Background: The role of protein kinase D (PKD) signaling in human epidermis is unclear.
Results: PKD isoforms have distinct and opposing growth regulatory functions in human keratinocytes. PKD3 is down-regulated upon differentiation and PKD3 silencing results in loss of keratinocyte proliferative potential.
Conclusion: PKD signaling is essential for maintaining human epidermal homeostasis.
Significance: PKD3 represents a potential drug target in hyperproliferative skin disorders.
Keywords: cell cycle, cell proliferation, differentiation, epidermis, protein kinase D (PKD), keratinocytes
Abstract
PKD is a family of three serine/threonine kinases (PKD-1, -2, and -3) involved in the regulation of diverse biological processes including proliferation, migration, secretion, and cell survival. We have previously shown that despite expression of all three isoforms in mouse epidermis, PKD1 plays a unique and critical role in wound healing, phorbol ester-induced hyperplasia, and tumor development. In translating our findings to the human, we discovered that PKD1 is not expressed in human keratinocytes (KCs) and there is a divergence in the expression and function of other PKD isoforms. Contrary to mouse KCs, treatment of cultured human KCs with pharmacological inhibitors of PKDs resulted in growth arrest. We found that PKD2 and PKD3 are expressed differentially in proliferating and differentiating human KCs, with the former uniformly present in both compartments whereas the latter is predominantly expressed in the proliferating compartment. Knockdown of individual PKD isoforms in human KCs revealed contrasting growth regulatory roles for PKD2 and PKD3. Loss of PKD2 enhanced KC proliferative potential while loss of PKD3 resulted in a progressive proliferation defect, loss of clonogenicity and diminished tissue regenerative ability. This proliferation defect was correlated with up-regulation of CDK4/6 inhibitor p15INK4B and induction of a p53-independent G1 cell cycle arrest. Simultaneous silencing of PKD isoforms resulted in a more pronounced proliferation defect consistent with a predominant role for PKD3 in proliferating KCs. These data underline the importance and complexity of PKD signaling in human epidermis and suggest a central role for PKD3 signaling in maintaining human epidermal homeostasis.
Introduction
Protein kinase D (PKD)2 is a novel family of three stress-responsive serine/threonine kinases (PKD-1, -2, and -3) involved in the regulation of diverse biological and pathological processes including cell proliferation and differentiation, adhesion and migration, protein transport, cell survival, angiogenesis, and tumor cell proliferation and metastasis (1, 2). Aberrant PKD activity and expression have been shown in a number of tumors including breast, skin, prostate, and pancreatic cancers (3–5). Most of our current understanding of PKD function, however, is based on characterization of PKD1, the founding member of this family; less is known about the function of other two isoforms (1, 6, 7). The PKD isoforms share highly homologous regulatory subdomains and can be activated by the same stimuli, however, recent studies in cancer cell lines have shown distinct but often coordinated functions for PKD isoforms (8, 9). The functional differences between these isoforms are likely based on their structural differences, level of expression, tissue specificity, and their interacting proteins (6). PKD activation generally involves phosphorylation of two conserved serine residues in the activation loop by diacylglycerol (DAG)-responsive novel PKC isozymes, followed by autophosphorylation to confer full activation (10). PKDs mediate the actions of a multitude of stimuli, including growth factors, G protein-coupled receptor agonists, phorbol esters, neuro-peptides, and oxidative stress (1); and can be found localized to the Golgi, plasma membrane, nucleus, or mitochondria, where they may have specialized functions.
In skin, earlier biochemical studies showed a correlation between PKD1 activity and proliferation status of cultured mouse KCs suggesting a pro-proliferative role for PKD1 signaling in epidermis (11, 12). In addition, PKD1 expression was shown to be up-regulated in mouse carcinomas and human hyperplastic disorders including basal cell carcinomas and psoriasis (4, 12). More recently, PKD1 was implicated in radiation-induced apoptosis in mouse KCs (13). Our own genetic in vitro and in vivo studies in mouse skin did not support a role for PKD1 in epidermal homeostasis. These studies however, revealed a critical and unique pro-proliferative role for PKD1 signaling in epidermal adaptive responses including stress-induced de-differentiation, wound re-epithelialization, and tumor promotion and development (14, 15). Interestingly, despite expression of all PKD isoforms in mouse KCs, PKD2, and PKD3 could not fully compensate for the loss of PKD1 signaling in stress-induced responses of mouse KCs. In translating our findings in mouse to human skin, we aimed to validate the importance of PKD1 signaling in human KCs. Surprisingly, we found a significant divergence in expression and function of PKD isoforms between mouse and human epidermis. Contrary to mouse KCs, PKD1 is undetectable in human KCs. In addition, PKD signaling is essential for normal proliferation and growth of human KCs. This is at least partly related to the divergence in expression and function of PKD3 between mouse and human KC. Moreover, our data showed distinct and even opposing growth regulatory functions for PKD2 and PKD3 in human KCs.
EXPERIMENTAL PROCEDURES
Epidermal Cultures
Human neonatal KCs were isolated from foreskins and grown in submerged cultures in the presence of irradiated 3T3 fibroblasts (16) using KC medium described by Wu et al. (17). Normal human KCs (NHKCs) were used between passages 2–6 in all experiments. To promote differentiation of NHKCs, cultures were grown to confluence and maintained for 4 days to induce stratification and expression of late markers of terminal differentiation. Primary cultures of mouse epidermis were established and induced to differentiate as described previously (15). For colony forming assay, 50 cells were seeded on lethally irradiated fibroblasts in a 60 mm culture dish, grown for 14 days and stained with Rhodamine B to visualize and quantify the number and size of colonies. Construction of the organotypic cultures has been described previously (18). Briefly, NHKCs were seeded onto the collagen matrix containing human dermal fibroblasts, submerged in the medium (19)and maintained for 4 days before raising to the air-liquid interface for an additional 8 days. The regenerated tissue was harvested and processed for routine histological analysis.
Analysis of Cell Proliferation
Assessment of DNA synthesis by thymidine incorporation has been described (15). When PKD inhibitors were used, cultures were treated for 20 h with [3H]thymidine added during the last 4 h. For cell cycle analysis, transduced NHKCs were fixed overnight with 70% ethanol, washed, and resuspended in PBS containing propidium iodide and RNaseA for 30 min. A total of 10,000 nuclei were examined in a FACSCalibur flow cytometer (BD Biosciences, NJ) and DNA histograms were analyzed by ModFit software.
