Background: No reliable treatment exists for cancer-related muscle loss.
Results: In muscles of mice with cancer, p-Stat3 stimulates proteolysis by activating caspase-3 and the ubiquitin-proteasome system through a C/EBPδ and myostatin pathway.
Conclusion: Inhibition of Stat3 suppresses cancer-induced muscle losses.
Significance: A small-molecule Stat3 inhibitor could be integrated into therapeutic strategies for preventing cancer-induced muscle losses.
Keywords: Cancer Biology, Caspase, Caspase-3, Muscle Atrophy, STAT3, Ubiquitin, Cancer Cachexia
Abstract
Cachexia occurs in patients with advanced cancers. Despite the adverse clinical impact of cancer-induced muscle wasting, pathways causing cachexia are controversial, and clinically reliable therapies are not available. A trigger of muscle protein loss is the Jak/Stat pathway, and indeed, we found that conditioned medium from C26 colon carcinoma (C26) or Lewis lung carcinoma cells activates Stat3 (p-Stat3) in C2C12 myotubes. We identified two proteolytic pathways that are activated in muscle by p-Stat3; one is activation of caspase-3, and the other is p-Stat3 to myostatin, MAFbx/Atrogin-1, and MuRF-1 via CAAT/enhancer-binding protein δ (C/EBPδ). Using sequential deletions of the caspase-3 promoter and CHIP assays, we determined that Stat3 activation increases caspase-3 expression in C2C12 cells. Caspase-3 expression and proteolytic activity were stimulated by p-Stat3 in muscles of tumor-bearing mice. In mice with cachexia caused by Lewis lung carcinoma or C26 tumors, knock-out of p-Stat3 in muscle or with a small chemical inhibitor of p-Stat3 suppressed muscle mass losses, improved protein synthesis and degradation in muscle, and increased body weight and grip strength. Activation of p-Stat3 stimulates a pathway from C/EBPδ to myostatin and expression of MAFbx/Atrogin-1 and increases the ubiquitin-proteasome system. Indeed, C/EBPδ KO decreases the expression of MAFbx/Atrogin-1 and myostatin, while increasing muscle mass and grip strength. In conclusion, cancer stimulates p-Stat3 in muscle, activating protein loss by stimulating caspase-3, myostatin, and the ubiquitin-proteasome system. These results could lead to novel strategies for preventing cancer-induced muscle wasting.
Introduction
In patients with advanced cancer, cachexia refers to the progressive loss of skeletal muscle and fat, resulting in weakness, diminished quality of life, poor responses to chemotherapy, and susceptibility to catabolic conditions (1). Cancer also causes early satiety and anorexia resulting in weight loss that is not reversed by improving nutritional intake because even those patients eating the recommended diets still lose weight (2). Alternatively, weight loss could develop because of competition for metabolic fuels between tumor and muscle. However, competition does not explain the development of cachexia because small, “pinpoint tumors” can produce muscle wasting, whereas large human tumors (>500 g) do not always induce wasting (3). In addition, there are no approved, regularly effective treatments that overcome the muscle wasting that is induced by cancer. Understanding the mechanisms underlying cancer cachexia should provide insights into potential treatment strategies. Our goal is to identify the mechanisms that could be manipulated so as to minimize the development of cachexia.
Factors that could contribute to the development of cancer cachexia include systemic inflammation. Inflammatory pathways are suggested because tumor necrosis factor-α (TNF-α), interleukin-1α (IL-1α), interferon-γ (IFN-γ), and IL-6 have been implicated in the pathogenesis of muscle wasting in mice (3, 4). However, the mechanisms and results have proven to be controversial. For example, some studies indicate that IL-6 administration to mice induces progressive wasting of muscle and fat stores, and ultimately, death (5–7). On the other hand, Zhou et al. (8) treated mice with pharmacological doses of IL-6 and failed to demonstrate significant changes in body weight or muscle mass. Moreover, some studies indicate that IL-6 alone does not cause muscle atrophy unless there is a second catabolic condition (9).
Pathways for the development of muscle wasting include activation of NF-κB or p53 (10, 11). Alternatively, the Jak2/Stat3 system could be active as it is reported to stimulate cancer cachexia (12–14). The Stat3 signaling pathway could be involved because IL-6 and other members of the IL-6 family (e.g. oncostatin M, IL-11, leukemia inhibitory factor (LIF), and ciliary neurotrophic factor (CNTF)) have each been reported to cause loss of muscle mass (15, 16). These factors can induce Stat3 activation. In fact, Bonetto et al. (12) analyzed an mRNA microarray system and concluded that Jak/Stat3 signaling is increased in muscles of mice bearing colon 26 tumors. They also overexpressed a dominant-negative Stat3 in muscles of mice that were treated with IL-6 or with Lewis lung carcinoma (LLC)3 or C26 cancer cells and found increased sizes of myofibers (17). In mice with pancreatic cancer, the Jak2 inhibitor, AG490, can suppress muscle losses (13). In addition, Stat3 is activated in muscles of ApcMin/+ mice that develop cancer cachexia (18). Activation of Stat3 has also been associated with the development of muscle atrophy in obesity (19), age-induced sarcopenia (20), inflammatory myopathies (21), or burns (22). Finally, we find that uremia activates p-Stat3 in muscle, initiating a signaling pathway that causes muscle protein loss (23). Thus, the loss of muscle protein that occurs in several catabolic conditions might result from activation of Stat3. Potentially, the identification of methods of targeting and suppressing Stat3 activity might prevent muscle wasting in catabolic conditions.
We identified a signaling pathway that proceeds from p-Stat3 to increased C/EBPδ expression that stimulates myostatin expression and loss of muscle mass (23). Myostatin is a negative growth factor in muscle and is increased in certain mouse models of muscle wasting, including cancer (24, 25). Potential mediators that increase myostatin expression include activated glucocorticoid receptors, forkhead transcription factors, and members of the C/EBP family of transcription factors (25–27).
Potential mechanisms accounting for loss of muscle mass in cancer cachexia include activation of proteases, such as the ubiquitin-proteasome system (UPS), an intracellular proteolytic system that degrades muscle proteins in several catabolic conditions including cancer cachexia(8, 28, 29). Another protease, caspase-3, interacts with the UPS to augment muscle protein degradation in two ways. Firstly, caspase-3 cleaves actomyosin, providing substrates for degradation by the UPS. In conditions such as uremia, the cleavage of actomyosin can be detected in muscles of mice or patients (30, 31). A second mechanism by which caspase-3 accelerates the loss of muscle mass is by stimulating protein degradation in the 26 S proteasome. This occurs when specific subunits of the 19 S proteasome are cleaved by caspase-3 (30–33). How these proteases were activated in cancer-induced loss of muscle mass has not been fully established. One possibility is that cancer cells secrete mediators that stimulate loss of muscle cell protein.
We confirmed that cancer cells secrete mediators that stimulate p-Stat3 in muscle cells resulting in loss of proteins. Secondly, we studied mice with cachexia induced by subcutaneous inoculation of isogenic, LLC, or C26 cells into C57/BL6 or CD2F1 mice, respectively. In these mouse models, we tested whether caspase-3 is activated to degrade actomyosin as occurs in the catabolic responses to uremia or diabetes (30, 31). Thirdly, we investigated whether activation of caspase-3 and the pathway from Stat3 activation (p-Stat3) to muscle wasting can be suppressed in mice lacking C/EBPδ or the ability to inhibit p-Stat3 in muscle. Our results suggest that inhibition of p-Stat3 might be developed into a strategy that blocks muscle wasting induced by certain cancers.
EXPERIMENTAL PROCEDURES
Reagents
C188-9 is a small-molecule inhibitor of Stat3 that targets the phosphotyrosyl peptide binding site within the Stat3 Src homology 2 (SH2) domain with Ki = 136 nm. It does not inhibit upstream Jak or Src kinases (34) and is well tolerated in mice when administered to mice at 12.5 mg/kg daily for 14 days (23, 34, 35). It was obtained from StemMed, Ltd. (Houston, TX).
Cell Cultures
Mouse C2C12 myoblasts (ATCC, Manassas, VA) and LLC cells (a gift of Dr. Yi-Ping Li; University of Texas Health Sciences Center, Houston, TX) were cultured in DMEM (Cellgro Mediatech, Manassas, VA), supplemented with 10% FBS (Invitrogen) plus penicillin-streptomycin (100 units/ml). C26 cells (a gift from Dr. V. Baracos, University of Alberta, Edmonton, Alberta, Canada) were cultured in RPMI 1640 medium (Sigma-Aldrich), supplemented with 10% FBS and with penicillin-streptomycin (100 units/ml).
