Abstract
Statistical analysis was carried out on large set of naturally occurring human amino acid variations and it was demonstrated that there is a preference for some amino acid substitutions to be associated with diseases. At an amino acid sequence level, it was shown that the disease-causing variants frequently involve drastic changes of amino acid physico-chemical properties of proteins such as charge, hydrophobicity and geometry. Structural analysis of variants involved in diseases and being frequently observed in human population showed similar trends: disease-causing variants tend to cause more changes of hydrogen bond network and salt bridges as compared with harmless amino acid mutations. Analysis of thermodynamics data reported in literature, both experimental and computational, indicated that disease-causing variants tend to destabilize proteins and their interactions, which prompted us to investigate the effects of amino acid mutations on large databases of experimentally measured energy changes in unrelated proteins. Although the experimental datasets were linked neither to diseases nor exclusory to human proteins, the observed trends were the same: amino acid mutations tend to destabilize proteins and their interactions. Having in mind that structural and thermodynamics properties are interrelated, it is pointed out that any large change of any of them is anticipated to cause a disease.
Keywords: amino acid variations, disease mutations, structure, hydrogen bond
Introduction
Humans are genetically similar to each other sharing about 99.9% of DNA code, while the rest 0.1% results in natural differences between individuals and their predisposition to diseases. With the rapid progress of whole genome sequencing, it is anticipated that genetic testing will become a routine procedure and therefore the ability to discriminate disease-causing and harmless variants are of crucial importance (Burgess, 2014; Orr and Chanock, 2008; Plon, et al., 2008). However, predicting the phenotype and disease association, especially for novel variants or variants occurring in genes never implicated in diseases, is not a trivial problem (Kucukkal, et al., 2014b; Stefl, et al., 2013; Zhang, et al., 2012a).
Significant efforts were invested in developing methods to predict disease associated SNPs (see (Alexov and Sternberg, 2013)). Some methods are sequence-based (Chen, et al., 2013; Dehouck, et al., 2009; Dehouck, et al., 2011; Dehouck, et al., 2013; Pires, et al., 2014; Schymkowitz, et al., 2005a; Schymkowitz, et al., 2005b; Yang and Zhou, 2008a; Yang and Zhou, 2008b), while other combine sequence information with physico-chemical and structural characteristics (Capriotti, et al., 2005; De Baets, et al., 2012; Ryan, et al., 2009; Schwarz, et al., 2010; Yue, et al., 2006). In parallel, several approaches were developed to predict the effects of variants on thermodynamic properties such as folding and binding free energies. Some of them focus on assessing the direction (the sign) of the energy change (Cheng, et al., 2006; Masso and Vaisman, 2014; Shihab, et al., 2013), while others calculate its magnitude as well (Capriotti, et al., 2004; Capriotti, et al., 2005; Chen, et al., 2013; Cheng, et al., 2006; Dehouck, et al., 2009; Dehouck, et al., 2011; Dehouck, et al., 2013; Pires, et al., 2014; Schymkowitz, et al., 2005a; Schymkowitz, et al., 2005b; Yang and Zhou, 2008a; Yang and Zhou, 2008b). These developments indicate that researchers have searched for features and patterns to discriminate disease-causing from harmless variants on several levels: sequence, structure, physico-chemical and thermodynamics levels (Kucukkal, et al., 2014b; Stefl, et al., 2013). The performance among the best methods utilizing one or several of the abovementioned characteristics, in terms of detecting disease-causing variants or predicting the free energy change, was found to be quite similar (Khan and Vihinen, 2010; Thusberg, et al., 2011), although direct comparison should be done with care (Nair and Vihinen, 2013; Vihinen, 2013; Vihinen, 2014). Perhaps this indicates that all relevant information is available at each of these levels and simply has to be properly used.
Even if we assume that these different levels contain the same amount of information with regard to being able to discriminate disease-causing and harmless variations, one still wants to deliver not only the predictions but also to provide detailed analysis of effects induced by the amino acid mutations (Kucukkal, et al., 2014a). Depending on the goals of a particular study, this additional information may include the observation that the variation occurs at highly conserved sequence position and therefore is predicted to be disease-causing, including non-coding conserved position (D'Haene, et al., 2009). For the purposes of another study, one may be interested in figuring out whether the mutation involves alteration of the physico-chemical properties of mutation site (Chen, et al., 2010). Furthermore, specific studies could be focusing on revealing the changes of the hydrogen bonds and salt bridges induced by the amino acid mutation (Boccuto, et al., 2014; Dolzhanskaya, et al., 2014; Takano, et al., 2012; Zhang, et al., 2013). Along the same line, in many cases, researchers will be interested to find out the effect of amino acid mutation on thermodynamic properties of the corresponding protein (Li, et al., 2014; Nishi, et al., 2013; Petukh, et al., 2014; Zhang, et al., 2012b). It should be mentioned that the last several quantities, changes of the folding and binding free energy, hydrogen bonds and salt bridges, can be measured experimentally and thus providing ultimate feedback for the in silico modeling (Takano, et al., 2012; Zhang, et al., 2011). Here, we are particularly interested in investigating the difference between disease-causing and harmless variants manifested at these four levels (sequence, structure, physico-chemical properties and thermodynamics) without invoking predictions, but utilizing mainly experimental data taken from various databases.
In this study, we provide a statistical and structural evaluation of amino acid variants currently seen in humans. Our goal is four-fold. First, following the original work of Casadio and colleagues (Casadio, et al., 2011) we aim to decipher global patterns (if any) between types of variations and their corresponding degrees of harmfulness together with their observed frequency in human population. For this purpose, we used the HumVar database (Release: 2014_07 of 09-Jul-2014, containing 69240 entries, out of which 37935 termed polymorphism, 24685 disease and 6578 unclassified (Capriotti, et al., 2006)), which is a comprehensive database of all disease-causing SNPs taken from UniProtKB and also SNPs without annotated involvement in disease termed as polymorphism (see also Varisnp database (Nair and Vihinen, 2013)). The second goal is to decipher the role of physico-chemical properties of the mutation site with respect to disease association. The third one is revealing structural origins of disease associated and polymorphic types of variations. In this context, we performed local structural evaluation of the proteins with such variations using HumanDisease (disease associated) and HumanPoly (polymorphism) datasets (Wei and Dunbrack Jr, 2013). Finally, the fourth goal is to investigate the effect of amino acid substitutions on thermodynamics of corresponding proteins and interactions. Here, we adopt the original definitions for disease and polymorphism provided in the corresponding databases mentioned above, but the polymorphism is referred in the text as harmless variant of simply a variant.
Materials and Methods
Secondary structure element assignment
The secondary structure element (SSE) at the mutated residue position was assigned with STRIDE (Heinig and Frishman, 2004). The algorithm utilizes hydrogen bond energy in a combination with information from statistically delivered backbone torsion angles to make prediction of SSE. Default parameters were used. STRIDE outputs six different categories of SSEs, which were combined into three classes in this study as: (a) alpha-helix and 310-helix; (b) strand; and (c) the rest of SSEs being coil, turn and bridge.
Surface exposure assignment
The solvent exposure of all residues in the protein was classified in three types with respect to their exposure to the solvent (adopted from (Levy, 2010)). Relative solvent accessible surface area (SAS) (the ratio between SAS of a residue in protein and in water (rSAS); rSASA=1 corresponding to totally exposed residue in the protein), calculated with NACCESS software (Hubbard and Thornton, 1993) determines the following categories:
Buried (rSAS = 0), if the side chain of the amino acid is completely buried inside the protein;
Partially Exposed (rSAS > 0 and rSAS ≤ 25 %), describes the partially exposed to the solvent residue;
Exposed (rSAS > 25 %), stands for the residue being on the protein surface.
Location of mutation sites within protein-protein complexes
We assigned the location of mutated residues in the protein-protein complex based on five categories (COR, SUP, RIM, INT and SUR) as previously described (Levy, 2010) by computing the rSAS of the residue in the monomeric (rSASAm) and complex (rSASAc) states, as well as their mutual difference (ΔrSAS=rSASm-rSASc). Thus, residues are considered to be at the interface if they are in COR (ΔrSAS>0 & rSASm>0.25 & rSASc<0.25), SUP (ΔrSAS>0 & rSASm<0.25) and RIM (ΔrSAS>0 & rSASc>0.25) regions; and are away from the interface if they are in SUR (ΔrSAS=0 & rSASc>0.25) and INT (ΔrSAS=0 & rSASc<0.25) regions. RIM and SUR locations indicate that the residue is exposed to the water solvent when the complex is formed. The solvent accessible surface area of a residue was calculated with NACCESS software (Hubbard and Thornton, 1993).