Antibodies and Western Blot Analysis
Antibodies against p63 (SC-8431), p53 (SC-126), p15 (SC-612), p21 (SC-397), PKD1 (SC-935 and SC-639), and β-actin (SC-1615) were from Santa Cruz Biotechnology (Santa Cruz, CA), loricrin (PRB-145P), and involucrin (PRB-140C) were from Covance (Berkeley, CA), PKD1/2 (CS-2052), and PKD3 (CS-5655) were from Cell Signaling Technologies (Danver, MA). Antibody against PKD2 was from Bethyl Laboratories (Montgomery, TX) and cyclin D1 was from BD Biosciences (San Jose, CA). For immunoblotting, NHKCs were lysed in lysis buffer (50 mm Tris/HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 10 mm NaF, 1 mm Na3VO4 plus Protease Inhibitor Mixture), and cleared cell lysates (30 μg) were separated by SDS-PAGE, transferred to nitrocellulose and blotted as described previously (15).
RNA Expression Analysis
Relative transcript levels of PKDs and cyclin-dependent kinase inhibitors in total RNA isolated from subconfluent cultures of NHKCs were determined by real time RT-qPCR using a 7300 Real Time System (Applied Biosystem, Foster City, CA). Total RNA (1 μg) was first reverse transcribed and the PCR reaction was run with Syber green Taq polymerase (Quantitect Kit, Qiagen, Valencia, CA) for 40 cycles of: 95 °C, 15 s, 55 °C, 30 s, and 72 °C, 30 s. All samples were run in triplicate and non-template controls were included in each run. The RNA levels of the target genes were normalized against GAPDH transcript levels and the comparative CT (2−ΔΔCT) method was used for calculating relative cytokine mRNA expression.
shRNA Knockdowns and Lentiviral Transduction
For lentiviral-mediated shRNA knockdowns several shRNAs were designed and subcloned in the pLKO1 vectors, packaged by triple transfection of 293T cells and used to transduce NHKCs at a multiplicity of infection of 2 as described previously (15). Following 2–4 days of drug selection, transduced NHKC were screened for knock-down by immunoblotting. For combinatorial knockdown of both PKD isozymes, shRNAs targeted to PKD2 and PKD3 were cloned in PLK01Puro and PLK01Neo, respectively. Low passage cultures were transduced with both viruses and were selected in medium containing both puromycin (0.7 μg/ml) and G418 (0.8 μg/ml) starting at 30 h post-transduction. The following shRNAs induced efficient knockdown of target proteins and were used in our studies: GACATACACGACCAAATTC targeted to human PKD1, AAGACTGCAAGTTTAACTGTC targeted to PKD2, CGGGAAACTGAATAATA AGAA (sh-1) and CCAGGAACCAAGTAAGA GAAT (sh-2) targeted to human PKD3. The oligonucleotide TGTAGGACTTACAGAACGT (scrambled) was used as a control. Transduced human KCs were used within two passages after transduction and drug selection.
Statistical Analyses
Differences among means were evaluated by Student's 2-tailed un-paired t test when comparing two groups and 2-way analysis of variance and the Tukey's HSD post hoc comparison when comparing more than 2 groups, using SPSS software (SPSS, Chicago, IL). Values with p < 0.05 were accepted as significant.
RESULTS
Divergence in PKD Expression and Function between Mouse and Human KCs
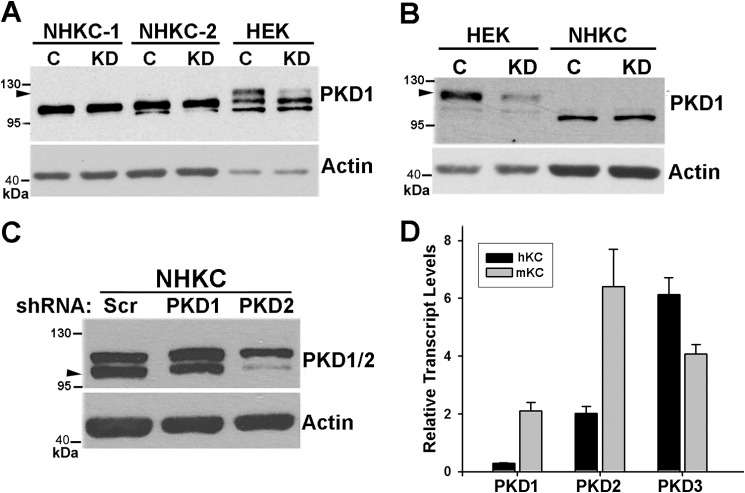
Previous studies have indicated that PKD1 is expressed in human skin and is up-regulated in hyperproliferative skin disorders (4, 20). To further investigate the role of PKD1 in human epidermis, we used lentivirus-mediated shRNA to knockdown PKD1 in NHKCs. Western blot analysis of cell lysates obtained from several preparations of NHKC or a human KC cell line (HaCat) using three different antibodies to PKD1 revealed immunoreactive proteins with similar molecular weights as PKDs (100 to 120 kDa) (Fig. 1 and data not shown). However, shRNA-mediated knockdown of PKD1 did not alter the intensity of these bands suggesting that none may represent PKD1 (Fig. 1, A–C). Similar analysis in 293-HEK cell line detected an immunoreactive protein with a slightly higher molecular weight than that in human KCs and this protein was efficiently knocked down by lentiviral transduction of shRNA targeted to PKD1 (Fig. 1, A and B). One of the antibodies (CS-2052) that cross-reacts with both PKD1 and PKD2 detected a doublet in NHKCs (Fig. 1C). To verify the efficiency of lentivirus-mediated shRNA knockdown in NHKCs, cells were transduced with lentiviruses encoding shRNAs targeted to either PKD1 or PKD2 and subjected to Western blot analysis using CS-2052. Knockdown of PKD1 did not alter intensities of any of the reactive proteins when compared with that of controls. However, PKD2 knockdown resulted in a marked reduction in the intensity of the lower band confirming the identity of one of the immunoreactive proteins as PKD2 (Fig. 1C). Given that transcripts of all PKD isoforms are detected in mouse KCs (14), transcript levels of PKD isoforms in total RNA isolated from mouse and human KCs were analyzed by RT-qPCR. As expected all three PKD isoforms were expressed in mouse KCs, albeit at variable levels (Fig. 1D). In human KCs however, the PKD1 transcript was barely detectable whereas the other two isoforms were expressed at moderate to high levels (Fig. 1D). These data indicated divergence in PKD1 expression between mouse and human KCs and identified PKD2 and PKD3 as major PKD isoforms in the latter.
FIGURE 1.
Expression of PKD isozymes in KCs. A and B, normal human KC (NHKC) or 293HEK cells were transduced with lentiviral vectors encoding either scrambled shRNA (C) or PKD1-specific shRNA (KD). Two different strains of human KC were used in A. After drug selection, cells were lysed, and 10–30 μg of cell lysates were analyzed by Western blotting using PKD1-specific antibodies SC-639 (A) or SC-935 (B). Actin was used as a loading control. Arrowheads point to immunoreactive band representing PKD1. The position of molecular weight markers on the blot is indicated on the left. C, NHKCs were transduced with lentiviral vectors encoding shRNA against PKD1, PKD2, or a scrambled control (Scr). Protein lysates (30 μg) were analyzed by immunoblotting using an antibody cross-reacting with both PKD1 and PKD2 (CS-2052). Arrowhead points to PKD2. Actin was used as a loading control. D, quantitative RT-PCR analysis of total RNA isolated from NHKC (black bars) and primary cultures of mouse epidermis (mKC; gray bars) using primer sets specific for human and mouse PKD isoforms. Relative transcript levels after normalized to GAPDH levels are shown as mean ± S.E. of triplicates.
