Abstract
p21-activated kinase 2 (Pak2), a serine/threonine kinase, has been previously shown to be essential for hematopoietic stem cell (HSC) engraftment. However, Pak2 modulation of long-term hematopoiesis and lineage commitment remain unreported. Utilizing a conditional Pak2 knock out (KO) mouse model, we found that disruption of Pak2 in HSCs induced profound leukopenia and a mild macrocytic anemia. Although loss of Pak2 in HSCs leads to less efficient short- and long-term competitive hematopoiesis than wild type (WT) cells, it does not affect HSC self-renewal per se. Pak2 disruption decreased the survival and proliferation of multi-cytokine stimulated immature progenitors. Loss of Pak2 skewed lineage differentiation toward granulocytopoiesis and monocytopoiesis in mice as evidenced by 1) a three to six-fold increase in the percentage of peripheral blood granulocytes and a significant increase in the percentage of granulocyte-monocyte progenitors (GMPs) in mice transplanted with Pak2-disrupted BM; 2) Pak2-disrupted BM and c-kit+ cells yielded higher numbers of more mature subsets of granulocyte-monocyte colonies and polymophonuclear neutrophils (PMNs), respectively, when cultured in the presence of granulocyte-macrophage colony stimulating factor (GM-CSF). Pak2 disruption resulted respectively in decreased and increased gene expression of transcription factors JunB and c-Myc, which may suggest underlying mechanisms by which Pak2 regulates granulocyte-monocyte lineage commitment. Furthermore, Pak2 disruption led to 1) higher percentage of CD4+CD8+ double positive T cells and lower percentages of CD4+CD8− or CD4−CD8+ single positive T cells in thymus and 2) decreased numbers of mature B cells and increased numbers of Pre-Pro B cells in BM, suggesting defects in lymphopoiesis.
Keywords: Pak2, hematopoietic progenitor cell, myelopoiesis, lymphopoiesis
Introduction
p21-activated kinases (Paks) are serine/threonine kinases that regulate diverse cellular activities including cytoskeletal remodeling, cell motility, cell proliferation, apoptosis and mitosis.1-3 Based on structural and functional similarities, Paks are divided into two groups. Group I consists of Pak1, Pak2, and Pak3 and group II consists of Pak4, Pak5, and Pak6.2 Among the six members of the Paks, Pak1, the most characterized isoform, is expressed in muscle, spleen, and brain.4 Pak2 is expressed ubiquitously while Pak3 is expressed principally in brain.5 Upregulation of Pak1 has been identified in a variety of human cancers, particularly hormone-dependent solid tumors, and is linked to tumor progression and poor survival in patients.5 Although structurally highly homologous to Pak1,6 recent studies suggest that Pak2 has shared and distinct functions in regulating cellular functions in different cell types.7-10
Few studies have examined the role of Paks in modulation of normal hematopoietic cell function and the pathogenesis of hematological malignancies.2,3,11-13 Previous work in our laboratory demonstrated that Pak1 knockout (Pak1-KO) mice have normal BM cellularity and complete blood count (CBC) profiles,14 and our unpublished data demonstrate that Pak1-KO hematopoietic stem cells (HSCs) reconstitute lethally irradiated mice as efficiently as wild type (WT) HSCs (D.W.C., unpublished). Thus, while Pak1 appears dispensable for steady-state hematopoiesis, a recent report demonstrated that Pak2 regulates HSC migration and engraftment.15 However, Pak2's regulation of long-term hematopoiesis and lineage commitment remains unreported.
In this study, utilizing novel lentiviral vectors and a conditional Pak2-KO murine model, we show that Pak2 disruption reduces proliferation and survival of hematopoietic progenitor cells (HPCs) in vitro, and in vivo leads to profound peripheral blood leukopenia while contributing to granulocyte/monocyte skewing and T and B cell differentiation/maturation defects. Pak2 disruption does not compromise HSC self-renewal per se. Bone marrow (BM) analyses revealed normal phenotypic HSC (defined as Lin−Sca1+c-Kit+CD150+CD48/41−) numbers, reduced Lin−Sca1+c-Kit+ cell (enriched for HPC) numbers, and an increased frequency of granulocyte-monocyte progenitors (GMPs), suggesting that a proliferation deficiency and myeloid lineage bias occurs at the progenitor cell level.
Materials And Methods
Mice and genotyping
To generate the conditional Pak2-KO mice, Pak2flox/flox mice (construction to be described elsewhere) were bred to Mx1Cre transgenic mice. Pak2flox/floxMx1Cre+ and Pak2flox/floxMx1Cre− mice were treated with 20mg/g body weight of polyIC every other day for total of three doses. Within 24 hours following final dose, BM cells were collected and Pak2 expression determined by Western blotting. Mice were maintained at Indiana University School of Medicine. All studies were reviewed and approved by the Institutional Animal Care and Use Committee and the Institutional Review Board.
Isolation of BM cells and c-kit positive cells
Low density mononuclear cells (LDMNCs) were isolated by density gradient centrifugation as described.16 LDMNCs were pooled from two femurs per mouse and enumerated using a hemocytometer. The absolute numbers of LDMNCs per femur were reported. C-kit+ cells were isolated by magnetic cell sorting using a CD117 isolation kit (Miltenyi Biotec, Auburn, CA, USA), according to manufacturer's protocol, with >90% selection purity. Unless otherwise mentioned, cells were cultured in Iscove modified Dulbecco medium (IMDM, from Lift Technologies, USA) with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% L-glutamine (Lonza, Walkersville, MD).
Transduction of cells with Lentiviral vectors
Lentiviral vectors encoding either enhanced green fluorescent protein (LVeGFP) or Cre recombinase-eGFP fusion protein (LVCre-eGFP) were used. The lentiviral eGFP backbone, helper plasmid, envelope plasmid and packaging 293T cells were generously provided by Dr. Helmut Hanenberg (Indiana University, Indianapolis). Cre recombinase cDNA was cloned into lentiviral eGFP backbone and virus generated using 293T cells, as described17. C-kit+ BM LDMNCs were transduced with lentivirus at a multiplicity of infection of 100:1 in the presence of murine interleukin-6 (mIL-6, 200 U/mL), Flt3 ligand (100 ng/mL), and murine stem cell factor (mSCF, 100 ng/mL)(all cytokines from Peprotech, Rocky Hill, NJ), as described, with minor modification.17 Four days post-transduction, GFP+ cells were sorted by FACS and used for in vitro assays. Experiments involving recombinant DNA were conducted following the National Institute of Health guidelines.
Colony assays
Methylcellulose-based colony assays were performed using LDMNCs, as described.16,18,19 Briefly, 20,000 sorted CD45.2+ BM LDMNCs were cultured in methylcellulose (MethoCult™ H4100, Stemcell Technologies, Vancouver, Canada) containing FBS (30%), β-mercaptoethanol, glutamine (1%), mSCF (100 ng/mL), murine granulocyte-macrophage-colony stimulating factor (mGM-CSF, 10 ng/mL), murine interleukin-3 (mIL-3, 5 ng/mL) and erythropoietin (4 units/ml) in 35mm Nunclon gridded dishes (Thermo Scientific). This assay measures multi-cytokine stimulated immature progenitors 20,21. All colonies including CFU-GM, CFU-GEMM and BFU-E in each dish were scored and the sum was shown in figures. Each condition was plated in triplicate or six repeats. Cell cultures were incubated in a 37°C humidified incubator with 5% CO2. In some experiments, individual colonies were collected and cytospin preparations of the progenies were subjected to Wright-Giemsa staining. For colony assays with mGM-CSF, mM-SCF, mG-CSF, mSCF or mIL-3 alone, 25,000 sorted CD45.2+ BM LDMNCs were plated in 0.3% agar culture medium containing 10% FBS and respective single cytokines 20. The absolute number of colonies was adjusted to reflect that contained per femur.
For multi-cytokine stimulated immature progenitor cell survival assays, equal numbers (2000 cells/35 mm dish) of lentiviral vector transduced GFP+Pak2flox/flox c-kit+ BM LDMNCs were serum starved in IMDM+1% BSA+100ng/ml mSCF for 0, 24, 48 and 72 hours prior to being plated for colony assay with mGM-CSF, mSCF, mIL-3 and EPO added to the culture as described above. All colonies including CFU-GM, CFU-GEMM and BFU-E in each dish were scored and the sum was used to calculate the percentage of survival.
