-surface contact with plant biomass. Previous proteomics data also had detected the presence of S-layer-associated GHs and substrate-binding proteins bound to cellulose (24), implicating the tāpirins and other cell surface-associated proteins in maintaining attachment to insoluble carbohydrates. Only the strongly cellulolytic Caldicellulosiruptor species were identified as deploying both classes of tāpirins, and those proteins were enriched on Avicel (Fig. 2). However, other hypothetical proteins downstream of the type IV locus in weakly cellulolytic Caldicellulosiruptor species were expressed upon growth on Avicel, enriched in the culture supernatant, and potentially are mediating cell-to-cell adherence in a community of both strongly cellulolytic and predominantly xylanolytic members. Based on the detection of signal peptides and transmembrane domains, the tāpirins are likely displayed on the cell membrane. The exact subcellular location of these proteins, however, remains to be determined and is the subject of ongoing experiments.
This study further confirmed and quantified the cellulose-binding function of the two orthologous classes of tāpirins. Based on yeast surface display (Fig. 4, G and H), the tāpirins can mediate attachment to cellulose without any association with other bacterial protein structures, such as the type IV pilus. In this case, S. cerevisiae is an attractive host for surface display of cellulose-binding proteins, because the species does not naturally adhere to cellulose. The affinity for cellulose appears to be relatively specific, as binding assays that included insoluble xylan failed to detect any of the tāpirins bound to this polysaccharide (Fig. 5). Using adsorption isotherms, the association constant for representatives from both classes of tāpirins could be estimated (Fig. 6). Both tāpirins bound to various forms of cellulose with affinities similar to those reported for CBMs (60, 62, 63, 72), fungal swollenin (64), and bacterial expansin (61). Taken together, the tāpirin proteins bind to cellulose with high affinity over a range of temperatures from 25 °C (trans vivo) to 70 °C (in vitro), confirming their hypothesized function as a new class of cellulose-binding proteins.
Based on amino acid sequence homology, the tāpirins are unique proteins currently only found in the genus Caldicellulosiruptor. Because there are no previously defined functional protein domain signatures in the tāpirin proteins, structural analysis from one class was completed. Here, we provide structural analysis of a truncated peptide derived from Calkro_0844 (“Calkro_0844_C”), a representative member of the tāpirins (Fig. 7). The solved structure of Calkro_0844_C indicates that it is a right-handed β-helix comprised of 11 turns that are held together by a long loop, which shields the hydrophobic face of the helix. Because of the unique features of these proteins, structural homology to other classes of proteins could not be assigned, indicating that this is truly a new class of biomolecules. Although no structural similarity could be assigned, the overall function of β-helix-containing proteins in cellular adhesion to glycoproteins has been described for Gram-negative pathogenic bacteria. Some examples of β-helix-containing proteins involved in attachment include larger adhesins from Bordetella pertussis (73, 74) and Haemophilus influenzae (75). In contrast however, the tāpirins are formed by nonpathogenic Gram-positive bacteria and are not known to be part of a type V secretion system. Overall, the tāpirins are significantly larger than currently known polysaccharide-binding modules, such as CBMs (3) or the pectate lyase-like protein from C. thermocellum (69), and establish a new paradigm by which lignocellulose-degrading microbes can attach to plant biomass. The possible exploitation of these novel proteins, or tāpirins, to improve plant biomass degradation for biofuels production is being pursued.
Supplementary Material
Acknowledgment
We thank B. M. Rao (North Carolina State University) for supplying yeast strain EBY100 and plasmid vector pCTCON for the yeast surface display experiments.
This work was supported by the BioEnergy Science Center and the Oak Ridge National Laboratory, a United States Department of Energy Bioenergy Research Center funded by the Office of Biological and Environmental Research in the Department of Energy Bioenergy Office of Science.

This article contains supplemental Table S1.
The atomic coordinates and structure factors (code 4WA0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase
- CBM
- carbohydrate-binding module
- DAP
- dilute acid-pretreated
- PDB
- Protein Data Bank
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane.
