Background: New structures of apolipoprotein (apo)A-IV reveal aligned, conserved proline residues of unknown function.
Results: Increasing deletion of prolines stabilizes apoA-IV and increases self-association but also increases lipid affinity and cholesterol efflux.
Conclusion: Proline residues play a concerted role in destabilizing the apoA-IV structure and modulate its function.
Significance: This is the first detailed study of the structural role of proline and of a stable trimeric exchangeable apolipoprotein.
Keywords: Apolipoprotein, Lipid-binding Protein, Mass Spectrometry (MS), Mutagenesis, Protein Cross-linking, Structure, Lipid Binding, Lipid-free, Oligomerization, Self-association, Apolipoprotein A-IV, Small Angle X-ray Scattering
Abstract
Apolipoprotein (apo)A-IV is a lipid emulsifying protein linked to a range of protective roles in obesity, diabetes, and cardiovascular disease. It exists in several states in plasma including lipid-bound in HDL and chylomicrons and as monomeric and dimeric lipid-free/poor forms. Our recent x-ray crystal structure of the central domain of apoA-IV shows that it adopts an elongated helical structure that dimerizes via two long reciprocating helices. A striking feature is the alignment of conserved proline residues across the dimer interface. We speculated that this plays important roles in the structure of the lipid-free protein and its ability to bind lipid. Here we show that the systematic conversion of these prolines to alanine increased the thermodynamic stability of apoA-IV and its propensity to oligomerize. Despite the structural stabilization, we noted an increase in the ability to bind and reorganize lipids and to promote cholesterol efflux from cells. The novel properties of these mutants allowed us to isolate the first trimeric form of an exchangeable apolipoprotein and characterize it by small-angle x-ray scattering and chemical cross-linking. The results suggest that the reciprocating helix interaction is a common feature of all apoA-IV oligomers. We propose a model of how self-association of apoA-IV can result in spherical lipoprotein particles, a model that may have broader applications to other exchangeable apolipoprotein family members.
Introduction
Apolipoproteins, well known for their lipid binding properties, function as critical transport molecules that mobilize and deliver lipids throughout the body. Apolipoprotein (apo)A-IV,3 the largest member of the exchangeable apolipoproteins family at 44 kDa, is a major component of HDL and is present on freshly secreted chylomicrons. Despite being the third most abundant protein associated with HDL (1), its biological role is unsettled. ApoA-IV production is increased during food intake, supporting a possible role in lipid absorption and satiety (2, 3). In addition, apoA-IV has been shown to prevent oxidation of lipids and suppress inflammation (4, 5). More recent studies have implied a potentially important role in altering the glucose sensitivity of pancreatic islets to alter insulin secretion (6). However, the mechanistic details for these functions remain unclear, in part because of a lack of structural understanding.
In general, high-resolution structural studies of lipid-free apolipoproteins have been challenging due to their ability to interconvert between multiple self-associated oligomeric states. To aid in this process, several studies have identified stable apolipoprotein point mutants and truncations that are more amenable to characterization. We identified a stable, truncated version of apoA-IV through limited proteolysis that retained the ability to form reconstituted lipoprotein particles (7) and resolved its x-ray crystal structure at 2.4 Å (8). As expected, apoA-IV is highly helical, consisting of a large helical bundle domain. Interestingly, the dimeric structure revealed that self-association occurs through a “helix swap” mechanism (described in detail in Deng et al. (8)), which integrates one helix from one chain with three helices from another chain to form tandem, head-to-tail four-helix bundle motifs (see Fig. 1). A similar dimerization mechanism was also observed in the crystal structure of an apoA-I truncation mutant (9), suggesting a common mechanism of self-association among apolipoproteins. These structures have been enlightening with respect to extrapolating to the lipid-bound forms of these apolipoproteins, at least in nascent discoidal HDL particles. Both models can be envisioned to open two helical hairpin “doors” to wrap around phospholipid acyl chains. This offers the first straightforward explanation for how two apolipoprotein molecules can interact while forming discoidal particles composed of antiparallel bi-helical belts as originally proposed by Segrest et al. (10) for apoA-I.
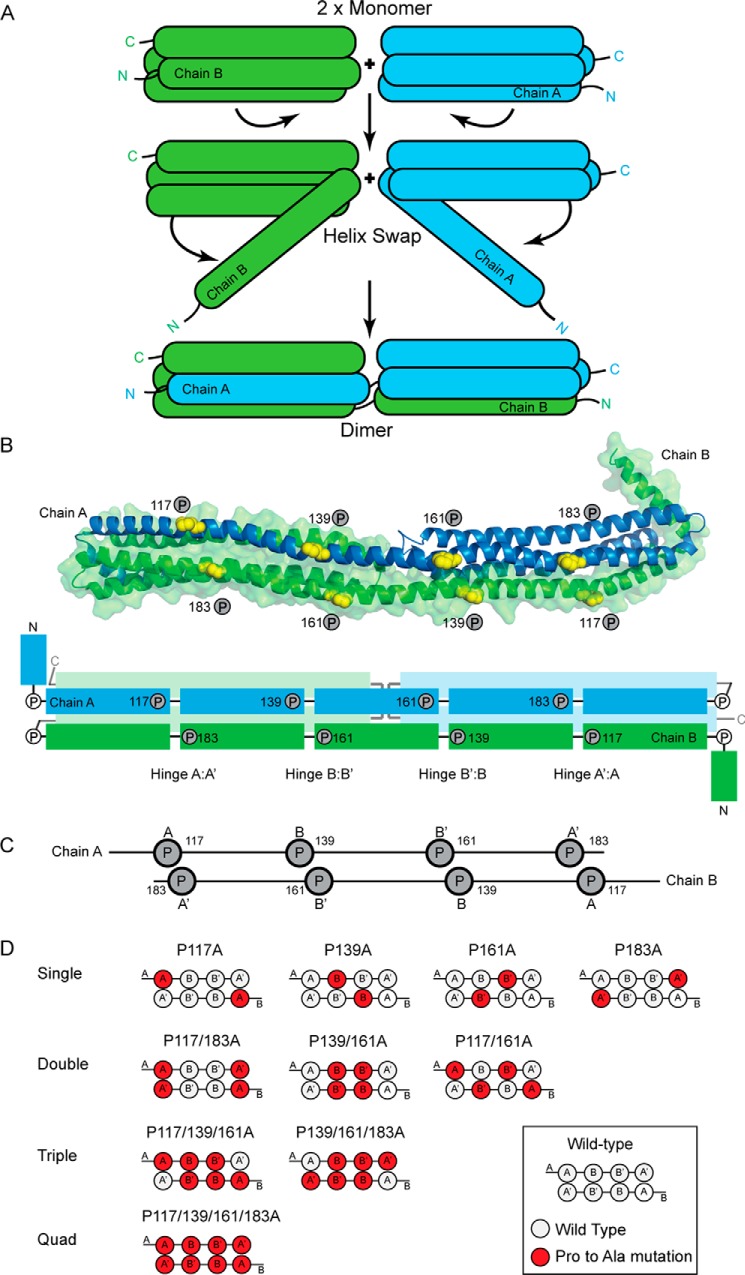
FIGURE 1.
Helix swap mechanism and mutant design. A, schematic representation of the helix swap responsible for dimerization of apoA-IV. The N terminus from each monomer dissociates from the four-helical bundle and forms a four-helical bundle with the adjacent monomer to form a dimer. B, an apoA-IV dimer as shown in our previously reported crystal structure (Protein Data Bank (PDB) ID 3S84) (8). Two monomers of apoA-IV that participate in a dimeric interaction are shown in graphic form in blue (Chain A) and green (Chain B). The prolines targeted for mutagenesis are shown as yellow spheres. The panel also has a schematic of the apoA-IV dimer. The proline-punctuated helical segments on the long shared helical domain are shown in darker color with conserved proline residues indicated. C, simplified diagram showing the location of proline residues in the dimer conformation. D, schematic representations of the various single, double, triple, and quadruple (Quad) proline to alanine mutations in the dimer conformation.
Although the apolipoproteins are indeed helical as suspected from their primary structure, they are punctuated by highly conserved proline residues, often spaced 22 amino acids apart (11, 12). This segmentation is particularly notable in apoA-IV. Biologically, proline mutations in apolipoproteins have been related to numerous disease states. Lipoprotein glomerulopathy can result from numerous proline mutations in apoE (13–15). Natural proline mutations in apoA-I have been related to low HDL cholesterol, premature coronary heart disease (16), lower level of lecithin:cholesterol acyltransferase (LCAT) activation (17), and renal amyloidosis (18). Additionally, proline mutation in apoA-II also causes amyloidosis in mice (19).
