Background: The architecture of spines in pyramidal neurons is critical for function.
Results: We identified δ-catenin as a critical regulator of spine architecture in hippocampal neurons.
Conclusion: δ-Catenin regulates spine architecture via its ability to interact with cadherin and PDZ domain-containing proteins.
Significance: The identification of molecular mechanisms underlying spine architecture is crucial for understanding the molecular and cellular basis of higher order brain functions.
Keywords: Cadherin, Catenin, Dendritic Spine, Hippocampus, Neuron, PDZ, Spine Architecture, Spine Dynamic
Abstract
The ability of neurons to maintain spine architecture and modulate it in response to synaptic activity is a crucial component of the cellular machinery that underlies information storage in pyramidal neurons of the hippocampus. Here we show a critical role for δ-catenin, a component of the cadherin-catenin cell adhesion complex, in regulating spine head width and length in pyramidal neurons of the hippocampus. The loss of Ctnnd2, the gene encoding δ-catenin, has been associated with the intellectual disability observed in the cri du chat syndrome, suggesting that the functional roles of δ-catenin are vital for neuronal integrity and higher order functions. We demonstrate that loss of δ-catenin in a mouse model or knockdown of δ-catenin in pyramidal neurons compromises spine head width and length, without altering spine dynamics. This is accompanied by a reduction in the levels of synaptic N-cadherin. The ability of δ-catenin to modulate spine architecture is critically dependent on its ability to interact with cadherin and PDZ domain-containing proteins. We propose that loss of δ-catenin during development perturbs synaptic architecture leading to developmental aberrations in neural circuit formation that contribute to the learning disabilities in a mouse model and humans with cri du chat syndrome.
Introduction
In hippocampal pyramidal neurons, the majority of excitatory synapses are localized on the heads of spines. There is a strong correlation between the size of the spine and synaptic strength (1, 2). The neck of spines allows biochemical compartmentalization and the architecture of the spine neck influences signaling in response to synaptic activity (3, 4). Spines are not static structures, but exhibit morphological plasticity in response to synaptic activity (5). Long-term potentiation inducing stimuli promote enlargement of spine heads, whereas stimuli that correlate with long-term depression induce spine shrinkage (6, 7). The ability of pyramidal neurons to generate appropriate spine and synaptic architecture and modulate it in response to synaptic activity is critical for the integrity of the cellular machinery that underlies information storage (8–10). Consistently, aberrations in spine architecture are associated with a variety of neurodevelopmental disorders (11, 12), including those associated with intellectual disability (13, 14). Our understanding of the molecular machinery that regulates synaptic architecture remains incomplete.
δ-Catenin (15) is a component of the cadherin-catenin cell adhesion complex (16). The cri du chat syndrome is associated with deletions of chromosome 5p. Loss of Ctnnd2, encoding δ-catenin, correlates with intellectual disability (17–19) in the cri du chat syndrome. Interestingly, a partial duplication of Ctnnd2 in an individual with the cri du chat syndrome with deletion of Ctnnd2 in the chromosomal deletion associated with the syndrome leads to a milder cognitive phenotype, further supporting a key role for δ-catenin in the intellectual disability associated with this syndrome (20). In addition, copy number variations of Ctnnd2 have been implicated in schizophrenia (21) and autism (22, 23). Furthermore, a mouse model of δ-catenin has deficits in spatial learning, Pavlovian fear conditioning, and hippocampal synaptic plasticity (24), suggesting that the functional roles of δ-catenin are vital in maintaining neuronal integrity necessary for higher order brain functions.
We have previously identified a critical role for δ-catenin in regulating spine density and architecture in developing hippocampal pyramidal neurons. Here, we demonstrate that the functional role of δ-catenin in regulating spine architecture extends to more mature synapses. Furthermore, we identified a molecular pathway that allows δ-catenin to mediate this functional role. Our data demonstrate that loss of δ-catenin in mature neurons of a mouse model leads to a significant reduction in spine head width and length. By taking advantage of shRNA-mediated knockdown in rat hippocampal neurons, we demonstrate that knockdown of δ-catenin during development leads to a similar phenotype. This functional role of δ-catenin is critically dependent on its ability to interact with cadherin and PDZ domain-containing proteins. Our results provide evidence for a key signaling pathway in regulating spine architecture during development in hippocampal pyramidal neurons. The compromise in synaptic architecture with loss of δ-catenin may underlie the behavioral phenotypes observed in the δ-catenin mouse model and individuals with cri du chat syndrome.
EXPERIMENTAL PROCEDURES
Animals
All experiments were approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee.
Synaptosome Preparation
Cortical/hippocampal tissue from P21/22 male mice was homogenized in 10 volumes of sucrose buffer containing 0.32 m sucrose, 5 mm HEPES, 0.1 mm EDTA solution and protease inhibitor mixture (Pierce, EDTA-free). The homogenate was centrifuged for 10 min at 300 × g. The resultant supernatant was centrifuged for 20 min at 12,000 × g to obtain synaptosome-enriched pellet. The pellet was resuspended in the same sucrose buffer and layered onto a tube with 0.6/0.8/1.2 m sucrose layers. After centrifugation for 90 min at 75,600 × g, the synaptosome-enriched interface between 0.8 and 1.2 m was collected and diluted in the sucrose buffer. The sample was centrifuged again for 60 min at 200,000× g to pellet the synaptosomes. The isolated synaptosomes were resuspended in the sucrose buffer and frozen immediately and Western blotted as described previously.
δ-Catenin N-term Mice
δ-Catenin N-term mice (also referred to as δ-catenin null mice) have been previously described (24).
Primary Neuron Culture
Primary rat neuronal cultures were generated from E18–19 rat hippocampi or P0 mice as previously described (25, 26). Briefly, dissociated neurons were plated at a density of 75,000 (150,000 for high density) on poly-l-lysine-coated 18-mm glass coverslips. Cultures are maintained at 37 ºC with 5% CO2. Ara-C (cytosine β-d-arabinofuranoside, Sigma) was added to 5 μm on DIV 2 and all the media were changed on DIV 4. Half of the media were changed twice every week. Lipofectamine 2000 (Life Technologies) was used for transfection as recommended by the manufacturer. Half of the medium was changed 1 day after transfection. Neurons were plated at 75,000 neurons per coverslip (12 well) for data shown in all figures except Fig. 4. For experiments in Fig. 4, neurons were plated at a higher density of 150,000 per coverslip (12 well).
FIGURE 4.