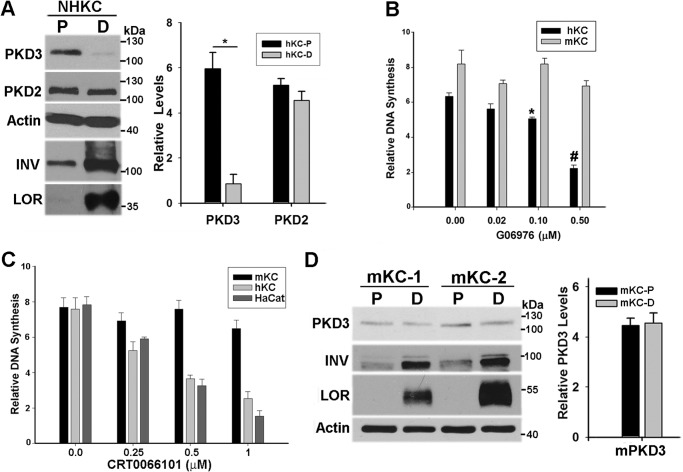
In the absence of a reliable anti-PKD3 antibody suitable for immunohistochemistry of intact skin, we next used Western blot analysis of cultures of NHKC grown under proliferative (sub-confluent) or differentiating conditions (post-confluent) to analyze distribution pattern of PKD2 and PKD3 in epidermis. In sub-confluent cultures, NHKCs are proliferative and express high levels of ΔNp63, a p53-related transcription factor with an essential role in maintenance of KC proliferation (21). In addition, low levels of early markers of differentiation such as involucrin (INV) are detected in these cultures. As cultures become confluent, the level of p63 is markedly reduced coinciding with the induction of markers of terminal differentiation such as loricrin (LOR) and profilaggrin/filaggrin (FIL) (Fig. 2A and Fig. 4). Using isotype-specific antibodies, both PKD2 and PKD3 were shown to be expressed in proliferating cultures of NHKCs. In post-confluent cultures, PKD2 levels remained relatively unchanged, whereas there was a marked down-regulation (>5 fold) of PKD3 (Fig. 2A). This is consistent with the restriction of PKD3 expression to the proliferative compartment of epidermis. Given the tight balance between KC proliferation and differentiation, down-regulation of PKD3 in differentiated NHKCs suggested a pro-proliferative role for PKD3 signaling.
FIGURE 2.
Divergence of PKD signaling between mouse and human KCs. A, protein lysates of subconfluent (P) and postconfluent (D) cultures of NHKC were analyzed by immunoblotting using antibodies specific to PKD3, PKD2, and markers of KC differentiation including involucrin (INV) and loricrin (LOR). Actin was used as a loading control. The graph shows the relative levels of PKD2 and PKD3 in P and D cultures. Band intensities from three Western blots quantified and normalized against actin levels and represented as mean ± S.E. *, p < 0.001. B and C, relative DNA synthesis measured by [3H]thymidine incorporation in subconfluent cultures of human and mouse KCs treated with increasing concentrations of Go6976 (B) or CRT0066101(C) for a period of 20 h. In C, both normal (hKC) and transformed (HaCat) human KCs were used. During the last 4 h, cultures were pulsed with 1 μCi [3H]thymidine and values expressed as mean ± S.E. (n = 8) from two independent experiments. Data were analyzed by ANOVA with significance at p < 0.001 for human cells and p = 0.5 for mouse cells. D, Western blot analysis of two independent primary cultures of mouse epidermis grown under proliferating (P) and differentiating (D) conditions using indicated antibodies. Graph shows PKD3 band intensities from three independent cell preparations presented as mean ± S.E.
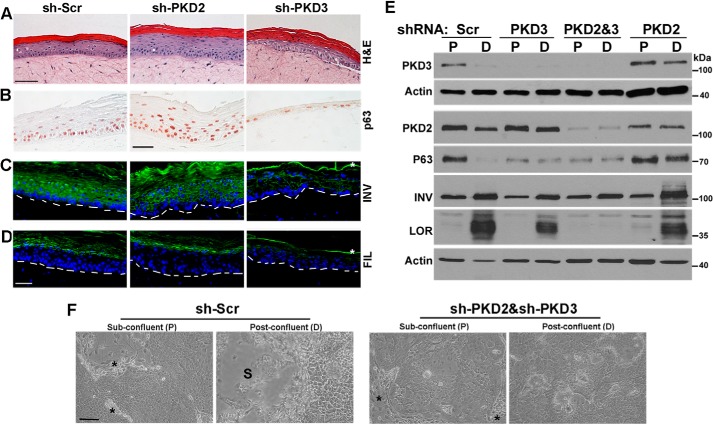
FIGURE 4.
PKD3 is required for normal proliferation and differentiation in regenerating epidermis. A–D, organotypic epidermal tissue regenerated from NHKCs expressing shRNA against PKD2, PKD3, and a Scr control. Tissue sections were analyzed by histology (A) or immunostaining with antibodies that recognize pan-p63 (B; brown nuclei) or differentiation markers including involucrin (INV) and filaggrin (FIL), both in green. Blue nuclear staining is DAPI (C and D). Dashed lines in C-D indicate the position of basement membrane, and * indicates nonspecific staining of cornified layer. Scale bar, 50 μm. E, Western blot analysis of proliferating (P) and differentiating (D) cultures of NHKC stably expressing Scr-shRNA or those targeting PKD2 or PKD3, or both isoforms (PKD2&3) with antibodies that recognize PKD3, PKD2, p63, and differentiation markers involucrin (INV) and loricrin (LOR). Actin is used as a loading control. A representative of three independent experiments is shown. F, NHKCs were co-transduced with two lentiviruses encoding sh-Scr or shRNA targeted to PKD2 and PKD3 (sh-PKD2/sh-PKD3), selected in medium containing both G418 and puromycin, and grown to confluence. Phase contrast images of cultures at sub-confluence (P) and 3 days post-confluence (D) are shown. * indicates irradiated 3T3 fibroblasts in sub-confluent cultures, and S marks the area where post-confluent cultures are stratified. Bar, 100 μm.