Hydrogen bonds (HB)
The missing hydrogen atoms on both the wild type and the mutant proteins were added to the structures with VMD software (version 1.9.1, topology file from CHARMM27 force field) (Yahyavi, et al., 2014). The possibility of a given residue to form HB with other residues in the protein was estimated based on the distance (D ≤ 2.4 Å) between oxygen acceptor and hydrogen (except HA and HB bound to Cα and Cβ respectively) atoms. Hydrogen bond angle constrains (Torshin, et al., 2002) were not taken into account – this was done to take into consideration plausible structural imperfections introduced by in silico made mutations. Only polar (S, T, N, Q, Y) and charged (R, H, K, D, E) amino acids were taken into account.
Salt bridges (SB)
In order to estimate the number of SB the particular residue might form with other protein residues, we calculated the distance between hydrogen atom (that is bound to nitrogen in positively charged residue) and oxygen atom (that is part of COO− group in negatively charged residues). The distance cut-off was set to be 4 Å. We also applied the rule that the pair of residues can form no more than one SB.
ProTherm and Skempi databases
The experimentally measured changes of the folding and binding free energy were taken from ProTherm (Bava, et al., 2004) and Skempi (Moal and Fernandez-Recio, 2012) databases, respectively. The ProTherm database is a collection of numerical data of thermodynamic parameters such as Gibbs free energy change for example, which is linked to Protein Data Bank (PDB) entries. As of February 2013, it contains 12561 amino acid mutations with available structural information. The Skempi database contains data of the changes in thermodynamic parameters for protein-protein interactions for which a structure of the complex has been solved and is available in the PDB. The last update of March 2012 consists of 3047 cases. In addition, we appended Skempi with non-redundant cases taken from other databases (Benedix, et al., 2009; Spassov and Yan, 2013). In both databases we collapsed the redundant entries (same protein, same mutation) into a single entry. However, if the experimental energy change for the same protein and same mutation differed more than 1.5 kcal/mol, the case was removed from databases. Otherwise, we used averaged value. In addition, cases for which the mutation site cannot be found in the wild type (WT) structure were removed as well. Since the initial crystal structures might have amino acids with missing atoms, we used the profix module from Jackal package to rebuild these missing atoms (Petrey, et al., 2003). It was done using default parameters and selecting “heavy atoms model” option. At the next step we applied scap module from the same Jackal package to substitute wild-type residue with the mutant to generate the mutant (MT) protein. To run scap we applied the following parameters: (a) CHARMM22 force field parameters, (b) large side-chain Jackal rotamer library was selected for the side-chain generation, and (c) predictions of the side chain orientation were made by applying scap option utilizing 3 initial structures. Finally for statistical analysis we used 2040 entries (81 proteins) from Skempi database and 2218 (128 proteins) from ProTherm database.
Results
Identification of frequently occurring mutations in the human population
First, following the original work of Casadio and colleagues (Casadio, et al., 2011) we statistically analyzed the HumVar dataset for any apparent patterns in disease-causing and harmless mutations. It was done by considering every possible amino acid mutation by taking all 380 different combinations of 20 natural amino acids. Then, we analyzed the database to decipher their frequency in the whole database, the frequency in the disease database, and the propensity of being the most harmful and the least harmful. It should be mentioned that the frequency of naturally occurring amino acid variations in HumVar depends on multiple factors, including genetic one, and therefore is a very complex phenomena. In this study the genetic factors are not taken into account and the corresponding amino acid variation frequencies are investigated as described below.
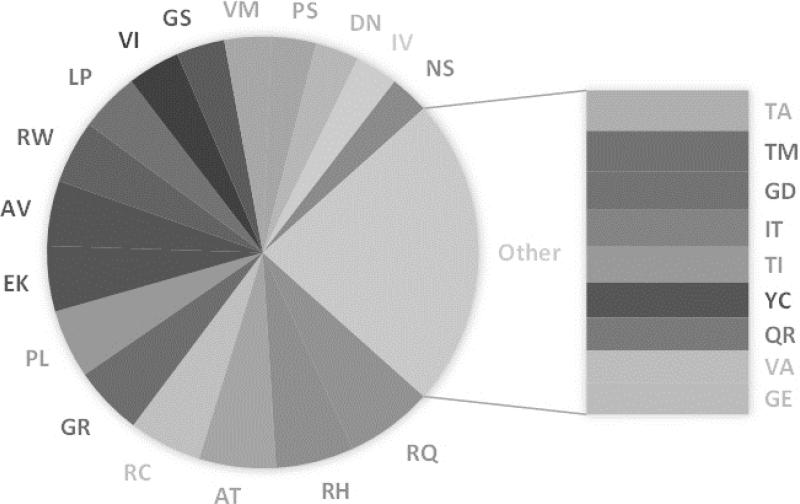
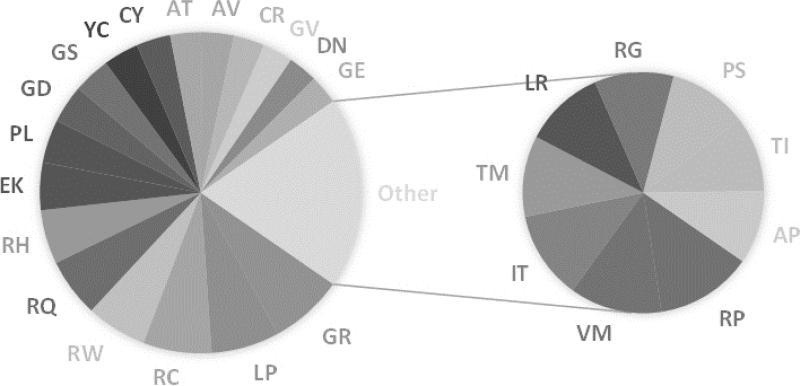
First of all, out of 380 combinations, 108 were never observed so far in the HumVar database and also 60 were observed only once. In contrast, the top 26 most frequent variants were found to make up to 46% of the whole database (disease and polymorphism/harmless). They are shown in Figure 1. The frequency of pair substitutions in the disease database also shows a quite interesting pattern as well, where the top 27 most frequent disease-causing variants make up to 53% of the disease database as indicated in Figure 2. About half of these turn out to be the same in both most frequent lists, while the other half represent high frequency types of variations seen mostly either in disease-causing or polymorphism/harmless databases. Particular examples are V→I (VI) and I→V (IV) amino acid mutations, which are seen much more frequently in polymorphism/harmless database.
Fig. 1.
Most frequent mutations. Frequency of top 26 mutations that make up 46% of HumVar database (disease and polymorphism). Rest of the 54% is made up by 246 mutations with 60 of them were observed only once.
Fig. 2.
Most frequent disease-causing mutations. Frequency of top 27 mutations that make up 53% of HumVar disease database. Rest of the 47% is composed of 245 mutations with 60 of them were observed only once.
For the purposes of further analysis, we use Casadio's definition of Disease Index (Pd) (Casadio, et al., 2011), which is a statistical measure for a particular mutation to be harmful. In this work we term this quantity “degree of harmfulness”, rather than Disease Index in order to be able to introduce other relevant definitions below. It is calculated as the ratio of the number of times a specific variation was found to be disease-causing and the number of times it was observed in the whole database (disease and polymorphism/harmless), i.e.
| (1) |
The first observation is that out of 380 plausible types of amino acid mutations, 87 cases were found to be only harmless variants. However, the frequencies of these variations in the entire HumVar database are very low, i.e. they were only seen 1-7 times, which is statistically insignificant and therefore they cannot be classified as harmless. Because of that, all variation types except the 108 that were never observed can be considered to be potentially disease- causing. Similarly, we checked if there are any variations that were always harmful (never seen in polymorphism/harmless database) and 13 of the variations seemed to be always harmful, however this is also statistically insignificant because they are observed only 1-2 times. Taking into account cases with very low frequency may overestimate or underestimate the degree of harmfulness for some underrepresented types of variations. Because of that, we calculated the degree of harmfulness via eq. (1) for variations that are observed at least 0.5% of the cases or more than 300 times (the most frequent mutation is observed 2090 times resulting in 3% of the cases). The results are presented in Table 1. Two types of variations are listed: (a) the most harmful variations with the largest score (degree of harmfulness > 0.5, eq. (1)) and the least harmful variations with the smallest score (degree of harmfulness < 0.2, eq. (1)). This analysis provides interesting findings about the nature of the amino acid being mutated and the degree of harmfulness.
Table 1.