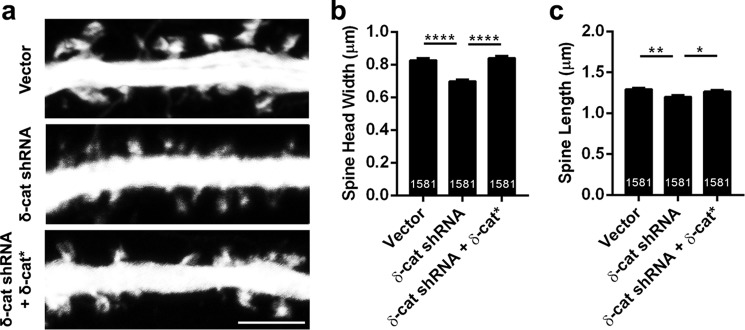
δ-Catenin knockdown effects on spine architecture persist with alterations in network activity. a, representative images of dendrites expressing vector, shRNA, or shRNA + δ-catenin*, transfected at DIV 11 and fixed at DIV 17 in neurons plated at high density (density, 150,000 per coverslip). b, spine head width and c, spine length in neurons expressing vector, shRNA, or shRNA + δ-catenin* (p values: *, <0.05; **, <0.005; ***, <0.0005, one-way ANOVA analysis and Dunnett's multiple comparison test with single pooled variance; scale, 5 μm).
cDNA Constructs
The shRNA for knockdown of δ-catenin has been previously described and validated. All constructs of δ-catenin have been previously described (26). The EGFP3-δ-catenin and K581M* constructs of δ-catenin were a kind gift from Dr. Shernaz Bamji (University of British Columbia) (27). EGFP-δ-catenin represents the XM_006520032.1. We note that there was a change in amino acid 942 (Ala to Val) relative to the GenBankTM sequence.
Antibodies
The mouse monoclonal anti-δ-catenin antibody was purchased from BD Transduction Labs (catalog number 611537, 1:800) and PROGEN Biotechnik GmbH (J19, catalog number 651152, 1:500). The N-cadherin antibody was purchased from BD Transduction Laboratories (catalog number 610920, 1:5000 for Western blot and 1:150 for immunocytochemistry). Antibody to PSD-95 was from EMD Millipore (catalog number MAB1598, 1:1000). Secondary antibodies were peroxidase anti-mouse IgG (Jackson ImmunoResearch, catalog number 715-035-151 1:8000) for Western blot, Alexa Fluor 555 (Life Technologies, catalog number A-21424, 1:200).
Confocal Microscopy
Images were taken on an inverted Zeiss Pascal LSM (26) or LSM 700 confocal microscope using a ×40 objective and a digital zoom of ×4 and Z-stack projection for measuring spine head width and length.
Time-lapse Analysis
Rat primary neurons were plated at a density of 50,000 on poly-l-lysine-coated glass bottom dish (14-mm microwell, MatTek catalog number P35G-0-14-C); Ara-C was added and the culture was maintained as previously described. Neurons were transfected with psuperEGFP vector and δ-catenin shRNA on DIV 11 and imaged on DIV 16–18. Dishes are maintained at air plus 5% CO2, in a 37 °C environment (Okolab control unit) in an imaging chamber (Warner Instruments) during imaging. Each sample was imaged for 5–8 h and 10–14 images of primary dendrite within 100 mm from the cell body of different individual neurons has been acquired. Time-lapse images were acquired using the following parameters: ×40, zoom 4, 512 × 512 frame size; 12 bit depth; Z-stack of 7 slices with 0.43-mm interval; 5 images were taken with a 5-min internal; definite focus was used during imaging. Images were analyzed blindly; ImageJ was used to measure the spine head width and length of dendritic protrusions within 100 mm from the cell body. Spines that could not be unambiguously identified at all time points were excluded from analysis. 510–660 dendritic protrusions were analyzed from each sample. 5 separate experiments were analyzed. Numbers of spines are: T1 to T3, n = 512 for spines ≤ 2 μm and n = 147 for spines >2 μm in the vector group; n = 505 for spines ≤2 μm and n = 143 for spines >2 μm in the δ-catenin shRNA group. For T4, n = 476 for spines ≤2 μm and n = 143 for spines >2 μm in the vector group; n = 458 for spines ≤2 μm and n = 137 for spines >2 μm in the δ-catenin shRNA group. For T5, n = 399 for spines ≤2 μm and n = 117 for spines >2 μm in the vector group; n = 391 for spines ≤2 μm and n = 125 for spines >2 μm in the δ-catenin shRNA group. Data were analyzed using Student's t test.
Spine Architecture Quantitation
Image analysis was done using NIH ImageJ. Images were minimally processed using Adobe Photoshop CS6 if required. Only contrast and brightness of overall image were adjusted if necessary. Spine head widths and lengths were assessed using ImageJ. These measurements were obtained from spines that were on dendrites within 80 μm from the cell body. For each set of experiments data were obtained from 3–4 independent experiments. The number of spines for each condition is indicated on the bar graph. For the mouse data, these numbers were obtained from secondary dendrites. Neurons from 4 to 5 mice for each genotype were analyzed.
Statistical Analysis
All data are shown as mean ± S.E. Data were analyzed in Prism 6 (GraphPad Software) by one-way ANOVA and Dunnett's multiple comparison test with single pooled variance. For data in Figs. 2 and 5, data were analyzed using two-tailed Student's unpaired t test assuming unequal variances. p < 0.05 was considered significant.
FIGURE 2.
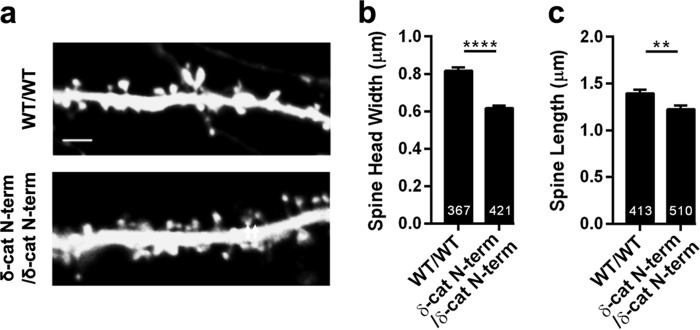
Spine architecture is perturbed in mature neurons from the δ-catenin N-term mice. a, representative images of dendrites expressing GFP from wild type and δ-catenin N-term mice (DIV 21). b, spine head width, and c, spine length in neurons from wild type and δ-catenin N-term mice expressing GFP.
FIGURE 5.
