Abstract
Invariant natural killer T (iNKT)-cell development is controlled by many polymorphic genes present in commonly used mouse inbred strains. Development of type 1 diabetes (T1D) in NOD mice partly results from their production of fewer iNKT-cells compared to non-autoimmune prone control strains including ICR. We previously identified several iNKT-cell quantitative trait genetic loci co-localized with known mouse and human T1D regions in a (NOD × ICR)F2 cross. To further dissect the mechanisms underlying the impaired iNKT-cell compartment in NOD mice, we carried out a series of bone marrow transplantation as well as additional genetic mapping studies. We found that impaired iNKT-cell development in NOD mice was mainly due to the inability of their double-positive (DP) thymocytes to efficiently select this T-cell population. Interestingly, we observed higher levels of CD1d expression by NOD than ICR DP thymocytes. The genetic control of the inverse relationship between the CD1d expression level on DP thymocytes and the frequency of thymic iNKT-cells was further mapped to a region on Chromosome 13 between 60.12 Mb and 70.59 Mb. The NOD allele was found to promote CD1d expression and suppress iNKT-cell development. Our results indicate that genetically controlled physiological variation of CD1d expression levels modulates iNKT-cell development.
Keywords: iNKT-cell, NOD mouse, CD1d, Linkage analysis, thymus
Introduction
CD1d-restricted invariant natural killer T (iNKT)-cells are a unique population with diverse immunoregulatory functions1. iNKT-cells can promote immune responses against tumors and infectious organisms, but they are also paradoxically capable of suppressing autoimmunity2. Distinct from conventional CD4 and CD8 T-cells, iNKT-cells are selected by CD1d-expressing CD4/CD8 double positive (DP) thymocytes1. We previously analyzed a large panel of classical and wild-derived inbred mouse strains and demonstrated a substantial strain-dependent variation in the frequencies of thymic and splenic iNKT cells3. The frequencies of iNKT-cells also vary significantly in humans4–8. Although the extent to which environmental factors may alter the frequency of iNKT-cells in humans is unknown, it appears that this phenotype is largely under polygenetic control8. Taken together, these results indicate that the development of iNKT-cells is a complex trait controlled by multiple genetic variants in both humans and mice.
We previously showed that the frequencies of iNKT-cells in NOD mice were at the lower end of the spectrum in both the thymus and spleen compared to most other inbred strains3. Defects in iNKT-cell development and function in NOD mice have been linked to autoimmune type 1 diabetes (T1D) development in this strain9–21. Despite sharing the same H2g7 major histocompatibility complex that is the primary genetic contributor to T1D development in NOD mice, the ICR/HaJ strain is completely resistant to this disease. Both NOD and ICR/HaJ (hereafter ICR) are related Swiss-derived inbred strains originating from an Ha/ICR outbred stock22, but differ significantly in their iNKT-cell frequencies3. To further understand the genetic basis of iNKT-cell development, we outcrossed the NOD mouse to the ICR strain and employed an F2 mapping strategy to identify multiple quantitative trait loci (QTL) that regulate the frequencies of thymic and splenic iNKT-cells23. We reported that several iNKT-cell QTL co-localized with previously known mouse and human T1D regions. These included a Chromosome (Chr) 12 QTL that overlapped with a syntenic human T1D locus on Chr 1423. While NOD mice have lower frequencies and numbers of iNKT-cells compared to the ICR strain, our F2 mapping study also identified several loci where NOD alleles promoted rather than suppressed iNKT-cell development23. These results indicate that in the context of the NOD genome, alleles that normally enhance iNKT-cell development are masked by other defects in this strain.
To gain further insight into the cellular mechanisms contributing to iNKT-cell deficiency in NOD mice and to aid in the eventual identification of the causative genes, we carried out a series of bone marrow (BM) chimerism experiments. These studies revealed that the iNKT-cell developmental defect in NOD mice was not cell intrinsic but was largely due to the inability of the DP thymocytes to efficiently select this T-cell subset. Unexpectedly, NOD DP thymocytes expressed higher levels of CD1d molecules compared to the ICR counterpart. Using a first backcross (BC1) mapping approach, we further showed that the inverse relationship between the CD1d expression level on DP thymocytes and the frequency of iNKT-cells was controlled by a locus on Chr 13 where the NOD allele enhanced CD1d expression and suppressed iNKT-cell development.
Results
Hematopoietic cell intrinsic but iNKT-cell extrinsic factors contribute to impaired iNKT-cell development in NOD mice
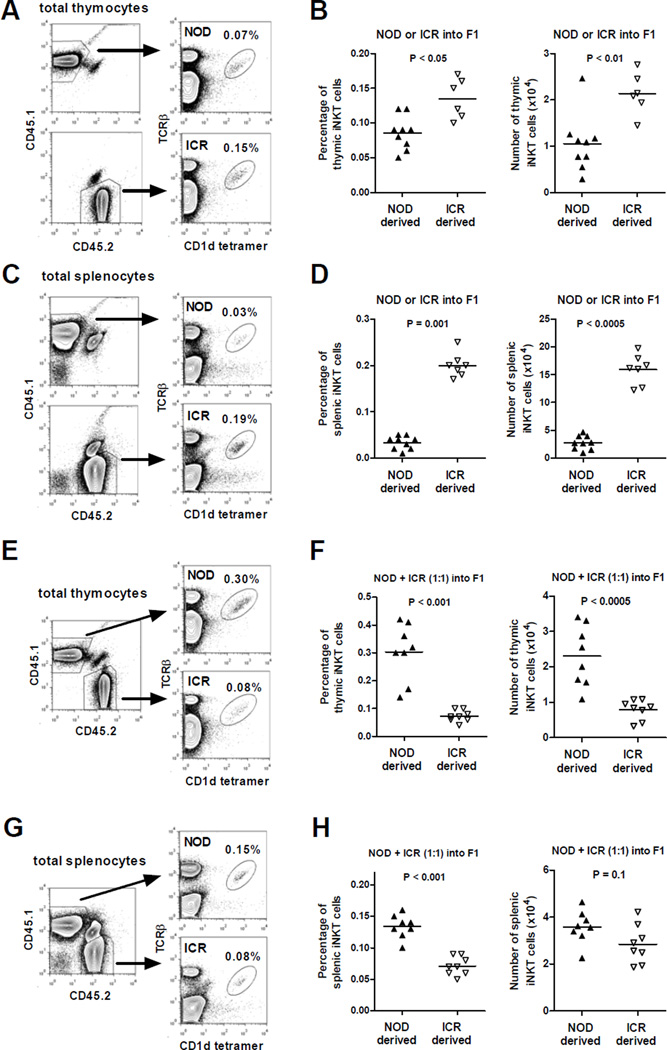
NOD and ICR mice have significantly different frequencies and numbers of thymic and splenic iNKT-cells as a result of genetic variations at multiple loci3, 23. We generated bone marrow (BM) chimeras to ask if factors intrinsic to hematopoietic cells respectively suppress and promote iNKT-cell development in NOD and ICR mice. To test this, we transferred T-cell depleted NOD (CD45.1+) or ICR (CD45.2+) BM cells into lethally irradiated (NOD × ICR)F1 recipients. Between 8 to 10 weeks post-BM reconstitution, we analyzed the frequency and number of donor-derived iNKT-cells in the thymus and spleen. As shown in Figure 1, ICR BM cells gave rise to higher frequencies and numbers of thymic (panels A and B) and splenic (panels C and D) iNKT-cells than those from NOD hematopoietic precursors in the reconstituted F1 recipients. We next determined if factors intrinsic or extrinsic to iNKT-cells control their varying differentiation from NOD and ICR BM cells. This was done by infusing T-cell depleted NOD and ICR BM cells mixed at a 1:1 ratio to chimerically reconstitute lethally irradiated (NOD × ICR)F1 mice. At the time of analyses, the respective reconstitution levels of NOD and ICR derived thymocytes in the F1 recipients were 41.8 ± 2.3 and 57.5 ± 2.2 (percentages, mean ± se). The respective reconstitution levels of NOD and ICR derived splenocytes in the F1 recipients were 35.1 ± 1.6 and 51.7 ± 1.8 (percentages, mean ± se). Unexpectedly, more thymic iNKT-cells (both the percentage and absolute number) were derived from NOD than ICR BM in the reconstituted F1 recipients (Fig. 1E and 1F). Similar results were also observed in the spleen (Fig. 1G and 1H) although the numerical difference between NOD and ICR derived iNKT-cells did not reach statistical significance. Collectively, these results indicate that when developing in isolation, the impaired differentiation of iNKT-cells in NOD mice is mainly due to defects extrinsic to this T-cell population but intrinsic to BM derived hematopoietic cells. We cannot rule out that hematopoietic cell extrinsic factors also contribute to the different levels of iNKT-cells between NOD and ICR strains as the design of the BM chimerism experiments did not specifically test this question.