We have previously shown that treatment of mouse epidermal cultures with a pan-PKD inhibitor has no adverse effect on normal cell proliferation in culture (15). To investigate whether PKD signaling plays a distinct role in NHKC proliferation, DNA synthesis in growing cultures of mouse and human KCs treated with either Go6976, a selective inhibitor of PKCα and PKDs, or CRT0066101, a more selective inhibitor of PKDs (22) were determined. As shown in Fig. 2, treatment of cultured KCs with either inhibitor resulted in a dose-dependent suppression of DNA synthesis in human but not mouse KCs (Fig. 2, B and C). To determine if these differences are due to differential regulation of PKD3 in human and mouse epidermis, proliferative, and differentiated cultures of mouse KC were subjected to Western blot analysis using an antibody that cross-reacts with both human and mouse PKD3. Analysis of primary cultures of mouse epidermis failed to show differentiation-induced suppression of PKD3 (Fig. 2D) suggesting differential regulation of PKD3 between mouse and human epidermis.
Overall, these data show significant divergence in expression and function of PKD isoforms between human and mouse KCs and suggest a role for PKD3 signaling in normal proliferation and differentiation of human KCs.
Contrasting Effects of PKD2 and PKD3 on the Human KC Growth and Proliferation
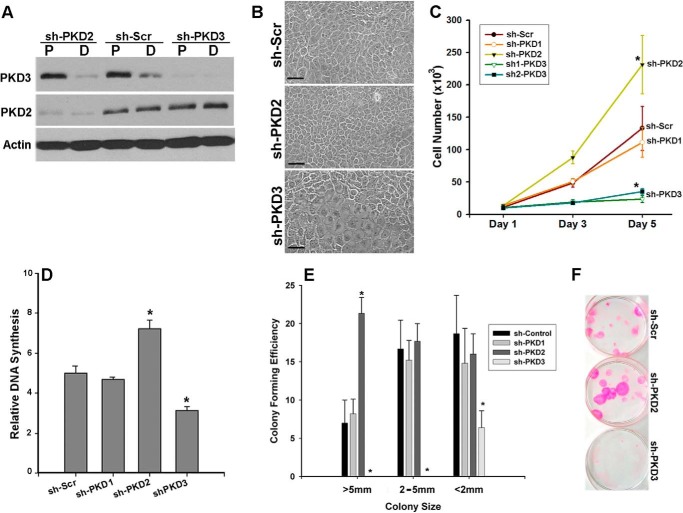
To investigate the role of PKD isoforms in regulation of NHKC proliferation, lentiviruses encoding isoform-specific shRNAs were used to stably knockdown individual PKD isoforms. Western blot analysis of proliferating and differentiated cultures of PKD2 (sh-PKD2), or PKD3 (sh-PKD3) knockdown NHKCs verified the specificity and efficiency of shRNA-mediated silencing, and showed that knockdown of one PKD isoform did not induce compensatory up-regulation of the other isoform (Fig. 3A). Interestingly, within 4–6 days after transduction, there were significant changes in the morphology and the growth behavior of NHKCs deficient in either PKD2 or PKD3 when compared with cultures expressing scrambled (Scr) shRNA (Fig. 3B). We then used a number of different parameters to assess the effects of PKD2- or PKD3-silencing on NHKC growth and proliferation. Monitoring the growth rate of these cultures for a period of 5 days revealed a significant deviation from control KCs expressing either Scr- or PKD1-shRNA (Fig. 3C). PKD3-deficient cells grew at a significantly slower rate than controls, whereas those deficient in PKD2 showed a marked enhancement in growth rate. The differences in cell number were not related to the altered plating efficiency of PKD2- or PKD3-deficient KCs as the initial number of cells adhered at 6 h post-seeding (Day 1) were comparable to that in control cultures. Moreover, the adverse effects of PKD3-shRNA on growth were not related to off-target effects as another shRNAs targeted to different regions of PKD3 transcript showed similar effects (Fig. 3C).
FIGURE 3.
Opposing growth regulatory functions of PKD2 and PKD3 in human KCs. A, NHKCs were transduced with lentiviruses encoding shRNA targeted to PKD2, PKD3, or a scramble (Scr) shRNA. Cell lysates were prepared from subconfluent (P) and postconfluent (D) cultures of transduced NHKCs and analyzed by immunoblotting using PKD isozyme-specific antibodies. Actin was used as a loading control. B, representative phase contrast images of transduced cultures of NHKCs showing morphology of KCs stably expressing control shRNA (Scr) or those targeted to PKD2 (sh-PKD2) or PKD3 (sh-PKD3). Images were taken at 200× magnification and scale bars, 50 μm. C, for growth curve, equal number of transduced KCs expressing shRNAs as indicated in the figure were plated, and total number of adhered live cells at 6 h (day 1), 3- and 5-days were determined. Values represent mean ± S.D. (n = 6) from three independent experiments. D, relative DNA synthesis in subconfluent cultures of PKD knockdown NHKCs were measured by [3H]thymidine incorporation after a 4-h pulse. The bars represent the mean ± S.E. (n = 3). A representative of three experiments is shown. E and F, colony forming efficiency of transduced NHKC expressing isoform-specific shRNA seeded at 2 cells/cm2, grown for 14 days, and stained with Rhodamine (F). Graph in E shows quantification of colonies based on their size. Values (mean ± S.E.) are based on triplicates from two independent experiments. Data were analyzed by ANOVA with significance at p < 0.001 (*).
Analysis of DNA synthesis by [3H]thymidine incorporation indicated a significant reduction in the proliferation rate of PKD3-deficient NHKCs when compared with the control (Fig. 3D). The effects of PKD3 knockdown on cell growth and proliferation was even more pronounced when cells were plated at clonal densities. Under these conditions, only a few abortive colonies measuring less than 2 mm in diameter were formed suggesting loss of proliferative potential of PKD3-deficient KCs (Fig. 3, E and F). On the contrary, knockdown of PKD2 resulted in a significantly higher rate of [3H]thymidine incorporation when compared with NHKCs expressing Scr-shRNA (Fig. 3D). Moreover, PKD2 silencing in NHKCs resulted in enhanced colony forming efficiency with a marked increase in the number of colonies larger than 5 mm in diameter consistent with the growth inhibitory role for PKD2 in human epidermis (Fig. 3, E and F). The differential effects of PKD2- and PKD3-silencing on NHKC proliferation, growth and clonogenicity revealed unique and contrasting growth regulatory roles for PKD isoforms in maintaining human epidermal homeostasis.