Cell cycle analysis of functional progenitor cells that form colonies in vitro
In vitro high specific activity tritiated 3H thymidine suicide assays were performed as previously described.22 Briefly, CD45.2+ BM LDMNCs were pulse-treated with control medium or with medium containing high specific activity 3H thymidine (50 mCi/mL, specific activity = 20 Ci/mmol; New England Nuclear, Boston, MA) for 30 minutes at 37°C. Cells were then washed three times with control medium before plating in clonogenic assays with mGM-CSF, mSCF, mIL-3 and EPO added to the culture as described above. The percentage of colony forming progenitors in S phase was determined by the following formula: Control colonies minus 3H thymidine colonies, divided by control colonies. All colonies including CFU-GM, CFU-GEMM and BFU-E in each dish were scored and the sum was used for the calculation.
Western blotting
Whole cell lysates were generated from equivalent number of cells and applied to Western blotting as described.23 Primary antibody was rabbit anti-Pak2 polyclonal antibody (Cell signaling Technology, Danvers, MA). Equal protein loading was monitored by using β-actin as an internal control.
Proliferation assay
C-kit+ BM cells were cultured in IMDM+10% FBS without cytokines overnight before plating into 96 well round bottom plates and adding mSCF (100 ng/mL, Peprotech, Rocky Hill, NJ) for 18 hours. 3H thymidine (1 mCi/well) (Amersham, Les Ulis, France) was added, and measured 6 hours later.
Annexin V staining
C-kit+ BM cells were cultured in IMDM+2% bovine serum albumin (BSA) and 100ng/ml mSCF overnight. Cells were then stained for 7-Amino-Actinomycin (7-AAD) and Annexin V-PE using PE Annexin V Apoptosis Detection kit, following manufacturer's instructions (BD Pharmingen).
Cell cycle analysis of lentiviral vector transduced c-kit+ cells
Cell cycle status of test cells was determined using propidium iodide (PI) staining, as previously described.24 Briefly, cells were washed with cold phosphate-buffered saline (PBS) twice then stained with equal volume of PBS containing 0.1mg/ml PI (Behring Diagnositics, La Jolla, CA), 0.6% Nonidet P-40 (NP-40), and PBS containing 2mg/ml RNAse (Sigma) for 30 minutes. Sample were analyzed on a FACScan (BDIS) and data analyzed using ModFit software (Verity Software House).
Flow cytometry
Peripheral white blood cells, BM LDMNCs, splenocytes or thymocytes were incubated with fluorochrome conjugated anti-mouse antibodies in ∼100 μL of PBS with 2% FBS at 4°C for 40 minutes, as previously described.19 One μg antibody was used per one million cells for each antibody. All antibodies were from BD Biosciences, BioLegend or eBiosciences. For lineage analysis, the following antibody panel was used: anti-CD45.1-APC, anti-CD45.2-PerCP-Cy5.5, anti-CD3e-FITC, anti-B220-HorizonV500, anti-CD8-PacificBlue, anti-CD4-APC eFluor 780 (eBiosciences), anti-Ly6G-PE-Cy7, and anti-11b/Mac1-PE. For BM HSC and HPC analysis, the following panel was used: FITC-conjugated anti-lineage markers (CD3, CD4, CD8, B220, Mac-1, Ly6G, Ter119), anti-Sca1-APC-Cy7, anti-c-Kit-PE, anti-CD150-APC, anti-CD41-PE-Cy 7, anti-CD48-PE-Cy 7, anti-CD135 (Flt3)-APC and anti-CD127(IL-7Ra)-PE-Cy7. SLAM-LSK cells, which represent long-term multi-lineage repopulating HSCs 25, are defined as Lin−Sca1+c-Kit+CD150+CD48/41− cells. BM common lymphoid progenitor cells (CLPs) are defined as Lin−IL7Rα+c-KitlowSca1low. For BM B cell flow, the following panel was used: anti-CD45.2-APC-Cy7, anti-B220-BV510, anti-IgM-e450, anti-IgD-APC, anti-CD43-PE and anti-CD24-FITC. BM mature B cells are defined as B220+IgM+IgD+, immature B cells are B220+IgM+IgD−, Pre B cells are B220+IgM−IgD−CD43−CD24high, Pro B cells are B220+IgM−IgD− CD43+CD24med and Pre-Pro B cells are B220+IgM−IgD−CD43+CD24− 26. For spleen B cell flow, anti-CD45.2-APC-Cy7, anti-B220-BV510, anti-CD93 FITC, anti-CD23-e450 and anti-CD21-PE were used. Splenic mature B cells are defined as B220+CD93−, immature B cells are B220+CD93+, marginal zone B cells are B220+CD93−CD21highCD23−, and follicular B cells are B220+CD93−CD21medCD23+ 26. For spleen T cell and polymorphonuclear neutrophils (PMNs) flow, anti-CD4.2-APC-Cy7, anti-CD3e-PECF594, anti-B220-BV510, anti-Mac-1-e450 and anti-Gr-1-FITC were used. For thymic T cell flow, anti-CD45.1-APC, anti-CD45.2-PacBlue, anti-CD3e-PECF594, anti-CD4-FITC and anti-CD8-PE-Cy7 were used. Data were acquired on an LSR II 407 flow cytometer outfitted with red (633 nm, 2 detectors), blue (488 nm, 5 detectors), and violet lasers (407 nm, 2 detectors) (BD Biosciences). Single color compensation controls were included in each experiment using polystyrene microbeads (BD Biosciences). Data analysis was performed using FlowJo 7.6.3 software (TreeStar, WA). Gates were determined using fluorescence minus-one controls. The absolute number of phenotypically defined CD45.2+ cell population in femur, spleen or thymus was obtained by multiplying the percentage of indicated cell population by absolute number of BM LDMNCs per femur, splenocytes or thymocytes respectively.
Transplantation, Mx1Cre induction, and blood isolation
For competitive repopulation assays, CD45.2+Pak2flox//floxMx1Cre+ or Pak2flox//floxMx1Cre− BM LDMNCs were mixed with CD45.1+ WT BoyJ cells at 1:1 ratios and injected I.V. into 8-10 week old lethally-irradiated (1100 cGy, divided into two doses 5-hours apart) CD45.1+/CD45.2+ WT recipient mice. For non-competitive transplantation experiment, CD45.2+Pak2flox//floxMx1Cre+ or Pak2flox//floxMx1Cre− BM LDMNCs were injected into CD45.1+ WT BoyJ mice. In all experiments, each recipient mouse received 3×106 total LDMNCs in 200 μL PBS. Two months following transplantation, Cre expression was induced by intraperitoneal (i.p.) injections of poly I poly C (polyIC, Sigma) dissolved in sterile PBS. Each transplanted mouse received a total of 6 escalating doses of polyIC starting at 10 μg/g body weight and ending with 25 μg/g (cumulative dose = 100 μg/g body weight). Peripheral blood was drawn from tail vein using di-potassium EDTA (ethylenediaminetetraacetic acid) salt as an anticoagulant prior to first dose polyIC administration and at the indicated time points after transplantation. Peripheral white blood cells were isolated by incubating (15 minutes, 23 C) 30-50 μL of blood in RBC lysis buffer (Qiagen), followed by washing twice in PBS.
For secondary transplantation, six months following polyIC administrations, 4×106 BM LDMNCs from primary recipients in the competitive transplantation experiments were injected into lethally irradiated CD45.1+/CD45.2+ WT recipient mice. BM LDMNCs from each donor were injected into 3 or 4 secondary recipients.
Peripheral blood complete blood cell count
Peripheral blood was collected from mice in the non-competitive transplant cohort prior to and post polyIC injections. Complete blood cell count (CBC) was determined using Hemavet 950 machine (Drew Scientific) following manufacturer's instruction. The absolute number of CD45.2+ WBCs was generated by multiplying total WBC number with the percentage of CD45.2+ cells in the peripheral blood.
Histologic analysis
Eight months post polyIC administrations, tibiae from the non-competitively transplanted cohort were collected and fixed in 1% formaldehyde at room temperature for 6 hours and then embedded in paraffin. Hematoxylin-eosin (H&E) were performed using routine methods.