Lists of most frequent, most harmful variations (gray columns, with degree of harmfulness > 50% and frequency > 0.5%) and most frequent, least harmful variations (with degree of harmfulness less than 20% and frequency > 0.5%)
| Variation | Frequency (%) | Degree of Harmfulness | Variation | Frequency (%) | Degree of Harmfulness |
|---|---|---|---|---|---|
| RC | 2.52 | 0.52 | VI | 1.82 | 0.10 |
| GR | 2.39 | 0.59 | IV | 1.47 | 0.08 |
| RW | 2.18 | 0.53 | TA | 1.33 | 0.13 |
| LP | 2.06 | 0.63 | VA | 1.07 | 0.17 |
| GD | 1.27 | 0.57 | LV | 0.93 | 0.19 |
| YC | 1.13 | 0.59 | SN | 0.90 | 0.19 |
| GE | 1.04 | 0.53 | KR | 0.82 | 0.16 |
| CY | 0.96 | 0.67 | ED | 0.81 | 0.16 |
| GV | 0.95 | 0.61 | DE | 0.69 | 0.19 |
| CR | 0.93 | 0.63 | SG | 0.64 | 0.15 |
| RP | 0.74 | 0.63 | AS | 0.62 | 0.16 |
| LR | 0.65 | 0.60 | RK | 0.58 | 0.12 |
| WR | 0.56 | 0.56 | EQ | 0.58 | 0.18 |
| FS | 0.54 | 0.52 | TS | 0.55 | 0.10 |
Degree of harmfulness and physico-chemical properties of amino acids
One apparent observation in the most disease-causing variations list is that 8 out of 14 amino acid mutation types involve Cys mutations (Table 1). Cys has a complex nature and it can be considered as hydrophilic (due to the thiol group), but also generally considered hydrophobic in the hydrophobicity scales (Betts and Russell, 2003). More importantly, the ability of forming disulfide bonds increases the role that Cys residues play in protein folding and stability. Another essential function of Cys residues is binding to various metal ions and in particular Zn2+ (Pace and Weerapana, 2014) among other functional roles (Pace and Weerapana, 2013). If a Cys involved in a disulfide bond is mutated, then it will certainly have a great impact on the structure and function as no other amino acid can fully compensate in that case. In the case of Zn2+ binding, the only amino acid that potentially can compensate would be His. However, there always will be cases involving substitution of Cys residue which does not participate in any specific interactions and such a substitution may not affect protein properties. Another important pattern in the most harmful variation list is the Arg and Pro substitutions (note that the high frequency of Arg substitution is due to highly mutated 5’-CpG dinucleotide). It is not surprising to see the Arg-involving variations such as RC, RP, and RW, are disease-causing. Such variations involve change of the charge and the entropic properties of the amino acid involved and this was extensively investigated in various proteins systems (Chiariotti, et al., 2012; Doss, et al., 2013; Doss, et al., 2014; George Priya Doss and Rajith, 2012; Kamaraj and Purohit, 2013; Kumar and Purohit, 2012; Kumar and Purohit, 2014; Kumar, et al., 2013; Pirolli, et al., 2014; Rajendran, et al., 2011). It was found that the majority of these amino acid mutations alter the protein flexibility as well as intra- or intermolecular hydrogen bonding network, resulting in significant deviation away from the wild-type characteristics of the proteins and that probably causing diseases, which is consistent with our statistical observations.
Not surprisingly, the least harmful variations list also reveals the importance of the wild-type residue physico-chemical characteristics being partially compensated by the mutant residue properties. With that being said, the variations that tend to be less harmful were found to introduce minimal changes of amino acid physico-chemical properties. For example, if the amino acid substitutions are hydrophobic to hydrophobic (IV, VI), have similar size (SA, TA, YF, LM, KR, ED) and also if they have the same charge (ED, KR), then they tend to be less harmful. In a recent experimental study, several cancer-relevant variations in Tin-1 kinase were studied to reveal their impact on folding free energy (Lori, et al., 2013). The amino acid mutations studied for the same protein are Y53H, E124E, E135K, E142D, which were found to be destabilizing by 0.38-4.89 kcal/mol. Consistent with our statistical and structural analyses (below), the E142D mutation was found to have a minimum effect on the stability of the protein while the E135K mutation was found to be the most destabilizing. Similarly, another experimental study found that a breast cancer-associated D67E mutation in breast cancer susceptibility protein 1 has retained the ligase and metal binding activity of the protein, barely perturbed its native global structure, and only slightly reduced its thermal stability (Atipairin, et al., 2011). This amino acid mutation was suggested to be either neutral or be involved in the disease development by other mechanisms.
Structural characterization
Next, we proceed with the evaluation of structural characteristics of the variations listed in Table 1. For this purpose we used HumanDisease (disease associated database, 1405 entries) and HumanPoly (database of polymorphic amino acid mutations, 1367 entries) (Wei and Dunbrack Jr, 2013) to determine whether any structural information exists in the PDB for wild type proteins involving these specific variation types. We collected four distinct categories of wild type PDB structures with corresponding information of mutation site (see below for categories definitions). Note that for significant fraction of cases listed in HumVar database, there is no corresponding structure in the HumanDisease and HumanPoly datasets, which results in fewer cases to examine and may shift the ratio between disease-causing and harmless cases associated with the amino acid mutation. For example, the most harmful variation in Table 1, the CY mutation, with degree of harmfulness 0.67, is seen in 15 structures from HumanDisease and 2 structures from HumanPoly sets, resulting in different harmful/harmless ratio in the structural databases. Being aware of this, we grouped the variations into four categories:
-
(a)
MD: The most harmful variation types (Table 1, first set) for which we can find structure in HumanDisease database. In other words, these are the cases identified by the statistical analysis of HumVar database as most harmful variation types and at the same time the variation type can be found in structural database HumanDisease (marked with gray in Table 1).
-
(b)
MV: The most harmful disease-causing variations (Table 1, first set) for which we can find structure in HumanPoly database. These represent cases which were identified in HumVar to be most harmful variation types, but belong to the polymorphic/harmless structural database HumanPoly.
-
(c)
LD: The least harmful disease-causing variations (Table 1, second set) for which we can find structure in HumanDisease database. This classification is made of case identified in HumVar to be least harmful variation types, but such variation is found in the disease structural database HumanDisease.
-
(d)
LV: The least harmful disease-causing variations (Table 1, second set) for which we can find structure in HumanPoly database. These cases are representing least harmful variation types found in HumVar which at the same time are seen in polymorphic/harmless structural database HumanPoly.
Such a grouping will allow us to make two types of analysis of physico-chemical characteristics at the amino acid mutation site: (a) differentiate disease from polymorphism/harmless variations (comparing MD and LD with MV and LV); and (b) compare most harmful (MD and MV) with least harmful (LD and LV) mutation types.
The PDB structures from this search were subjected to further structural analysis. The structural properties that we considered for the wild-type and mutant residue in question are the changes in salt bridges and hydrogen bonding network upon mutation, as well as change in solvent accessible surface area of amino acid mutation site. In addition, we examined each altered mutation site in terms of secondary structure, i.e. helix, sheet or coil for each of four groups of variation types.
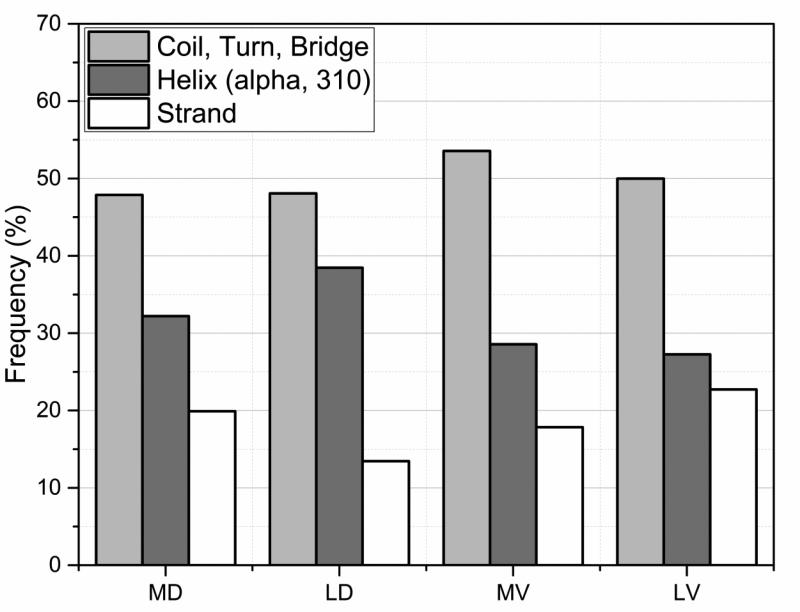
First of all, this later assessment indicates that the disease and polymorphism/harmless cases were found to show similar location in secondary structure elements (SSE) as seen in Fig. 3. One observation is that these mutation types (listed in Table 1) in general tend to be located more often in a helix or a coil/turn/bridge rather than in a strand. The fact that significant fraction of disease-causing mutations are located in coil/turn/bridge regions, presumably the most flexible regions, can be explained by their frequent involvement in catalytic functions and molecular recognition. In addition, this is consistent with previous findings, where 21.7% of disease-causing mutations were found to be in intrinsically disordered regions (Vacic, et al., 2012). The same work indicated that 20% of these mutations were found to induce disorder-to-order transitions (Vacic, et al., 2012). The observation that there is little difference in the distribution of disease and polymorphic/harmless variations with SSE indicates that SSE location is not important factor for disease association. Indeed, the variation types that were found responsible for 46% of these transitions are: RW, RC, EK, RH, RQ. Specifically the first two Arg substitutions are one of the top disease-causing variations (Table 1) and also all of these variations introduce severe changes in terms of amino acid properties. Therefore, the degree of harmfulness of an amino acid mutation may be more dependent on the type of mutation than its location in SSEs. In other words, the other factors that play a role in the degree of harmfulness of a variation might be more important than its location in SSEs.
