Abstract
Numerous reports have suggested that immunogenetic factors may influence HIV-1 acquisition, yet replicated findings that translate between study cohorts remain elusive. Our work aimed to test several hypotheses about genetic variants within the IL10-IL24 gene cluster that encodes interleukin (IL)-10, IL-19, IL-20, and IL-24. In aggregated data from 515 Rwandans and 762 Zambians with up to 12 years of follow-up, 190 single nucleotide polymorphisms (SNPs) passed quality control procedures. When HIV-1-exposed seronegative subjects (n = 486) were compared with newly seroconverted individuals (n = 313) and seroprevalent subjects (n = 478) who were already infected at enrollment, rs12407485 (G>A) in IL19 showed a robust association signal in adjusted logistic regression models (odds ratio = 0.64, P = 1.7 × 10−4, and q = 0.033). Sensitivity analyses demonstrated that (i) results from both cohorts and subgroups within each cohort were highly consistent; (ii) verification of HIV-1 infection status after enrollment was critical; and (iii) supporting evidence was readily obtained from Cox proportional hazards models. Data from public databases indicate that rs12407485 is part of an enhancer element for three transcription factors. Overall, these findings suggest that molecular features at the IL19 locus may modestly alter the establishment of HIV-1 infection.
Keywords: Africa, HIV-1 infection, IL19, SNP, association, validation
INTRODUCTION
Sexual transmission of HIV-1 infection is an inefficient process1 that is limited in part by a clear selection bias for the transmitted/founder viruses.2, 3 Studies in the past two decades have revealed various host genetic factors as probable determinants of HIV-1 acquisition (seroconversion) after high-risk sexual exposure.4–6 However, with rare exceptions, these findings have proved to be difficult to confirm or replicate. Indeed, recent genome-wide association studies (GWAS) have repeatedly failed to generate consistent findings when HIV-1-exposed seronegative (HESN) subjects are compared with HIV-1-infected individuals in a typical case-control study design.7–11 Notably, the only confirmed variant to confer resistance to HIV-1 seroconversion, a 32-bp deletion in the CCR5 open reading frame (CCR5-Δ32), is not applicable to populations of African and Asian ancestries.4
Several investigations with partially consistent results have highlighted single nucleotide polymorphisms (SNPs) within the interleukin (IL)-10 gene (IL10) on chromosome 1q32.1. In particular, -592A>C (rs1800872T>G), -819T>C (rs1800871A>G), and -1082A>G (rs1800896T>C) in the IL10 promoter have been implicated individually or collectively (as haplotypes) in several cohorts.12–19 As IL-10 is a potent anti-inflammatory cytokine that regulates viral clearance,20 a biological role in HIV-1 acquisition is quite plausible. Fine mapping for IL10 and neighboring loci, including IL19, IL20, and IL24, has been attempted in an African American cohort,19 but causal variants have not been identified. Our work here aimed to close this gap through dense coverage of the IL10-IL24 gene cluster in two distinct African cohorts, under the assumption that fine mapping based on African populations with relatively short haplotype blocks 21–23 can resolve ambiguities and offer new insights.
RESULTS
Overall characteristics of two prospective cohorts available to this study
Within two cohorts of HIV-1 serodiscordant couples from Lusaka, Zambia (1995 to 2012) and Kigali, Rwanda (2005 to 2012),24–28 our work focused on the analyses of three subgroups, i.e., HIV-1-exposed seronegative (HESN) subjects who had sufficient follow-up (≥9 months) for repeated verification of HIV-1 transmission status, HIV-1 seroconverters (SCs) identified during quarterly follow-up visits, as well as HIV-1 seroprevalent subjects (SPs) who were already infected at the time of enrollment. After several layers of quality control (QC), a total of 515 Rwandans and 762 Zambians were available for this study (Table 1). Within each cohort, the three subgroups shared similar demographic features (age and sex), but HESNs had substantially longer follow-up than SCs (P <0.001 in both cohorts) (Table 1), suggesting that their categorization as HESNs was not an artifact of infrequent testing. On the other hand, genital ulcer/inflammation (GUI) as a well-known risk for HIV-1 acquisition28–30 was more frequent in SCs than HESNs (P <0.001 in both cohorts). In subsequent analyses of host genetic variants, statistical models were adjusted for potential confounders whenever possible.
Table 1.
Overall characteristics of 1,277 eligible subjects from two African cohorts.
| Subjects from Kigali, Rwandaa | Subjects from Lusaka, Zambiaa | |||||
|---|---|---|---|---|---|---|
| Baseline characteristics | HESNs | SCs | SPs | HESNs | SCs | SPs |
| No. of subjects | 228 | 61 | 226 | 258 | 252 | 252 |
| Sex ratio (M/F) | 0.97 (112/116) | 1.03 (31/30) | 0.93 (109/117) | 1.12 (136/122) | 0.65 (99/153) | 0.84 (115/137) |
| Earliest enrollment date | Aug. 2002 | Jan. 2002 | Aug. 2002 | Mar. 1995 | Mar. 1995 | Mar. 1995 |
| Latest enrollment date | Mar. 2005 | Feb. 2011 | Jan. 2005 | Jan. 2006 | Jun. 2009 | Jan. 2006 |
| Age at enrollment: mean ± SD | 31.8 ± 7.3 | 30.7 ± 8.9 | 32.5 ± 7.4 | 32.2 ± 8.4 | 28.6 ± 7.5 | 31.8 ± 8.0 |
| Follow-up visitsa: median (IQR) | 16 (12–20) | 4 (3–9) | 14 (10–17) | 18 (10–28) | 8 (4–13) | 13 (6–22) |
| Follow-up time (months)b: median (IQR) | 48 (35–60) | 10 (6–23) | 48 (34–60) | 53 (27–81) | 18 (9–39) | 51 (27–75) |
| Genital ulcer/inflammation (GUI)c: no. (%) | 26 (11.4) | 19 (31.2) | 49 (21.7) | 43 (16.7) | 105 (41.7) | 81 (32.1) |
For this study, each cohort is divided into three subgroups. Abbreviations and the last eligible visit are defined in the text (see Methods).
P <0.001 between HESNs and SCs in each cohort.
As recorded in the last 12 months of follow-up before HIV-1 transmission or end of last visits.
