Abstract
The crystal structure of a fully glycosylated HIV-1 gp120 core in complex with CD4 receptor and Fab 17b at 4.5 Å resolution reveals 9 of the 15 N-linked glycans of core gp120 to be partially ordered. The glycan at position Asn262 had the most extensive and well-ordered electron density, and a GlcNAc2Man7 was modeled. The GlcNAc stem of this glycan is largely buried in a cleft in gp120, suggesting a role in gp120 folding and stability. Its arms interact with the stems of neighboring glycans from the oligomannose patch, which is a major target for broadly neutralizing antibodies.
Keywords: HIV-1 gp120, glycan shield, role of N262
Introduction
The HIV-1 envelope glycoprotein gp160 (Env) mediates viral entry and consists of a trimeric oligomer containing the receptor binding subunit gp120 and the membrane-fusion subunit gp41. As Env is the only viral protein on the surface of HIV-1 virions, it is the primary target of the antibody response. The majority of neutralizing antibodies are directed towards gp120 as it is more distal to the membrane than gp41 and consequently more accessible for immune recognition. Accordingly, gp120 contains features that facilitate evasion of the immune response such as sequence-variable loops (V1–V5) and a dense network of N-linked glycans (81 on average per trimer).
Although studies with patient sera indicate N-linked glycosylation sites evolve in parallel with the adaptive immune response1, a physical mechanism for their role in immune evasion is poorly understood. One hypothesis proposes that N-linked glycans form a ‘glycan shield’, which provides a barrier that prevents B-cell receptor recognition of the underlying protein surface. The gp120 glycans are weakly immunogenic, because they are host-derived, and therefore can only be recognized in non-self contexts, such as unusual clusters or as glycopeptides2. Nonetheless, a substantial number of broadly neutralizing antibodies (bNAbs) have now been isolated3–6 that recognize clusters of N-linked glycans centered around Asn332 (N332) and Asn160 (N160). Our understanding of glycan shielding is complicated by allosteric effects in which deletion of glycans can affect antibody binding to distal epitopes, likely as a result of changes in glycan processing or packing on the viral surface.1,7 Thus, the interest in structural characterization of an intact glycan shield to elucidate its physical properties as well as its role in immune evasion and recognition.
Unfortunately, crystallization of glycoproteins can be challenging due to the heterogeneity and conformational flexibility of glycans, especially when the glycans make up a large proportion of the protein mass, as here where gp120 is almost ~50% carbohydrate. While crystal structures of glycan-dependent bNAbs bound to partially glycosylated gp120 have been published, a crystal structure of a fully glycosylated HIV-1 gp120 is not available, although a cryo-EM structure of fully glycosylated BG505 Env trimer was recently determined at 5.8 Å 8. Here, we present the crystal structure at 4.5 Å resolution of a fully glycosylated YU2 gp120 core bound to two-domains (D1D2) of CD4 and the antigen-binding fragment (Fab) of antibody 17b. The structure reveals a role for the glycan at residue 262 in both protein folding and stabilization as well as in immune evasion.
Materials and Methods
Protein production and crystallization
The YU2 gp120 core construct used for crystallization contains variable loop deletions, and N- and C-terminal truncations as previously described9. The codon optimized gp120 core sequence was cloned into a phCMV vector and was expressed in 293 Freestyle™ (293F) cells in the presence of 50 μM kifunensine. The supernatant was passed over a F105 affinity column and the eluted protein purified by size exclusion chromatography (Hiload 26/60 Superdex S200 prep grade, Amersham). The two-domain CD4 was produced in Chinese hamster ovarian cells (CHO) and purified following a previously described protocol10. To isolate the Fab fragment, 17b IgG was enzymatically digested with activated Lys-C beads following the vendor’s standard protocol (Pierce). The Fab was further purified through a protein A affinity column followed by size-exclusion chromatography. Protein complexes were assembled by mixing gp120 with ligands at 1:1.2 molar ratio for an hour at room temperature before purification by size exclusion chromatography. Ternary complexes were assembled by sequential addition of CD4 followed by 17b Fab with purification by size exclusion chromatography after addition of each ligand.