Figure 1. Hematopoietic cell intrinsic but iNKT-cell extrinsic factors contribute to impaired iNKT-cell development in NOD mice.
T-cell depleted BM cells (5×106) isolated from NOD or ICR were transferred alone or co-injected at a 1:1 ratio into lethally irradiated (1100 Rads) (NOD × ICR)F1 recipients. The frequency and number of iNKT-cells were determined in the thymus and spleen at 8–10 weeks post BM reconstitution. (A and C) Representative flow cytometry profiles of the thymus (A) or spleen (C) cells isolated from an F1 recipient reconstituted with either NOD (CD45.1) or ICR (CD45.2) BM cells. (B and D) Summarized results of the frequency and number of thymic (B) and splenic (D) iNKT-cells in F1 recipients reconstituted with either NOD (CD45.1) or ICR (CD45.2) BM cells. (E and G) Representative flow cytometry profiles of the thymus (E) or spleen (G) isolated from an F1 recipient reconstituted with equal number of NOD (CD45.1) and ICR (CD45.2) BM cells. (F and H) Summarized results of the frequency and number of thymic (F) and splenic (H) iNKT-cells in F1 recipients reconstituted with equal number of NOD (CD45.1) and ICR (CD45.2) BM cells. Each symbol represents one BM recipient. The horizontal bar indicates the mean. Results are pooled from 2 independent experiments. Statistical analysis was performed using the Mann Whitney test.
ICR DP thymocytes are more capable than those from NOD mice to support iNKT-cell development
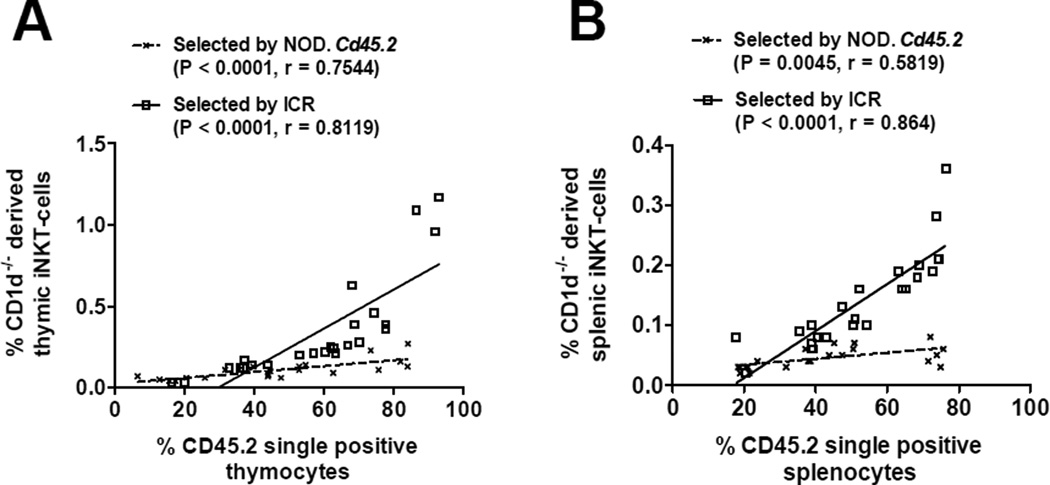
The results described above suggested a possibility that the ability of NOD and ICR DP thymocytes to select and support the development of iNKT-cells is different. To directly test this, we generated BM chimeras by transferring T-cell depleted NOD.Cd1d−/− (CD45.1+) BM cells admixed at different ratios with NOD.Cd45.2 or ICR BM cells into lethally irradiated (NOD × ICR)F1 recipients. CD1d-deficient DP thymocytes lack the ability to select but can become iNKT-cells in the presence of NOD.Cd45.2 or ICR DP thymocytes. As shown in Figure 2A, there was a positive correlation between the frequency of NOD.Cd1d−/− originated iNKT-cells found in the thymus and the proportion of NOD.Cd45.2 or ICR derived thymocytes. We also observed similar results in the spleens of the BM chimeras (Fig. 2B). However, it was evident that ICR-derived DP thymocytes were more capable of selecting iNKT-cells compared to the NOD.Cd45.2 derived counterpart. This conclusion is based on the observation that the degree of increase in CD1d-deficient BM derived iNKT-cells as a function of the gradually elevated proportion of CD1d-sufficient DP thymocytes was far greater when developing in the co-recipients of ICR than NOD.Cd45.2 BM.
Figure 2. ICR DP thymocytes are more capable than those from NOD mice to support the development of iNKT-cells.
NOD.Cd1d−/− BM cells admixed with those from NOD.Cd45.2 or ICR at different ratios (from 4:1 to 1:4) were transferred into lethally irradiated (NOD × ICR)F1 recipients. The frequencies of NOD.Cd1d−/− derived iNKT-cells were determined in the thymus (A) and spleen (B) of the BM chimeras at 8–10 weeks post BM reconstitution. Gating strategy was similar to that described in Figure 1. The percentage of iNKT-cells is presented as the proportion of CD45.1+/CD45.2− cells. Statistically significant correlations (Pearson correlation, P < 0.05) between the percentages of NOD.Cd1d−/− derived iNKT-cells and the proportions of CD45.2+ cells were found for those selected by NOD.Cd45.2 or ICR in both the thymus and spleen. The solid and dotted lines represent the best fit linear regression lines.