Immunogenetic findings based on both cohorts (screening models)
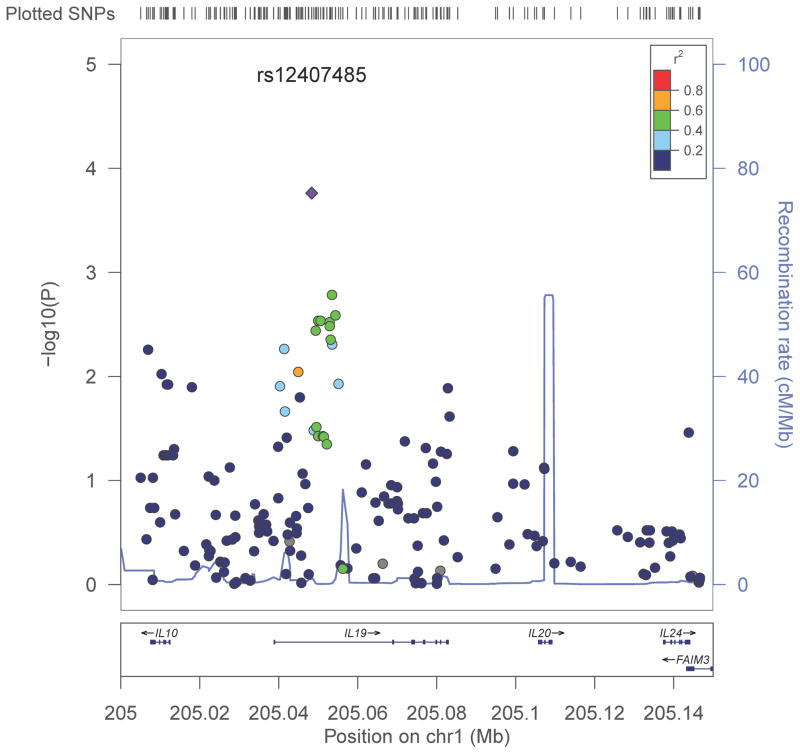
For the genomic region spanning IL10, IL19, IL21, and IL24 (nucleotide positions 205,007,570 to 205,144,106, plus a 3-kb extra region on each side), genotyping using a fine-mapping assay (the ImmunoChip) yielded 189 candidate SNPs in Rwandans and 191 candidate SNPs in Zambians (Tables S1 and S2 in Supplemental Materials), which more than tripled the SNP coverage provided by a gene-centric, custom bead array that helped with generating hypotheses.19 In joint logistic regression models adjusted for sex, age, cohort, and genetic ancestry, QQ plot (Figure S1), Manhattan regional plot (Figure 1), and other summary statistics (Table S3) for the combined dataset (190 candidate SNPs and 1,277 subjects) revealed multiple SNPs of interest. When the P values were corrected for 190 candidate SNPs (assuming dominant effects for minor alleles) or 95 independent tests (eliminating 95 tagging SNPs), only rs12407485, an intronic SNP within IL19, retained statistical significance (Pc = 0.031 or 0.016): the minor allele of rs12407485 was less prevalent in HIV-1-infected subjects (SCs + SPs) than in HESNs (odds ratio = 0.64, P = 1.7 × 10−4, and false discovery rate (FDR) = 0.033) (Tables S3 and S4).
Figure 1.
Manhattan regional plot based on analyses of 190 candidate SNPs in the IL10-IL24 gene cluster. For the aggregated dataset (515 Rwandans and 762 Zambians combined), P values (the Y-axis on the left) are derived from screening models that assume dominant effects for minor alleles (see text). Recombination rates (Y-axis on the right) and linkage disequilibrium measures (color-coded r2 values) correspond to data from Yoruban Africans (the 1000 Genomes Project). The rs12407485 SNP in IL19 is represented by a diamond.
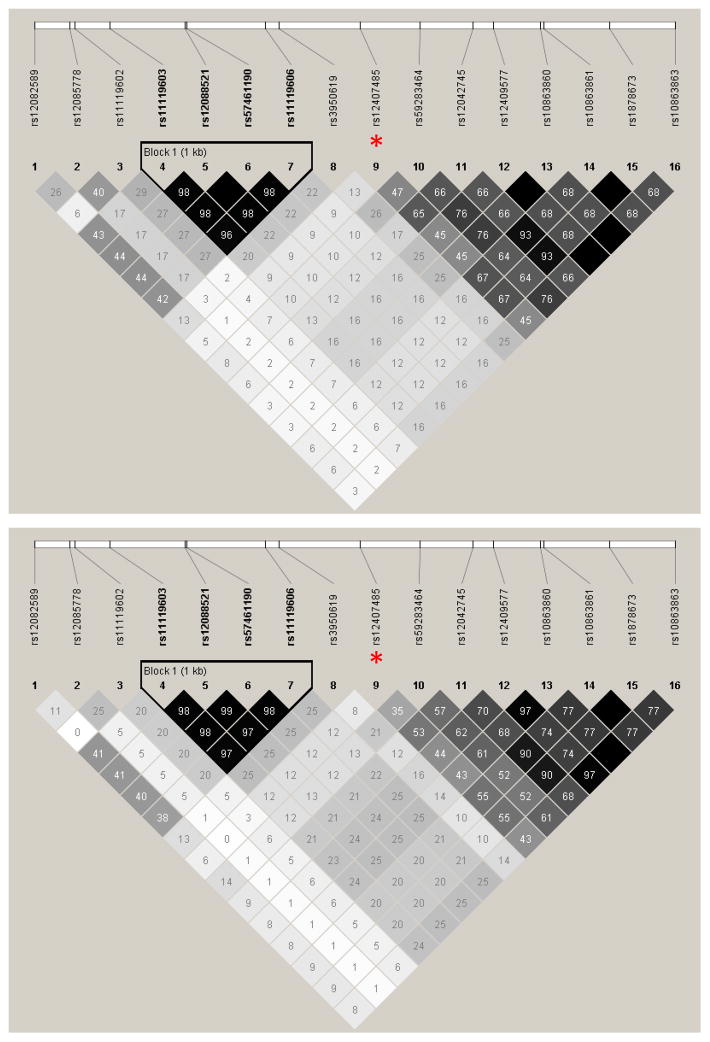
Local haplotype structures
The candidate SNPs used for association analyses did not form any extended haplotypes in either cohort (data not shown). In a 6-kb subregion around rs12407485, the haplotype structures were quite similar between Rwandans and Zambians, regardless of HIV-1 infection status (Figure 2). A single haplotype block containing four SNPs was 871-bp upstream from rs12407485, while the downstream SNPs did not form any blocks. In both cohorts, SNPs that could partially (r2 >0.50) tag rs12407485 included rs12042745, rs10863861, and rs1878673 (r2 = 0.53–0.67) (Figure 2), which largely accounted for the detection of multiple secondary association signals (Table S3). Among frequent haplotypes encompassing rs12407485 and 10 other SNPs (five in each direction), only one had the rs12407485-A allele (Table S5).
