Abstract
Genome-wide association studies (GWAS) hold tremendous promise to improve our understanding of human biology. Recent GWAS have revealed over 75 loci associated with erythroid traits, including the 4q27 locus that is associated with red blood cell size (mean corpuscular volume, MCV). The close linkage disequilibrium block at this locus harbors the CCNA2 gene that encodes cyclin A2. CCNA2 mRNA is highly expressed in human and murine erythroid progenitor cells and regulated by the essential erythroid transcription factor GATA1. To understand the role of cyclin A2 in erythropoiesis, we have reduced expression of this gene using short hairpin RNAs in a primary murine erythroid culture system. We demonstrate that cyclin A2 levels affect erythroid cell size by regulating the passage through cytokinesis during the final cell division of terminal erythropoiesis. Our study provides new insight into cell cycle regulation during terminal erythropoiesis and more generally illustrates the value of functional GWAS follow-up to gain mechanistic insight into hematopoiesis.
Keywords: Erythropoiesis, Cyclin A2, Cell cycle, Genome-wide association study, Genetics
INTRODUCTION
Genome-wide associations studies (GWAS) have been successful at identifying thousands of common genetic variants associated with human diseases and traits [1, 2]. GWAS of hematologic traits hold promise for gaining an improved understanding of human hematopoiesis [2]. We and others have previously used such follow-up studies to identify BCL11A as a silencer of fetal hemoglobin expression, cyclin D3 as a key regulator of the number of cell divisions during erythropoiesis, and TRIM58 as a ubiquitin ligase necessary for terminal erythropoiesis [3-5]. In the case of cyclin D3, we found that variation in the number of cell divisions resulting from variable levels of cyclin D3 could impact both erythroid cell size and number [3]. Recent GWAS of erythroid traits have revealed a locus associated with red cell size (mean corpuscular volume, MCV) at 4q27 [6]. The close linkage disequilibrium block at the 4q27 locus harboring the most significantly associated genetic variants contains the CCNA2 gene, which encodes cyclin A2 [6, 7].
Given our prior studies of cell cycle regulation during terminal erythropoiesis, we reasoned that studying the role of cyclin A2 during erythropoiesis would provide insight into how this protein could result in natural variation in MCV. The absence of cyclin A2 causes embryonic lethality and its absence from the hematopoietic compartment results in stem cell depletion and consequent pancytopenia [8], which contrasts with our ability to study cyclin D3 in vivo using viable knockout mice [3]. Therefore, we reasoned that reducing the level of cyclin A2 in a primary murine fetal liver erythroid culture system with synchronous differentiation would be ideal to specifically study its dosage-dependent role in erythropoiesis. Since cyclin A2 is degraded at mitosis during each cell division [8], we postulated that this knockdown strategy would be successful and occur soon after introduction of short hairpin RNAs (shRNAs). Importantly, studies have suggested that a cis-regulatory element may harbor the causal non-coding variant at this locus, which is predicted to alter expression of CCNA2, and therefore such knockdown should mimic the situation observed with natural human variation [6, 7, 9].
MATERIALS AND METHODS
Cell culture
293T cells were maintained in DMEM with 10 % fetal bovine serum, 2 mM L-glutamine, and 1 % penicillin/streptomycin. For production of retrovirus, 293T cells were transfected with the appropriate viral packaging and genomic vectors using FuGene 6 reagent (Promega) according to the manufacturer’s protocol. Culture of primary mouse cells is described below.
Constructs
The shRNA sequences targeting mouse Ccna2 were obtained from the RNAi Consortium of the Broad Institute (http://www.broadinstitute.org/rnai/trc) and had the following sequences:
sh4 – AAAAGTTAATGAAGTACCTGACTATGTCGACATAGTCAGGTACTTCATTAAC
sh5 – AAAAGCTTCGAAGTTTGAAGAAATAGTCGACTATTTCTTCAAACTTCGAAGC
These sequences were cloned into the BbsI restriction sites of the linearized MSCV-pgkGFP-U3-U6P retroviral vector, which co-expresses GFP driven by the PGK promoter.