Comparison of SLAM protein expression in NOD and ICR mice
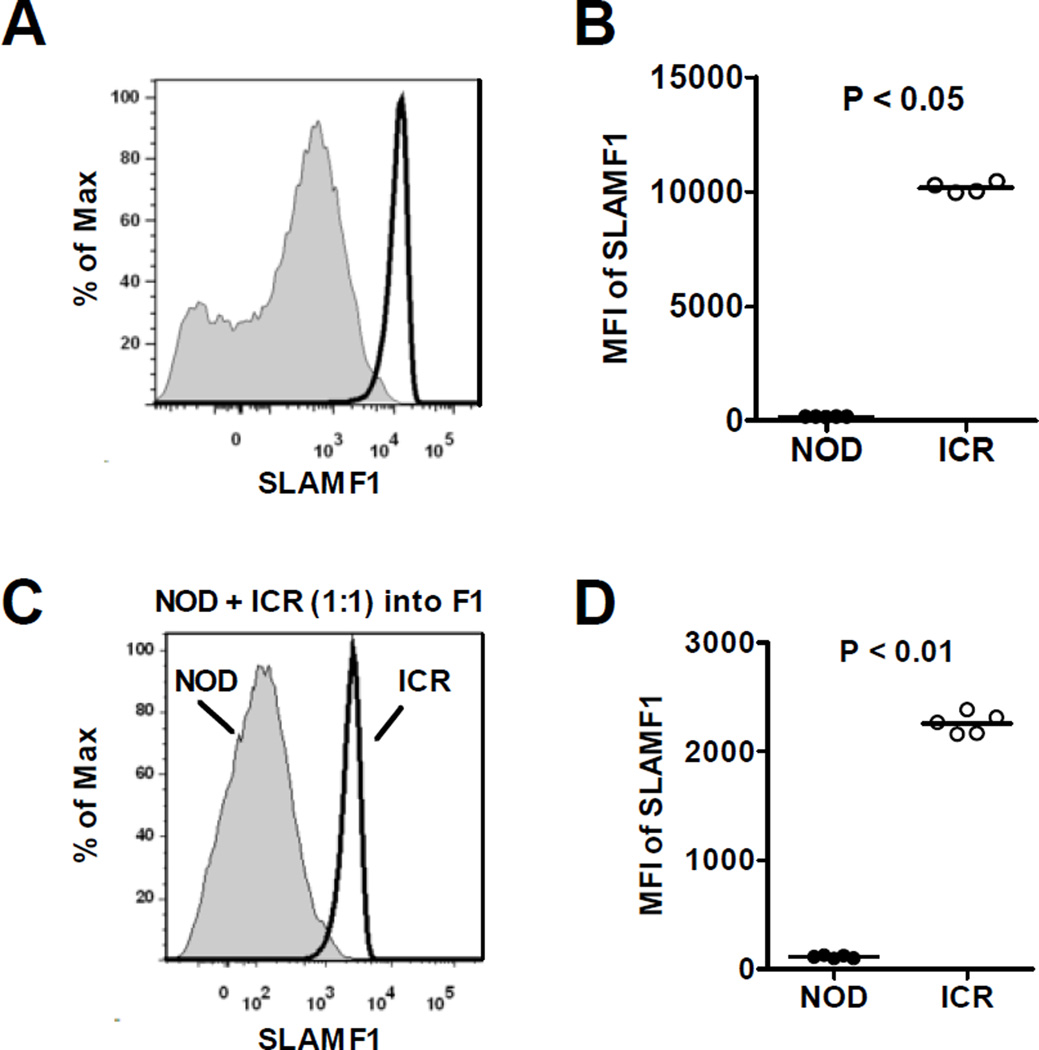
Several SLAM family proteins, including SLAMF1 (CD150), SLAMF3 (CD229/Ly9), and SLAMF6 (Ly108), have been shown to play important roles in the development of iNKT-cells24–28. SLAMF1 and SLAMF6 promote normal iNKT cell development24. On the other hand, SLAMF3 suppresses iNKT-cell development28. Previous studies have also documented reduced expression level of SLAMF1 in NOD mice that in part contributes to the numerical defect of iNKT-cells in this strain29, 30. Thus, we determined if differential expression of SLAMF1, SLAMF3, and SLAMF6 on NOD and ICR DP thymocytes could represent the iNKT-cell extrinsic factors that distinguish these two strains. SLAMF1 was expressed at a significantly lower level on DP thymocytes in NOD mice compared to the ICR strain (Fig. 3A and 3B). Overall expression levels of SLAMF3 and SLAMF6 were found to be comparable between NOD and ICR mice (data not shown). We also observed that SLAMF1 expression remained significantly lower on NOD-derived DP thymocytes compared to those of the ICR origin in the mixed BM chimeras (Fig. 3C and 3D).
Figure 3. Analyses of SLAMF1 expression.
(A and B) Expression levels of SLAMF1 on DP thymocytes of 7-week-old NOD and ICR males were analyzed by flow cytometry. Representative histograms of SLAMF1 on CD4+/CD8+ gated cells are shown in (A). NOD and ICR mice are respectively indicated by shaded areas and solid lines. Results from 4–5 mice per strain are summarized in (B). (C and D) Expression of SLAMF1 was analyzed on DP thymocytes in NOD/ICR mixed BM chimeras. (C) Representative histograms of SLAMF1 expression (gated on NOD CD45.1+ CD45.2− or ICR CD45.1− CD45.2+ DP thymocytes). (D) The summarized results from 5 BM chimeras are shown. Each symbol represents one mouse. The horizontal bar dictates the mean. Statistical analysis was performed using the Mann Whitney test.
CD1d expression level inversely correlates with the frequency of iNKT-cells
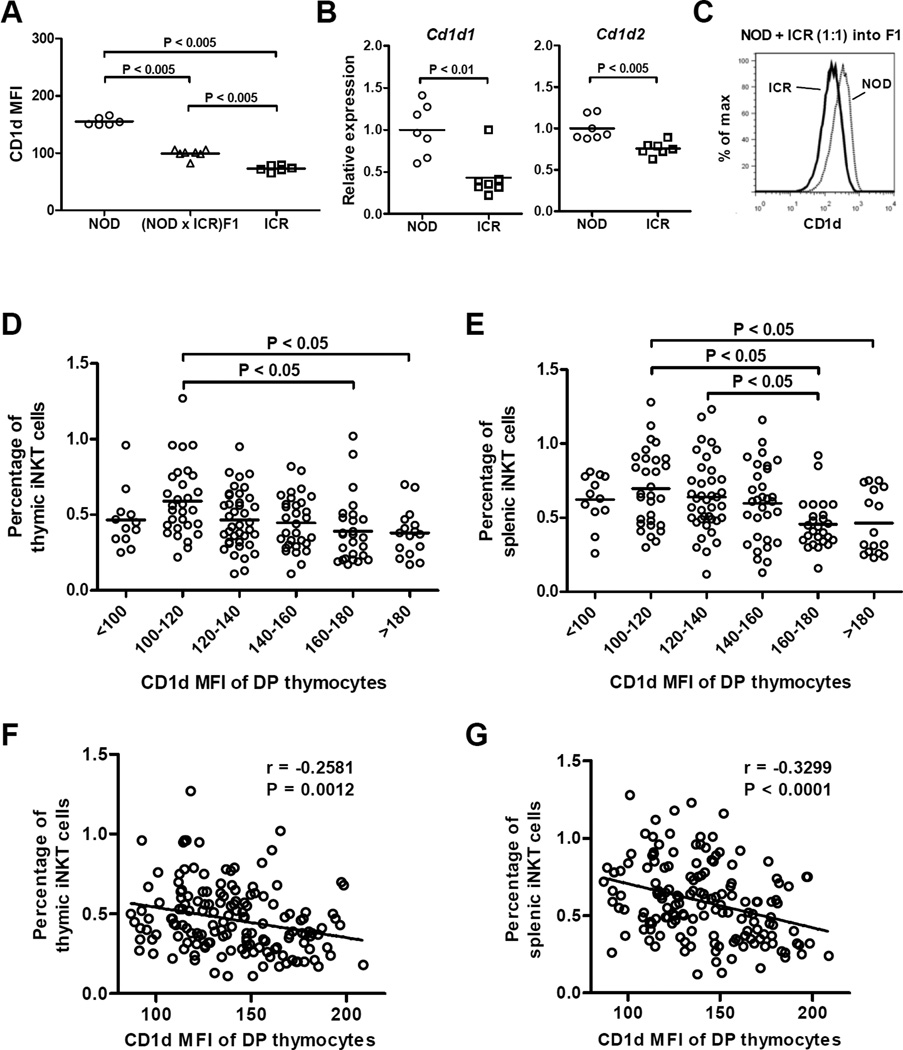
CD1d expression on DP thymocytes is essential and sufficient to support the development of iNKT-cells31–35. Reduced expression of CD1d molecules on DP thymocytes could represent another possible mechanism underlying impaired iNKT-cell extrinsic factors limiting their development in NOD mice. Therefore, we compared the levels of CD1d protein expression on DP thymocytes in NOD and ICR mice. To our surprise, NOD mice were found to express a higher level of CD1d than that in the ICR strain (Fig. 4A). (NOD × ICR)F1 mice showed an intermediate level of CD1d expression, indicating this trait is genetically controlled in a semi-dominant manner (Fig. 4A). To determine if the difference is regulated at the mRNA level, we independently analyzed Cd1d1 and Cd1d2 transcripts and confirmed that both genes were expressed at higher levels in the DP thymocytes of NOD mice than in the ICR strain (Fig. 4B). It was also evident that the difference was greater in the Cd1d1 than Cd1d2 gene. The differential levels of CD1d molecules on the DP thymocytes of NOD and ICR mice are cell autonomously controlled as the same respectively high and low expression patterns were observed in the mixed BM chimeras (Fig. 4C).
Figure 4. CD1d expression level inversely correlates with the frequency of iNKT-cells.