At >80% confluence, the medium bathing C2C12 myoblasts was changed to DMEM supplemented with 2% horse serum (Sigma-Aldrich) to induce C2C12 myoblast differentiation into myotubes (36). The conditioned medium from cultured C26 or LLC cells (cells cultured in medium for 36 h) was collected and centrifuged (450 × g, 5 min, 4 °C); the medium was diluted 1:5 with 2% horse serum before adding to C2C12 myotubes (37). The area of C2C12 myotubes was measured using the National Institutes of Health ImageJ program.
Mice
All animal experiments and procedures were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC). CD2F1 female mice at 8–10 weeks of age (Charles River; New York, NY) received subcutaneous injections of isogenic C26 tumor cells (5 × 106 cells in 500 μl of medium). After 5 days, tumor-bearing mice received daily intraperitoneal injections of the diluent, D5W (5 g of dextrose in 100 ml of water) or 12.5 mg/kg of body weight of C188-9 in the diluent for 14 days. Control and C188-9-treated tumor-bearing mice were pair-fed (food eaten by the C188-9-treated cancer-bearing mouse was fed to the control, tumor-bearing mouse the following day). Body weights and tumor sizes were measured daily.
Mice with muscle-specific knock-out of Stat3 (Stat3 KO) were created by breeding transgenic mice expressing Stat3flox/flox with mice expressing muscle creatine kinase Cre (MCK-Cre) (23). C/EBPδ heterozygous mice were a gift of Dr. Esta Sterneck (Center for Cancer Research, NCI, National Institutes of Health, Frederick, MD). These mice were cross-bred to produce homozygous C/EBPδ mice. Both muscle-specific Stat3 KO and muscle-specific C/EBPδ KO mice are fertile and exhibit normal growth. Muscle-specific Stat3 KO or C/EBPδ KO mice and control mice at 8–10 weeks were inoculated with isogenic LLC cancer cells, and body weights were measured over 18 days during pair feeding. Subsequently, tibialis anterior and gastrocnemius muscles, plus predominantly red-fiber soleus and predominantly white-fiber extensor digitorum longus muscles, were dissected, weighed, immediately frozen in liquid nitrogen, and stored at −80 °C.
Real-time PCR
Primer sequences and methods have been described (23).
Western Blotting and 14-kDa Actin Detection
Gastrocnemius muscles were homogenized in radioimmunoprecipitation assay buffer plus a phosphatase inhibitor and a Complete mini protease inhibitor (Roche Applied Science, 1 mg of protein per 20 μl of radioimmunoprecipitation assay buffer). Muscle lysates were evaluated by Western blotting as described (5, 36, 38). For detection of the 14-kDa actin fragment, 30 mg of gastrocnemius muscle was placed in PBS that contained the protease inhibitor in a ratio of 1:30 (31). After homogenization, the lysates (∼100 μl) were centrifuged at 3300 × g for 10 min at 4 °C. Pellets were resuspended in 2× Laemmli sample buffer, boiled for 20 min, and separated on a 15% SDS gel and subjected to Western blotting with anti-actin antibody (1:500 dilution). This antibody recognizes the carboxyl-terminal 11 amino acids of α-actin (Sigma-Aldrich). The antibody will recognize the full length of actin at 42 kDa and the cleaved 14-kDa actin fragment (30).
Protein Synthesis and Degradation
Isolated soleus and extensor digitorum longus muscles were used to measure the rate of protein synthesis or degradation as described (36, 39).
Caspase-3 Promoter Activity Assay
Dr. Sabbagh (Montreal, Quebec, Canada) kindly provided a series of deletions of the caspase-3 promoter in a luciferase reporter construct. The luciferase reporter constructs contained 0 (−178 + 14), 1 (−1368 + 14), 2 (−1776 + 14), or 3 (−2245 + 14) putative Stat3 binding sites, respectively (Fig. 2C). The negative control was the reverse orientation of the −2245 + 14-bp DNA fragment cloned in a luciferase reporter construct. Each caspase-3 promoter-luciferase reporter construct was electroporated into C2C12 cells with or without plasmids that express Stat3 using the Invitrogen Neon transfection system. In addition, a pRSV-βgal vector was transfected into the cells to serve as an internal control. At 24 h after transfection, cells containing the each construct were incubated with or without 100 ng/ml IL-6 for 6 h, and luciferase and βgal activities were assessed. IL-6 was added to activate Stat3.
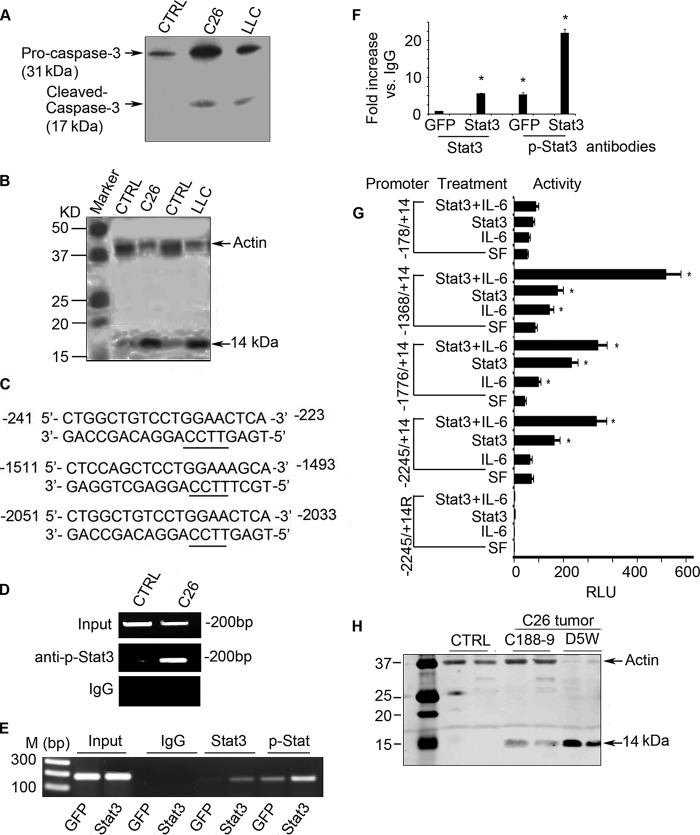
FIGURE 2.
p-Stat3 stimulates caspase-3 transcription, augmenting cancer-induced muscle loss. A, representative Western blot of caspase-3 and cleaved (activated) caspase-3 in muscles of mice bearing C26 or LLC tumors. CTRL, control. B, representative Western blots demonstrating increased caspase-3 activity in muscles of mice bearing C26 or LLC tumors. Activity was assessed from the cleavage of actomyosin, which produces a 14-kDa actin fragment in muscle found in cancer and other catabolic conditions. C, the three putative Stat3 binding sites in the caspase-3 promoter. D, a CHIP assay revealed that p-Stat3 binds to the caspase-3 promoter in C2C12 myotubes that had been treated with conditioned medium from C26 cells. E, C2C12 myotubes were infected with an adenovirus expressing GFP or Stat3. After 24 h, cells expressing Stat3 were stimulated by adding IL-6. Results of the CHIP assay revealed binding of p-Stat3 to the caspase-3 promoter. M indicates molecular marker. F, immunoprecipitated DNA was obtained as in panel E and subjected to RT-PCR analysis. The -fold change of Stat3 or p-Stat3 that is associated with DNA when compared with the value obtained with anti-IgG is shown (*, p < 0.05 versus GFP expressing cells). G, C2C12 cells were transfected with plasmids expressing different deletions of the caspase-3 promoter plus a plasmid that expresses constitutively active Stat3. Cells were then treated with or without IL-6 for 6 h, and luciferase activity was measured to assess caspase-3 promoter activity. (Results are mean ± S.E.; *, p < 0.05 versus results obtained in cells cultured in serum-free (SF) medium.) H, representative Western blots demonstrating caspase-3 proteolytic activity as an increase in the 14-kDa actin fragment in muscles of mice bearing tumor or being treated with or without C188-9.
CHIP Analysis
For CHIP assays, we used the Millipore kit according to the manufacturer's instructions. Briefly, DNA-protein complexes from C2C12 myotubes were cross-linked by adding 1% formaldehyde (Sigma-Aldrich) for 10 min. Cells were washed three times with ice-cold PBS containing a protease inhibitor (Sigma-Aldrich). Myotubes were lysed in buffer, vortexed, and sonicated (VibraCell Sonicator) for 10 s at power 4; this sonicate procedure was repeated four times to obtain DNA fragments ranging between 300 and 800 bp. After centrifugation, the protein-DNA lysate was diluted 10-fold in CHIP buffer and precleared using salmon sperm DNA and protein A/G-agarose beads for 1 h at 4 °C. Each 100 μl of protein-DNA lysate was used as an input control.