Figure 2.
Patterns of linkage disequilibrium (LD) for SNPs around rs12407485 (a non-coding SNP in IL19). Candidate SNPs within a 3-kb region beyond rs12407485 (indicated by asterisks) are analyzed for HIV-1-infected Rwandans (top panel) and Zambians (bottom panel). The pairwise r2 values are boxed. Almost identical results are seen in analyses of HIV-1-exposed seronegative (HESN) subjects.
Confirmatory findings from several sensitivity analyses and multivariable models
The negative association between rs12407485-A and HIV-1 acquisition was highly consistent when analyses were done for separate cohorts and subgroups within each cohort, with ORs ranging from 0.59 to 0.71 (P = 0.007–0.243) when age, sex, and genetic ancestry were treated as covariates (Table 2). The magnitude of effect size attributable to rs12407485-A was persistently modest in both cohorts, with a 39% protection against seroconversion for Rwandans and 36% for Zambians. Statistical power for detecting such minor effects in the study cohorts was adequate (Table S6).
Table 2.
Association of rs12407485 minor allele A with resistance to HIV-1 acquisition, as revealed by adjusted logistic regression models in two separate cohorts.
| rs12407485-A in Rwandans | rs12407485-A in Zambians | |||||||
|---|---|---|---|---|---|---|---|---|
| Subgroups in modela | n | OR | 95% CI | P | n | OR | 95% CI | P |
| HESNs vs. (SCs + SPs) | 515 | 0.61 | 0.43–0.88 | 0.008 | 762 | 0.64 | 0.47–0.88 | 0.007 |
| HESNs vs. SCs | 289 | 0.71 | 0.39–1.27 | 0.243 | 510 | 0.65 | 0.45–0.95 | 0.025 |
| HESNs vs. SPs | 454 | 0.59 | 0.40–0.86 | 0.007 | 510 | 0.63 | 0.44–0.92 | 0.016 |
The three subgroups are defined in Table 1 and text; odds ratio (OR) and 95% confidence interval (CI) have been adjusted for age, sex, and regional genetic ancestry (MDS1-MDS4, see text). OR >1.0 is unfavorable (being seropositive for HIV-1 infection).
Two additional analyses revealed that the negative association of rs12407485-A with HIV-1 acquisition was not apparent if analyses relied on subjects identified as HIV-1 seronegative (HIV−) or seropositive (HIV+) at the time of enrollment (no follow-up) or after two years of regular testing for HIV-1 infection (Table 3). For example, when 478 HIV+ individuals were compared with 799 HIV− subjects at baseline, rs12407485-A was slightly less frequent in the HIV+ group (adjusted OR = 0.71), with a corrected P value of 0.475 (0.005 × 95). The relevance of demographic features (age and sex) and geography to HIV-1 acquisition was not apparent either (P = 0.015–0.742) when the interim analyses were conducted at baseline or two years into follow-up.
Table 3.
Sensitivity analyses: alternative logistic regression models for 1,277 subjects (515 Rwandans and 762 Zambians).
| Baseline model: 478 seropositives vs. 799 seronegativesb | Interim model: 678 seropositives vs. 599 seronegatives-c | |||||
|---|---|---|---|---|---|---|
| Factorsa in model | OR | 95% CI | Adjusted P | OR | 95% CI | Adjusted P |
| Age >40 years | 1.18 | 0.84–1.68 | 0.343 | 0.85 | 0.61–1.20 | 0.363 |
| Female sex | 1.07 | 0.85–1.37 | 0.557 | 1.20 | 0.95–1.51 | 0.129 |
| Country (Rwanda) | 3.50 | 1.27–9.62 | 0.015 | 1.18 | 0.44–3.13 | 0.742 |
| rs12407485-A | 0.71 | 0.55–0.90 | 0.005d | 0.69 | 0.55–0.87 | 0.002d |
As defined in Table 1 and text. Odds ratio (OR) and confidence interval (CI) are further adjusted for genetic ancestry (MDS1-MDS4, see text). OR >1.0 is unfavorable (being seropositive).
At enrollment, only SPs (n = 478) are HIV-1 seropositive, while the other subjects (n = 799) are seronegative.
Up to two years of follow-up after enrollment, with 200 confirmed seroconversion events (as in Figure 3).
P ≥0.19 when corrected for 95 independent tests.
In analyses restricted to HESNs and SCs with confirmed transmission source partners, an expanded multivariable model further indicated that the negative association of rs12407485-A with HIV-1 acquisition was independent of other factors (adjusted OR = 0.52, 95% CI = 0.37–0.75) (Table 4). Indeed the relative effect size (OR) for rs12407485-A improved when viral and host factors applicable to both cohorts were evaluated simultaneously (Table 4).
Table 4.
Multivariable logistic regression model: inclusion of other covariates that are applicable to 708 subjects (HESNs and SCs) from both cohorts.
| Factorsa in model | n | OR | 95% CI | Adjusted P |
|---|---|---|---|---|
| Age >40 years | 91 | 0.53 | 0.30–0.95 | 0.031 |
| Female sex | 370 | 0.94 | 0.65–1.34 | 0.721 |
| Region (Rwanda) | 222 | 0.34 | 0.14–0.86 | 0.023 |
| GUI | 171 | 3.70 | 2.48–5.51 | <0.0001 |
| Donor VL: >100,000 copies/mLb | 257 | 2.08 | 1.42–3.03 | <0.0001 |
| Donor VL: <10,000 copies/mLb | 166 | 0.37 | 0.22–0.61 | <0.0001 |
| HLA-B, P2-Metc | 314 | 1.28 | 0.91–1.81 | 0.152 |
| rs12407485-A | 263 | 0.52 | 0.37–0.75 | <0.001 |
As defined in Table 1 and excluding 24 subjects with missing information for GUI or HIV-1 viral load (VL) in cohabiting index (donor) partners. Odds ratio (OR) and confidence interval (CI) are further adjusted for genetic ancestry (MDS1-MDS4, see text). OR >1.0 is unfavorable (being HIV-1 seropositive).