Structurally, these residues are commonly thought to terminate helical secondary structure, creating segmentation because they disrupt the hydrogen-bonding pattern of a helix. In fact, the Picket Fence model (20), the Belt model along with its variations (10, 21–24), and the Double-Super Helix model (25) for apoA-I in HDL all feature proline residues for segmentation and/or induction of curvature in the structures. Initial observations on apoA-I revealed that the prolines were not simply involved in turns, but could occur in the middle of a helix, providing a significant bend to its direction (26). In our crystal structure of apoA-IV, some of the conserved prolines cause helical turns, whereas others cause “kinks” or slight bends in an otherwise continuous helix (8). Interestingly, the prolines tended to “line up” in direct opposition across the dimer interface (see Fig. 1, B and C), indicating an important functional role.
Despite their high conservation in the exchangeable apolipoproteins, the roles of prolines in terms of structure, stability, function, and oligomerization have not been systematically investigated. Taking advantage of the detailed structural understanding of the apoA-IV core helical domain from the crystal structure, we took the opportunity to carefully characterize the role of these aligned proline residues on the structural and functional properties of apoA-IV.
EXPERIMENTAL PROCEDURES
Mutagenesis
Point mutations to convert proline to alanine residues were done following a previously published protocol based on the QuikChangeTM site-directed mutagenesis protocol (27). Mutations were confirmed by DNA sequencing.
Protein Expression and Purification
Recombinant human apoA-IV WT and various proline mutants were produced and purified in Escherichia coli as described previously (8). Size exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 200 (GE Healthcare) column was performed to isolate oligomeric species for the various constructs. The isolated oligomeric species were concentrated at 4 °C using Amicon Ultra-15 centrifuge filter units.
Sedimentation Velocity
Analytical ultracentrifugation experiments were performed using a Beckman XL-I equipped with absorbance optics and a four-hole rotor. Sedimentation velocity was performed in a two-channel carbon-filled Epon centerpiece at 48,000 rpm, 20 °C with protein concentration at 0.15 mg/ml that had been dialyzed into 20 mm NaPO4, pH 7.4, 100 mm NaF. Absorbance at 215 nm was used to monitor the protein, and data were analyzed using Sedfit (28).
SEC
To determine oligomeric redistribution over time, purified protein was diluted to 0.15 mg/ml, a physiological concentration, in PBS with 1 mm EDTA and incubated at 37 °C. At varying time points, 300 μl of sample was collected and examined using the Superdex 200 10/300 (GE Healthcare) gel filtration column. The elution process was monitored with absorbance at 235 nm.
Chemical Cross-linking
The isolated trimer of P117/P139A/P161A/P183A apoA-IV was cross-linked with bis(sulfosuccinimidyl) suberate as reported previously (25). Freshly solubilized bis(sulfosuccinimidyl) suberate (spacer arm of 11.4 Å) was added to the protein solutions at molar ratios of cross-linker to protein of 50:1. The samples were incubated at 4 °C for 12 h. After quenching with an excess of Tris-HCl, the cross-linked proteins were exhaustively digested with trypsin at a 1:20 mass ratio of trypsin to apoA-IV for 2 h at 37 °C followed by a second spike of trypsin and another 2-h incubation. The resulting peptides were then lyophilized to dryness and stored at −80 °C until analyzed by MS.
Mass spectrometry and Interpretation
Nano-LC-MS/MS analyses were performed on a TripleTOF® 5600+ (AB SCIEX, Toronto, Ontario, Canada) coupled to an Eksigent (Dublin, CA) NanoLC-Ultra® nanoflow system. Dried samples were reconstituted in formic acid/H2O 0.1/99.9 (v/v), and 5 μl (containing 1–3 μg of digest) was loaded onto C18 IntegraFritTM trap column (outer diameter of 360 μm, inner diameter of 100, and 25-μm packed bed) from New Objective, Inc. (Woburn, MA) at 2 μl/min in formic acid/H2O 0.1/99.9 (v/v) for 15 min to desalt and concentrate the samples. For the chromatographic separation, the trap column was switched to align with the analytical column, Acclaim® PepMap100 (inner diameter of 75 μm, length of 15 cm, C18 particle sizes of 3 μm, and pore sizes of 100 Å) from Dionex-Thermo Fisher Scientific. The protein was eluted at 300 nl/min using a varying mobile phase gradient from 95% phase A (formic acid/H2O 0.1/99.9, v/v) to 40% phase B (formic acid/acetonitrile 0.1/99.9, v/v) for 35 min (1% per min) and then from 40% B to 85% B in 5 min with re-equilibration. The effluent from the nano-LC was introduced into the mass spectrometer using a NanoSpray® III source (AB SCIEX). The instrument was operated in positive ion mode for 65 min, where each cycle consisted of one TOF-MS scan (0.25-s accumulation time, in a 350–1500 m/z window) followed by 30 information-dependent acquisition mode MS/MS scans on the most intense candidate ions selected from an initially performed TOF-MS scan during each cycle. Each product ion scan was operated under vender-specified high-sensitivity mode with an accumulation time of 0.075 s and CE of 43 with an 8-unit scan range. The .wiff files were converted to Mascot generic files using a file converting algorithm embedded in the PeakView® v1.2.0.3 software (AB SCIEX) and uploaded into Cross-ID. Given the sequence of apoA-IV, the cross-linker used, the isotopic possibilities, and the enzyme used for peptide generation, the program first identifies all possible cross-link interactions based on a user-defined window of error for each mass detected in the experiment (20 ppm for this instrument). Once candidate identifications are determined, the program then compares theoretical MS/MS fragmentation patterns to the actual data, generating a probability score that reflects the confidence of the identification. These identifications are then visually verified by experienced personnel before the cross-link is declared valid.
DMPC Clearance Assay
The ability for apoA-IV and various mutants to emulsify suspended 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) (Avanti Polar Lipids) was tested using a DMPC clearance microplate protocol based on previously published methods (7). DMPC was prepared in buffer, and protein concentration was 0.21 mg/ml, with a 3:1 ratio of DMPC:protein. In a 96-well plate (Fisher catalog number 12-565-501), 25.4 μl of DMPC and 7.6 μl of Tris buffer (25 mm Tris, pH 8.5, 150 mm NaCl, and 1 mm EDTA) were pipetted into one well, and 200 μl of protein sample was pipetted into another well. The plate was then incubated for 10 min in a BioTek SynergyTM HT microplate reader that was preheated to 26 °C. After 10 min, protein sample was transferred to the well containing DMPC and Tris buffer, and the well was monitored for 325 nm absorbance in 30-s intervals for 30 min.
Cholesterol Efflux Assay
Fatty acid-free BSA was acquired from EMD Millipore Calbiochem. 8-bromo-3′-5′cAMP was from Sigma-Aldrich. Fetal bovine serum was from was from Gemini Bio-Products (West Sacramento, CA). PBS was from Thermo Fisher Scientific. RAW 264.7 mouse macrophages were from the American Type Culture Collection (Manassas, VA). The Amplex Red cholesterol assay kit was from Life Technologies.
The transformed mouse macrophage cell line RAW 264.7 (ATCC) was maintained in Dulbecco's modified eagle medium-high-glucose (DMEM) supplemented with 10% FBS. Cells were grown to 80% confluency in a 48-well plate, and then growth medium (DMEM/10% FBS) was replaced with that containing 1.0 μCi/ml [1,2-3H]cholesterol and 0.3 mm 8-bromo-3′-5′cAMP for 16 h to stimulate ABCA-I expression. Following a PBS wash, treatment medium (DMEM with 0.2% fatty acid-free BSA and 0.3 mm 8-bromo-3′-5′ cAMP) with and without lipid-free apolipoprotein acceptors at 10 μg/ml (apoA-I, apoA-IV, or apoA-IV P117/P139A/P161A/P183A trimer) was added at t = 0 h and incubated at 37 °C. After 6 h, the efflux medium was removed and spun at 1,000 × g for 5 min to remove any cells from the medium. The medium was analyzed by liquid scintillation counting.