Crystallization trials were carried out in 96-well vapour diffusion sitting drop trays (Hampton Research) by mixing 0.1μl of protein with an equal volume of well solution, using the Cartesian Honeybee robotic system (Honeybee Robotics). The Hampton Crystal Screen (Hampton Research), Precipitant Synergy Screen (Emerald BioSystems) and Wizard Screen (Emerald BioSystems) were initially used for crystal screening. Optimization of condition 54 from the Precipitant Synergy Screen yielded diffraction quality crystals in 1.4 M sodium formate, 16.6% PEG 3350, 0.1 M CaCl2, 0.1 M Acetate pH 4.5, at 20°C.
Data collection, structure solution and refinement
Crystals were cryoprotected by brief immersion in 20–30% PEG 3350, with either 20% glycerol, 25–35% ethylene glycol or 15% 2R,3R-butanediol, before being flash-cooled in liquid nitrogen. Data were collected at the ID-22 beamline (SER-CAT) at the Advanced Photon Source, and processed with HKL-200011. The best diffraction to 4.5 Å resolution was obtained for a crystal soaked in 2R,3R-butanediol (Table 1). The data were indexed in space group P43212. The calculated Matthews’ coefficient is 2.53 Å3/Dalton with a 51% solvent content with one gp120/Fab/CD4 complex (MW~115,000 Da) per asymmetric unit.
Table 1.
Data collection and refinement statistics
| Data collection
| |
|---|---|
| Beamline | APS 22ID |
| Wavelength (Å) | 1.000 |
| Space group | P43212 |
| Unit cell parameters (Å) | a = b=171.3 c = 151.9 |
| Resolution (Å) | 47.5-4.5 (4.7-4.5) a |
| Observations | 60,950 |
| Unique reflections | 13,317 (1,321) a |
| Redundancy | 4.6 (4.6) a |
| Completeness (%) | 95.2 (97.3) a |
| <I>/<σI> | 9.0 (2.4) a |
| Rsymb | 0.13 (0.92) a,b |
| Rpimc | 0.07 (0.36) a,c |
|
| |
| Refinement statistics | |
|
| |
| Resolution (Å) | 47.5-4.5 (4.8-4.5) |
| Reflections (work) | 12,621 (2,473) |
| Reflections (test) | 666 (143) |
| Rcryst(%)d | 27.7 (30.1) |
| Rfree(%)e | 32.2 (34.0) |
| Average B-values (Å2) | |
| All proteins | 219 |
| gp120 | 169 |
| 17b Fab | 236 |
| CD4 D1D2 | 177 |
| Glycans | 223 |
| Wilson B-value (Å2) | 249 |
|
| |
| RMSD from ideal geometry | |
|
| |
| Bond length (Å) | 0.003 |
| Bond angles (°) | 0.757 |
|
| |
| Ramachandran statistics (%)f | |
|
| |
| Favored | 97.7 |
| Outliers | 0.0 |
|
| |
| PDB ID | 4RQS |
Numbers in parentheses refer to the highest resolution shell.
Rsym = Σhkl Σi | Ihkl,i − <Ihkl> |/Σhkl Σi Ihkl,I, where Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l, <Ihkl> is the average intensity for that reflection, and n is the redundancy.
Rpim is a redundancy-independent measure of the quality of intensity measurements. Rpim = Σhkl (1/(n−1))1/2 Σi | Ihkl,i − <Ihkl> |/Σhkl Σi Ihkl,I, where Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l, <Ihkl> is the average intensity for that reflection, and n is the redundancy.
Rcryst = Σhkl | Fo − Fc |/Σhkl | Fo | x 100
Rfree was calculated as for Rcryst, but on a test set comprising 5% of the data excluded from refinement.