Quantitative Real-Time PCR
CD45.2+Lin−kit+ cells were sorted from LDMNCs from CD45.1+ BoyJ mice that were transplanted with CD45.2+Pak2flox/floxMx1Cre+ or Pak2flox/floxMx1Cre− marrow by FACS. mRNA was isolated using Miltenyi μMACS mRNA isolation kit (Miltenyi) and then reverse transcribed to cDNA using iScript reverse transcription supermix kit (Bio-Rad, Hercules, CA). Quantitative Real-Time PCR (qRT-PCR) was performed using SsoAdvanced universal probes supermix (Bio-Rad) and TaqMan probes against murine JunB, c-Myc, PU.1 and GAPDH (Appplied Biosystems). qRT-PCR detection was done on a Bio-Rad CFX96 thermocycler (Bio-Rad). 2-ΔΔCT method was used to analyze the gene expression levels as previously described. 27
Statistics
Statistical analyses were performed with GraphPad Prism 5.0 or Excel. Data were reported as mean ± SE and were analyzed using unpaired two-tailed student's t tests or ANOVA with appropriate post-hoc comparisons. Differences yielding p<0.05 were defined as statistically significant.
Results
Conditional disruption of Pak2 in hematopoietic cells
Pak2 nullizygous mice die before embryonic age 8.5 days. Thus, to study Pak2 deletion in the marrow compartment in vivo, Pak2flox/flox mice were crossed with Mx1Cre mice. Although we successfully deleted Pak2 in hematopoietic cells following polyIC administration in Pak2flox/floxMx1Cre+ mice (Supplementary Figure S.1A), all mice died within 14 days of polyIC treatment, for reasons unrelated to hematopoiesis. Thus, to better understand a role for Pak2 in hematopoiesis, we isolated and transduced Pak2flox/flox c-kit+ BM cells with lentiviral vectors encoding either enhanced green fluorescent protein (LVeGFP) or Cre recombinase- enhanced green fluorescent protein fusion protein (LVCre-eGFP). Transduction of the c-kit+ cells with either LVCre-eGFP or LVeGFP caused less than 20% cell death. As shown in supplementary figure S.1B, transduction with LVCre-eGFP successfully reduced Pak2 expression in c-kit+ cells.
Pak2 regulates HPC proliferation, survival and apoptosis
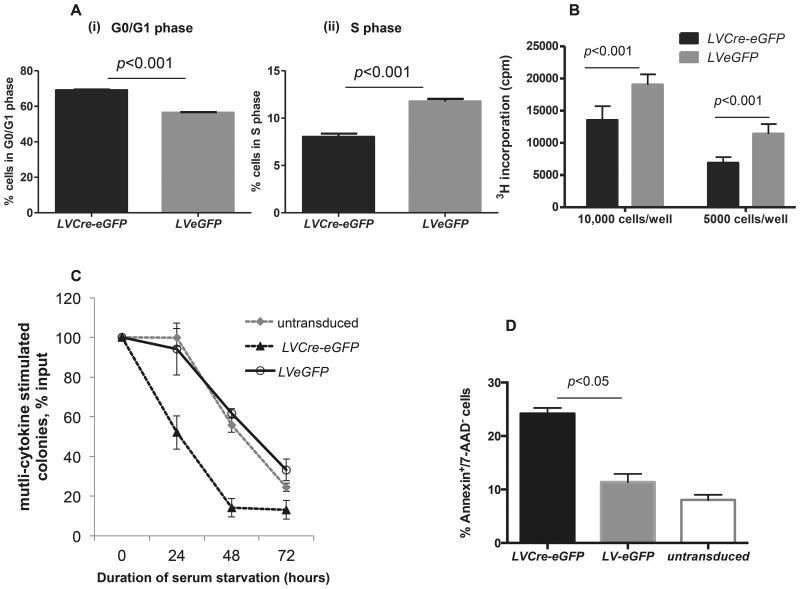
Prior studies suggested group I Paks regulate G1 to S transition in certain cell lines 28 as well as proliferation and survival in HPCs.15 To further study Pak2 in HPCs, we cultured LVCre-eGFP or LVeGFP transduced GFP+Pak2flox/flox c-kit+ cells (enrich for HPCs) with SCF overnight to allow the cells to recover from the stress associated with flow cytometric sorting before their use in functional assays. Although most active as a co-stimulating factor for HPC and HSC proliferation, SCF has modest stimulating activity by itself 29,30 and was therefore used in cell cycle and proliferation assays. Pak2-disrupted cells showed an ∼15% increase in the frequency of cells in the G0/G1 phase, a corresponding 14% decrease in S phase (Figure 1A), and an approximately 30-50% decrease in SCF-induced proliferation in Pak2-disrupted HPCs (Figure 1B), consistent with impaired transition from G0/G1 to S phase. Despite the survival signal provided by SCF, serum starved Pak2-disrupted progenitors demonstrated substantial impairment in multi-cytokine stimulated colony formation as evidenced by the more significant decrease in the sum of CFU-GM, CFU-GEMM and BFU-E when compared to WT cells, indicating a survival disadvantage (Figure 1C). Following serum starvation, Pak2-disrupted c-kit+ cells rapidly underwent apoptosis, as compared to WT (Figure 1D).
Figure 1. Pak2 regulates hematopoietic progenitor cell proliferation, survival and apoptosis.
Pak2flox/flox c-kit+ BM cells were untransduced or transduced with lentiviral vectors encoding either eGFP (LVeGFP) or Cre-eGFP (LVCre-eGFP). GFP+ cells were sorted and used. (A) Cells were cultured in IMDM+10% FBS+100ng/ml of mSCF for 24 hours before cell cycle analysis. Panels (i) and (ii) demonstrate the pooled G0/G1 phase and S phase data from at least 3 mice, respectively. Student t test, p<0.05. (B) Cells were cultured overnight in IMDM+10% FBS without cytokines before being plated in IMDM+1%BSA and stimulated with 100ng/ml SCF for 18 hours. 3H incorporation was measured. Hest, p<0.001. (C) Cells were serum starved in IMDM+1% BSA+100ng/ml mSCF for indicated time period prior to being plated for multi-cytokine stimulated colony assay with IL-3, SCF, GM-CSF and EPO added to the culture. All colonies including CFU-GM, CFU-GEMM and BFU-E in each dish were scored and the sum was used to calculate the percentage of survival. Y axis indicates colony formation as percentage of that at hour 0. Untransduced vs LVeGFP, p>0.05, LVCre-eGFP vs LVeGFP, p<0.05 at hour 24, 48 and 72. Student t test. (D) Cells were serum starved in IMDM+1% BSA+100ng/ml mSCF for overnight, apoptotic death of cells was determined by Annenxin/7-AAD staining. One-way ANOVA, p<0.05 between LVCre-eGFP and LVeGFP. Representative data from 2-3 independent experiments are shown.
Loss of Pak2 in HSCs leads to less efficient short- and long-term competitive hematopoiesis than WT cells
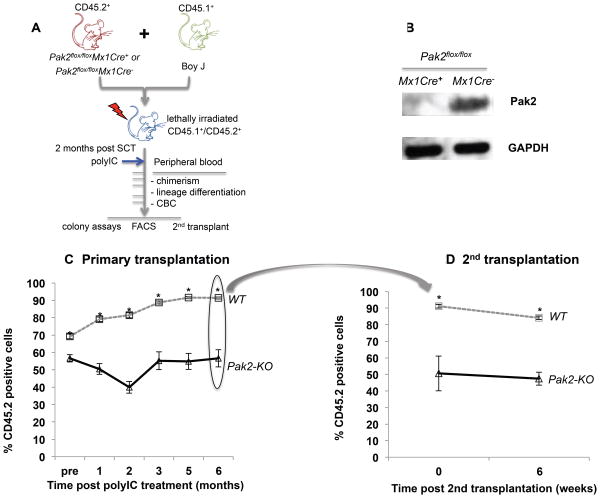
We performed competitive repopulation assays by isolating CD45.2+Pak2flox/floxMx1Cre+ or Pak2flox/floxMx1Cre− BM LDMNCs, mixing them with CD45.1+ WT BoyJ cells at 1:1 ratio, and transplanting them into lethally-irradiated CD45.1+/CD45.2+ hosts (Figure 2A). Following stable hematopoietic reconstitution (2 months post-transplantation), we injected control and experimental mice with polyIC, thus inducing Cre expression in Mx1Cre+ mice. This design allows hematopoietic cell specific deletion of Pak2, using WT as internal controls (Pak2flox/flox cells vs. WT BoyJ cells) and the ability to track Pak2-KO HSC-derived short- and long-term hematopoiesis in vivo. The role of Pak2 on HSC homing was not examined in this assay as Pak2 deletion was induced after HSC homing and engraftment. Loss of Pak2 gene expression in transplanted CD45.2+ cells is shown Figure 2B.