Small-angle X-ray Scattering
Small-angle x-ray diffraction (SAXS) data on the quadruple proline mutant was collected at beam line 12-ID-B at the Advance Photon Source located in Argonne National Laboratory (see Table 1). A flow cell was used to reduce radiation damage. 120 μl of sample was loaded into the flow cell and oscillated during the course of data collection. Data were collected at room temperature, with an exposure time of 1.0 s, and a total of 30 images were taken for each sample. Small angle diffraction images were captured using a Pilatus 2M detector. Data collected were subtracted, examined, and averaged using PRIMUS (29). Also in PRIMUS, Rg was determined, and Dmax was calculated using AutoGNOM (see Table 1) (30). GNOM was used for indirect transformation, and GASBOR was used for ab initio reconstruction (31). Depending on the data set, dimer (P2) or trimer symmetry (P3) was used as a constraint in GASBOR. For the trimer reconstruction, P1 as a symmetry constraint was also tested, and a similar trefoil was observed as with P3. Ten independent ab initio reconstructions for each oligomeric form of apoA-IV were averaged using the DAMAVER package (32). The superposition of our full-length model of the apoA-IV trimer was performed using SUPCOMB (33).
TABLE 1.
SAXS parameters derived from scattering curves
| PR calculated |
Guinier calculated |
||||
|---|---|---|---|---|---|
| Rg | I(0) | Dmax | Rg | I(0) | |
| Å | Å | Å | |||
| Dimer | 56.4 | 0.14 | 195 | 56.19 | 0.15 |
| Trimer | 66.9 | 0.21 | 225 | 67.34 | 0.23 |
RESULTS
Upon resolution of the apoA-IV crystal structure, it was apparent that dimerization resulted from extensive contacts between monomers that buried a substantial hydrophobic core. However, the relatively modest melting temperature of 54 °C indicated that apoA-IV was more “fluid” than implied from the static crystal structure. Therefore, we hypothesized that one possible function of the conserved prolines might be to destabilize the bundle, which in turn would help promote lipid access to the hydrophobic core. In contrast, we predicted that a more stable and rigid four-helix bundle might have a reduced affinity for lipids because access to the hydrophobic core would be limited and the protein would favor its soluble, lipid-free state.
To investigate this possibility, we concentrated on the conserved prolines residing within the extended dimerization helix (helix B) that is interchanged or “swapped” between dimer-participating chains. The B helices from each chain run anti-parallel to each other and contain 4 proline residues (117, 139, 161, and 183) equally spaced 22 residues apart (Fig. 1,B and C). Although the helices run anti-parallel, the nature of the proline spacing invokes a fascinating symmetry that aligns them across the helices from the different chains. This creates a set of four paired prolines that we speculated could form a series of unidirectional hinges (all bending toward the bundle core) (8). To test their function, we systematically converted these proline residues to alanine. Although there is no ideal amino acid substitution for proline, we selected alanine because of its small size, its propensity over proline to form an α-helical structure, its reasonable similarity in terms of polarity, and the relatively high degree of evolutionary substitution interchange between proline and alanine (34). Our expectation was that, without the “kinking” induced by proline, helix B would have a stronger helical tendency, become less segmented, and hence generate a more stable helix bundle with weaker lipid affinity.
The panel of mutants tested is outlined in Fig. 1D. We began by replacing each proline individually (single mutants). This results in two mutations per dimer, one for each apoA-IV molecule (P117A, P139A, P161A, P183A). We also generated double mutants in which opposing prolines were targeted (P117A/P183A and P139A/P161A) and one in which non-opposing prolines were mutated (P117A/P161A). Two triple mutants were generated (P117A/P139A/P161A and P139A/P161A/P183A) along with one quadruple mutant with all four prolines substituted.
ApoA-IV Oligomeric Distribution Is Altered by Proline Mutagenesis
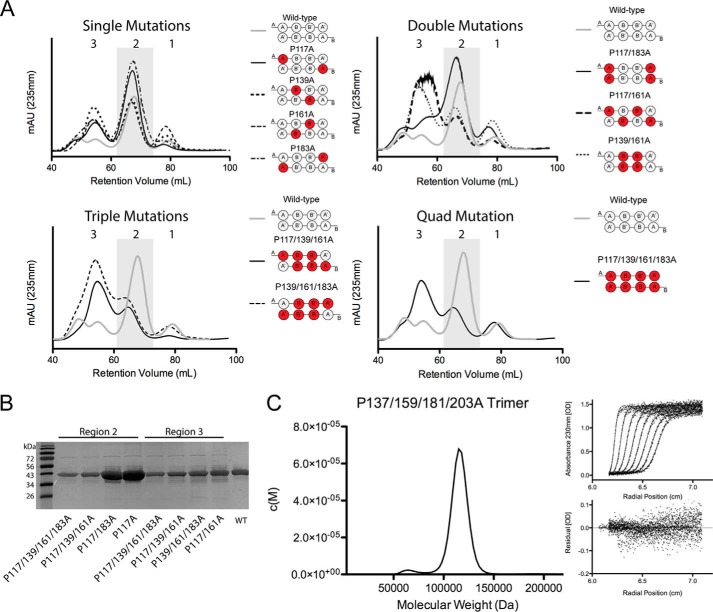
We first analyzed each mutant after purification by SEC to determine the degree to which they self-associate (8). As we have previously described, at 4 °C WT apoA-IV distributes primarily as a dimer with small amounts of monomer immediately after purification (8). We also noticed a larger molecular weight species that eluted after the void volume (Fig. 2A). We previously established that the dominant peak (region 2) corresponded to dimeric apoA-IV and that region 1 corresponded to the monomeric protein by sedimentation velocity (8). However, the higher molecular weight species in region 3 were not characterized because they tended to dissociate into the lower molecular weight forms when isolated. This is reminiscent of apoA-I (35) and apoE (36) whose higher self-associated states can be observed under equilibrium conditions in solution, but quickly redistribute to lower self-associated forms during purification.
FIGURE 2.
Extensive proline to alanine substitution in apoA-IV alters the oligomeric distribution. A, size exclusion chromatography was used to separate different apoA-IV oligomeric forms that are stable in solution. The UV absorbance trace for apoA-IV WT is compared with single, double, triple, and quadruple (Quad) point mutants. The trace is divided into three regions based on retention volume (region 1, 75–85 ml; region 2, 60–75 ml; and region 3, 50–60 ml). The scale of the UV traces among the different panels are comparable. mAU, milliabsorbance units. B, SDS-PAGE analysis of protein isolated from regions 3 and 2 of the proline mutants and compared with apoA-IV WT. C, a sedimentation velocity was used to determine the molecular mass of the protein isolated from region 3 from SEC of the quadruple mutant. Eight representative data fits and a residual of fit are shown.
The single proline mutants also primarily distributed as dimer and monomer, although the contribution of the larger oligomers tended to increase slightly versus WT (Fig. 2A, top left). However, the oligomeric pattern was more profoundly affected in the case of the double proline mutations. Although P117A/P183A largely resembled WT apoA-IV, P117A/P161A and P139A/P161A exhibited significant increases in the high molecular weight oligomers (Fig. 2A, top right). A similar shift was observed for the triple and quadruple apoA-IV mutants (Fig. 2A, bottom right and left). An SDS-PAGE analysis of the material eluting in regions 2 and 3 showed intact apoA-IV protein at its correct molecular mass of 44.5 kDa, thus ruling out contributions of contaminating proteins or covalent oligomers to the results (Fig. 2B).
The increased fractional distribution of the multiple proline mutants in region 3 when compared with WT apoA-IV suggested that this species may have been stabilized by the mutations. To test this, we collected the fractions in region 3 from the quadruple mutant P117/P139A/P161A/P183A apoA-IV, concentrated them by ultrafiltration at room temperature, and then reapplied the sample to the SEC column. We found that this isolated species underwent minimal redistribution after 24 h (data not shown), indicating a stable association. This sample was characterized by sedimentation velocity (Fig. 2C), revealing a single dominant peak with a calculated molecular mass of 115 ± 9 kDa, consistent with a trimer of apoA-IV.