Mouse fetal liver erythroid progenitor purification, retrovirus infection and in vitro culture
E14.5-15.5 fetal liver cells were homogenized in PBS supplemented with 2 % FBS and 100 µM EDTA. Mature erythrocytes were lysed by the addition of ammonium chloride solution (StemCell Technologies, Inc.) at a 1:4 ratio and incubation on ice for 10 min. After washing, the remaining cells were incubated with a cocktail of biotin-conjugated antibodies, including Lineage Cocktail (BD 559971), Ter119 (eBioscience 13-5921-85), CD16/32 (Abcam 25249), Sca-1 (BD 553334), CD34 (MCA1825B), CD41 (MCA2245B). After magnetic depletion with streptavidin beads (BD 557812) a pure fetal liver Ter119-negative erythroid progenitor population was obtained [10].
For retroviral infection, 293T cells were transfected with retroviral construct described above along with the pCL-eco packaging vector. Media was changed the day after transfection. After 24 hours, this media was collected and filtered at 0.45 µm immediately prior to infection of purified erythroid progenitor cells. The cells were mixed with viral supernatant and polybrene (filtered 4 mg/ml stock) was added to the mixture at a final concentration of 0.4 µl/ml of media in a 24-well plate, at a density of 100,000 cells per well. The cells were spun at approximately 32 °C for 90 minutes at 2000 rpm.
Subsequently for differentiation, cells were resuspended in IMDM containing 15 % fetal bovine serum and 0.5 U/ml erythropoietin (EPO, Amgen) for up to 66 h at 37°C, 5 % CO2.
May Grϋnwald-Giemsa Staining
Approximately 50,000-200,000 cells were centrifuged on to poly-L-lysine coated slides and stained with May-Grϋnwald-Giemsa as described previously [3]. Then slides were mounted with coverslips and examined. Stained cells were captured, processed and analyzed using Axiovision Microscopy Software (Carl Zeiss).
Cell cycle analyses, phospho-Histone H3 staining and PKH labeling
In vitro cultured erythroid cells were pulsed with 10µM 5-ethynyl-2’-deoxyuridine (EdU) for 30 min and EdU incorporation was detected using an EdU flow kit (Invitrogen C10418) at indicated time points as described by the manufacturer’s protocol. Propidium iodide (PI) was added to stain for DNA content after RNAse digestion. The PI signal data was acquired on a linear scale.
For phospho-Histone H3 staining erythroid cells were fixed and permeabilized using reagents from an EdU flow kit (Invitrogen C10418). Incubation of anti-phospho-Histone H3 rabbit monoclonal antibody (Ser10, clone MC463, Millipore) was conducted at room temperature for 45 minutes at a 1:400 dilution. After washing, incubation with secondary antibody donkey anti-rabbit AlexaFluor647 (Jackson Labs 711-605-152) was conducted for 30min in the dark at a 1:200 dilution. Anti-GFP-FITC antibody (Abcam ab6662) was used to identify GFP-infected cells at a 1:200 dilution. Propidium iodide (PI) was added to stain for DNA content after RNAse digestion. The PI signal data was acquired on a linear scale.
To allow tracking of the number of cell divisions cells were labeled with the PKH26 red fluorescent cell linker kit (Sigma-Aldrich, PKH26GL-1KT) as described previously [3]. An aliquot of the labeled cells was used to measure the mean fluorescence intensity (MFI) of PKH26 immediately after labeling (0 h). The number of cell divisions was calculated as described previously [3].
Flow cytometry analysis and sorting
For flow cytometry analysis, in vitro cultured erythroid cells were washed in PBS and stained with 7AAD or 1 µg/ml Propidium Iodide (PI), 1:100 APC-conjugated Ter119 (eBioscience 17-5921-83), 1:300 PE-conjugated CD71 (eBioscience 12-0711-83) and 1 µg/ml Hoechst and followed by FACS analysis (BD LSR II flow cytometer) [11]. Data was analyzed using FlowJo v10 (Tree Star).
