Abstract
Mitochondrial DNA (mtDNA) is normally present at thousands of copies per cell and is packaged into several hundred higher-order structures termed nucleoids1. The abundant mtDNA-binding protein, transcription factor A mitochondrial (TFAM), regulates nucleoid architecture, abundance, and segregation2. Complete mtDNA depletion profoundly impairs oxidative phosphorylation (OXPHOS), triggering calcium-dependent stress signaling and adaptive metabolic responses3. However, the cellular responses to mtDNA instability, a physiologically relevant stress observed in many human diseases and aging, remain ill-defined4. Here we show that moderate mtDNA stress elicited by TFAM deficiency engages cytosolic antiviral signaling to enhance the expression of a subset of interferon-stimulated genes (ISG). Mechanistically, we have found that aberrant mtDNA packaging promotes escape of mtDNA into the cytosol, where it engages the DNA sensor cGAS and promotes STING-IRF3-dependent signaling to elevate ISG expression, potentiate type I interferon responses, and confer broad viral resistance. Furthermore, we demonstrate that herpesviruses induce mtDNA stress, which potentiates antiviral signaling and type I interferon responses during infection. Our results further demonstrate that mitochondria are central participants in innate immunity, identify mtDNA stress as a cell-intrinsic trigger of antiviral signaling, and suggest that cellular monitoring of mtDNA homeostasis cooperates with canonical virus sensing mechanisms to fully license antiviral innate immunity.
To explore the cellular responses to mtDNA stress in the absence of OXPHOS deficiency, we employed a TFAM heterozygous knockout (Tfam+/−) mouse model. Cells and tissues from these animals exhibit modest or no significant differences in mtDNA-encoded transcripts and oxygen consumption rates, despite ~50% depletion of mtDNA (Extended Data Fig. 1a–c)5,6. In addition to mtDNA depletion, Tfam+/− mouse embryonic fibroblasts (MEFs) have reduced oxidative mtDNA damage repair capacity and markedly altered mtDNA packaging, organization, and distribution (Fig. 1a)6. Nucleoids in Tfam+/− MEFs were less numerous and exhibited a larger size distribution (Fig. 1a and Extended Data Fig. 1d). Thus, Tfam+/− cells provide a robust model to characterize cellular responses triggered by moderate mtDNA stress.
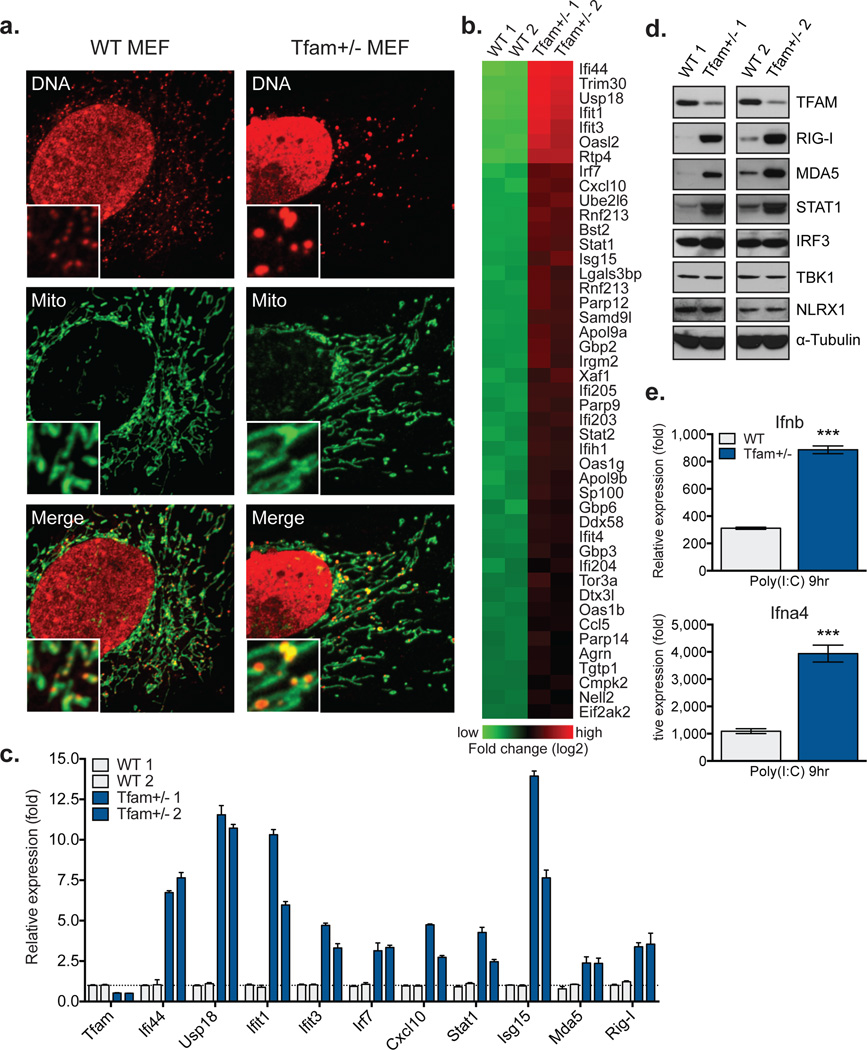
Figure 1. Tfam+/− cells exhibit mtDNA stress, elevated ISG expression, and augmented type I interferon responses.
a, Confocal microscopy images of MEFs stained with anti-DNA (DNA) and anti-HSP60 (Mito) antibodies. b, Heatmaps of microarray analyses. Genes in Tfam+/− MEFs exhibiting statistically significant (p<0.05), two-fold or greater increases over WT are shown. c-d, qRT-PCR (c) and western blots (d) of basal ISG expression in two littermate WT and Tfam+/− MEF lines. e, qRT-PCR analysis of type I interferon expression in MEFs 9 hrs post cytosolic delivery of poly(I:C). Error bars indicate ± s.e.m. of triplicates and are representative of three independent experiments. ***=p<0.001.
Gene expression profiling of Tfam+/− MEFs revealed an unexpected enrichment of interferon-stimulated genes (ISGs) and antiviral signaling factors (Fig. 1b). Of the 45 most over expressed genes, 39 were ISGs, including many with direct antiviral activity (i.e. Ifi44, Ifit1, Ifit3, Oasl2, Rtp4)7,8. There was also increased expression of cytoplasmic RNA and DNA sensors, such as Ddx58 (RIG-I), Ifih1 (MDA5), and p200 family proteins Ifi203, Ifi204, and Ifi205, as well as transcription factors Irf7, Stat1, and Stat2, ISGs that function to positively reinforce the antiviral response. Direct measurement of basal ISG mRNA and protein expression in Tfam+/− MEFs validated the microarray results (Fig. 1c–d). Finally, Tfam+/− MEFs expressed 3–4 fold more Ifnb and Ifna4 upon transfection with the MDA5 agonist poly(I:C) (Fig. 1e), consistent with enhanced type I interferon responses.
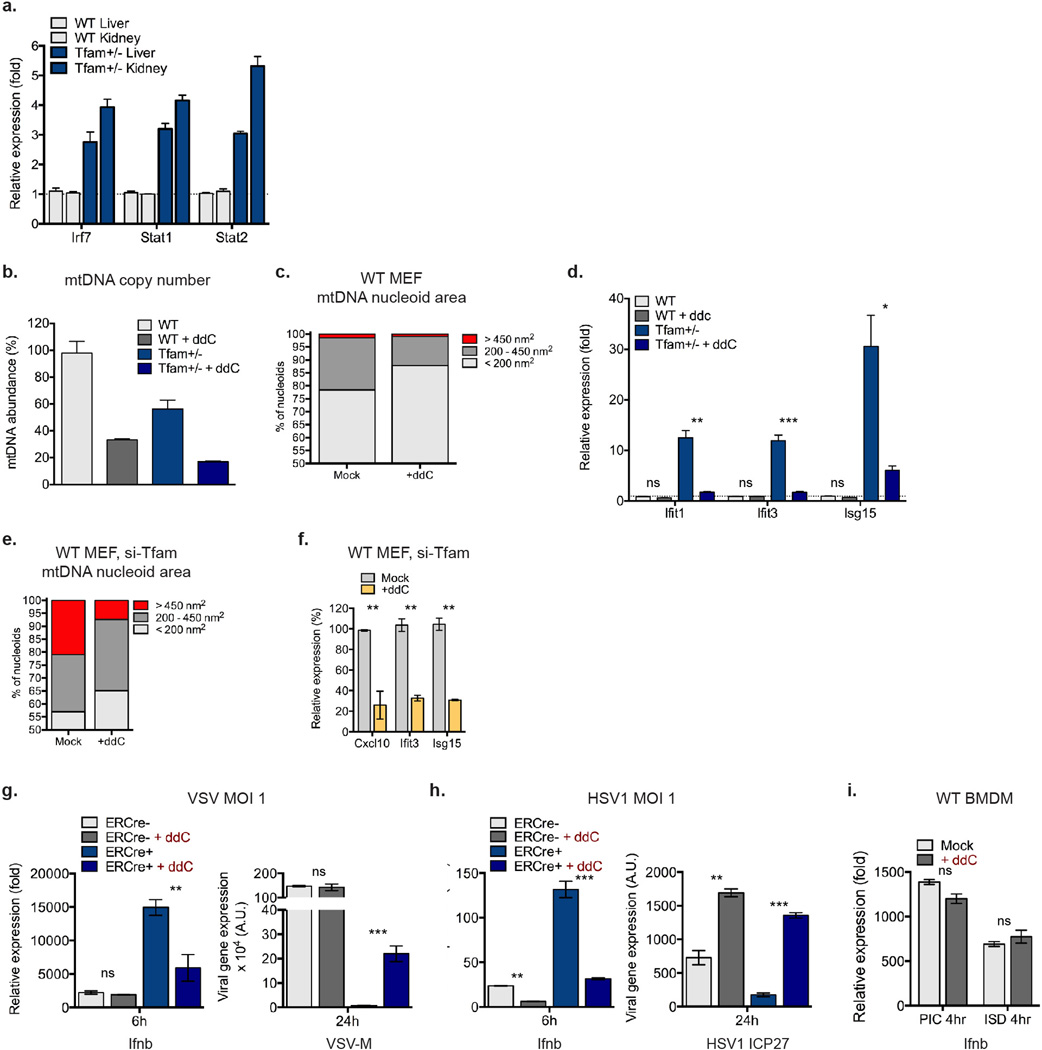
To ensure that the mtDNA stress and ISG expression phenotypes were not unique to Tfam+/− MEFs, we employed inducible TFAM depletion models (TFD). Analogous to Tfam+/− cells, TFD MEFs and bone marrow-derived macrophages (BMDM) displayed mtDNA stress phenotypes, augmented ISG expression, and heightened type I interferon responses to poly(I:C) (Extended Data Fig. 1d–i). Collectively, these data indicate that TFAM depletion induces mtDNA nucleoid stress that triggers antiviral ‘priming,’ characterized by basally elevated ISG expression and potentiated type I interferon production.