(A) Flow cytometry analysis of CD1d expression on DP thymocytes of 7-week-old NOD, ICR, and (NOD × ICR)F1 males. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the Mann Whitney test. (B) The relative levels of Cd1d1 (left) and Cd1d2 (right) transcripts in DP thymocytes isolated from NOD and ICR mice. Total RNA was isolated from sorted DP thymocytes and reverse transcribed into cDNA. The abundance of Cd1d1 and Cd1d2 cDNA was determined by qPCR and normalized to that of Gapdh. The results are presented as the fold change relative to the mean of all analyzed NOD mice. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the Mann Whitney test. (C) CD1d expression difference in NOD and ICR DP thymocytes is cell autonomously controlled. NOD and ICR BM cells were mixed at a 1:1 ratio and transferred into lethally irradiated (1100 Rads) (NOD × ICR)F1 recipients. CD1d expression on NOD or ICR derived DP thymocytes was determined by flow cytometry at 9 weeks post BM reconstitution. The representative result of 3 BM recipients is shown. (D and E) A cohort of 163 first backcross (BC1) males were analyzed to test if there is an inverse relationship between CD1d expression level on DP thymocytes and the frequency of iNKT-cells. BC1 mice are grouped based on the expression level of CD1d and the frequencies of thymic (D) and splenic (E) iNKT-cells are statistically compared by one way analysis of variance followed by the Tukey’s multiple comparison test to compare all possible pairs. Each symbol represents one mouse. The horizontal bar indicates the mean. Overall negative correlations were also found between CD1d expression levels on DP thymocytes and iNKT-cell frequencies in the thymus (F) and spleen (G). The corresponding Pearson correlation r and the P value are indicated.
It is possible that the respectively higher and lower expression of CD1d molecules in NOD and ICR mice is a strain specific characteristic but not a factor that contributes to the difference of their iNKT-cell compartment. Thus, we outcrossed NOD mice to the ICR strain to generate F1 females that were subsequently mated to NOD males to generate a cohort of first backcross (BC1) mice where multiple loci capable of modulating iNKT-cell development were segregating. We analyzed the expression level of CD1d molecules on DP thymocytes as well as the frequencies of thymic and splenic iNKT-cells in the BC1 progeny. The BC1 mice were arbitrarily grouped based on the expression levels of CD1d on their DP thymocytes. As respectively shown in Figures 4D and 4E, lower expression of CD1d molecules was associated with higher percentages of thymic and splenic iNKT-cells. In addition, there was an overall negative correlation between CD1d expression levels on DP thymocytes and iNKT-cell frequencies in the thymus and spleen (Fig. 4F and 4G). Both CD4+ and DN iNKT-cells were similarly affected (data not shown). Thus, these results suggest that in a mixed NOD/ICR genetic background, variation in the physiological level of CD1d expression could inversely affect iNKT-cell development.
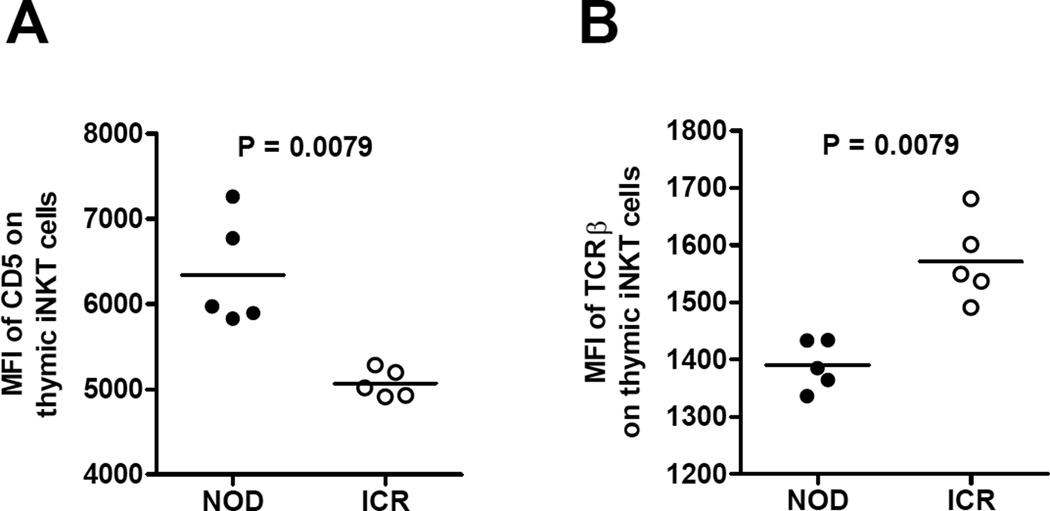
We also analyzed CD5 expression on thymic iNKT-cells to find evidence of altered TCR signals in these cells between NOD and ICR mice as a result of engaging different levels of CD1d molecules on DP thymocytes. As shown in Figure 5A, we observed higher expression levels of CD5 on NOD thymic iNKT-cells compared to those in the ICR mice. On the other hand, the overall fluorescent intensity of TCRβ staining was lower on NOD thymic iNKT-cells than those in the ICR mice (Fig. 5B). These results suggest that NOD iNKT-cells received stronger signals from the CD1d-TCR interaction and those expressing higher levels of TCR may have been deleted. We did not observe a difference in the fluorescent intensity of CD1d tetramer staining, indicating that the remaining iNKT-cells have similar TCR affinity in NOD and ICR mice (data not shown).
Figure 5. Evidence of differential CD1d-TCR interaction strength on thymic iNKT-cells between NOD and ICR mice.
The MFI of (A) CD5 and (B) TCRβ staining on NOD and ICR thymic iNKT-cells (gated on TCRβ+/CD1d tetramer+). Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the Mann Whitney test.
Genetic mapping of CD1d expression
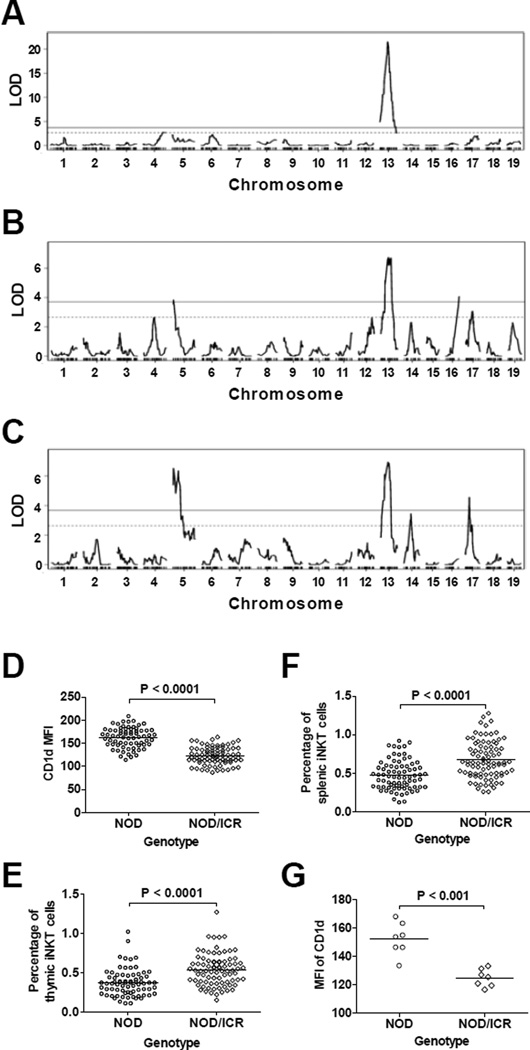
We previously identified multiple QTL regulating the development of iNKT-cells in a (NOD × ICR)F2 cross23. We reasoned that if the expression level of CD1d is a factor that differentially regulates iNKT-cell development in NOD and ICR mice, then a common genetic locus would be identified to control both traits. To test this possibility, we conducted a QTL analysis for the level of CD1d expression on DP thymocytes using the BC1 mice described in Figures 4D and 4E. We identified one highly significant QTL (LOD score > 20, explaining 48% of the variance) on Chr 13 regulating CD1d expression on DP thymocytes (Fig. 6A). The peak LOD score was marked by rs29566823 (62.59Mb) with a 95% confidence interval defined by rs46755152 (60.12 Mb) and rs13481868 (70.59 Mb). The Chr 13 QTL was not found to interact with other genetic loci. The NOD allele at the Chr 13 locus enhanced the overall expression level of CD1d on DP thymocytes in the BC1 mice (Fig. 6D). In our previously reported (NOD × ICR)F2 cross studies23, the same Chr 13 locus overlapping with Idd14 was identified as a suggestive QTL for the percentage of splenic iNKT-cells in the genome scan for main effect analysis, and as a significant locus for both thymic and splenic iNKT-cell frequencies in the multiple regression analyses. In these prior analyses, the NOD allele at this Chr 13 locus reduced the percentage of iNKT-cells in the F2 progeny. To further test the inverse relationship between the level of CD1d expression and the percentage of iNKT-cells, we also conducted QTL analysis for the frequency of thymic and splenic iNKT-cells in the BC1 progeny generated in the current study. Consistent with the previous F2 cross results, a significant QTL was found at the same Chr 13 locus (Figs. 6B and 6C) where the NOD allele lowered the frequency of thymic and splenic iNKT-cells (Figs. 6E and 6F respectively). Also consistent with the previous F2 cross, we identified a similar linkage on Chr 17. The location of the Chr 17 QTL is 5–20 Mb distal to the H2 locus. Taken together, our results suggest that a gene(s) within the Chr 13 locus modulates the development of iNKT-cells through reciprocally altering the expression level of CD1d, although the possibility that these two phenotypes are regulated by distinct genetic variants cannot be completely ruled out.