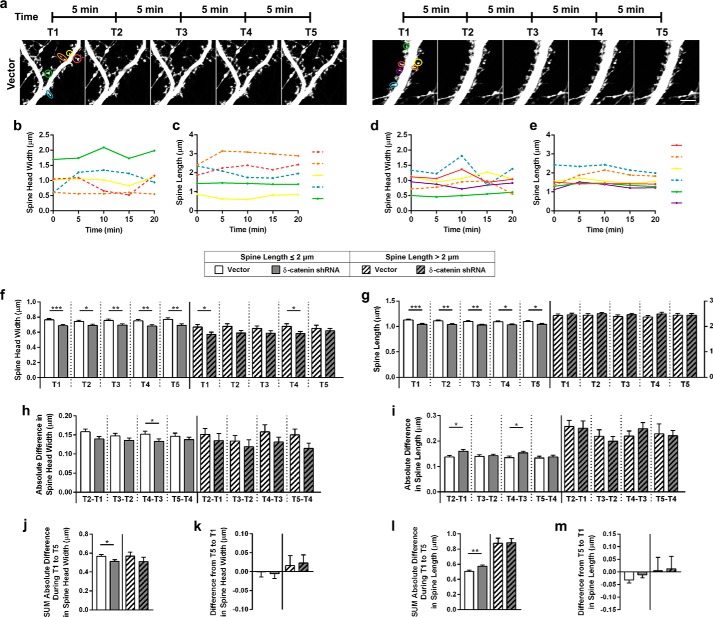
Knockdown of δ-catenin alters spine dynamics. a, representative images of dendritic segments from neurons expressing vector and shRNA at indicated time points. b, spine head widths from spines, expressing vector; c, spine lengths from spines, expressing vector; d, spine head widths from spines, expressing shRNA; e, spine lengths from spines, expressing shRNA, at time points indicated for color coded spines as indicated in first panel of a. Solid lines, spine lengths of spines less than or equal to 2 μm in length and dashed lines for spines greater than 2 μm in length. f, spine head widths of spines less than or equal to 2 μm in length and greater than 2 μm in length at the time points indicated. g, spine lengths of spines less than or equal to 2 μm in length and greater than 2 μm in length at the time points indicated. The scale on the right is for the group with lengths greater than 2 μm. h, absolute differences in spine head width for the two groups at the time points indicated. i, absolute differences in spine length for the two groups at the time points indicated. j, sum of absolute differences in spine head width from T1 (0 min) to T5 (20 min) in spines from the two groups. k, difference in spine head width from T5 (20 min) to T1 (0 min) in spines from the two groups. l, sum of absolute differences in spine head length from T1 (0 min) to T5 (20 min) in spines from the two groups. m, difference in spine length from T5 (20 min) to T1 (0 min) in spines from the two groups (p values: *, <0.05; **, <0.005; ***, <0.0005, Student's t test; scale, 5 μm).
RESULTS
δ-Catenin Is Enriched in Synaptosomes
δ-Catenin is a component of the cadherin-catenin cell adhesion complex and we have previously demonstrated a role for δ-catenin in regulating spine and synapse density and dendrite morphogenesis (26, 28).
Hippocampal development in the mouse proceeds through distinct stages in which differentiation and synapse formation occurs between E19 and P12 and synapse formation and maturation between P10 and P30 (29). δ-Catenin is well expressed at all stages of development, however, its cellular distribution is altered during development. We examined the distribution of δ-catenin in cortical synaptosomes from P21/22 mouse brains. Mouse cortical tissue was homogenized and processed as described under “Experimental Procedures” to obtain synaptosomes. Total homogenate, low speed pellet, high speed supernatant, and synaptosome fractions were immunoblotted with two separate commercial antibodies to δ-catenin. Enrichment of the synaptosome fraction was confirmed by immunoblotting with an antibody to PSD-95, an excitatory postsynaptic marker. Consistent with a functional role at synapses, δ-catenin was enriched in synaptosomes, similar to the enrichment observed for PSD-95 (Fig. 1). These results confirm that δ-catenin is enriched in synaptosomes during development and is consistent with a synaptic functional role for δ-catenin at more advanced stages of hippocampal development.
FIGURE 1.
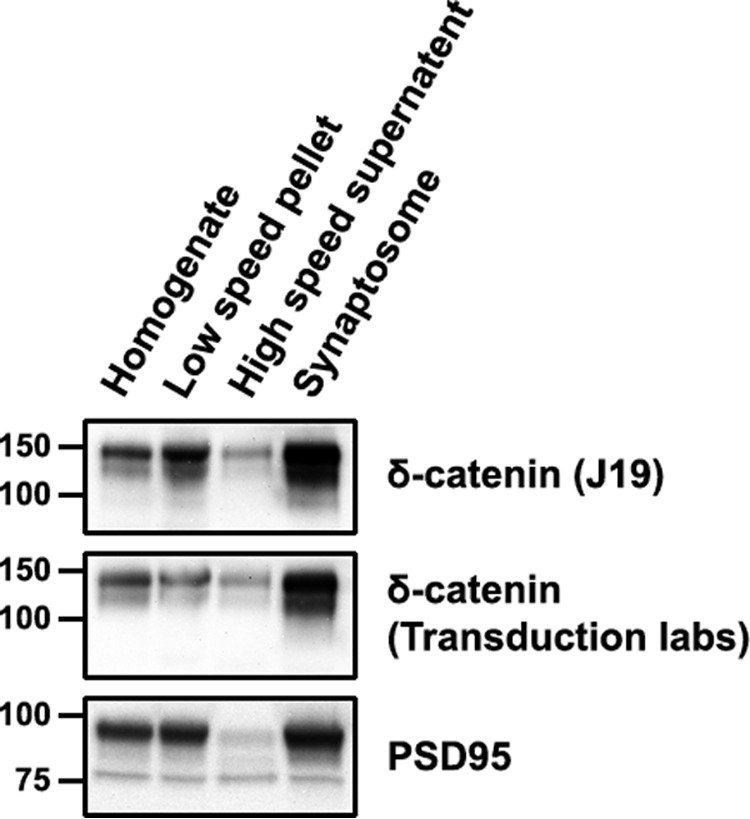
δ-Catenin is enriched in developing synaptosomes. Western blots of the indicated fractions from synaptosome preparations from P21/P22 mouse cortical tissue probed with two separate antibodies to δ-catenin and PSD-95.
Loss of δ-Catenin in Mature Neurons in a Mouse Model Perturbs Synaptic Architecture
We have previously demonstrated that loss of δ-catenin in a mouse model results in a reduction in spine head width and length during development (26). It is known that some proteins perturb synaptic structure and function during development, but not in the adult. For example, loss of SynGAP in a mouse model perturbs spine densities when eliminated before development, but not after development (30). To determine the effects of loss of δ-catenin in the mouse model, we examined the spine head width and length in primary cultured neurons from control and δ-catenin N-term mice (Fig. 2) at DIV 21. Loss of δ-catenin in this mouse model leads to a reduction in the head width and length of spines. Thus δ-catenin is a critical regulator of spine architecture in both developing and mature neurons. However, these effects may also be a result of developmental alterations that occur in the absence of δ-catenin.
Knockdown of δ-Catenin Perturbs Synaptic Architecture in Mature Neurons
To examine if acute loss of δ-catenin in mature neurons has similar effects, we took advantage of shRNA-mediated knockdown of δ-catenin in cultured rat neurons (75,000 neurons per coverslip). We have previously validated this shRNA (26, 28). Rat primary neurons in culture were transfected with constructs expressing vector, shRNA, or shRNA, and a full-length cDNA of δ-catenin encoding an shRNA-resistant protein (δ-catenin*) at DIV 14. The plasmids for these also express EGFP. Neurons were fixed at DIV 21 and imaged by confocal microscopy, and spine head width and lengths were examined (Fig. 3a). Knockdown of δ-catenin resulted in a significant decrease in the spine length and spine head width (Fig. 3b), which was rescued by the expression of the shRNA-resistant form of δ-catenin. Taken together with our previous studies, these results indicate that (a) δ-catenin is a critical regulator of spine architecture in pyramidal neurons, (b) this functional role extends to different stages during development, and (c) this functional role of δ-catenin is cell autonomous.
FIGURE 3.