Since mitochondrial stress can trigger release of mtDNA into the cytosol to engage the NLRP3 inflammasome, we assayed for extra-mitochondrial mtDNA in TFD cells9,10 . Analysis of pure cytosolic extracts revealed a 3–4 fold increase of specific mtDNA fragments from the D-loop regulatory region, indicating liberation of immunostimulatory mtDNA into the cytosol (Extended Data Fig. 2)11. Confocal and electron microscopy of TFD cells revealed significantly elongated, interconnected mitochondrial networks consistent with a hyperfused phenotype (Fig. 1a and Extended Data Figs. 1e, g, 3a–b). Since mitochondrial fission facilitates proper nucleoid distribution and removal of damaged mtDNA, we examined whether mitochondrial hyperfusion in TFD cells governed mtDNA stress-induced ISG expression12,13. Knockdown of mitofusin-1 (Mfn1) induced fission and largely abrogated ISG expression in TFD MEFs (Extended Data Fig. 3c–e). Moreover, depletion of the mtDNA quality-control enzyme endonuclease G-like 1 (EXOG) exacerbated ISG expression in Tfam+/− MEFs (Extended Data Fig. 3f)14. Collectively, these data indicate that TFAM depletion promotes accumulation of aberrant mtDNA, which accesses the cytosol to engage innate immune signaling.
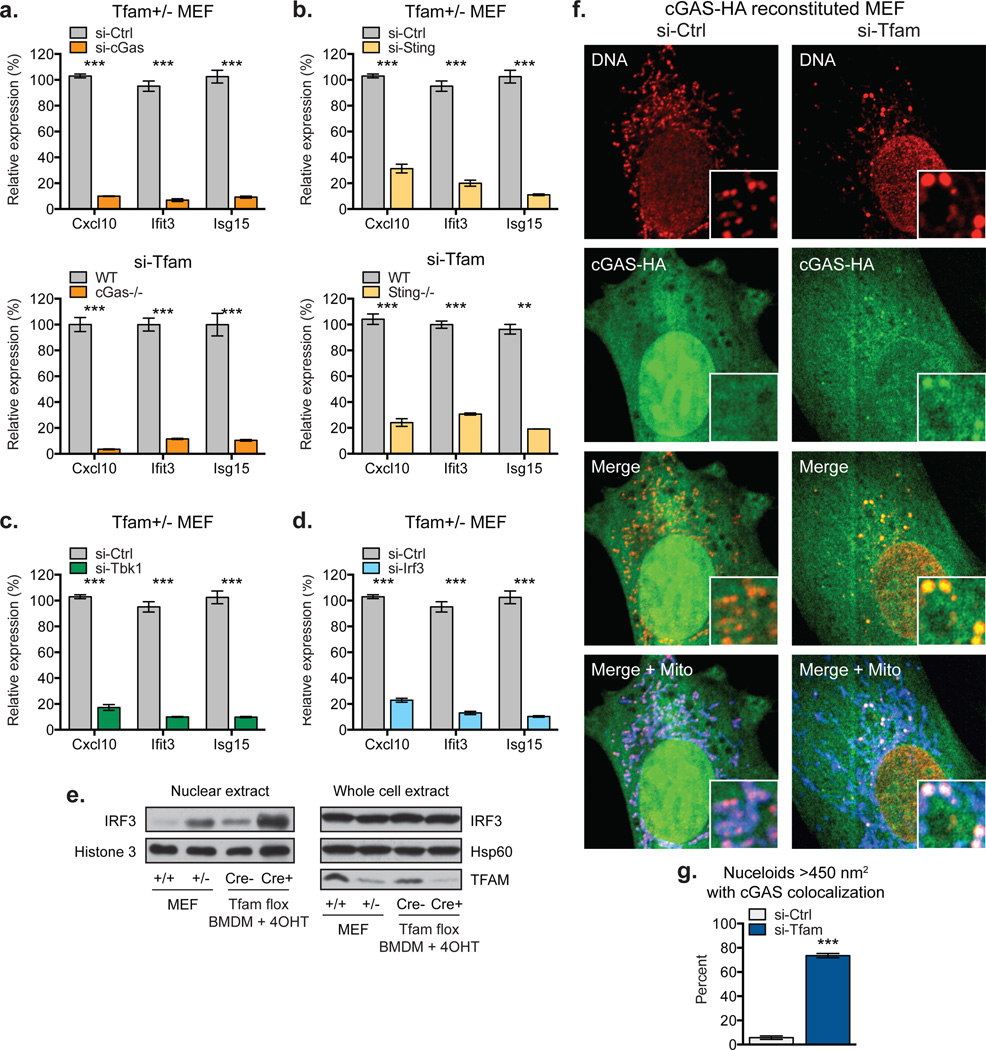
We next examined the involvement of the cytosolic DNA sensor cGAS in mtDNA stress signaling, as it mediates ISG expression in response to exogenous and endogenous immunostimulatory DNA species15–17. Knockdown of cGAS in Tfam+/− MEFs or TFAM depletion in cGas−/− MEFs largely abrogated ISG expression (Fig. 2a). Furthermore, ISG mRNAs in TFD cells were reduced 70–90% in the absence of STING, indicating cGAS-STING signaling is the predominant driver of mtDNA stress-induced ISG expression (Fig. 2b). STING signals via the TBK1-IRF3/7 axis to trigger antiviral gene expression, and knockdown of either TBK1 or IRF3 robustly blocked ISG expression in Tfam+/− MEFs (Fig. 2c–d)18,19. This was accompanied by enhanced nuclear accumulation of IRF3, consistent with IRF3 activating ISG transcription (Fig. 2e). Finally, using murine cGAS reconstituted cGAS−/− MEFs, we observed prominent re-localization of cGAS from nuclear and/or cytoplasmic pools to the vicinity of aberrant mtDNA nucleoids in TFD MEFs (Fig. 2f–g). Taken together, these results indicate that mtDNA stress facilitates cGAS-dependent sensing of cytoplasmic mtDNA, resulting in STING-TBK1-IRF3 signaling to trigger ISG expression.
Figure 2. mtDNA stress triggers ISG expression in a cGAS- and STING-dependent fashion.
a-b, ISG expression in Tfam+/− MEFs transfected with the indicated siRNAs (top panels), or WT, cGas−/− (a), and Sting−/− (b) MEFs transfected with TFAM siRNAs (bottom panels). c-d, ISG expression in Tfam+/− MEFs transfected with the indicated siRNAs for 96 hrs. e, Western blots of whole cell and nuclear extracts of WT and Tfam+/− MEFs or Tfam flox ERCre−/+ BMDM exposed to 4OHT for 96 hrs. f-gcGAS−/− MEFs reconstituted with cGAS-HA were transfected with the indicated siRNAs for 96 hrs, then stained with anti-DNA, anti-Hsp60 [Mito], and anti-HA antibodies and imaged. cGAS colocalization scoring was performed as described in the Methods. Error bars indicate ± s.e.m. of triplicates and are representative of three independent experiments. *=p<0.05, **=p<0.01, ***=p<0.001.
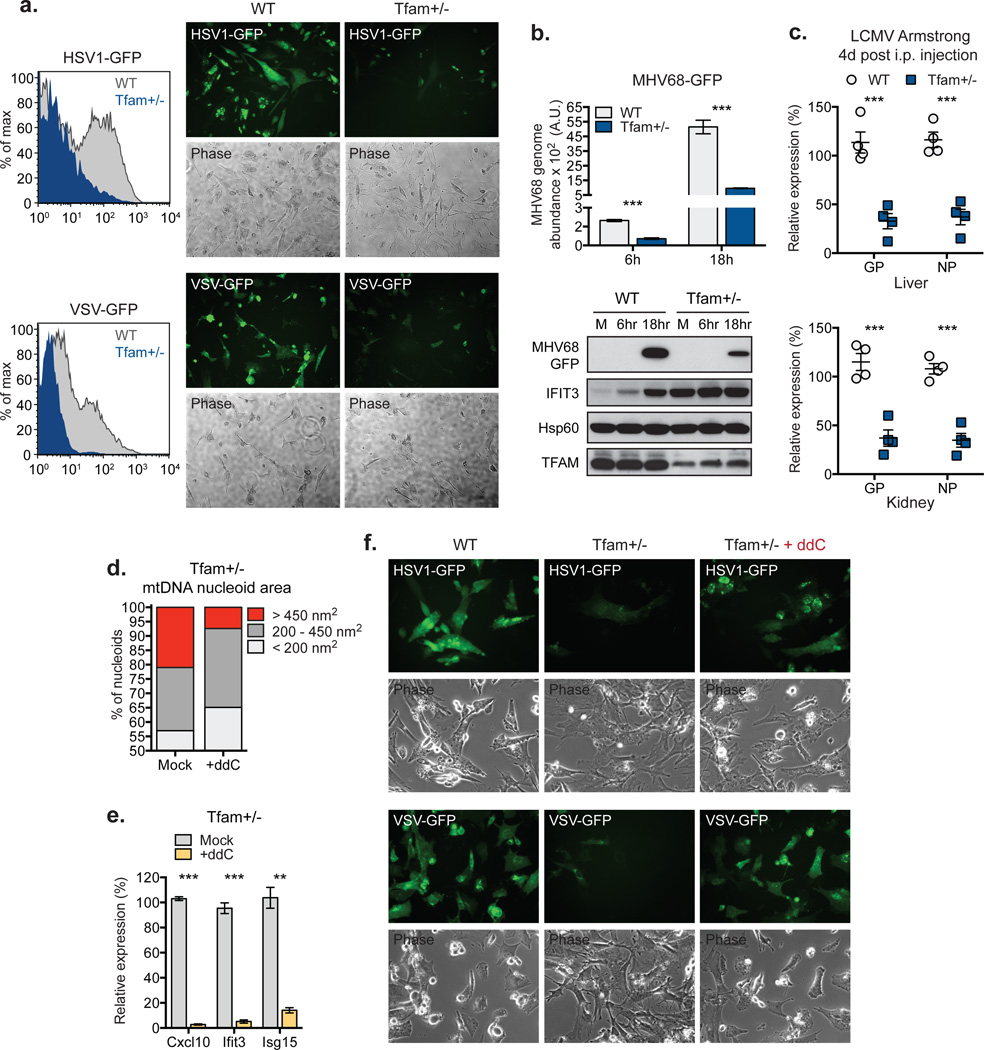
To establish functional significance of mtDNA stress-induced antiviral priming, we challenged MEFs with Herpes Simplex Virus-1 (HSV-1) or Vesicular Stomatitis virus (VSV) that express GFP for easy detection. In contrast to WT cells, which displayed robust viral GFP expression post-infection, Tfam+/− MEFs were markedly resistant to HSV-1 and VSV (Fig. 3a). In addition, Tfam+/− MEFs exhibited heightened type I interferon and ISG expression upon viral challenge, consistent with potentiated type I interferon responses in these cells (Extended Data Fig. 4a). Similar results were obtained upon challenge with the rodent gamma-herpesvirus MHV-68 (Fig. 3b and Extended Data Fig. 4b). Furthermore, TFD BMDM displayed augmented antiviral gene expression and markedly lower HSV-1- and VSV-encoded mRNA and protein 6–24 hours post infection (Extended Data Fig. 5c–f). Finally, we found that Tfam+/− mice exhibit basally elevated ISG expression, which confers resistance to acute infection by lymphocytic choriomeningitis virus (LCMV) Armstrong (Extended Data Fig. 5a and Fig. 3c).