Proline Mutagenesis Increases the Oligomeric Stability of ApoA-IV
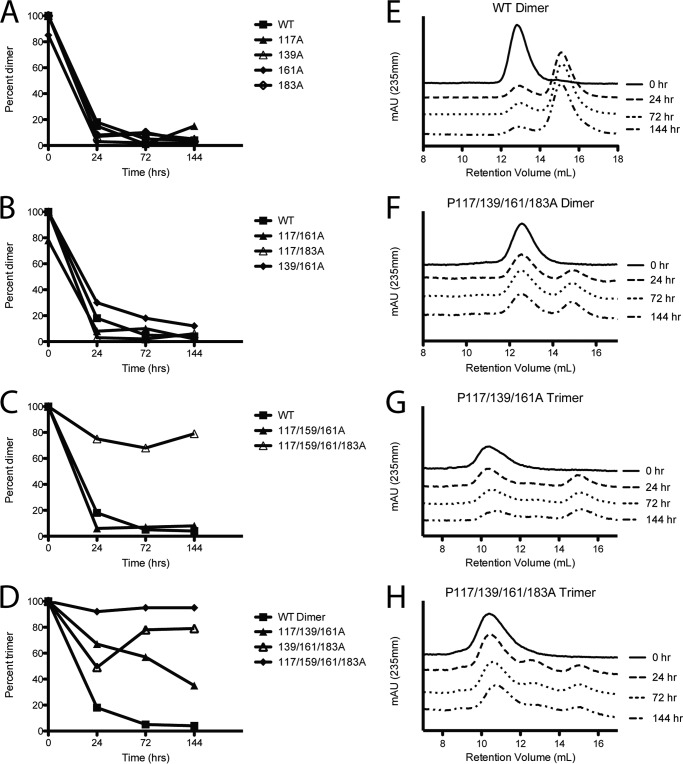
Using purified oligomeric species and SEC, we monitored the redistribution of the mutant apoA-IV samples over time at 37 °C. Previously, we established that apoA-IV is highly stable for days in its dimeric and monomeric forms when incubated below 25 °C (37). However, at 37 °C, isolated dimers redistribute to a 30:70 dimer:monomer mixture within 24 h (Fig. 3, A and E) (37). To assess the stability of the mutants, we isolated the most dominant oligomeric forms adopted by the mutants and demonstrated that they were stable at 4 °C (shown as the 0 h trace in Fig. 3, E–H). The proline mutants were then incubated at 37 °C for 24, 72, and 144 h and analyzed by SEC. Similar to WT, the isolated dimer of single, double, and triple proline mutants readily distributed into the monomer and dimer forms by 24 h (Fig. 3, A–C). However, when the dimer of the quadruple proline mutation was tested, the distribution resulted in significantly more dimer than WT after 144 h with an 60:40 dimer:monomer mix (Fig. 3, C and F). For those mutants that showed prominent trimers, we performed a similar experiment on isolated trimers. In each case, the mutants showed some degree of redistribution after 144 h. However, there were interesting differences. For example, the trimer for P117A/P139A/P161A redistributed between the trimer and monomer bands with a near complete absence of dimer (Fig. 3G). By contrast, trimeric P117A/P139A/P161A/P183A remained largely trimeric with minimal redistribution to dimer and monomer (Fig. 3, D and H).
FIGURE 3.
Oligomeric stability of isolated apoA-IV triple and quadruple proline mutants. For each protein, the sample was first isolated at 4 °C by SEC. Purified oligomeric species of ApoA-IV and proline mutants were incubated at 37 °C for the indicated time and reanalyzed by SEC. The ratio is calculated as the area under the desired species divided by the total area of regions 1–3. A–C, stability of the purified WT dimer when compared with the single proline mutants (A), the double proline mutants (B), and the triple and quadrupole proline mutants (C). D, stability of purified trimer showing the WT dimer for comparison. Note: WT trimer cannot be isolated. E–H, individual SEC UV absorbance traces showing the distribution of oligomeric species over time for WT (E), P117/P139A/P161A/P183A dimer (F), P117A/P139A/P161A trimer (G), and P117/P139A/P161A/P183A trimer (H). mAU, milliabsorbance units.
Proline Mutagenesis Increases the Thermal Stability of ApoA-IV
We next determined the effect of the proline mutations on the overall secondary structure of apoA-IV in its various oligomeric states using circular dichroism. All mutants exhibited the classical shape of a highly helical protein with characteristic minima at 208 and 222 nm similar to WT apoA-IV, with minor fluctuations in the magnitude of the peaks at 208 and 222 nm (data not shown).
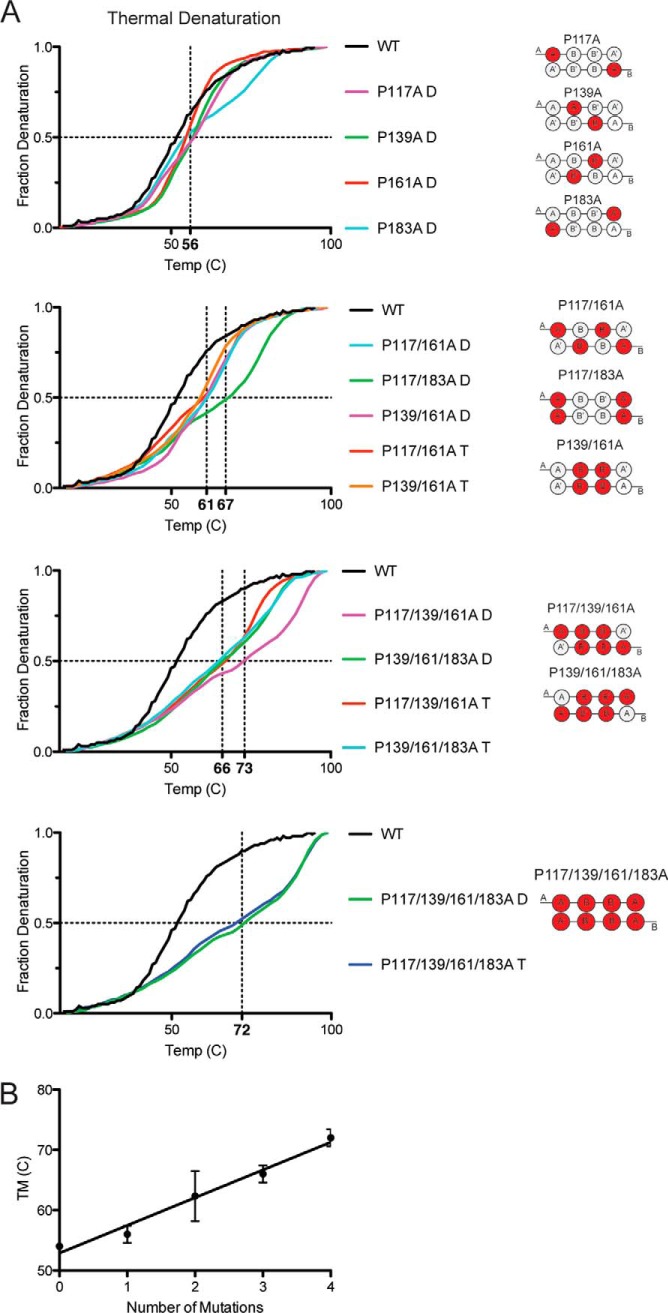
We next determined the effects of the proline mutations on protein stability using thermal denaturation experiments as monitored by circular dichroism. The temperature melting curve for apoA-IV is not fully reversible (data not shown), which limits the determination of thermodynamic unfolding parameters (38). However, information can still be gleaned about the relative stability of the WT when compared with the mutant proteins by comparing the melting temperature (Tm). The melting curve for the WT apoA-IV dimer was sigmoidal with a Tm of 54 °C, matching previous studies (8) (Fig. 4A). Analysis of the single proline mutations showed minor increases in Tm (ranging from 55 to 57 °C); however, as more proline residues were mutated to alanine, a significant increase in the Tm was observed (Fig. 4A). Most of the double mutants exhibited a Tm in the range of 61–67 °C with the triple mutants ranging from 66 to 73 °C. The Tm of the quadruple mutant was near 72 °C, an increase of 18 °C versus WT. We noticed that the increases in Tm seemed to be more related to the number of proline substitutions in a given protein and not necessarily related to their position along the shared helix. This is illustrated in Fig. 4B in which a simple linear regression analysis suggested that each proline to alanine mutation increased the Tm on average 4.6 °C ± 0.4. We also noted that the shape of the unfolding curve changed from the classic sigmoidal progression of WT apoA-I to a multiphasic pattern as more proline residues were substituted. This indicates that introduction of alanine for proline resulted in complicated unfolding states that cannot be explained by a single state transition.
FIGURE 4.
Thermal stability of proline mutants by CD. A, protein stability was measured by thermal denaturation and monitored for helical content at 222 nm with the midpoint of denaturation (i.e. Tm) indicated by the dotted vertical lines. Each trace is representative of three scans run on independently prepared samples. B, averaged Tm plotted versus number of proline mutations showing general stabilizing trend of proline substitutions. Error bars represent one sample standard deviation calculated from averaging the data from each mutant containing that number of mutations.
The thermal denaturation curves also allowed us to compare the stabilizing forces between the oligomeric forms (i.e. dimers and trimers of the same mutant). With the exception of one mutant, P117A/P139A/P161A, the dimer and trimer forms of four out of five mutants shared similar denaturation curves. This suggests that the forces stabilizing the trimer appear to be similar to the dimer. This is consistent with previous findings that the monomer and dimer of apoA-IV exhibited similar melting curves (8). However, it should be noted that we cannot rule out the possibility that the starting protein is redistributing during the experiment.