Fig. 3.
The distribution of mutations sites within secondary structure elements (SSEs) for the four types of mutations sites MD, MV, LD and LV (see text for details).
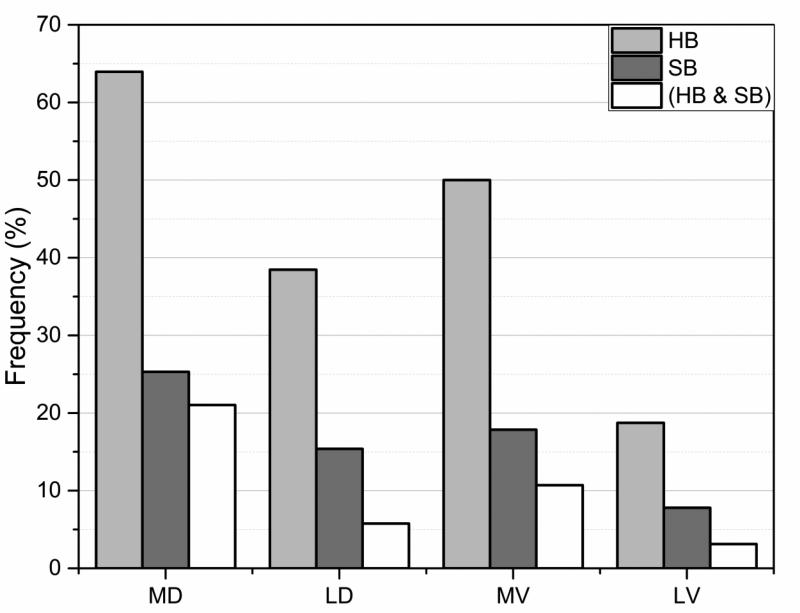
Turning to the structural impact of the variation types (listed in Table 1), the most harmful variation types (MD and MV) were found to cause the largest changes in the hydrogen bonding network at the vicinity of mutation site as shown in Fig. 4. Among the most harmful variation types in Table 1, more than 60% involve alteration of wild type hydrogen bonds. The changes in number of salt bridges also indicate the same trend. Similar observations were made in many case studies showing that disease-causing variations either affect structural integrity (Boccuto, et al., 2014; Takano, et al., 2012) or catalytic properties (Zhang, et al., 2013; Zhang, et al., 2010) by altering hydrogen bonds or salt bridges (Yue, et al., 2005; Yue and Moult, 2006). On the other hand, the harmless variation types (noted as LV) cause the least amount of change in hydrogen bonding and number of salt bridges (Fig. 4). The next paragraph suggests plausible explanation of this observation.
Fig. 4.
Percentage of cases with changed (lost/gain of) hydrogen bonds (blue) and salt bridges (red) for the four types of mutations sites MD, MV, LD and LV (see text for details). The vertical axis indicates percentage of the cases the effect was found.
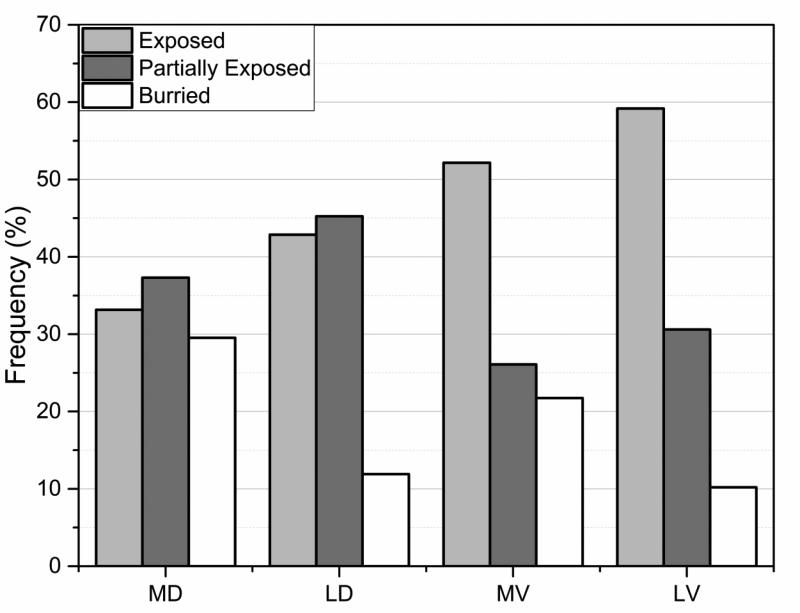
Quite interesting patterns were observed with regards to degree of burial of the mutated residues. First of all, as indicated in Fig. 5, the least harmful variation types in the polymorphism/harmless database (LV group) are found to be mostly exposed in WT structures. For more than 55% of cases the wild type side chain of a residue being involved in LV amino acid mutation are found to be totally surface exposed. Perhaps this explains why LV amino acid mutations were found to cause the least alterations of hydrogen bonds and salt bridges (Fig. 4). Since their wild type side chains are totally exposed to the water phase, these residues are not involved in any specific interactions (if they participate in a hydrogen bond or salt bridge, their side chain will be partially obscured from the water). Substitution of such side chains by another, especially if it is a residue having similar physico-chemical properties, is expected not to have impact on structural integrity of the corresponding protein. At the other side of the spectrum are harmful variations (MV and MD) (Fig. 5). The most harmful variation types in the disease list (noted as MD) are found to have the highest amount of burial degree compared to other three types, an observation consistent with previous works (Yue, et al., 2005; Yue and Moult, 2006). If such an amino acid mutation involves charged or polar group, most probably the group will be involved in specific interactions in the wild type protein, such as hydrogen bond or salt bridge, in order to gain favorable energy to compensate for the unfavorable desolvation penalty (caused by the burial). If mutated, these wild type hydrogen bonds and salt bridges will be lost, resulting is the observed trend that disease-causing amino acid mutations cause more changes in hydrogen bond and salt bridges than polymorphic/harmless ones.
Fig. 5.
Degree of burial of WT residues classified three categories: exposed, partially exposed and buried. See text for more details for the burial classification and for MD, MV, LD and LV.
Effects of mutations on thermodynamic characteristics of proteins
Amino acid mutations almost always affect the thermodynamic properties of the corresponding macromolecules, such as folding and binding free energies. In this section, we first briefly outline the general trend of the effects of disease-causing amino acid mutations on stability and interactions as reported in literature. Then, we use two experimental databases, ProTherm(Bava, et al., 2004) and Skempi (Moal and Fernandez-Recio, 2012) extended with cases taken from other databases (as previously described in Method section) to deliver statistics about the distribution of the change of folding and binding free energy caused by amino acid sunstitutions.
Observations taken from literature
Experimentally, it was repeatedly shown that most of the disease-causing amino acid mutations tend to be destabilizing, i.e. lowering the folding free energy (Grothe, et al., 2013; Khan, et al., 2013; Lori, et al., 2013; Tokuriki and Tawfik, 2009). The degree of destabilization was found to be elevated for amino acid mutations that introduce drastic changes of physico-chemical properties at the mutation site. For most of the cases the destabilization was accompanied by structural changes as well (Khan, et al., 2013; Lori, et al., 2013). The same trend was found in in silico modeling studies utilizing structural information (Boccuto, et al., 2014; Dolzhanskaya, et al., 2014; Tokuriki and Tawfik, 2009; Yue, et al., 2005; Yue and Moult, 2006). However, some amino acid mutations can be disease-causing and at the same time stabilizing the corresponding protein (Takano, et al., 2012; Witham, et al., 2011; Zhang, et al., 2011).
Similar trend, but not so pronounced, was reported in the literature for the effects of amino acid mutations on protein binding. It was indicated that any deviation of the wild type properties caused by amino acid mutations, enhanced or reduced affinity for example, possesses high risk of developing a disease. Experimentally it was shown that disease-causing amino acid mutations alter macromolecular interactions (Domoszlai, et al., 2014; Patel, et al., 2011; Placone, et al., 2014; Placone and Hristova, 2012; Wu, et al., 2010; Yang, et al., 2013; Zhang, et al., 2011). Computational studies indicated that amino acid mutations alter binding affinity of proteins and make the electrostatic component of the binding energy less favorable (Nishi, et al., 2013; Teng, et al., 2009). Genomic-scale investigations were also carried out with combined efforts of machine learning and statistical potentials and showed that macromolecular interactions are affected by amino acid mutations (Berliner, et al., 2014).
Statistics of experimental data
Currently there is no large database providing both an extensive list of human amino acid variations and experimentally determined changes of the folding and binding free energies. However, such databases exist for investigator-introduced amino acid mutations: ProTherm (Bava, et al., 2004) and Skempi (Moal and Fernandez-Recio, 2012). The ProTherm database (Bava, et al., 2004) contains the measured folding free energy changes for investigator-made amino acid mutations, and extended Skempi database (Moal and Fernandez-Recio, 2012) – for binding free energy changes. Although these amino acid mutations are not naturally occurring in human population (and most of the proteins are not human proteins), but were introduced by investigators for various reasons and therefore are biased, still one wonders if similar observation can be made as for naturally occurring human variations.