Figure 6. Genetic mapping of CD1d expression and the iNKT-cell frequency.
QTL analysis was performed to map genetic loci that regulate the expression level of CD1d on DP thymocytes and the frequencies of thymic and splenic iNKT-cells. (A) The LOD score plot of CD1d expression on DP thymocytes. (B) The LOD score plot of thymic iNKT-cell frequency among total thymocytes. (C) The LOD score plot of splenic iNKT-cell frequency among total splenocytes. The solid and dashed lines respectively represent the genome-wide adjusted significant (P < 0.01) and suggestive (P < 0.1) LOD score thresholds. (D) The single ICR allele at the Chr 13 peak marker rs29566823 reduced the expression of CD1d on DP thymocytes. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the unpaired t test. (E) The single ICR allele at the Chr 13 peak marker rs46755152 increased the frequency of thymic iNKT-cells. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the unpaired t test. (F) The single ICR allele at the Chr 13 peak marker rs46755152 increased the frequency of splenic iNKT-cells. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the unpaired t test. (G) Independent confirmation of genotype-phenotype (CD1d expression level) association in 14 BC1 mice. The homozygous NOD genotypes at Chr 13 markers rs46755152 and rs13481868 significantly enhanced the expression of CD1d on DP thymocytes. Each symbol represents one mouse. The horizontal bar indicates the mean. Statistical analysis was performed using the Mann Whitney test.
One caveat of our CD1d expression analysis in the mapping study is that not all BC1 mice were analyzed on the same day. While we considered it unlikely that the highly significant QTL identified on Chr 13 was due to day-to-day variations in the instrument and the antibody staining process, this remained a possibility. Thus, we generated a separate cohort of 14 BC1 mice and determined the CD1d expression level on their DP thymocytes within the same experiment. All 14 BC1 mice were also genotyped for two SNP markers that define the 95% confidence interval of the Chr 13 QTL. In all of the 14 mice, these two SNP markers were perfectly linked (same NOD or NOD/ICR genotypes). As shown in Figure 6G, the BC1 mice that carried homozygous NOD alleles at both SNPs expressed significantly higher levels of CD1d than those that were heterozygous at both markers. Therefore, we have identified a Chr 13 locus that inversely regulates the level of CD1d expression on DP thymocytes and the frequency of thymic and splenic iNKT-cells.
Analyses of thymic iNKT-cell frequencies and DP thymocyte CD1d expression levels in various inbred strains
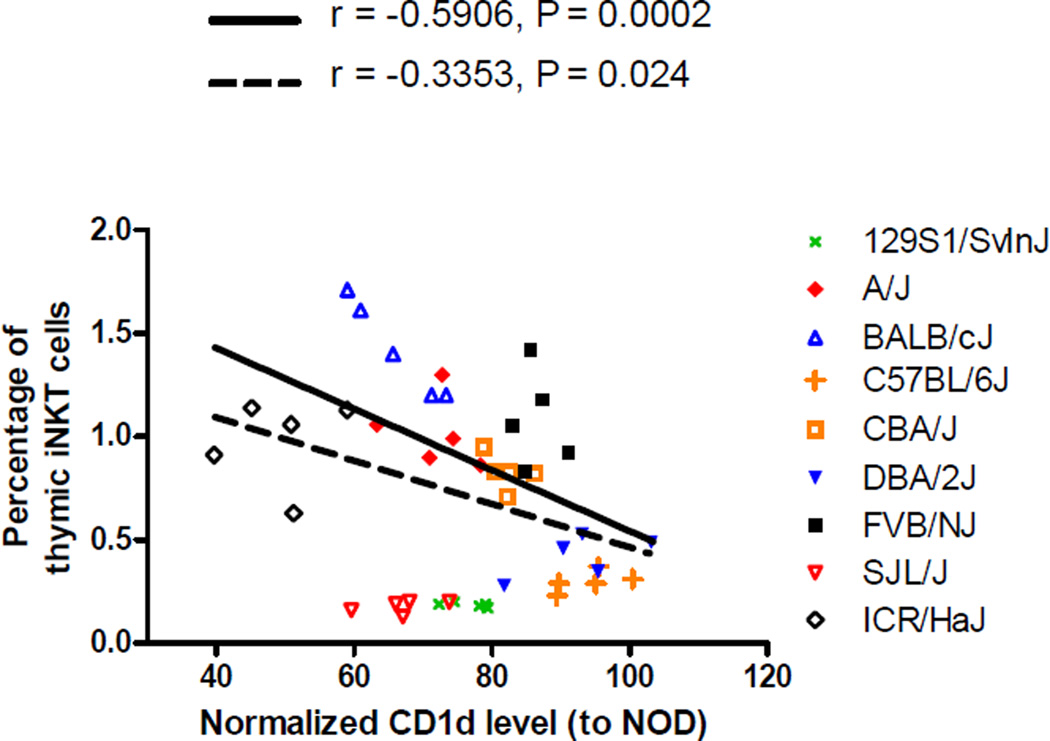
We wished to test if the inverse relationship between CD1d expression levels on DP thymocytes and the frequency of thymic iNKT-cells observed in NOD and ICR mice could be extended to other mouse strains. To test this, we analyzed a panel of classical inbred strains, including CBA/J, BALB/cJ, 129/SvInJ, A/J, FVB/J, SJL/J, C57BL6/J, and DBA2/J. We also include ICR/HaJ mice for comparison. We normalized the mean fluorescent intensity level of each individual mouse to the mean of 3 NOD/ShiLtJ mice analyzed on the same days to control for day-to-day variation in the analysis of CD1d expression on DP thymocytes. When all analyzed strains were considered, there was a significant negative correlation (P = 0.024) between the CD1d expression level and the frequency of thymic iNKT-cells (Fig. 7). Notably, 129/SvInJ and SJL/J mice appeared to fall outside of the linear relationship when compared to other strains (Fig. 7). When 129/SvInJ and SJL/J mice were excluded from the statistical analysis, the negative correlation becomes even more significant (P = 0.0002) between the CD1d expression level and the percentage of thymic iNKT-cells (Fig. 7). These results suggest that the presence of the inverse relationship between the level of CD1d expression on DP thymocytes and the frequency of thymic iNKT-cells is genetic background dependent.
Figure 7. Negative correlation of thymic iNKT-cell frequencies and DP thymocyte CD1d expression levels in a panel of inbred strains.
The proportion of thymic iNKT-cells and the level of CD1d expression on DP thymocytes were determined in 7-week-old males of the indicated strains. For each strain, 5 mice were analyzed side by side with 3 NOD males in the same experiment. The CD1d expression level was then normalized to the mean of the 3 NOD mice. Each symbol represents one mouse. The dotted line represents the best fit linear regression line when all strains are considered. The solid line denotes the best fit liner regression line when 129S1/SvInJ and SJL/J are excluded. The corresponding Pearson correlation r and the P value are also indicated.