These values were calculated using Molprobity (http://molprobity.biochem.duke.edu/)
The structure was determined by molecular replacement (MR) using Phaser with high-resolution structures of YU2 core gp120 (PDBID: 4JZW), 2-domain CD4 (PDBID: 2NY1) and 17b Fab (PDBID: 2NY1) as the MR models. Model building was carried out using Coot-0.7 (www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) and refinement was implemented in using Phenix 1.8.1-1168 (www.phenix-online.org). Ramachandran restraints were used during refinement due to the low resolution of the dataset. Final Rcryst and Rfree values are 27.7% and 32.2%. Energy minimized rotamers of high mannose N-linked glycan models were obtained from http://glycam.ccrc.uga.edu/glylib and used as initial models during refinement. Glycan nomenclature and geometry were monitored throughout the refinement with the PDB CArbohydrate REsidue check (PDBCARE) online tool (www.glycosciences.de). Buried molecular surface areas were analyzed with the Molecular Surface Package (http://www.csb.yale.edu/userguides/graphics/msp/msu_local.html) using a 1.7 Å probe radius and standard van der Waals radii12. Fab residues were numbered according to Kabat nomenclature13 and gp120 is numbered using the standard HxBC2 convention.
Results and Discussion
Production and optimization of crystals containing fully glycosylated gp120
Although variable loops V1, V2, and V3 were removed from gp120 core constructs, 15 N-linked glycosylation sites are present on the YU2 gp120 core. These glycans can introduce considerable heterogeneity in the glycoprotein due either to diversity in the glycoforms that can be added by the glycan processing enzymes within the Golgi 14, or to conformational flexibility of the glycans15. To counteract glycoform variability, we expressed gp120 in the presence of 50 μM kifunensine in human embryonic kidney 293 (293T) cells, which yields predominantly GlcNAc2Man9 at the glycosylation sites, in human embryonic kidney 293 GnTI−/− (293S) cells, which results in GlcNAc2Man5-9 glycans, and in Drosophila Schneider 2 (S2) cells, which results in paucimannose glycans. Extensive crystal screening of gp120 from these three systems yielded gp120-cocrystals only when the gp120 was expressed in 293T cells in the presence of kifunensine.
Complexes were assembled to facilitate crystallization, as we sought to minimize the effect of glycan heterogeneity and flexibility by introducing potential crystal packing contacts through the addition of bulky protein ligands. In principle, this approach may also constrain glycan flexibility by limiting motion around the glycan sites proximal to the ligands. Thus, a panel of antibody Fabs that recognize the CD4-binding site (CD4bs) and the CD4-induced (CD4i) epitope, in addition to two-domain CD4, was used to assemble gp120 complexes for crystal screening. Ultimately, diffraction quality crystals grew from a complex containing fully glycosylated YU2 gp120 core (produced in 293T cells in the presence of kifunensine) bound to 17b Fab and D1D2 CD4.
Initial diffraction experiments on tetragonal crystals yielded lower than 6 Å data with streaking of diffraction spots and severe anisotropy along the c* direction that made indexing difficult. For some crystals, five minutes of dehydration extended the diffraction limit to 4–5 Å resolution. One crystal that was briefly soaked in 15% 2R,3R-butanediol after five minutes of dehydration gave the best diffraction and was used for structural analysis. Unfortunately, longer dehydration times, annealing, or chemical cross-linking with glutaraldehyde, did not improve resolution or diffraction quality.
Structure of fully glycosylated HIV-1 gp120
The overall protein structure of fully glycosylated HIV-1 gp120 bound to 17b Fab and D1D2 CD4 closely resembled previously published structures of deglycosylated complexes containing the same ligands (PDBID: 1RZK), although small differences would not be discernable at this low resolution. The main differences were subtle shifts in interdomain dispositions in CD4 and 17b, which could be influenced by crystal packing. As anticipated, these ligands, particularly CD4, participated in numerous protein-protein and protein-glycan crystal contacts around the glycosylated face of gp120 (Fig. S1). Glycan-glycan crystal contacts, previously reported in a fully glycosylated SIV gp120 structure (PDBID: 2BF1), were absent here.