REFERENCES
- 1. Gilkes N. R., Warren R. A., Miller R. C., Jr., Kilburn D. G. (1988) Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 263, 10401–10407 [PubMed] [Google Scholar]
- 2. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. (1988) Studies of the cellulolytic system of Trichoderma reesei QM 9414. Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 3. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brulc J. M., Antonopoulos D. A., Miller M. E., Wilson M. K., Yannarell A. C., Dinsdale E. A., Edwards R. E., Frank E. D., Emerson J. B., Wacklin P., Coutinho P. M., Henrissat B., Nelson K. E., White B. A. (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 1948–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hess M., Sczyrba A., Egan R., Kim T. W., Chokhawala H., Schroth G., Luo S., Clark D. S., Chen F., Zhang T., Mackie R. I., Pennacchio L. A., Tringe S. G., Visel A., Woyke T., et al. (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 [DOI] [PubMed] [Google Scholar]
- 7. Warnecke F., Luginbühl P., Ivanova N., Ghassemian M., Richardson T. H., Stege J. T., Cayouette M., McHardy A. C., Djordjevic G., Aboushadi N., Sorek R., Tringe S. G., Podar M., Martin H. G., Kunin V., et al. (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565 [DOI] [PubMed] [Google Scholar]
- 8. Mello L. V., Chen X., Rigden D. J. (2010) Mining metagenomic data for novel domains: BACON, a new carbohydrate-binding module. FEBS Lett. 584, 2421–2426 [DOI] [PubMed] [Google Scholar]
- 9. Rigden D. J., Eberhardt R. Y., Gilbert H. J., Xu Q., Chang Y., Godzik A. (2014) Structure- and context-based analysis of the GxGYxYP family reveals a new putative class of glycoside hydrolase. BMC Bioinformatics 15, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suen G., Weimer P. J., Stevenson D. M., Aylward F. O., Boyum J., Deneke J., Drinkwater C., Ivanova N. N., Mikhailova N., Chertkov O., Goodwin L. A., Currie C. R., Mead D., Brumm P. J. (2011) The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 6, e18814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jun H. S., Qi M., Gong J., Egbosimba E. E., Forsberg C. W. (2007) Outer membrane proteins of Fibrobacter succinogenes with potential roles in adhesion to cellulose and in cellulose digestion. J. Bacteriol. 189, 6806–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gong J., Egbosimba E. E., Forsberg C. W. (1996) Cellulose-binding proteins of Fibrobacter succinogenes and the possible role of a 180-kDa cellulose-binding glycoprotein in adhesion to cellulose. Can. J. Microbiol. 42, 453–460 [Google Scholar]
- 13. Ezer A., Matalon E., Jindou S., Borovok I., Atamna N., Yu Z., Morrison M., Bayer E. A., Lamed R. (2008) Cell surface enzyme attachment is mediated by family 37 carbohydrate-binding modules, unique to Ruminococcus albus. J. Bacteriol. 190, 8220–8222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Q., Morrison M., Nelson K. E., Bayer E. A., Atamna N., Lamed R. (2004) A novel family of carbohydrate-binding modules identified with Ruminococcus albus proteins. FEBS Lett. 566, 11–16 [DOI] [PubMed] [Google Scholar]
- 15. Rakotoarivonina H., Larson M. A., Morrison M., Girardeau J. P., Gaillard-Martinie B., Forano E., Mosoni P. (2005) The Ruminococcus albus pilA1-pilA2 locus: expression and putative role of two adjacent pil genes in pilus formation and bacterial adhesion to cellulose. Microbiology 151, 1291–1299 [DOI] [PubMed] [Google Scholar]
- 16. Pegden R. S., Larson M. A., Grant R. J., Morrison M. (1998) Adherence of the Gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins. J. Bacteriol. 180, 5921–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mosoni P., Gaillard-Martinie B. (2001) Characterization of a spontaneous adhesion-defective mutant of Ruminococcus albus strain 20. Arch. Microbiol. 176, 52–61 [DOI] [PubMed] [Google Scholar]
- 18. Vodovnik M., Duncan S. H., Reid M. D., Cantlay L., Turner K., Parkhill J., Lamed R., Yeoman C. J., Berg Miller M. E., White B. A., Bayer E. A., Marinšek-Logar R., Flint H. J. (2013) Expression of cellulosome components and type IV pili within the extracellular proteome of Ruminococcus flavefaciens 007. PLoS ONE 8, e65333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Te'o V. S., Saul D. J., Bergquist P. L. (1995) celA, another gene coding for a multidomain cellulase from the extreme thermophile Caldocellum saccharolyticum. Appl. Microbiol. Biotechnol. 43, 291–296 [DOI] [PubMed] [Google Scholar]
- 20. Gibbs M. D., Saul D. J., Lüthi E., Bergquist P. L. (1992) The β-mannanase from “Caldocellum saccharolyticum” is part of a multidomain enzyme. Appl. Environ. Microbiol. 58, 3864–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brunecky R., Alahuhta M., Xu Q., Donohoe B. S., Crowley M. F., Kataeva I. A., Yang S. J., Resch M. G., Adams M. W., Lunin V. V., Himmel M. E., Bomble Y. J. (2013) Revealing nature's cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science 342, 1513–1516 [DOI] [PubMed] [Google Scholar]