Discussion
In the current study, we used mixed BM chimeras to demonstrate that iNKT-cell extrinsic but hematopietic cell intrinsic factors contribute to their developmental defects in NOD mice. In addition, we showed that CD1d expression levels on DP thymocytes inversely correlated with the frequencies of thymic and splenic iNKT-cells in the BC1 progeny. The genetic control of CD1d expression was further mapped by linkage analysis to a Chr 13 region that explained 48% of the variance. This indicates that a polymorphic gene(s) within the Chr 13 QTL plays a major role in modulating the CD1d expression level although other loci with minor effects may also be involved in this regulation. The same Chr 13 locus was also found to significantly regulate iNKT-cell frequencies in the BC1 mice. Collectively, our results suggest that a gene within the Chr 13 linkage interval regulates iNKT-cell development through reciprocally modulating CD1d expression on DP thymocytes. The Chr 13 locus was identified as a suggestive QTL that regulated iNKT-cell frequency in our previous (NOD × ICR)F2 cross. Thus, our current study also demonstrates that analyses of sub-phenotypes potentially linked to iNKT-cell development are a useful approach to further confirm and define the biological contribution of previously identified loci.
Thymic iNKT-cells have been shown to exert an effect on other developing T-cells36, 37. It is possible that the number of iNKT-cells in the thymus could modulate the expression of CD1d on DP thymocytes. Thus, the inverse relationship between the level of CD1d expression and the frequency of iNKT-cells could be due to an iNKT-cell derived factor that suppresses the expression of CD1d molecules. However, we consider this explanation unlikely. This conclusion is based on the results presented in Figure 4. In the BM chimeras where a 1:1 ratio of NOD and ICR BM cells were transferred, NOD and ICR derived DP thymocytes respectively expressed high and low levels of CD1d even when they were exposed to the same iNKT-cells (Fig. 4C). In addition, in our BC1 progeny, various iNKT-cell frequencies were observed in mice having similar levels of CD1d expression. Therefore, our results suggest that a gene(s) on Chr 13 controls the level of CD1d expression which in turn modulates iNKT-cell development. There are 214 protein coding and non-coding genes listed at Mouse Genome Informatics (www.informatics.jax.org) within the Chr 13 QTL confidence interval (Supplementary Table 1). Among these, there are several protease cathepsin protein coding genes, including Ctsl. Thymic expression of cathepsin L (encoded by Ctsl) is essential for iNKT-cell development38. However, a possibility of Ctsl as the underlying gene of the Chr 13 QTL can be ruled out because cathepsin L deficient mice express normal levels of CD1d on their DP thymocytes38. Our gene expression results indicate that the level of CD1d expression is regulated at the transcript level by a gene(s) within the Chr 13 linkage region. Within this QTL confidence interval, there are also several zinc finger protein and miRNA coding genes, which could potentially regulate CD1d expression at the transcriptional level. Future mapping and gene expression studies will test this possibility.
CD1d expression in the thymus is essential to support the development of iNKT-cells. However, the mechanisms controlling CD1d expression on DP thymocytes remain largely unknown. Unique polymorphisms present in the CD1d promoter of PWD/PhJ and PWK/PhJ strains were demonstrated to decrease CD1d expression on DP thymocytes39. The same report showed that reduced CD1d expression was associated with the lower frequency of iNKT-cells also observed in PWD/PhJ and PWK/PhJ mice when compared to the C57BL/6 strain. On the other hand, we showed that there was an inverse relationship between the level of CD1d expression on DP thymocytes and the frequency of iNKT-cells in NOD and ICR strains and their BC1 progeny. Interestingly, the frequency of iNKT-cells was not found to be further increased in our BC1 mice showing the lowest CD1d expression level (Fig. 4D and 4E). We also attempted to determine if the inverse relationship observed between NOD and ICR mice could be applied to other commonly used classical inbred strains. Our results showed that only a subset but not all the strains analyzed here exhibited a negative correlation between CD1d expression levels and iNKT-cell frequencies. Development of iNKT-cells is a complex trait controlled by multiple genes. Previous mouse genetic studies, including ours, have identified more than 10 loci by congenic mapping approaches or crosses between two strains9, 10, 15, 19, 21, 23, 40, 41. Thus, it is reasonable to consider that the effect of CD1d expression level on iNKT-cell development could be masked by other dominant allelic variants in certain genetic backgrounds. We have also compared the frequencies of iNKT-cells between wild-type NOD and NOD.CD1d+/− mice but did not observe a significant difference (data not shown), albeit the CD1d expression level was reduced by about 50% in the latter strain. Thus, these results indicate that the physiological variation in CD1d expression plays an important role in controlling the development of iNKT-cells, but reducing its level alone is not sufficient to increase the frequency of these T-cells in NOD mice.
Previous studies utilizing NOD congenic strains or a cross between NOD and B6 mice did not identify the Chr 13 locus for the frequency of iNKT-cells. Congenic mapping studies have not tested the Chr 13 region. One explanation to why the Chr 13 locus was not mapped in the previous NOD and B6 cross is that the underlying gene is not polymorphic between NOD and B6 strain. This possibility is supported by the similar CD1d expression level observed between NOD and B6 strains (Figure 7).
The exact mechanism underlying the inverse relationship between CD1d expression level on DP thymocytes and the frequency of iNKT-cells remains to be elucidated. Lck promoter driven transgenic over-expression of CD1d molecules exclusively on thymocytes did not lead to reduced iNKT-cells34, 35. On the other hand, when driven by a MHC class I gene promoter, over-expression of CD1d caused a significant reduction of iNKT-cells as a result of enhanced dendritic cell (DC) mediated negative selection42. We also observed higher level of CD1d expression on NOD thymic DC (CD11c+) compared to their ICR counterparts (data not shown). Future studies will determine if differential expression of CD1d molecules on thymic DC contributes to the respectively low and high iNKT-cell frequencies in NOD and ICR mice as a result of negative selection.
Homotypic interactions of SLAMF1 and SLAMF6, presumably at the DP stage, are important for the development of iNKT-cells24. NOD and ICR have different haplotypes at the Slam/Cd2 locus on the distal end of Chr 1 (based on available SNP genotyping results at the Mouse Phenome Database, http://phenome.jax.org/) that most likely modulate their expression levels in cis as shown previously29, 30, 43. Although lower SLAMF1 expression on NOD DP thymocytes could explain their reduced ability to select and support the development of iNKT-cells, we do not consider it the major contributor to the observed difference in the frequency and number of this unique T-cell subset in NOD and ICR mice. This interpretation is based on the results of our previously reported F2 mapping studies23 and the BC1 genetic analyses described here where the Slam/Cd2 locus was not identified as a QTL for the frequency of thymic iNKT-cells. Since SLAMF1 expression remained lower on NOD-derived DP thymocytes compared to those of the ICR origin in mixed BM chimeras, our results indicate that ICR derived signals do not promote NOD iNKT-cell development through normalizing SLAMF1 expression on their precursors. Our results also demonstrate that low expression of SLAMF1 on NOD DP thymocytes do not significantly prevent them from becoming iNKT-cells when present in a permissive environment. Although both SLAMF1 and SLAMF6 contribute to iNKT-cell development in the thymus, the latter appears to play a more important role as SLAMF6 deficient mice have fewer iNKT-cells compared to Slamf1 knockout mutants24. Therefore, the level of SLAMF6 in NOD mice was able to compensate for their low SLAMF1 expression to support iNKT-cell development in the presence of ICR DP thymocytes in the mixed BM chimeras. Nonetheless, our results do not argue against a role of low SLAMF1 expression in defective iNKT-cell development in NOD mice as previously indicated29, 30. Optimal development of iNKT-cells is most likely finely tuned by the combined effects of the expression levels of CD1d and SLAMF family proteins, including SLAMF1, SLAMF3, and SLAMF6.
Our mixed BM chimerism experiments also demonstrate that the developmental defect of NOD iNKT-cells is not cell-intrinsic. Indeed, in mixed BM chimeras more thymic iNKT-cells were derived from NOD than ICR hematopoietic precursors. This indicates that the normally impaired development of iNKT-cells in NOD mice is due to non-cell autonomous factors. We previously identified a major QTL on Chr 17 regulating iNKT-cell frequencies in a (NOD × ICR)F2 cross23. The same locus was also identified in the BC1 cross. The NOD but not the ICR allele of the Chr 17 QTL promoted iNKT-cell development. Future studies will determine if a gene(s) within this region regulates iNKT-cell development in a cell-intrinsic manner.
