Abstract
Antiretroviral therapy (ART) potently suppresses HIV-1 replication, but the virus persists in quiescent infected CD4+T cells as a latent integrated provirus, and patients must indefinitely remain on therapy. If ART is terminated, these integrated proviruses can reactivate, driving new rounds of infection. A functional cure for HIV requires eliminating low-level ongoing viral replication that persists in certain tissue sanctuaries and preventing viral reactivation. The HIV Tat protein plays an essential role in HIV transcription by recruiting the kinase activity of the P-TEFb complex to the viral mRNA’s stem–bulge–loop structure, TAR, activating transcriptional elongation. Because the Tat-mediated transactivation cascade is critical for robust HIV replication, the Tat/TAR/P-TEFb complex is one of the most attractive targets for drug development. Importantly, compounds that interfere with transcription could impair viral reactivation, low-level ongoing replication, and replenishment of the latent reservoir, thereby reducing the size of the latent reservoir pool. Here, we discuss the potential importance of transcriptional inhibitors in the treatment of latent HIV-1 disease and review recent findings on targeting Tat, TAR, and P-TEFb individually or as part of a complex. Finally, we discuss the impact of extracellular Tat in HIV-associated neurocognitive disorders and cancers.
1 Introduction
Antiretroviral therapy (ART) potently suppresses replication of human immunodeficiency virus (HIV) driving viral loads to undetectable levels (<50 copies/ml), but fails to permanently eradicate the virus (Chun et al. 1997; Finzi et al. 1997; Wong et al. 1997). Unfortunately, HIV still persists mostly in latently infected memory CD4+T cells in individuals on suppressive ART, and these cells represent a long-lasting source of resurgent virus upon the interruption of ART (Finzi et al. 1999). The long half-life of infected memory CD4+T cells is partly responsible for the lifelong persistence of HIV (Finzi et al. 1999; Siliciano et al. 2003). In addition to latently infected cells, persistence can also be attributed to ongoing low levels of viral replication in infected subjects on ART (Fletcher et al. 2014; Palmer et al. 2008). Cell-associated viral RNA can be detected in gut and lymph nodes, suggesting continuous viral production in these compartments during ART and these anatomical reservoirs may constitute viral sanctuaries (Yukl et al. 2010).
As current anti-HIV drugs do not inhibit transcription from integrated viral genomes and do not prevent viral particle release from stable cellular reservoirs, novel classes of antiretrovirals (ARVs) are needed to inhibit these processes. An ideal drug candidate should be able to inhibit viral production from integrated viral genomes and permanently silence HIV transcription.
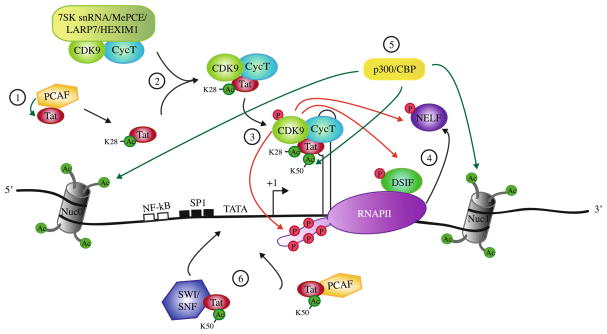
In newly infected cells, cellular transcription factors such as NF-κB initiate HIV basal transcription at the 5′ long-terminal repeat (LTR) but result in short, abortive viral transcripts due to RNA polymerase II (RNAPII) pausing shortly after promoter clearance (Toohey and Jones 1989). An RNA stem–loop structure called transactivation response element (TAR) spontaneously forms within the first 59 nucleotides of each viral transcript. The viral protein Tat, a 101 amino acid protein, is initially expressed from rare full-length transcripts that are multiply spliced. After acetylation of Tat at lysine 28 by the p300/CBP-associated factor (PCAF), Tat recruits the positive transcription elongation factor b (P-TEFb) [composed of cyclin T1 and cyclin-dependent kinase 9 (CDK9)] from a large inactive complex composed of 7SK snRNA, the methylphosphate capping enzyme, MePCE, the La-related protein, LARP7, and HEXIM1 proteins (Fig. 1) (Barboric et al. 2007; Krueger et al. 2008; Sedore et al. 2007). Tat binds to P-TEFb, and the complex binds the TAR RNA (D’Orso and Frankel 2010). Tat binds to TAR by a specific arginine-rich basic domain between residues 49 and 57. Once in close proximity to the pre-initiation complex, autophosphorylated CDK9 (Garber et al. 2000) phosphorylates negative elongation factors DSIF and NELF, converting DSIF into a positive elongation factor and causing NELF to release from the complex. In addition, CDK9 phosphorylates serine 2 of the RNAPII C-terminal domain (CTD) heptapeptide repeat, allowing the interaction of RNAPII with additional factors involved in productive transcription elongation (Fig. 1) [Reviewed in (Ott et al. 2011)]. Tat is released from TAR and P-TEFb after being acetylated at lysine 50 by p300/CBP and hGCN5. Freed Tat can then recruit factors such as PCAF and SWI/SNF leading to further chromatin remodeling enhancing HIV transcription elongation. Studies based on chromatin immunoprecipitation and fluorescence recovery after photobleaching suggested that Tat and P-TEFb could stay on the elongating RNAPII throughout the transcription of the entire HIV gene and could undergo several cycles of association/dissociation during the elongation process (Bres et al. 2005; Molle et al. 2007). The elongation complex is then converted into a highly processive unit and promotes the synthesis of full-length viral transcripts by more than 100-fold (Cullen 1986).