Plasma VL in known index partners (the reference group has medium plasma VL, i.e., between 10,000 and 100,000 copies/mL).
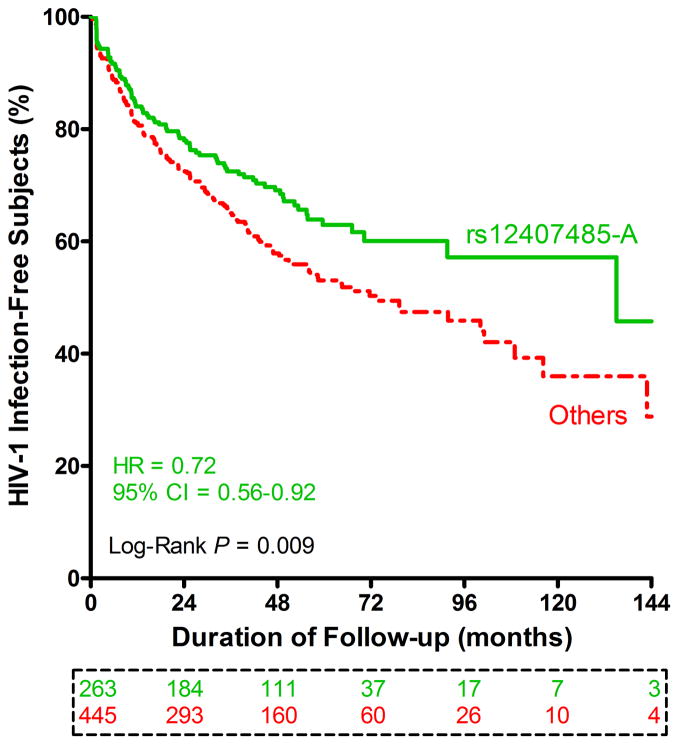
Results from alternative analyses: rs12407485 genotypes and time to seroconversion
In univariable analyses restricted to 222 Rwandans and 486 Zambians who were HIV-1 seronegative at enrollment, individuals with rs12407485-A displayed a slower rate of seroconversion when compared with other subjects (Figure 3). A multivariable Cox proportional hazards model (Table 5) confirmed the independent association of rs12407485-A with delayed HIV-1 acquisition (adjusted hazard ratio (HR) = 0.65, P = 0.001). Previously identified factors associated with HIV-1 seroconversion,28, 30 including genital ulcer/inflammation (GUI) and HIV-1 viral load (VL) in known (cohabiting) index partners, were also independent factors in this select group of subjects (Table 5). Overall, GUI was the most prominent predictor of HIV-1 acquisition (HR = 2.75, P <0.0001), while low VL (<10,000 copies/mL) in index partners was the most favorable factor in the model (HR = 0.42, P <0.0001).
Figure 3.
Immunogenetic influences on HIV-1 seroconversion. Kaplan-Meier curves are based on high-risk subjects (222 Rwandans and 486 Zambians) who are seronegative at enrollment, with stratification by rs12407485 genotypes (GG homozygosity versus GA + AA). Results beyond the first 12 years of quarterly follow-up visits are censored (for lack of remaining subjects). Crude hazard ratio (HR) and 95% confidence intervals (CI) are estimates from a univariable Cox proportional hazards model (no adjustment for other factors). Subjects available at seven major visit intervals are boxed and color coded.
Table 5.
Timing of multifactorial influences on HIV-1 acquisition: Alternative model for at-risk subjects in both cohorts (222 Rwandans and 486 Zambians).
| Factorsa in Cox model | n | HR | 95% CI | Adjusted P |
|---|---|---|---|---|
| Age >40 years | 91 | 0.58 | 0.37–0.90 | 0.016 |
| Female sex | 370 | 0.94 | 0.73–1.21 | 0.648 |
| Region (Rwanda) | 222 | 0.50 | 0.24–1.02 | 0.057 |
| GUI | 171 | 2.75 | 2.15–3.52 | <0.0001 |
| Donor VL: >100,000 copies/mL | 257 | 1.68 | 1.27–2.12 | <0.001 |
| Donor VL: <10,000 copies/mL | 166 | 0.42 | 0.28–0.65 | <0.0001 |
| HLA-B, P2-Met | 314 | 1.27 | 1.00–1.61 | 0.051 |
| rs12407485-A | 263 | 0.65 | 0.50–0.84 | 0.001 |
Insights from bioinformatics analyses
Data from HaploReg revealed that rs12407485 is within an enhancer element with predicted binding affinity for three transcription factors (Table S7 in Supplemental materials). A single tagging SNP, rs17016339 (not covered by the ImmunoChip or by the Illumina 1M Duo chip (Illumina, San Diego, CA, USA)), has been seen in Africans (r2 = 0.99). This intronic SNP is also relevant to gene transcription (Table S7). A second SNP, rs4347211, can partially tag rs17016339 (r2=0.77). Although rs4347211 is covered by the Illumina 1M Duo chip, existing datasets at dbGaP (https://www.ncbi.nlm.nih.gov/gap, last accessed on December 19, 2014) did not include two GWAS based on African natives.
The NCBI Global Cross-database has no existing entries for IL19-related GWAS data or gene expression QTLs (eQTLs), but the SNP and CNV Annotation (SCAN) database indicates that both rs12407485 and rs17016339 are probable trans-acting eQTLs (P = 8 × 10−5 in Yoruban samples) for GARS (chromosome 7p15), a gene that encodes glycyl-tRNA synthetase (Table S8).
The newly established Finemapping Data Portal (also based on the ImmunoChip)31 lists five IL10 SNPs (rs1518110, rs1518111, rs3024493, rs3024495, and rs3024490) that are predicted to be causal variants previously associated with four autoimmune disorders, but neither rs12407485 nor rs17016339 is in strong LD with these IL10 SNPs of interest.