Figure 2. Loss of Pak2 in HSCs leads to less efficient short- and long-term competitive hematopoiesis than WT cells.
(A) Schematic of competitive repopulation transplantation. (B) Pak2 expression in sorted CD45.2+ BM cells at 6 months post-polylC administrations. The percentage of peripheral blood CD45.2+ cells in lethally irradiated (C) primary and (D) secondary recipients. N=6-8 and n=9-11 mice/group in primary and secondary transplantation experiments, respectively. Representative data from 3 competitive transplantation experiments are shown. Student t test, * indicates p<0.05 between Pak2-KO and WT. In secondary transplantation experiment, 0 vs 6 weeks: p>0.05 in both WT and Pak2-KO cohorts.
Over time, mice transplanted with WT CD45.2+ BM cells showed increased percentage of CD45.2+ peripheral blood cells, as CD45.2+ WT HSCs outcompete CD45.1+ BoyJ HSCs.31 In contrast, CD45.2+ Pak2-KO HSCs failed to outcompete CD45.1+ BoyJ cells, showing stable chimerism by six months post Pak2 deletion. At six months post-polyIC, Pak-KO CD45.2+ chimerism decreased to less than 60% (57+/- 5% vs 91+/- 0.6%) of WT (Figure 2C). Thus, while capable of hematopoietic reconstitution, Pak2 disruption in CD45.2+ HSCs led to less efficient short- and long-term competitive hematopoiesis as compared to WT cells. Prior to polyIC administration, mice reconstituted with Pak2-KO BM had lower chimerism than those transplanted with WT BM from causes that are unclear to us at this time.
To evaluate the role of Pak2 in regulating HSC long-term BM repopulation, we performed secondary transplantation at six months post Pak2 deletion in primary recipients. At six weeks following secondary transplantation, chimerism in secondary recipients that were reconstituted with either WT or Pak2-KO HSCs was stable (Figure 2D), suggesting that Pak2 disruption does not compromise HSC self-renewal per se. The decreased chimerism in peripheral blood (Figure 2C) may be a result of reduced survival and proliferation of HPCs and terminally differentiated mature lineages.
Pak2 disruption results in severe leukopenia and mild macrocytic anemia
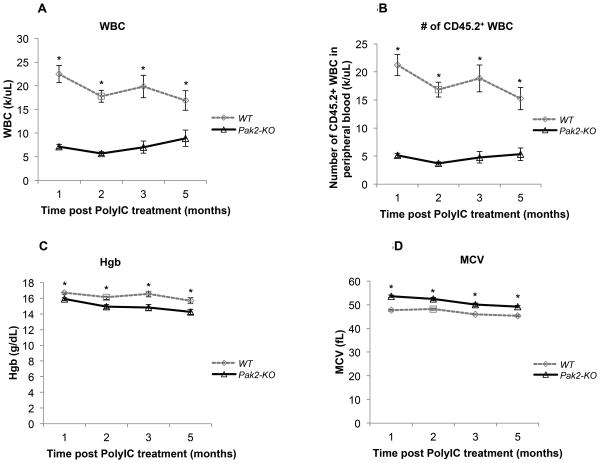
To study the ability of CD45.2+ Pak2-KO HSCs to maintain long-term hematopoiesis without contribution from CD45.1+ WT BoyJ cells, we also transplanted a cohort of CD45.1+ WT BoyJ mice non-competitively with CD45.2+Pak2flox/floxMx1Cre+ or Pak2flox/floxMx1Cre− marrow, again inducing Cre expression by polyIC injection two months post transplantation. Peripheral blood from mice reconstituted with Pak2-KO HSCs displayed severe leukopenia with a near 75% reduction in total (Figure 3A) and CD45.2+ peripheral blood white blood cell (WBC) count (Figure 3B) when compared to mice transplanted with WT HSCs starting at one month post Pak2 deletion. These mice also developed a very mild but statistically significant macrocytic anemia (Figure 3C and 3D).
Figure 3. Pak2 disruption results in severe leukopenia and mild macrocytic anemia.
CD45.1+ WT BoyJ mice were non-competitively transplanted with CD45.2+Pak2flox/floxMx1Cre+ or Pak2flox/floxMx1Cre− BM cells. Peripheral blood was collected at indicated time points post polylC treatment for complete blood cell count (CBC) and flow cytometry. (A) total white blood cell (WBC) count, (B) the absolute number of CD45.2+ WBCs in the peripheral blood (C) hemoglobin (Hgb) level, (D) mean corpuscular volume (MCV) of red blood cells. N=6-8/group. Representative data from 3 experiments are shown. Student t test, * indicates p<0.05 between Pak2-KO and WT.
Pak2 disruption diminishes phenotypic HPC but preserves phenotypic HSC population
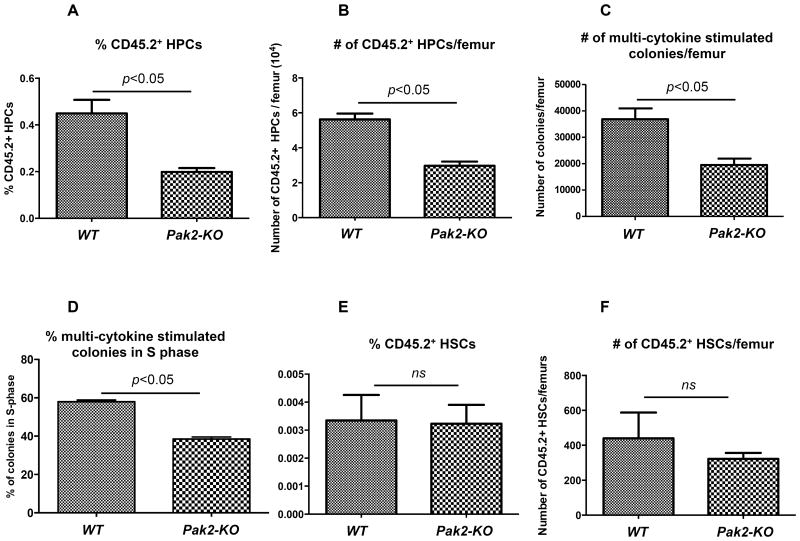
Given the striking leukopenia in mice reconstituted with Pak2-KO BM cells in the competitive and non-competitive transplant cohorts, we assessed phenotypically-defined HSC and HPC frequency and numbers in the BM of these mice. Despite the significant difference in peripheral blood WBC counts, we found no difference in the number of BM LDMNCs per femur between mice transplanted with Pak2-KO and WT BM cells (Supplementary Figure S.2A). Histological examination also revealed no gross difference in total cellularity (Supplementary Figure S.2B). However, Pak2-KO BM showed a 50% diminution in the frequency and total number of Lin−Sca1+c-Kit+ cell population that is enriched for HPCs (Figures 4A-B, representative flow gating shown in Supplementary Figure S.3). When compared to mice transplanted with WT cells, those transplanted with Pak2-KO BM cells had approximately 50% less multi-cytokine stimulated colonies (the sum of CFU-GM, CFU-GEMM and BFU-E) per femur as assessed by colony assays containing GM-CSF, SCF, IL-3 and EPO (Figure 4C).
Figure 4. Pak2 disruption diminishes phenotypic HPC but preserves phenotypic HSC population.