Figure 3. mtDNA stress potentiates viral resistance.
a, Viral GFP expression in MEFs infected with HSV1-GFP or VSV-GFP at MOI 0.5 for 24 hrs. b, MHV68-GFP abundance in MEFs infected at MOI 0.5. c, LCMV Armstrong gene expression 4d after i.v. infection of WT and Tfam+/− mice; n=4. d-f, Nucleoid area (d) or ISG expression (e) of MEFs exposed to ddC for 96 hrs. ddC-exposed MEFs were infected with HSV1-GFP or VSV-GFP at MOI 0.1 and imaged after 24 hrs. Error bars represent ± s.e.m. of triplicates (b,e) or quadruplicates (c) and all panels are representative of three independent experiments. **=p<0.01, ***=p<0.001.
To probe a direct requirement for mtDNA stress in antiviral priming in TFAM deficient cells, we utilized dideoxycytidine (ddC), a deoxyribonucleoside analog that specifically inhibits mtDNA replication and decreases mtDNA nucleoid size2,20. Treatment of WT MEFs with ddC resulted in reduced mtDNA copy number and decreased average nucleoid size without altering basal ISG expression (Extended Data Fig. 5b–d). In contrast, ddC drastically diminished mtDNA stress (i.e. enlarged nucleoids measuring greater than 450nm2) in Tfam+/− and TFD MEFs (Fig. 3d and Extended Data Fig. 5e), which was accompanied by attenuation of antiviral priming and basal ISG expression (Fig. 3e and Extended Data Fig. 5d, f). Moreover, ddC ablated the viral resistance phenotype of Tfam+/− MEFs (Fig. 3f). We observed similar decreases in type I interferon production and a reduction in the viral resistance phenotype in ddC-treated TFD BMDM (Extended Data Fig. 5g–h, blue bars). These results demonstrate that mtDNA stress directly potentiates antiviral innate immunity.
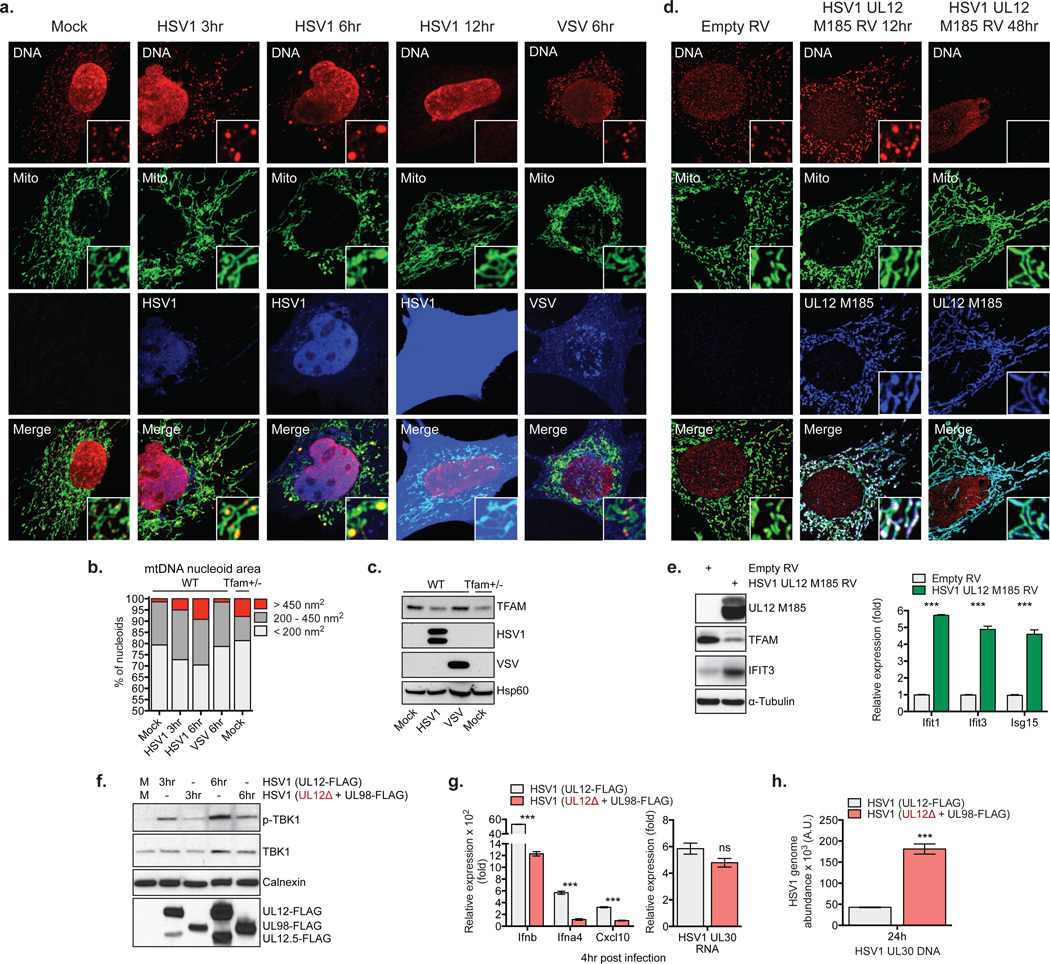
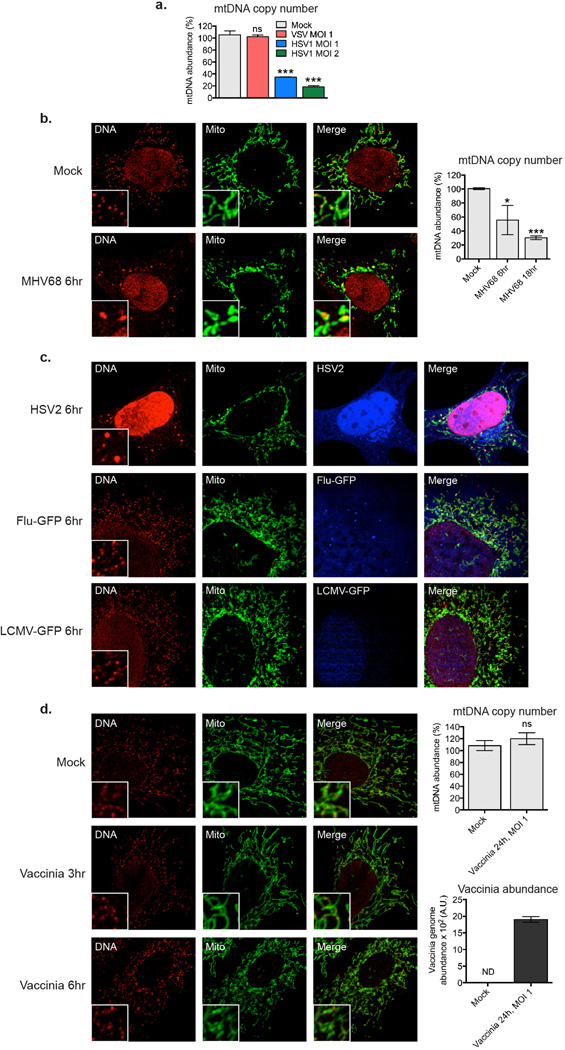
The observation that ddC treated WT BMDM displayed reduced Ifnb and increased viral gene expression upon challenge with HSV-1, despite normal responses to cytosolic nucleic acids (Extended Data Fig. 5h–i, gray bars), indicated to us that virus-induced mtDNA stress may boost host antiviral responses, consistent with reports linking viral infection to mtDNA dysregulation21,22. The alpha-herpesvirus protein UL12.5 encoded by HSV-1 and HSV-2 localizes to mitochondria and promotes rapid mtDNA depletion in human cells, which we confirmed in MEFs (Extended Data Fig. 6a)22–24. Since mtDNA depletion and nucleoid stress are often coupled, we explored nucleoid architecture and abundance kinetically during HSV-1 infection. Remarkably, 3 hours after challenge with HSV-1, mtDNA stress was readily apparent, with nucleoids less evenly distributed and enlarged (Fig. 4a). After 6 hours, ~10% of nucleoids measured larger than 450 nm2, and there was a significant decrease in total nucleoid intensity. After 12 hours, we observed pronounced mtDNA depletion. The mtDNA stress observed 3 to 6 hours after HSV-1 challenge closely mirrored that of TFAM deficient cells (Fig. 4b), as did TFAM protein levels (Fig. 4c). MHV-68 and HSV-2 triggered mtDNA stress similar to HSV-1, indicating that mtDNA stress is a common cellular perturbation during herpesvirus infection (Extended Data Fig. 6b–c). However, induction of mtDNA stress and TFAM depletion were not a general consequence of viral infection, as cells infected with VSV, Influenza, LCMV, and Vaccinia possessed normal mtDNA architecture, TFAM expression, and copy number (Fig. 4a–c and Extended Data Fig. 6c–d).
Figure 4. HSV-1 induces mtDNA stress and TFAM depletion sufficient to trigger ISG expression and necessary to fully engage antiviral immunity.
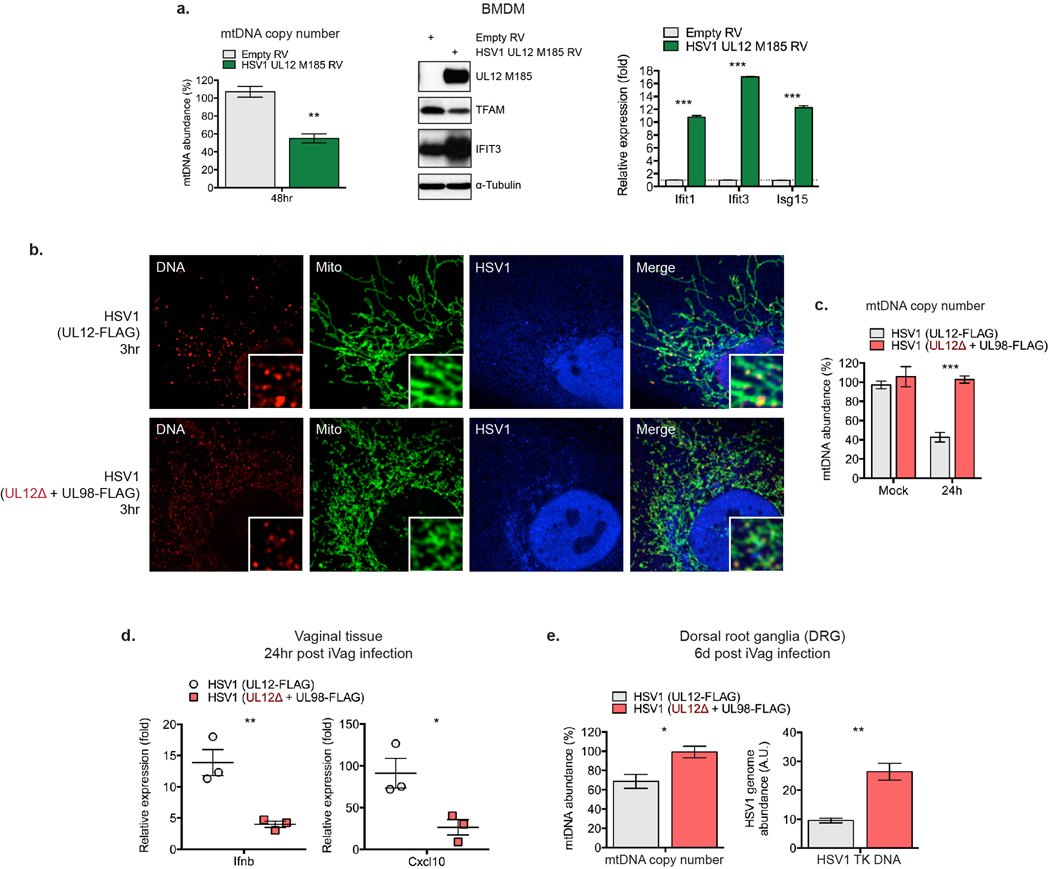
a-c, WT MEFs were mock infected or infected with HSV1-GFP or VSV-GFP at MOI 10 for the indicated times, and imaged after staining with anti-DNA, anti-Hsp60 [Mito], and anti-HSV [HSV1] or GFP [VSV] antibodies (a). mtDNA nucleoid area was calculated as described in the Methods (b). Extracts were blotted as indicated (c). d-e, WT MEFs were transduced with HSV1 UL12 M185-FLAG-expressing or empty retroviruses (RV) and cells were stained with anti-DNA, anti-Hsp60 [Mito] and anti-FLAG antibody [UL12 M185] (d), and protein or ISG expression examined after 24 hrs (e). f-g, Protein and RNA expression in BMDM infected with HSV1 (UL12-FLAG) or UL12-deficient HSV1 (UL12Δ + UL98-FLAG) at MOI 2 for the indicated times,. h, HSV1 genome abundance in L929 cells were infected as in f-g. Error bars indicate ± s.e.m. of triplicates and are representative of three independent experiments. ***=p<0.001.