Other observations
Among other SNPs analyzed in this study, rs1800896 (-1082 G>A) and rs1800872 (-592C>A) in IL10 and rs2981572 in IL20 have been highlighted in various studies related to HIV-1 acquisition or disease progression.12, 13, 15–19 The two IL10 SNPs, rs1800896 and rs1800872 (tagged by rs1518111, rs1800871, and three other SNPs in Africans) had borderline statistical significance (P = 0.050 and 0.057, respectively) in screening models (Table S3); these minor associations were readily dismissed by the FDR values (0.242 for both) and by inconsistencies with the directions of reported associations. The IL20 SNP, rs2981572, could not be confirmed either (P = 0.340). Meanwhile, several cohort-specific observations (adjusted P <0.01) were noted, but again the false discovery rates (q ≥0.37) were too high to warrant further attention.
DISCUSSION
In contrast with earlier reports, new data from two African cohorts failed to confirm the potential roles of several IL10 and IL20 SNPs in HIV-1 acquisition. Instead, promising evidence from primary and secondary models persistently and consistently suggested that a non-coding SNP (rs12407485) in IL19 (1q32.2) might be an independent host factor negatively associated with incident and prevalent HIV-1 infection. Although these epidemiologic findings still fell short of a genome-wide significance, the consensus results and their underlying biology are promising in light of existing data from several public databases.
By extending the usual comparison of SCs (incident cases) versus HESNs (contemporary controls)28, 30 to further analyses of HESNs against seroprevalent subjects (SPs) alone or SPs combined with SCs, we were able to demonstrate that the immunogenetic relationships seen with rs12407485 in IL19 were remarkably similar in all analyses (e.g., Table 2 and Table 4). These separate and joint analyses of seroincident and seroprevalent HIV-1 infection may benefit future studies as identification of pathogenetic mechanisms relevant to HIV-1 prevention remains critical in many parts of Africa where treatment as prevention32 is still unaffordable and where internal and external validation of immunogenetic relationships is quite rare.
Our findings here can attest to the daunting challenges presented by an evolving and relative trait such as resistance to HIV-1 acquisition. In populations with high rates of HIV-1 transmission, the HIV-1 infection status of seronegative subjects can change rapidly (in months), which should mandate the need for longitudinal testing before resistance to HIV-1 infection is ascertained.33 Unfortunately, most studies can only compare HIV-1-infected subjects with HIV-1 seronegative (HIV-) individuals in a typical case-control study design, often without reference to (i) the characteristics of source/donor partners, (ii) assessment of exposure based on follow-up data (especially biological evidence), and (iii) verification of HIV-1 infection status in the initially HIV- subjects. Confirmation of HIV-1 infection through repeated testing is particularly important as positive serology took months to develop after acute infection.34 Our study was able to overcome these apparent deficiencies and further established that even an extra two years of follow-up after enrollment could have led to false negative findings (Table 3). Findings based on the Multicenter AIDS Cohort Study also suggested that ascertainment of high risk for HIV-1 infection after enrollment is critical to the identification of host genetic factors that delays or prevents HIV-1 acquisition.35
Lack of supporting evidence form earlier efforts7–10 might also reflect their inability to target enough IL19 SNPs or infer IL19 genotypes based on known genetic structure (Table S7). The rs12407485 SNP in IL19 is a case in point: it was not covered by previous GWAS arrays; its only tagging SNP (rs17016339) was overlooked by GWAS as well (Table S3 and Table S7). Indeed, many causal SNPs identified by the ImmunoChip are beyond the scope of routine genotyping for GWAS because they are typically distant (~14-kb away) from coding sequences.31 By focusing on immune response genes, especially regions with unambiguous genetic associations from independent genome-wide scans and meta-analyses, the ImmunoChip is advantageous in terms of locus-specific SNP coverage and fine mapping.31
Non-genetic factors (e.g., co-infections) that mediate HIV-1 acquisition36 are expected to obscure modest, immunogenetic contributions, especially since risk for exposure can evolve (e.g., the motivation for child-bearing diminishes as couples age). A prerequisite to any claim for a determinant of HIV-1 resistance is the proof of adequate exposure.3, 37 Frequent diagnosis of genital ulcer/inflammation in our cohorts provided clear evidence that high-risk factors did exist in HESNs and their partners (Table 1). In index partners and confirmed transmission source partners, HIV-1 viral load was another important parameter that is known to improve statistical modeling (Table 5), although not all subjects (e.g., SPs) could be analyzed this way. These practical issues can be addressed in cohorts with frequent testing and adequate exposure.
As far as rs12407485 (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=12407485) in IL19 is concerned, its novel association with resistance to HIV-1 acquisition may reflect the importance of various immunoregulatory cytokines, including IL-10 that controls the clearance of viral, bacterial, and parasitic infections.20, 38 Many pathogens produce IL-10-like molecules to subvert host immunity and maintain latency.39 IL-19 itself is closely related to IL-10 in terms of amino acid sequence homology and function, and both are expressed by a wide range of cell lineages to suppress inflammation and cell adhesion40, 41 or to facilitate tissue repair,42, 43 usually in the contraction phase of immune responses. A connection between rs12407485 and cytokine-mediated, immunoregulatory functions is probable because rs12407485 variants are predicted to alter an enhancer-like sequence motif.
The rs12407485 locus is already known to be a trans-acting eQTL for the glycyl-tRNA synthetase gene (GARS). In African populations, the lone tagging SNP (rs17016339) for rs12407485 is another probable trans-acting eQTL for GARS, with multiple functional properties in cell lines. These eQTL relationships can be more complex than expected, as the tRNA synthetase gene family is notorious for splicing products that commercial gene expression arrays cannot distinguish.44 Follow-up studies may need to take advantage of high-resolution techniques, especially RNA-Seq, to fully elucidate the potential function of rs12407485 or its tagging SNP (r17016399) as a trans-acting eQTL for GARS (http://www.ncbi.nlm.nih.gov/gene/2617).