Quantitative RT-PCR
Isolation of RNA was performed using the miRNeasy Mini Kit (Qiagen). An on-column DNase (Qiagen) digestion was performed according to manufacturer’s instructions. RNA was quantified by a NanoDrop spectrophotometer (Thermo Scientific). Reverse transcription was carried out using the iScript cDNA Synthesis Kit (Bio-Rad). Realtime PCR was performed using the ABI 7900 Machine Real-Time PCR System and SYBR Green PCR Master Mix (Applied Biosystems). The following primers were used for quantitative RT-PCR:
Ccna2 forward 5’-TGGATGGCAGTTTTGAATCACC-3’
Ccna2 reverse 5’-CCCTAAGGTACGTGTGAATGTC-3’
Ubc forward 5’- GAGTTCCGTCTGCTGTGTGA-3’
Ubc reverse 5’- CCTCCAGGGTGATGGTCTTA-3’
Western Blotting
Cells were harvested at indicated timepoints and processed as previously described [3]. After SDS gel electrophoresis and western blotting, membranes were blocked with 3% BSA-PBST and probed with cyclin A rabbit polyclonal antibody (H-432: sc-751, Santa Cruz) at a 1:1000 dilution or ACTB mouse monoclonal antibody (AC-15, Sigma) at a 1:2500 dilution. Membranes were washed, incubated with sheep-anti-mouse or donkey-anti-rabbit peroxidase-coupled secondary antibodies (NA931 and NA934, GE Healthcare) and incubated for 1 min with Western Lightning Plus-ECL substrate (Perkin Elmer). Proteins were visualized by exposure to scientific imaging film (Kodak).
In silico analyses of cyclin A2 gene regulation and expression
Human and murine cyclin A2 mRNA expression patterns were obtained from publicly available microarray and RNA sequencing data [12-15]. Compiled GATA1 occupancy and nucleosome depleted region (NDR) data were obtained and analyzed as described [16, 17]. Expression data was analyzed as previously described [12-15, 17].
RESULTS
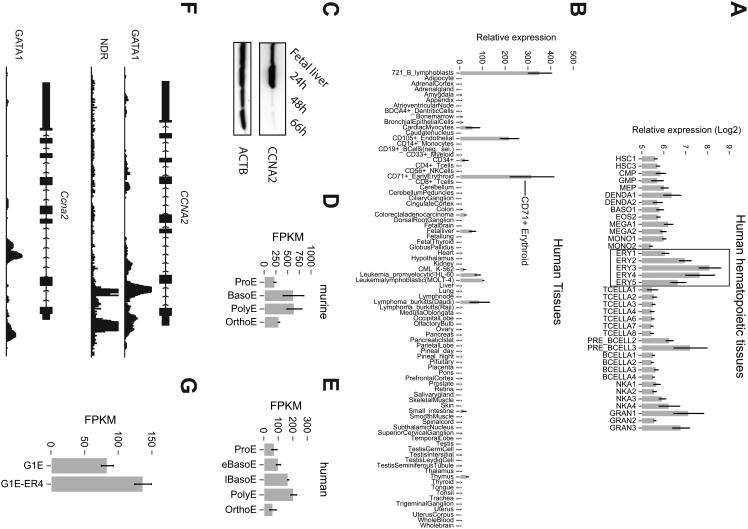
Cyclin A2 was highly expressed in erythroid cells (Figure 1A, B) and downregulated during the late stages of terminal maturation concomitant with the cell cycle exit that temporally occurs at the same time (Figure 1C, D, E) [3]. The key erythroid transcription factor GATA1 occupied chromatin within the CCNA2 locus and Ccna2 mRNA is induced upon reactivation of Gata1 in the murine erythroid cell line G1E-ER4 (1F, G). Thus, cyclin A2 demonstrates similar expression in human and murine erythroid cells and is potentially regulated by the key erythroid transcription factor GATA1.
Figure 1. Cyclin A2 expression and regulation in human and murine erythroid tissues.