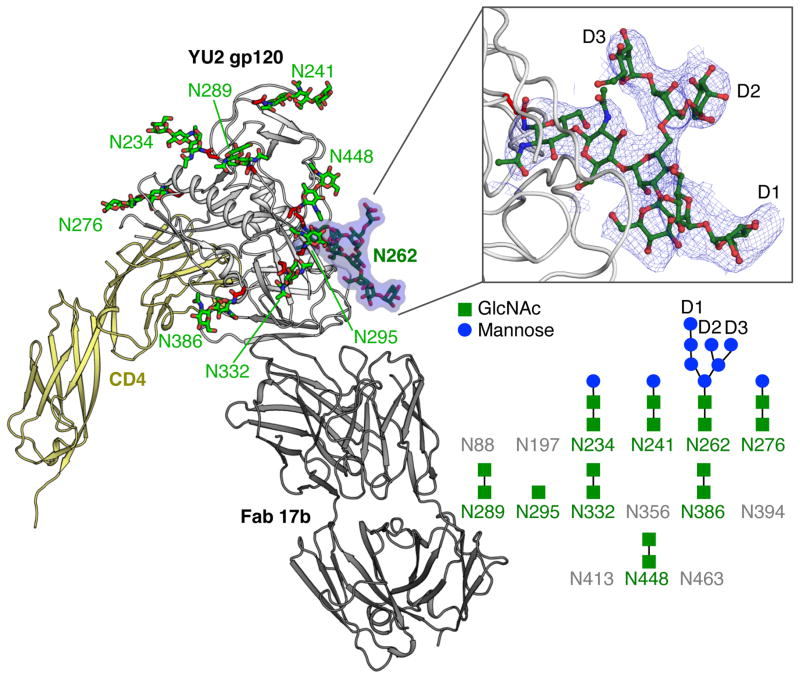
Although details at the side-chain level are not generally well resolved at 4.5 Å resolution, other than perhaps for large aromatic side chains, the bulky glycan core (two N-acetylglucosamines attached to the Asn) from 9 N-linked glycans were visible, as well as most of the glycan attached to Asn262 (Fig. 1). Greater definition of glycan Asn262 was possible because it was wedged into a cleft in gp120 and engaged in crystal contacts with a neighboring symmetry mate. Ring stacking interaction is observed between Pro212 and the first GlcNAc of glycan Asn262. Lack of electron density at other N-linked glycosylation sites is likely due to disorder or heterogeneity, or perhaps from lower levels of glycosylation at these positions.
Figure 1.
Crystal structure of fully glycosylated HIV-1 YU2 gp120. The crystal structure of fully glycosylated YU2 gp120 bound to two-domain CD4 and 17b Fab is shown in cartoon representation. N-linked glycans are displayed in ball-and-stick representation and labelled according to sequence position. The glycan at Asn262 (N262) is highlighted and displayed in more detail in the upper right inset. The blue mesh in the inset represents the 2Fobs-Fcalc electron density map at a 1σ contour level. The glycans on the N-linked glycosylation sites of YU2 gp120 core that can clearly observed in the electron density maps are represented in the lower right. The sites with observable density for glycans are colored green with N-acetyl-glucosamine in green and mannose in blue. Glycosylation sites without any observable density are colored gray. The observed glycan structures are shown schematically. D1-3 indicates the terminal arms of high mannose glycans.
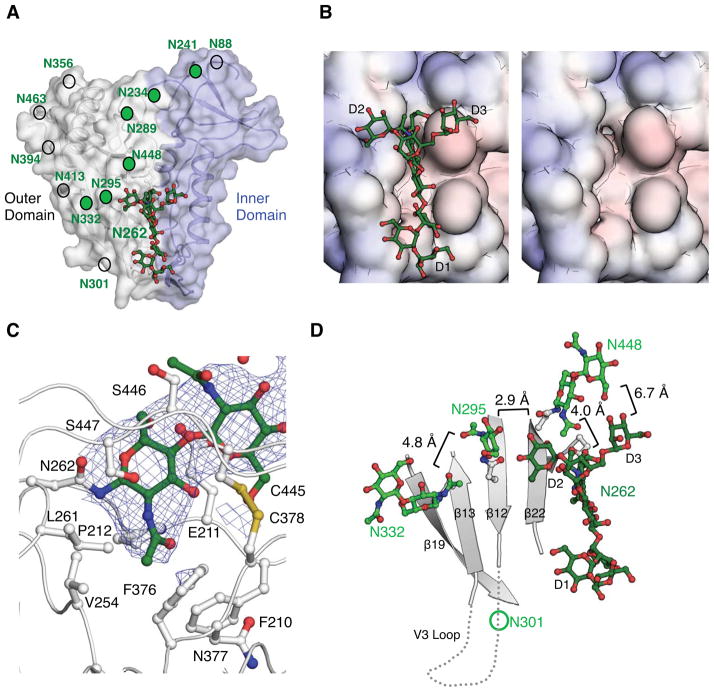
The glycosylation sites on the outer domain of gp120 were distributed on average ~15 Å from each other, as measured from attachment to the Asn residue [Fig. 2(A)]. The oligomannose patch around the glycan site Asn332 exhibited a higher density of glycans, with glycans distributed on average ~10 Å from each other. Considering that the distance between the glycan core to the α-mannose tips of the arms is ~10 Å, some glycan-glycan interactions would certainly possible within this oligomannose patch. However, lack of ordered electron density for these interactions suggested they were mostly weak or absent, with the outer hydrophilic glycan rings likely interacting with solvent16.