In other epidemiologic studies, IL19 is one of the 139 genes in the human genome that have been associated with virus diversity in human populations,45 and several IL19 gene variants have been associated with ulcerative colitis, a disease that is characterized by excessive inflammation of intestinal mucosa.46 The two SNPs identified in a Japanese cohort, rs2243188 (intronic) and rs2243193 (3′ UTR), have minor alleles that appear to be protective, but both actually tag many functionally relevant loci in Asians, some of which contribute to enhancer-related sequence motifs as well. A second study47 based on analyses of Europeans has identified an IL19 haplotype as being protective against palmoplantar pustulosis, a skin disease caused by chronic inflammation. If the importance of IL-19 to skin and mucosal health is generalizable, manipulation of IL-19 expression could be an attractive option for intervention.
PATIENTS AND METHODS
Study populations
Subjects available to this study were members of HIV-1 discordant couples assembled by the Rwanda-Zambia HIV-1 Research Group between 1995 and 2012 for voluntary testing and counseling (VTC). The overall study design and quarterly follow-up strategies have been described in detail elsewhere.24–26, 30, 48 For this study, analyses focused on comparing HESNs versus SCs and SPs with adequate follow-up and DNA samples for genotyping (Table 1). To minimize the potential bias for phylogenetically related viruses24, 25 among the HIV-1 infected subjects (SCs and SPs), SPs known as transmission source partners (donor partners for SCs) were excluded, although their characteristics, especially GUI and VL,30, 48 were treated as covariates in multivariable models whenever possible. A small number of SCs with unknown source partners (viruses not confirmed in suspected source partners) were not analyzed for timing of HIV-1 infection after enrollment, as several factors in donor partners have strong influences and thus must be treated as covariates.30, 48
Ethical aspects
The research outlined in this study was approved annually by the institutional review boards at clinical sites in Africa (Lusaka, Zambia and Kigali, Rwanda) and two collaborating institutions in the United States (Emory University and University of Alabama at Birmingham). All subjects (legally married adults) gave written informed consent for participation in VTC and related research activities. Interventional strategies for combination antiretroviral therapy (cART), medical male circumcision, use of condoms, and other applicable measures (e.g., diagnosis and treatment of sexually transmitted infections) followed national or regional guidelines whenever possible. All visits beyond cART initiation, either self-reported or confirmed by presence of antiretrovirals in blood samples, were excluded from analyses.
Factors already known to mediate HIV-1 transmission
Various interim analyses demonstrated that heterosexual transmission in both cohorts was associated with several donor (index) and recipient (initially seronegative) characteristics,24, 27, 29, 30, 48 including: (i) GUI in both partners, (ii) plasma VL in index partners (SPs), and (iii) male circumcision. Several host genetic factors from classic human leukocyte antigen (HLA) genes and the KIR2DS4 locus have been reported for the Zambian cohort,27, 28, 30, 49 but so far only the dimorphism for position 2 (P2) of the 9mer HLA-B leader peptide has been shown to impact HIV-1 acquisition in both cohorts when time to HIV-1 acquisition was assessed in Cox proportional hazard models.28 Potential confounding by these known factors was considered in multivariable modeling, whenever applicable.
Genotyping using the ImmunoChip (Illumina, San Diego, CA)
Genomic DNA samples from eligible subjects were used for SNP genotyping at a genomics core facility (University of Alabama at Birmingham). SNP alleles were assigned using the joint calling and haplotype phasing algorithm implemented in BEAGLECALL 50. For each cohort, we completed a series of data cleaning and quality control procedures for ~195,000 SNPs on the ImmunoChip, excluding loci based on the following criteria: (i) known duplication, (ii) missingness (call rate <98.5%), (iii) minor allele frequency (MAF) <0.02 in either cohort, or (iv) deviation from Hardy-Weinberg equilibrium (HWE) (P <10−6). In the end, 190 candidate SNPs spanning the IL10-IL24 gene cluster (± a 3-kb region at each end of the cluster) were retained for analyses in both cohorts, while 78 monomorphic loci and ~40 rare SNPs were excluded (Tables S1 and S2 in Supplemental Materials).
Additional data trimming based on ImmunoChip results
Subjects with ImmunoChip data were further evaluated against three additional (and occasionally overlapping) quality control criteria: (i) overall call rates <95%, (ii) failing sex determination (n=0); or (iii) inferred familial relatedness based on kinship coefficients estimated using the KING software package.51, 52 Subjects were selectively excluded in a pair-wise fashion for up to third degree kinship.
Population stratification
To control for population stratification within the study cohorts, we performed a multidimensional scaling analysis (MDS) using a subset of the ImmunoChip SNPs and the KING software package.51 When SNPs mapped to known regions of extended LD within European populations52, 53 were excluded, 34,390 SNPs with pairwise r2 <0.20 were retained for MDS. The top four MDS vectors (MDS1-MDS4) were then used as covariates in subsequent association analyses.
Assessment of linkage disequilibrium (LD) and haplotype structures in study populations
For each cohort stratified by HIV-1 infection status (positive or negative), local haplotypes in the IL10-IL24 gene cluster were inferred using HaploView.54 The ability of individual SNPs to tag adjacent loci was determined by r2 values. To facilitate an alternative strategy for Bonferroni correction of p values and for assessment of statistical power in various analytical approaches (Table S6), the number of independent SNPs within the candidate loci was calculated using SimpleM.55
Screening and statistical modeling of genetic associations with HIV-1 acquisition
To maximize statistical power (Table S6) and to minimize the number of random testing, association analyses began with the analyses of aggregated SNP data from both cohorts, in which HESNs (controls) were compared with SCs + SPs (cases). The PLINK program (v1.07)56 was used to test all candidate SNPs that passed the QC procedures, with an emphasis on consensus results. The accompanying QQ plot and Manhattan regional plot were created using the R package (qqman at http://cran.r-project.org/web/packages/qqman/index.html) and LocusZoom (http://csg.sph.umich.edu/locuszoom/),57 respectively. Recombination rates in the Manhattan regional plot were based on SNP data from Africans (Yorubans) in the International HapMap Project.22 In these screening models, the P values and OR estimates (including 95% CI) for individual SNPs were adjusted for four potential confounders (sex, age, country of origin, and regional genetic ancestry) that were applicable to all subjects (Table S3). Preliminary association signals (P <0.05 after Bonferroni correction) were subjected to three sets of sensitivity analyses, i.e., (i) HESNs versus SCs alone in each cohort, (ii) HESNs versus SPs alone in each cohort, and (iii) multivariable models that further accounted for previously established covariates, including three categories of index partners’ plasma VLs (low, <10,000 copies/mL, medium, 10,000–100,000 copies/mL, and high, >100,000 copies/mL) (not applicable to SCs with unknown transmission source partners).24, 27, 30 Moreover, Kaplan-Meier curves were used to compare time-dependent events of HIV-1 seroconversion during the study interval (a secondary outcome). Using SAS version 9.3 (SAS Institute, Cary, NC). The HRs (mean and 95% confidence interval) for HIV-1 acquisition were based on Cox proportional hazards models, with statistical adjustments for other covariates. Candidate SNPs already reported in the literature, especially in studies of Africans7, 8 and African Americans19 or in fine-mapping for causal variants,31 were also noted, so were SNPs that had FDR <0.20 in the initial screening models (Table S3).
