Background: FoF1 synthesizes ATP by ion gradient-powered Fo c-ring CW rotation viewed from the periplasm.
Results: An electrostatic Fo-cR50/aE196 leash and proton gate promotes CW rotation against ATPase-driven CCW rotation by as much as one c subunit.
Conclusion: A subunit a “grab and push” mechanism rotates the Fo c-ring CW.
Significance: How Fo drives CW rotation for ATP synthesis against F1 ATPase-dependent CCW torque is a major unresolved question.
Keywords: ATP Synthase, F1Fo ATPase, Molecular Motor, Proton Transport, Single-molecule Biophysics, Nanodiscs
Abstract
Living organisms rely on the FoF1 ATP synthase to maintain the non-equilibrium chemical gradient of ATP to ADP and phosphate that provides the primary energy source for cellular processes. How the Fo motor uses a transmembrane electrochemical ion gradient to create clockwise torque that overcomes F1 ATPase-driven counterclockwise torque at high ATP is a major unresolved question. Using single FoF1 molecules embedded in lipid bilayer nanodiscs, we now report the observation of Fo-dependent rotation of the c10 ring in the ATP synthase (clockwise) direction against the counterclockwise force of ATPase-driven rotation that occurs upon formation of a leash with Fo stator subunit a. Mutational studies indicate that the leash is important for ATP synthase activity and support a mechanism in which residues aGlu-196 and cArg-50 participate in the cytoplasmic proton half-channel to promote leash formation.
Introduction
Most living organisms rely primarily on the FoF1 ATP synthase to maintain the non-equilibrium concentration gradient of ATP to ADP and Pi that provides the main source of energy for cellular processes (1). The ATP synthase is composed of two rotary motors, Fo and F1, joined by their rotor and stator components (Fig. 1A). To synthesize ATP, the Fo motor translocates H+ or, in some species Na+ ions across the membrane at the expense of a non-equilibrium electrochemical potential as a means to drive rotation of a ring of c subunits relative to subunit a. When viewed from the periplasm of Escherichia coli, protons move into the cytoplasm concurrent with clockwise (CW)4 rotation. This rotation forces conformational changes in the catalytic sites of F1 that drive ATP synthesis. Conversely, the F1 motor can hydrolyze ATP via a mechanism that alternates successively between its three catalytic sites to drive counterclockwise (CCW) rotation in a manner that forces the Fo motor to pump protons to the periplasm.
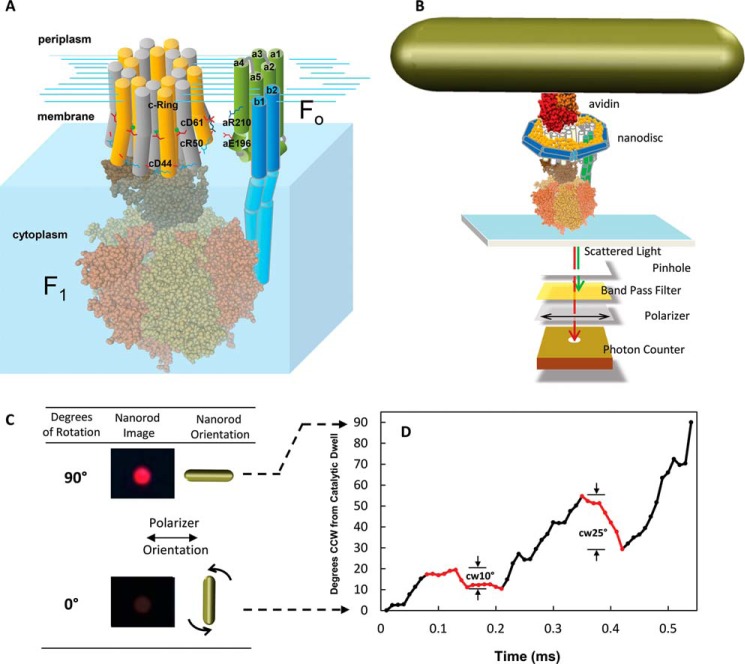
FIGURE 1.
A, subunit components of the FoF1 ATP synthase. The E. coli Fo motor shown as a schematic includes subunits a (gray) and b1 and b2 (green) and the c10-ring, where individual c subunits are distinguished in yellow and white. The F1 ATPase motor (PDB code 3OAA) subunits include α (orange), β (tan), γ (light brown), and ϵ (dark brown). B, single molecule measurements of rotation using gold nanorods as a probe. The FoF1 complexes were incorporated into lipid bilayer nanodiscs (n-FoF1) that each contained a bilayer of phospholipid molecules surrounded by the membrane scaffold protein to stabilize Fo (27). The n-FoF1 molecules were attached to the microscope slide via His6 tags on the N termini of the F1 β subunits. The c2▿C mutation to E. coli FoF1 was biotinylated to attach the avidin-coated gold nanorod (78 × 34 nm). The intensity of red light scattered from a single nanorod was measured as a function of time through a polarizing filter that was aligned for minimum intensity at one of the three ATPase-dependent catalytic dwells. C, scattered light intensity when the long axis of a nanorod is perpendicular and parallel to the direction of polarization. D, transient dwells that rotate the c-ring CW (red data) against the force of ATPase-driven CCW rotation (black data) observed during a single F1ATPase power stroke. The rotational positions for the first 90° of single power strokes as a function of time were determined from the arcsine (31) where minimum and maximum intensity values of light scattered from a nanorod in the presence of 1 mm Mg2+ATP represent 0°, the end of the F1 catalytic dwell, and 90°.
Each c subunit in the ring that comprises the Fo rotor is in the form of a hairpin of two transmembrane helices (TMH) connected by a loop on the side that docks with F1 (2). The size of the c-ring is specific to the organism and has, to date, been found to vary from as small as eight copies in bovine mitochondria (3) to as large as 15 copies in cyanobacteria (4). A highly conserved carboxyl group (cAsp-61 in E. coli) located on the outer TMH of subunit c in the middle of the membrane is essential for Fo-dependent ion translocation (5, 6). Because the E. coli ATP synthase contains a c10 ring, 10 protons are transferred across the membrane during each revolution that involves a net of three ATPs made or consumed.
There is currently no high-resolution structure of Fo that includes subunit a, which is directly involved in Fo-dependent torque generation. Subunit a provides half-channels for ions to access cAsp-61 from the membrane surface and contains an essential and highly conserved arginine residue (aArg-210 in E. coli) that faces cD61 (7–10). As the result of extensive mutational analysis and cross-linking studies (11–14), subunit a is believed to fold into five TMHs, of which TMHs 2–5 form a four-helix bundle (15, 16). The center of TMHs 2–5 provides an aqueous accessible half-channel in subunit a for ions to enter from the periplasm to protonate cAsp-61 during ATP synthesis (17). Cross-linking evidence shows that protonation of this half-channel causes TMHs 4 and 5 to swivel in a manner thought to facilitate proton transfer to cAsp-61 (18).
Crystal structures of c-rings demonstrate that the carboxyl side chain equivalent to cAsp-61 in E. coli provides an ion-binding site at each of the subunit c-c interfaces, known as the ion-locked conformation, that stabilizes the ion in the membrane upon dehydration (19). Recent c-ring structures determined at low pH show that the essential carboxylate can adopt an open extended conformation that would facilitate ion loading (20). When a c-subunit completes a rotation, the change from a closed to an open conformation is anticipated to promote the displacement of the ion as the carboxylate rotates into the proximity of aArg-210.