Cellular protein-DNA lysates were immunoprecipitated overnight at 4 °C with antibodies against Stat3, p-Stat3, or normal rabbit IgG (Santa Cruz Biotechnology). Subsequently, the lysates were incubated with protein A/G-agarose beads (Santa Cruz Biotechnology) for 1 h at 4 °C. The complexes were washed as described by the manufacturer. Immunoprecipitated DNA was then reverse cross-linked at 65 °C for 4 h in the presence of 0.2 m NaCl and purified using phenol/chloroform/isoamyl alcohol. A total of 5 μl of the purified DNA was subjected to PCR amplification of a 190-bp fragment using primers that were derived from the caspase-3 promoter: forward 5′-TGGGTATCTTCCTCAATCCC-3′and reverse 5′-GTGACACATGGCTTTAGTCC-3′). The amplified DNA includes the predicted Stat3 binding site of −241 to −223.
Proteasome Activity
Proteasomes were partially purified by differential centrifugation; proteasome activity was measured as the release of 7-amino-4-methylcoumarin (AMC) from the fluorogenic peptide substrate LLVY-AMC (N-Suc-Leu-Leu-Val-Tyr-AMC) (33).
Muscle Force
Mouse grip strength was measured as described (23). Briefly, grip strength was assessed five times at 1-min intervals using a grip strength meter (Columbus Instruments, Columbus, Ohio). The average grip strength over 4 days was calculated.
Statistical Analysis
Student's t test was used when two experimental groups were compared, and analysis of variance was used when data from three or four groups were studied. After variance analyses, pairwise comparisons were made by the Student-Newman-Keuls test. The data are presented as means ± S.E.
RESULTS
Conditioned Medium from Cultured C26 Cancer Cells Activates Stat3, Causing Myotube Atrophy
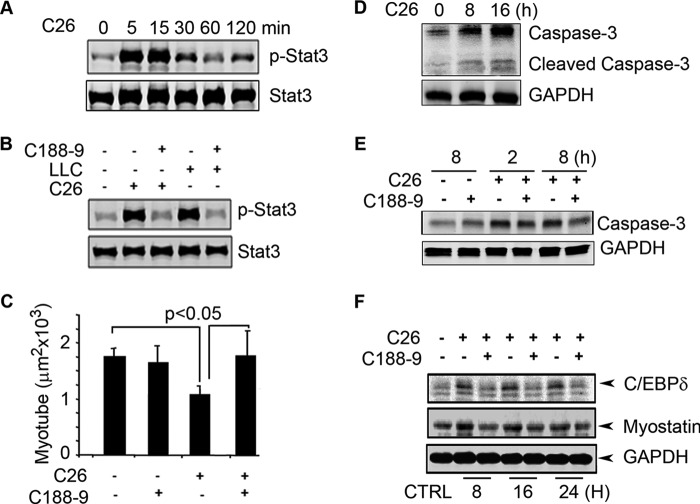
Evidence from patients with cancer cachexia or from rodent models of cancer indicates that cancer cells release factors that stimulate the loss of muscle mass (40). To confirm this result, we added conditioned medium from cultures of C26 cells to C2C12 myotubes. Within 5 min, the conditioned medium stimulated a >10-fold increase in p-Stat3 (Fig. 1A). When we added the Stat3 inhibitor, C188-9, to C2C12 myotubes, the stimulation of p-Stat3 by conditioned medium from cultured C26 cells was blocked (similar results were present in C2C12 myotubes treated with conditioned medium from LLC cells) (Fig. 1B). Notably, conditioned medium from C26 cells also reduced the size of myotubes, and C188-9 prevented the decrease in myotube size (Fig. 1C). These results indicate that activation of p-Stat3 stimulates muscle protein losses and that blocking p-Stat3 can prevent loss of muscle cell proteins.
FIGURE 1.
Conditioned medium from C26 cancer cells activates p-Stat3 in C2C12 myotubes, a model of skeletal muscle. A, representative Western blots for p-Stat3 and Stat3 exposed to the conditioned medium of C26 for different times. B, representative Western blots for p-Stat3 or Stat3 with or without C188-9 in conditioned medium for 15 min. C, the average sizes of C2C12 myotubes following a 72-h incubation in C26-conditioned medium with or without C188-9. Error bars indicate means ± S.E. D, representative Western blots of caspase-3 and cleaved caspase-3 of C2C12 incubated in C26-conditioned medium. E, representative Western blots of pro-caspase-3 in C2C12 myotubes treated with C26-conditioned medium with or without C188-9. F, representative Western blots of C/EBPδ and myostatin in C2C12 myotubes treated with C26-conditioned medium with or without C188-9. CTRL, control.
In mice with chronic kidney disease or diabetes, we have found that caspase-3 activation stimulates muscle proteolysis (30, 32, 41). To test whether the conditioned medium increases caspase-3 activity, we measured the cleaved (activated) form of caspase-3 in C2C12 myotubes following their incubation with conditioned medium from C26 cancer cells. There was an increase in both the expression of caspase-3 and activation of caspase-3 (Fig. 1D). When we added C188-9 to C26-conditioned medium in C2C12 myotubes, there was decreased expression of caspase-3 (Fig. 1E), suggesting that p-Stat3 stimulates caspase-3 expression to participate in the loss of muscle cell protein.
Because p-Stat3 activation by uremia will stimulate the expression of C/EBPδ and myostatin in muscles (23), we examined whether conditioned medium from C26 cells will stimulate these components of the intracellular signaling pathway (23). Incubation of C2C12 myotubes with conditioned medium from C26 cells increased the expression of both C/EBPδ and myostatin. These responses were blocked by C188-9 (Fig. 1F). Thus, cultured C26 cells release a factor that stimulates p-Stat3 and activates caspase-3 plus loss of protein from C2C12 muscle cells.
In Cancer Cachexia, p-Stat3 Stimulates Caspase-3 Transcription in Muscle
The UPS by itself degrades actomyosin and myofibrillar proteins slowly (42), but when caspase-3 is activated, it cleaves actomyosin and the myofibrillar proteins to provide substrates for degradation in the UPS (5, 30, 32). Caspase-3 also can cleave specific subunits of the 19 S proteasome particle, which stimulates the proteolytic activity of the 26 S proteasome (33). Because we find that conditioned medium from cultured C26 cancer cells stimulates caspase-3 expression and activation in skeletal muscle cells (Fig. 1), we examined whether C26 or LLC tumors inoculated in mice will stimulate caspase-3 expression and activation in muscle. Fig. 2A indicated that the levels of pro-caspase-3 and cleaved caspase-3 (activated caspase-3) are increased in muscles of mice bearing C26 or LLC tumors. To assess whether there is also increased proteolysis, we examined muscle for 14-kDa fragments of actin in the insoluble fraction of muscle biopsies, as occurs in animals and patients with catabolic conditions (30, 31). There was an increase in the 14-kDa actin fragment in muscles of mice with loss of muscle mass from LLC or C26 tumors when compared with results from pair-fed, control mice without tumors (Fig. 2B). These results indicate that caspase-3 participates in the muscle proteolysis that is present in tumor-bearing mice.
To determine whether the increase in caspase-3 expression in muscles of tumor-bearing mice is linked to Stat3 activation, we used the MatInspector program and found that there are three putative Stat3 binding sites within the 3-kb caspase-3 promoter (Fig. 2C). We then tested whether an increase in p-Stat3 would stimulate caspase-3 expression. C2C12 myotubes were exposed to conditioned medium from cultured C26 cells for 24 h. We performed a CHIP assay using lysates of C2C12 myotubes. The lysates were immunoprecipitated with anti-p-Stat3 antibody, and DNA from the immunocomplex was subjected to PCR analysis. We found evidence that p-Stat3 binds to the caspase-3 promoter (Fig. 2D). To examine whether p-Stat3 interacts with the caspase-3 promoter, we infected C2C12 myotubes with an adenovirus that expresses Stat3 or with a control adenovirus that expresses GFP. We also added 100 ng/ml IL-6 to the C2C12 myotubes to activate Stat3. Lysates of cells treated with IL-6 or Stat3 were immunoprecipitated with anti-Stat3 or anti-p-Stat3 antibodies. The immunoprecipitated DNA was analyzed by PCR or RT-PCR. Again, the results indicate that p-Stat3 binds to the caspase-3 promoter (Fig. 2, E and F).