Bioinformatics: surveys of four public databases (last accessed on December 19, 2014)
SNPs displaying association signals in our analyses were first queried in HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) for known LD with SNPs uncovered by the 1000 Genomes Project (regardless of coverage by the ImmunoChip) or annotated by the ENCODE project.58, 59 Additional surveys took advantage of the NCBI Global Cross-database (http://www.ncbi.nlm.nih.gov/), which captures SNPs associated with various human traits, diseases and/or cellular gene expression profiles.60 Further information from the SNP and CNV Annotation (SCAN) Database (http://www.scandb.org/newinterface/index.html) and the Finemapping Data Portal http://www.broadinstitute.org/pubs/finemapping/)31 helped to identify cis- and trans-acting SNPs of functional importance. Overall, these searches were expected to provide a broad and comprehensive view of local genetic structure and SNP functions.
Supplementary Material
Acknowledgments
This work was supported primarily by the National Institute of Allergy and Infectious Disease (NIAID), through R01 AI071906 (and its supplemental award) to R.A.K./J.T. and R01 AI064060 to E.H.. S.A. and E.H. received additional funding from the International AIDS Vaccine Initiative (Protocol C), with further support (UL1 RR025008) from the Clinical Translational Science Award program, National Center for Research Resources. We are grateful to members of the Rwanda-Zambia HIV-1 Research Group for their valuable contributions to patient enrollment. We also thank Paul Farmer, Naw Htee Khu, Hailin Lu, and Travis R. Porter for assistance with sample inventories, genotyping, and data management.
References
- 1.CDC. National Center for HIV/AIDS VH, STD, and TB Prevention. HIV transmission risk. 2014 http://www.cdc.gov/hiv/pdf/policies_transmission_risk_factsheet.pdf.
- 2.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, et al. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191(Suppl 1):S68–77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 5.Lederman MM, Alter G, Daskalakis DC, Rodriguez B, Sieg SF, Hardy G, et al. Determinants of protection among HIV-exposed seronegative persons: an overview. J Infect Dis. 2010;202 (Suppl 3):S333–8. doi: 10.1086/655967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telenti A, McLaren P. Genomic approaches to the study of HIV-1 acquisition. J Infect Dis. 2010;202 (Suppl 3):S382–6. doi: 10.1086/655969. [DOI] [PubMed] [Google Scholar]
- 7.Petrovski S, Fellay J, Shianna KV, Carpenetti N, Kumwenda J, Kamanga G, et al. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS. 2011;25(4):513–8. doi: 10.1097/QAD.0b013e328343817b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lingappa JR, Petrovski S, Kahle E, Fellay J, Shianna K, McElrath MJ, et al. Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS ONE. 2011;6(12):e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limou S, Delaneau O, van Manen D, An P, Sezgin E, Le Clerc S, et al. Multicohort genomewide association study reveals a new signal of protection against HIV-1 acquisition. J Infect Dis. 2012;205(7):1155–62. doi: 10.1093/infdis/jis028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lane J, McLaren PJ, Dorrell L, Shianna KV, Stemke A, Pelak K, et al. A genome-wide association study of resistance to HIV infection in highly exposed uninfected individuals with hemophilia A. Hum Mol Genet. 2013;22(9):1903–10. doi: 10.1093/hmg/ddt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLaren PJ, Coulonges C, Ripke S, van den Berg L, Buchbinder S, Carrington M, et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 2013;9(7):e1003515. doi: 10.1371/journal.ppat.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci U S A. 2000;97(26):14467–72. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasilescu A, Heath SC, Ivanova R, Hendel H, Do H, Mazoyer A, et al. Genomic analysis of Th1-Th2 cytokine genes in an AIDS cohort: identification of IL4 and IL10 haplotypes associated with the disease progression. Genes Immun. 2003;4(6):441–9. doi: 10.1038/sj.gene.6363983. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Song W, Lobashevsky E, Wilson CM, Douglas SD, Mytilineos J, et al. Cytokine and chemokine gene polymorphisms among ethnically diverse North Americans with HIV-1 infection. J Acquir Immune Defic Syndr. 2004;35(5):446–54. doi: 10.1097/00126334-200404150-00002. [DOI] [PubMed] [Google Scholar]
- 15.Shrestha S, Strathdee SA, Galai N, Oleksyk T, Fallin MD, Mehta S, et al. Behavioral risk exposure and host genetics of susceptibility to HIV-1 infection. J Infect Dis. 2006;193(1):16–26. doi: 10.1086/498532. [DOI] [PubMed] [Google Scholar]
- 16.Erikstrup C, Kallestrup P, Zinyama-Gutsire RB, Gomo E, Butterworth AE, Pedersen BK, et al. Reduced mortality and CD4 cell loss among carriers of the interleukin-10 -1082G allele in a Zimbabwean cohort of HIV-1-infected adults. AIDS. 2007;1(17):2283–91. doi: 10.1097/QAD.0b013e3282f153ed. [DOI] [PubMed] [Google Scholar]
- 17.Oleksyk TK, Shrestha S, Truelove AL, Goedert JJ, Donfield SM, Phair J, et al. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10(4):309–22. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naicker DD, Werner L, Kormuth E, Passmore JA, Mlisana K, Karim SA, et al. Interleukin-10 promoter polymorphisms influence HIV-1 susceptibility and primary HIV-1 pathogenesis. J Infect Dis. 2009;200(3):448–52. doi: 10.1086/600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shrestha S, Wiener HW, Aissani B, Song W, Shendre A, Wilson CM, et al. Interleukin-10 (IL-10) pathway: genetic variants and outcomes of HIV-1 infection in African American adolescents. PLoS ONE. 2010;5(10):e13384. doi: 10.1371/journal.pone.0013384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12(11):1301–9. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prentice HA, Pajewski NM, He D, Zhang K, Brown EE, Kilembe W, et al. Host genetics and immune control of HIV-1 infection: fine mapping for the extended human MHC region in an African cohort. Genes Immun. 2014;15(5):275–81. doi: 10.1038/gene.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, et al. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17(10):901–10. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76(1):397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kempf MC, Allen S, Zulu I, Kancheya N, Stephenson R, Brill I, et al. Enrollment and retention of HIV discordant couples in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2008;47(1):116–25. doi: 10.1097/QAI.0b013e31815d2f3f. [DOI] [PubMed] [Google Scholar]