Figure 2.
Location and interactions of the Asn262 glycan in gp120. (A) The Asn262 glycan (N262) is shown as green ball-and-sticks in the context of the inner and outer domains of the gp120 core. Visible N-linked glycosylation sites are indicated by green circles and locations of other potential glycosylation sites by non-filled circles. (B) The solvent accessible surface of the weakly negatively charged cleft into which the Asn262 glycan inserts is displayed and colored according to surface electrostatic potential [−10 kT/e (red) to 10 kT/e (blue)] as calculated by the Adaptive Poisson-Boltzmann Solver (APBS). The The cleft is shown with and without glycan. (C) The interaction of Asn262 glycan with the cleft in gp120 around the largely buried GlcNAc core. The blue mesh represents the 2Fobs-Fcalc electron density map at a 1σ contour level. The side chains of protein residues that constitute the cleft around the glycan base are shown in ball-and-stick representation and labelled. (D) The organization of other gp120 glycans in proximity to the Asn262 glycan. The closest distances between the glycans are labelled. The positions of the V3 loop and the Asn301 glycan that are absent in the YU2 gp120 core construct are approximated.
Structure of the N262 glycan
The glycosylation sequon at N262 is 99% conserved, and its removal by mutation significantly reduces HIV-1 viral infectivity17 and gp120 protein expression18, suggesting that the carbohydrate at this site plays an important role in protein folding. These observations were supported by the structure, which revealed the core of the Asn262 glycan is largely buried (155 Å2) within a weakly negatively charged protein cleft [Fig. 2(B)]. This cleft extends nearly all the way along one face of the gp120 protein surface, demarking the border between the inner and outer domains (Fig. 2(A). The cleft is particularly deep around the Asn262 glycan, where the N-acetyl group of the first GlcNAc inserts into the protein surface [Fig. 2(C)]. Thus, the glycan at position Asn262 appears to stabilize gp120 by bridging the inner and outer domains of gp120 and by blocking solvent access to the protein core. Presumably, protein side chains could have evolved to fill this space, but the glycan may play an additional role in allowing potential motions between the inner and outer domains during the membrane fusion process.
The terminal arms of the Asn262 glycan protrude from the protein cleft with the D1 arm extending along but reaching outside the cleft, the D2 arm reaching towards glycan Asn295, and the D3 arm facing the inner domain [Fig. 2(D)]. The glycan conformation may be stabilized by its extensive interaction with gp120 protein, which buries 440 Å2 of the glycan surface. A neighboring symmetry mate makes a few contacts with the tips of the N262 glycan (Fig. S1), but may not be influencing the overall conformation since a similar glycan conformation is observed at N262 for other partially glycosylated gp120 structures19,20. However, the core of the glycan at Asn448 within gp120 is positioned only 4 Å from the stem of the Asn262 glycan [Fig. 2(D)], which likely stabilizes the stem and orientation of the Asn262 glycan in the absence of crystal contacts. Notably, the D2 arm of the Asn262 glycan is only 2.9 Å from the first GlcNAc of the Asn295 glycan, which in turn is only 4.8 Å away from the Asn332 glycan stem. Thus, these glycans can have long range stabilizing effects on each other.