To test whether binding of p-Stat3 to the caspase-3 promoter stimulates caspase-3 transcription, we transfected C2C12 myoblasts with a luciferase plasmid that expresses the caspase-3 promoter (a serial deletion of the promoter was described under “Experimental Procedures”). These cells were also co-transfected with a plasmid that expresses constitutively active Stat3 (Stat3C) or with an “empty” plasmid. The pRSV-βgal plasmid was added to each transfection reaction as a transfection control. In these experiments, we activated Stat3 with IL-6. In C2C12 cells treated with IL-6 (to activate p-Stat3 or cells that were expressing Stat3C), there was increased activity of the caspase-3 promoter. The highest caspase-3 promoter activity was found in IL-6-treated cells that expressed Stat3C (Fig. 2G). As noted, the negative control was the promoter fragment (−2245 + 14), cloned in the reverse orientation. Cells expressing this construct exhibited virtually no luciferase expression. In cells containing the truncated caspase-3 promoter (−178/+14), luciferase activity was absent and remained absent even in cells treated with IL-6 or transfected with Stat3C or transfected with Stat3 and treated with IL-6 (Fig. 2G). Interestingly, the full-length caspase-3 promoter exhibited relatively low luciferase activity, suggesting the presence of a negative regulatory element within the full-length promoter.
To identify whether p-Stat3 initiates caspase-3 proteolytic activity in muscle, we measured the 14-kDa actin fragment in the insoluble fraction of muscles of tumor-bearing mice. The level of the 14-kDa actin fragment was increased in muscles of C26 tumor-bearing mice. This increase was blocked by treating mice bearing C26 tumors with C188-9 (Fig. 2H).
Muscle-specific Stat3 KO in Mice with LLC Tumors Improves Skeletal Muscle Metabolism
Our in vitro results from cultured cells suggest that C/EBPδ and myostatin are “downstream” from Stat3 activation (Fig. 1) (23). An alternative explanation would be that C188-9 inhibits either C/EBPδ or myostatin in addition to p-Stat3. To evaluate these two possibilities, we created mice with muscle-specific Stat3 KO. These mice are fertile and develop normally (23). At 8–10 weeks of age, Stat3 KO and Stat3flox/flox male mice (controls) were injected subcutaneously with LLC and pair-fed for 18 days. In the control, the body weight of Stat3flox/flox mice bearing LLC tumors declined significantly. In contrast, mice with muscle-specific, Stat3 KO experienced improved growth despite the presence of LLC tumors (Fig. 3A). The greater gain in body weight was due, at least in part, to larger masses of muscle including the gastrocnemius (Fig. 3B). Consistent with the increased muscle mass, muscle-specific Stat3 KO improved myofiber sizes despite the presence of the LLC tumor (Fig. 3, C and D). These differences in the growth of muscle were independent of changes in tumor size (Fig. 3E). We also found that the greater muscle mass in mice with muscle-specific KO of Stat3 was accompanied by a greater grip strength despite the presence of LLC tumors (Fig. 3F). Similar results were observed in male and female mice with muscle-specific Stat3 KO (only results from male mice are shown).
FIGURE 3.
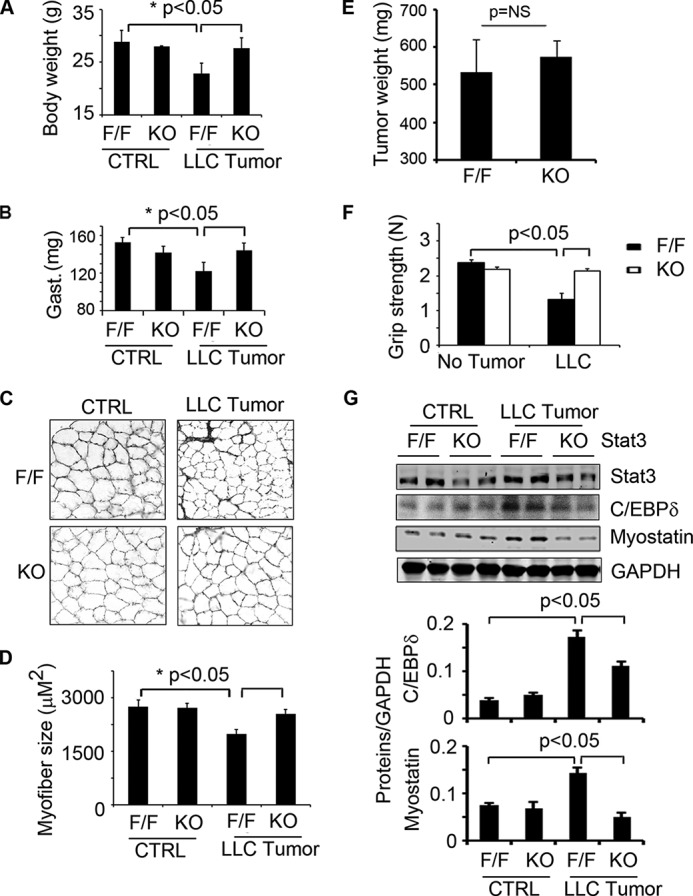
Muscle-specific Stat3 KO in mice suppresses LLC-induced loss of muscle mass. Mice with muscle-specific Stat3 KO (KO) or control mice (Stat3flox/flox (F/F)) were used; 10 mice in each group were injected with LLC cells. A–G, after 18 days, we assessed: body weight (A); weights of gastrocnemius (Gast.) muscles (B); differences in cross-sections of tibialis anterior muscles that were immunostained with anti-laminin (C); the average sizes of myofibers (D); weights of tumors (E); muscle grip (mean values ± S.E.) (F); and representative Western blots of Stat3, C/EBPδ, and myostatin in gastrocnemius muscles of Stat3 KO mice versus control, floxed Stat3 (F/F mice) that had been treated with and without LLC tumors (G). CTRL, control. Densities of these Western blots were corrected for GAPDH and quantified (bar graphs).
Muscle-specific Stat3 KO also led to suppression of the expression of both C/EBPδ and myostatin despite LLC tumors (Fig. 3G). Notably, growth of control mice without tumors was not significantly different from the growth rates of non-tumor-bearing mice with muscle-specific Stat3 KO or the Stat3flox/flox mice (Fig. 3A). Thus, muscle-specific KO of Stat3 neither improved nor interfered with the growth of mice. We conclude that genetic inhibition of p-Stat3 in vivo yields results similar to those achieved by chemical inhibition of p-Stat3 in cultured cells (Figs. 1 and 3).
C/EBPδ KO in Mice Suppresses LLC Tumor-induced Cachexia
Because the loss of muscle mass in mice with LLC cancer is associated with increased expressions of p-Stat3, C/EBPδ, and myostatin in muscles (Fig. 3), we examined whether C/EBPδ is necessary for LLC-induced muscle wasting. C/EBPδ KO mice are fertile and develop normally (23, 43), and the absence of C/EBPδ did not affect the growth of LLC tumors (data not shown). In C/EBPδ KO mice bearing LLC tumors, body and muscle mass increased versus the loss of body and muscle mass that occurs in control mice bearing LLC tumors (Fig. 4, A and B). Consistent with the increase in muscle weight, C/EBPδ KO preserved the myofiber sizes despite the presence of LLC tumors (Fig. 4, C and D).
FIGURE 4.
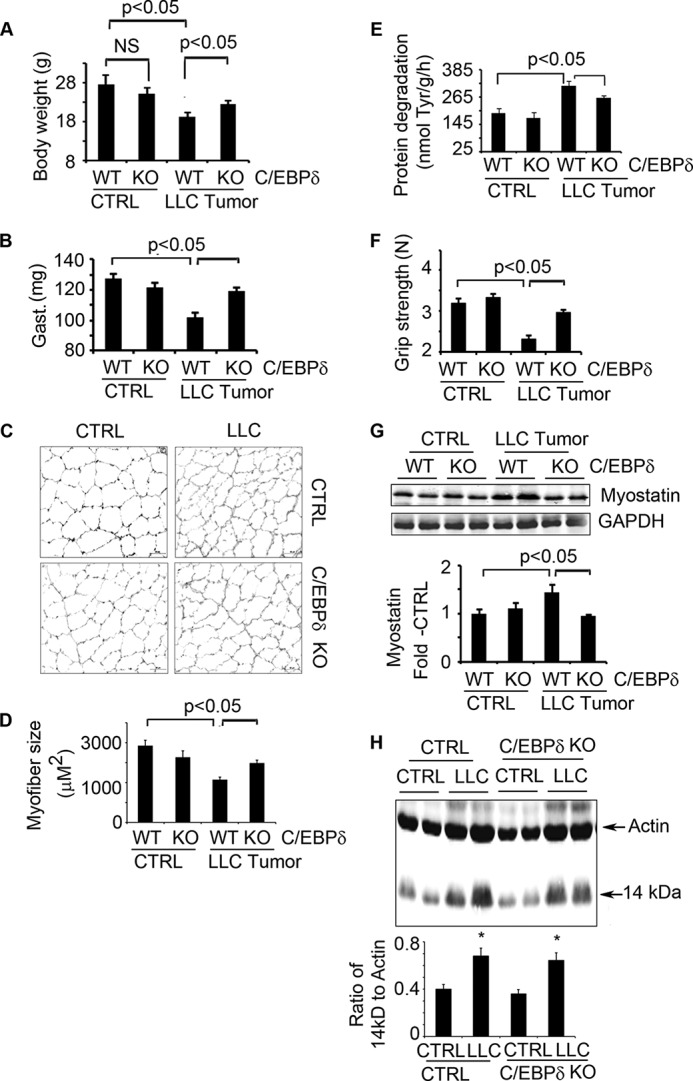
Elimination of C/EBPδ in mice suppresses LLC-induced cachexia. A–F, C/EBPδ KO and control mice were injected with LLC cells. Eighteen days later, we measured: body weight (A); weights of gastrocnemius (Gast.) (B); cross-sections of tibialis anterior muscles following immunostaining with anti-laminin (C); average myofiber sizes (D); rates of muscle protein degradation (E); and muscle grip strength (F). CTRL, control. G, representative Western blots of myostatin in muscles of C/EBPδ KO or control mice inoculated with or without LLC (upper panel) are shown. The -fold changes in myostatin expression when compared with results in control mice are shown (lower panel, quantification of myostatin levels corrected for GAPDH). H, representative Western blots of 14-kDa actin fragment in muscles of wild type or C/EBPδ KO mice bearing tumors. The -fold changes in 14 kDa to total actin are shown (lower panel,* p < 0.05 versus non-tumor mice). Results are reported as mean ± S.E. NS, not significant.