- 27.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008;181(4):2626–35. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merino AM, Song W, He D, Mulenga J, Allen S, Hunter E, et al. HLA-B signal peptide polymorphism influences the rate of HIV-1 acquisition but not viral load. J Infect Dis. 2012;205(12):1797–805. doi: 10.1093/infdis/jis275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song W, He D, Brill I, Malhotra R, Mulenga J, Allen S, et al. Disparate associations of HLA class I markers with HIV-1 acquisition and control of viremia in an African population. PLoS ONE. 2011;6(8):e23469. doi: 10.1371/journal.pone.0023469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015 doi: 10.1038/nature13835. in press (e-Publication ahead of print: October 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young JM, Turpin JA, Musib R, Sharma OK. Outcomes of a National Institute of Allergy and Infectious Diseases Workshop on understanding HIV-exposed but seronegative individuals. AIDS Res Hum Retroviruses. 2011;27(7):737–43. doi: 10.1089/aid.2010.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384(9939):258–71. doi: 10.1016/S0140-6736(14)60164-1. [DOI] [PubMed] [Google Scholar]
- 35.Tang J, Shelton B, Makhatadze NJ, Zhang Y, Schaen M, Louie LG, et al. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J Virol. 2002;76(2):662–72. doi: 10.1128/JVI.76.2.662-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corey L, Wald A, Celum CL, Quinn TC. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr. 2004;35(5):435–45. doi: 10.1097/00126334-200404150-00001. [DOI] [PubMed] [Google Scholar]
- 37.Vince N, Bashirova AA, Lied A, Gao X, Dorrell L, McLaren PJ, et al. HLA class I and KIR genes do not protect against HIV type 1 infection in highly exposed uninfected individuals with hemophilia A. J Infect Dis. 2014;210(7):1047–51. doi: 10.1093/infdis/jiu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couper KN, Blount DG, Wilson MS, Hafalla JC, Belkaid Y, Kamanaka M, et al. IL-10 from CD4CD25Foxp3CD127 adaptive regulatory T cells modulates parasite clearance and pathology during malaria infection. PLoS Pathog. 2008;4(2):e1000004. doi: 10.1371/journal.ppat.1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001;9(2):86–92. doi: 10.1016/s0966-842x(00)01919-3. [DOI] [PubMed] [Google Scholar]
- 40.Romano MF, Lamberti A, Petrella A, Bisogni R, Tassone PF, Formisano S, et al. IL-10 inhibits nuclear factor-kappa B/Rel nuclear activity in CD3-stimulated human peripheral T lymphocytes. J Immunol. 1996;156(6):2119–23. [PubMed] [Google Scholar]
- 41.England RN, Preston KJ, Scalia R, Autieri MV. Interleukin-19 decreases leukocyte-endothelial cell interactions by reduction in endothelial cell adhesion molecule mRNA stability. Am J Physiol Cell Physiol. 2013;305(3):C255–65. doi: 10.1152/ajpcell.00069.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann SR, Rösen-Wolff A, Tsokos GC, Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin Immunol. 2012;143(2):116–27. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Zhou JJ, Wang F, Xu Z, Lo WS, Lau CF, Chiang KP, et al. Secreted histidyl-tRNA synthetase splice variants elaborate major epitopes for autoantibodies in inflammatory myositis. J Biol Chem. 2014;289(28):19269–19275. doi: 10.1074/jbc.C114.571026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Bresolin N, Clerici M, et al. Genome-wide identification of susceptibility alleles for viral infections through a population genetics approach. PLoS Genet. 2010;6(2):e1000849. doi: 10.1371/journal.pgen.1000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto-Furusho JK, Alvarez-Leon E, Fragoso JM, Gozalishvilli A, Vallejo M, Vargas-Alarcon G. Protective role of interleukin-19 gene polymorphisms in patients with ulcerative colitis. Hum Immunol. 2011;72(11):1029–32. doi: 10.1016/j.humimm.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 47.Kingo K, Mossner R, Koks S, Ratsep R, Kruger U, Vasar E, et al. Association analysis of IL19, IL20 and IL24 genes in palmoplantar pustulosis. Brit J Dermatol. 2007;156(4):646–52. doi: 10.1111/j.1365-2133.2006.07731.x. [DOI] [PubMed] [Google Scholar]
- 48.Merino AM, Sabbaj S, Easlick J, Goepfert P, Kaslow RA, Tang J. Dimorphic HLA-B signal peptides differentially influence HLA-E- and natural killer cell-mediated cytolysis of HIV-1-infected target cells. Clin Exp Immunol. 2013;174(3):414–423. doi: 10.1111/cei.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merino A, Malhotra R, Morton M, Mulenga J, Allen S, Hunter E, et al. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis. 2011;203(4):487–95. doi: 10.1093/infdis/jiq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85(6):847–61. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, et al. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet. 2012;8(4):e1002640. doi: 10.1371/journal.pgen.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian C, Kosoy R, Nassir R, Lee A, Villoslada P, Klareskog L, et al. European population genetic substructure: further definition of ancestry informative markers for distinguishing among diverse European ethnic groups. Mol Med. 2009;15(11–12):371–83. doi: 10.2119/molmed.2009.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 55.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32(4):361–9. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38(Database issue):D620–5. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44(5):502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.