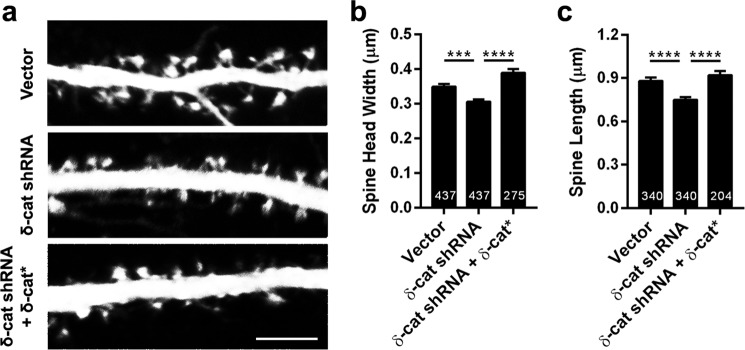
Knockdown of δ-catenin perturbs spine architecture in mature neurons. a, representative images of dendrites expressing vector, shRNA, or shRNA + δ-catenin*, transfected at DIV 14 and fixed at DIV 21 (density, 75,000 neurons per coverslip). b, spine head width and c, spine length in neurons expressing vector, shRNA, or shRNA + δ-catenin* (p values: *, <0.05; **, <0.005; ***, <0.0005, one-way ANOVA analysis and Dunnett's multiple comparison test with single pooled variance; scale, 5 μm).
It is well established that plating primary neurons at different cell densities affects network activity and alters dendrites and spine significantly (31–33). We examined if the δ-catenin loss-induced perturbation of spine architecture would be altered by changes in network activity. To this end, we plated neurons at a higher density (150,000 neurons per coverslip) and examined if knockdown of δ-catenin perturbs spine architecture (Fig. 4, a and b). High-density cultured neurons expressing shRNA to δ-catenin had a significant reduction in the head width and length that could be rescued by co-expression of the shRNA-resistant form of δ-catenin. These results suggest that loss of δ-catenin leads to an inherent compromise of spine architecture and this is independent of neuronal network activity.
Alterations in Spine Dynamics in the Absence of δ-Catenin
Because the architecture of spines was compromised in the absence of δ-catenin, we chose to examine if the dynamics of spine head width and length were altered in the absence of δ-catenin. To this end, neurons in culture were transfected with vector or shRNA at DIV 11 and examined by time-lapse microscopy on DIV 16–18 over a total of 20 min with images obtained every 5 min (Fig. 5a). Spine head widths and lengths were obtained at each time point. For the purpose of analysis, we split the spines into two groups, those with spine lengths less than or equal to 2 μm and those with spine lengths greater than 2 μm. Fig. 5, b–e, demonstrate head widths and lengths at different time points for a few spines as indicated in Fig. 5a. We examined spine head width and length parameters from similar experiments for larger groups of neurons to define average behavior. In the less than or equal to 2-μm head width group, the spine head widths in the shRNA group were smaller at all time points examined, whereas in the greater than 2-μm group, a significant difference in head width was only observed at some time points (Fig. 5f). We also performed a similar analysis for spine lengths. In the group of spines with lengths less than or equal to 2 μm, the lengths of the spines in the shRNA group were significantly smaller at each time point. However, in the greater than 2-μm group, the spine lengths between the vector and shRNA group showed no significant differences (Fig. 5g). In the less than or equal to 2-μm group, the absolute differences in spine head widths were not significantly different between the vector and the shRNA except at one time point. The greater than 2-μm group showed a trend toward alterations in the absolute difference in spine head width between the vector and shRNA, however, these differences were not significant (Fig. 5h). Similar results were obtained for the spine length (Fig. 5i). The sum of the absolute differences from T1 to T5 were significantly reduced in the less than or equal to 2-μm shRNA group, but not the greater than 2-μm group, in comparison to the vector group (Fig. 5j). Similar results were observed for spine length (Fig. 5k). The difference in spine head widths and spine lengths from T5 to T1 were not significantly different for any of the groups (Fig. 5, l and m). Taken together, these results suggest that δ-catenin may regulate the spine head width and length of the less than or equal to 2 μm and greater than 2 μm head width/length differentially. The effects of loss of δ-catenin on the smaller head width and length population are more prominent. In this group, the loss of δ-catenin leads to an inherent sustained decrease in spine head width and length. Interestingly, in this group of spines, the head width is less dynamic, whereas the spine length is more dynamic in the shRNA group in comparison to the vector control group.
Taken together, these results suggest that loss of δ-catenin has a more prominent effect on spines with smaller head width and length. In this population of spines, loss of δ-catenin leads to a net decrease in spine dynamics associated with head width and an increase in spine dynamics associated with spine length.
Role of ARM Domains of δ-Catenin in Regulation of Spine Architecture
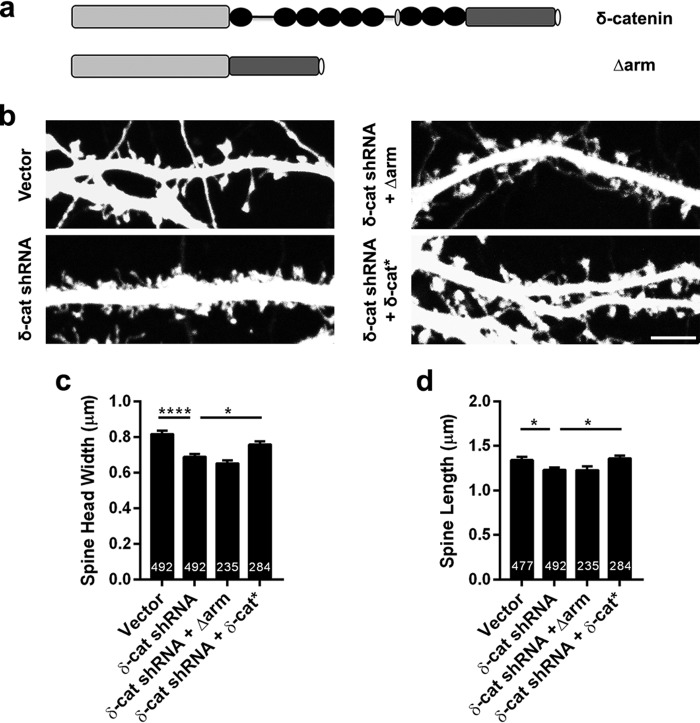
We sought to examine the mechanism by which δ-catenin regulates synaptic architecture. The structure of δ-catenin includes a series of armadillo (ARM) repeats within the central region of the protein (34). Because ARM domains have been implicated in protein-protein interactions and the ARM domains of δ-catenin have been demonstrated to mediate interaction with presenilin (35) and N-cadherin (36), we chose to examine if the ARM repeats are critical for the ability of δ-catenin to regulate spine architecture. To this end, cultured rat primary neurons were transfected at DIV 11 with vector, shRNA to δ-catenin, shRNA to δ-catenin plus a construct of δ-catenin lacking the ARM domains and shRNA plus δ-catenin* (Fig. 6, a and b). Neurons were fixed at DIV 17 and examined by confocal microscopy and spine parameters were obtained. The expression of the shRNA to δ-catenin caused a significant reduction in the spine head width (Fig. 6, b and c). These parameters could be rescued by the shRNA-resistant full-length δ-catenin, but not the construct of δ-catenin lacking the ARM domains (Fig. 6c). Similar results were observed for spine length (Fig. 6d). This suggests that ARM domain interactions are necessary for the ability of δ-catenin to regulate both spine length and head width during development.