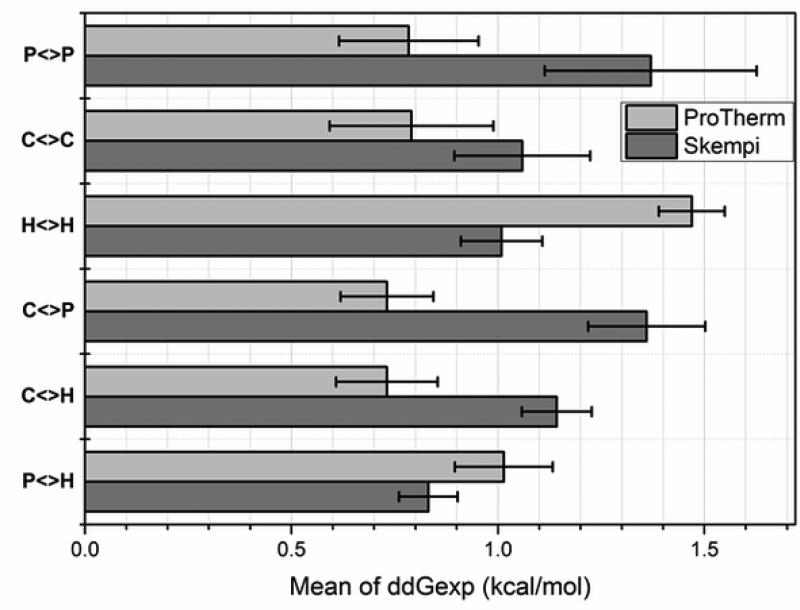
The first question that we would like to address is what are the average changes of the folding and binding free energies seen in experimental databases. The change is taken as the difference between mutant and the wild type free energy, and thus a positive value indicates that amino acid mutation reduces the free energy. We group the results according to the wild type amino acid physico-chemical characteristics (Fig. 6). It can be seen that the mean value of the change of both binding and folding free energies is a positive number indicating that on average amino acid substitutions tend to destabilize proteins and their interactions (see Supp. Table S1a and S1b for individual energies of all available amino acid substitutions). Perhaps this implies that wild type proteins and their complexes are nearly energetically optimized as suggested by a computational study (Brock, et al., 2007). This should not be considered as a statement that more stable proteins and better protein interactions cannot be engineered (successful re-engineerings were reported in the past (Baxa, et al., 1999; Fu, et al., 2009)), but rather as a suggestion that random amino acid substitutions tend to have no effect or to destabilize protein's thermodynamics properties.
Fig. 6.
The mean of the experimental folding and binding free energy change caused by mutations taken from ProTherm and Skempi databases and grouped by physico-chemical property as: P<>P – polar to polar side chain substitution; C<>C – charged to charged; H<>H – hydrophobic to hydrophobic, C<>P – charged to polar and polar to charged; C<>H – charged to hydrophobic and hydrophobic to charged; P<>H – polar to hydrophobic and hydrophobic to polar. Charged residues (C): R, K, D and E; polar residues (P): S, T, N, and Q; hydrophobic residues (H): A, V, I, and L. Standard error is provided for each bar.
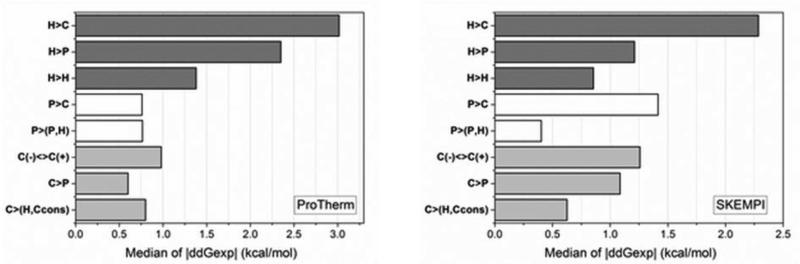
The second question in this section is how the absolute magnitude of the change of binding and folding free energies is related to the physico-chemical properties of the amino acid side chains being involved (Fig. 7). It should be clarified that we are seeking for the magnitude of the change not the direction, since it was previously demonstrated in the literature that disease-causing amino acid mutations result in both stabilization or destabilization of proteins and their complexes (Alexov and Sternberg, 2013; Yates and Sternberg, 2013). Here we ask what types of amino acid substitutions cause the largest changes of the folding and binding free energies in ProTherm (Bava, et al., 2004) and extended Skempi (Moal and Fernandez-Recio, 2012) databases respectively and are these types of substitutions similar to the patterns reported above for disease-causing variations? To address such questions, the median of the absolute value of free energy change for both databases is shown in Fig. 7a,b. The side chains are grouped according to their physico-chemical nature as charged, polar and hydrophobic. Then, we grouped the type of substitution(s) (maximum two categories) which results in the largest median of absolute free energy change. For example, in case of folding (Fig. 7a) and binding free energy (Fig. 7b), the largest change was found to occur for substitutions from hydrophobic to charged side chains. The second largest change within the same group was found for mutations from hydrophobic to polar amino acids. The changes associated with the rest of substitutions are smaller. The trends in Fig. 7 can be summarized as following: the largest changes of free energy are seen for cases involving drastic changes of the physico-chemical properties of the wild type residue. Thus, investigator made amino acid substitutions in ProTherm (Bava, et al., 2004) and extended Skempi (Moal and Fernandez-Recio, 2012) databases show the same trend as disease-causing variations seen in human population.
Fig. 7.
The median of experimental folding and binding free energy absolute change caused by mutations taken from ProTherm and Skempi databases and grouped according to largest effect and the rest of cases. Single arrow represents substitution for one type residue to another, as for example H>C is mutation from wild type hydrophobic to charged amino acid in the mutant. Charged residues (C): R, K, D, and E; polar residues (P): S, T, N, and Q; hydrophobic residues (H): A, V, I, and L. R and K are positively charged residues noted as C(+); while D and E – negatively charged residues noted as C(-). During C<>Ccons amino acid substitutions the charge of the residue conserves, that includes C(+)>C(+) and C(-)>C(-) groups of mutations.
Comparison of effects for monomeric proteins and protein complexes
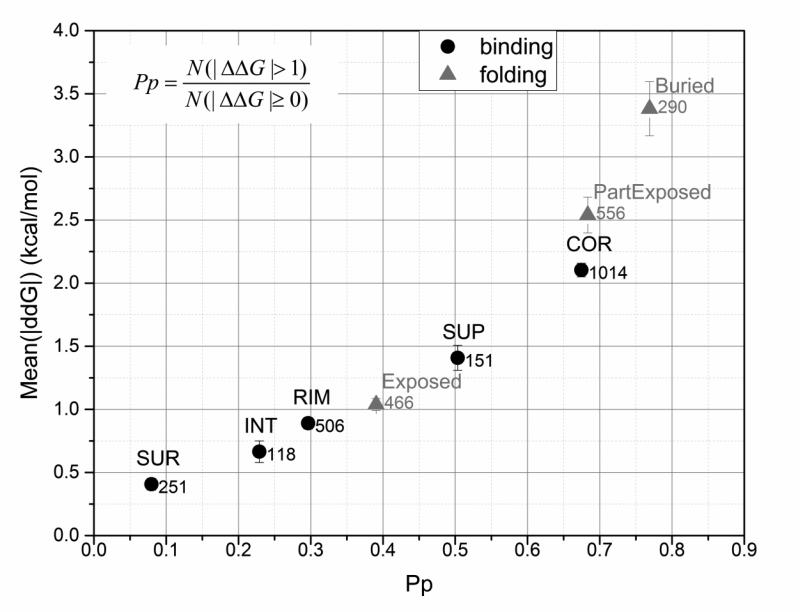
The above analysis was done putting monomeric proteins and protein complexes on the same footage. However, the folding and the binding may be affected by amino acid substitutions differently. Thus, using ProTherm and extended Skempi databases, along with structural information as explained in the Method section, we investigate plausible correlation between degree of burial, magnitude of the energy change and probability index Pp (perturbation index as defined by Casadio and colleagues (Casadio, et al., 2011) and calculated as probability of given amino acid mutation to cause significant change in free energy, eq. (2)).
| (2) |
Results are shown in Fig. 8, where the mutation sites are grouped according to the definitions of burial (exposed, partially exposed and buried) and in case of protein complexes, adopting the five classification scheme (SUR,INT, RIM, SUP and COR) (see Method section). One can see that there is practically linear dependence of the mean of the corresponding energy change as a function of Pp. Consistently with the analysis above, the amino acid mutations involving sites buried in the core of the monomeric protein or in the core of the protein complex interface cause largest energy change and are associated with largest Pp value.