Finally we sought to determine whether HSV-1-induced mtDNA dysregulation is necessary and sufficient to potentiate antiviral signaling. Transduction of MEFs and BMDM with replication-incompetent retroviruses encoding the mitochondria-targeted HSV-1 UL12 M185 gene product was sufficient to cause mitochondrial hyperfusion, nucleoid enlargement, and mtDNA loss, indicative of mtDNA stress (Fig. 4d and Extended Data Fig. 7a)24. UL12 M185 expression was also sufficient to trigger TFAM depletion and antiviral priming (i.e. augmented ISG mRNA and protein expression) (Fig. 4e and Extended Data Fig. 7a). To explore the effect of HSV-1-induced mtDNA stress on innate antiviral responses, we employed a recombinant, UL12-deficient HSV-1 strain (UL12Δ + UL98-FLAG) that is severely impaired in its ability to induce mtDNA stress but replicates similarly to a matched UL12-sufficient strain (Extended Data Fig. 7b–c)25. Infection with UL12Δ HSV-1 resulted in attenuated phospho-TBK1, type I interferon and ISG expression between 3 to 6 hours post infection, despite comparable early HSV-1 gene expression (Fig. 4f–g). However, after 24 hours, UL12Δ HSV-1 genome abundance was roughly three fold higher compared to the UL12-suficient control, consistent with impaired antiviral innate immunity (Fig. 4h). Finally, UL12Δ HSV-1 elicited less robust antiviral innate immune responses in the vagina and more readily spread to dorsal root ganglia of WT mice due to a deficit in mtDNA stress-dependent antiviral priming (Extended Data Fig. 7d–e). These results reveal that herpesvirus-induced mtDNA stress is necessary to effectively engage ISG expression and antiviral priming, and suggest that cellular monitoring of mtDNA homeostasis may represent an additional sensory mechanism to robustly engage antiviral innate immunity.
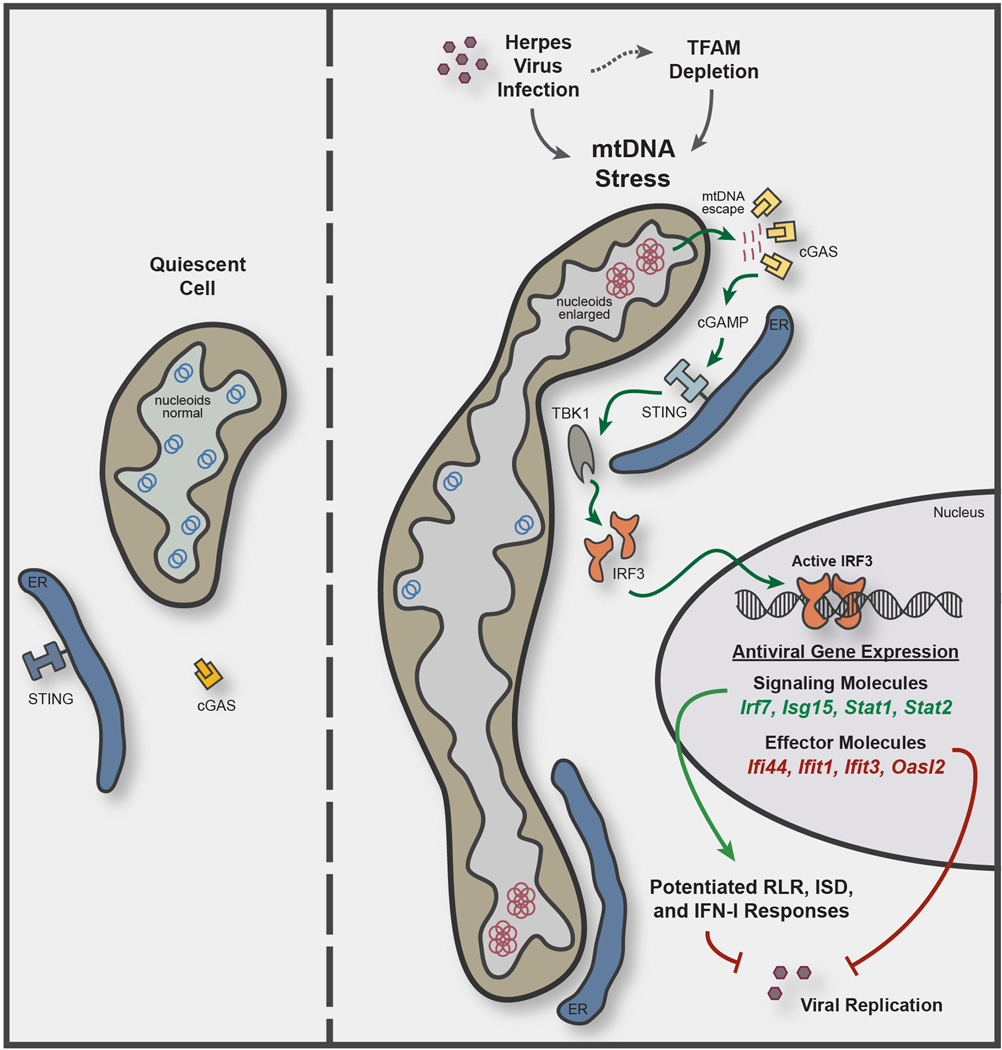
In closing, our work uncovers a novel cellular response to mtDNA stress that engages the antiviral innate immune response. Specifically, we show that mtDNA stress, induced by herpesvirus infection and mediated by loss of the mtDNA packaging protein TFAM, triggers a cGAS-STING-IRF3-dependent pathway to upregulate ISGs and potentiate type I interferon responses to viral infection (Extended Data Fig. 8). Our results support a model whereby viral-mediated disruption of mtDNA homeostasis serves as a cell-intrinsic indicator of infection that works in parallel with canonical virus sensing to fully license antiviral innate immunity. Conversely, pathologic type I interferon signatures promote autoimmune diseases such as systemic lupus erythematosus (SLE), and altered ISG expression correlates with radiation-resistant and metastatic phenotypes in some cancers26,27. Mitochondrial and mtDNA dysregulation have been noted in SLE, and perturbations in TFAM and/or mtDNA homeostasis are frequently observed in cancer28–30. Therefore, further investigation into this pathway will not only expand our knowledge of innate antiviral defense, but it may also broaden our understanding of how mitochondria contribute to the pathogenesis of human diseases and aging beyond their well-characterized roles in metabolism, apoptosis, and reactive oxygen species production.
On-line only Methods
Animal Strains
Tfam+/− and Tfamflox/flox mice were previously described and maintained on a C57BL/6 background6,31. Tfamflox/flox mice were bred to ERCre transgenic mice from Jackson (stock # 004682) for inducible, 4OHT-mediated deletion. All animal experiments were conducted in compliance with guidelines established by the Yale University Animal Care and Use Committee.
Antibodies and Reagents
Rabbit anti-mouse TFAM polyclonal anti-sera was previously described6, rabbit anti-VSV polyclonal anti-sera was a gift of John Rose at Yale University, mouse anti-Viperin was a gift of Peter Cresswell at Yale University, and rabbit-anti IFIT3 was a gift of Ganes Sen at Cleveland Clinic. The following antibodies were obtained commercially: goat anti-Hsp60 (N-20) and rabbit anti-Calnexin (H-70) (Santa Cruz Biotechnology); mouse and rabbit anti-FLAG (F1804, F7425) (Sigma); mouse anti-DNA (CBL186) (Millipore); mouse anti-GFP (JL-8) (BD Biosciences); rabbit anti-HSV1/2 (ab9533) and anti-Histone 3 (ab1791) (Abcam); rat anti-HA-FITC (11988506001) (Roche); rabbit anti-NLRX1 (17215-1-AP) (Proteintech); mouse anti-α-tubulin (DM1A) (Neomarkers); mouse anti-GAPDH (6C5) (Ambion); and rabbit anti-RIG-I (D14G6), -MDA5 (D74E4), -STAT1 (9172), -IRF3 (D83B9), -TBK1 (D1B4), and -phospho-TBK1 (D52C2) (Cell Signaling Technology). mIFNα ELISA and recombinant mIFNβ was from PBL Assay Science, and mIL-6 ELISA was from eBioscience. All primer sequences and siRNAs utilized are found in Extended Data Tables 1–2.
Cell Culture
Primary WT, Tfam+/−, Sting−/−, and cGas−/− MEFs were generated from E12.5–14.5 embryos, maintained in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biological), and subcultured no more than five passages before experiments. Sting−/− MEFs were kindly provided by Dr. Glen Barber at the University of Miami32. L929 cells were obtained from ATCC and maintained in DMEM supplemented with 10% FBS. siRNA transfection of MEFs was performed with 25 nM siRNA duplexes (IDT) and Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions. ddC (Sigma) was resuspended in PBS, added to MEFs or BMDM at a final concentration of 10–20 µM, and replenished every 48 hours. BMDM were generated from bone marrow of 8–12 week old littermate Tfamfl/flERCre− and Tfamfl/flERCre+ mice and cultured on Petri plates in DMEM containing 10% FBS plus 30% L929 culture media. To induce Cre-mediated deletion, 1 µM 4OHT dissolved in DMSO (Sigma) was added to BMDM cultures on day 6 and incubated for an additional 2–3 days. Cells were then lifted from plates by incubating in cold PBS containing 1 mM EDTA, re-plated in fresh media containing 10% L929 conditioned media, and allowed to rest overnight before experimentation (for a total of 72 or 96hrs 4OHT exposure). Transfection of ISD33 and Poly(I:C) (Sigma) into the cytosol of BMDM was performed using Lipofectamine 2000 (Invitrogen). Briefly, 1×106 BMDM were seeded in 6-well dishes after 4OHT treatment, and transfected the next day with 4µg ISD/well or 2.5µg/well of Poly(I:C) complexed at a ratio of 2:1 Lipofectamine 2000 to nucleic acid. Poly(I:C) transfection into the cytosol of MEFs was performed as described34.