FIGURE 6.
The ARM region of δ-catenin regulates its ability to influence spine architecture. a, schematic of constructs used. b, representative images of dendrites from neurons expressing vector, shRNA, or shRNA + δ-catenin *, transfected at DIV 11 and imaged at DIV 17. c, quantitation of spine head width, and d, spine length in neurons expressing vector, shRNA, or shRNA + δ-catenin* (p values: *, <0.05; **, <0.005; ***, <0.0005, one-way ANOVA analysis and Dunnett's multiple comparison test with single pooled variance; scale, 5 μm).
Cadherin Dependence of the Ability of δ-Catenin to Regulate Synaptic Architecture
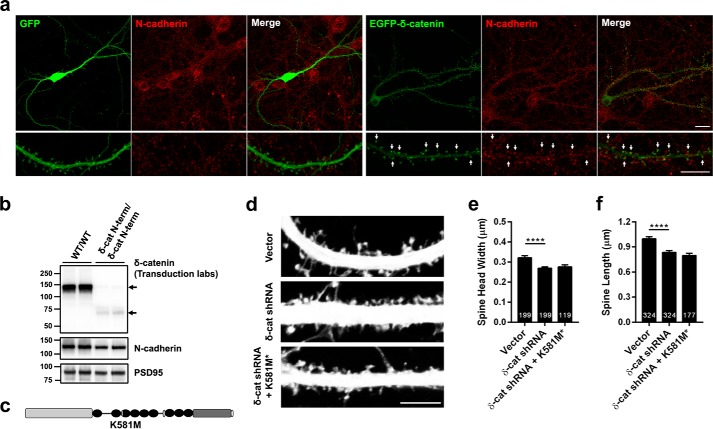
One of the proteins that are known to bind to δ-catenin within the ARM repeat domains is N-cadherin. Given the known role of N-cadherin in regulating synaptic architecture (37–39) and spine plasticity (40–42), we examined if δ-catenin cooperates with N-cadherin to regulate spine architecture. We examined the colocalization of endogenous N-cadherin with EGFP and EGFP-tagged δ-catenin in cultured rat hippocampal neurons by transfecting EGFP or EGFP-δ-catenin, immunostaining with N-cadherin and confocal microscopy. Note that in cells that were expressing extremely high levels of EGFP-δ-catenin, the morphology appeared to be aberrant, so neurons expressing low to medium levels of EGFP δ-catenin were used for imaging and analysis. As expected, N-cadherin was localized to spines labeled by EGFP. A significant amount of EGFP-δ-catenin colocalized with N-cadherin on spine heads (Fig. 7a), suggesting that the two proteins may cooperate to regulate synaptic structure.
FIGURE 7.
The interaction of δ-catenin with N-cadherin is necessary for its ability to regulate spine architecture. a, confocal images of DIV 19–20, transfected at DIV 17, rat hippocampal neurons expressing EGFP or EGFP δ-catenin and immunostained for N-cadherin. N-cadherin is localized at spines and a significant amount of EGFP-δ-catenin and N-cadherin colocalize at spine heads (indicated by arrows). (Scale, top panel, 20 μm; bottom, 10 μm.) b, Western blot analysis of synaptosome from control and δ-catenin N-term mice with antibodies to δ-catenin and PSD-95. N-cadherin levels are significantly reduced in the δ-catenin N-term mouse synaptosomes (p = 0.023, Student's t test). c, schematic of construct used. d, representative images of dendrites from neurons expressing vector, shRNA, or shRNA + δ-catenin K518M* mutant that abolishes its interaction with N-cadherin. e, quantitation of spine head width, and f, spine length in neurons expressing vector, shRNA, or shRNA + K581M* (p values: *, <0.05; **, <0.005; ***, <0.0005, one-way ANOVA and Dunnett's multiple comparison test with single pooled variance; scale, 5 μm).
We have previously demonstrated that the δ-catenin N-term mice have reduced global levels of N-cadherin (26). To examine if this reflects specific alterations in the levels of N-cadherin at synapses, we examined the levels of N-cadherin in synaptosomes from wild type and δ-catenin N-term mice during development (P21/22) by Western blot analysis relative to the levels of PSD-95. δ-Catenin N-term mice had a significant reduction (by about 25%) in the levels of N-cadherin in synaptosomes (Fig. 7b), suggesting that loss of δ-catenin significantly reduces the synaptic pool of N-cadherin (values are in arbitrary units, control, 1.738 ± 0.114; δ-catenin N-term, 1.296 ± 0.048, p = 0.023, Student's t test).
In addition, these results suggested that the reduction of the synaptic pool of N-cadherin in the absence of δ-catenin might be linked to the functional cooperation of δ-catenin and N-cadherin in regulating synaptic architecture. To further examine this cooperation, we took advantage of a point mutation in δ-catenin, K581M* (Fig. 7c), which has been demonstrated to significantly abolish its interaction with N-cadherin (27). Primary rat cultured neurons were transfected with vector or shRNA or shRNA plus δ-catenin-K581M* at DIV 11–12. Neurons were fixed and imaged by confocal microscopy at DIV 17–18 to estimate spine parameters (Fig. 7d). Although the expression of the shRNA caused a significant reduction in the head width of spines, the expression of the δ-catenin-K581M* did not rescue the phenotype (Fig. 7e). Similar results were obtained for the spine length (Fig. 7f). This suggests that the interaction of δ-catenin and N-cadherin is critical for the ability of δ-catenin to regulate spine architecture.
δ-Catenin Regulates Spine Architecture via PDZ-dependent Interactions
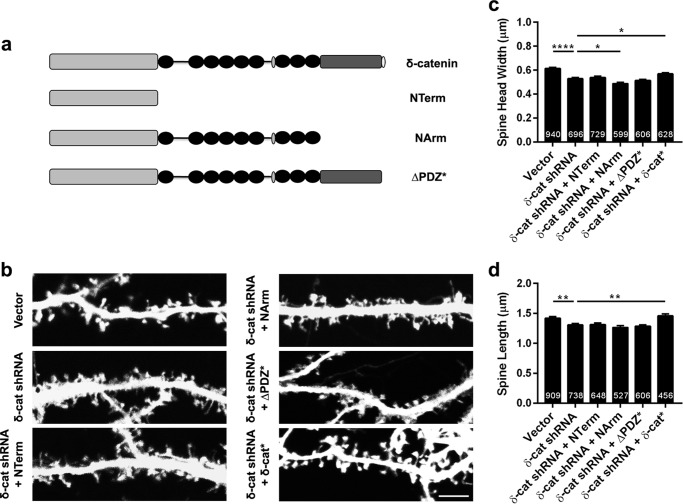
In addition to the ARM domains, δ-catenin includes an N-terminal region and a C-terminal region that includes a PDZ binding motif. To examine if the ability of δ-catenin to regulate spine architecture was dependent on other domains of δ-catenin, we examined the ability of δ-catenin constructs encoding different regions of δ-catenin. To this end, rat primary neurons in culture were transfected (DIV 11) with vector, shRNA, or shRNA plus constructs encoding the N-terminal region, the N-terminal, and ARM repeats or the full-length protein lacking the PDZ motif (Fig. 8a). In addition, neurons were transfected with shRNA and full-length δ-catenin*. Neurons were fixed at DIV 17, and spine parameters were examined by confocal microscopy (Fig. 8b). The expression of the shRNA significantly reduced spine head width and this could only be rescued by the expression of the full-length δ-catenin* (Fig. 8c). Similar results were obtained for the spine length (Fig. 8d). Because the Δ PDZ construct includes the entire protein but the PDZ motif, we conclude that the PDZ motif is necessary for the ability of δ-catenin to regulate spine architecture. Taken together, these studies indicate that both cadherin and PDZ-dependent interactions are necessary for the ability of δ-catenin to regulate spine architecture in developing neurons.