Fig. 8.
Effect of the location of the mutated site on the change in binding/folding free energies. On the x-axis: the probability of the wild type residue being in the given location to cause large change in binding/folding free energy (perturbation index, Pp) due to mutation. On the y-axis: the averaged absolute value of experimentally obtained binding/folding free energies provided with standard error of mean as an error bar and the total number of cases across appropriate databases. Experimentally determined values of binding/folding free energies were obtained from extended Skempi and ProTherm databases respectively. Five location types of mutation sites were considered for binding energy change analysis (COR, RIM, SUP, SUR, and INT), and three – for folding free energy analysis (Buried, Partially Buried, and Exposed). For details see Method section.
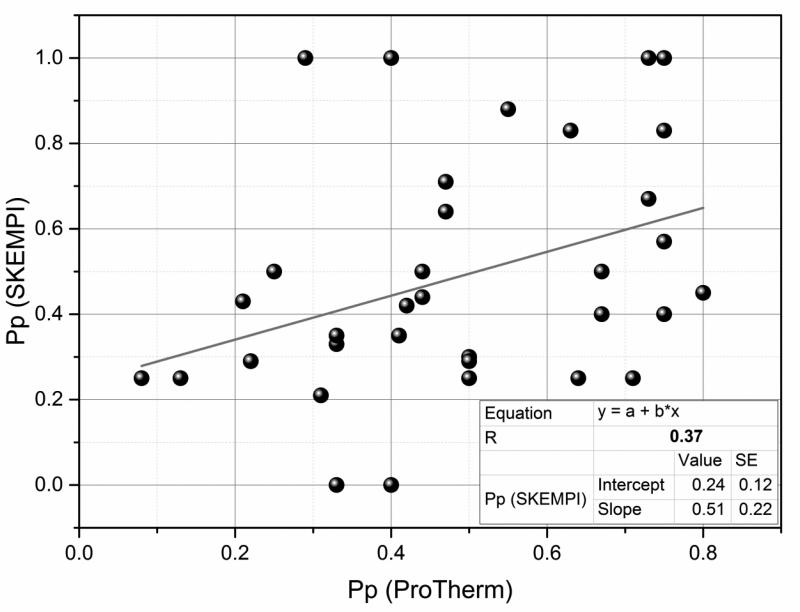
While the effect of relative burial on the magnitude of the corresponding energy change was found to be quite similar for monomeric proteins and protein complexes, it is tempting to investigate if the same similarity exists at level of types of amino acid substitutions. For this purpose we plot the Pp of individual mutation types found in ProThem against the same types in extended Skempi databases (Fig. 9). It can be seen that there is practically no correlation. This observation reflects various factors as the difference in the physico-chemical properties of protein interfaces and protein interior, different distributions of mutation sites (see Fig. 8) and difference in the specific interactions.
Fig. 9.
Correlation between perturbation indexes (Pp) of the same mutation types causing change in binding and free energy (experimental values were obtained from extended Skempi and ProTherm databases respectively).
Discussion and Conclusion
Our findings provide a relation between variation types and their degree of harmfulness and follow the original work of Casadio and colleagues (Casadio, et al., 2011), but on much larger dataset. It is interesting to compare both works in terms of the delivered Pd's or degree of harmfulness to check the sensitivity of the results with respect to the expansion of HumVar database. The original work (Casadio, et al., 2011) was done on about 21,000 amino acid variations, while in this paper we deal with more than 69,000 cases. The Pd's taken from Casadio and colleagues (Casadio, et al., 2011) were plotted against the Pd's delivered in this work (see Supp. Fig. S1) and very good correlation was obtained (R=0.90). However, the y-intercept of the fitting line is shifted by 0.2 points, indicating clear tendency that our Pd indexes are smaller than those of Casadio and colleagues (Casadio, et al., 2011). The reason for this tendency stems from the different distribution of disease and polymorphic variations in the old and newest versions of HumVar database. In the old HumVar database the ratio between disease and polymorphic variations is 1.36, while it is significantly lower (0.65) in the current release, and thus resulting in smaller Pd values. Despite of such difference, our results generally agree with the results of Casadio and colleagues (Casadio, et al., 2011). Similarly, following Casadio and colleagues, we plotted Pp versus Pd values, but including protein complexes as well (Supp. Fig. S2a,b). The obtained correlation coefficients in both cases, ProTherm and extended Skempi databases, are very similar to those reported by Casadio and colleagues (Casadio, et al., 2011). The relatively low correlation coefficients (Supp. Fig. S2a,b) indicate that disease causing effects are not exclusory associated with changes of the thermodynamic properties, but may invoke many other changes of the wild type characteristics of the corresponding proteins and protein complexes.
We would like to mention that although the data is taken largely within broad amino acid classifications, it is crucial not to over-interpret. It is important to stress on the meaning of our statistical classifications. For example, the least harmful classification does not mean that those variations are completely harmless but it rather encapsulates the observation in the HumVar database that their probability of being harmful is relatively low (less than 20%). Therefore, it is quite possible that some of those amino acid mutation types may be found harmful in future studies, but this would not contradict with our current statistical evaluations. Also, we are well aware that the disease-causing effects come from a combination of various properties such as: variation types, whether the amino acid mutation introduces a change in salt bridges or hydrogen bonding network or degree of burial, the nature of the mutation site meaning that if the amino acid mutation is taking place in the active site or whether it is in an allosteric path. Therefore, the same variation type may be disease-causing in one protein but completely harmless in another. Because of that, our analysis should be considered as providing general trend in understanding the variation type effects and their association with harmfulness in conjunction with local structural features implicated in disease.
The statistical analysis indicated that disease-causing variations, especially those referred here as the most harmful variation types (MP and MD), tend to invoke drastic changes of physico-chemical properties of amino acid mutation site and native hydrogen and salt bridge networks. The most harmful variation sites were also found to be less surface exposed as compared with polymorphic/harmless sites. These observations are in accordance with previous works of Moult and colleagues (Yue, et al., 2005; Yue and Moult, 2006). Indeed, they have shown that the protein structure stability changes caused by single residue substitutions are associated mostly with changes in hydrophobic area, overpacking, backbone strain and loss of electrostatic interactions (Wang and Moult, 2001). Taking all these observations together, it can be concluded that the disease-causing amino acid mutations are typically associated with large change of the native properties of the corresponding proteins and their interactions. From thermodynamics perspective, most of the experimental and computational results reported in the literature indicate that disease-causing amino acid mutations tend to destabilize proteins and their complexes. Such an observation was previously made by Tawfik and colleagues (Tokuriki, et al., 2007; Tokuriki and Tawfik, 2009) and was attributed as a major constraint on protein evolvability. The larger the change – the higher is the probability that amino acid mutation is disease-causing, although the disease-effect will depend on the overall characteristics of the wild type protein. Small changes of the folding or binding free energy of a relatively unstable protein or a protein involved in weak interactions may have drastic effect of its function as well.
It was pointed out that any significant deviation of the wild type characteristics (as stability, hydrogen bonds, salt bridges, and interactions) may lead to diseases. Both over-stabilizing and destabilizing changes might be equally crucial for the protein functionality. An amino acid mutation that makes a protein very rigid while flexibility is important for its function or a amino acid mutation that makes a transient interaction almost permanent - both should be causing a large alteration of protein function and perhaps will be disease-causing. As mentioned above, large-scale studies reported in the literature indicate that the disease-causing variations tend to destabilize proteins (Casadio, et al., 2011; Wang and Moult, 2001; Yue, et al., 2005; Yue and Moult, 2006) and their interactions (Nishi, et al., 2013; Teng, et al., 2009). Why there is such a preference? Is it a result of wild type protein thermodynamic properties? Based on the statistics obtained from investigator made amino acid substitutions and their experimentally measured free energy changes as reported in ProTherm (Bava, et al., 2004) and extended Skempi (Moal and Fernandez-Recio, 2012), we speculate that wild type proteins and interactions are near optimized (as it was demonstrated for the electrostatic component of the binding free energy (Brock, et al., 2007)). Thus most of amino acid mutations (unless purposely engineered) tend to have no effect or to destabilize proteins and their interactions.
The linkage between different characteristics such as physico-chemical properties, ability to form hydrogen bonds and salt bridges and thermodynamics (folding and binding free energy changes) with respect to harmfulness is also clear. The most harmful variation types were found to be associated with drastic changes of physico-chemical properties, hydrogen bonds and salt bridges networks of the wild type. Similarly, the largest changes of the free energy were found experimentally to occur upon amino acid substitutions involving large change of the physico-chemical properties of the wild type. Thus, the disease-causing effect is demonstrated at various levels, which are interconnected. A large change of the free energy is frequently associated with a buried amino acid mutation site, where the amino acid substitution results in radical change of the physico-chemical properties including removal/addition of hydrogen bonds and salt bridges. Such interconnectivity, perhaps, is the reason why methods utilizing different features have similar performance. However, from point of view of understanding the molecular mechanism of diseases, one is interested to reveal all details of the changes induced by the amino acid mutation, not just some. Knowing the details will facilitate development of therapeutic solutions.