Flow cytometric and colonogenic analysis of BM in mice that were competitively transplanted with the 1:1 mix of CD45.1+ BoyJ and CD45.2+Pak2flox/floxMx1Cre+ (Pak2-KO) or Pak2flox/floxMx1Cre− (WT) BM cells. (A) The percentage and (B) absolute number of CD45.2+ phenotypic HPCs (Lin−Sca1+c-Kit+) in the BM. (C) The absolute number of CD45.2+ multi-cytokine stimulated colonies which represent the sum of CFU-GM, CFU-GEMM and BFU-E per femur. (D) The percentage of CD45.2+ multi-cytokine stimulated colonies that were in S phase as determined by high specific activity tritiated thymidine suicide assays. (E) The percentage and (F) absolute number of phenotypic CD45.2+ HSCs (Lin−Sca1+c-Kit+CD150+CD48/41−) in the BM. Representative data from 2-4 independent experiments are shown, 3 or more mice per genotype in each experiment. Student t test. ns=non-significant, p>0.05. Data was collected at 6 to 8 months post PIPC administration.
We reasoned that the reduction in the phenotypic HPC population might be the result of decreased proliferation of Pak2 deficient progenitors. Using high specific activity tritiated thymidine killing assays with GM-CSF, SCF, IL-3 and EPO added in the culture, we revealed a near 35% decrease in the percentage of Pak2-disrupted multi-cytokine stimulated colonies in S phase (Figure 4D), consistent with decreased numbers of these colonies (Figure 4C).
We then examined SLAM-LSK cells (Lin−Sca1+c-Kit+CD150+CD48/41−), which represent long-term multi-lineage repopulating HSCs 25 (representative flow gating shown in Supplementary Figure S.3). We found no difference in the frequency (Figure 4E) or absolute number (Figure 4F) of phenotypically identified long-term repopulating HSCs between mice transplanted with Pak2-KO and WT marrow.
Pak2 disruption induces myeloid skewing
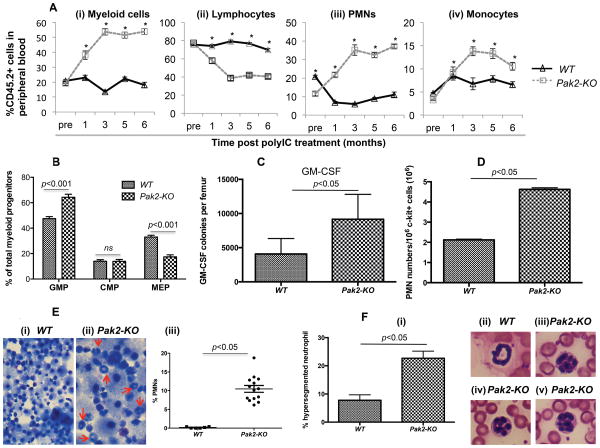
Using eight-parameter flow cytometry (representative flow gating shown in Supplementary Figure S.4), we evaluated multi-lineage reconstitution in each recipient mouse from both the competitive and the non-competitive transplantation cohorts. In the competitive transplantation cohort, starting at 3 months post Pak2 deletion, the percentage of CD45.2+ peripheral blood myeloid cells showed a greater than 2.5-fold increase in mice reconstituted with Pak2-KO BM cells (Figure 5A - i). The skewed lineage differentiation was most dramatic in the granulocytic lineage, as demonstrated by greater than 3-fold increase in the percentage of polymorphonuclear neutrophils (PMNs) in these mice (Figure 5A-iii). There was a small but significant increase in the percentage of monocytes (Figure 5A-iv). Accordingly, these mice showed a 30-40% decrease in the percentage of lymphoid cells (Figure 5A-ii). We observed the lineage skewing as early as one month following deletion of Pak2 in vivo. Despite the dramatic increase in the percentage of myeloid cells, the severe leukopenia in Pak2-KO BM transplanted mice resulted in significantly lower absolute myeloid counts in peripheral blood, as compared to mice transplanted with WT cells (Supplementary Figure S.5A-C).
Figure 5. Pak2 disruption induces myeloid skewing.
(A) The percentage of peripheral blood CD45.2+ myeloid cells (B220−CD3−), lymphocytes (combining CD3+ and B220+ gates), PMNs (B220−CD3−Mac-1highLy6Ghigh) and monocytes (B220−CD3−Mac-1+Ly6Glow) is illustrated in panel (i), (ii), (iii) and (iv), respectively. N=6-10 mice/group. Student t test, * indicates p<0.001 between Pak2-KO and WT. (B) The percentage of granulocyte-monocyte progenitors (GMP: Lin−Sca1−c-Kit+CD34+FcγRII/III+), megakaryocyte-erythroid progenitors (MEP: Lin−Sca1−c-Kit+CD34TcγRII/III−) and common myeloid progenitors (CMP: Lin−Sca1−c-Kit+CD34+FcγRII/III−/low) within the myeloid progenitor (MP: Lin−Sca1−c-Kit+) population. N=5-6 mice/group. Two way ANOVA, p<0.001. (C) Colony formation of sorted CD45.2+ BM LDMNCs in the presence of GM-CSF. N=6-10 mice/group. Student t test, p<0.05 between Pak2-KO and WT. (D) 106 lentiviral vector transduced Pak2flox/flox c-kit+ cells were cultured in murine GM-CSF for 7 days prior to Mac-1 and Ly6G staining. Absolute number of PMNs (Mac-1highLy6Ghigh) is shown. Student t test, p<0.05. (E) cytospin preparations of the progenies from the multi-cytokine stimulated colony assays shown in figure 4C. Arrows indicate PMNs. Panel (i) and (ii) illustrate representative images under 100× magnification; panel (iii) shows the percentage of PMNs. Total of 1093 and 837 progenies were counted from WT and Pak2-KO colonies, respectively. N=6. Student t test, p<0.05 (F) Peripheral blood smears from mice transplanted with Pak2-KO ad WT BM (magnification 100×). (i) Percentage of hypersegmented neutrophils (n=3 mice/group), (ii) A normal neutrophil with a typical doughnut-shaped nucleus, (iii) to (v) Representative Pak2-KO neutrophils with hypersegmented nucleus. Representative data from at least 2 experiments are shown.
Approximately six-fold higher numbers of CD45.2+ PMNs were observed in the spleens of mice transplanted with Pak2-KO BM than in those reconstituted with WT cells (Supplementary Figure S.5D). These data indirectly suggest that 1) granulocytopenia in peripheral blood is not because Pak2-KO PMNs are not capable of migrating out of the BM; 2) there may be a defect in Pak2-KO PMN circulation.
To directly assess the consequence of Pak2 disruption on myeloid differentiation, we performed several experiments. First, we evaluated the composition of myeloid progenitor cells in BM phenotypically using flow cytometry. As shown in figure 5B, mice reconstituted with Pak2-KO BM cells showed a 17% increase in the percentage of phenotypic granulocyte-monocyte progenitors (GMP: Lin−Sca1−c-Kit+CD34+FcγRII/III+) and a 16% decrease in the percentage of phenotypic megakaryocyte-erythroid progenitors (MEP: Lin−Sca1−c-Kit+CD34−FcγRII/III−) within the myeloid progenitor (MP: Lin−Sca1−c-Kit+) population. WT and Pak2-KO marrow reconstituted mice showed equivalent percentage of common myeloid progenitors (CMP: Lin−Sca1−c-Kit+CD34+FcγRII/III−/low).
We then performed granulocyte-macrophage colony forming unit assays by culturing CD45.2+ BM LDMNCs cells in agar culture medium in the presence of GM-CSF only. Pak2-KO BM cells contained greater than 2-fold more granulocyte-macrophage progenitors than WT cells both as a frequency (Supplementary Figure S.6A) and in total numbers per femur (Figure 5C). In contrast, when cultured in the presence of G-CSF, SCF or IL-3 alone, Pak2-KO BM cells yielded significantly lower numbers of colonies than WT control (Supplementary Figure S.6B-D). There was a trend towards fewer M-CSF colonies in Pak2-KO than in WT BM (Supplementary Figure S.6E). To further confirm that loss of Pak2 induces granulocyte differentiation, lentiviral vector transduced GFP+Pak2flox/flox c-kit+ cells were maintained in liquid culture in the presence of GM-CSF. Seven days later, cells were stained for CD11b and Ly6G. Cells transduced with LVCre-eGFP yielded a significantly higher frequency (Supplementary Figure S.7) and total number (Figure 5D) of PMNs than those transduced with LVeGFP. Cytospin preparations of the progenies from the multi-cytokine stimulated colony assays shown in Figure 4C were subjected to Wright-Giemsa staining and morphological evaluation. We observed a significantly higher percentage of PMNs in the Pak2-KO progenies (Figure 5E-i to iii). In summary, we have provided several lines of evidence that Pak2 disruption in HSCs induces myeloid skewing, particularly increased granulocytopoiesis. Interestingly, higher frequency of neutrophil nuclear hypersegmentation (greater than 5 lobes) was observed in the peripheral blood from mice transplanted with Pak2-KO BM, suggesting granulocyte dysplasia (Figure 5F-i to v).