In conclusion, we identified a Chr 13 locus that inversely regulates CD1d expression on DP thymocytes and the frequency of iNKT-cells. In addition, our results indicate that the reduced frequency of iNKT-cells in NOD mice is not cell autonomously controlled but is mainly due to the impaired ability of their BM derived cells, most likely DP thymocytes, to support the development of this T-cell subset.
Materials and Methods
Mice
NOD/LtDvs (hereafter NOD) mice are maintained at the Medical College of Wisconsin (MCW) by brother-sister mating. ICR/HaJ (ICR), NOD.Cd45.2, and NOD.Cd1d−/− mice were purchased from The Jackson Laboratory (TJL) (stock numbers 009122, 014149, and 006330 respectively) and maintained by brother-sister mating at the MCW. Five-week-old male NOD/ShiLtJ, CBA/J, BALB/cJ, 129/SvInJ, A/J, FVB/J, SJL/J, C57BL6/J, and DBA2/J were purchased from TJL and housed in the MCW vivarium for two weeks before the experiments. NOD mice were outcrossed to ICR mice to generate reciprocal F1 females that were subsequently mated to NOD males to generate first backcross (BC1) progeny. A total of 163 male BC1 mice were generated for analysis. Results from all BC1 males derived from both directions of the F1 crosses were combined for statistical analyses. All BC1 mice were analyzed at 7 weeks old. Previous studies showed sex difference in the frequency of thymic iNKT-cells10. Based on this reason, male mice were used in all analyses. All animal protocols were approved by the MCW Institutional Animal Care and Use Committees.
Generation of bone marrow chimeras
Bone marrow (BM) cells were harvested from both femurs and tibias of 6–10 week-old donors. T-cells were depleted using anti-CD3 microbeads and LD columns from Miltenyi Biotec (San Diego, CA). NOD and ICR BM cells (5×106 total) were intravenously transferred alone or co-injected at a 1:1 ratio into lethally (1100 Rads) irradiated 4–8 week-old (NOD × ICR)F1 mice. In separate experiments, T-cell depleted NOD.Cd1d−/− BM cells admixed at different ratios with those from NOD.Cd45.2 or ICR mice were used as donor cells. BM chimeras were analyzed at 8–10 weeks post reconstitution. The F1 mice were used as recipients to avoid potential rejection of the BM cells, and to distinguish NOD and ICR derived cells.
Flow cytometry
Red blood cell depleted single cell suspensions were prepared from the thymus and the spleen. Cells were Fc-blocked at room temperature with anti-CD16/CD32 (clone 2.4G2, Bio×Cell) for 10 min followed by staining with an antibody cocktail containing CD1d/α-galactosylceramide analog (PBS-57) loaded tetramers, anti-CD4 (clone RM4-5), and anti-TCRβ (clone H57-597) for 30 min at 4C. Thymocytes were also stained with anti-CD8 (clone 53-6.7) and anti-CD4 as well as anti-CD1d (clone 1B1), anti-CD5 (clone 53-7.3), anti-SLAMF1 (clone TC15-12F12.2), anti-SLAMF3 (clone Ly9ab3), or anti-SLAMF6 (clone 330-AJ) as indicated. For analyzing BM chimeras, the thymus and spleen were similarly processed and cells were stained with CD1d tetramers, anti-CD4, and anti-TCRβ, anti-CD45.1 (clone A20) and anti-CD45.2 (clone 104). CD1d tetramers were provided by the NIH tetramer core facility. All fluorochrome conjugated antibodies were purchased from BD Bioscience (San Jose, CA), BioLegend (San Diego, CA), or eBioscience (San Diego, CA). Stained cells were washed, run on a FACSCalibur or a LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and analyzed using the FlowJo software (Tree Star, Ashland, OR). Dead cells were excluded by propidium iodide or 7-AAD staining. Both dyes were purchased from Sigma-Aldrich (St. Louis, MO).
Single nucleotide polymorphism genotyping
Tail genomic DNA was purified by the Wizard Genomic DNA Purification kit (Promega, Madison, WI). Purified tail DNA samples of BC1 mice used for mapping studies were sent to GeneSeek Illumina Genotyping Services (Lincoln, NE) and genotyped with the Mouse Universal Genotyping Array (MUGA) as previously described23. This platform analyzes 7,851 single nucleotide polymorphism (SNP) markers throughout the mouse genome with an average spacing of 325 Kb. NOD, ICR and (NOD × ICR)F1 DNA samples were also included for quality control purposes. Genotypes on these control samples were used to filter out non-polymorphic markers as well as markers that were technical failures based on inconsistent genotypes between parental strains and the F1 sample. A total of 1,528 markers on autosomes passed the filtering and were used in the QTL mapping analysis. Genetic map positions of these markers (cM) were updated to the new mouse genetic map using the web tool mouse map converter at http://cgd.jax.org44. The new map resolves inconsistencies between the physical and genetic maps.
Quantitative trait loci (QTL) analysis
The distributions of measured phenotypes were skewed so all traits were log-transformed before analysis. A three-stage QTL mapping approach was performed on each trait using R/qtl v1.25–15 (http://www.rqtl.org/)45 as described46, 47. Briefly, the first stage is a single-locus genome scan to identify single loci associated with the CD1d expression, thymic and splenic iNKT-cell traits (main effect QTL). The respective genome-wide adjusted thresholds for significant (P < 0.01) and suggestive (P < 0.1) LOD scores were based on 10,000 permutations of the observed data. QTL confidence intervals (CIs) were determined by the posterior probability as previously described46. In stage 2 of the analysis, simultaneous pair-wise scans to detect additive and epistatic effects were performed. The third stage was to collect QTL detected at previous 2 stages to construct a multiple-QTL model to determine the combined effects of all QTL detected on the traits.
Quantitative real time polymerase chain reaction (qPCR)
NOD and ICR thymocytes isolated form 7 week-old males were stained with anti-CD4 and anti-CD8, and DP cells were sorted on a FACS Aria IIu (BD Biosciences). The purity was higher than 99%. Total RNA was extracted using the RNeasy Plus Mini kit (Qiagen) as recommended by the manufacturer. First strand cDNA was synthesized with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instruction. qPCR was performed on a 7900HT Sequence Detection System (Applied Biosystems) using TaqMan® gene expression assays for Gapdh (assay number: Mm99999915_g1), Cd1d1 (assay number: Mm00783541_s1), and Cd1d2 (assay number: Mm00776138_gh). Relative expression of Cd1d1 and Cd1d2 was determined using Gapdh as the internal control using the comparative Ct method.
Statistical analysis
General statistical analysis was performed using Prism 5 (GraphPad Software). The specific statistics used to calculate P values for different comparisons are indicated in the corresponding Figure Legends.
Supplementary Material
Acknowledgements
We are also grateful to the NIH tetramer core facility for providing us CD1d tetramers. This work was supported by National Institutes of Health grants DK077443 and AI110963 (to Y.-G.C.), DK46266 and DK95735 (to D.V.S.), as well as by grants from the Helmsley Charitable Trust (2014PG-T1D048 to D.V.S.), The Juvenile Diabetes Research Foundation, The American Diabetes Association, and the Children’s Hospital of Wisconsin Foundation.