Supplementary Material
Acknowledgments
Funding:
E.A and M.P. were supported by a grant from NIH, NIGMS, grant number R01GN093937.
T.G.K was supported by a grant from Clemson University Provost office.
Footnotes
All authors declare no conflict of interests.
References
- Alexov E, Sternberg M. Understanding molecular effects of naturally occurring genetic differences. J Mol Biol. 2013;425(21):3911–3. doi: 10.1016/j.jmb.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Atipairin A, Canyuk B, Ratanaphan A. Substitution of aspartic acid with glutamic acid at position 67 of the BRCA1 RING domain retains ubiquitin ligase activity and zinc (II) binding with a reduced transition temperature. JBIC Journal of Biological Inorganic Chemistry. 2011;16(2):217–226. doi: 10.1007/s00775-010-0718-y. [DOI] [PubMed] [Google Scholar]
- Bava KA, Gromiha MM, Uedaira H, Kitajima K, Sarai A. ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004;32(Database issue):D120–1. doi: 10.1093/nar/gkh082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Steinbacher S, Weintraub A, Huber R, Seckler R. Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity. J Mol Biol. 1999;293(3):693–701. doi: 10.1006/jmbi.1999.3165. [DOI] [PubMed] [Google Scholar]
- Benedix A, Becker CM, de Groot BL, Caflisch A, Bockmann RA. Predicting free energy changes using structural ensembles. Nat Methods. 2009;6(1):3–4. doi: 10.1038/nmeth0109-3. [DOI] [PubMed] [Google Scholar]
- Berliner N, Teyra J, Colak R, Garcia Lopez S, Kim PM. Combining structural modeling with ensemble machine learning to accurately predict protein fold stability and binding affinity effects upon mutation. PLoS One. 2014;9(9):e107353. doi: 10.1371/journal.pone.0107353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. Bioinformatics for geneticists. 2003;317:289. [Google Scholar]
- Boccuto L, Aoki K, Flanagan-Steet H, Chen CF, Fan X, Bartel F, Petukh M, Pittman A, Saul R, Chaubey A. A mutation in a ganglioside biosynthetic enzyme, ST3GAL5, results in salt & pepper syndrome, a neurocutaneous disorder with altered glycolipid and glycoprotein glycosylation. Hum Mol Genet. 2014;23(2):418–33. doi: 10.1093/hmg/ddt434. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock K, Talley K, Coley K, Kundrotas P, Alexov E. Optimization of electrostatic interactions in protein-protein complexes. Biophys J. 2007;93(10):3340–52. doi: 10.1529/biophysj.107.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DJ. Disease genetics: all together now for variant interpretation. Nat Rev Genet. 2014;15(4):216. doi: 10.1038/nrg3702. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20:63–68. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33(Web Server issue):W306–10. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio R, Vassura M, Tiwari S, Fariselli P, Luigi Martelli P. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum Mutat. 2011;32(10):1161–70. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- Chen CW, Lin J, Chu YW. iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinformatics 14 Suppl. 2013;2:S5. doi: 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Salem RM, Rao F, Fung MM, Bhatnagar V, Pandey B, Mahata M, Waalen J, Nievergelt CM, Lipkowitz MS. Common charge-shift mutation Glu65Lys in K+ channel beta(1)-Subunit KCNMB1: pleiotropic consequences for glomerular filtration rate and progressive renal disease. Am J Nephrol. 2010;32(5):414–24. doi: 10.1159/000320131. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JL, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins-Structure Function and Bioinformatics. 2006;62(4):1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- Chiariotti L, Doss CGP, NagaSundaram N. Investigating the Structural Impacts of I64T and P311S Mutations in APE1-DNA Complex: A Molecular Dynamics Approach. PLoS ONE. 2012;7(2):e31677. doi: 10.1371/journal.pone.0031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haene B, Attanasio C, Beysen D, Dostie J, Lemire E, Bouchard P, Field M, Jones K, Lorenz B, Menten B. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: implications for mutation screening. PLoS Genet. 2009;5(6):e1000522. doi: 10.1371/journal.pgen.1000522. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baets G, Van Durme J, Reumers J, Maurer-Stroh S, Vanhee P, Dopazo J, Schymkowitz J, Rousseau F. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Research. 2012;40(D1):D935–D939. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y, Grosfils A, Folch B, Gilis D, Bogaerts P, Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25(19):2537–2543. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. Bmc Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Rooman M, Gilis D. BeAtMuSiC: prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Research. 2013;41(W1):W333–W339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhanskaya N, Gonzalez MA, Sperziani F, Stefl S, Messing J, Wen GY, Alexov E, Zuchner S, Velinov M. A novel p.Leu(381)Phe mutation in presenilin 1 is associated with very early onset and unusually fast progressing dementia as well as lysosomal inclusions typically seen in Kufs disease. J Alzheimers Dis. 2014;39(1):23–7. doi: 10.3233/JAD-131340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domoszlai T, Martincuks A, Fahrenkamp D, Schmitz-Van de Leur H, Kuster A, Muller-Newen G. Consequences of the disease-related L78R mutation for dimerization and activity of STAT3. J Cell Sci. 2014;127(Pt 9):1899–910. doi: 10.1242/jcs.137422. [DOI] [PubMed] [Google Scholar]
- Doss CGP, Nagasundaram N, Chakraborty C, Chen L, Zhu H. Extrapolating the effect of deleterious nsSNPs in the binding adaptability of flavopiridol with CDK7 protein: a molecular dynamics approach. Human genomics. 2013;7(1):10. doi: 10.1186/1479-7364-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss CGP, Rajith B, Magesh R, Kumar AA. Influence of the SNPs on the structural stability of CBS protein: Insight from molecular dynamics simulations. Frontiers in Biology. 2014;9(6):504–518. [Google Scholar]
- Downward J. RAS's Cloak of Invincibility Slips at Last? Cancer cell. 2014;25(1):5–6. doi: 10.1016/j.ccr.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Fu H, Grimsley GR, Razvi A, Scholtz JM, Pace CN. Increasing protein stability by improving beta-turns. Proteins. 2009;77(3):491–8. doi: 10.1002/prot.22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George Priya Doss C, Rajith B. A New Insight into Structural and Functional Impact of Single-Nucleotide Polymorphisms in PTEN Gene. Cell Biochemistry and Biophysics. 2012;66(2):249–263. doi: 10.1007/s12013-012-9472-9. [DOI] [PubMed] [Google Scholar]
- Grothe HL, Little MR, Sjogren PP, Chang AA, Nelson EF, Yuan C. Altered protein conformation and lower stability of the dystrophic transforming growth factor beta-induced protein mutants. Molecular vision. 2013;19:593. [PMC free article] [PubMed] [Google Scholar]
- Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32(Web Server issue):W500–2. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. Naccess. Computer Program. Department of Biochemistry and Molecular Biology, University College; London: 1993. [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kamaraj B, Purohit R. Computational Screening of Disease-Associated Mutations in OCA2 Gene. Cell Biochemistry and Biophysics. 2013;68(1):97–109. doi: 10.1007/s12013-013-9697-2. [DOI] [PubMed] [Google Scholar]
- Khan RH, Chaturvedi D, Mahalakshmi R. Methionine Mutations of Outer Membrane Protein X Influence Structural Stability and Beta-Barrel Unfolding. PLoS ONE. 2013;8(11):e79351. doi: 10.1371/journal.pone.0079351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31(6):675–84. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Kucukkal TG, Petukh M, Li L, Alexov E. Structural and Physico-Chemical Effects of Disease and Non-Disease nsSNPs on Proteins. Curr Opin Struct. Biol. 2014a doi: 10.1016/j.sbi.2015.01.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukkal TG, Yang Y, Chapman SC, Cao W, Alexov E. Computational and experimental approaches to reveal the effects of single nucleotide polymorphisms with respect to disease diagnostics. Int J Mol Sci. 2014b;15(6):9670–717. doi: 10.3390/ijms15069670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Purohit R. Computational investigation of pathogenic nsSNPs in CEP63 protein. Gene. 2012;503(1):75–82. doi: 10.1016/j.gene.2012.04.032. [DOI] [PubMed] [Google Scholar]
- Kumar A, Purohit R. Use of Long Term Molecular Dynamics Simulation in Predicting Cancer Associated SNPs. PLoS computational biology. 2014;10(4):e1003318. doi: 10.1371/journal.pcbi.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Rajendran V, Sethumadhavan R, Purohit R. Molecular Dynamic Simulation Reveals Damaging Impact of RAC1 F28L Mutation in the Switch I Region. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED. A simple definition of structural regions in proteins and its use in analyzing interface evolution. J Mol Biol. 2010;403(4):660–670. doi: 10.1016/j.jmb.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Li M, Petukh M, Alexov E, Panchenko AR. Predicting the Impact of Missense Mutations on Protein-Protein Binding Affinity. J Chem Theory Comput. 2014;10(4):1770–1780. doi: 10.1021/ct401022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori C, Lantella A, Pasquo A, Alexander LT, Knapp S, Chiaraluce R, Consalvi V. Effect of Single Amino Acid Substitution Observed in Cancer on Pim-1 Kinase Thermodynamic Stability and Structure. PLoS One. 2013;8(6):e64824. doi: 10.1371/journal.pone.0064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masso M, Vaisman II. AUTO-MUTE 2.0: A portable framework with enhanced capabilities for predicting protein functional consequences upon mutation. Advances in bioinformatics 2014. 2014 doi: 10.1155/2014/278385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moal IH, Fernandez-Recio J. SKEMPI: a Structural Kinetic and Energetic database of Mutant Protein Interactions and its use in empirical models. Bioinformatics. 2012;28(20):2600–7. doi: 10.1093/bioinformatics/bts489. [DOI] [PubMed] [Google Scholar]