Altered myeloid transcription factor gene expression in Pak2-KO HSCs and HPCs
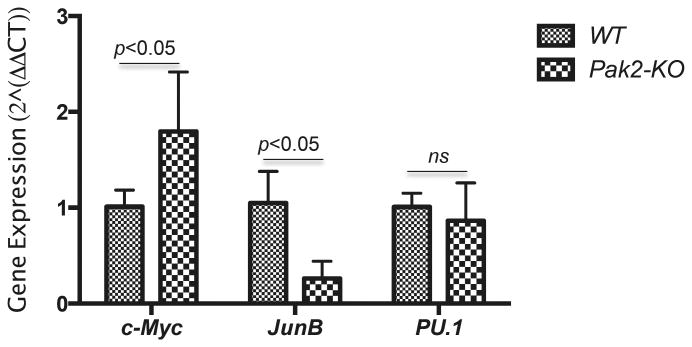
To begin to understand possible underlying mechanisms by which Pak2 regulates myeloid lineage differentiation, we examined gene expression levels of several transcription factors that are known to regulate myelopoiesis. Pak2 disruption led respectively to an approximately 3-fold decrease and 2-fold increase in JunB and Myc expression levels in Lin−kit+ cells, a population of cells that are enriched for HSCs and HPCs (Figure 6). There was no difference in PU.1 expression between the two groups.
Figure 6. Altered myeloid transcription factor gene expression in Pak2-KO HSCs and HPCs.
Seven months post polylC injection, CD45.2+Lin−kit+ cells were sorted from LDMNCs from CD45.1+ BoyJ mice transplanted with CD45.2+Pak2flox/floxMx1Cre+ or Pak2flox/floxMx1Cre− marrow, their JunB, c-Myc, PU.1 expression was determined by qRT-PCR. N=3-4 mice/group. Representative data from 2 experiments are shown. Two-way ANOVA.
Loss of Pak2 leads to deficient lymphopoiesis
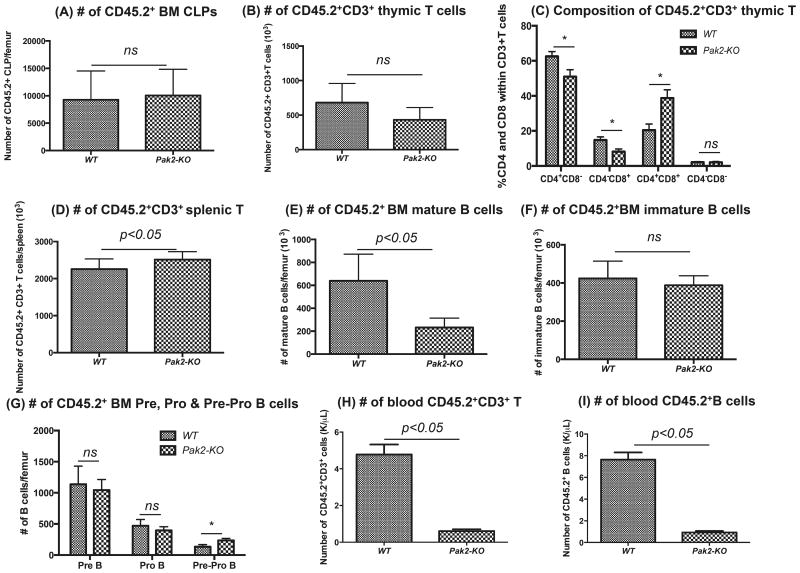
One may argue that the observed myeloid skewing is simply due to more profound defect in lymphoipoiesis. We next evaluated the lymphoid lineages in the BM, thymus, spleen and blood from mice transplanted with Pak2-KO BM. We found no differences in the numbers of phenotypic common lymphoid progenitors (CLPs: Lin−IL7Rα+c-KitlowSca1low) between WT and Pak2-KO BM (Figure 7A). Mice transplanted with Pak2-KO BM had comparable numbers of CD45.2+CD3+ T cells in the thymus (Figure 7B). Interestingly, within the CD45.2+CD3+ thymic T cell population, there was significantly higher percentage of CD4+CD8+ double positive T cells and lower percentages of CD4+CD8− or CD4−CD8+ single positive T cells in Pak2-KO mice when compared to WT controls, suggesting a possible differentiation/maturation defect in Pak2-KO T cells in thymus (Figure 7C). No difference in the numbers of splenic CD45.2+CD3+ T cells was found (Figure 7D). Loss of Pak2 led to significantly lower numbers of mature B cells and higher numbers of Pre-Pro B cells in BM, respectively. There was no difference in the numbers of immature B cells, Pre B cells and Pro B cells in the BM between Pak2-KO and WT groups (Figure 7E-G, representative flow gating shown in Supplementary Figure S.8A). These data suggest a possible differentiation/maturation defect in Pak2-KO B cells in BM. The numbers of splenic mature B, immature B, marginal zone B and follicular B cells were lower in mice with Pak2-KO BM when compared to WT controls (Supplementary Figure S.8B-C), indicating a possible survival and/or proliferation defect in Pak2-KO B cells in spleen. In peripheral blood, mice with Pak2-KO BM had significantly lower numbers of CD3+ T cells and B cells than WT controls (Figure 7H-I), consistent with severe leukopenia in these mice.
Figure 7. Loss of Pak2 leads to deficient lymphopoiesis.
A) Absolute number of CD45.2+ common lymphoid progenitors (CLPs: Lin−IL7Ra+c-KitlowSca1low), (B) number of CD45.2+CD3+ T cells in thymus, (C) percentage of CD4+CD8+, CD4+CD8−, CD4−CD8+ and CD4−CD8− cells within the CD45.2+CD3+ thymic T cell population, (D) number of CD45.2+CD3+ T cells in spleen, (E) numbers of CD45.2+ BM mature B cells (B220+lgM+lgD+), (F) numbers of CD45.2+ BM immature B cells (B220+lgM+lgD−), (G) numbers of CD45.2+ BM Pre B (B220+lgM−lgD−CD43−CD24high), Pro B (B220+lgM−lgD−CD43+CD24med) and Pre-Pro B cells (B220+lgM−lgD−CD43+CD24−), (H) number of peripheral blood CD45.2+CD3+ T cells and (I) number of peripheral blood CD45.2+B220+ B cells was determined by flow cytometry. Absolute number of each cell population was obtained by multiplying the percentage with cellularity of thymus or spleen or WBC count. N=4-6 mice/group. Student t test, * indicates p<0.05 between Pak2-KO and WT. Representative data from 2-3 experiments.
Discussion
Here, utilizing a Pak2flox/flox mouse model, we assessed the functional consequence of HSC-autonomous Pak2 disruption in vivo. Transplanted Pak2-disrupted HSCs produced profound peripheral leukopenia, mild macrocytic anemia, myeloid skewing and T and B cell differentiation/maturation defects. Although loss of Pak2 in HSCs leads to less efficient short- and long-term competitive hematopoiesis than wild type (WT) cells, it does not affect HSC self-renewal per se. Pak2-disrupted BM maintains its cellularity and phenotypic HSCs. However, Pak2 disruption diminished numbers of phenotypically-defined HPCs (Figure 4A-B) and multi-cytokine stimulated immature progenitors (Figure 4C) in BM. Our data suggest that Pak2 regulates HPC function. As Pak2 is a major downstream effector of the Rho GTPases Cdc42 and Rac, our finding agrees with the reports that Rac GTPases regulate HSC and HPC proliferation, survival, and apoptosis.32-34
Our finding that Pak2 disruption leads to severe leukopenia in mice (Figure 3A-B) is consistent with decreased numbers of HPCs (Figure 4A-B) and multi-cytokine stimulated progenitors in Pak2-KO BM (Figure 4C), which may be explained by impaired survival (Figure 1A, S.6D) and proliferation of Pak2-KO HPCs (Figure 4D). When specific progenitors were examined, we found that Pak2 disruption selectively increases GM-CSF colony forming progenitors (Figure 5C-D) while decreasing G-CSF, M-CSF and IL-3 stimulated progenitors in BM (Supplementary Figure S6). This data clearly demonstrates Pak2's selective regulation of GM-CSF responding colony forming progenitors. Since Pak1, a structurally highly homologous isoform of Pak2, is known to regulate Ras signaling35, which mediates GM-CSF signaling transduction36, it is possible that Pak2 may modulate GM-CSF signaling. Work is ongoing to examine how loss of Pak2 alters GM-CSF signaling pathway in HPCs.