A contributor to the loss of muscle mass occurring in LLC-bearing mice was an increase in protein degradation in muscles. Notably, the increased rate of muscle protein degradation was prevented in C/EBPδ KO mice (Fig. 4E). Moreover, larger muscles in these mice were associated with an increase in their grip strength (Fig. 4F). C/EBPδ KO also suppressed tumor-induced expression of myostatin in mouse muscles (Fig. 4G). Taken together, these results demonstrate that C/EBPδ is required in the signaling pathway that links activation of p-Stat3 to myostatin and loss of muscle mass.
When we incubated C2C12 myotubes with conditioned medium from cultured LLC cells, we found that caspase-3 is activated and that C/EBPδ expression is increased (Fig. 1, D and F). Likewise, we found that there is increased p-Stat3, caspase-3, and C/EBPδ (Fig. 3G) in muscles of mice bearing LLC tumors. To determine whether caspase-3 activation depends on C/EBPδ, we examined muscles from wild type and C/EBPδ KO mice that were or were not bearing LLC tumors. Western blotting revealed that the presence of LLC tumors increased the 14-kDa actin fragment in muscles of control and C/EBPδ KO mice (Fig. 4H). This result indicates that Stat3 activates caspase-3 independent of C/EBPδ.
Chemical Inhibition of p-Stat3 by C188-9 Improves Cancer-induced Muscle Wasting
The muscle mass loss induces by LLC tumors was also present in another model of cancer cachexia, created by inoculating 8–10-week-old CD2F1 female mice with isogenic C26 cells. Our preliminary studies indicate the latter model produces uniform and severe cachexia phenotype in mice. Therefore, we selected this model for Stat3 inhibitor treatment. At 5 days after injecting C26 tumor cells, the mice began treatment with C188-9. It suppressed activation of Stat3 and the expression of C/EBPδ and myostatin in muscles (Fig. 5A). In addition, C188-9 treatment led to an increase in body weights versus tumor-bearing mice treated with the diluent (Fig. 5B). As with results in mice with LLC tumors, C188-9 administration blocked the decrease in muscle mass and in the sizes of muscle fibers (Fig. 5, C and D). Importantly, the increase in muscle mass in mice bearing C26 tumors treated with C188-9 was associated with an improvement in their grip strength (Fig. 5E). Finally, we found that the increase in muscle mass resulted from increases in protein synthesis and decreases in protein degradation detected in soleus and extensor digitorum longus muscles (Fig. 5, F and G).
FIGURE 5.
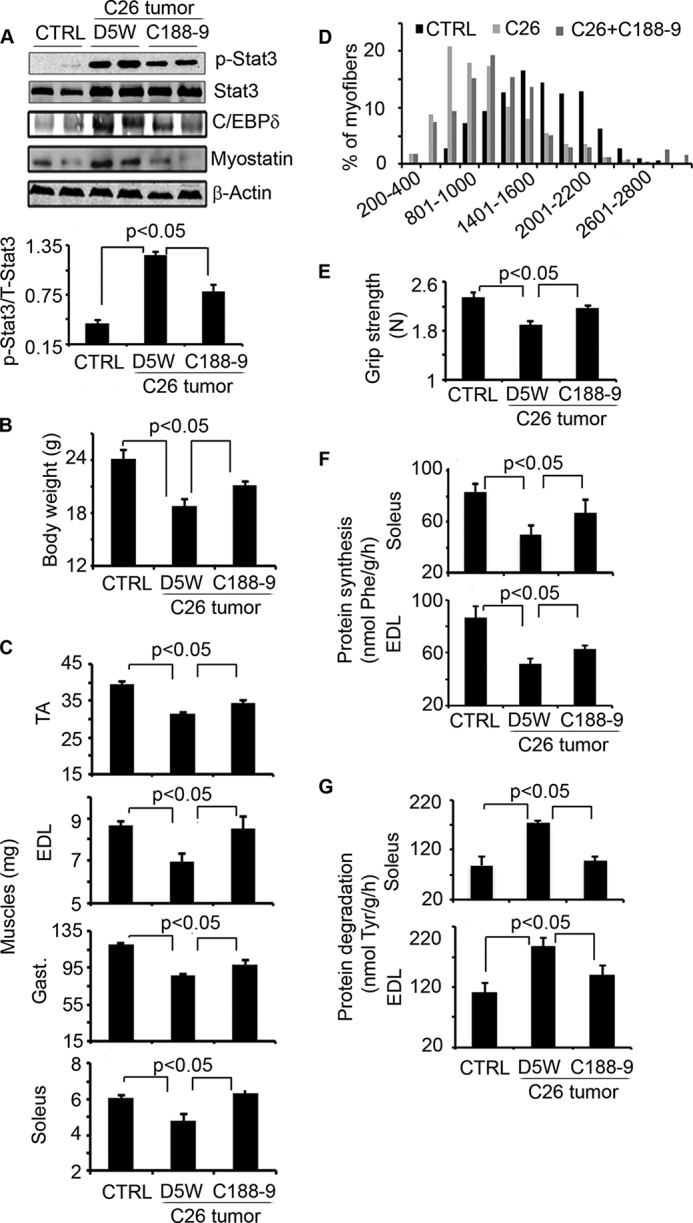
Blocking Stat3 activation with C188-9, a Stat3 inhibitor, suppresses cancer cachexia. CD2F1 mice bearing C26 tumors for 5 days were subsequently treated with C188-9 or D5W twice daily for 14 days. CD2F1 mice without C26 tumors served as controls (CTRL). Twelve mice were in each group. A, upper panel, representative Western blots of different proteins from lysates of gastrocnemius muscles (Gast.). Lower panel, quantification was corrected for GAPDH levels. B–G, we also measured: body weights (B); muscle mass (C); TA, tibialis anterior; the distribution of myofiber sizes in tibialis anterior muscles of the three groups of mice (D).; muscle grip strength (E); the rate of protein synthesis (F); and the rate of protein degradation in soleus and extensor digitorum longus (EDL) muscles (G). Results are mean ± S.E.
Cancer-induced Cachexia Stimulates Activation of Stat3 and Proteolysis by the UPS
To examine how cancer influences the mechanisms of proteolysis, we added conditioned medium from C26 cells to C2C12 myotubes and assessed the breakdown of myofibrillar proteins over 72 h. Conditioned medium increased p-Stat3 and decreased the presence of the myosin heavy chain protein (MyHC). The loss of the myofibrillar protein, MyHC, was blocked by C188-9 (Fig. 6A). In C2C12 muscle cells, the conditioned medium from C26 cells increased mRNA levels of both MuRF-1 and MAFbx/Atrogin-1, and treatment with C188-9 blocked these responses (Fig. 6B). Similar results were observed in vivo; mRNA and protein levels of MAFbx/Atrogin-1 and MuRF-1 were increased in muscles of tumor-bearing mice. These responses were not present in mice with muscle-specific Stat3 KO or in mice with C/EBPδ KO. Activation of the UPS was blocked by administration of C188-9 (Fig. 6, C–F).
FIGURE 6.