The free energy of cellular ATP is directly proportional to the logQ, where Q = [ATP] / [ADP][Pi]. Under steady-state conditions, the cytoplasm of E. coli typically contains 3 mm ATP, 0.4 mm ADP, and 6 mm Pi so that the logQ ≅ 0.1 (21). For the chloroplast FoF1, a logQ ≅ 0.1 results at steady state from a modest ΔpH ≅ 1.5 in contrast to a logQ ≅ −4.9 in the absence of an electrochemical potential (22). Rat and mouse skeletal muscles can maintain a logQ ≅ 6.0 (8 mm ATP, 0.008 mm ADP (23), and 0.8 mm Pi), whereas cat skeletal muscle at rest has a logQ ≅ 0.2 (7.8 mm ATP, 1.3 mm ADP, and 3.8 mm Pi) that drops to −2.3 after exercise (24).
A major unresolved question is how the Fo motor uses the electrochemical gradient to generate enough torque so that the c-ring will undergo CW rotation even when the logQ is high. A Brownian ratchet mechanism has been postulated to power Fo rotation (25, 26). Two noncolinear ion access half-channels from each side of the membrane, like those identified in subunit a that lead to the cAsp-61 carboxylate, must be present for a Brownian ratchet to function. In addition, rotational diffusion of the c-ring relative to subunit a must be periodically restricted in some manner.
Recently, a periodic restriction in rotation lasting 50–175 μs that results from stepping between adjacent c subunits in the c-ring has been observed in single-molecule experiments of E. coli FoF1 incorporated into lipid bilayer nanodiscs (n-FoF1) to stabilize Fo (27). This 36° stepping, observed as a transient dwell, occurs as the result of an interaction between subunit a and subunit c known as a leash. However, the molecular basis for the interaction responsible for the leash and its relevance to the mechanism of ATP synthesis remains undetermined. Mutations to either cAsp-61 or aArg-210, the only known interaction between these subunits, have been found to decrease the ability to form the leash to only a small extent. Although proton gradient-driven CW rotation in the ATP synthase direction has been observed with single molecules of FoF1 embedded in liposomes (28) and extended lipid bilayers (29), the time resolution of these experiments was not sufficient to resolve the presence of the leash.
Using single-molecule measurements with a time resolution of 10 μs, we now show that, upon formation of the leash, the Fo complex overcomes the force of F1 ATP hydrolysis-driven CCW rotation to rotate the c-ring CW in the ATP synthase direction by as much as one c subunit. Mutational studies indicate that the leash is important for ATP synthase activity and support a mechanism in which residues aGlu-196 and cArg-50 participate in the cytoplasmic proton half-channel to promote leash formation. The results presented here provide a framework for a mechanism in which this Fo-dependent CW rotation results from protonation-dependent conformational changes in subunit a that push the van der Waals surface of TMH 4 against the surface of subunit c.
EXPERIMENTAL PROCEDURES
Preparations of n-FoF1 and Inverted E. coli Membrane Vesicles
Mutations were created from the pFV2 plasmid construct as described by Ishmukhametov et al. (10). The detergent-solubilized FoF1 purified from both the wild type and mutants was immediately incorporated into nanodiscs as described previously (27). Inverted membrane vesicles (IMVs) of E. coli containing FoF1 were isolated as reported previously (30).
Disulfide Cross-linking
Samples of detergent-solubilized FoF1 were isolated as described previously (27) and divided into aliquots of 50 μg of protein, which were incubated for 1 h at 25 °C in a final volume of 40 μl that contained TMG buffer (50 mm Tris, 5 mm MgCl2, 10% (v/v) glycerol (pH 7.5)/HCl) (sample 1), 1.5 mm βMSH in TMG buffer (sample 2), 2.0 mm CuCl2 and 5.5 mm O-phenanthroline in TMG (sample 3), or 0.5 mm M2M (1,2-ethanediyl bismethanethiosulfonate) (sample 4). Subsequently, 5 μl of 500 mm EDTA (pH 8.0) was added to samples 3 and 4 and incubated for 15 min prior to the addition of 1 μl of 10 mm fluorescein maleimide to all samples. After a 2-h incubation, unreacted fluorescein maleimide was removed by 10 successive concentration/dilution cycles using 7 ml of Pierce spin concentrators with a 150 kDa cutoff. The final sample concentrations were normalized, and 40 μg of protein was loaded onto a premade SDS 8–16% gradient denaturing gel (Bio-Rad). Fluorescent bands were imaged using UV light prior to Coomassie staining.
ATP Synthase Assays
The ATP synthesis activity of IMVs was measured using the ATP bioluminescence assay kit CLS II luciferin/luciferase system (Roche Diagnostics) as the result of a pH gradient using buffer 1 (pH 8.0) and buffer 2 (pH 4.4), which were each composed of 20 mm succinate, 20 mm Tricine, 50 mm MES, 200 mm NaCl, 5 mm NaP, 5 mm MgSO4, and buffer 3, which was composed of 200 mm Tricine, 158 mm KCl, and 242 mm KOH (pH 8.9). Prior to use, the lyophilized luciferin/luciferase mixture was dissolved in 10 ml of buffer 3 according to the directions of the vendor. Freshly prepared IMVs were suspended in buffer 1 at 52.73 mg of protein/ml as a stock for use in the assays. For each assay, 27.5 μl of the IMV stock was added to a cuvette that contained 135.5 μl of buffer 2 and 4 μl of a 1 μg/ml solution of valinomycin, incubated for 5 min at 25 °C, and then mixed rapidly in a cuvette containing 155 μl of the buffer 3-luciferin/luciferase solution and 8 μl of 2 mm ADP. The rate of increase of 562 nm luminescence intensity was monitored immediately upon mixing in a Cary Eclipse spectrometer at 28 °C with the photomultiplier tube detector at 800 V and the slit width at 20 nm.
ATP Hydrolysis Assay
The rate of ATP hydrolysis of membrane vesicles was measured in a buffer containing 50 mm Tris-HCl (pH 8.0), 20 mm KCl, 1 mm MgCl2, and 2 mm ATP using a coupled enzyme assay with pyruvate kinase and lactic dehydrogenase as described in Ref. 31.
Proton Translocation-dependent Quenching of ACMA Fluorescence
Fluorescence of ACMA was excited at 410 nm and monitored at 480 nm using a Cary Eclipse fluorescence spectrometer. Fluorescence quenching was initiated by the addition of a final concentration of either 5 mm ATP or 1 mm NADH to IMVs suspended in a buffer containing 10 mm HEPES (pH 7.5), 100 mm KCl, 10 mm MgCl2, and 0.25 mg/ml of ACMA. The reaction was terminated by addition of carbonyl cyanide p-chlorophenylhydrazone to a final concentration of 3 μm.
Single-molecule Studies
Assembly of n-FoF1 molecules on a microscope slide and the attachment of gold nanorods to the c-ring of Fo were carried out as described in Ref. 27. Rotation measurements of single molecules of n-FoF1 were made as described previously (27, 32).
MATLAB Algorithms for Transient Dwell Identification
Transient dwells during transition events were identified using custom programs developed in MATLAB R2013b. The program first ascertained the minimum and maximum intensities for each data set, and transitions were selected for further analysis that changed from within 5% of the minimum at the end of a catalytic dwell to 5% of the maximum values. Earlier studies showed that this rotation is CCW as a function of increasing intensity (32). The rotational position was then calculated from the observed intensity values using the arcsine of the light intensity scattered from a nanorod as determined previously (27, 31).
Transient dwells during a transition were identified when the net rotation was either CW or less than 1.5° for a period of more than 15 μs, equivalent to ≥3 consecutive data points at 200 kHz. These criteria limited the resolution of CW rotation to a minimum of >3°. To minimize the inclusion of pauses unrelated to transient dwells, transitions were then classified as containing transient dwells when they were observed to have two or more transient dwell events occur. The percentage of transitions that contain transient dwells was then calculated for each data set. To identify the extent of CW rotation during a transient dwell, the start and end points of CW rotation were determined by the identifying the rotational positions at which angular velocity approached zero.