Pak2's apparent negative regulation of granulocyte and monocyte lineage commitment is intriguing. Myeloid differentiation requires coordination of multiple transcription factors exerting their functions at different levels of the HSC and HPC hierarchy.37 JunB, a component of the Activator Protein-1 (AP-1) transcription factor, is a transcriptional regulator of myelopoiesis.38,39 Loss of JunB in myeloid lineage leads to increased numbers of granulocyte progenitors and ultimately myeloproliferative disease in mice.38,39 Our findings that Pak2 disruption in HSCs leads to increased percentages of phenotypically defined GMP (Figure 5B) and increased responsiveness to GM-CSF (Figure 5C-D) partially overlaps with the phenotype in mice with JunB deficient myeloid lineage. The fact that Pak2 disruption results in down-regulation of JunB suggests that Pak2 may regulate myelopoiesis through JunB. Indeed, Pak1, an isoform of Pak2, regulates gene transcription through interaction with c-Jun40, another component of AP-1.
Johnasen et al reported that c-Myc must be negatively regulated for myeloblasts to differentiate into neutrophils,41 suggesting an essential role of c-Myc in myelopoiesis and granulocytopoiesis. Furthermore, Pak1 knockdown in human colorectal cancer cell lines inhibits the expression of c-Myc,42,43 indicating that Pak1 may positively regulate c-Myc expression. Considering that Pak2 negatively regulates myelopoiesis in our study (Figure 5), we expected a decrease in c-Myc expression in Pak2-KO HSCs and HPCs. In contrast, we observed a near 2-fold increase in c-Myc expression in these cells (Figure 6). Although required for the formation of the earliest myeloid transcriptional network, 44,45 PU.1 expression is not altered in Pak2-KO HSCs and HPCs (Figure 6). These data suggest that Pak2 regulates later stage of myelopoiesis downstream of CMP, consistent with the finding that Pak2 disruption does not affect phenotypic CMPs in BM (Figure 5B). Together, our data indicate that Pak2 may regulate myelopoiesis possibly through its regulation of multiple transcription factors.
In addition to myeloid skewing, mice transplanted with Pak2-KO BM also develop hypersegmented neutrophils, sharing some of the phenotype found in mice with STAT3 deficient hematopoietic cells.46 Given that STAT3 directly activates Rac1 47, as a downstream target of Rac1, Pak2 may be regulated by STAT3.
In addition to the direct effects of Pak2 on HPC granulocyte lineage commitment (Figure 5B-F), defect in lymphopoiesis may further contribute to peripheral blood myeloid lineage skewing in mice transplanted with Pak2-KO BM. Increased percentage of CD4+CD8+ T cells but decreased percentages of CD4 or CD8 single positive cells in thymus suggest that Pak2-KO T cells may have differentiation/maturation defect in thymus (Figure 7B-C). Lower numbers of mature B cells and higher numbers of Pre-Pro B cells in BM suggests a possible B cell differentiation/maturation defect in BM (Figure 7E-G). Lower numbers of splenic mature B, immature B, marginal zone B and follicular B cells indicates a possible survival and/or proliferation defect in Pak2-KO B cells in spleen. (Figure S.8B-C). Furthermore, it is also possible that loss of Pak2 reduces the survival and proliferation of terminally differentiated mature T and B cells in blood as evidenced by reduced numbers of peripheral blood T and B cells (Figure 7H-I). Considering the role of group I Paks in cytoskeleton dynamics and cell-cell interaction, defect in T and B cell differentiation/maturation may be the result of deficient interaction between Pak2-KO T and B precursors with other cells in the thymus and BM microenvironment.
A number of Pak inhibitors are under development as novel agents particularly in hormone dependent solid cancers.48-50 However, few studies have addressed the implication of Paks regulation of normal and leukemic HPC functions. Our findings that Pak2 disruption in HSCs induces severe leukopenia, granulocyte-monocyte skewing and T and B cell differentiation/maturation defects may have potential implications in targeting Pak2 to treat both hematological and non-hematological malignancies. Furthermore, our observation indicates that the potential hematological side effects following Pak2 inhibition may warrant consideration.
Conclusion
Pak2 displays different functions at different points in hematopoiesis. Pak2 negatively regulates granulocyte-monocyte lineage commitment while positively regulating survival and proliferation of HPCs that are not committed to granulocyte-monocyte lineage. Furthermore, Pak2 may act at other aspects of hematopoiesis including PMN segmentation and circulation, as well as T and B cell differentiation/maturation in thymus and BM.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Emmanuel Katsanis and Dr. Mary Dinauer for helpful discussions, Michal Jander for help maintaining mouse colonies, Audrey McGuire for assisting the histological analysis, Shi Chen, Jessica Stokes and Emely Hoffman for technical assistance and Heather Daniel, Kim Carpenter and Vanessa Frisinger for administrative assistance.
Grant support: Y.Z. was supported by Morris Green Scholarship, American Cancer Society IRG-74-001-35 and a Lymphoma SPORE Career Development Award; D.W.C. was supported by National Institutes of Health grant R01 CA074177-15; J.C. was supported by R01 CA142928 and R01 CA148805; K.S. was supported by a predoctoral HHMI fellowship and a fellowship from National Institutes of Health Grant T32 CA111198; H.E.B. was supported by R01 HL56416, R01 HL67384, and HL112669; H.E.B. and E.F.S. were supported by P01 DK090948. This work was supported in part by the Indiana Center for Excellence in Molecular Hematology (NIDDK P30 DK090948). The Flow Cytometry Research Facility is partially funded by NCI P30 CA082709.
Footnotes
Author contributions: Yi Zeng: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing and final approval of manuscript.
Hal E. Broxmeyer: Conception and design, collection and assembly of data, data analysis and interpretation and manuscript editing.
Karl Staser: Collection and assembly of data, data analysis and interpretation and manuscript editing.
Brahmananda Reddy Chitteti, Su-Jung Park, Seongmin Hahn, Scott Cooper, Zejin Sun, Li Jiang, XianLin Yang, Jin Yuan and Rachelle Kosoff: Collection and assembly of data.
Edward F. Srour: Data analysis and interpretation, manuscript editing.
George Sandusky: Collection and assembly of data, data interpretation.
Jonathan Chernoff: Provision of study material and manuscript editing.
Wade Clapp: Conception and design, data interpretation, manuscript editing and final approval of manuscript.
Conflict-of-interest disclosure: The authors have no conflicts of interest to disclose.