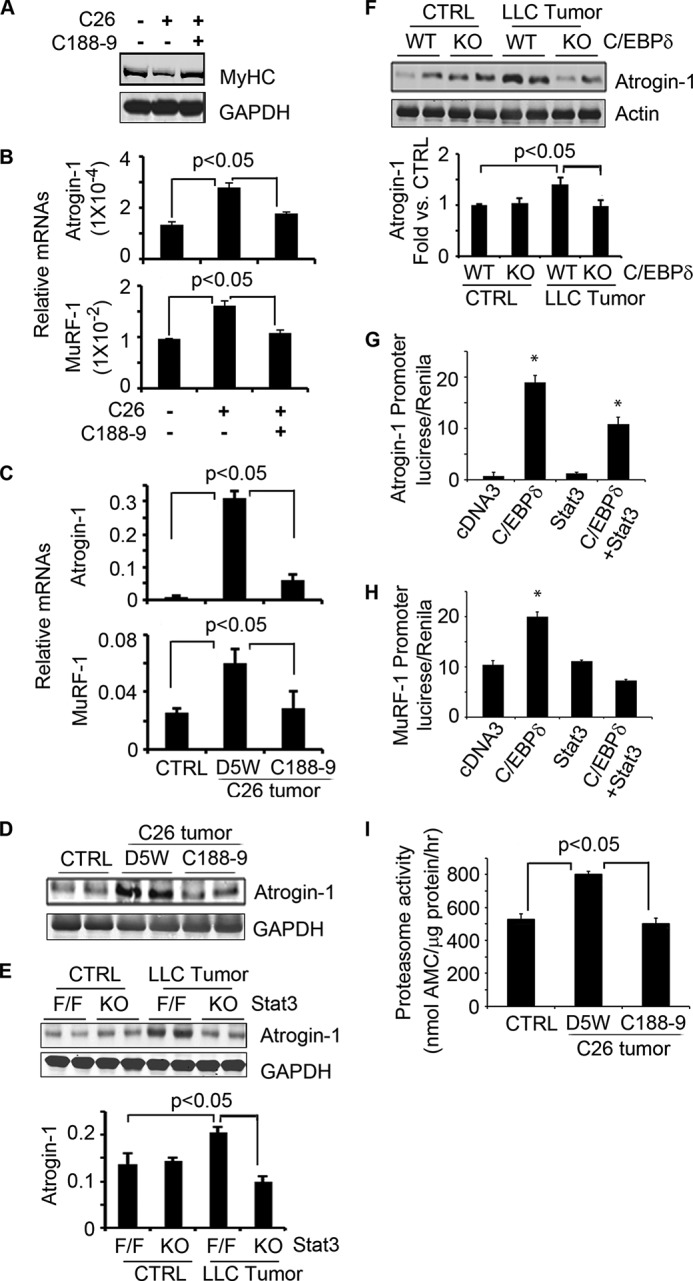
Activation of C/EBPδ induces proteolysis in the UPS in cancer cachexia. A, C2C12 myotubes were treated with conditioned medium from C26 cells with or without C188-9 for 72 h. A representative Western blot shows a decrease in myosin heavy chain protein that was blocked by C188-9. B, C2C12 myotubes were incubated in conditioned medium from C26 cells for 24 h with or without C188-9. Levels of mRNAs of MAFbx/Atrogin-1 and MuRF-1 are shown. C and D, CD2F1 mice bearing C26 tumors were treated with C188-9 for 2 weeks. mRNAs of MAFbx/Atrogin-1 and MuRF-1 and representative Western blots of MAFbx/Atrogin-1 in gastrocnemius muscle are shown. CTRL, control. E, LLC tumors were injected into mice with muscle-specific KO of Stat3 or Stat3flox/flox (F/F). After 18 days, representative Western blots from muscle show an increase in the MAFbx/Atrogin-1 protein corrected for GAPDH (mean ± S.E.). F, LLC cells were injected into C/EBPδ KO mice, and 14 days later, a representative Western blot from muscle shows the MAFbx/Atrogin-1 protein. Results corrected for GAPDH are presented as mean ± S.E. G, MAFbx/Atrogin-1 promoter activity was increased in cells that overexpress C/EBPδ. H, MuRF-1 promoter activity was increased in cells that overexpress C/EBPδ. I, proteasomes isolated from muscles of mice with or without tumors were used to measure the proteasome activity using the fluorogenic peptide, LLVY-AMC, as a substrate. Results are mean ± S.E.
What stimulates expression of MAFbx/Atrogin-1 or MuRF-1? Using the MatInspector program, we found that there are C/EBPδ binding sites within the promoter of both MAFbx/Atrogin-1 and MuRF-1. Thus, we overexpressed C/EBPδ in C2C12 myoblasts and found that the increase in C/EBPδ promoted the activity of the MAFbx/Atrogin-1 and MuRF-1 promoters; expression of constitutively active Stat3C did not change promoter activities. Interestingly, overexpression of both Stat3C and C/EBPδ in C2C12 muscle cells decreased MAFbx/Atrogin-1 promoter activity when compared with results in C2C12 cells that only expressed C/EBPδ. These results suggest that there is a negative regulatory response that is stimulated when both Stat3C and C/EBPδ are expressed (Fig. 6G). A similar response of MuRF-1 promoter to Stat3C or C/EBPδ was obtained (Fig. 6H). We conclude that C/EBPδ up-regulates MAFbx/Atrogin-1 or MuRF-1 mRNA levels, resulting in degradation of muscle proteins in mice bearing C26 tumors. We also assessed proteolytic activity of the 26 S proteasome in muscles of mice bearing C26 tumors. There was increased activity of the proteasome, which was suppressed by C188-9 as was the expression of the E3 ubiquitin ligases (Fig. 6I).
DISCUSSION
The development of cachexia is a dreaded complication of cancers not only because cachexia is debilitating but also because it is responsible for cancer-related death (44). Despite intensive investigation, the mechanisms underlying cachexia are controversial, and clinically reliable methods of combating or preventing cancer cachexia are inadequate. Our novel discoveries are as follows. First, we studied two models of cancer cachexia, produced by C26 and LLC tumors. We found that tumors activate Stat3 in skeletal muscle and initiate two pathways of proteolysis, the UPS and caspase-3. Second, we uncovered a new pathway from activated Stat3 to caspase-3, and we identified that Stat3 will bind to the caspase-3 promoter, increasing the expression of pro-caspase-3. This is relevant because we have shown that caspase-3 cleaves the complex structure of muscle proteins, providing substrates for the UPS. Caspase-3 also cleaves subunits of the proteasome, leading to increased proteolysis in the 26 S proteasome. Third, we found that a similar proteolytic pathway is activated in muscles of mice with cancer cachexia or chronic kidney disease; activated Stat3 stimulates expression of C/EBPδ, leading to MAFbx/Atrogin-1 and myostatin expression. This is important because there is no evidence that the pathway that is activated in chronic kidney disease would be stimulated in cancer-induced muscle wasting. Finally, the small-molecule Stat3 inhibitor exhibits specificity for p-Stat3 and was successful in blocking the muscle wasting induced by cancer.
How is Stat3 activated in muscles of mice bearing C26 or LLC cancers? Potential explanations include responses to inflammatory cytokines (6, 45). Results in Fig. 1 show that conditioned medium from C26 or LLC tumors stimulates p-Stat3 in C2C12 myotubes, which is in a pattern similar to that we encountered when we treated C2C12 myotubes with IL-6 (5). Because it is known that C26 (46) or LLC tumor cells (47) secrete IL-6, we speculate that IL-6 is at least one of the factors in conditioned medium stimulating p-Stat3 in muscle cells. Inclusion of the stat3 inhibitor, C188-9, in C2C12 cells that were treated with conditioned medium from the cancer cells resulted in suppression of p-Stat3, caspase-3, and the UPS, leading to larger sizes of the C2C12 myotubes. This is consistent with the study of Bonetto et al. (17) about activation of Stat3 in muscles of tumor-bearing mice. In other studies as well, muscle wasting has been associated with higher levels of circulating IL-6 especially when a second catabolic condition is present (9). In addition, Zhang et al. (37) reported that cancers secrete TNF-α, which stimulates phospho-p38 and phospho-C/EBPβ, resulting in increased expression of MAFbx/Atrogin-1. In our experiments, C188-9 blocked Stat3 phosphorylation but not the phosphorylation of p38 (data not shown), indicating that our results cannot be explained by phosphorylation of p38.
How does Stat3 activation cause muscle wasting? Previously, we found that muscle wasting in a model of chronic uremia that activates Stat3 initiates a pathway to C/EBPδ and to myostatin, resulting in loss of muscle mass (23). In the mouse model of cancer cachexia, we found that this signaling pathway contributes to tumor-induced cancer cachexia (Figs. 2 and 7). In addition, in the cancer cachexia model, we found that p-Stat3 stimulates caspase-3 transcription. The mechanism involves Stat3 binding to the caspase-3 promoter, resulting in increased expression of pro-caspase-3. The participation of caspase-3 in the loss of muscle protein was identified when we found a higher level of the 14-kDa actin fragment in muscles of mice bearing cancers. The involvement of caspase-3 is important because caspase-3 promotes muscle protein losses in two ways. First, it cleaves the complex structure of actomyosin and myofibrillar proteins to produce substrates for the UPS, and second, caspase-3 stimulates 26 S proteasome activity by cleaving regulatory subunits of 19 S (30, 31, 33).