RESULTS
Fo-dependent Rotation in the ATP Synthase (CW) Direction
We determined changes in the rotational position of individual 78 × 34 nm gold nanorods attached to the c subunit ring of single n-FoF1 molecules powered by ATP hydrolysis using dark field microscopy with a polarizing filter and a bandpass filter to eliminate all but the red light scattered from the nanorod (Fig. 1B). A rotating nanorod may appear to be stationary when its rotation is not eccentric. However, the scattered red light intensity will change in a sinusoidal manner (Fig. 2C) as the nanorod rotates relative to the direction of the polarizer (supplemental Movie 1). To measure rotational position as a function of time during individual 120° F1 ATPase power strokes, the polarizing filter was aligned with each nanorod to minimize the scattered light intensity during one of the three catalytic dwells. During the CCW rotation of the 120° F1 ATPase power stroke subsequent to that dwell, the nanorod will undergo a “transition” in which the light intensity passes from a minimum through a maximum when the nanorod has rotated 90° (27, 33). Transitions were collected from 5-s data sets from each molecule at data acquisition rates between 100–200 kHz in 10-kHz increments for a total collection time of 55 s using a single photon-counting avalanche photodiode. The rotational position as a function of time for each transition (Fig. 1D) was determined from the arcsine of the intensity as described under “Experimental Procedures.” Each data set yielded in excess of 300 transitions that were analyzed in this manner.
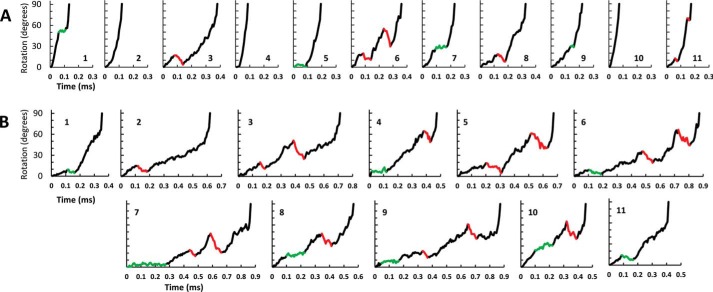
FIGURE 2.
A and B, examples of successive F1 ATPase transitions observed during single-molecule data acquisition data sets of n-FoF1 at a viscosity of (A) 0. 9 cP (aqueous buffer) or (B) 3.0 cP (25% PEG-400). Examples of transient dwells where rotation was halted or contained CW rotation are indicated in green and red, respectively. Transient dwells were not observed in transitions A2, A4, and A10.
Fig. 1D shows an example of a transition collected from a single n-FoF1 ATPase molecule during ATPase-driven rotation in the presence of 1 mm MgATP. Under these conditions, the logQ ≅ 5.6 on the basis of estimated MgADP and Pi concentrations of ∼50 μm each (34). The two transient dwells (red data), which are evidence of the leash, initially formed after the c-ring had rotated 18° and 55° from the end of the F1 ATPase catalytic dwell. Consequently, the net rotation between transient dwells was 37°, consistent with the CCW advancement of one c subunit in the c10 ring, as reported previously (27). During the transient dwells shown in Fig. 1D, Fo was able to rotate the c-ring CW by 10° and 25°, respectively, against the force of ATPase-driven rotation by F1.
Occurrence and Extent of Fo-dependent Rotation in the ATP Synthase Direction
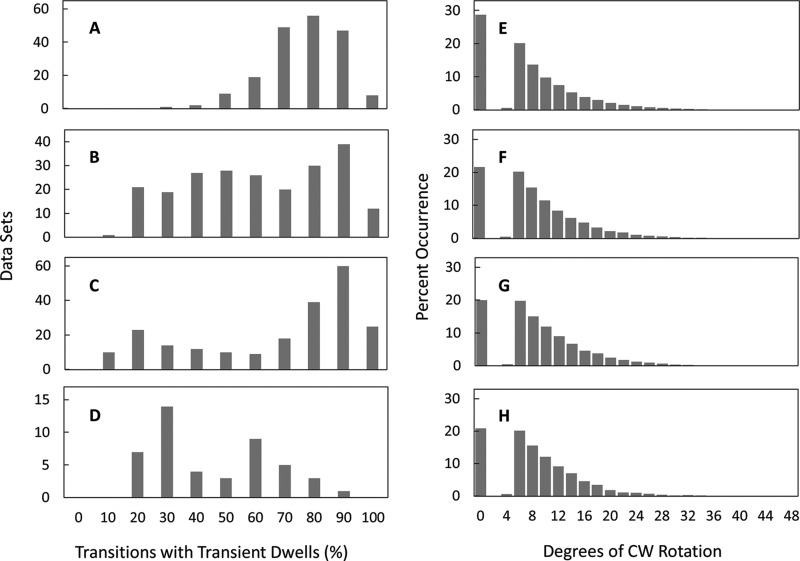
Examples of successive transitions from single molecules (Fig. 2) show transitions that lack transient dwells as well as those where rotation is halted or where CW rotation occurs. Custom software was written to identify the occurrence of transient dwells during 90° rotational transitions of ATPase-driven n-FoF1 power strokes and to determine the extent of CW rotation when it exceeded 3°. Rotation of the c-ring in the CW direction was observed during 72% of the 225,767 transient dwells examined from 207 molecules, whereas no net rotation was observed during the remaining 28% (Fig. 3, A–G). Of the former, almost all (99.88%) rotated CW ≤36° with an average CW rotation of ∼11°. These results indicate that when the leash between subunit a and the c-ring forms, Fo can force the c-ring to rotate in the ATP synthase direction by as much as one c subunit against the force of F1 ATPase-driven rotation.
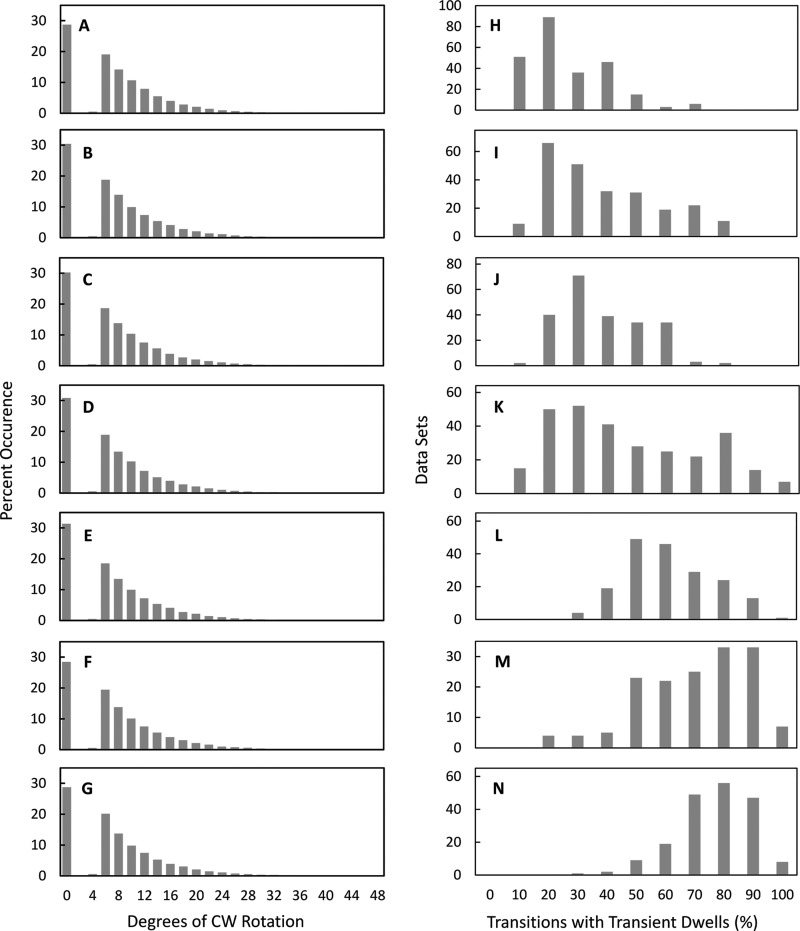
FIGURE 3.