Viral Stocks and Infections
VSV-G-GFP35, HSV1-GFP36, MHV-68-GFP, HSV237, Vaccinia virus38, Influenza A PR8 NS1-GFP39, HSV1 (UL12-FLAG) and HSV1 (UL12Δ + UL98-FLAG)25 were maintained as described34,40,41. MEFs or BMDM were infected at the indicated multiplicity of infection (MOI) in serum-free DMEM for one hour, washed, and incubated for various times. Cells were then fixed and stained for microscopy, lysed for western blot, solubilized in RLT plus (Qiagen) for RNA extraction, or prepared for FACS analysis. FACS was performed by first trypsinizing MEFs, followed by labeling with LIVE/DEAD Fixable Far Red stain (Molecular Probes). Cells were then fixed with 4% paraformaldehyde, washed, and analyzed on a FACSCalibur flow cytometry machine (BD). FACS plots were first gated on live cells before analyzing viral GFP fluorescence. Viral gene expression in BMDM was determined using qRT-PCR as described below, except that after values were normalized against GAPDH cDNA using the 2−ΔΔCt method, all data points were subtracted by one to center on zero.
LCMV ARM infection of WT and Tfam+/− mice was performed as described42. Briefly, 10-week-old female mice were infected with 2 × 105 p.f.u. of virus i.p., and four days post infection, mice were sacrificed, tissues isolated, and total RNA prepared using RNeasy Plus kits (QIAGEN). After generating cDNA, samples were subjected to qPCR analysis as described below using published methods43,44.
In Vivo HSV-1 Infection, DRG isolation, and Viral Titration
6-week old female mice were purchased from Charles River Laboratories and treated with Depo Provera (GE Healthcare) five days before infection45. The vaginal canals of Depo Provera treated mice were swabbed with a Calginate swab (Fisher) and 106 plaque forming units were delivered via pipette tip into the vagina. One day post infection, vaginal tissue was isolated for RNA extraction. Infected mice were euthanized at indicated time points and dorsal root ganglia (DRG) were dissected as previously described46. DRG were homogenized using a motorized pestle and total DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. Relative HSV-1 genome abundance was determined using primers specific for nuclear Tert and HSV-1 TK.
cGAS-HA and UL12 M185 Cloning and Retroviral Expression
A plasmid encoding HSV1 UL12 M185 SPA containing a 3X FLAG tag at the C-terminus was described previously24. This construct, or a plasmid encoding murine cGAS-HA (Invivogen), was subcloned into the pMXs-IRES-Puro vector and replication incompetent retroviruses were packaged using plat-E cells according to the manufacturer’s instructions (Cell Biolabs). SV40 large T immortalized cGAS−/− MEFs were exposed to supernatants containing cGAS-HA retroviruses and incubated overnight. Two days post transduction, 3 µg/ml puromycin was added to select a stable population of cells expressing cGAS-HA. Supernatants containing empty or UL12 M185 SPA retroviruses and 4 µg/ml polybrene were incubated with cells 5 × 104 MEFs or 2 × 105 BMDM in 12 well dishes for a period of 8 hours. Viral supernatants were then washed off, fresh media was added to wells, and the cells were incubated for the duration of the experiment until lysis.
Quantitative PCR
To measure relative gene expression by qRT-PCR, total cellular RNA was isolated using RNeasy Plus RNA extraction kit (QIAGEN). Approximately 400–2000ng RNA was normalized across samples and cDNA was generated using the High Capacity cDNA RT kit (Applied Biosystems). cDNA was then subjected to qPCR using Fast SYBR Green Master Mix (Applied Biosystems) and primers as indicated on the ViiA7 Real Time PCR system (Life Technologies). Three technical replicates were performed for each biological sample, and expression values of each replicate were normalized against GAPDH cDNA using the 2−ΔΔCt method. For relative expression (fold), control samples were centered at 1; for relative expression (%), control samples were centered at 100%. mtDNA copy number analysis was performed as described using primers specific to nuclear Tert and the D-loop region of mtDNA (listed in Extended Data Table 1)6. Relative HSV-1 genome abundance was determined using primers specific for nuclear Tert and HSV-1 UL30 or TK. Relative MHV68 genome abundance was determined using primers specific for nuclear Tert and MHV68 ORF40. Relative Vaccinia genome abundance was determined using primers specific for nuclear Tert and VV DNApol E9L.
Immunofluorescence Microscopy
For all microscopy images containing mtDNA nucleoids and associated panels, cells were grown on coverslips and transfected, treated, and/or infected as described. After washing in PBS, cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min, blocked with PBS-10% FBS for 30 min, stained with primary antibodies for 60 min, and stained with secondary antibodies for 60 min. Cells were washed with PBS between each step. Coverslips were mounted with Prolong Gold anti-fade reagent containing Dapi (Molecular Probes). Cells were imaged on a Zeiss LSM 510 META with a 63X water corrected objective. A digital scan zoom of 3.0 was used to enhance magnification. Images were pseudo-colored and merged using ImageJ software (NIH). For microscopy images in Fig. 3, MEFs were infected as described and fixed with 4% paraformaldehyde for 20 min. Viral GFP fluorescence and phase contrast images were captured using an Olympus IX-71 inverted scope with a 10X (Fig. 3a) or 20X (Fig. 3e) objective. Viral GFP images were pseudo-colored using ImageJ.
For nucleoid area quantification, approximately 10–15 unique fields of view from 10 distinct confocal images, comprising between 200–400 nucleoids, were captured at random. After incorporating scale information obtained from the LSM Image Browser (Zeiss), images were made binary and the area of each nucleoid was determined using the ‘Analyze Particles’ feature of ImageJ. Nucleoids were divided into the three size cutoffs, less than 200nm2, between 200–450nm2, and greater than 450nm2, and the percentage of nucleoids falling within each category was plotted. Percent of nucleoids >450 nm2 displaying cGAS colocalization was scored by calculating nucleoid area from 5 distinct images of si-Ctrl and si-Tfam transfected cGAS-HA reconstituted MEFs as described above. Nucleoids greater than 450 nm2 with a substantial cGAS co-localization signal were scored as positive.
Electron Microscopy
MEFs grown in petri dishes and on coverslips for orientation were fixed in 2.5% gluteraldehyde in 0.1M sodium cacodylate buffer pH 7.4 for 1 hour. The cells were rinsed in sodium cacodylate and those in petri dishes were scraped and spun down in 2% agar. All samples were fixed in 1% osmium tetroxide for 1 hour, en bloc stained in 2% uranyl acetate in maleate buffer pH 5.2 for a further hour, rinsed and dehydrated in an ethanol series, and infiltrated with resin (Embed812 EMS) and baked over night at 60°C. Hardened blocks were cut using a Leica UltraCut UCT. 60nm sections were collected on formver/carbon coated grids and contrast stained using 2% uranyl acetate and lead citrate. Samples were viewed on an FEI Tencai Biotwin TEM at 80Kv. Images were taken using Morada CCD and iTEM (Olympus) software.
For mitochondrial perimeter quantification, approximately 10–15 unique EM images of each genotype were captured at random. After incorporating scale information from iTEM software, the perimeter along the outer membrane of each mitochondrion was traced and quantified using ImageJ. Mitochondria were divided into the three size cutoffs, less than 2µm, between 2–5µM, and greater than 5µm, and the percentage of mitochondria falling within each category was plotted.
Oxygen Consumption Analysis
Cells were plated in XF96 plates (SeaHorse Biosciences) at 10,000 cells/well and the next day cellular O2 consumption was determined in a SeaHorse Bioscience XF96 extracellular flux analyzer according to manufacturer instructions. Cells were maintained at 37 degrees C in normal growth medium without serum.
Nuclear Fractionation and Western Blotting
Whole cell extracts were solubilized in SDS lysis buffer (20 mM Tris-HCl, 1% SDS, pH 7.5, containing protease and phosphatase inhibitors), boiled for 5 minutes, and DNA was sheared by sonication. For nuclear extraction, PBS-washed cell pellets were resuspended in 10 pellet volumes of RSB buffer (10 mM NaCl, 1.5 mM CaCl2, 10 mM Tris-HCl pH 7.5), swelled on ice for 10 min, homogenized with a motorized Teflon pestle, and the homogenate was centrifuged at 980 g for 10 min to pellet nuclei. Pellets were washed 5X in PBS, SDS was then added to a final concentration of 1%, and extracts were boiled for 5 min before sonicating to shear DNA and normalizing protein concentration. Western blotting was performed using standard protocols, and Hsp60 was used as whole cell extract loading controls, while Histone 3 was probed as a nuclear loading control.
Detection of mtDNA in Cytosolic Extracts
Digitonin extracts from MEFs and BMDM were generated largely as described47. WT and Tfam+/− MEFs (7 × 106) or Tflox ERCre−/+ BMDM exposed to 4OHT for 72 hours (1 × 107) were each divided into two equal aliquots, and one aliquot was resuspended in 500 µl of 50 µM NaOH and boiled for 30 minutes to solubilize DNA. 50µl 1M Tris-HCl pH 8 was added to neutralize the pH, and these extracts served normalization controls for total mtDNA. The second equal aliquots were resuspended in roughly 500 µl buffer containing 150 mM NaCl, 50 mM HEPES, pH 7.4, and 15–25 µg/ml digitonin (EMD Chemicals). The homogenates were incubated end over end for 10 minutes to allow selective plasma membrane permeabilization, then centrifuged at 980 g for 3 min three times to pellet intact cells. The first pellet was saved as the ‘Pel’ fraction for western blotting. The cytosolic supernatants were transferred to fresh tubes and spun at 17000 g for 10 min to pellet any remaining cellular debris, yielding cytosolic preps free of nuclear, mitochondrial, and ER contamination. DNA was then isolated from these pure cytosolic fractions using QIAQuick Nucleotide Removal Columns (QIAGEN). qPCR was performed on both whole cell extracts and cytosolic fractions using nuclear DNA primers (Tert) and mtDNA primers (Dloop1-3, Cytb, 16S, Nd4), and the Ct values obtained for mtDNA abundance for whole cell extracts served as normalization controls for the mtDNA values obtained from the cytosolic fractions. This allowed effective standardization among samples and controlled for any variations in the total amount of mtDNA in control and TFAM-deficient samples. Using this digitonin method, no nuclear Tert DNA was detected in the cytosolic fractions, indicating nuclear lysis did not occur.
Bioinformatic Analyses
Total cellular RNA from WT and Tfam+/− littermate MEFs was prepared using RNeasy Plus RNA extraction kits (QIAGEN) and used for the expression microarray procedure in conjunction with the Emory University Integrated Genomics Core. RNA integrity was first verified by an Agilent Bioanalyzer and then amplified, labeled, and hybridized onto Mouse Gene 1.0 ST arrays (Affymetrix) using standard protocols recommended by the manufacturer, starting from 2 µg of total RNA. Data were normalized by the RMA method using GeneSpring software (Agilent) for each biological sample in duplicate. The student’s t test was used to determine statistically significant changes in expression in Tfam+/− MEFs relative to WTs, with a cutoff p value of 0.0548. Heatmaps were generated using MultiExperiment Viewer49.