FIGURE 7.
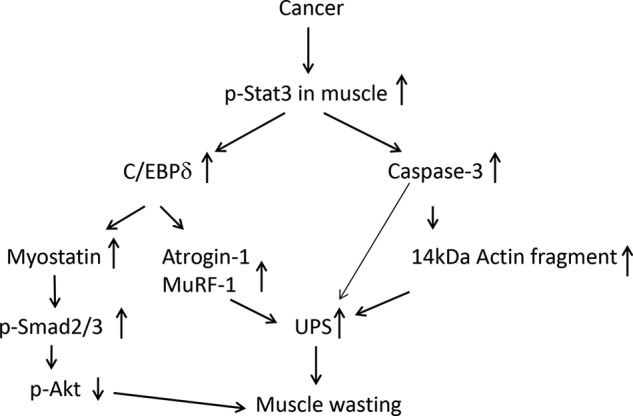
Cancer activates Stat3 in muscle to stimulate loss of muscle mass via two signaling pathways. In one pathway, p-Stat3 stimulates caspase-3 transcription and activity. In a second pathway, p-Stat3 stimulates C/EBPδ expression and activity, which increases myostatin and MAFbx/Atrogin-1 and MuRF-1. Both pathways result in protein losses in muscle.
The mechanisms by which C188-9 increased muscle mass in tumor-bearing mice included improvements in muscle protein synthesis and suppression of protein degradation. We and others report that increases in the mRNAs of the muscle-specific, E3 ubiquitin ligases, MAFbx/Atrogin-1 and MuRF-1, are associated with muscle wasting in catabolic conditions (48–50). Notably, we found that C188-9 treatment not only reduces the mRNAs of MAFbx/Atrogin-1 and MuRF1 but also improves both protein synthesis and degradation in muscles of mice bearing tumors (Fig. 4). We also uncovered insights into the mechanisms by which cancer induces muscle expression of MAFbx/Atrogin-1; we found C/EBPδ binding sites in the MAFbx/Atrogin-1 promoter and showed that overexpression of C/EBPδ in C2C12 cells increased MAFbx/Atrogin-1 promoter activity (Fig. 6). Likewise, there are C/EBPδ binding elements within the MuRF-1 promoter, indicating common responses of MuRF-1 and MAFbx/Atrogin-1.
Myostatin was increased in muscles of mice with cancer cachexia, and this response was suppressed by genetic deletion of either Stat3 or C/EBPδ KO or by treatment of cancer-bearing mice with C188-9. We conclude there is a key role for myostatin in the cancer-induced signaling pathway, leading to muscle protein losses, because C/EBPδ KO mice exhibited suppression of myostatin while preserving muscle mass despite the presence of LLC tumors. This conclusion is consistent with results reported by Zhou et al. (8). They demonstrated that blocking myostatin with a soluble ActRIIB receptor reversed cancer cachexia and improved survival. Other evidence for an important role of myostatin in muscle wasting is our report that inhibiting myostatin with an anti-myostatin peptibody blocks the muscle wasting of chronic uremia (36). Taken together, our results and those of others indicate that myostatin is a negative regulator of muscle mass in cachexia.
Treatment strategies for patients with cancer cachexia include eliminating the tumor and optimizing nutritional status. Tumor elimination is difficult to achieve, in part because loss of muscle mass can reduce a patient's ability to tolerate chemotherapy. Presently, there are no clinically available drugs that directly target Stat3. We identified that C188-9 targets the phosphotyrosyl peptide binding site within the Stat3 SH2 domain (34, 35). C188-9 directly binds to Stat3 with high affinity (KD = 4.7 nm) and blocks Stat3 binding to its phosphotyrosyl-peptide ligand with a Ki = 136 nm (34). C188-9 does not inhibit upstream Jak or Src kinases (34) and is well tolerated in mice in doses up to 100 mg/kg daily (23).4 Our results of treating mouse models of cancer cachexia with C188-9 suggest that it has promise to treat or prevent cachexia in patients. Alternatively, the results suggest that potential approaches to treat cancer-induced muscle wasting, including methods targeting muscle-specific caspase-3, suppress C/EBPδ or block myostatin.
Acknowledgment
We thank Dr. Laurant Sabbagh (Montreal, Quebec, Canada) for kindly providing us with deletions of the caspase-3 promoter in a luciferase reporter construct.
The project was supported by the generous support of Dr. and Mrs. Harold Selzman, the Norman S. Coplon extramural research grants from Satellite Health, American Diabetic Association (1-11-BS-194), and the Pilot/Feasibility award of the Diabetes Research Center (P30-DK079638) at Baylor College of Medicine (to L. Z.). This work was also supported by National Institutes of Health Grants R37 DK37175 (to W. E. M.) and P50 CA058183, P50 CA097007, and R21 CA149783, Cancer Prevention and Research Institute of Texas Grant RP100421, and Grants from the Alkek Foundation and the Dan L. Duncan Cancer Center (to D. T.), and by a fellowship from the Sao Paulo Research Foundation (FAPESP 2012/03142-8) (to K. A. S. S.).
D. J. Tweardy, L. Zhang, and W. E. Mitch, unpublished data.
- LLC
- Lewis lung carcinoma
- p-Stat3
- phosphorylated Stat3
- C/EBP
- CCAAT/enhancer-binding protein
- UPS
- ubiquitin-proteasome system
- AMC
- 7-amino-4-methylcoumarin
- Stat3C
- constitutively active Stat3.
REFERENCES
- 1. Tisdale M. J. (2009) Mechanisms of cancer cachexia. Physiol. Rev. 89, 381–410 [DOI] [PubMed] [Google Scholar]
- 2. Evans W. J., Morley J. E., Argilés J., Bales C., Baracos V., Guttridge D., Jatoi A., Kalantar-Zadeh K., Lochs H., Mantovani G., Marks D., Mitch W. E., Muscaritoli M., Najand A., Ponikowski P., Rossi Fanelli F., Schambelan M., Schols A., Schuster M., Thomas D., Wolfe R., Anker S. D. (2008) Cachexia: a new definition. Clin. Nutr. 27, 793–799 [DOI] [PubMed] [Google Scholar]
- 3. Morrison S. D. (1976) Control of food intake in cancer cachexia: a challenge and a tool. Physiol Behav. 17, 705–714 [DOI] [PubMed] [Google Scholar]
- 4. Tisdale M. J. (2002) Cachexia in cancer patients. Nat. Rev. Cancer 2, 862–871 [DOI] [PubMed] [Google Scholar]
- 5. Zhang L., Du J., Hu Z., Han G., Delafontaine P., Garcia G., Mitch W. E. (2009) IL-6 and serum amyloid A synergy mediates angiotensin II-induced muscle wasting. J. Am. Soc. Nephrol. 20, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodman M. N. (1994) Interleukin-6 induces skeletal muscle protein breakdown in rats. Proc. Soc. Exp. Biol. Med. 205, 182–185 [DOI] [PubMed] [Google Scholar]
- 7. Tsujinaka T., Fujita J., Ebisui C., Yano M., Kominami E., Suzuki K., Tanaka K., Katsume A., Ohsugi Y., Shiozaki H., Monden M. (1996) Interleukin 6 receptor antibody inhibits muscle atrophy and modulates proteolytic systems in interleukin 6 transgenic mice. J. Clin. Invest. 97, 244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou X., Wang J. L., Lu J., Song Y., Kwak K. S., Jiao Q., Rosenfeld R., Chen Q., Boone T., Simonet W. S., Lacey D. L., Goldberg A. L., Han H. Q. (2010) Reversal of cancer cachexia and muscle wasting by ActRIIB Antagonism leads to prolonged survival. Cell 142, 531–543 [DOI] [PubMed] [Google Scholar]
- 9. White J. P., Puppa M. J., Sato S., Gao S., Price R. L., Baynes J. W., Kostek M. C., Matesic L. E., Carson J. A. (2012) IL-6 regulation on skeletal muscle mitochondrial remodeling during cancer cachexia in the ApcMin/+ mouse. Skelet. Muscle 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Op den Kamp C. M., Langen R. C., Snepvangers F. J., de Theije C. C., Schellekens J. M., Laugs F., Dingemans A. M., Schols A. M. (2013) Nuclear transcription factor κB activation and protein turnover adaptations in skeletal muscle of patients with progressive stages of lung cancer cachexia. Am. J. Clin. Nutr. 98, 738–748 [DOI] [PubMed] [Google Scholar]
- 11. Schwarzkopf M., Coletti D., Sassoon D., Marazzi G. (2006) Muscle cachexia is regulated by a p53-PW1/Peg3-dependent pathway. Genes Dev. 20, 3440–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonetto A., Aydogdu T., Kunzevitzky N., Guttridge D. C., Khuri S., Koniaris L. G., Zimmers T. A. (2011) STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS One. 6, e22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilabert M., Calvo E., Airoldi A., Hamidi T., Moutardier V., Turrini O., Iovanna J. (2014) Pancreatic cancer-induced cachexia is Jak2-dependent in mice. J. Cell. Physiol. 229, 1437–1443 [DOI] [PubMed] [Google Scholar]