Effects of PKD Silencing on Epidermal Differentiation
In epidermis, proliferative KCs residing in the basal layer undergo cell cycle arrest, migrate outward and commit to terminal differentiation to generate the skin permeability barrier (23). To study the effects of PKD silencing in the context of human epidermal tissue, control or PKD isoform knockdown NHKC were used to regenerate epidermal tissue in organotypic cultures. In this system, KCs are seeded onto a dermal equivalent and raised to the air/liquid interface to produce correctly polarized epidermis with spatially organized expression of marker proteins for proliferation and differentiation (24). As shown in Fig. 4, tissue regenerated from NHKCs expressing control shRNA (sh-Scr) was well-polarized and composed of all epidermal layers, including the stratum granulosum and corneum (Fig. 4A). As expected, nuclear p63 protein was mainly restricted to basal layer KCs (Fig. 4B), consistent with its expression pattern in human epidermis (21, 25). Moreover, expression of the early differentiation proteins like INV preceded that of the late differentiation proteins like FIL (Fig. 4, C and D).
Epidermal tissue regenerated from PKD2-deficient KCs (sh-PKD2) showed a significant increase in epidermal thickness, although all differentiated layers of epidermis including granular and cornified layers were present. Immunostaining of tissue sections verified normal distribution of markers of early (INV) and late (FIL) epidermal differentiation (Fig. 4, A–C); however, p63 staining was extended to upper spinous layers (Fig. 4D). On the contrary, epidermal tissues regenerated from PKD3-deficient NHKCs were hypoplastic showing a marked differentiation defect as indicated by a lack of distinct spinous and granular layers and retention of nuclei in cornified layers (parakeratosis) (Fig. 4A). Immunostaining revealed a marked reduction in p63 staining consistent with the loss of KC proliferative potential. Moreover, INV and FIL were expressed at markedly reduced levels indicating a differentiation defect. Analysis of tissue formed from NHKC transduced with another PKD3-specific shRNAs showed a similar phenotype (data not shown). To quantify alterations in protein markers of proliferation and differentiation induced by PKD-silencing, lysates of PKD2- or PKD3-knockdown NHKCs grown in subconfluent (P) or post-confluent (D) cultures were analyzed by immunoblotting. Western blot analysis of subconfluent cultures of PKD3-deficient NHKCs showed a marked decline in p63 levels when compared with control cultures (Fig. 4E). However, loss of PKD3 did not induce premature induction of markers of differentiation including INV and LOR. These proteins were induced in post-confluent cultures, although at a significantly lower levels than those in control KCs (Fig. 4E). This was consistent with the differentiation defect observed in PKD3-deficient epidermis (Fig. 4, C and D). In PKD2-deficient cultures, p63 levels in differentiated cultures were elevated when compared with controls (Fig. 4E), consistent with p63 distribution in PKD2-deficient epidermis (Fig. 4B).
Given the opposing effects of PKD2 and PKD3 on KC proliferation, we next determined whether loss of both isoforms may partially reverse the proliferation or differentiation defect induced by PKD3 silencing. NHKC were co-transduced with lentiviruses encoding PKD2-shRNA/Puro and those encoding PKD3-shRNA/Neo and selected in media containing puromycin and G418 to silence both PKD2 and PKD3 (Fig. 4E). As a control, NHKCs were co-transduced with lentiviruses encoding Scr-shRNA/Puro and Scr-shRNA/Neo. Subconfluent cultures of NHKCs deficient in both PKD isoforms displayed morphological features and growth behavior similar to that observed for PKD3 knockdown KCs (Fig. 4F). However, when grown to post-confluence to induce differentiation, double PKD knockdown cultures failed to stratified and differentiate (Fig. 4F). Consistent with these observations, Western blot analysis of sub-confluent and post-confluent cultures of PKD2&3-deficient KCs verified comparable levels of markers of proliferation (p63) and early differentiation (INV) to those of PKD3 knockdown KCs. However, expression of a marker of terminal differentiation (LOR) was further suppressed in double PKD knockdown KCs (Fig. 4E). These data indicated a predominant growth regulatory role for PKD3 in epidermal progenitor population, and suggested some overlapping or compensatory functions for PKD2 and PKD3 in proliferating compartment of human epidermis. Overall, the progressive nature of proliferation defect, loss of clonogenicity, diminished tissue regenerative potential and the low levels of p63 in PKD3-deficient cells are consistent with failure of self-renewal of KC progenitor cells and suggest an essential pro-proliferative role for PKD3 signaling in human epidermis.
PKD3 Silencing Induces p15INK4B Expression and a G1 Cell Cycle Arrest
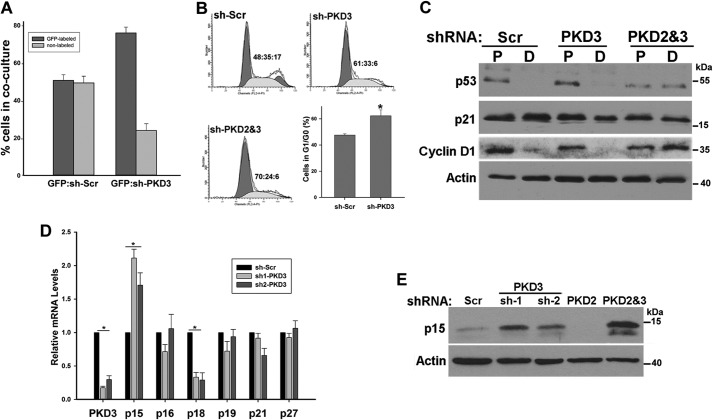
PKDs are involved in regulation of vesicle transport (26, 27). More recently, PKD3 has been shown to regulate prostate cancer cell growth through modulation of secretion of key tumor promoting factors (28). Therefore, the requirement for PKD3 in NHKC proliferation may be due to a paracrine signaling defect mediated by PKD3-dependent secreted factors. To address this question, we determined whether NHKC could rescue the growth defect of PKD3-deficient NHKCs by establishing co-cultures. GFP-labeled NHKC were admixed at a 1:1 ratio with either PKD3-deficient or control NHKCs (non-labeled) and alterations in the ratio of GFPpos:GFPneg cells over a period of 3 days were determined by flow cytometry. After 3 days of culture, the proportion of GFP-labeled cells was at ∼50 and 75% in control and PKD3-deficient co-cultures, respectively (Fig. 5A). The rapid decline in the number of PKD3-deficient KCs indicated that secretion from the normal neighboring KCs could not fully rescue their growth defect.
FIGURE 5.
PKD3 silencing leads to p15INK4B up-regulation and G1 cell cycle arrest. A, 1 × 105 GFP-labeled NHKCs were mixed with equal number of transduced NHKCs expressing either Scr-shRNA or PKD3-shRNA and grown in culture for 3 days. Cells were collected, and the percentage of GFP-labeled cells in each culture was determined by flow cytometry. B, FACS cell cycle profiles of control or PKD3 or PKD2&3 knockdown NHKCs after selection at 7 days post-transduction. Percent of cells in G1:S:G2/M are indicated in the graph. Bar graph shows the proportion of cells in G1/G0 in the control and PKD3-deficient culture (n = 4). C, Western blot analysis of lysates prepared from transduced NHKCs described in Fig. 4 grown as subconfluent (P) and postconfluent (D) cultures using antibodies recognizing p53, p21, and cyclin D1. D, NHKC transduced with lentiviruses encoding Scr-shRNA or one of the two shRNA targeting different region of PKD3. Total RNA from subconfluent cultures was isolated and analyzed by real time RT-qPCR using primers sets specific to PKD3 or CKIs as indicated at the bottom of the graph. E, cell lysates from two NHKCs stably expressing an shRNA targeted to PKD3 or those targeting to PKD2, and a combination of PKD2 and PKD3-sh1 (PKD2&3) were subjected to Western blot analysis using antibodies specific to p15. Actin is used as a loading control. All data were expressed as mean ± S.D. from at least three independent experiments. *, p < 0.001 versus control.