Human CCNA2 mRNA expression levels in (A) purified human hematopoietic tissues [13] and (B) a panel of 79 human tissues [14]. Expression data was analyzed as previously described, except microarray gene expression was not normalized to 0 in (A). (C) CCNA2 protein levels of whole fetal liver and cultured fetal liver cells at indicated timepoints as assessed by western blot. ACTB was used as a loading control. (D, E) CCNA2 mRNA expression levels of primary human and murine stage-fractionated human erythroblasts cultured from blood-derived progenitors. Expression peaks in both species at the polychromatophilic erythroblast stage. RNA-seq data was obtained and analyzed as previously described [12, 17]. FKPM = fragments per kilobase of transcript per million; ProE = proerythroblast; BasoE = basophilic erythroblast; eBasoE = early basophilic erythroblast; lBasoE = late basophilic erythroblast; PolyE = polychromatophillic erythroblast; OrthoE = orthochromatophillic erythroblast. (F) Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) analysis of transcription factor GATA1 occupancy at the human (top) and murine cyclin A2 locus (bottom). GATA1 occupancy is observed both intronically and in the proximal promoter region. Nucleosome depleted regions (NDR) are shown across the human CCNA2 locus and overlap with GATA1 occupancy. (G) Ccna2 mRNA expression as measured in murine erythroid cell line G1E-ER4 before and after reactivation of GATA1 [15, 26].
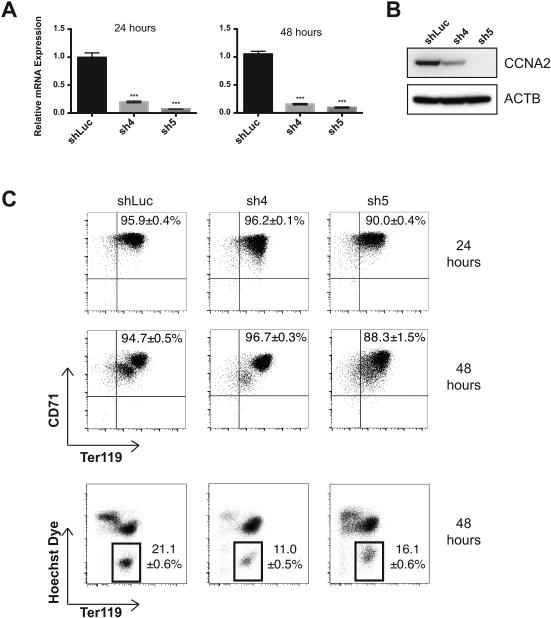
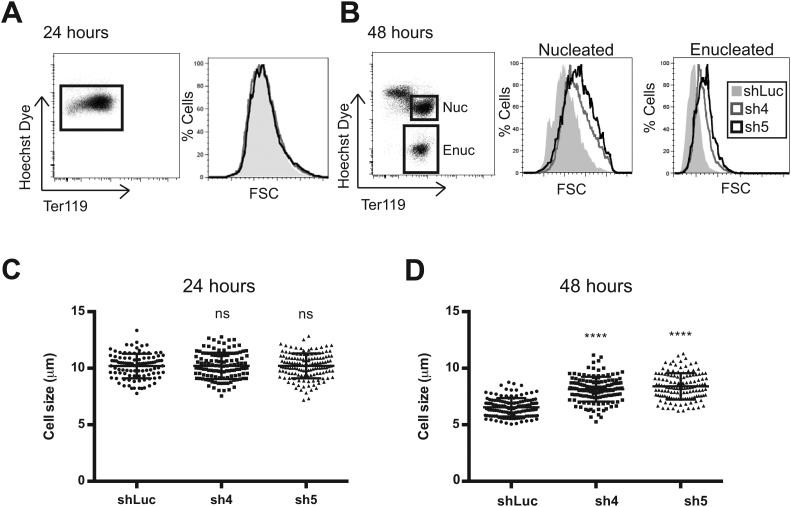
To examine the role of cyclin A2 in erythropoiesis, its level was reduced using shRNAs in primary murine erythroid cells. This reduction in cyclin A2 levels occurred soon after infection of the cells, as assessed by a knockdown of >80% at the mRNA level at both 24 and 48 hours and western blot at 24 hours (Figure 2A, B). As judged by induction of expression of the cell surface markers Ter119 and CD71 (transferrin receptor) [3, 18, 19], knockdown of cyclin A2 did not significantly perturb differentiation at 24 hours (Figure 2C). We did note a mild decrease (1.4 – 2-fold) in enucleation of these cells at 48 hours as assessed using Hoechst 33342 dye, indicating some perturbation at the stages of differentiation that immediately precede enucleation (Figure 2C). We additionally observed that the erythroid cells had no major change in size at 24 hours, as assessed by both forward scatter (FSC) using flow cytometry and by direct measurement of cell diameter using phase contrast microscopy (Figures 3A, C). However, at 48 hours, we did observe a significant increase in both FSC and cell diameter among both the nucleated and enucleated fractions (Figures 3B, D). Interestingly, at 48 hours, the control cells were on average 6.5 μm in diameter (close to the value observed for mature murine red blood cells) [20], while sh4-infected cells were 8.1 μm and sh5-infected cells were 8.4 μm (Figure 3D). This is consistent with the observation that common genetic variation in potential regulatory elements of the cyclin A2 locus appears to affect red cell size in humans [6, 7].