Fig. 1.
HIV-1 transcription elongation. 1 Upon Tat acetylation on Lys28 by PCAF, 2 Tat recruits P-TEFb (CDK9/cyclin T1) from a large inactive complex with 7SK snRNA/MePCE/LARP7/HEXIM1. 3 Tat/P-TEFb complex binds to TAR. CDK9 phosphorylates Ser2 of the RNAPII CTD, stalled shortly after transcription initiation. 4 CDK9 phosphorylates the negative elongation factor NELF, which is released from RNAPII, and DSIF that becomes a positive transcription elongation factor. 5 Tat is acetylated at Lys50 by p300/CBP, resulting in the release of the Tat/P-TEFb complex from TAR. p300/CBP acetylates Nuc0 and Nuc1 allowing for chromatin remodeling. 6 Tat recruits to the initiation start site SWI/SNF, PCAF, and additional factors not depicted here to further promote transcription. Green arrow acetylation; red arrow phosphorylation
During ART, HIV replication in a subset of memory CD4+T cells is progressively silenced and the HIV-1 provirus is maintained in a latent transcriptional state by a multitude of molecular mechanisms. These include low levels of Tat or P-TEFb, absence of cellular transcription factors NF-κB and NFAT, presence of repressors such as CBF-1 and YY1 as well as transcriptional interference [reviewed in (Margolis 2010; Mbonye and Karn 2011)]. Entry into latency is also regulated by specific epigenetic chromatin modifications of nucleosomes present at the HIV promoter, notably, deacetylation and methylation of histone N-terminal tails by specific enzymes such as histone deacetylase 1 (HDAC1) and Suv39H1 (Margolis 2010; Mbonye and Karn 2011). During transcriptional reactivation, Tat recruits chromatin remodeling factors, such as SWI/SNF, that are responsible for changes in the local chromatin structure (Easley et al. 2010; Mahmoudi 2012) and histone acetyl transferases (HAT) such as p300/CBP, which can reverse the effects of histone deacetylation (Fig. 1) (Benkirane et al. 1998; Lusic et al. 2003).
2 HIV Transcription Inhibitors and Deep-Latency
Tat is an attractive target for therapeutic intervention because it has no cellular homologs and it is expressed early in the viral life cycle. Direct inhibition of Tat blocks the feedback loop that drives exponential production of viral progeny. An ideal anti-HIV small-molecule candidate should have a drug-like structure, be soluble in physiologic conditions, inexpensive, used at low posology and able to penetrate sanctuary sites such as gut, lymph nodes and the brain. In the specific case of a transcription inhibitor, it is essential that the compound inhibits only Tat-dependent transcription without affecting cellular transcription. This can be accomplished by directly targeting Tat/TAR interaction or the recruitment of P-TEFb by Tat to the transcriptionally paused RNAPII. A specific Tat-dependent transcription inhibitor should not disrupt HIV basal transcription, which is promoted by cellular transcription factors shared with cellular genes (e.g., NF-κB, Sp1, NFAT, etc) (Cullen 1991; Jeang et al. 1993; Pessler and Cron 2004), and this explains why anti-Tat molecules are unable to fully inhibit acute HIV replication, unlike ARVs targeting other viral proteins (Mousseau et al. 2012). However, in contrast with ARVs that have no activity on viral expression from cells containing an integrated provirus and can only block de novo infection, an HIV transcription inhibitor mediates complete transcriptional inhibition in these cells (Mousseau et al. 2012).
Several strategies have been employed for eradicating the latent HIV reservoir including ART regimen intensification, vaccines, HDAC inhibitors, gene therapy, and stem cell transplantation (Deeks et al. 2012). The most commonly explored strategy focuses on purging the reservoirs using anti-latency agents such as HDAC inhibitors, while preventing novel infections by maintaining ART (Deeks 2012). This approach is based on the premise that reactivation of HIV from latency will kill the infected cells or allow the immune system to eliminate them. However, a clinical trial using the HDAC inhibitor Vorinostat demonstrated that the reactivation of latently infected CD4+T cells did not result in a measurable decrease in the size of the latent reservoir (Archin et al. 2012). Moreover, Siliciano and colleagues showed that the current methods to measure the latent reservoir (viral outgrowth assay) could have underestimated its size by up to 60-fold, and multiple reactivation events would be necessary for the required viral reactivation of every single latently infected cell (Ho et al. 2013). Each eradication approach has its own inherent challenges and none have demonstrated definitive success. The countless failures in obtaining total HIV eradication have brought forth the concept of a “functional cure”—defined by the persistence of HIV genetic material in the body without detectable viral replication in the absence of ART, without loss of CD4+T cells, no clinical progression, and lack of HIV transmission.