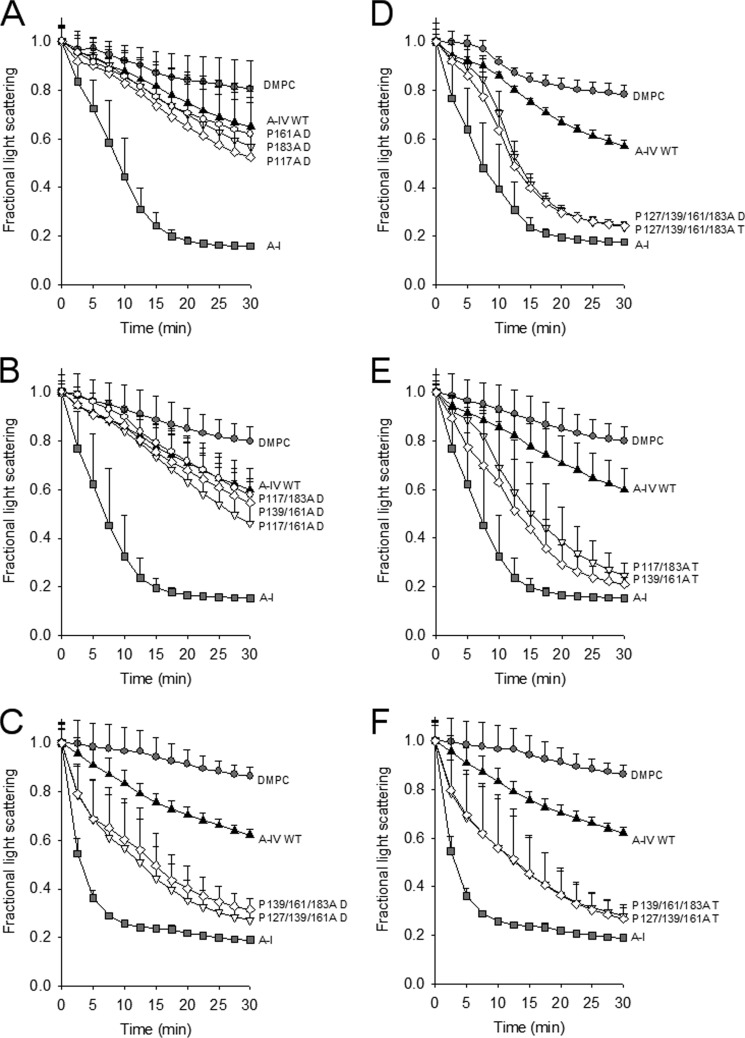
Effect of Proline Substitution on Lipid Binding and Cholesterol Efflux Functionality
To analyze the functional implications of the proline substitutions, we performed a lipid clearance assay using DMPC. This assay measures both the ability of a given protein to associate with lipid and its ability to reorganize large multilamellar liposomes into small lipoprotein-like particles analogous to those found in plasma. This is reflected as a time-dependent decrease in light scattering from the liposomes. Fig. 5 shows that WT lipid-free apoA-IV cleared DMPC liposomes to a moderate extent versus the lipid alone, whereas apoA-I (included here as a positive control) was highly effective over the 30 min of the assay. This is consistent with numerous previous studies (39, 40). The single proline mutants (present as dimers) showed no significant difference in their ability to clear DMPC when compared with WT (also present as a dimer), although they trended toward being slightly more effective (Fig. 5A). The dimeric forms of the double proline mutants were also similar to WT (Fig. 5B). Surprisingly, the trimeric forms of P117A/P183A and P139A/P161A showed a marked increase in the ability to clear DMPC, even approaching that of apoA-I (Fig. 5E). Both the dimeric and the trimeric forms of the triple proline mutants were also significantly more effective than WT (Fig. 5, C and F) as was both dimer and trimer of the quadruple mutant (Fig. 5D).
FIGURE 5.
Ability of apoA-IV proline mutants to bind and reorganize DMPC liposomes in the dimeric and trimeric states. A, dimers of WT apoA-IV and mutant dimers. DMPC liposomes were prepared and incubated with equal masses of isolated dimers for each mutant at 24.5 °C for 30 min in a visible spectrophotometer. DMPC without protein and human apoA-I were included as negative and positive controls, respectively. B and C, same experiment with isolated dimers of the double proline mutants (B) and isolated dimers of the triple proline mutants (C). D, isolated dimer and trimer from the quadruple proline mutant. E, isolated trimers from the double proline mutants. F, isolated trimers from the triple proline mutants. For all panels, the data are normalized to t = 0 for each experiment (set to 1.0) and represent averaged data from three independent experiments performed on the same day. The error bars represent one sample standard deviation.
We have previously shown that apoA-IV is capable of generating HDL particles via the ATP binding cassette A1 cell surface transporter in macrophages, although not as effectively as apoA-I or apoA-II (39). Thus, we determined the cholesterol efflux ability of the trimeric quadruple mutant and compared this with WT apoA-I and apoA-IV (Fig. 6). This mutant was selected because of its remarkable oligomeric stability and, based on the thermodynamic studies outlined above, was predicted to remain in its trimeric state for the length of this bioassay. Interestingly, the proline substitutions enhanced the ability of apoA-IV to promote cholesterol efflux, becoming similar to that of apoA-I.
FIGURE 6.
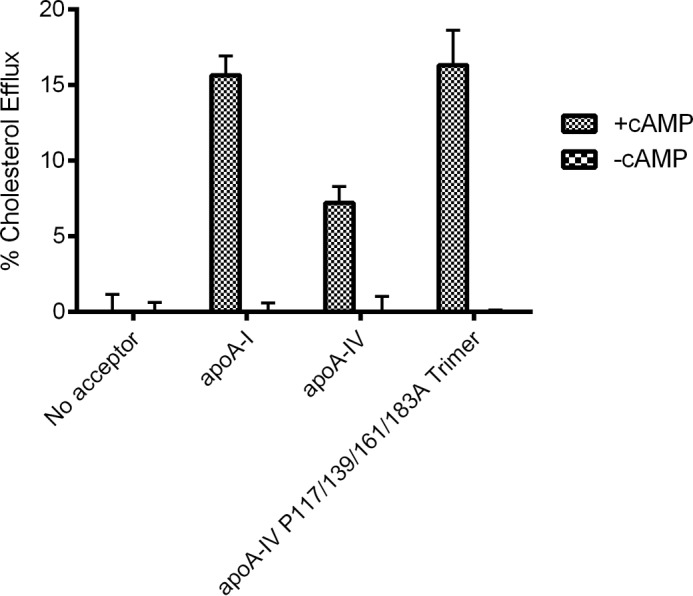
Cholesterol efflux comparison of wild-type dimer apoA-IV, apoA-I, and trimer form of quadruple apoA-IV mutant. RAW 264.7 mouse macrophages were grown to confluency, exchange-labeled with tritiated free cholesterol, and treated with cAMP to up-regulate ABCA1 (see “Experimental Procedures”). After washing, test medium was applied containing 10 μg/ml of each of the indicated lipid-free proteins for 6 h at 37 °C. The appearance of cholesterol-based radioactivity in the medium was determined by liquid scintillation counting. Cells lacking cAMP, and therefore ABCA1 expression, were included as controls to verify ABCA1 specificity. The reported efflux values were adjusted for the minor efflux to medium containing no acceptor protein. The data represent triplicate analyses, and error bars show one sample standard deviation. The asterisk represents a significant difference from WT apoA-IV by a two-tailed Student's t test (p < 0.05).