Footnotes
Conflict of interest
The authors have no financial conflicts of interest.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol. 2008;20(3):358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YG, Tsaih SW, Serreze DV. Genetic control of murine invariant natural killer T-cell development dynamically differs dependent on the examined tissue type. Genes Immun. 2012;13(2):164–174. doi: 10.1038/gene.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan AC, Serwecinska L, Cochrane A, Harrison LC, Godfrey DI, Berzins SP. Immune characterization of an individual with an exceptionally high natural killer T cell frequency and her immediate family. Clin Exp Immunol. 2009;156(2):238–245. doi: 10.1111/j.1365-2249.2009.03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montoya CJ, Pollard D, Martinson J, Kumari K, Wasserfall C, Mulder CB, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. 2007;122(1):1–14. doi: 10.1111/j.1365-2567.2007.02647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kis J, Engelmann P, Farkas K, Richman G, Eck S, Lolley J, et al. Reduced CD4+ subset and Th1 bias of the human iNKT cells in Type 1 diabetes mellitus. J Leukoc Biol. 2007;81(3):654–662. doi: 10.1189/jlb.1106654. [DOI] [PubMed] [Google Scholar]
- 7.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. 2005;35(5):1399–1407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 8.Lee PT, Putnam A, Benlagha K, Teyton L, Gottlieb PA, Bendelac A. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest. 2002;110(6):793–800. doi: 10.1172/JCI15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YG, Driver JP, Silveira PA, Serreze DV. Subcongenic analysis of genetic basis for impaired development of invariant NKT cells in NOD mice. Immunogenetics. 2007;59(9):705–712. doi: 10.1007/s00251-007-0236-4. [DOI] [PubMed] [Google Scholar]
- 10.Esteban LM, Tsoutsman T, Jordan MA, Roach D, Poulton LD, Brooks A, et al. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol. 2003;171(6):2873–2878. doi: 10.4049/jimmunol.171.6.2873. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher JM, Jordan MA, Snelgrove SL, Slattery RM, Dufour FD, Kyparissoudis K, et al. Congenic analysis of the NKT cell control gene Nkt2 implicates the peroxisomal protein Pxmp4. J Immunol. 2008;181(5):3400–3412. doi: 10.4049/jimmunol.181.5.3400. [DOI] [PubMed] [Google Scholar]
- 12.Hammond KJ, Poulton LD, Palmisano LJ, Silveira PA, Godfrey DI, Baxter AG. alpha/beta-T cell receptor (TCR)+CD4-CD8- (NKT) thymocytes prevent insulin-dependent diabetes mellitus in nonobese diabetic (NOD)/Lt mice by the influence of interleukin (IL)-4 and/or IL-10. J Exp Med. 1998;187(7):1047–1056. doi: 10.1084/jem.187.7.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong S, Wilson MT, Serizawa I, Wu L, Singh N, Naidenko OV, et al. The natural killer T-cell ligand alpha-galactosylceramide prevents autoimmune diabetes in non-obese diabetic mice. Nat Med. 2001;7(9):1052–1056. doi: 10.1038/nm0901-1052. [DOI] [PubMed] [Google Scholar]
- 14.Lehuen A, Lantz O, Beaudoin L, Laloux V, Carnaud C, Bendelac A, et al. Overexpression of natural killer T cells protects Valpha14- Jalpha281 transgenic nonobese diabetic mice against diabetes. J Exp Med. 1998;188(10):1831–1839. doi: 10.1084/jem.188.10.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuki N, Stanic AK, Embers ME, Van Kaer L, Morel L, Joyce S. Genetic dissection of V alpha 14J alpha 18 natural T cell number and function in autoimmune-prone mice. J Immunol. 2003;170(11):5429–5437. doi: 10.4049/jimmunol.170.11.5429. [DOI] [PubMed] [Google Scholar]
- 16.Naumov YN, Bahjat KS, Gausling R, Abraham R, Exley MA, Koezuka Y, et al. Activation of CD1d-restricted T cells protects NOD mice from developing diabetes by regulating dendritic cell subsets. Proc Natl Acad Sci U S A. 2001;98(24):13838–13843. doi: 10.1073/pnas.251531798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV, et al. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med. 2001;7(9):1057–1062. doi: 10.1038/nm0901-1057. [DOI] [PubMed] [Google Scholar]
- 18.Shi FD, Flodstrom M, Balasa B, Kim SH, Van Gunst K, Strominger JL, et al. Germ line deletion of the CD1 locus exacerbates diabetes in the NOD mouse. Proc Natl Acad Sci U S A. 2001;98(12):6777–6782. doi: 10.1073/pnas.121169698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno A, Wang J, Cheng L, Im JS, Shi Y, Porcelli SA, et al. Enhanced early expansion and maturation of semi-invariant NK T cells inhibited autoimmune pathogenesis in congenic nonobese diabetic mice. J Immunol. 2008;181(10):6789–6796. doi: 10.4049/jimmunol.181.10.6789. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Geng YB, Wang CR. CD1-restricted NK T cells protect nonobese diabetic mice from developing diabetes. J Exp Med. 2001;194(3):313–320. doi: 10.1084/jem.194.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zekavat G, Mozaffari R, Arias VJ, Rostami SY, Badkerhanian A, Tenner AJ, et al. A novel CD93 polymorphism in non-obese diabetic (NOD) and NZB/W F1 mice is linked to a CD4+ iNKT cell deficient state. Immunogenetics. 2010;62(6):397–407. doi: 10.1007/s00251-010-0442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leiter E, Atkinson M, editors. NOD mice and related strains: research applications in diabetes, AIDS, cancer and other diseases. R.G. Lands Company; 1998. [Google Scholar]
- 23.Tsaih SW, Khaja S, Ciecko AE, MacKinney E, Chen YG. Genetic control of murine invariant natural killer T cells maps to multiple type 1 diabetes regions. Genes Immun. 2013;14(6):380–386. doi: 10.1038/gene.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27(5):751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11(5):435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, et al. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36(6):986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang SH, Cannons JL, Cheng J, et al. A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes. J Immunol. 2013;190(5):2121–2128. doi: 10.4049/jimmunol.1202145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sintes J, Cuenca M, Romero X, Bastos R, Terhorst C, Angulo A, et al. Cutting edge: Ly9 (CD229), a SLAM family receptor, negatively regulates the development of thymic innate memory-like CD8+ T and invariant NKT cells. J Immunol. 2013;190(1):21–26. doi: 10.4049/jimmunol.1202435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178(3):1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 30.Jordan MA, Fletcher JM, Jose R, Chowdhury S, Gerlach N, Allison J, et al. Role of SLAM in NKT Cell Development Revealed by Transgenic Complementation in NOD Mice. J Immunol. 2011;186(7):3953–3965. doi: 10.4049/jimmunol.1003305. [DOI] [PubMed] [Google Scholar]
- 31.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol. 2000;164(5):2412–2418. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 33.Schumann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol. 2005;175(11):7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- 34.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202(2):239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmer MI, Colmone A, Felio K, Xu H, Ma A, Wang CR. A cell-type specific CD1d expression program modulates invariant NKT cell development and function. J Immunol. 2006;176(3):1421–1430. doi: 10.4049/jimmunol.176.3.1421. [DOI] [PubMed] [Google Scholar]
- 36.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11(8):709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14(11):1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honey K, Benlagha K, Beers C, Forbush K, Teyton L, Kleijmeer MJ, et al. Thymocyte expression of cathepsin L is essential for NKT cell development. Nat Immunol. 2002;3(11):1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 39.Borg ZD, Benoit PJ, Lilley GW, Aktan I, Chant A, DeVault VL, et al. Polymorphisms in the CD1d promoter that regulate CD1d gene expression are associated with impaired NKT cell development. J Immunol. 2014;192(1):189–199. doi: 10.4049/jimmunol.1301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Liang ZY, Matsuki N, Van Kaer L, Joyce S, Wakeland EK, et al. A murine locus on chromosome 18 controls NKT cell homeostasis and Th cell differentiation. Journal of Immunology. 2003;171(9):4613–4620. doi: 10.4049/jimmunol.171.9.4613. [DOI] [PubMed] [Google Scholar]
- 41.Tsukamoto K, Ohtsuji M, Shiroiwa W, Lin Q, Nakamura K, Tsurui H, et al. Aberrant genetic control of invariant TCR-bearing NKT cell function in New Zealand mouse strains: possible involvement in systemic lupus erythematosus pathogenesis. J Immunol. 2008;180(7):4530–4539. doi: 10.4049/jimmunol.180.7.4530. [DOI] [PubMed] [Google Scholar]
- 42.Chun T, Page MJ, Gapin L, Matsuda JL, Xu H, Nguyen H, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003;197(7):907–918. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, et al. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, et al. A new standard genetic map for the laboratory mouse. Genetics. 2009;182(4):1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 46.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159(1):371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wergedal JE, Ackert-Bicknell CL, Tsaih SW, Sheng MH, Li R, Mohan S, et al. Femur mechanical properties in the F2 progeny of an NZB/B1NJ × RF/J cross are regulated predominantly by genetic loci that regulate bone geometry. J Bone Miner Res. 2006;21(8):1256–1266. doi: 10.1359/jbmr.060510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.