A–G, the extent of Fo-dependent CW rotation against the force of F1 ATPase-driven rotation. Shown is the distribution of transient dwells as a function of the degrees of CW rotation observed against the force of ATPase-driven CCW rotation. H–N, distribution of single molecule n-FoF1 transition data sets as a function of the percentage of occurrence of transient dwells during ATPase-dependent power strokes. Viscosities of 0.9 cP (A and H), 1.2 cP (B and I), 1.5 cP (C and J), 1.8 cP (D and K), 2.3 cP (E and L), 3.0 cP (F and M), and 4.3 cP (G and N) were obtained by the presence of 0%, 5%, 10%, 15%, 20%, 25%, and 30% PEG-400 in the buffer, respectively.
Fig. 3, H—N, shows the distribution of 5-s data sets for single n-FoF1 molecules as a function of the percentage of transitions in each data set that contain transient dwells. The occurrence of transient dwells increased with the viscosity of the medium because of the time constant for leash formation relative to the velocity of F1 ATPase-driven CCW rotation reported previously (27). At viscosities of 3.0 and 4.2 centiPoise, transient dwells were present an average of 65 ± 1.5% (all reported errors are S.E.) and 72 ± 0.9%, respectively. The algorithm used here revealed that transient dwells were present an average of 21.7 ± 0.9% of the time in aqueous buffer (0.9 cP), indicating that leash formation is not limited to rotation at high viscosities. It is noteworthy that, although leash formation occurred more frequently at higher viscosities, the fraction of transient dwells that rotated CW and the average extent of CW rotation observed were independent of viscosity (Fig. 3, A–G).
Juxtaposition of aGlu-196 and cArg-50 in Fo
Subunit c residues cArg-50 and cAsp-61 and subunit a residues aGlu-196 and aArg-210 were selected as candidates that potentially contribute to the interaction responsible for the formation of the leash in Fo. Although a high-resolution structure of the E. coli c-ring has not been determined, the crystal structures from the c-ring of spinach (35) shows that cArg-50 faces the exterior of the c-ring at the cytoplasm-membrane interface (Fig. 4B), which also exists in the c-ring structures from spirulina (4) and pea chloroplasts (36). Crystallographic information is not currently available for subunit a. However, covalent modification studies with membrane-impermeant maleimides map aGlu-196 to the cytoplasmic end of TMH4, the same TMH that contains aArg-210 (15). The possible involvement of aGlu-196 in an electrostatic protein interaction at the membrane surface has been suggested by the chemical reactivity of aE196C to maleimides (11, 12, 37).
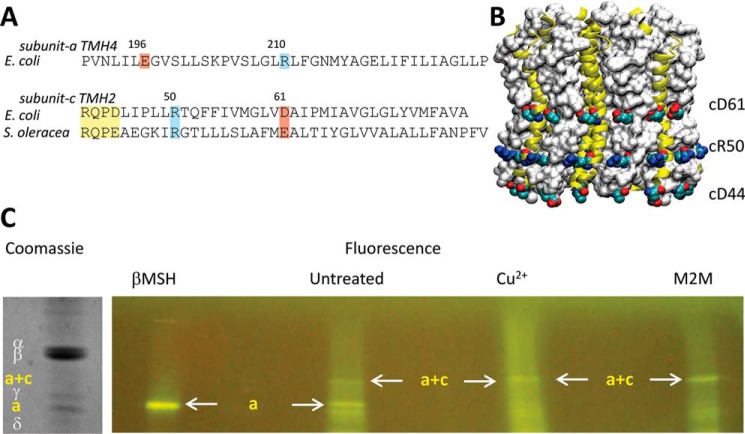
FIGURE 4.
A, sequence alignments of subunit a TMH4 with subunit c TMH2, where subunit c residues in the F1 docking loop, are shown in yellow. B, the c14-ring from Pisum sativum Fo (PDB code 3V3C). C, verification that residues aGlu-196 and cArg-50 are juxtaposed in Fo at a distance capable of forming an electrostatic interaction. Following addition of either βMSH, 1,2-ethanediyl bismethanethiosulfonate, or CuCl2, FoF1-aE196C/cR50C/c2▿C, available sulfhydryl groups were fluorescently labeled with fluorescein maleimide. The subunits were separated by SDS-PAGE and visualized by fluorescence and Coomassie stain.
To determine whether cArg-50 and aGlu-196 are in close enough proximity to form an electrostatic interaction that could be responsible for the leash, the extent of cross-linking of the E. coli Fo cR50C/aE196C double mutant was measured These cysteines were reduced either by addition of βMSH or were cross-linked via 1,2-ethanediyl bismethanethiosulfonate, which forms a bridge that is the same distance as a salt bridge between arginine and glutamic acid side chains, or by Cu2+-mediated disulfide formation. Before the FoF1 subunits were separated by SDS-PAGE, the treated samples were exposed to fluorescein maleimide to modify remaining free sulfhydryl groups. The only other cysteine present in FoF1 used for these experiments was the subunit c 2▿C insertion that placed a cysteine on the opposite side of the membrane from the Cys pair to be cross-linked. Consequently, the formation of a cR50C/aE196C disulfide will be evident as a loss of fluorescence from subunit a and because of fluorescein maleimide modification of the subunit c 2▿C insertion by the appearance of a fluorescent band corresponding to the combined molecular mass of subunits a and c.
In the 1,2-ethanediyl bismethanethiosulfonate-treated sample (Fig. 4C), the prominent fluorescent band observed had a molecular weight consistent with the cross-linked product of subunit a and subunit c. In contrast, fluorescence from subunit a was most intense in the βMSH-treated sample. The intensity of this subunit a band was inversely proportional to the fluorescence intensity of the subunit a/c-cross-linked product band, which was present even in the absence of an oxidizing or cross-linking agent. These data indicate that residues cArg-50 and aGlu-196 are juxtaposed at a distance capable of forming a salt bridge.
Electrostatic Interactions Promote Leash Formation but Not Fo-dependent CW Rotation
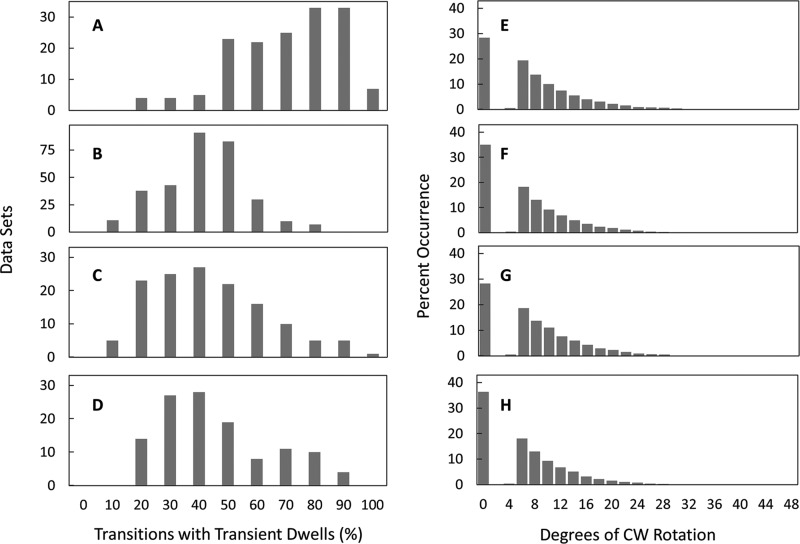
To assess the extent to which specific residues contribute to the ability of Fo to form the leash, mutation-dependent changes in the distribution of transient dwells per molecule were examined at viscosities of 3.0–4.25 cP, when they occur most frequently. In wild-type n-FoF1 at 4.25 cP, transient dwells formed an average of 72 ± 0.9% of the time (Fig. 5A). Under the same conditions, the ability of n-FoF1 aR210G as well as n-FoF1 cD61G to form transient dwells decreased <1.3-fold to an average of 56 ± 1.7% (Fig. 5B) and 62 ± 1.9% (Fig. 5C), respectively. In contrast, n-FoF1 aE196Q molecules decreased the occurrence of transient dwells ∼2-fold to an average of 40 ± 3.1% of the transitions (Fig. 5D). Although the aR210G, cD61G, and aE196Q mutations decreased the average occurrence at which transient dwells formed, the extent of the Fo-dependent CW rotation against the force of ATPase-driven rotation was essentially unchanged and averaged ∼11° (Fig. 5, E–H). In fact, the fraction of transient in which Fo pushed the c-ring in the CW direction increased to a total of about 80% of transient dwells observed.