Small-angle X-ray Scattering Analysis of the Quadruple Proline Mutant
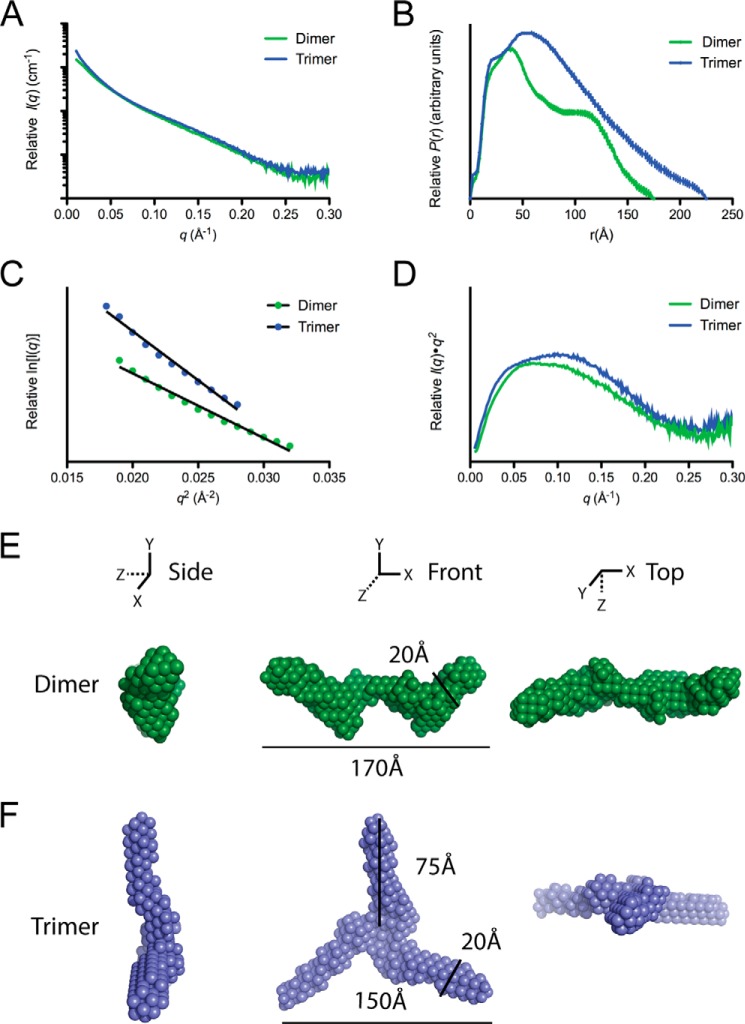
We have previously utilized small-angle x-ray diffraction (SAXS) to characterize the overall molecular shapes in solution of both the monomer and the dimer forms of WT apoA-IV. As SAXS requires a highly homogenous sample, the high stability of the quadruple proline mutant (i.e. extremely slow interconversion rate) allowed for the characterization of both the trimer and the dimer forms (Fig. 7, A–D). Analysis of the scattering profile shows that the dimeric form has a calculated radius of gyration (Rg) of 56.7 ± 1.5 Å, with a Dmax of 195 Å and a bimodal pairwise distance-distribution (P(r)) curve, indicating a semi-modular, rod-like structure comparable with the WT protein (Table 1). Interestingly, the trimeric form had a significantly larger Rg of 67.5 ± 1.1 Å, and a Dmax of 225 Å. The trimer P(r) curve resembled an elongated rod with a spherical peak, as indicated by a single peak with a shoulder, significantly different from the dimer (Fig. 7B). For both the dimer and the trimer forms, the Rg, and I0 calculated from Guinier analysis matched closely to that of P(r) curve (Table 1), and the Kratky plots indicated that the proteins are structured (Fig. 7D).
FIGURE 7.
Small angle x-ray scattering of dimeric and trimeric forms of the quadruple proline mutant. A and B, scattering profile (A) and P(r) function (B). C and D, Guinier plot (C) and Kratky plot (D) of dimer (green) and trimer (blue) form of the quadruple proline mutant. E and F, averaged ab initio reconstructions of dimer (E) and trimer (F).
Based on the SAXS data, three-dimensional envelopes for both the dimer and the trimer forms were created using ab initio reconstruction (Fig. 7, E and F). In each case, 10 independent reconstructions were generated and then averaged (Table 1). The dimeric envelope revealed a long rod, similar to WT apoA-IV (37, 41), but was more segmented or kinked along the length. Surprisingly, this feature resembled a previously characterized point mutation, F334A, which also cleared DMPC effectively (7). However, unlike F334A, the quadruple proline mutant dimer is more structured when comparing their Kratky plots (37). Strikingly, the trimer envelope showed a propeller structure with three rods radiating from a central point related by 3-fold symmetry (Fig. 7F). The dimensions of each rod are roughly 20 Å wide and 75 Å in length, as measured from the central point to the outer tip.
Chemical Cross-linking Analysis of the Quadruple Proline Mutant
Given the high structural similarity between the apoA-IV monomer and its dimer, it was reasonable to expect that the trimer formed by the quadruple mutant might also exhibit similar intermolecular contacts. To test this, we performed a chemical cross-linking experiment on the isolated trimeric form of the quadruple proline mutant. We obtained 69 cross-linked peptides that reported about 46 distance constraints (i.e. Lys pairs) within the trimer. Most of these cross-links were also observed in our previous study of WT ApoA-IV dimer (37). Fig. 8 shows a contact plot of the WT apoA-IV dimer rendered from our previously proposed structure (41). The plot shows the proximity of each residue with respect to all other residues in a two-dimensional format. The colored areas show where the residues indicated on each axis come within 20 Å of each other (i.e. close enough to be cross-linked by the reagent we used). The experimental cross-links derived in the quadruple mutant trimer are superimposed on the plot. For the most part, these fall within the colored regions and are thus highly consistent with the WT dimer structure, even when the residues are far apart in the sequence. However, there were a few cross-links that fell outside a colored region, but these could easily be consistent with the model with minor adjustments of the structure. These data strongly suggest that the intermolecular contacts within the stabilized apoA-IV trimer are similar to those in the stable WT apoA-IV dimer and its monomer.
FIGURE 8.
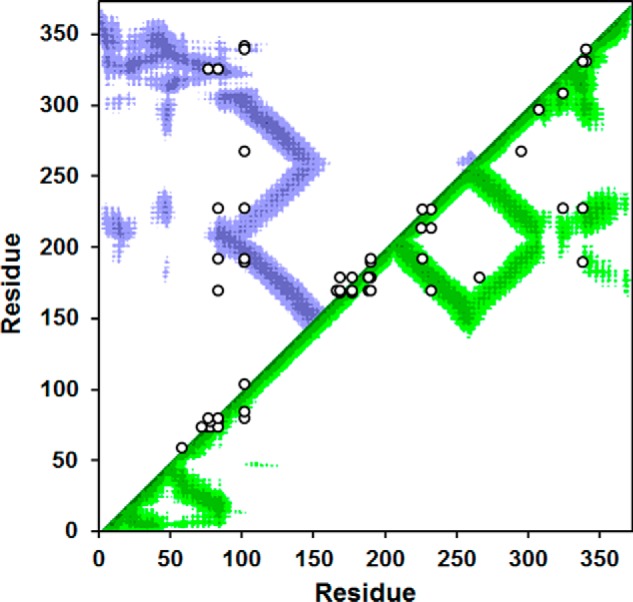
Cross-linking data from the stable trimer of the quadruple proline mutant superimposed on a contact plot of the latest model of dimeric WT apoA-IV. The x and y axes of the plot indicate the residue number of apoA-IV from 1 to 376. The diagonal line bisects the figure, and the top/left is used to evaluate intermolecular interactions, whereas the bottom/right shows intramolecular interactions. Using our latest structure of the human WT apoA-IV dimer (8), we produced a contact map for each amino acid. The regions of the darkest shade represent proximity of 1–10 Å, and the lighter gradations represent 11–15, 16–20, and 21–24 Å, respectively. White indicates greater than 24 Å. For example, residue 100 of one molecule (x axis) is within 11–15 Å of residue 200 (y axis) of the other molecule in the dimer structure as indicated by the purple color at their intersection on the graph. On the other hand, residue 100 is quite far away from residue 250 of the other molecule (white where they intersect). The cross-links between Lys residues identified in trimeric P117/P139A/P161A/P183A apoA-IV are superimposed in white on the figure. In cases where Lys assignments in cross-links were ambiguous, the Lys residues with the shortest separation distance were plotted.
DISCUSSION
Highly conserved proline residues have long been thought to play an important structural role in the exchangeable apolipoproteins (i.e. initiation and termination of helices); however, limited structure-function studies have been performed to explore these ideas. The reluctance of mutating proline residues likely stems from a concern of inducing large overall structural perturbations, making data interpretation difficult. These concerns, while valid, have been partially mitigated in this study by targeting specific proline residues based on the crystal structure of apoA-IV.
Given the remarkable alignment of proline residues in the long swapped helix in dimeric apoA-IV, we hypothesized that these proline residues might function as hinges and help to destabilize the helical bundle to facilitate lipid binding. Indeed, our data clearly showed that substitution of these proline residues with alanine increased the overall protein stability as measured by an increase in melting temperature, supporting that the proline residues destabilize the bundle. In addition, we also observed an increase in the preference of the dimer form and higher order species versus the monomer. Our results indicate that these effects were not strictly dependent on the position of a particular proline mutation. For example, we did not find that substitution of any one proline (or even a particular pair of prolines) always resulted in changes to oligomerization or stability. Instead, these observations correlated best with the overall number of proline residues mutated in helix B. This suggests that all of these prolines work together to have a concerted effect on apoA-IV structure. However, contrary to our hypothesis, extensive proline disruption led to an unexpected increase in the ability of apoA-IV to bind to and reorganize phospholipid membranes despite the increased stability of the proteins in the thermodynamic sense.