To provide insights into the nature of proliferation defect induced by PKD3 silencing, cell cycle profiles of NHKC stably expressing either Scr- or PKD3-shRNA were analyzed using flow cytometry. Cell cycle analysis revealed a significant increase in the percentage of cells in G0/G1 and reduced number of cells in S phase and G2/M in PKD3 knockdown cells (Fig. 5B), consistent with a G1 cell cycle arrest. In addition Western blot analysis of PKD3-deficient KCs indicated a marked reduction in cyclin D1 levels consistent with a G1 arrest (Fig. 5C). As expected, combinatorial silencing of both isozymes resulted in accumulation of a larger fraction of cells in G0/G1. However, even in these cultures, there was no sub-G1 peak indicating that loss of PKD signaling did not induce apoptosis in NHKCs suggesting that PKD3 silencing induces growth arrest without affecting cell death (Fig. 5B). Previous studies have shown that knockdown of p63 in NHKC induces a G1 cell cycle arrest that is dependent on p53 function and its target, cyclin-dependent kinase inhibitor (CKI) p21 (29). Although p63 levels in PKD3-deficient KC are significantly reduced, Western blot analysis of PKD3-deficient cells for p53 and p21 did not show significant alteration in levels of these two proteins suggesting a different mechanism (Fig. 5C). In addition, PKD3 silencing in HaCat cells, a human KC cell line with two mutated p53 alleles, which are unable to regulate p21 expression (30) induced a similar growth defect as those observed in NHKCs, suggesting a p53-independent mechanism (data not shown).
To determine the molecular basis for G1-arrest induced by PKD3 silencing, we analyzed the expression profiles of CKIs in NHKCs depleted of PKD3. For these experiments we generated two PKD3 knockdown NHKCs using two independent shRNA targeted to various regions of PKD3 transcript. Total RNA from transduced NHKC stably expressing sh-Scr or either of two sh-PKD3 were isolated and transcript levels of various CKIs were examined by real time RT-qPCR. As shown in Fig. 5D, PKD3 silencing in NHKC using two independent shRNA resulted in a reproducible and significant up-regulation of p15INK4B (Fig. 5D). p15INK4B is known to form a complex with CDK4 or CDK6, and prevents G1 progression (31). Western blot analysis of lysates prepared from subconfluent cultures of PKD3-deficient KCs confirmed up-regulation of p15Ink4B at protein levels (Fig. 5E). P15 levels were highest when both PKD2 and PKD3 were silenced, consistent with a more pronounced growth defect of these cultures. These results identified p15INK4B as a potential mediator of human KC growth arrest induced by PKD3 silencing.
DISCUSSION
Most of our current understanding of PKD function in skin is based on characterization of PKD1 in mouse KCs. We have previously shown that although all PKD isoforms are expressed in mouse epidermis, PKD1 plays a unique and critical role in epidermal adaptive responses. In translating these findings to human skin, we discovered that PKD1 is not detectable in human KCs and that there are major differences in PKD isoforms expression, regulation, and function between mouse and human epidermis. Clearly, mouse skin is not a useful model to study PKD functions in human epidermis. These differences are likely reflective of major structural and functional differences between mouse and human skin as well as the context-dependent functions of PKDs (2, 6). Contrary to mouse skin, we found PKD signaling to be essential for maintaining human epidermal homeostasis. This is at least partly related to the divergence in regulation and function of PKD3 between mouse and human KC. Moreover, our findings indicate that PKD-2 and -3 in human KCs have contrasting growth regulatory roles, possibly interacting with different signaling pathways or operating in different KC subpopulations. Our studies underline the importance and complexity of PKD signaling in human KCs and suggest a central role for PKD3 signaling in regulating human epidermal homeostasis.
Previous studies have reported a correlation between PKD1 expression and proliferation status of human epidermis suggesting a proproliferative and anti-differentiating role for this PKD isoform (4). Our studies suggest that these immune-histochemical studies may have been misinterpreted due to the cross-reactivity of anti-PKD1 antibodies to other proteins in human KCs. Despite this, our work supports a proproliferative role for PKD signaling in human epidermis; however, this role is complex and mediated specifically by PKD3, a lesser known member of PKD family. Although little is known about PKD3 signaling in normal tissues, PKD3 has recently been shown to contribute to prostate and breast cancer cell growth and survival, at least partly through regulation of secretion of key tumor promoting factors (28, 32, 33). Even though the precise mechanism of growth arrest induced by PKD3 silencing in NHKCs remains to be elucidated, the inability of normal KC to rescue this proliferation defect in co-cultures suggests that PKD3 regulates KC proliferation through cell-autonomous mechanisms likely by transducing growth promoting signals from neighboring cells or the growth medium. The progressive nature of proliferation defect, lack of clonogenicity and the low tissue regenerative potential of PKD3-knockdown NHKCs are consistent with a critical role for PKD3 signaling in maintaining self-renewal of progenitor cells (25, 29). More importantly, PKD3 is down-regulated in differentiated human KCs consistent with a pro-proliferative role for this isoform. In epidermis, when basal layer KCs commit to differentiation, they undergo cell cycle arrest and migrate outward to differentiate. However, given that PKD silencing does not induce premature differentiation, down-regulation of PKD3 is likely required for the KCs to exit the proliferative compartment, not to induce differentiation.