FIGURE 5.
A–D, effects of site-directed mutations on the distribution of single-molecule n-FoF1 transition data sets as a function of the percentage of occurrence of transient dwells during F1 ATPase-dependent power strokes. E–H, the effects of mutations on the extent of Fo-dependent CW rotation. Shown is the distribution of transient dwells as a function of the degrees of CW rotation observed against the force of ATPase-driven CCW rotation. Data were collected at a viscosity of 4.3 cP for WT (A and E), aR210G (B and F), cD61G (C and G), and aE196Q (D and H) mutants of FoF1.
As shown in Fig. 6B, n-FoF1 cR50L and n-FoF1 aE196L molecules formed transient dwells at 3.0 cP almost 2-fold less often (37 ± 0.8% and 39 ± 1.7%, respectively) than observed with wild-type n-FoF1 (Fig. 6A, 65 ± 1.5%). This was about the same magnitude as that observed with n-FoF1 aE196Q, which is consistent with the evidence in Fig. 3C showing that aGlu-196 and cArg-50 can form a salt bridge. However, none of the single mutants examined completely eliminated the ability to form the leash. The cD44A/R50L/D61G triple mutant was examined to determine whether the effects of eliminating all of the charged residues exposed to the exterior of the c-ring had an additive effect on the ability to form transient dwells. The triple mutant decreased the average occurrence of transient dwells formed to 41 ± 1.7%, approximately a 2-fold decrease from the wild type (Fig. 6C), which was comparable with that of n-FoF1 cR50L. Consequently, the effects of the single-site mutations on the ability to form the leash were not additive but were dominated by the cArg-50 mutation.
FIGURE 6.
A–C, effects of site-directed mutations on the distribution of single-molecule n-FoF1 transition data sets as a function of the percentage of occurrence of transient dwells during F1 ATPase-dependent power strokes. D–F, the effects of mutations on the extent of Fo-dependent CW rotation. Shown is the distribution of transient dwells as a function of the degrees of CW rotation observed against the force of ATPase-driven CCW rotation. Data were collected at a viscosity of 3.0 cP for WT (A and D), cR50L (B and E), and cD44A/cR50L/cD61G (C and F) mutants of FoF1.
The extent of Fo-dependent CW rotation that occurred upon formation of the leash was not affected by either the cR50L mutation or the cD44A/R50L/D61G triple mutant (Fig. 6, D–F). The fact that removal of all charges on the outer surface of the c-ring did not eliminate the ability of Fo to push the c-ring CW against the force of the F1 ATPase indicates that this CW rotation results when the van der Waals surface of subunit a TMH4 pushes against the surface of each c subunit in the c-ring rather than from electrostatic interactions between these proteins.
Leash Formation Is Important to ATP Synthesis
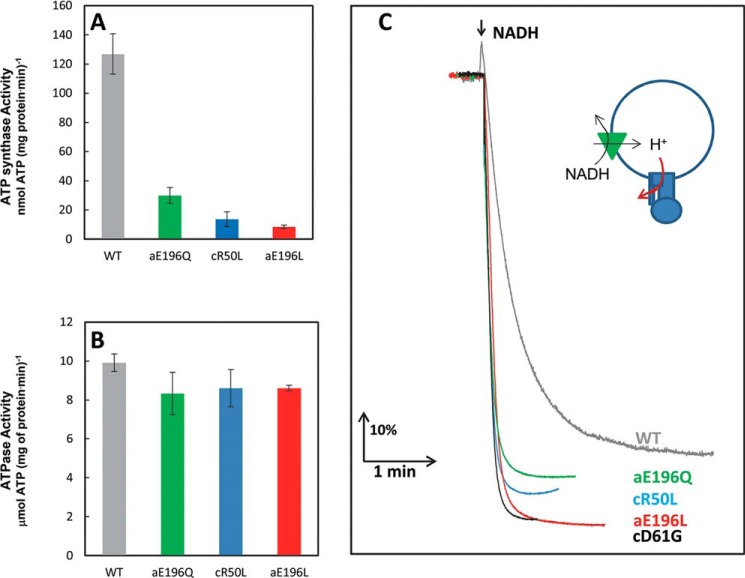
Mutations that decreased the ability to form the leash were found to decrease ATP synthase activity to a greater extent than ATPase activity of FoF1 in isolated inverted membrane vesicles. As shown in Fig. 7A, the aE196Q mutation, which could still form a hydrogen bond to cArg-50, decreased ATP synthase activity 4.7-fold. Elimination of the ability to form an electrostatic interaction between cArg-50 and aGlu-196 by mutations to a leucine decreased ATP synthase activity 11-fold and 15-fold, respectively. In contrast, membrane ATPase activity of the cR50L, aE196L, and aE196Q mutants decreased only by about 20% (Fig. 7B). Therefore, the ability to form an electrostatic interaction between cArg-50 and aGlu-196 is important to the mechanism of ATP synthesis.
FIGURE 7.
A–C, effects of the FoF1 mutations cR50L, aE196L, and aE196Q using inverted membrane vesicles on ATP synthesis (A) and ATP hydrolysis (B) activity measured by ensemble coupled enzyme assays and on NADH-driven ACMA quenching (C). The green triangle represents electron transfer complexes that pump protons in response to NADH, and the red arrow indicates the mutation-dependent restriction of Fo-dependent proton flow that results in an increase in ACMA quenching.
The cR50L, aE196L, and aE196Q mutations that decreased the occurrence of leash formation also significantly decreased the ability of protons to escape from inverted E. coli membrane vesicles, as measured by the extent of NADH-dependent proton pumping using isolated IMVs of E. coli (Fig. 7C). Because the F1 complex of the ATP synthase is on the outside of IMVs, the size of the proton gradient formed from NADH-dependent proton pumping can be measured by the extent of ACMA fluorescence quenching. The cD61G mutant, known to block proton translocation through Fo (6), and the ability of the uncoupler carbonyl cyanide p-chlorophenylhydrazone to collapse the gradient served as controls. The cR50L and aE196L mutants increased the extent of NADH-dependent ACMA quenching in a similar manner to that observed with the cD61G mutant (Fig. 7C). The extent of ACMA quenching observed with the aE196Q mutant was intermediate between that of the WT and the other mutants, consistent with ACMA quenching results reported previously for mutations of aGlu-196 by Vik et al. (38). The results shown in Fig. 7 are consistent with the periodic formation of an electrostatic interaction between cArg-50 and aGlu-196 that serves as a proton gate that is important for ATP synthesis but not ATP hydrolysis-dependent proton translocation.