References
- 1.Ong CC, Jubb AM, Zhou W, et al. p21-activated kinase 1: PAK'ed with potential. Oncotarget. 2011;2(6):491–496. doi: 10.18632/oncotarget.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28(28):2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28(1-2):51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radu M, Semenova G, Kosoff R, Chernoff J. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14(1):13–25. doi: 10.1038/nrc3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6(6):459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 6.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 7.Coniglio SJ, Zavarella S, Symons MH. Pak1 and Pak2 mediate tumor cell invasion through distinct signaling mechanisms. Mol Cell Biol. 2008;28(12):4162–4172. doi: 10.1128/MCB.01532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bright MD, Garner AP, Ridley AJ. PAK1 and PAK2 have different roles in HGF-induced morphological responses. Cell Signal. 2009;21(12):1738–1747. doi: 10.1016/j.cellsig.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 9.Siu MK, Wong ES, Chan HY, et al. Differential expression and phosphorylation of Pak1 and Pak2 in ovarian cancer: effects on prognosis and cell invasion. Int J Cancer. 2010;127(1):21–31. doi: 10.1002/ijc.25005. [DOI] [PubMed] [Google Scholar]
- 10.Kosoff R, Chow HY, Radu M, Chernoff J. Pak2 kinase restrains mast cell FcepsilonRI receptor signaling through modulation of Rho protein guanine nucleotide exchange factor (GEF) activity. J Biol Chem. 2013;288(2):974–983. doi: 10.1074/jbc.M112.422295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Liu F, Li F. PAK as a therapeutic target in gastric cancer. Expert Opin Ther Targets. 2010;14(4):419–433. doi: 10.1517/14728221003642019. [DOI] [PubMed] [Google Scholar]
- 12.Carter JH, Douglass LE, Deddens JA, et al. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10(10):3448–3456. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 13.Mao X, Onadim Z, Price EA, et al. Genomic alterations in blastic natural killer/extranodal natural killer-like T cell lymphoma with cutaneous involvement. J Invest Dermatol. 2003;121(3):618–627. doi: 10.1046/j.1523-1747.2003.12406.x. [DOI] [PubMed] [Google Scholar]
- 14.Allen JD, Jaffer ZM, Park SJ, et al. p21-activated kinase regulates mast cell degranulation via effects on calcium mobilization and cytoskeletal dynamics. Blood. 2009;113(12):2695–2705. doi: 10.1182/blood-2008-06-160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorrance AM, De Vita S, Radu M, et al. The Rac GTPase effector p21-activated kinase is essential for hematopoietic stem/progenitor cell migration and engraftment. Blood. 2013;121(13):2474–2482. doi: 10.1182/blood-2012-10-460709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YY, Vik TA, Ryder JW, et al. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J Exp Med. 1998;187(11):1893–1902. doi: 10.1084/jem.187.11.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Si Y, Pulliam AC, Linka Y, et al. Overnight transduction with foamyviral vectors restores the long-term repopulating activity of Fancc-/- stem cells. Blood. 2008;112(12):4458–4465. doi: 10.1182/blood-2007-07-102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DE, Hangoc G, Cooper S, et al. The effects of purified recombinant murine interleukin-3 and/or purified natural murine CSF-1 in vivo on the proliferation of murine high- and low-proliferative potential colony-forming cells: demonstration of in vivo synergism. Blood. 1987;70(2):401–403. [PubMed] [Google Scholar]
- 19.Staser K, Park SJ, Rhodes SD, et al. Normal hematopoiesis and neurofibromin-deficient myeloproliferative disease require Erk. J Clin Invest. 2013;123(1):329–334. doi: 10.1172/JCI66167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broxmeyer HE, Hoggatt J, O'Leary HA, et al. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nat Med. 2012;18(12):1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broxmeyer HE, Orschell CM, Clapp DW, et al. Rapid mobilization of murine and human hematopoietic stem and progenitor cells with AMD3100, a CXCR4 antagonist. J Exp Med. 2005;201(8):1307–1318. doi: 10.1084/jem.20041385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broxmeyer HE, Williams DE, Cooper S, et al. Comparative effects in vivo of recombinant murine interleukin 3, natural murine colony-stimulating factor-1, and recombinant murine granulocyte-macrophage colony-stimulating factor on myelopoiesis in mice. J Clin Invest. 1987;79(3):721–730. doi: 10.1172/JCI112877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SJ, Beck BD, Saadatzadeh MR, Haneline LS, Clapp DW, Lee SH. Fanconi anemia D2 protein is an apoptotic target mediated by caspases. J Cell Biochem. 2011;112(9):2383–2391. doi: 10.1002/jcb.23161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jetmore A, Plett PA, Tong X, et al. Homing efficiency, cell cycle kinetics, and survival of quiescent and cycling human CD34(+) cells transplanted into conditioned NOD/SCID recipients. Blood. 2002;99(5):1585–1593. doi: 10.1182/blood.v99.5.1585. [DOI] [PubMed] [Google Scholar]
- 25.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 26.Yabas M, Teh CE, Frankenreiter S, et al. ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat Immunol. 2011;12(5):441–449. doi: 10.1038/ni.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Nheu T, He H, Hirokawa Y, Walker F, Wood J, Maruta H. PAK is essential for RAS-induced upregulation of cyclin D1 during the G1 to S transition. Cell Cycle. 2004;3(1):71–74. [PubMed] [Google Scholar]
- 29.Tong J, Gordon MS, Srour EF, et al. In vivo administration of recombinant methionyl human stem cell factor expands the number of human marrow hematopoietic stem cells. Blood. 1993;82(3):784–791. [PubMed] [Google Scholar]
- 30.Hoffman R, Tong J, Brandt J, et al. The in vitro and in vivo effects of stem cell factor on human hematopoiesis. Stem Cells. 1993;11(Suppl 2):76–82. doi: 10.1002/stem.5530110813. [DOI] [PubMed] [Google Scholar]
- 31.Waterstrat A, Liang Y, Swiderski CF, Shelton BJ, Van Zant G. Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood. 2010;115(2):408–417. doi: 10.1182/blood-2008-03-143370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancelas JA. On how Rac controls hematopoietic stem cell activity. Transfusion. 2011;51(Suppl 4):153S–159S. doi: 10.1111/j.1537-2995.2011.03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang X, Cancelas JA, Li L, et al. R-Ras and Rac GTPase cross-talk regulates hematopoietic progenitor cell migration, homing, and mobilization. J Biol Chem. 2011;286(27):24068–24078. doi: 10.1074/jbc.M111.226951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengupta A, Arnett J, Dunn S, Williams DA, Cancelas JA. Rac2 GTPase deficiency depletes BCR-ABL+ leukemic stem cells and progenitors in vivo. Blood. 2010;116(1):81–84. doi: 10.1182/blood-2009-10-247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel AS, Allen JD, Park SJ, et al. Pak1 regulates multiple c-Kit mediated Ras-MAPK gain-in-function phenotypes in Nf1+/− mast cells. Blood. 2008;112(12):4646–4654. doi: 10.1182/blood-2008-04-155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emanuel PD. Juvenile myelomonocytic leukemia and chronic myelomonocytic leukemia. Leukemia. 2008;22(7):1335–1342. doi: 10.1038/leu.2008.162. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 38.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119(3):431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell. 2001;104(1):21–32. doi: 10.1016/s0092-8674(01)00188-x. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Solana B, Motwani M, Li DQ, Eswaran J, Kumar R. p21-activated kinase-1 signaling regulates transcription of tissue factor and tissue factor pathway inhibitor. J Biol Chem. 2012;287(47):39291–39302. doi: 10.1074/jbc.M112.404061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansen LM, Iwama A, Lodie TA, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol. 2001;21(11):3789–3806. doi: 10.1128/MCB.21.11.3789-3806.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He H, Huynh N, Liu KH, et al. P-21 activated kinase 1 knockdown inhibits beta-catenin signalling and blocks colorectal cancer growth. Cancer Lett. 2012;317(1):65–71. doi: 10.1016/j.canlet.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 43.He H, Shulkes A, Baldwin GS. PAK1 interacts with beta-catenin and is required for the regulation of the beta-catenin signalling pathway by gastrins. Biochim Biophys Acta. 2008;1783(10):1943–1954. doi: 10.1016/j.bbamcr.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 45.Scott EW, Fisher RC, Olson MC, Kehrli EW, Simon MC, Singh H. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6(4):437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 46.Mantel C, Messina-Graham S, Moh A, et al. Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood. 2012 doi: 10.1182/blood-2012-01-404004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran BH, Guan KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290(5489):144–147. doi: 10.1126/science.290.5489.144. [DOI] [PubMed] [Google Scholar]
- 48.Porchia LM, Guerra M, Wang YC, et al. 2-amino-N-{4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]-phenyl} acetamide (OSU-03012), a celecoxib derivative, directly targets p21-activated kinase. Mol Pharmacol. 2007;72(5):1124–1131. doi: 10.1124/mol.107.037556. [DOI] [PubMed] [Google Scholar]
- 49.Nheu TV, He H, Hirokawa Y, et al. The K252a derivatives, inhibitors for the PAK/MLK kinase family selectively block the growth of RAS transformants. Cancer J. 2002;8(4):328–336. doi: 10.1097/00130404-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 50.Deacon SW, Beeser A, Fukui JA, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15(4):322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.