Our findings indicated that PKD3 silencing in NHKCs coincide with a marked reduction in p63, a major regulator of proliferation and differentiation in epidermis (29, 34). Similar to p63, PKD3 is predominantly expressed in the proliferative compartment of epidermis and is down-regulated in differentiated KCs. Additionally, knock down of either PKD3 or p63 leads to G1 cell cycle arrest. However, p21 has been identified as a major mediator of G1 arrest induced by p63 silencing (29), whereas p21 transcript or protein levels are not altered in PKD3 knockdown NHKCs. We have identified p15INK4B as a potential mediator of growth arrest induced by PKD3 silencing. p15INK4B is known to inhibit cyclinD-CDK4/6 complex formation and RB phosphorylation, thus resulting in G1 cell-cycle arrest (31). p15INK4B is expressed at very low levels in proliferating KC, is up-regulated upon differentiation (35) (data not shown) and is a target of TGFβ-induced growth arrest (36). More recent studies have shown up-regulation of both p15 INK4B and p21 in response to the knockdown of p63 or Myc in human KCs (37) suggesting a critical role for p15INK4B in regulating KC growth arrest. Therefore, mitogenic signaling transduced by PKD3 is likely to be critical in suppressing p15INK4B expression, hence maintaining KCs in the proliferative compartment of epidermis. In addition, dramatic reduction in p63 levels that follows PKD3 silencing suggests that a role for PKD3 signaling in p63 stabilization. Because of key roles of p63 in cell growth and proliferation, apoptosis and differentiation, p63 levels are tightly regulated, mainly by the ubiquitin-dependent proteasomal degradation pathway (38). Recent studies have shown that degradation of p63 during KC differentiation by E3 ubiquitin ligase requires GSK3 kinase activity (39). Given that GSK3 is a potential substrate for PKD signaling (40), it is tempting to speculate that PKD3 signaling may stabilize p63 in proliferative KCs through negative regulation of GSK3.
A major finding of our studies is contrasting growth regulatory roles of PKD2 and PKD3. Despite sharing high sequence homology, PKDs show isoform-specific functions based on their structural differences, levels of expression, tissue specificity, and their interacting proteins (6). Although PKD2 and PKD3 are both expressed in proliferating NHKCs, our loss-of-function studies indicated distinct growth regulatory functions for these two isoforms. This is likely due to structural differences between PKD2 and PKD3 and their interactions with distinct signaling pathways regulating proliferation in human KCs. PKD1 and PKD2 share higher homology in their structure than with PKD3, which lacks some regulatory elements including the N terminus hydrophobic domain or the C terminus PDZ binding motif (41) and contains divergent pleckstrin homology and C1 domain (42, 43). Contrary to our findings in normal cells, recent studies in cancer cell lines have suggested coordinated functions of PKD2 and PKD3 in promoting cancer cell growth and invasion (33, 44). This apparent contradiction in PKD2 signaling in normal and cancer cells could be explained by the fact that cancer cells resist inhibitory signals that might otherwise stop their growth (45). It is worth noting however, that despite opposing growth regulatory roles of PKD2 and PKD3 in NHKCs, the higher magnitude of growth inhibition in these cells following pharmacological inhibition of pan PKD signaling or combinatorial knockdown of PKD2 and PKD3 suggests some overlapping or compensatory functions for PKD2 and PKD3 in proliferating compartment of human epidermis.
In summary, we have identified a complex and critical role for PKD signaling in maintaining homeostasis in human epidermis. Given the importance of PKD1 signaling in mediating adaptive responses of mouse epidermis, we expect unique functions for PKD2 and PKD3 in modulating adaptive responses of human epidermis. Determining the interacting signaling pathways and direct downstream effectors of these PKD isoforms in regulating growth and differentiation of human epidermis is of great interest for future studies and will have therapeutic value in treating skin hyperproliferative disorders. It is therefore of high importance that we make clear the role of PKD isoforms in a relevant model to human skin early on, so that future translational strategies will be more effective.
Acknowledgments
We thank Dr. M. Simon for constructive discussion and criticism of the study, and Kamil Alzayady, Megan Smead, and Ninche Alston for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R21AR056013 (to S. G.).
- PKD
- protein kinase D
- KC
- keratinocyte
- NHKC
- normal human keratinocytes
- Scr
- scrambled
- INV
- involucrin
- FIL
- filaggrin
- LOR
- loricrin
- CKI
- cyclin-dependent kinase inhibitor.
REFERENCES
- 1. Rozengurt E. (2011) Protein Kinase D Signaling: Multiple Biological Functions in Health and Disease. Physiology 26, 23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ellwanger K., Hausser A. (2013) Physiological functions of protein kinase D in vivo. IUBMB Life 65, 98–107 [DOI] [PubMed] [Google Scholar]
- 3. LaValle C. R., George K. M., Sharlow E. R., Lazo J. S., Wipf P., Wang Q. J. (2010) Protein kinase D as a potential new target for cancer therapy. Biochim. Biophys. Acta 1806, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ristich V. L., Bowman P. H., Dodd M. E., Bollag W. B. (2006) Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br. J. Dermatol. 154, 586–593 [DOI] [PubMed] [Google Scholar]
- 5. Guha S., Tanasanvimon S., Sinnett-Smith J., Rozengurt E. (2010) Role of protein kinase D signaling in pancreatic cancer. Biochem. Pharmacol. 80, 1946–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fu Y., Rubin C. S. (2011) Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 12, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steinberg S. F. (2012) Regulation of Protein Kinase D1 Activity. Mol. Pharmacol. 81, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borges S., Storz P. (2013) Protein kinase D isoforms: new targets for therapy in invasive breast cancers? Expert review of anticancer therapy 13, 895–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wille C., Seufferlein T., Eiseler T. (2014) Protein Kinase D family kinases: roads start to segregate. Bioarchitecture 4, 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rozengurt E., Rey O., Waldron R. T. (2005) Protein kinase D signaling. J. Biol. Chem. 280, 13205–13208 [DOI] [PubMed] [Google Scholar]
- 11. Ernest Dodd M., Ristich V. L., Ray S., Lober R. M., Bollag W. B. (2005) Regulation of Protein Kinase D During Differentiation and Proliferation of Primary Mouse Keratinocytes. J. Invest. Dermatol. 125, 294–306 [DOI] [PubMed] [Google Scholar]
- 12. Rennecke J., Rehberger P. A., Fürstenberger G., Johannes F. J., Stöhr M., Marks F., Richter K. H. (1999) Protein-kinase-Cmu expression correlates with enhanced keratinocyte proliferation in normal and neoplastic mouse epidermis and in cell culture. Int. J. Cancer 80, 98–103 [DOI] [PubMed] [Google Scholar]