Figure 2. Effects on differentiation from reduction of cyclin A2 levels in erythroid progenitors.
(A) Relative mRNA levels of Ccna2 in cultured fetal liver cells at 24 and 48 hours as assessed by quantitative RT-PCR (n ≥ 3 per group, results are shown as +/− standard deviation, *** = p < 0.001 using two-sided Student t-test). (B) Ccna2 protein levels of cultured fetal liver cells at 24 hours as assessed by western blot. (C) Representative flow cytometry plots of murine erythroid surface markers CD71 vs. Ter119 at 24 hours (top) and Ter119 vs. Hoechst 33342 staining at 48 hours (bottom) are shown. Results are representative of at least 3 independent experiments. Differences in enucleation frequency between control and knockdown cells are statistically significant, p < 0.05 using the two-sided Student t-test.
Figure 3. Increased erythrocyte size resulting from reduction of cyclin A2 levels in erythroid progenitors.
(A, B) Forward scatter (FSC) profiles for a control shRNA (shLuc) or for shRNAs targeting Ccna2 (sh4, sh5) are shown for the nucleated and enucleated fractions at 24 and 48 hours, respectively. Results are representative of at least 3 independent experiments. (C, D) Cell size measurements using phase contrast microscopy images are shown as measured in micrometers (μm). Enucleated and nucleated cells were difficult to separate using phase contrast microscopy and therefore all cells were counted together. At least 100 cells were measured per group and the results are shown at 24 and 48 hours, respectively (ns = non-significant difference, **** = p < 0.0001 using two-sided Student t-test).
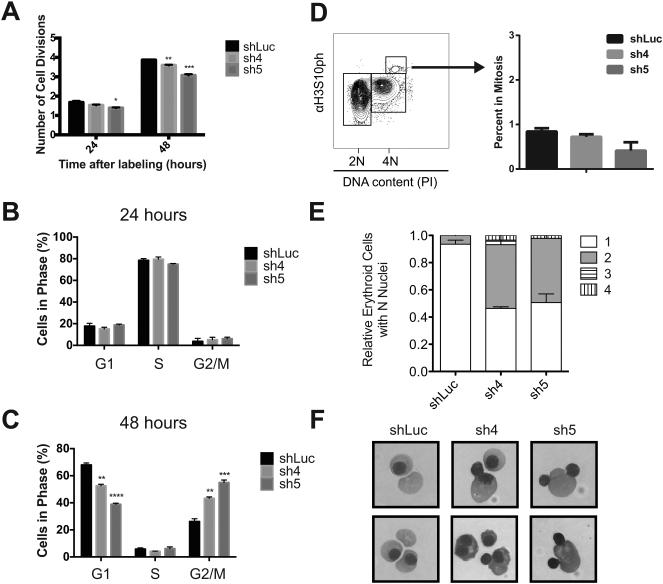
To understand the mechanisms underlying this observation, we initially examined the number of cell divisions during terminal erythroid maturation, using PKH26 labeling [3]. Both in control and knockdown cells, approximately 2 cell divisions had occurred by 24 hours. In contrast, at 48 hours the cyclin A2 knockdown cells demonstrated a reduction in the average number of cell divisions (Figure 4A); control cells had undergone an average of 3.9 cell divisions, whereas the two groups of cyclin A2 knockdown cells (sh4 and sh5 cultures) completed 3.6 and 3.1 divisions, respectively (Figure 4A). This observation suggests that the terminal cell division is impaired in cells with reduced levels of cyclin A2. Next, we pulse-labeled the cultured cells with 5-ethynyl-2’ deoxyuridine (EdU), a modified nucleoside that gets incorporated into newly synthesized DNA, to assess cell cycle progression. There was no significant difference between knockdown and control cells at 24 hours (Figure 4B); as expected the majority of cells were in S phase [3]. At 48 hours ~70% of control cells had exited the cell cycle and were in the G0/G1 state and ~25 % were in G2/M. In contrast, there was an increase (20-30% of total cells) in G2/M with a concomitant reduction in the fraction of cells in G1 (Figure 4C) 48 hours following the knockdown of cyclin A2. We stained for phosphorylated histone H3 (phospho-histone H3), a well-known marker of mitosis (prior to anaphase), to assess the stage of G2/M at which this accumulation occurs [21]. The fraction of cells with phospho-histone H3 – about 1% - was not affected by a knockdown of Ccna2 (Figure 4D). This demonstrates that the accumulation of cells in G2/M is not attributable to an accumulation of cells in the early stages of mitosis, since dephosphorylation of histone H3 only occurs during anaphase. This observation suggests that erythroid cells with reduced cyclin A2 levels either accumulate in G2 phase prior to entry into mitosis or fail to undergo cytokinesis appropriately after progression through anaphase.