Statistical Analyses
Error bars displayed throughout the manuscript represent standard error of the mean (s.e.m.) unless indicated, and were calculated from triplicate or quadruplicate technical replicates of each biological sample. For in vivo experiments, error bars were calculated from the average of triplicate technical replicates of 3–4 mice per point. Sample sizes were chosen by standard methods to ensure adequate power, and no randomization or blinding was used for animal studies. Statistical significance was determined using the unpaired student’s t test; * = p<0.05; ** = p<0.01; *** = p<0.001; n.s., not significant (p>0.05). Data shown are representative of two to three independent experiments, including microscopy images, western blots, and viral challenges.
Extended Data
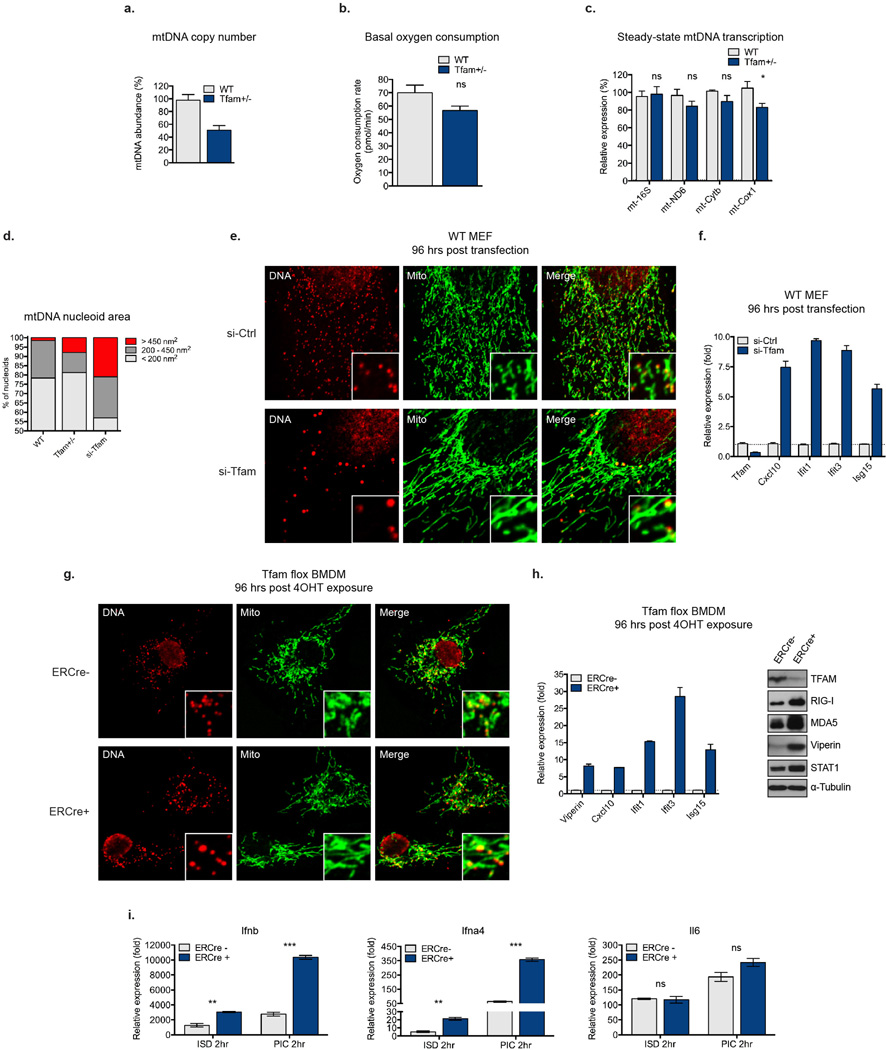
Extended Data Figure 1. TFAM deficiency induces mtDNA depletion, nucleoid stress, elevated ISG expression, and augmented type I interferon responses, but does not drastically alter oxygen consumption and mt-transcription rates.
a, qPCR analysis of relative mtDNA copy number from WT and Tfam+/− MEFs. b, Basal oxygen consumption rate of WT and Tfam+/− MEFs as determined by Seahorse Bioscience XF96 Extracellular Flux assay. c, qRT-PCR of mtDNA-encoded rRNA (16s) and mRNA (ND6, Cytb, Cox1) transcripts in WT and Tfam+/− MEFs. d-f, Untransfected Tfam+/− (d) or WT MEFs transfected with control [si-Ctrl] or Tfam [si-Tfam] siRNAs (d-f) were stained with anti-Hsp60 [Mito] and anti-DNA [DNA] antibodies. Nucleoid area from multiple independent images was calculated, stratified into groups, and graphed as percentage of the total number of nucleoids counted for each sample (d). Inset panels are magnified 3X to enhance viewing of mitochondrial network and nucleoid architecture (e). TFAM and ISG mRNA expression were measured by qRT-PCR (f). g-i, Tfamflox/flox ERCre− or Tfamflox/flox ERCre+ BMDM were incubated in 4OHT for 96hrs to induce TFAM depletion. Immunofluorescence staining was performed as described above (g). ISG mRNA and protein expression was monitored by qRT-PCR and western blotting (h). qRT-PCR analysis of type I interferon and Il6 expression in 4OHT-treated Tfam flox ERCre−/+ BMDM two hours post cytosolic delivery of interferon-stimulatory DNA [ISD] or poly(I:C) [PIC] (i). Error bars indicate ± s.e.m. of triplicates and data are representative of three independent experiments.. *=p<0.05, **=p<0.01, ***=p<0.001, ns=not significant. Unless noted p<0.05.
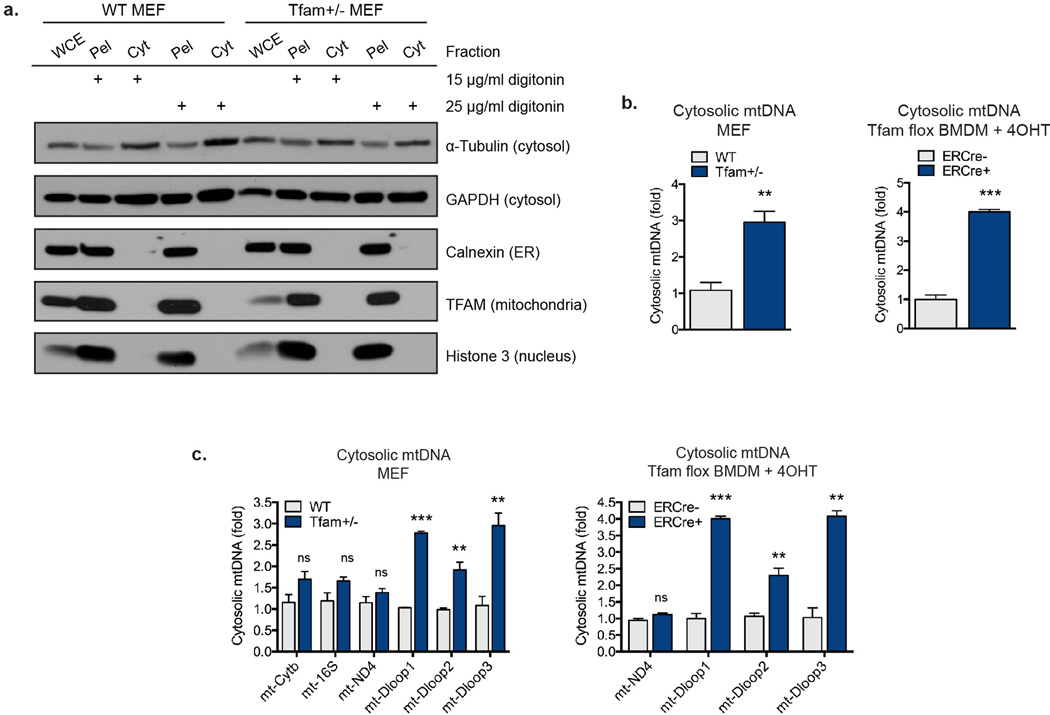
Extended Data Figure 2. TFAM deficiency promotes accumulation of cytosolic mtDNA.
a, WT or Tfam+/− MEFs were subjected to digitonin fractionation as described in the Methods and whole cell extracts [WCE], pellets [Pel], or cytosolic extracts [Cyt] were blotted using the indicated antibodies. b, DNA was extracted from digitonin extracts of WT and Tfam+/− MEFs or Tfamflox/flox ERCre− or Tfamflox/flox ERCre+ BMDM incubated in 4OHT for 72hrs. Cytosolic mtDNA was quantitated via qPCR using the mt-Dloop3 primer set. Normalization was performed as described in the Methods. c, Samples were prepared as described in b, and cytosolic mtDNA was quantitated via qPCR using the indicated primer sets. Error bars indicate ± s.e.m. of triplicates and data are representative of three independent experiments.**=p<0.01, ***=p<0.001.
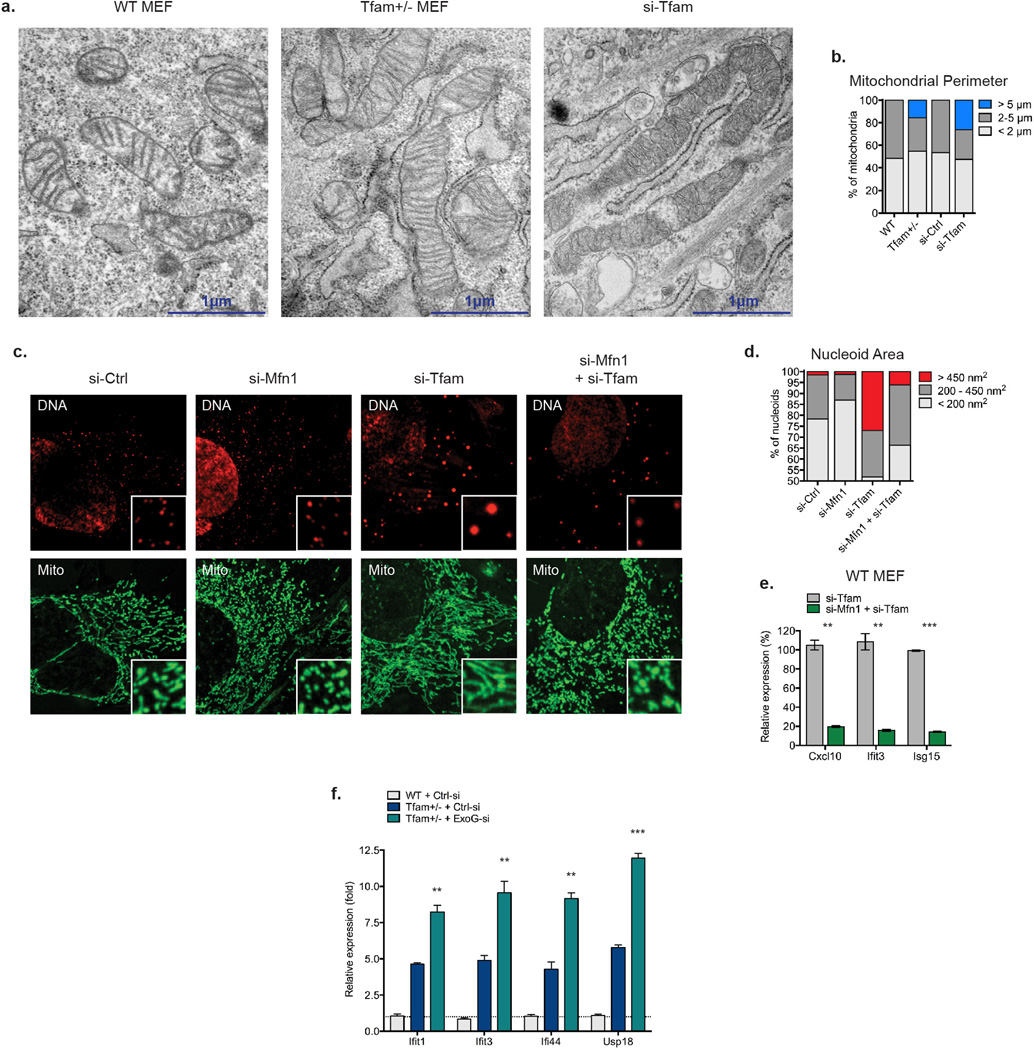
Extended Data Figure 3. Mitochondrial hyperfusion regulates the accumulation of mtDNA nucleoid stress in TFD MEFs.