- 13. Arun S. N., Kaddour-Djebbar I., Shapiro B. A., Bollag W. B. (2011) Ultraviolet B irradiation and activation of protein kinase D in primary mouse epidermal keratinocytes. Oncogene 30, 1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rashel M., Alston N., Ghazizadeh S. (2014) Protein kinase D1 has a key role in wound healing and skin carcinogenesis. J. Invest. Dermatol. 134, 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jadali A., Ghazizadeh S. (2010) Protein kinase D is implicated in the reversible commitment to differentiation in primary cultures of mouse keratinocytes. J. Biol. Chem. 285, 23387–23397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rheinwald J. G., Green H. (1975) Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinocyte colonies from single cells. Cell 6, 331–343 [DOI] [PubMed] [Google Scholar]
- 17. Wu Y. J., Parker L. M., Binder N. E., Beckett M. A., Sinard J. H., Griffiths C. T., Rheinwald J. G. (1982) The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells nonkeratinizing epithelia. Cell 31, 693–703 [DOI] [PubMed] [Google Scholar]
- 18. Ghazizadeh S., Katz A. B., Harrington R., Taichman L. B. (2004) Lentivirus-Mediated Gene Transfer to Human epidermis. J. Invest. Dermatol. Symp. Proc. 9, 269–275 [DOI] [PubMed] [Google Scholar]
- 19. Egles C., Garlick J., Shamis Y. (2010) Three-Dimensional Human Tissue Models of Wounded Skin. in Epidermal Cells (Turksen K., ed), pp 345–359, Humana Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivanova P., Atanasova G., Poumay Y., Mitev V. (2008) Knockdown of PKD1 in normal human epidermal keratinocytes increases mRNA expression of keratin 10 and involucrin: early markers of keratinocyte differentiation. Arch. Dermatol. Res. 300, 139–145 [DOI] [PubMed] [Google Scholar]
- 21. Parsa R., Yang A., McKeon F., Green H. (1999) Association of p63 with Proliferative Potential in Normal and Neoplastic Human Keratinocytes. J. Invest. Dermatol. 113, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 22. Harikumar K. B., Kunnumakkara A. B., Ochi N., Tong Z., Deorukhkar A., Sung B., Kelland L., Jamieson S., Sutherland R., Raynham T., Charles M., Bagherazadeh A., Foxton C., Boakes A., Farooq M., Maru D., Diagaradjane P., Matsuo Y., Sinnett-Smith J., Gelovani J., Krishnan S., Aggarwal B. B., Rozengurt E., Ireson C. R., Guha S. (2010) A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol. Cancer Ther. 9, 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watt F. M. (2014) Mammalian skin cell biology: at the interface between laboratory and clinic. Science 346, 937–940 [DOI] [PubMed] [Google Scholar]
- 24. Carlson M. W., Alt-Holland A., Egles C., Garlick J. A. (2001) Three-Dimensional Tissue Models of Normal and Diseased Skin in Current Protocols in Cell Biology, Chapter 19, Unit 19.9, John Wiley & Sons, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Senoo M., Pinto F., Crum C. P., McKeon F. (2007) p63 Is Essential for the Proliferative Potential of Stem Cells in Stratified Epithelia. Cell 129, 523–536 [DOI] [PubMed] [Google Scholar]
- 26. Hausser A., Storz P., Märtens S., Link G., Toker A., Pfizenmaier K. (2005) Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIβ at the Golgi complex. Nat. Cell Biol. 7, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bossard C., Bresson D., Polishchuk R. S., Malhotra V. (2007) Dimeric PKD regulates membrane fission to form transport carriers at the TGN. J. Cell Biol. 179, 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LaValle C. R., Zhang L., Xu S., Eiseman J. L., Wang Q. J. (2012) Inducible Silencing of Protein Kinase D3 Inhibits Secretion of Tumor-Promoting Factors in Prostate Cancer. Mol. Cancer Therap. 11, 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Truong A. B., Kretz M., Ridky T. W., Kimmel R., Khavari P. A. (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20, 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lehman T. A., Modali R., Boukamp P., Stanek J., Bennett W. P., Welsh J. A., Metcalf R. A., Stampfer M. R., Fusenig N., Rogan E. M. (1993) p53 mutations in human immortalized epithelial cell lines. Carcinogenesis 14, 833–839 [DOI] [PubMed] [Google Scholar]
- 31. Cánepa E. T., Scassa M. E., Ceruti J. M., Marazita M. C., Carcagno A. L., Sirkin P. F., Ogara M. F. (2007) INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 59, 419–426 [DOI] [PubMed] [Google Scholar]
- 32. Chen J., Deng F., Singh S. V., Wang Q. J. (2008) Protein Kinase D3 (PKD3) Contributes to Prostate Cancer Cell Growth and Survival Through a PKCϵ/PKD3 Pathway Downstream of Akt and ERK 1/2. Cancer Res. 68, 3844–3853 [DOI] [PubMed] [Google Scholar]
- 33. Hao Q., McKenzie R., Gan H., Tang H. (2013) Protein Kinases D2 and D3 Are Novel Growth Regulators in HCC1806 Triple-negative Breast Cancer Cells. Anticancer Res. 33, 393–399 [PubMed] [Google Scholar]
- 34. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 35. Alani R. M., Hasskarl J., Münger K. (1998) Alterations in cyclin-dependent kinase 2 function during differentiation of primary human keratinocytes. Mol. Carcinog. 23, 226–233 [DOI] [PubMed] [Google Scholar]
- 36. Hannon G. J., Beach D. (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371, 257–261 [DOI] [PubMed] [Google Scholar]
- 37. Wu N., Rollin J., Masse I., Lamartine J., Gidrol X. (2012) p63 Regulates Human Keratinocyte Proliferation via MYC-regulated Gene Network and Differentiation Commitment through Cell Adhesion-related Gene Network. J. Biol. Chem. 287, 5627–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li C., Xiao Z. X. (2014) Regulation of p63 protein stability via ubiquitin-proteasome pathway. Biomed. Res. Int. 2014, 175721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galli F., Rossi M., D'Alessandra Y., De Simone M., Lopardo T., Haupt Y., Alsheich-Bartok O., Anzi S., Shaulian E., Calabrò V., La Mantia G., Guerrini L. (2010) MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell Sci. 123, 2423–2433 [DOI] [PubMed] [Google Scholar]
- 40. Shin S., Wolgamott L., Yoon S.-O. (2012) Regulation of endothelial cell morphogenesis by the protein kinase D (PKD)/glycogen synthase kinase 3 (GSK3)β pathway, Am. J. Physiol. Cell Physiol. 303, 743–756 [DOI] [PubMed] [Google Scholar]
- 41. Sánchez-Ruiloba L., Cabrera-Poch N., Rodríguez-Martínez M., López-Menéndez C., Jean-Mairet R. M., Higuero A. M., Iglesias T. (2006) Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J. Biol. Chem. 281, 18888–18900 [DOI] [PubMed] [Google Scholar]
- 42. Rey O., Papazyan R., Waldron R. T., Young S. H., Lippincott-Schwartz J., Jacamo R., Rozengurt E. (2006) The Nuclear Import of Protein Kinase D3 Requires Its Catalytic Activity. J. Biol. Chem. 281, 5149–5157 [DOI] [PubMed] [Google Scholar]
- 43. Anderson G., Chen J., Wang Q. J. (2005) Individual C1 domains of PKD3 in phorbol ester-induced plasma membrane translocation of PKD3 in intact cells. Cell Signal. 17, 1397–1411 [DOI] [PubMed] [Google Scholar]
- 44. Zou Z., Zeng F., Xu W., Wang C., Ke Z., Wang Q. J., Deng F. (2012) PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J. Cell Sci. 125, 4800–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]