Previously, we have proposed that apoA-IV lipid affinity is regulated by a clasp mechanism where the N and C termini both interact with each other and the core bundle to limit lipid binding (39, 40, 42). Disruption of the interaction leads to avid lipid association and the ability to solubilize phospholipid liposomes. Further insight into this mechanism was recently gained by SAXS analysis of the fast lipid-binding mutant F334A, which showed structural changes to the core helical bundle domain (37). Interestingly, our SAXS analysis revealed apparently similar conformational changes for the quadruple proline mutant (i.e. an overall kinking of the long rod-like structure characteristic of WT apoA-IV). Therefore, it is possible that structural changes in the core could indirectly influence the clasp mechanism and increase lipid affinity despite a more stable hydrophobic core in the helical bundle. Increased lipid affinity might also arise from the stability and accumulation of the larger oligomer, which could increase the local concentration of apoA-IV to promote protein-lipid particle formation. Although the functional role of apoA-IV clasp interaction and why it has evolved to bind lipids weakly has not been resolved, it is interesting that the conserved proline residues are needed to maintain this characteristic.
Following the determination of the apoA-IV dimer structure, we suggested that the transition to the monomer was analogous to closing the blade of a pocketknife (8). Indeed, the introduction of glycine residues into the middle of helix B (essentially oiling the hinge of the pocketknife) favors distribution to the monomeric form. In contrast, we now show that replacing the helix-disrupting proline residues with helix-promoting alanine residues favored the formation of the higher oligomeric forms. We would argue that this results from the stabilization of helix B, which can stay more extended in the dimer and trimer forms versus the monomer. The reason for preference for the trimer over the dimer for several of the mutants is currently unclear, but is most likely concentration-dependent.
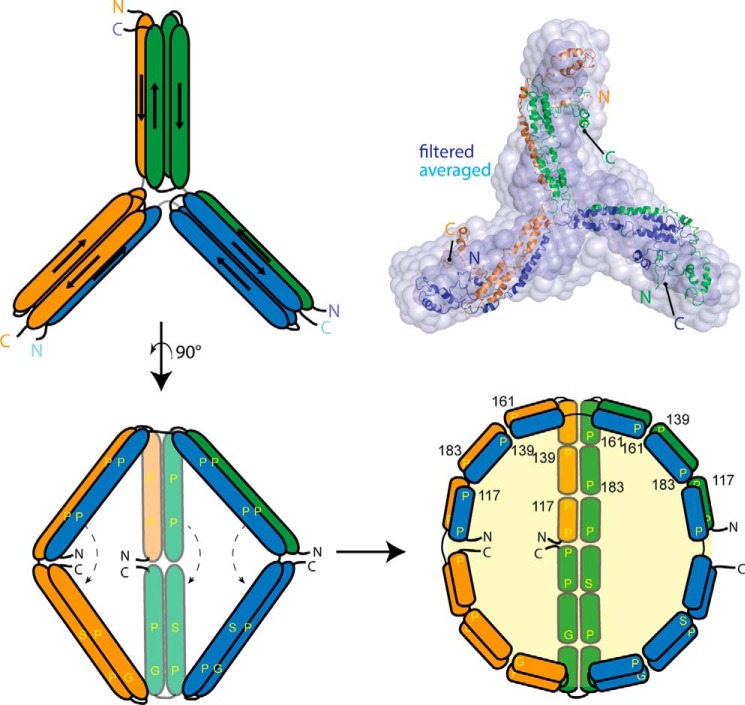
The increase in stability of the proline mutants, particularly the quadruple mutant, provided an opportunity to characterize a stable trimeric apolipoprotein species for the first time. The SAXS data indicated a propeller-like structure, whereas the cross-linking data were strikingly similar to the intra- and intermolecular patterns we previously observed for the WT dimer and similar to the intramolecular patterns found in isolated WT monomers (37). A logical interpretation is that all three oligomeric states are built upon similar molecular interactions. Because both the monomer and the dimer form a robust four-helix bundle, we propose that each blade of the trimer propeller is a four-helix bundle. Furthermore, because apoA-IV dimerization is achieved by swapping one of the helices in the bundle (8), it follows that a similar domain-swap mechanism accounts for the trimer. Here, instead of two chains reciprocally swapping a B helix in the dimer, each chain accepts and donates a helix from a different polypeptide in a daisy chain-like fashion. The propeller structure could be generated by a slight bend in the center of the shared helix such that each apoA-IV molecule is bent 120°. This concept is illustrated in Fig. 9 where the extended helix of chain 1 (orange) swaps with chain 2 (green), chain 2 swaps with chain 3 (blue), and chain 3 swaps with chain 1 to close the formation. Note that all intermolecular interactions occurring in the trimer also occur in an identical fashion in the dimer, except that each molecule is swapping with two partners versus one. This concept allows for the straightforward visualization of higher order oligomers such as tetramers in which an additional molecule is added to the chain, requiring only a minor adjustment in the bend angle of the participating polypeptides. Interestingly, this is reminiscent of the Trefoil model of apoA-I in spherical HDL particles, which can account for up to five molecules on the surface of a lipoprotein through a similar helix-swapping paradigm (43). If our ideas are correct, these helix-swapping connections may represent a common mode of interaction that may apply to apolipoproteins beyond apoA-IV and apoA-I.
FIGURE 9.
Trimeric model apoA-IV based on the SAX data from the quadruple mutant, and possible mode of lipid accumulation. Utilizing the ab initio reconstruction, our previously generated model for the full-length monomer of apoA-IV (41), and the crystal structure (3S84) (8) of apoA-IV, we propose that the domain swap mechanism responsible for dimerization can also result in a trimer. The atomic trimer model was superimposed onto both the averaged and the filtered ab initio molecular envelopes using SUPCOMB (33) with an normalized spatial discrepancy value of 5.4 and 2.5, respectively. Invoking a similar mechanism as described previously (8), swinging away the helical arms (indicated by the dashed line) would expose the hydrophobic core to promote lipid sequestration. Due to the segmentation of proline, glycine, and serine residues, which could provide curvature, it is possible to envision a spherical particle resulting.
Three-dimensionally, the trimer model also suggests a mechanism for how HDL can transition from a disc to a sphere through additional self-association. Previously, we proposed a mechanism for how dimeric apoA-IV can emulsify lipids to create a disk-like particle by swinging open a pair of helical arms that fold across the swapped helical backbone (8). In the case of a trimer, the same swinging action can be applied to open the hydrophobic core and allow for lipid binding. This creates a spherical cavity for lipid without changing the molecular interactions within and between apoA-IV molecules (Fig. 9). Although speculative, preliminary experiments in our laboratory and the results of others substantiate the existence of spherical HDL particles that exclusively contain apoA-IV in human plasma. Future studies will be directed at isolating and studying the structure of these particles. It will also be interesting to determine whether similar self-association assemblies also occur for apoA-I as a similar helix-swapping theme was also identified with apoA-I crystal structure of the N-terminal fragment (9) or whether the proline residues in apoA-I also play a role in regulating its oligomeric state. Furthermore, as HDL particles can contain a large number of apolipoprotein chains, it will be important to determine whether the “swapping mechanism” is a driving force that facilitates the accumulation of additional chains to an HDL particle.
Acknowledgments
We thank Drs. Yun-Xing Wang and Xianyang Fan from the Protein-Nucleic Acid Interaction Section at the Structural Biophysical Laboratory of the NCI, National Institutes of Health for providing us with help in data collection and beam time to perform the SAXS experiments. We also thanks the X-ray Science Division at Argonne National Laboratory for technical support at the beamline 12-ID-B and the Proteomics Core at the University of Cincinnati. We gratefully acknowledge Martin Jones from the University of Alabama at Birmingham for development of the contact plots used herein.
This work was supported by National Institutes of Health R01 Grants GM098458 (to T. B. T. and W. S. D.) and HL67093 (to W. S. D.) and by National Institutes of Health Training Grant T32 HL007382 (to X. D.).
- apoA-IV
- apolipoprotein A-IV
- DMPC
- 1,2-dimyristoyl-sn-glycero-3-phosphocholine
- SAXS
- small angle x-ray scattering
- SEC
- size exclusion chromatography
- P(r)
- pairwise distance-distribution
- 8-bromo-3′-5′cAMP
- 8-bromoadenosine 3′-5′-cAMP.