Overall, we observed dense packing of glycans in the oligomannose patch centered on the Asn332 glycan, a target of bNAbs7. The dense packing may affect the orientation of the glycans, and it has been proposed that such unusually dense packing may enhance immune recognition7. Surprisingly, despite the close proximity to this bNAb target around N332 and its high conservation across HIV-1 strains, the Asn262 glycan does not appear to be involved in any epitope recognized by currently available bNAbs.
Conclusion
The 4.5 Å structure of a fully glycosylated gp120 core reveals molecular details of how the glycan shield is organized and uncovers a role for the glycan at Asn262 in stabilizing the gp120 structure. The results described here are consistent with ordered Asn262 glycans observed in the partially deglycosylated x-ray and fully glycosylated EM structures of pre-fusion trimeric HIV-1 Env8,19,20. The study further confirms that crystallization of highly glycosylated proteins can be facilitated by controlling the expression system to produce limited glycan types and by introducing ligands to increase non-glycan surfaces for crystal lattice formation.
Supplementary Material
Acknowledgments
Grant Sponsor: NIH grants RO1 AI084817 (I.A.W), Scripps CHAVI-ID UM1 AI100663 (I.A.W.), GM U54 GM94586 (I.A.W.), and the Intramural Research Program of the Vaccine Research Center, NIAID/NIH.
We are grateful to A. Kim for assistance with core gp120 production and to members of the Wilson lab for help, especially R.L. Stanfield for help with X-ray data processing, A. Sarkar for useful discussions on glycan chemistry and conformation, and J.P. Verenini for help in manuscript formatting. We thank the AIDS Reagent Database for CD4 and J. Robinson for 17b antibody. X-ray data sets were collected at the Advanced Photon Source beamline 22-ID. Use of sector 22 (Southeast Region Collaborative Access Team) was supported by the US Department of Energy, Basic Energy Sciences, Office of Science under contract number W-31-109-Eng-38. This work is supported by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH, and by grants from the Skaggs Institute (I.A.W.), from the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium, and from the NIH: GM U54 GM94586 (to I.A.W.), R01 AI084817 (to I.A.W.), and the Scripps CHAVI-ID UM1 AI100663 (I.A.W.). L.K. is grateful to the American Foundation for AIDS Research for a Mathilde Krim Fellowship in Basic Biomedical Research. The content is the responsibility of the authors and does not necessarily reflect the official views of the NIGMS, NIAID or NIH. This is manuscript #XXX from The Scripps Research Institute. The authors declare no competing financial interests.
Footnotes
Accession Numbers
The structure factors and atomic coordinates of fully glycosylated YU2 gp120 core bound to 17b Fab and 2-domain CD4 have been deposited in the RCSB Protein Data Bank (http://www.rcsb.org/) under PDBID 4RQS.
References
- 1.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 3.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O’Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Program NCS, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Protocol GPI, Koff WC, Wilson IA, Burton DR, Poignard P. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol GPI, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, Scheid JF, Eden C, Mouquet H, Nussenzweig MC, Schwartz O. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med. 2013;210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct & Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 10.Ryu SE, Kwong PD, Truneh A, Porter TG, Arthos J, Rosenberg M, Dai XP, Xuong NH, Axel R, Sweet RW, et al. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature. 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinsowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Connolly ML. The molecular surface package. Journal of molecular graphics. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 13.Martin AC. Accessing the Kabat antibody sequence database by computer. Proteins. 1996;25:130–133. doi: 10.1002/(SICI)1097-0134(199605)25:1<130::AID-PROT11>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Woods RJ, Tessier MB. Computational glycoscience: characterizing the spatial and temporal properties of glycans and glycan-protein complexes. Curr Opin Struct Biol. 2010;20:575–583. doi: 10.1016/j.sbi.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bundle DR, Young NM. Carbohydrate-protein interactions in antibodies and lectins. Curr Opin Struct Biol. 1992;2:666–673. [Google Scholar]
- 17.Huang P, Yu S, Wu C, Liang L. Highly conserved antigenic epitope regions of hemagglutinin and neuraminidase genes between 2009 H1N1 and seasonal H1N1 influenza: vaccine considerations. J Transl Med. 2013;11:47. doi: 10.1186/1479-5876-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.