- 22. Blumer-Schuette S. E., Brown S. D., Sander K. B., Bayer E. A., Kataeva I., Zurawski J. V., Conway J. M., Adams M. W., Kelly R. M. (2014) Thermophilic lignocellulose deconstruction. FEMS Microbiol. Rev. 38, 393–448 [DOI] [PubMed] [Google Scholar]
- 23. Gibbs M. D., Reeves R. A., Farrington G. K., Anderson P., Williams D. P., Bergquist P. L. (2000) Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40, 333–340 [DOI] [PubMed] [Google Scholar]
- 24. Blumer-Schuette S. E., Giannone R. J., Zurawski J. V., Ozdemir I., Ma Q., Yin Y., Xu Y., Kataeva I., Poole F. L., 2nd, Adams M. W., Hamilton-Brehm S. D., Elkins J. G., Larimer F. W., Land M. L., Hauser L. J., et al. (2012) Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J. Bacteriol. 194, 4015–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blumer-Schuette S. E., Lewis D. L., Kelly R. M. (2010) Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl. Environ. Microbiol. 76, 8084–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Young J., Chung D., Bomble Y. J., Himmel M. E., Westpheling J. (2014) Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass. Biotechnol. Biofuels 7, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vanfossen A. L., Lewis D. L., Nichols J. D., Kelly R. M. (2008) Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann. N.Y. Acad. Sci. 1125, 322–337 [DOI] [PubMed] [Google Scholar]
- 28. Dam P., Kataeva I., Yang S. J., Zhou F., Yin Y., Chou W., Poole F. L., 2nd, Westpheling J., Hettich R., Giannone R., Lewis D. L., Kelly R., Gilbert H. J., Henrissat B., Xu Y., Adams M. W. (2011) Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 39, 3240–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z. W., Lee S. H., Elkins J. G., Morrell-Falvey J. L. (2011) fSpatial and temporal dynamics of cellulose degradation and biofilm formation by Caldicellulosiruptor obsidiansis and Clostridium thermocellum. AMB Express 1, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozdemir I., Blumer-Schuette S. E., Kelly R. M. (2012) S-layer homology domain proteins Csac_0678 and Csac_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 78, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanfossen A. L., Verhaart M. R., Kengen S. M., Kelly R. M. (2009) Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75, 7718–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yokoyama H., Yamashita T., Morioka R., Ohmori H. (2014) Extracellular secretion of noncatalytic plant cell wall-binding proteins by the cellulolytic thermophile Caldicellulosiruptor bescii. J. Bacteriol. 196, 3784–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 37. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., Sangrador-Vegas A., Scheremetjew M., Rato C., Yong S. Y., Bateman A., et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Studier F. W. (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 40. Boder E. T., Wittrup K. D. (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 [DOI] [PubMed] [Google Scholar]
- 41. Nam J.-M., Fujita Y., Arai T., Kondo A., Morikawa Y., Okada H., Ueda M., Tanaka A. (2002) Construction of engineered yeast with the ability of binding to cellulose. J. Mol. Catal. B Enzym. 17, 197–202 [Google Scholar]
- 42. Sreenath H. K., Bingman C. A., Buchan B. W., Seder K. D., Burns B. T., Geetha H. V., Jeon W. B., Vojtik F. C., Aceti D. J., Frederick R. O., Phillips G. N., Jr., Fox B. G. (2005) Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr. Purif. 40, 256–267 [DOI] [PubMed] [Google Scholar]
- 43. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grosse-Kunstleve R. W., Adams P. D. (2003) Substructure search procedures for macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 59, 1966–1973 [DOI] [PubMed] [Google Scholar]
- 45. McCoy A. J., Storoni L. C., Read R. J. (2004) Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr. D Biol. Crystallogr. 60, 1220–1228 [DOI] [PubMed] [Google Scholar]
- 46. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Skubák P., Pannu N. S. (2013) Automatic protein structure solution from weak X-ray data. Nat. Commun. 10.1038/ncomms3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Engh R. A., Huber R. (1991) Accurate bond and angle parameters for x-ray protein-structure refinement. Acta Crystallogr. A Found. Crystallogr. 47, 392–400 [Google Scholar]
- 52. VanFossen A. L., Ozdemir I., Zelin S. L., Kelly R. M. (2011) Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 108, 1559–1569 [DOI] [PubMed] [Google Scholar]
- 53. Onyenwoke R. U., Lee Y. J., Dabrowski S., Ahring B. K., Wiegel J. (2006) Reclassification of Thermoanaerobium acetigenum as Caldicellulosiruptor acetigenus comb. nov and emendation of the genus description. Int. J. Syst. Evol. Microbiol. 56, 1391–1395 [DOI] [PubMed] [Google Scholar]