a-b, WT MEFs were transfected with control or TFAM siRNAs for 96hrs. Cells were fixed and processed for EM analysis (a). Mitochondrial perimeter measurements were obtained from multiple independent images, stratified into groups, and graphed as percentage of the total number of mitochondria counted for each sample (b). c-e, WT MEFs were transfected with control, Mfn1, and/or TFAM siRNAs for 96hrs. Cells were fixed and stained with anti-Hsp60 antibody [Mito] and anti-DNA antibody [DNA] for confocal microscopy (c). Nucleoid area from multiple independent images was calculated as previously described (d). RNA was extracted for ISG expression analysis by qRT-PCR (e). f, WT and Tfam+/− MEFs were transfected with the indicated siRNAs for 96 hours and ISG expression analyzed by qRT-PCR. Error bars indicate ± s.e.m. of triplicates and data are representative of two independent experiments. **=p<0.01, ***=p<0.001.
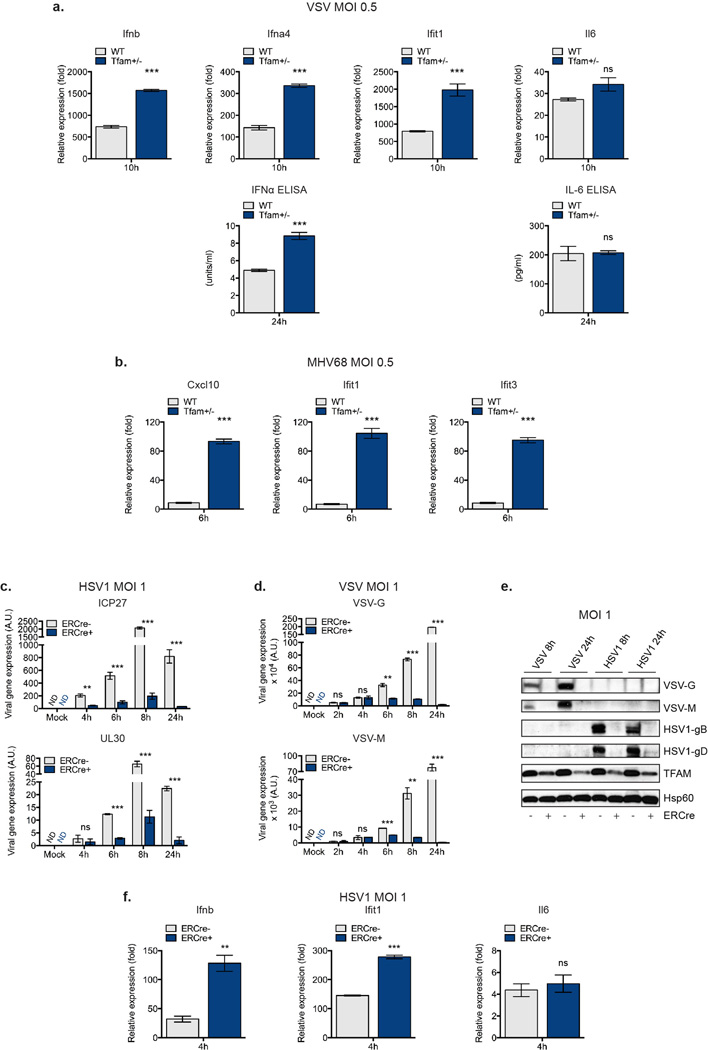
Extended Data Figure 4. mtDNA stress in TFD MEFs and BMDM potentiates type I interferon responses to viral infection and enhances viral clearance.
a-b, WT and Tfam+/− MEFs were infected with VSV-GFP (a) or MHV68-GFP (b) and after the indicated times, cytokine and ISG mRNA expression determined by qRT-PCR, or cytokine secretion determined by ELISA. c-f, Tfamflox/flox ERCre− or Tfamflox/flox ERCre+ BMDM were incubated in 4OHT for 96hrs to induce TFAM depletion. Cells were infected with HSV1-GFP (c, e-f) or VSV-GFP (d-e), incubated for the indicated times, and viral gene expression was determined by qRT-PCR (c-d) and western blotting (e), or cytokine and ISG mRNA expression determined by qRT-PCR (f). Error bars indicate ± s.e.m. of triplicates and data are representative of two independent experiments. **=p<0.01, ***=p<0.001, ns=not significant.
Extended Data Figure 5. Tissues fromTfam+/− mice display elevated ISG expression, and ddC abrogates mtDNA stress, ISG expression and viral resistance phenotypes of TFD cells.
a, RNA was extracted from the liver and kidneys of 8 week old WT and Tfam+/− mice (n=2 each) and subjected to qRT-PCR analysis for basal ISG expression. b-d, Relative mtDNA copy number (b), mtDNA nucleoid area (c) and ISG expression (d) of WT and Tfam+/− MEFs exposed to ddC for 96hrs. e-f, mtDNA nucleoid area (e) and ISG expression (f) of WT MEFs transfected with control or TFAM siRNAs for 96 hrs in the presence or absence of ddC. g-i, Tfamflox/flox ERCre− or Tfamflox/flox ERCre+ BMDM were incubated in 4OHT for 96hrs to induce TFAM depletion in the presence of ddC. ddC was washed out and cells allowed to recover overnight before infection. Cells were infected with VSV-GFP (g) or HSV1-GFP (h) at MOI 1, or WT BMDM transfected with poly(I:C) or ISD (i), and incubated for the indicated times. Ifnb expression or viral gene expression was determined by qRT-PCR. Error bars indicate ± s.e.m. of triplicates and data are representative of two independent experiments. *=p<0.05, **=p<0.01, ***=p<0.001, ns=not significant. Unless noted p<0.05.
Extended Data Figure 6. Alpha- and gamma-herpesviruses induce mtDNA stress, but RNA viruses Influenza and LCMV do not.
a, Relative mtDNA copy number of WT MEFs 24 hours post infection with VSV-GFP, HSV1-GFP, or mock at the indicated MOIs. b, WT MEFs were infected with MHV68-GFP at a MOI 0.5 and after the indicated times, cells were stained and subjected to confocal microscopy or relative mtDNA copy number determined. c, WT MEFs were infected with HSV2, Flu-GFP, or LCMV-GFP, at a MOI 10, and after 6 hours, cells were stained and subjected to confocal microscopy. d, WT MEFs were infected with Vaccinia virus at a MOI 10 or 1, and after the indicated times, cells were stained and subjected to confocal microscopy or relative mtDNA copy number determined. Error bars indicate ± s.e.m. of triplicates and data are representative of two independent experiments. *=p<0.05, **=p<0.01, ***=p<0.001, ns=not significant.
Extended Data Figure 7. HSV1 UL12 M185 expression is sufficient to trigger mtDNA stress, TFAM depletion, and antiviral priming in BMDM; infection with UL12-deficient HSV-1 fails to induce mtDNA stress, elicits lower vaginal type I interferon responses, and spreads more readily to DRG.
a, WT BMDM were transduced with HSV1 UL12 M185 expressing or empty retroviruses (RV) and relative mtDNA abundance, protein expression, and ISG mRNA expression determined. b, WT MEFs were infected HSV1 (UL12-FLAG) or UL12-deficient HSV1 (UL12Δ + UL98-FLAG) at MOI 10 for 3 hours and analyzed by confocal microscopy. c, WT MEFs were infected HSV1 (UL12-FLAG) or UL12-deficient HSV1 (UL12Δ + UL98-FLAG) at MOI 2 for 24 hours and mtDNA abundance was determined by qPCR. d, The vaginas of WT mice (n=3 per condition) were inoculated with 106 p.f.u. of HSV1 (UL12-FLAG) or UL12-deficient HSV1 (UL12Δ + UL98-FLAG), and 24 hours post infection, vaginal RNA was extracted and gene expression analyzed by qRT-PCR. e, Mice (n=3 per condition) were infected as previously described, and 10 days post infection, DNA from DRG was isolated for mtDNA and HSV-1 genome abundance measurements by qPCR. Error bars indicate ± s.e.m. of triplicates and data are representative of two independent experiments. *=p<0.05, **=p<0.01, ***=p<0.001; unless noted p<0.05, ns=not significant.
Extended Data Figure 8. Model illustrating mtDNA stress-dependent antiviral priming.
TFAM depletion, induced genetically or during Herpes virus infection, triggers mtDNA stress, characterized by nucleoid loss and enlargement. This results in the release of fragmented mtDNA that recruits and activates peri-mitochondrial cGAS to generate the second messenger cyclic GMP-AMP (cGAMP) and activate ER-resident STING. STING then activates TBK1, which phosphorylates IRF3 to induce dimerization and nuclear translocation. Active IRF3 elevates basal gene expression of ISGs with antiviral signaling and effector functions. Signaling ISGs, such as IRF7, ISG15, STAT1, and STAT2 cooperate with IRF3 to potentiate RIG-I-like receptor (RLR), interferon-stimulatory DNA (ISD), and type I interferon (IFN-I) responses, and effector ISGs, such as IFI44, IFIT1, IFIT3, and OASL2, augment viral resistance. Both outcomes collectively and robustly boost innate antiviral defenses to dampen viral replication.