DISCUSSION
The results presented here show that when the leash has formed, Fo rotates the c-ring in the ATP synthesis CW direction by as much as one c subunit against the CCW force of F1 ATPase-driven rotation >70% of the time. Although the occurrence of leash formation increases with the viscosity-dependent decrease in F1 ATPase-driven angular velocity, leash formation and the associated Fo-dependent CW rotation do occur at low viscosities of aqueous buffer. The data presented here show that leash formation is strongly facilitated by the formation of a salt bridge between aGlu-196 and cArg-50. This is supported by the cross-linking results shown in Fig. 4 and by the observation that the aE196L and cR50L mutations decreased the occurrence of leash formation by about the same extent (Figs. 5 and 6). The involvement of aGlu-196 in an electrostatic protein interaction is also supported by the observations that aE196C was the only cytoplasmic or periplasmic cysteine mutant whose reactivity with membrane-impermeant maleimides was dependent upon treatment with high salt (11, 12, 37). However, these experiments did not clearly demonstrate the participation of this residue in an electrostatic interaction because cysteine is not ionic.
It is clear from the data shown in Fig. 7 that residues aGlu-196 and cArg-50 not only facilitate leash formation but also have a significant role in a proton translocation gating mechanism related to ATP synthesis. One of the binding sites of the proton channel-blocking inhibitor oligomycin A, for which Fo is named, maps to a region of subunit a that includes aGlu-196 (39–41). This residue was also once proposed to contribute to the proton translocation pathway in early subunit a models that placed the residue in the hydrophobic portion of the membrane (42). On the basis of cross-linking and Ag+ sensitivity experiments involving Cys substitutions, the 194–199 region of subunit a was suggested to be somehow involved in a H+ translocation gating mechanism (43).
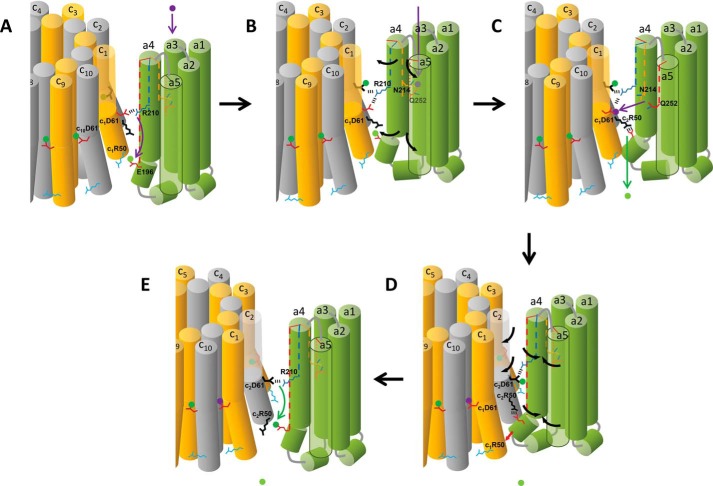
The ATP synthase mechanism shown in Fig. 8 incorporates the results presented here showing the importance of aGlu-196 and cArg-50 in ATP synthesis (Fig. 7A), proton gating (Fig. 7, C and D), and leash formation (Figs. 5 and 6) as well as the leash-dependent rotation in the ATP synthase (CW) direction against the force of F1 ATPase CCW rotation (Fig. 3). An animation of this mechanism is shown in supplemental Movie 2. Prior to forming the leash (Fig. 8A), aGlu-196 has become protonated by the proton displaced from c1Asp-61 by aArg-210.
FIGURE 8.
Model for the mechanism of Fo-dependent CW rotation in the ATP synthase direction. A, the deprotonated periplasmic channel conformation of subunit a, where the cytoplasmic end of aTMH4 is positioned between c-ring subunits c10 and c1. The position of aArg-210 on aTMH4 has displaced the proton (green dot) from c1Asp-61 to protonate aGlu-196. Vertical dotted lines in subunit a TMHs indicate the rotational position of residues. B, protonation of the periplasmic half-channel (purple dot) induces the subunit a conformational change where aTMH4 rotates relative to the c-ring. C, the PPC subunit a conformation. The cytoplasmic end of aTMH4 containing aGlu-196 is positioned between c-ring subunits c1 and c2 to form the leash. Leash formation is facilitated by a salt bridge between c2Arg-50 and aGlu-196, which moves the proton to the cytoplasm. The PPG conformation caused aR210 to be rotated from subunit c1 to c2 in the c-ring. This conformation also facilitates proton transfer from the periplasmic half channel to c1Asp-61. D, the reversion of subunit a from the PPG to the DPG conformation by van der Waals repulsion when the leash is engaged is responsible for CW c-ring rotation (E). Release of the leash occurs after CW rotation by 36°. As subunit a forms the DPG conformation, aArg-210 rotates into position to transfer the proton from c2Asp-61 to aGlu-196, which facilitates the release of the leash.
The interior of subunit a TMHs 2–5 has been shown to provide an aqueous accessible channel for ions to enter from the periplasm to protonate cAsp-61 during ATP synthesis (17). Cross-linking experiments (18, 44) indicate that protonation of this half-channel via aAsn-214 and aGln-252 promotes the rotation of TMHs 4 and 5 relative to other subunit a helices and also relative to the c-ring (Fig. 8B). We propose that this movement of aTMH4 from the deprotonated periplasmic channel conformation to the protonated periplasmic channel (PPC) conformation facilitates leash formation by allowing a juxtaposition of aGlu-196 with c2Arg-50 to form a salt bridge (Fig. 8C). Formation of the leash is represented by an interdigitation of the cytoplasmic end of aTMH4 containing aGlu-196 between subunits c1 and c2 of the c-ring. Formation of the electrostatic interaction with c2Arg-50 deprotonates aGlu-196 and moves the proton to the cytoplasm. It is noteworthy that the mutants examined here that removed charged residues at the interfacial surfaces of subunit a and subunit c decreased but did not eliminate leash formation altogether. Consequently, although the ionic interaction between aGlu-196 and cArg-50 significantly increases the ability of subunit a to interdigitate between two c subunits in the c-ring, it is not absolutely required for the leash to engage.
Formation of the PPC conformation as shown in Fig. 8C is believed to move aAsn-214 and aGln-252 into a position that enables the transfer of the proton to c1Asp-61 (17) so that c1Asp-61 reverts to the closed-locked position (19, 20). Because this half-channel also includes His-245, Glu-219, and perhaps other residues (1), there is probably a complex network for proton movement so that the proton transferred to c1Asp-61 is probably not the same one that entered the half-channel to induce formation of the PPC conformation.
We propose that the energy released as subunit a reverts from the PPC to the deprotonated periplasmic channel conformation is used to push the leash-engaged aTMH4 against subunit c1 to rotate the c-ring in the ATP synthase (CW) direction (Fig. 8D, red arrow). This push must result from steric interactions between subunit a and the c-ring rather than from specific electrostatic interactions because the mutants examined here, including the cD44A/cR50L/cD61G triple mutant that removed all of the charged residues on the outer surface of the c-ring, did not alter the occurrence of the Fo-dependent CW rotation when the leash had formed, nor did it decrease the extent of Fo-dependent CW rotation.
In Fig. 8E, the leash is disengaged from the c-ring when aGlu-196 becomes protonated as aArg-210 deprotonates c2Asp-61. The location of residues aGlu-196 and cArg-50 that facilitate leash formation at the membrane surface enables them to exist as independent species in a manner that disengages the leash. In comparison, molecular dynamics simulations predict that aArg-210 must maintain electrostatic interactions with one to two of the cAsp-61 side chains as the result of their positions in the hydrophobic core of the membrane (45).