Figure 4. Cyclin A2 reduction impairs the transition through cytokinesis during terminal erythropoiesis.
(A) PKH26 labeling was used to assess the number of cell divisions during the fetal liver cultures at 24 and 48 hours (n ≥ 3 per group, results are shown as +/− standard deviation; * = p < 0.05, ** = p < 0.01, *** = p < 0.001 using two-sided Student t-test in comparison with shLuc control). (B, C) EdU labeling of cells at 24 and 48 hours, respectively, with separation of various phases of the cell cycle (n ≥ 3 per group, results are shown as +/− standard deviation; ** = p < 0.01, *** = p < 0.001 using two-sided Student t-test in comparison with shLuc control). (D) Phospho-histone H3 staining is shown with the percentage of cells in mitosis (n ≥ 3 per group, results are shown as +/− standard deviation and did not reach statistical significance using a two-sided Student t-test, p > 0.05). (E) Distribution of relative amounts of cells with N nuclei is shown for N = 1-4 nuclei using cytocentrifuge specimens at 48 hours. Results are shown as +/− standard deviation, which were obtained by analysis of > 200 cells per group). (F) Representative examples of binuclear morphology in mature cells from cytocentrifuge specimens shown from images acquired with a 63X objective compared with normal orthochromatic erythroblasts and enucleated reticulocytes from the shLuc control.
We therefore analyzed cytocentrifuge slides at 24 and 48 hours to attempt to gain insight into the nature of this arrest during terminal erythropoiesis. While knockdown and control cells at 24 hours showed no appreciable morphological differences, at 48 hours there was a 7-fold increase in the number of binucleate orthochromatic erythroblasts in both cyclin A2 knockdown cultures (Figure 4E, F). Thus, cells with reduced cyclin A2 levels frequently fail to undergo cytokinesis during the terminal erythroid cell division, but nonetheless go on to enucleate, forming larger red blood cells than normal. We noted on 48 hour cytocentrifuge specimens from the cyclin A2 knockdown cells many instances of binucleate cells that were undergoing enucleation, suggesting that this phenomenon does indeed occur and explains the observation that such cells can still undergo enucleation and therefore end up larger in size (Figure 4F).
DISCUSSION
Our findings illustrate an important and previously unappreciated mechanism that regulates red cell size and that appears to underlie human inter-individual variation in this trait [2, 6]. By regulating the ability of cells to progress through the final cytokinesis during terminal erythropoiesis, cyclin A2 regulates red blood cell size. We have previously shown that the number of cell divisions during terminal erythropoiesis is critical for determining red cell size and number and is affected by variation in cyclin D3 levels [3]. In contrast, in the absence of cyclin A2, only passage through cytokinesis in the final division of erythropoiesis appears to be affected, while prior divisions occur without observable perturbations. This may explain why the regulatory variants upstream of cyclin A2 are only significantly associated with red blood cell size in humans. While A-type cyclins have previously been implicated in the regulation of entry into mitosis in other cell types [8, 22], our finding that this factor is necessary for the initiation of cytokinesis has not been previously described and may be unique to terminal erythropoiesis. We note that the phenotypic similarity between the cytokinesis defect with reduced cyclin A2 and that observed in the context of congenital dyserythropoietic anemias is striking and there may be molecular connections between these pathways [23-25]. Our studies defining the roles of cyclin A2, cyclin D3, and BCL11A in erythropoiesis illustrate how the genetic contribution to variation seen in GWAS provides only a minimal estimate of the biological contribution and major perturbations of these genes can show dramatic phenotypes that illuminate fundamental molecular mechanisms [2-4].