FIGURE 8.
The PDZ motif is critical for the ability of δ-catenin to regulate spine architecture. a, schematic of constructs used. b, representative images of dendrites from neurons expressing vector, shRNA, or shRNA + the indicated constructs. c, quantitation of spine head width and d, spine length in neurons expressing vector, shRNA, or shRNA + indicated constructs (p values: *, <0.05; **, <0.005; ***, <0.0005, one-way ANOVA and Dunnett's multiple comparison test with single pooled variance; scale, 5 μm).
DISCUSSION
In this study, we demonstrate that δ-catenin, a component of the cadherin-catenin cell adhesion complex (43), is localized in synaptosomes and is a critical regulator of spine architecture in developing and mature neurons. Mature neurons from mice that genetically lack δ-catenin have perturbations in synaptic architecture. This phenotype is replicated by shRNA-mediated knockdown suggesting that the functional role of δ-catenin in regulating spine architecture is cell autonomous. Furthermore, the phenotype persists with alterations in synaptic activity. Taken together, these results suggest that δ-catenin is a critical regulator of spine architecture in developing and mature neurons.
The alterations in spine lengths and head widths in the absence of δ-catenin are not reflected by a change in spine dynamics, suggesting that spines are inherently smaller in the absence of δ-catenin. Furthermore, we demonstrate that δ-catenin is co-localized with N-cadherin during development and loss of δ-catenin in a mouse model leads to a reduction in the synaptic pool of N-cadherin. Interestingly, the ability of δ-catenin to influence spine architecture is critically dependent on its ability to interact both with cadherin and PDZ domain-containing proteins (Fig. 9). We propose a model in which δ-catenin is anchored by N-cadherin and PDZ domain-containing proteins at synapses and regulates spine architecture via downstream effectors that ultimately link to the actin cytoskeleton.
FIGURE 9.
Model for δ-catenin-mediated regulation of synaptic architecture. A, δ-catenin promotes the trafficking and stabilization of N-cadherin at synapses. B, stabilization of the N-cadherin-δ-catenin complex allows δ-catenin to recruit and stabilize PDZ domain-containing proteins that modulate the spine actin cytoskeleton via effectors to influence spine architecture.
Previous studies suggest that p120ctn, a member of the δ-catenin family of proteins lacking the PDZ motif, influences spine architecture, suggesting that members of this family of proteins may have functions in synaptic regulation. In similarity to δ-catenin, loss of p120ctn leads to a reduction in the head width and length of spines (44). However, the ability of p120ctn to influence spine head width is dependent on its ability to interact with cadherin and regulate the activity of Rac, the small GTP-binding protein, whereas its ability to influence spine length is independent of its ability to bind cadherin, but dependent on its ability to influence Rho. This is in stark contrast to the mechanisms that allow δ-catenin to regulate spine architecture. We have previously demonstrated that loss of δ-catenin does not perturb the global levels of active Rac and Rho (26), suggesting that δ-catenin and p120ctn, whereas selectively requiring cadherin association, may influence the actin cytoskeleton to influence spine head width and modulate spine length through independent pathways. It would be interesting, in the future, to dissect out these signaling pathways and determine the mechanisms underlying the divergence of the two signaling pathways.
Our results are consistent with data (27) that demonstrate that knockdown of δ-catenin leads to a reduction in the activity induced stability of cadherin. Furthermore, Brigidi et al. (27) demonstrated that knockdown of δ-catenin perturbs chemically induced LTP-induced alterations in spine architecture in a cadherin-dependent manner. Our data demonstrate that the reduction in head width and length induced by knockdown of δ-catenin is independent of alterations in neural network activity. Taken together with our data, these results suggest that loss of δ-catenin leads to a compromise of both basal and activity-induced alterations in spine architecture.
Our studies are also consistent with other studies that suggest that δ-catenin may function in recruitment and stabilization of synaptic proteins and N-cadherin. δ-Catenin promotes trafficking of N-cadherin to membrane (45), masks an endocytic signal in cadherin (46), recruits and tethers (47) the presenilin 1 complex to cadherin (48), recruits AMPA receptor-binding protein and glutamate receptor interacting protein to AMPA receptor and promotes stabilization of surface GluR2 (36). We propose a model (Fig. 7) in which δ-catenin promotes the trafficking and stabilization of N-cadherin at the synapse. Once stabilized, δ-catenin, through its PDZ-binding motif recruits PDZ domain-containing proteins, which through other effector molecules modulate the actin cytoskeleton to influence spine architecture.
N-cadherin is key regulator of synaptic plasticity (40, 42, 49, 50). N-cadherin has a key role in plasticity induced spine stabilization and spine remodeling (51) and synaptic activity promotes lateral expansion of the spine head in an N-cadherin-dependent manner. Given our finding of a key role for δ-catenin in determining spine architecture in a cadherin-dependent manner, it is possible that δ-catenin is one of the critical mediators of the ability of N-cadherin to participate in spine and synaptic plasticity.
Although the classical studies in hippocampal neurons have been predominantly focused on N-cadherin, it is now clear that several other cadherin family molecules are expressed in pyramidal neurons. For example, knockdown of cadherin-9, a type II cadherin, disrupts synaptic integrity and promotes the formation of filopodia (52) in CA3 pyramidal neurons. Given our key finding that δ-catenin regulates spine architecture via interaction with cadherins, it is interesting to speculate that δ-catenin might partner with more than one type of cadherin to regulate synaptic architecture. Consistently with this, our data (Fig. 7a) indicates that a pool of δ-catenin is not associated with N-cadherin.
Our data demonstrates that interactions mediated via the type I PDZ motif of δ-catenin are critical in the ability of δ-catenin to regulate synaptic architecture. Several PDZ domain-containing proteins are known to bind to the PDZ motif of δ-catenin. These include Erbin, Densin-180, S-SCAM, Papin, AMPA receptor-binding protein, and glutamate receptor interacting protein (53–56). Given the diversity of PDZ domain-containing proteins at the synapse, it is likely that other PDZ domain proteins interacting with δ-catenin remain to be identified. It would be interesting in the future to determine which of these proteins cooperate with δ-catenin to regulate synaptic architecture and dissect the entire signaling pathway that allows δ-catenin to modulate the actin cytoskeleton.