The extent of the Fo-dependent CW rotation against the force of ATPase-driven CCW rotation was as much as 36°, which, for the c10-ring of E. coli, is equivalent to a movement to the adjacent c subunit, whereas the average was 11°. The CW rotation of 11° may then be the position at which the mechanical advantage of the leash has decreased to the point that it is, on average, overwhelmed by the F1 ATPase-driven torque. The CW rotation observed here may be evidence of the Fo power stroke because of the protonation-dependent conformational change of subunit a, shown in Fig. 8D, or elastic energy generated by F1ATPase CCW rotation when the leash is engaged. The Fo-dependent rotation in the ATP synthase (CW) direction occurred in the absence of the transmembrane proton gradient required for sustained CW rotation by the three to four successive c subunits in the c-ring necessary for net ATP synthesis. Therefore, during the CW rotation observed here, proton movement may progress through all of the steps shown in Fig. 8 but, in the case of the cD61G mutant, may only progress as far as shown in Fig. 7D before F1 ATPase CCW rotation reverses the steps shown in Fig. 8 to return the proton to the periplasmic side of Fo (Fig. 8A). Although we have been able to vary the occurrence of leash formation kinetically by varying the viscosity of the medium or by mutating aGlu-196 and cArg-50, we have not yet identified conditions that alter the extent of Fo-dependent CW rotation. Consequently, more work is required to distinguish whether the CW rotation results from a power stroke or elastic energy.
The experiments presented here show that the grab and push function of the leash in E. coli Fo is more effective when an electrostatic interaction can form between aGlu-196 and cArg-50. Sequence alignments of subunit a TMH4 and subunit c TMH2 from a variety of plant, animal, and bacterial organisms are shown in Fig. 9. In the subunit a sequences examined, aGlu-196 is either conserved or a carboxyl residue is present one helical turn away, where, in many cases, two adjacent carboxyl groups face the N-terminal end of subunit c TMH2.
FIGURE 9.
Sequence alignments of subunit a TMH4 and subunit c TMH2 aligned by organism. Sequence alignments of subunit c TMH2 are sorted by positively charged residues exposed to the outer surface of the c-ring near the cytoplasmic side of the membrane. Organisms for which a crystal structure of the c-ring is available are highlighted in yellow, and residues in which the c subunit side chains are not exposed to the outer surface are highlighted in gray.
All of the subunit c ring structures solved to date contain cArg-50 or a compensating positively charged residue that faces the N-terminal end of subunit a TMH4 (3, 46–51). Residues that are not exposed to the outer surface of the c-ring in available structures are highlighted in gray (Fig. 9). It is noteworthy that fewer residues are sequestered in the smaller c-rings (c8 and c10) than in the larger c14 and c15 rings from chloroplasts and cyanobacteria because of the curvature of the outer ring surface. Among the sequences examined, a positively charged residue is almost always found on the cytoplasmic side of the outer ring surface. In the bovine c8-ring, cLys-43 is the exposed positively charged residue that has a trimethylated ϵ-amino group, which suggests that the positive charge plays an important functional role (51). This residue is highly conserved in vertebrates and invertebrate phyla and has been found to be completely methylated in 29 metazoan species examined (52). When the lysine in this position is found in unicellular eukaryotes and prokaryotes, it is unmethylated. It is important to reiterate that the data presented here suggest that the grab and push mechanism results from an interaction of the van der Waals surfaces of subunit a and the c-ring that does not strictly depend on the presence of an electrostatic interaction.
Supplementary Material
This project was supported, in whole or in part, by National Institutes of Health Grant R01GM097510 (to W. D. F.).

This article contains supplemental Movies 1 and 2.
- CW
- clockwise
- CCW
- counterclockwise
- TMH
- transmembrane helix
- IMV
- inverted membrane vesicle
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- PPC
- protonated periplasmic channel
- ACMA
- 9-amino-6-chloro-2-methoxy acridine.
REFERENCES
- 1. Spetzler D., Ishmukhametov R., Hornung T., Martin J., York J., Jin-Day L., Frasch W. D. (eds) (2012) Photosynthesis, in Advances in Photosynthesis and Respiration (Eaton-Rye J., Tripathy B., Sharkey T., eds.), Vol. 34, pp. 561–590, Springer, Dordrecht, The Netherlands [Google Scholar]
- 2. Girvin M. E., Fillingame R. H. (1994) Hairpin folding of subunit c of F1Fo ATP synthase: 1H distance measurements to nitroxide-derivatized aspartyl-61. Biochemistry 33, 665–674 [DOI] [PubMed] [Google Scholar]
- 3. Watt I. N., Montgomery M. G., Runswick M. J., Leslie A. G., Walker J. E. (2010) Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 16823–16827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pogoryelov D., Yildiz O., Faraldo-Gómez J. D., Meier T. (2009) High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 16, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 5. Pogoryelov D., Krah A., Langer J. D., Yildiz Ö., Faraldo-Gómez J. D., Meier T. (2010) Microscopic rotary mechanism of ion translocation in the F-0 complex of ATP synthases. Nat. Chem. Biol. 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 6. Fillingame R. H., Peters L. K., White L. K., Mosher M. E., Paule C. R. (1984) Mutations altering aspartyl-61 of the ω subunit (uncE protein) of Escherichia coli H+-ATPase differ in effect on coupled ATP hydrolysis. J. Bacteriol. 158, 1078–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Angevine C. M., Fillingame R. H. (2003) Aqueous access channels in subunit a of rotary ATP synthase. J. Biol. Chem. 278, 6066–6074 [DOI] [PubMed] [Google Scholar]
- 8. Vik S. B., Antonio B. J. (1994) A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J. Biol. Chem. 269, 30364–30369 [PubMed] [Google Scholar]
- 9. Lightowlers R. N., Howitt S. M., Hatch L., Gibson F., Cox G. B. (1987) The proton pore in the Escherichia coli F0F1-ATPase: a requirement for arginine at position 210 of the a-subunit. Biochim. Biophys. Acta 894, 399–406 [DOI] [PubMed] [Google Scholar]
- 10. Ishmukhametov R. R., Pond J. B., Al-Huqail A., Galkin M. A., Vik S. B. (2008) ATP synthesis without R210 of subunit a in the Escherichia coli ATP synthase. Biochim. Biophys. Acta 1777, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long J. C., Wang S., Vik S. B. (1998) Membrane topology of subunit a of the F1F0 ATP synthase as determined by labeling of unique cysteine residues. J. Biol. Chem. 273, 16235–16240 [DOI] [PubMed] [Google Scholar]
- 12. Valiyaveetil F. I., Fillingame R. H. (1998) Transmembrane topography of subunit a in the Escherichia coli F1F0 ATP synthase. J. Biol. Chem. 273, 16241–16247 [DOI] [PubMed] [Google Scholar]
- 13. Wada T., Long J. C., Zhang D., Vik S. B. (1999) A novel labeling approach supports the five-transmembrane model of subunit a of the Escherichia coli ATP synthase. J. Biol. Chem. 274, 17353–17357 [DOI] [PubMed] [Google Scholar]
- 14. DeLeon-Rangel J., Ishmukhametov R. R., Jiang W., Fillingame R. H., Vik S. B. (2013) Interactions between subunits a and b in the rotary ATP synthase as determined by cross-linking. FEBS Lett. 587, 892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwem B. E., Fillingame R. H. (2006) Cross-linking between helices within subunit a of Escherichia coli ATP synthase defines the transmembrane packing of a four-helix bundle. J. Biol. Chem. 281, 37861–37867 [DOI] [PubMed] [Google Scholar]