ACKNOWLEDGEMENTS
We thank P. Sicinski, C. Walkley, O. Steinberg-Shemer, E. Traxler, J. Flygare, J. Shi, and members of the Sankaran and Lodish laboratories for helpful discussions and advice. This work was supported by the German National Academic Foundation (to L.S.L.) and National Institutes of Health grants P01 HL032262 (to H.F.L.), R21 HL120791, and R01 DK103794 (to V.G.S.).
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
REFERENCES
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.Sankaran VG, Orkin SH. Genome-wide association studies of hematologic phenotypes: a window into human hematopoiesis. Current opinion in genetics & development. 2013;23:339–344. doi: 10.1016/j.gde.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sankaran VG, Ludwig LS, Sicinska E, et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes & development. 2012;26:2075–2087. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science. 2008;322:1839–1842. doi: 10.1126/science.1165409. [DOI] [PubMed] [Google Scholar]
- 5.Thom CS, Traxler EA, Khandros E, et al. Trim58 degrades Dynein and regulates terminal erythropoiesis. Developmental cell. 2014;30:688–700. doi: 10.1016/j.devcel.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Harst P, Zhang W, Mateo Leach I, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul DS, Albers CA, Rendon A, et al. Maps of open chromatin highlight cell type-restricted patterns of regulatory sequence variation at hematological trait loci. Genome Res. 2013;23:1130–1141. doi: 10.1101/gr.155127.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalaszczynska I, Geng Y, Iino T, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Park SE, Kim M, et al. A functional single nucleotide polymorphism at the promoter region of cyclin A2 is associated with increased risk of colon, liver, and lung cancers. Cancer. 2011;117:4080–4091. doi: 10.1002/cncr.25930. [DOI] [PubMed] [Google Scholar]
- 10.Flygare J, Rayon Estrada V, Shin C, et al. HIF1alpha synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood. 2011;117:3435–3444. doi: 10.1182/blood-2010-07-295550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji P, Jayapal SR, Lodish HF. Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nature cell biology. 2008;10:314–321. doi: 10.1038/ncb1693. [DOI] [PubMed] [Google Scholar]
- 12.An X, Schulz VP, Li J, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome research. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci U S A. 2014;111:6131–6138. doi: 10.1073/pnas.1318948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulirsch JC, Lacy JN, An X, et al. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet. 2014;10:e1004890. doi: 10.1371/journal.pgen.1004890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes & development. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Socolovsky M, Gross AW, et al. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 20.Murdock RC, Reynolds C, Sarelius IH, et al. Adaptation and survival of surface-deprived red blood cells in mice. Am J Physiol Cell Physiol. 2000;279:C970–980. doi: 10.1152/ajpcell.2000.279.4.C970. [DOI] [PubMed] [Google Scholar]
- 21.Kadauke S, Udugama MI, Pawlicki JM, et al. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuno N, den Elzen N, Pines J. Human cyclin A is required for mitosis until mid prophase. J Cell Biol. 1999;147:295–306. doi: 10.1083/jcb.147.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traxler E, Weiss MJ. Congenital dyserythropoietic anemias: III's a charm. Blood. 2013;121:4614–4615. doi: 10.1182/blood-2013-05-497602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iolascon A, Heimpel H, Wahlin A, et al. Congenital dyserythropoietic anemias: molecular insights and diagnostic approach. Blood. 2013;122:2162–2166. doi: 10.1182/blood-2013-05-468223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liljeholm M, Irvine AF, Vikberg AL, et al. Congenital dyserythropoietic anemia type III (CDA III) is caused by a mutation in kinesin family member, KIF23. Blood. 2013;121:4791–4799. doi: 10.1182/blood-2012-10-461392. [DOI] [PubMed] [Google Scholar]
- 26.Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Molecular and cellular biology. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]