δ-Catenin N-term mice have severe cognitive abnormalities (24). Although the molecular basis of this is unclear, synaptic deficits are common in a number of neurodevelopmental disorders associated with intellectual disability (14). Our data, taken together with previous data that demonstrate that loss of δ-catenin enhances synaptic density and function, suggests that loss of δ-catenin leads to developmental synaptic aberrations that may contribute to deficits in the formation, stabilization, and function of neural circuits. These developmental aberrations may thus underlie the behavioral deficits observed in the δ-catenin N-term mouse model and humans with the cri du chat syndrome.
Acknowledgments
Some of the data presented in this manuscript was collected in the laboratory of Dr. Louis Reichardt at the University of California, San Francisco, supported by the Simon's Foundation. We gratefully acknowledge Dr. Reichardt's support.
Note Added in Proof
The n values under “Experimental Procedures” pertaining to the time lapse analysis shown in Fig. 5 were not correct in the version of this article that was published on February 27, 2015 as a Paper in Press. These errors have been corrected. The corrections do not change the interpretation of the results or the conclusions.
This work was supported, in whole or in part, by National Institutes of Health Institutional Development Award (IDeA) Grant 5P20GM103471-10 from the NIGMS, start up funds from the Munroe-Meyer Institute, and grants from the Alzheimer's Association, The Nebraska Research Initiative, and Nebraska EPSCoR Grant EPS-1004094.
- EGFP
- enhanced green fluorescent protein
- DIV
- days in vitro
- ARM
- armadillo
- ANOVA
- analysis of variance.
REFERENCES
- 1. Tada T., Sheng M. (2006) Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16, 95–101 [DOI] [PubMed] [Google Scholar]
- 2. Oh W. C., Hill T. C., Zito K. (2013) Synapse-specific and size-dependent mechanisms of spine structural plasticity accompanying synaptic weakening. Proc. Natl. Acad. Sci. U.S.A. 110, E305-E312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Araya R., Vogels T. P., Yuste R. (2014) Activity-dependent dendritic spine neck changes are correlated with synaptic strength. Proc. Natl. Acad. Sci. U.S.A. 111, E2895-E2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tønnesen J., Katona G., Rózsa B., Nägerl U. V. (2014) Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 17, 678–685 [DOI] [PubMed] [Google Scholar]
- 5. Bosch M., Castro J., Saneyoshi T., Matsuno H., Sur M., Hayashi Y. (2014) Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82, 444–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oe Y., Tominaga-Yoshino K., Hasegawa S., Ogura A. (2013) Dendritic spine dynamics in synaptogenesis after repeated LTP inductions: dependence on pre-existing spine density. Sci. Rep. 3, 1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheng M., Ertürk A. (2014) Long-term depression: a cell biological view. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saneyoshi T., Fortin D. A., Soderling T. R. (2010) Regulation of spine and synapse formation by activity-dependent intracellular signaling pathways. Curr. Opin. Neurobiol. 20, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vanleeuwen J. E., Penzes P. (2012) Long-term perturbation of spine plasticity results in distinct impairments of cognitive function. J. Neurochem. 123, 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kasai H., Fukuda M., Watanabe S., Hayashi-Takagi A., Noguchi J. (2010) Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci. 33, 121–129 [DOI] [PubMed] [Google Scholar]
- 11. Boda B., Mendez P., Boury-Jamot B., Magara F., Muller D. (2014) Reversal of activity-mediated spine dynamics and learning impairment in a mouse model of Fragile X syndrome. Eur. J. Neurosci. 39, 1130–1137 [DOI] [PubMed] [Google Scholar]
- 12. Kulkarni V. A., Firestein B. L. (2012) The dendritic tree and brain disorders. Mol. Cell. Neurosci. 50, 10–20 [DOI] [PubMed] [Google Scholar]
- 13. Verpelli C., Montani C., Vicidomini C., Heise C., Sala C. (2013) Mutations of the synapse genes and intellectual disability syndromes. Eur. J. Pharmacol. 719, 112–116 [DOI] [PubMed] [Google Scholar]
- 14. Pavlowsky A., Chelly J., Billuart P. (2012) Emerging major synaptic signaling pathways involved in intellectual disability. Mol. Psychiatry 17, 682–693 [DOI] [PubMed] [Google Scholar]
- 15. Kosik K. S., Donahue C. P., Israely I., Liu X., Ochiishi T. (2005) δ-Catenin at the synaptic-adherens junction. Trends Cell Biol. 15, 172–178 [DOI] [PubMed] [Google Scholar]
- 16. Arikkath J. (2009) Regulation of dendrite and spine morphogenesis and plasticity by catenins. Mol. Neurobiol. 40, 46–54 [DOI] [PubMed] [Google Scholar]
- 17. Medina M., Marinescu R. C., Overhauser J., Kosik K. S. (2000) Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics 63, 157–164 [DOI] [PubMed] [Google Scholar]
- 18. Zhang X., Snijders A., Segraves R., Zhang X., Niebuhr A., Albertson D., Yang H., Gray J., Niebuhr E., Bolund L., Pinkel D. (2005) High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am. J. Hum. Genet. 76, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiurazzi P., Oostra B. A. (2000) Genetics of mental retardation. Curr. Opin. Pediatr. 12, 529–535 [DOI] [PubMed] [Google Scholar]
- 20. Sardina J. M., Walters A. R., Singh K. E., Owen R. X., Kimonis V. E. (2014) Amelioration of the typical cognitive phenotype in a patient with the 5pter deletion associated with Cri-du-chat syndrome in addition to a partial duplication of CTNND2. Am. J. Med. Genet. A 164, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 21. Vrijenhoek T., Buizer-Voskamp J. E., van der Stelt I., Strengman E., Genetic Risk and Outcome in Psychosis (GROUP) Consortium, Sabatti C., Geurts van Kessel A., Brunner H. G., Ophoff R. A., Veltman J. A. (2008) Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am. J. Hum. Genet. 83, 504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gai X., Xie H. M., Perin J. C., Takahashi N., Murphy K., Wenocur A. S., D'arcy M., O'Hara R. J., Goldmuntz E., Grice D. E., Shaikh T. H., Hakonarson H., Buxbaum J. D., Elia J., White P. S. (2012) Rare structural variation of synapse and neurotransmission genes in autism. Mol. Psychiatry 17, 402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilman S. R., Iossifov I., Levy D., Ronemus M., Wigler M., Vitkup D. (2011) Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 70, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Israely I., Costa R. M., Xie C. W., Silva A. J., Kosik K. S., Liu X. (2004) Deletion of the neuron-specific protein δ-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 14, 1657–1663 [DOI] [PubMed] [Google Scholar]
- 25. Yuan Y., Singh D., Arikkath J. (2013) Mef2 promotes spine elimination in absence of δ-catenin. Neurosci. Lett. 536, 10–13 [DOI] [PubMed] [Google Scholar]