- 54. Gera N., Hussain M., Wright R. C., Rao B. M. (2011) Highly stable binding proteins derived from the hyperthermophilic Sso7d scaffold. J. Mol. Biol. 409, 601–616 [DOI] [PubMed] [Google Scholar]
- 55. Ito J., Fujita Y., Ueda M., Fukuda H., Kondo A. (2004) Improvement of cellulose-degrading ability of a yeast strain displaying Trichoderma reesei endoglucanase II by recombination of cellulose-binding domains. Biotechnol. Prog. 20, 688–691 [DOI] [PubMed] [Google Scholar]
- 56. Fan L.-H., Zhang Z.-J., Yu X.-Y., Xue Y.-X., Tan T.-W. (2012) Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. U.S.A. 109, 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baek S.-H., Kim S., Lee K., Lee J.-K., Hahn J.-S. (2012) Cellulosic ethanol production by combination of cellulase-displaying yeast cells. Enzyme Microb. Technol. 51, 366–372 [DOI] [PubMed] [Google Scholar]
- 58. Tsai S.-L., Goyal G., Chen W. (2010) Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 76, 7514–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wen F., Sun J., Zhao H. (2010) Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 76, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tomme P., Boraston A., McLean B., Kormos J., Creagh A. L., Sturch K., Gilkes N. R., Haynes C. A., Warren R. A., Kilburn D. G. (1998) Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B Biomed. Sci. Appl. 715, 283–296 [DOI] [PubMed] [Google Scholar]
- 61. Georgelis N., Yennawar N. H., Cosgrove D. J. (2012) Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc. Natl. Acad. Sci. U.S.A. 109, 14830–14835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Riedel K., Ritter J., Bauer S., Bronnenmeier K. (1998) The modular cellulase CelZ of the thermophilic bacterium Clostridium stercorarium contains a thermostabilizing domain. FEMS Microbiol. Lett. 164, 261–267 [DOI] [PubMed] [Google Scholar]
- 63. Din N., Forsythe I. J., Burtnick L. D., Gilkes N. R., Miller R. C., Jr., Warren R. A., Kilburn D. G. (1994) The cellulose-binding domain of endoglucanase A (CenA) from Cellulomonas fimi: evidence for the involvement of tryptophan residues in binding. Mol. Microbiol. 11, 747–755 [DOI] [PubMed] [Google Scholar]
- 64. Jäger G., Girfoglio M., Dollo F., Rinaldi R., Bongard H., Commandeur U., Fischer R., Spiess A. C., Büchs J. (2011) How recombinant swollenin from Kluyveromyces lactis affects cellulosic substrates and accelerates their hydrolysis. Biotechnol. Biofuels 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dong A., Xu X., Edwards A. M., Midwest Center for Structural Genomics, Structural Genomics Consortium, Chang C., Chruszcz M., Cuff M., Cymborowski M., Di Leo R., Egorova O., Evdokimova E., Filippova E., Gu J., Guthrie J., et al. (2007) In situ proteolysis for protein crystallization and structure determination. Nat. Methods 4, 1019–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wernimont A., Edwards A. (2009) In situ proteolysis to generate crystals for structure determination: An update. PLoS ONE 4, e5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lombard V., Bernard T., Rancurel C., Brumer H., Coutinho P. M., Henrissat B. (2010) A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 [DOI] [PubMed] [Google Scholar]
- 68. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 69. Close D. W., D'Angelo S., Bradbury A. R. (2014) A new family of β-helix proteins with similarities to the polysaccharide lyases. Acta Crystallogr. D Biol. Crystallogr. 70, 2583–2592 [DOI] [PubMed] [Google Scholar]
- 70. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 71. Rakotoarivonina H., Jubelin G., Hebraud M., Gaillard-Martinie B., Forano E., Mosoni P. (2002) Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiology 148, 1871–1880 [DOI] [PubMed] [Google Scholar]
- 72. Linder M., Salovuori I., Ruohonen L., Teeri T. T. (1996) Characterization of a double cellulose-binding domain: synergistic high affinity binding to crystalline cellulose. J. Biol. Chem. 271, 21268–21272 [DOI] [PubMed] [Google Scholar]
- 73. Emsley P., Charles I. G., Fairweather N. F., Isaacs N. W. (1996) Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381, 90–92 [DOI] [PubMed] [Google Scholar]
- 74. Clantin B., Hodak H., Willery E., Locht C., Jacob-Dubuisson F., Villeret V. (2004) The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc. Natl. Acad. Sci. U.S.A. 101, 6194–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yeo H.-J., Yokoyama T., Walkiewicz K., Kim Y., Grass S., Geme J. W., 3rd (2007) The structure of the Haemophilus influenzae HMW1 pro-piece reveals a structural domain essential for bacterial two-partner secretion. J. Biol. Chem. 282, 31076–31084 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.