- 16. Hakulinen J. K., Klyszejko A. L., Hoffmann J., Eckhardt-Strelau L., Brutschy B., Vonck J., Meier T. (2012) Structural study on the architecture of the bacterial ATP synthase Fo motor. Proc. Natl. Acad. Sci. U.S.A. 109, E2050–E2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fillingame R. H., Steed P. R. (2014) Half channels mediating H transport and the mechanism of gating in the F sector of Escherichia coli FF ATP synthase. Biochim. Biophys. Acta 1837, 1063–1068 [DOI] [PubMed] [Google Scholar]
- 18. Moore K. J., Fillingame R. H. (2008) Structural interactions between transmembrane helices 4 and 5 of subunit a and the subunit c-ring of Escherichia coli ATP synthase. J. Biol. Chem. 283, 31726–31735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meier T., Polzer P., Diederichs K., Welte W., Dimroth P. (2005) Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus. Science 308, 659–662 [DOI] [PubMed] [Google Scholar]
- 20. Symersky J., Pagadala V., Osowski D., Krah A., Meier T., Faraldo-Gómez J. D., Mueller D. M. (2012) Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 19, 485-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weber J., Senior A. E. (1997) Catalytic mechanism of F1-ATPase. Biochim. Biophys. Acta 1319, 19–58 [DOI] [PubMed] [Google Scholar]
- 22. Turina P., Samoray D., Gräber P. (2003) H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1-liposomes. EMBO J. 22, 418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kushmerick M. J., Moerland T. S., Wiseman R. W. (1992) Mammalian skeletal muscle fibers distinguished by contents of phosphocreatine, ATP, and Pi. Proc. Natl. Acad. Sci. U.S.A. 89, 7521–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kushmerick M. J., Meyer R. A., Brown T. R. (1992) Regulation of oxygen consumption in fast- and slow-twitch muscle. Am. J. Physiol. 263, C598–C606 [DOI] [PubMed] [Google Scholar]
- 25. Junge W., Lill H., Engelbrecht S. (1997) ATP synthase: an electrochemical transducer with rotatory mechanics. Trends Biochem. Sci. 22, 420–423 [DOI] [PubMed] [Google Scholar]
- 26. Oster G., Wang H., Grabe M. (2000) How Fo-ATPase generates rotary torque. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishmukhametov R., Hornung T., Spetzler D., Frasch W. D. (2010) Direct observation of stepped proteolipid ring rotation in E. coli FoF1-ATP synthase. EMBO J. 29, 3911–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Diez M., Zimmermann B., Börsch M., König M., Schweinberger E., Steigmiller S., Reuter R., Felekyan S., Kudryavtsev V., Seidel C. A., Gräber P. (2004) Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat. Struct. Mol. Biol. 11, 135–141 [DOI] [PubMed] [Google Scholar]
- 29. Watanabe R., Tabata K. V., Iino R., Ueno H., Iwamoto M., Oiki S., Noji H. (2013) Biased Brownian stepping rotation of FoF1-ATP synthase driven by proton motive force. Nat. Commun. 4, 1631. [DOI] [PubMed] [Google Scholar]
- 30. Lowry D. S., Frasch W. D. (2005) Interactions between β D372 and γ subunit N-terminus residues γ K9 and γ S12 are important to catalytic activity catalyzed by Escherichia coli F1F0-ATP synthase. Biochemistry 44, 7275–7281 [DOI] [PubMed] [Google Scholar]
- 31. Martin J. L., Ishmukhametov R., Hornung T., Ahmad Z., Frasch W. D. (2014) Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. U.S.A. 111, 3715–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hornung T., Martin J., Spetzler D., Ishmukhametov R., Frasch W. D. (2011) Microsecond resolution of single-molecule rotation catalyzed by molecular motors. Methods Mol. Biol. 778, 273–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spetzler D., York J., Daniel D., Fromme R., Lowry D., Frasch W. (2006) Microsecond time scale rotation measurements of single F(1)-ATPase Molecules. Biochemistry 45, 3117–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noji H., Bald D., Yasuda R., Itoh H., Yoshida M., Kinosita K., Jr. (2001) Purine but not pyrimidine nucleotides support rotation of F(1)-ATPase. J. Biol. Chem. 276, 25480–25486 [DOI] [PubMed] [Google Scholar]
- 35. Vollmar M., Schlieper D., Winn M., Büchner C., Groth G. (2009) Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase. J. Biol. Chem. 284, 18228–18235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saroussi S., Schushan M., Ben-Tal N., Junge W., Nelson N. (2012) Structure and flexibility of the C-ring in the electromotor of rotary F(0)F(1)-ATPase of pea chloroplasts. Plos ONE 7, e43045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang D., Vik S. B. (2003) Close proximity of a cytoplasmic loop of subunit a with c subunits of the ATP synthase from Escherichia coli. J. Biol. Chem. 278, 12319–12324 [DOI] [PubMed] [Google Scholar]
- 38. Vik S. B., Cain B. D., Chun K. T., Simoni R. D. (1988) Mutagenesis of the α subunit of the F1Fo-ATPase from Escherichia coli: mutations at Glu-196, Pro-190, and Ser-199. J. Biol. Chem. 263, 6599–6605 [PubMed] [Google Scholar]
- 39. John U. P., Nagley P. (1986) Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 207, 79–83 [DOI] [PubMed] [Google Scholar]
- 40. Breen G. A., Miller D. L., Holmans P. L., Welch G. (1986) Mitochondrial DNA of two independent oligomycin-resistant Chinese hamster ovary cell lines contains a single nucleotide change in the ATPase 6 gene. J. Biol. Chem. 261, 11680–11685 [PubMed] [Google Scholar]
- 41. Ray M. K., Connerton I. F., Griffiths D. E. (1988) DNA sequence analysis of the Olir2–76 and Ossr1–92 alleles of the Oli-2 region of the yeast Saccharomyces cerevisiae: analysis of related amino-acid substitutions and protein-antibiotic interaction. Biochim. Biophys. Acta 951, 213–219 [DOI] [PubMed] [Google Scholar]
- 42. Cox G. B., Fimmel A. L., Gibson F., Hatch L. (1986) The mechanism of ATP synthase: a reassessment of the functions of the b and a subunits. Biochim. Biophys. Acta 849, 62–69 [DOI] [PubMed] [Google Scholar]
- 43. Moore K. J., Angevine C. M., Vincent O. D., Schwem B. E., Fillingame R. H. (2008) The cytoplasmic loops of subunit a of Escherichia coli ATP synthase may participate in the proton translocating mechanism. J. Biol. Chem. 283, 13044–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moore K. J., Fillingame R. H. (2013) Obstruction of transmembrane helical movements in subunit a blocks proton pumping by F1Fo ATP synthase. J. Biol. Chem. 288, 25535–25541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aksimentiev A., Balabin I. A., Fillingame R. H., Schulten K. (2004) Insights into the molecular mechanism of rotation in the Fo sector of ATPs [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stock D., Leslie A. G., Walker J. E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 47. Dautant A., Velours J., Giraud M. F. (2010) Crystal structure of the Mg·ADP-inhibited state of the yeast F1c10-ATP synthase. J. Biol. Chem. 285, 29502–29510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meier T., Krah A., Bond P. J., Pogoryelov D., Diederichs K., Faraldo-Gómez J. D. (2009) Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 391, 498–507 [DOI] [PubMed] [Google Scholar]
- 49. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) A new type of proton coordination in an F(1)F(o)-ATP synthase rotor ring. PLoS Biol. 8, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schulz S., Iglesias-Cans M., Krah A., Yildiz O., Leone V., Matthies D., Cook G. M., Faraldo-Gómez J. D., Meier T. (2013) A new type of Na+-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biol. 11, e1001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen R., Fearnley I. M., Palmer D. N., Walker J. E. (2004) Lysine 43 is trimethylated in subunit C from bovine mitochondrial ATP synthase and in storage bodies associated with batten disease. J. Biol. Chem. 279, 21883–21887 [DOI] [PubMed] [Google Scholar]
- 52. Walpole T. B., Palmer D. N., Jiang H., Ding S., Fearnley I. M., Walker J. E. (2015) Conservation of complete trimethylation of lysine 43 in the rotor ring of c-subunits of metazoan ATP synthase. Mol. Cell. Proteomics, 10.1074/mcp.M114.047456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.