- 26. Arikkath J., Peng I. F., Ng Y. G., Israely I., Liu X., Ullian E. M., Reichardt L. F. (2009) δ-Catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J. Neurosci. 29, 5435–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brigidi G. S., Sun Y., Beccano-Kelly D., Pitman K., Mobasser M., Borgland S. L., Milnerwood A. J., Bamji S. X. (2014) Palmitoylation of δ-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat. Neurosci. 17, 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arikkath J., Israely I., Tao Y., Mei L., Liu X., Reichardt L. F. (2008) Erbin controls dendritic morphogenesis by regulating localization of δ-catenin. J. Neurosci. 28, 7047–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mody M., Cao Y., Cui Z., Tay K. Y., Shyong A., Shimizu E., Pham K., Schultz P., Welsh D., Tsien J. Z. (2001) Genome-wide gene expression profiles of the developing mouse hippocampus. Proc. Natl. Acad. Sci. U.S.A. 98, 8862–8867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clement J. P., Aceti M., Creson T. K., Ozkan E. D., Shi Y., Reish N. J., Almonte A. G., Miller B. H., Wiltgen B. J., Miller C. A., Xu X., Rumbaugh G. (2012) Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell 151, 709–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ivenshitz M., Segal M. (2010) Neuronal density determines network connectivity and spontaneous activity in cultured hippocampus. J. Neurophysiol. 104, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 32. Biffi E., Regalia G., Menegon A., Ferrigno G., Pedrocchi A. (2013) The influence of neuronal density and maturation on network activity of hippocampal cell cultures: a methodological study. PLoS One 8, e83899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cullen D. K., Gilroy M. E., Irons H. R., Laplaca M. C. (2010) Synapse-to-neuron ratio is inversely related to neuronal density in mature neuronal cultures. Brain Res. 1359, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paffenholz R., Kuhn C., Grund C., Stehr S., Franke W. W. (1999) The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp. Cell Res. 250, 452–464 [DOI] [PubMed] [Google Scholar]
- 35. Levesque G., Yu G., Nishimura M., Zhang D. M., Levesque L., Yu H., Xu D., Liang Y., Rogaeva E., Ikeda M., Duthie M., Murgolo N., Wang L., VanderVere P., Bayne M. L., Strader C. D., Rommens J. M., Fraser P. E., St. George-Hyslop P. (1999) Presenilins interact with armadillo proteins including neural-specific plakophilin-related protein and β-catenin. J. Neurochem. 72, 999–1008 [DOI] [PubMed] [Google Scholar]
- 36. Silverman J. B., Restituito S., Lu W., Lee-Edwards L., Khatri L., Ziff E. B. (2007) Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 27, 8505–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yam P. T., Pincus Z., Gupta G. D., Bashkurov M., Charron F., Pelletier L., Colman D. R. (2013) N-cadherin relocalizes from the periphery to the center of the synapse after transient synaptic stimulation in hippocampal neurons. PLoS One 8, e79679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pielarski K. N., van Stegen B., Andreyeva A., Nieweg K., Jüngling K., Redies C., Gottmann K. (2013) Asymmetric N-cadherin expression results in synapse dysfunction, synapse elimination, and axon retraction in cultured mouse neurons. PLoS One 8, e54105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reinés A., Bernier L. P., McAdam R., Belkaid W., Shan W., Koch A. W., Séguéla P., Colman D. R., Dhaunchak A. S. (2012) N-cadherin prodomain processing regulates synaptogenesis. J. Neurosci. 32, 6323–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mysore S. P., Tai C. Y., Schuman E. M. (2008) N-cadherin, spine dynamics, and synaptic function. Front. Neurosci. 2, 168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mysore S. P., Tai C. Y., Schuman E. M. (2007) Effects of N-cadherin disruption on spine morphological dynamics. Front. Cell Neurosci. 1, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mendez P., De Roo M., Poglia L., Klauser P., Muller D. (2010) N-cadherin mediates plasticity-induced long-term spine stabilization. J. Cell Biol. 189, 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arikkath J., Reichardt L. F. (2008) Cadherins and catenins at synapses: roles in synaptogenesis and synaptic plasticity. Trends Neurosci. 31, 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elia L. P., Yamamoto M., Zang K., Reichardt L. F. (2006) p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis M. A., Ireton R. C., Reynolds A. B. (2003) A core function for p120-catenin in cadherin turnover. J. Cell Biol. 163, 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nanes B. A., Chiasson-MacKenzie C., Lowery A. M., Ishiyama N., Faundez V., Ikura M., Vincent P. A., Kowalczyk A. P. (2012) p120-catenin binding masks an endocytic signal conserved in classical cadherins. J. Cell Biol. 199, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Restituito S., Khatri L., Ninan I., Mathews P. M., Liu X., Weinberg R. J., Ziff E. B. (2011) Synaptic autoregulation by metalloproteases and γ-secretase. J. Neurosci. 31, 12083–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim H., He Y., Yang I., Zeng Y., Kim Y., Seo Y. W., Murnane M. J., Jung C., Lee J. H., Min J. J., Kwon D. D., Kim K. K., Lu Q., Kim K. (2012) δ-Catenin promotes E-cadherin processing and activates β-catenin-mediated signaling: implications on human prostate cancer progression. Biochim. Biophys. Acta 1822, 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heisler F. F., Lee H. K., Gromova K. V., Pechmann Y., Schurek B., Ruschkies L., Schroeder M., Schweizer M., Kneussel M. (2014) GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. Proc. Natl. Acad. Sci. U.S.A. 111, 5030–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tai C. Y., Kim S. A., Schuman E. M. (2008) Cadherins and synaptic plasticity. Curr. Opin. Cell Biol. 20, 567–575 [DOI] [PubMed] [Google Scholar]
- 51. Okamura K., Tanaka H., Yagita Y., Saeki Y., Taguchi A., Hiraoka Y., Zeng L. H., Colman D. R., Miki N. (2004) Cadherin activity is required for activity-induced spine remodeling. J. Cell Biol. 167, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams M. E., Wilke S. A., Daggett A., Davis E., Otto S., Ravi D., Ripley B., Bushong E. A., Ellisman M. H., Klein G., Ghosh A. (2011) Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 71, 640–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ide N., Hata Y., Deguchi M., Hirao K., Yao I., Takai Y. (1999) Interaction of S-SCAM with neural plakophilin-related Armadillo-repeat protein/δ-catenin. Biochem. Biophys. Res. Commun. 256, 456–461 [DOI] [PubMed] [Google Scholar]
- 54. Deguchi M., Iizuka T., Hata Y., Nishimura W., Hirao K., Yao I., Kawabe H., Takai Y. (2000) PAPIN: a novel multiple PSD-95/Dlg-A/ZO-1 protein interacting with neural plakophilin-related armadillo repeat protein/δ-catenin and p0071. J. Biol. Chem. 275, 29875–29880 [DOI] [PubMed] [Google Scholar]
- 55. Laura R. P., Witt A. S., Held H. A., Gerstner R., Deshayes K., Koehler M. F., Kosik K. S., Sidhu S. S., Lasky L. A. (2002) The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of δ-catenin and ARVCF. J. Biol. Chem. 277, 12906–12914 [DOI] [PubMed] [Google Scholar]
- 56. Izawa I., Nishizawa M., Ohtakara K., Inagaki M. (2002) Densin-180 interacts with δ-catenin/neural plakophilin-related armadillo repeat protein at synapses. J. Biol. Chem. 277, 5345–5350 [DOI] [PubMed] [Google Scholar]