REFERENCES
- 1. Tabet F., Rye K.-A. (2009) High-density lipoproteins, inflammation and oxidative stress. Clin. Sci. 116, 87–98 [DOI] [PubMed] [Google Scholar]
- 2. Tso P., Liu M. (2004) Apolipoprotein A-IV, food intake, and obesity. Physiol. Behav. 83, 631–643 [DOI] [PubMed] [Google Scholar]
- 3. Wang F., Kohan A. B., Kindel T. L., Corbin K. L., Nunemaker C. S., Obici S., Woods S. C., Davidson W. S., Tso P. (2012) Apolipoprotein A-IV improves glucose homeostasis by enhancing insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 109, 9641–9646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qin X., Swertfeger D. K., Zheng S., Hui D. Y., Tso P. (1998) Apolipoprotein AIV: a potent endogenous inhibitor of lipid oxidation. Am. J. Physiol. 274, H1836–H1840 [DOI] [PubMed] [Google Scholar]
- 5. Vowinkel T., Mori M., Krieglstein C. F., Russell J., Saijo F., Bharwani S., Turnage R. H., Davidson W. S., Tso P., Granger D. N., Kalogeris T. J. (2004) Apolipoprotein A-IV inhibits experimental colitis. J. Clin. Invest. 114, 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li X., Xu M., Wang F., Kohan A. B., Haas M. K., Yang Q., Lou D., Obici S., Davidson W. S., Tso P. (2014) Apolipoprotein A-IV reduces hepatic gluconeogenesis through nuclear receptor NR1D1. J. Biol. Chem. 289, 2396–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tubb M. R., Silva R. A. G. D., Pearson K. J., Tso P., Liu M., Davidson W. S. (2007) Modulation of apolipoprotein A-IV lipid binding by an interaction between the N and C termini. J. Biol. Chem. 282, 28385–28394 [DOI] [PubMed] [Google Scholar]
- 8. Deng X., Morris J., Dressmen J., Tubb M. R., Tso P., Jerome W. G., Davidson W. S., Thompson T. B. (2012) The structure of dimeric apolipoprotein A-IV and its mechanism of self-association. Structure 20, 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mei X., Atkinson D. (2011) Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286, 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., De Loof H., Harvey S. C. (1999) A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274, 31755–31758 [DOI] [PubMed] [Google Scholar]
- 11. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 12. Elshourbagy N. A., Walker D. W., Boguski M. S., Gordon J. I., Taylor J. M. (1986) The nucleotide and derived amino acid sequence of human apolipoprotein A-IV mRNA and the close linkage of its gene to the genes of apolipoproteins A-I and C-III. J. Biol. Chem. 261, 1998–2002 [PubMed] [Google Scholar]
- 13. Mitani A., Ishigami M., Watase K., Minakata T., Yamamura T. (2011) A novel apolipoprotein E mutation, ApoE Osaka (Arg158 Pro), in a dyslipidemic patient with lipoprotein glomerulopathy. J. Atheroscler. Thromb. 18, 531–535 [DOI] [PubMed] [Google Scholar]
- 14. Luo B., Huang F., Liu Q., Li X., Chen W., Zhou S.-F., Yu X. (2008) Identification of apolipoprotein E Guangzhou (arginine 150 proline), a new variant associated with lipoprotein glomerulopathy. Am. J. Nephrol. 28, 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oikawa S., Matsunaga A., Saito T., Sato H., Seki T., Hoshi K., Hayasaka K., Kotake H., Midorikawa H., Sekikawa A., Hara S., Abe K., Toyota T., Jingami H., Nakamura H., Sasaki J. (1997) Apolipoprotein E Sendai (arginine 145→proline): a new variant associated with lipoprotein glomerulopathy. J. Am. Soc. Nephrol. 8, 820–823 [DOI] [PubMed] [Google Scholar]
- 16. Miller M., Aiello D., Pritchard H., Friel G., Zeller K. (1998) Apolipoprotein A-IZavalla (Leu159→Pro): HDL cholesterol deficiency in a kindred associated with premature coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 18, 1242–1247 [DOI] [PubMed] [Google Scholar]
- 17. Utermann G., Haas J., Steinmetz A., Paetzold R., Rall S. C., Jr., Weisgraber K. H., Mahley R. W. (1984) Apolipoprotein A-IGiessen (Pro143→Arg). Eur. J. Biochem. 144, 325–331 [DOI] [PubMed] [Google Scholar]
- 18. Murphy C. L., Wang S., Weaver K., Gertz M. A., Weiss D. T., Solomon A. (2004) Renal apolipoprotein A-I amyloidosis associated with a novel mutant Leu64Pro. Am. J. Kidney Dis. 44, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 19. Higuchi K., Yonezu T., Tsunasawa S., Sakiyama F., Takeda T. (1986) The single proline-glutamine substitution at position 5 enhances the potency of amyloid fibril formation of murine apo A-II. FEBS Lett. 207, 23–27 [DOI] [PubMed] [Google Scholar]
- 20. Phillips J. C., Wriggers W., Li Z., Jonas A., Schulten K. (1997) Predicting the structure of apolipoprotein A-I in reconstituted high-density lipoprotein disks. Biophys. J. 73, 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., Axelsen P. H. (1999) The structure of human lipoprotein A-I: evidence for the “belt” model. J. Biol. Chem. 274, 14541–14544 [DOI] [PubMed] [Google Scholar]
- 22. Bhat S., Sorci-Thomas M. G., Alexander E. T., Samuel M. P., Thomas M. J. (2005) Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J. Biol. Chem. 280, 33015–33025 [DOI] [PubMed] [Google Scholar]
- 23. Martin D. D. O., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. (2006) Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281, 20418–20426 [DOI] [PubMed] [Google Scholar]
- 24. Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., 3, Smith J. D., Gogonea V., Hazen S. L. (2007) The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol.14, 861–868 [DOI] [PubMed] [Google Scholar]
- 25. Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., Gutsche I., Zaccai G., Didonato J. A., Hazen S. L. (2009) Double superhelix model of high density lipoprotein. J. Biol. Chem. 284, 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. (1997) Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. U.S.A. 94, 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng L., Baumann U., Reymond J.-L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115–e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 [Google Scholar]
- 30. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 [Google Scholar]
- 31. Svergun D. I., Petoukhov M. V., Koch M. H. J. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkov V. V., Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozin M. B., Svergun D. I. (2001) Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 [Google Scholar]
- 34. Betts M. J., Russell R. B. (2007) Amino-acid properties and consequences of substitutions. in Bioinformatics for Geneticists (Barnes M.R., Gray I.C., eds), Vol. 2, pp. 311–339, John Wiley & Sons Ltd., Chichester, UK [Google Scholar]
- 35. Davidson W. S., Hazlett T., Mantulin W. W., Jonas A. (1996) The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. U.S.A. 93, 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garai K., Frieden C. (2010) The association-dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry 49, 9533–9541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng X., Morris J., Chaton C., Schröder G. F., Davidson W. S., Thompson T. B. (2013) Small-angle X-ray scattering of apolipoprotein A-IV reveals the importance of its termini for structural stability. J. Biol. Chem. 288, 4854–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Greenfield N. J. (2006) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pearson K., Saito H., Woods S. C., Lund-Katz S., Tso P., Phillips M. C., Davidson W. S. (2004) Structure of human apolipoprotein A-IV: a distinct domain architecture among exchangeable apolipoproteins with potential functional implications. Biochemistry 43, 10719–10729 [DOI] [PubMed] [Google Scholar]
- 40. Pearson K., Tubb M. R., Tanaka M., Zhang X. Q., Tso P., Weinberg R. B., Davidson W. S. (2005) Specific sequences in the N and C termini of apolipoprotein A-IV modulate its conformation and lipid association. J. Biol. Chem. 280, 38576–38582 [DOI] [PubMed] [Google Scholar]
- 41. Walker R. G., Deng X., Melchior J. T., Morris J., Tso P., Jones M. K., Segrest J. P., Thompson T. B., Davidson W. S. (2014) The structure of human apolipoprotein A-IV as revealed by stable is isotope-assisted cross-linking, molecular dynamics, and small angle x-ray scattering. J. Biol. Chem. 289, 5596–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tubb M. R., Silva R. A. G. D., Fang J., Tso P., Davidson W. S. (2008) A Three-dimensional homology model of lipid-free apolipoprotein A-IV using cross-linking and mass spectrometry. J. Biol. Chem. 283, 17314–17323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang R., Silva R. A. G. D., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. (2011) Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18, 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]