- 14. Watchorn T. M., Dowidar N., Dejong C. H., Waddell I. D., Garden O. J., Ross J. A. (2005) The cachectic mediator proteolysis inducing factor activates NF-κB and STAT3 in human Kupffer cells and monocytes. Int. J Oncol. 27, 1105–1111 [PubMed] [Google Scholar]
- 15. Hebenstreit D., Horejs-Hoeck J., Duschl A. (2005) JAK/STAT-dependent gene regulation by cytokines. Drug News Perspect. 18, 243–249 [DOI] [PubMed] [Google Scholar]
- 16. Tisdale M. J. (1998) New cachexic factors. Curr. Opin. Clin. Nutr. Metab Care 1, 253–256 [DOI] [PubMed] [Google Scholar]
- 17. Bonetto A., Aydogdu T., Jin X., Zhang Z., Zhan R., Puzis L., Koniaris L. G., Zimmers T. A. (2012) JAK/STAT3 pathway inhibition blocks skeletal muscle wasting downstream of IL-6 and in experimental cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 303, E410-E421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alvestrand A., Ahlberg M., Fürst P., Bergström J. (1983) Clinical results of long-term treatment with a low protein diet and a new amino acid preparation in patients with chronic uremia. Clin. Nephrol. 19, 67–73 [PubMed] [Google Scholar]
- 19. Klover P. J., Clementi A. H., Mooney R. A. (2005) Interleukin-6 depletion selectively improves hepatic insulin action in obesity. Endocrinology 146, 3417–3427 [DOI] [PubMed] [Google Scholar]
- 20. Glass D. J. (2007) Two tales concerning skeletal muscle. J. Clin. Invest. 117, 2388–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weil W. M., Glassner P. J., Bosco J. A., III (2007) High-altitude illness and muscle physiology. Bull. NYU. Hosp. Jt. Dis. 65, 72–77 [PubMed] [Google Scholar]
- 22. Jeschke M. G., Chinkes D. L., Finnerty C. C., Kulp G., Suman O. E., Norbury W. B., Branski L. K., Gauglitz G. G., Mlcak R. P., Herndon D. N. (2008) Pathophysiologic response to severe burn injury. Ann. Surg. 248, 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L., Pan J., Dong Y., Tweardy D. J., Dong Y., Garibotto G., Mitch W. E. (2013) Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab. 18, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carlson C. J., Booth F. W., Gordon S. E. (1999) Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am. J. Physiol. 277, R601–R606 [DOI] [PubMed] [Google Scholar]
- 25. Ma K., Mallidis C., Bhasin S., Mahabadi V., Artaza J., Gonzalez-Cadavid N., Arias J., Salehian B. (2003) Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am. J. Physiol. Endocrinol. Metab. 285, E363–E371 [DOI] [PubMed] [Google Scholar]
- 26. Ma K., Mallidis C., Artaza J., Taylor W., Gonzalez-Cadavid N., Bhasin S. (2001) Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am. J. Physiol. Endocrinol. Metab. 281, E1128–E1136 [DOI] [PubMed] [Google Scholar]
- 27. Allen D. L., Unterman T. G. (2007) Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am. J. Physiol. Cell Physiol 292, C188-C199 [DOI] [PubMed] [Google Scholar]
- 28. Han H. Q., Zhou X., Mitch W. E., Goldberg A. L. (2013) Myostatin/activin pathway antagonism: molecular basis and therapeutic potential. Int. J. Biochem. Cell Biol. 45, 2333–2347 [DOI] [PubMed] [Google Scholar]
- 29. Mitch W. E., Goldberg A. L. (1996) Mechanisms of muscle wasting: the role of the ubiquitin-proteasome system. N. Engl. J. Med. 335, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 30. Du J., Wang X., Meireles C. L., Bailey J. L., Debigare R., Zheng B., Price S. R., Mitch W. E. (2004) Activation of caspase 3 is an initial step triggering muscle proteolysis in catabolic conditions. J. Clin. Invest. 113, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Workeneh B., Rondon-Berrios H., Zhang L., Hu Z., Ayehu G., Ferrando A., Kopple J. D., Wang H., Storer T. W., Fournier M., Lee S. W., Du J., Mitch W. E. (2006) Development of a diagnostic method for detecting increased muscle protein degradation in patients with catabolic conditions. J. Am. Soc. Nephrol. 17, 3233–3239 [DOI] [PubMed] [Google Scholar]
- 32. Song Y.-H., Li Y., Du J., Mitch W. E., Rosenthal N., Delafontaine P. (2005) Muscle-specific expression of insulin-like growth factor-1 blocks angiotensin II-induced skeletal muscle wasting. J. Clin. Invest. 115, 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang X. H., Zhang L., Mitch W. E., LeDoux J. M., Hu J., Du J. (2010) Caspase-3 cleaves specific 19 S proteasome subunits in skeletal muscle stimulating proteasome activity. J. Biol. Chem. 285, 21249–21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Redell M. S., Ruiz M. J., Alonzo T. A., Gerbing R. B., Tweardy D. J. (2011) Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood 117, 5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu X., Kasembeli M. M., Jiang X., Tweardy B. J., Tweardy D. J. (2009) Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS. One. 4, e4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L., Rajan V., Lin E., Hu Z., Han H. Q., Zhou X., Song Y., Min H., Wang X., Du J., Mitch W. E. (2011) Pharmacological inhibition of myostatin suppresses systemic inflammation and muscle atrophy in mice with chronic kidney disease. FASEB J. 25, 1653–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang G., Jin B., Li Y. P. (2011) C/EBPβ mediates tumour-induced ubiquitin ligase atrogin1/MAFbx upregulation and muscle wasting. EMBO J. 30, 4323–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas S. S., Dong Y., Zhang L., Mitch W. E. (2013) Signal regulatory protein-α interacts with the insulin receptor contributing to muscle wasting in chronic kidney disease. Kidney Int. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clark A. S., Mitch W. E. (1983) Comparison of protein synthesis and degradation in incubated and perfused muscle. Biochem. J. 212, 649–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Todorov P., Cariuk P., McDevitt T., Coles B., Fearon K., Tisdale M. (1996) Characterization of a cancer cachectic factor. Nature 379, 739–742 [DOI] [PubMed] [Google Scholar]
- 41. Wang X., Hu Z., Hu J., Du J., Mitch W. E. (2006) Insulin resistance accelerates muscle protein degradation: activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology 147, 4160–4168 [DOI] [PubMed] [Google Scholar]
- 42. Solomon V., Goldberg A. L. (1996) Importance of the ATP-ubiquitin-proteasome pathway in degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem. 271, 26690–26697 [DOI] [PubMed] [Google Scholar]
- 43. Staiger J., Lueben M. J., Berrigan D., Malik R., Perkins S. N., Hursting S. D., Johnson P. F. (2009) C/EBPβ regulates body composition, energy balance-related hormones and tumor growth. Carcinogenesis 30, 832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tisdale M. J. (2010) Reversing cachexia. Cell 142, 511–512 [DOI] [PubMed] [Google Scholar]
- 45. Goodman M. N. (1991) Tumor necrosis factor induces skeletal muscle protein breakdown in rats. Am. J. Physiol. 260, E727-E730 [DOI] [PubMed] [Google Scholar]
- 46. Fujimoto-Ouchi K., Tamura S., Mori K., Tanaka Y., Ishitsuka H. (1995) Establishment and characterization of cachexia-inducing and -non-inducing clones of murine colon 26 carcinoma. Int. J. Cancer 61, 522–528 [DOI] [PubMed] [Google Scholar]
- 47. Puppa M. J., Gao S., Narsale A. A., Carson J. A. (2014) Skeletal muscle glycoprotein 130's role in Lewis lung carcinoma-induced cachexia. FASEB J. 28, 998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 49. Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee S. W., Dai G., Hu Z., Wang X., Du J., Mitch W. E. (2004) Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J. Am. Soc. Nephrol. 15, 1537–1545 [DOI] [PubMed] [Google Scholar]