- Nair P, Vihinen M. VariBench: a benchmark database for variations. Hum Mutat. 2013;34(1):42–9. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- Nishi H, Tyagi M, Teng S, Shoemaker BA, Hashimoto K, Alexov E, Wuchty S, Panchenko AR. Cancer missense mutations alter binding properties of proteins and their interaction networks. PLoS One. 2013;8(6):e66273. doi: 10.1371/journal.pone.0066273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr N, Chanock S. Common genetic variation and human disease. Adv Genet. 2008;62:1–32. doi: 10.1016/S0065-2660(08)00601-9. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N, Weerapana E. Zinc-Binding Cysteines: Diverse Functions and Structural Motifs. Biomolecules. 2014;4(2):419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NJ, Weerapana E. Diverse Functional Roles of Reactive Cysteines. ACS Chemical Biology. 2013;8(2):283–296. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- Patel G, Johnson DS, Sun B, Pandey M, Yu X, Egelman EH, Wang MD, Patel SS. A257T linker region mutant of T7 helicase-primase protein is defective in DNA loading and rescued by T7 DNA polymerase. J Biol Chem. 2011;286(23):20490–9. doi: 10.1074/jbc.M110.201657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey D, Xiang Z, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53 Suppl. 2003;6:430–5. doi: 10.1002/prot.10550. others. [DOI] [PubMed] [Google Scholar]
- Petukh M, Wu B, Stefl S, Smith N, Hyde-Volpe D, Wang L, Alexov E. Chronic Beryllium Disease: Revealing the Role of Beryllium Ion and Small Peptides Binding to HLA-DP2. PLoS One. 2014;9(11):e111604. doi: 10.1371/journal.pone.0111604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DE, Ascher DB, Blundell TL. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42(Web Server issue):W314–9. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirolli D, Sciandra F, Bozzi M, Giardina B, Brancaccio A, De Rosa MC. Insights from Molecular Dynamics Simulations: Structural Basis for the V567D Mutation-Induced Instability of Zebrafish Alpha-Dystroglycan and Comparison with the Murine Model. PLoS ONE. 2014;9(7):e103866. doi: 10.1371/journal.pone.0103866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placone J, He L, Del Piccolo N, Hristova K. Strong dimerization of wild-type ErbB2/Neu transmembrane domain and the oncogenic Val664Glu mutant in mammalian plasma membranes. Biochim Biophys Acta. 2014;1838(9):2326–30. doi: 10.1016/j.bbamem.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placone J, Hristova K. Direct assessment of the effect of the Gly380Arg achondroplasia mutation on FGFR3 dimerization using quantitative imaging FRET. PLoS One. 2012;7(10):e46678. doi: 10.1371/journal.pone.0046678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–91. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran V, Purohit R, Sethumadhavan R. In silico investigation of molecular mechanism of laminopathy caused by a point mutation (R482W) in lamin A/C protein. Amino Acids. 2011;43(2):603–615. doi: 10.1007/s00726-011-1108-7. [DOI] [PubMed] [Google Scholar]
- Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25(11):1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Roedelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods. 2010;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Research. 2005a;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz JWH, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proceedings of the National Academy of Sciences of the United States of America. 2005b;102(29):10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GL, Edwards KJ, Day IN, Gaunt TR. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human mutation. 2013;34(1):57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov VZ, Yan L. pH-selective mutagenesis of protein-protein interfaces: In silico design of therapeutic antibodies with prolonged half-life. Proteins. 2013;81(4):704–14. doi: 10.1002/prot.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl S, Nishi H, Petukh M, Panchenko AR, Alexov E. Molecular mechanisms of disease-causing missense mutations. J Mol Biol. 2013;425(21):3919–36. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K, Liu D, Tarpey P, Gallant E, Lam A, Witham S, Alexov E, Chaubey A, Stevenson RE, Schwartz CE. An X-linked channelopathy with cardiomegaly due to a CLIC2 mutation enhancing ryanodine receptor channel activity. Hum Mol Genet. 2012;21(20):4497–507. doi: 10.1093/hmg/dds292. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Madej T, Panchenko A, Alexov E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys J. 2009;96(6):2178–88. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369(5):1318–32. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19(5):596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Torshin IY, Weber IT, Harrison RW. Geometric criteria of hydrogen bonds in proteins and identification of “bifurcated” hydrogen bonds. Protein Eng. 2002;15(5):359–63. doi: 10.1093/protein/15.5.359. [DOI] [PubMed] [Google Scholar]
- Vacic V, Markwick PR, Oldfield CJ, Zhao X, Haynes C, Uversky VN, Iakoucheva LM. Disease-associated mutations disrupt functionally important regions of intrinsic protein disorder. PLoS computational biology. 2012;8(10):e1002709. doi: 10.1371/journal.pcbi.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M. Guidelines for reporting and using prediction tools for genetic variation analysis. Hum Mutat. 2013;34(2):275–82. doi: 10.1002/humu.22253. [DOI] [PubMed] [Google Scholar]
- Vihinen M. Proper reporting of predictor performance. Nat Methods. 2014;11(8):781. doi: 10.1038/nmeth.3032. [DOI] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17(4):263–70. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wei Q, Dunbrack RL., Jr The role of balanced training and testing data sets for binary classifiers in bioinformatics. PLoS ONE. 2013;8(7):e67863. doi: 10.1371/journal.pone.0067863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witham S, Takano K, Schwartz C, Alexov E. A missense mutation in CLIC2 associated with intellectual disability is predicted by in silico modeling to affect protein stability and dynamics. Proteins. 2011;79(8):2444–54. doi: 10.1002/prot.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sommers JA, Suhasini AN, Leonard T, Deakyne JS, Mazin AV, Shin-Ya K, Kitao H, Brosh RM., Jr Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010;116(19):3780–91. doi: 10.1182/blood-2009-11-256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyavi M, Falsafi-Zadeh S, Karimi Z, Kalatarian G, Galehdari H. VMD-SS: A graphical user interface plug-in to calculate the protein secondary structure in VMD program. Bioinformation. 2014;10(8):548–50. doi: 10.6026/97320630010548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Wu M, Li Y, Zheng GY, Cao HQ, Sun W, Yang R, Zhang H, Sheng YH, Kong XQ. Mutation p.S335X in GATA4 reduces its DNA binding affinity and enhances cell apoptosis associated with ventricular septal defect. Curr Mol Med. 2013;13(6):993–9. doi: 10.2174/15665240113139990053. others. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhou Y. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions. Protein Science. 2008a;17(7):1212–1219. doi: 10.1110/ps.033480.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhou Y. Specific interactions for ab initio folding of protein terminal regions with secondary structures. Proteins-Structure Function and Bioinformatics. 2008b;72(2):793–803. doi: 10.1002/prot.21968. [DOI] [PubMed] [Google Scholar]
- Yates CM, Sternberg MJ. The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J Mol Biol. 2013;425(21):3949–63. doi: 10.1016/j.jmb.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 2005;353(2):459–73. doi: 10.1016/j.jmb.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7(1):166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356(5):1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Miteva MA, Wang L, Alexov E. Analyzing effects of naturally occurring missense mutations. Comput Math Methods Med. 2012a;2012:805827. doi: 10.1155/2012/805827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Norris J, Kalscheuer V, Wood T, Wang L, Schwartz C, Alexov E, Van Esch H. A Y328C missense mutation in spermine synthase causes a mild form of Snyder-Robinson syndrome. Hum Mol Genet. 2013;22(18):3789–97. doi: 10.1093/hmg/ddt229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Norris J, Schwartz C, Alexov E. In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in spermine synthase. PLoS One. 2011;6(5):e20373. doi: 10.1371/journal.pone.0020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Teng S, Wang L, Schwartz CE, Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31(9):1043–9. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang L, Gao Y, Zhang J, Zhenirovskyy M, Alexov E. Predicting folding free energy changes upon single point mutations. Bioinformatics. 2012b;28(5):664–71. doi: 10.1093/bioinformatics/bts005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.