Extended Data Table 1. Oligonucleotides utilized in qPCR.
| Gene name | Forward and reverse oligo. sequences |
|---|---|
| mGapdh | GACTTCAACAGCAACTCCCAC |
| TCCACCACCCTGTTGCTGTA | |
| mTfam | AAGGATGATTCGGCTCAGG |
| GGCTTTGAGACCTAACTGG | |
| mmt-16S | GTTACCCTAGGGATAACAGCGC |
| GATCCAACATCGAGGTCGTAAACC | |
| mmt-ND6 | TTAGCATTAAAGCCTTCACC |
| CCAACAAACCCACTAACAAT | |
| mmt-Cytb | AGTAGACAAAGCCACCTTGA |
| CCGCGATAATAAATGGTAAG | |
| mmt-Cox1 | GCCCCAGATATAGCATTCCC |
| GTTCATCCTGTTCCTGCTCC | |
| mlfna4 | CTTTCCTCATGATCCTGGTAATGAT |
| AATCCAAAATCCTTCCTGTCCTTC | |
| mlfnb | CCCTATGGAGATGACGGAGA |
| CCCAGTGCTGGAGAAATTGT | |
| mll6 | TGATGCACTTGCAGAAAACA |
| ACCAGAGGAAATTTTCAATAGGC | |
| mViperin | ATAGTGAGCAATGGCAGCCT |
| AACCTGCTCATCGAAGCTGT | |
| mlfit1 | CAAGGCAGGTTTCTGAGGAG |
| GACCTGGTCACCATCAGCAT | |
| mlfit3 | TTCCCAGCAGCACAGAAAC |
| AAATTCCAGGTGAAATGGCA | |
| mlfi44 | CTGATTACAAAAGAAGACATGACAGAC |
| AGGCAAAACCAAAGACTCCA | |
| mlsg15 | CTAGAGCTAGAGCCTGCAG |
| AGTTAGTCACGGACACCAG | |
| mUsp18 | GAGAGGACCATGAAGAGGA |
| TAAACCAACCAGACCATGAG | |
| mlrf7 | CAATTCAGGGGATCCAGTTG |
| AGCATTGCTGAGGCTCACTT | |
| mCxcl10 | CCAAGTGCTGCCGTCATTTTC |
| GGCTCGCAGGGATGATTTCAA | |
| mStat1 | CGCGCATGCAACTGGCATATAACT |
| ATGCTTCCGTTCCCACGTAGACTT | |
| mStat2 | TGATCTCTAACAGACAGGTGG |
| CTGCATTCACTTCTAAGGACTC | |
| mMda5 | CGGAAGTTGGAGTCAAAGC |
| TTTGTTCAGTCTGAGTCATGG | |
| mRig-I | GAGTACCACTTAAAGCCAGAG |
| AATCCATTTCTTCAGAGCATCC | |
| HSV1 ICP27 RNA | TTTCTCCAGTGCTACCTGAAGG |
| TCAACTCGCAGACACGACTCG | |
| HSV1 UL30 RNA | CGCGCTTGGCGGGTATTAACAT |
| TGGGTGTCCGGCAGAATAAAGC | |
| VSV G | CAAGTCAAAATGCCCAAGAGTCACA |
| TTTCCTTGCATTGTTCTACAGATGG | |
| VSV M | TATGATCCGAATCAATTAAGATATG |
| GGGACGTTTCCCTGCCATTCCGATG | |
| LCMV GP | TGCCTGACCAAATGGATGATT |
| CTGCTGTGTTCCCGAAACACT | |
| LCMV NP | CAGAAATGTTGATGCTGGACTGC |
| CAGACCTTGGCTTGCTTTACACAG | |
| MHV68 gDNA ORF40 | TAGCCACACCTCCCACGC |
| ATTCAAGACCTGAACATAGTGC | |
| Vaccinia E9L | CGGCTAAGAGTTGCACATCCA |
| CTCTGCTCCATTTAGTACCGATTCT | |
| HSV1 gDNA TK | ATACCGACGATCTGCGACCT |
| TTATTGCCGTCATAGCGCGG | |
| HSV1 gDNA UL30 | ATCACCGACCCGGAGAG |
| CAGGCGCTTGTTGGTGT | |
| m.mtDNA Dloop 1 | AATCTACCATCCTCCGTGAAACC |
| TCAGTTTAGCTACCCCCAAGTTTAA | |
| m.mtDNA Dloop 2 | CCCTTCCCCATTTGGTCT |
| TGGTTTCACGGAGGATGG | |
| m.mtDNA Dloop 3 | TCCTCCGTGAAACCAACAA |
| AGCGAGAAGAGGGGCATT | |
| m.mtDNA CytB | GCTTTCCACTTCATCTTACCATTTA |
| TGTTGGGTTGTTTGATCCTG | |
| m.mtDNA 16S | CACTGCCTGCCCAGTGA |
| ATACCGCGGCCGTTAAA | |
| m.mtDNA ND4 | AACGGATCCACAGCCGTA |
| AGTCCTCGGGCCATGATT | |
| m.nucDNA Tert | CTAGCTCATGTGTCAAGACCCTCTT |
| GCCAGCACGTTTCTCTCGTT |
Extended Data Table 2. Dicer substrate siRNAs utilized.
All siRNAs were predesigned by IDT and transfected at 25nM final concentration.
| Gene name | IDT Duplex name |
|---|---|
| mTfam | MMC.RNAI.N009360.12.1 |
| mExoG | MMC.RNAI.N172456.12.1 |
| mSting | MMC.RNAI.N028261.12.1 |
| mcGas | MMC.RNAI.N173386.12.1 |
| mTbk1 | MMC.RNAI.N019786.12.1 |
| mlrf3 | MMC.RNAI.N016849.12.1 |
| mMfn1 | MMC.RNAI.N024200.12.1 |
| Firefly luciferase (si-Ctrl) | FLuc-S1 |
Acknowledgements
We thank Navdeep Chandel for TFAMflox/flox mice, John Schoggins and Skip Virgin for cGas−/− MEFs, Kapil Bahl and Jack Schell for advice with VSV infections, and Siyuan Ding for HSV-1 gene expression analysis. This work was supported by a joint grant from the United Mitochondrial Disease Foundation and Mitocon and NIH R01 AG047632 (G.S.S.), NIH. R01 AI054359 and R01 AI081884 (A.I.), Canadian Institutes for Health Research grant MOP37995 and a Canada Research Chair in Molecular Virology (J.R.S.), American Cancer Society Postdoctoral Fellowship PF-13-035-01-DMC (A.P.W.), United Mitochondrial Disease Foundation Postdoctoral Fellowship (N.R.), NIH F31 AG039163 (M.C.T.), NIH T32 AI055403 (W.K-H), and Alberta Innovates-Health Solutions and a Queen Elizabeth II Graduate Scholarship (B.A.D).
Footnotes
Author Contributions
A.P.W. designed and performed experiments, analyzed data, interpreted results and wrote the paper; W.K.H. provided viral stocks, advice on viral infection protocols, and performed in vivo HSV-1 infections; M.S. performed LCMV and Flu infections; M.C.T. aided in experimental design and assisted with viral infections; C.M.P. performed experiments and analyzed data; M.B. performed steady-state mitochondrial transcript analysis; N.R. assisted with gene expression array analysis; D.A.M. generated cGas−/− MEFs; B.A.D. and J.R.S. generated and provided HSV-1 UL12 constructs and HSV-1 ΔUL12 viruses; S.M.K. provided reagents and facilities for LCMV infections and interpreted results; S.M.L. and R.E.M. provided reagents and advice and help perform viral infections; A.I. supplied reagents, designed experiments, and interpreted results; GSS designed experiments, interpreted results and wrote the paper.
Author Information
Microarray data have been submitted to the NCBI (GEO #GSE63767).
The authors have no competing financial interests to declare.
References
- 1.Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 2.Kasashima K, Sumitani M, Endo H. Human mitochondrial transcription factor A is required for the segregation of mitochondrial DNA in cultured cells. Exp Cell Res. 2011;317:210–220. doi: 10.1016/j.yexcr.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–722. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 6.Woo DK, et al. Mitochondrial Genome Instability and ROS Enhance Intestinal Tumorigenesis in APC(Min/+) Mice. Am. J. Pathol. 2012;180:24–31. doi: 10.1016/j.ajpath.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rusinova I, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimada K, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholls TJ, Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 12.Ban-Ishihara R, Ishihara T, Sasaki N, Mihara K, Ishihara N. Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proceedings of the National Academy of Sciences. 2013;110:11863–11868. doi: 10.1073/pnas.1301951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malena A, Loro E, Di Re M, Holt IJ, Vergani L. Inhibition of mitochondrial fission favours mutant over wild-type mitochondrial DNA. Hum. Mol. Genet. 2009;18:3407–3416. doi: 10.1093/hmg/ddp281. [DOI] [PubMed] [Google Scholar]
- 14.Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. EXOG, a novel paralog of Endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ablasser A, et al. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 17.Cai X, Chiu Y-H, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 18.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. J Immunol. 2013;190:1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pohjoismäki JLO, et al. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic Acids Res. 2006;34:5815–5828. doi: 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiedmer A, et al. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J Virol. 2008;82:4647–4655. doi: 10.1128/JVI.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corcoran JA, Saffran HA, Duguay BA, Smiley JR. Herpes simplex virus UL12.5 targets mitochondria through a mitochondrial localization sequence proximal to the N terminus. J Virol. 2009;83:2601–2610. doi: 10.1128/JVI.02087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duguay BA, Smiley JR. Mitochondrial Nucleases ENDOG and EXOG Participate in Mitochondrial DNA Depletion Initiated by Herpes Simplex Virus 1 UL12.5. J Virol. 2013;87:11787–11797. doi: 10.1128/JVI.02306-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duguay BA, et al. Elimination of mitochondrial DNA is not required for herpes simplex virus 1 replication. J Virol. 2014;88:2967–2976. doi: 10.1128/JVI.03129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:541–547. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 27.Khodarev NN, et al. Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 2007;67:9214–9220. doi: 10.1158/0008-5472.CAN-07-1019. [DOI] [PubMed] [Google Scholar]
- 28.Lee H-T, et al. Leukocyte mitochondrial DNA alteration in systemic lupus erythematosus and its relevance to the susceptibility to lupus nephritis. Int J Mol Sci. 2012;13:8853–8868. doi: 10.3390/ijms13078853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H-M, Sugino H, Aoki C, Nishimoto N. Underexpression of mitochondrial-DNA encoded ATP synthesis-related genes and DNA repair genes in systemic lupus erythematosus. Arthritis Res. Ther. 2011;13:R63. doi: 10.1186/ar3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace DC. Mitochondria and cancer. Nat. Rev. Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods Section References
- 31.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Tal MC, et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalton KP, Rose JK. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology. 2001;279:414–421. doi: 10.1006/viro.2000.0736. [DOI] [PubMed] [Google Scholar]
- 36.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012 doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang IK, Pillai PS, Iwasaki A. Efficient influenza A virus replication in the respiratory tract requires signals from TLR7 and RIG-I. Proceedings of the National Academy of Sciences. 2013;110:13910–13915. doi: 10.1073/pnas.1303275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012;12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardenas I, et al. Placental viral infection sensitizes to endotoxin-induced pre-term labor: a double hit hypothesis. Am. J. Reprod. Immunol. 2011;65:110–117. doi: 10.1111/j.1600-0897.2010.00908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall HD, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh RM, Seedhom MO. Lymphocytic choriomeningitis virus (LCMV): propagation, quantitation, and storage. Curr Protoc Microbiol. 2008;Chapter 15 doi: 10.1002/9780471729259.mc15a01s8. Unit 15A.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCausland MM, Crotty S. Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods. 2008;147:167–176. doi: 10.1016/j.jviromet.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parr MB, et al. A mouse model for studies of mucosal immunity to vaginal infection by herpes simplex virus type 2. Lab. Invest. 1994;70:369–380. [PubMed] [Google Scholar]
- 46.Malin SA, Davis BM, Molliver DC. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc. 2007;2:152–160. doi: 10.1038/nprot.2006.461. [DOI] [PubMed] [Google Scholar]
- 47.Holden P, Horton WA. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res Notes. 2009;2:243. doi: 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raimundo N, et al. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Bio Techniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]