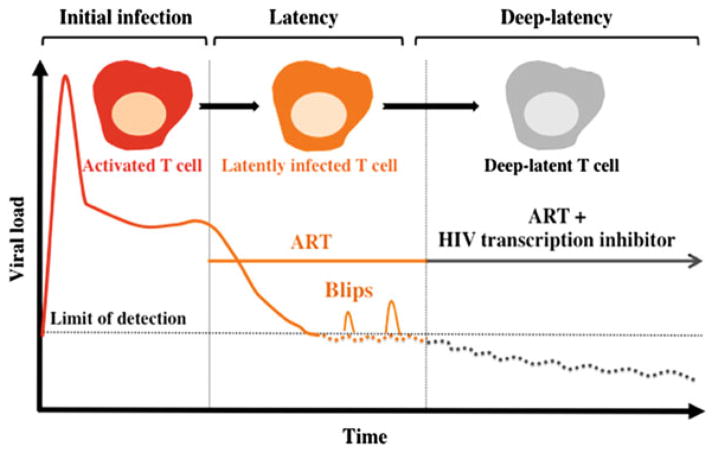
An alternative approach that represents a significant departure from established paradigms of eradicating latent reservoirs involves the use of a therapeutic agents targeting HIV transcription. These could potentially suppress residual levels of viral transcription in latently infected cells, thereby establishing a state of deep-latency refractory to viral reactivation. Specifically, rather than activating the endogenous latent reservoir, one would drive the residual transcription that occurs during ART into long-term deep-latency. Despite the persistence of the HIV genetic material in the body, a transcriptional inhibitor treatment combined with ART would be aimed at reducing the size of the latent reservoir pool by potentially blocking ongoing viral replication, as well as reactivation events or “blips” (Jones and Perelson 2007; Sklar et al. 2002) that replenish the viral latent reservoir, a key limitation of current ART (Fig. 2).
Fig. 2.
Hypothetically, a transcription inhibitor could promote deep-latency and block reactivation. High levels of circulating virus are observed in the blood of infected individuals upon HIV infection. Upon ART initiation, viral mRNA is reduced to below 50 copies/ml and memory CD4+T cells remain latently infected. If ART is halted, there is an immediate rebound of virus production that correlates with CD4+T-cell reactivation. Combining a transcription inhibitor to ART could induce a state of deep-latency that would lead to the reduction in size of the viral reservoir
Transcriptional inhibitors could also reduce morbidities associated with persistent levels of immune activation caused by low-level ongoing replication in subjects on suppressive ART (Hunt 2010). Moreover, by suppressing viral transcription to a state of deep-latency refractory to reactivation, ART could potentially be interrupted without viral rebound to alleviate patient’s side effects.
3 Extracellular Tat
ART curtails HIV replication; however, there is a paradoxical increase in the prevalence of HIV-associated neurocognitive disorders (HAND) (Bagashev and Sawaya 2013), which include a group of syndromes that range from undetectable neurocognitive impairments to severe forms of encephalitis/dementia (Gannon et al. 2011). A commonly accepted explanation for HAND is the inability of various ART regimens to pass through the blood–brain barrier (BBB), thus rendering the brain a viral sanctuary (Bagashev and Sawaya 2013). HIV-1 does not infect neurons; therefore while still debated, neurotoxicity has been postulated to be mediated by extracellular Tat upon its direct passage through the BBB or after secretion from infected glia cells (Li et al. 2009). Tat uptake by glial cells (King et al. 2006; Ma and Nath 1997) or interaction with certain cell receptors on a variety of cell types has been widely reported (Albini et al. 1996, 1998; Debaisieux et al. 2012; Ensoli et al. 1993; Fawell et al. 1994; Rusnati and Presta 2002). A high level of Tat mRNA has been found in the brain of HAND patients (Del Valle et al. 2000; Hofman et al. 1994; Hudson et al. 2000; Li et al. 2009; Wesselingh et al. 1993; Wiley et al. 1996), and anti-Tat antibodies were detected in their serum (Aldovini et al. 1986) or their cerebrospinal fluid (Bachani et al. 2013). Moreover, injection of Tat or stable expression of Tat in brain showed neurological symptoms similar to those associated with HAND (Gorantla et al. 2012; Kim et al. 2003; Li et al. 2009; Weeks et al. 1995; Zucchini et al. 2013).
Tat has been proposed to mediate neurotoxicity by several mechanisms: induction of oxidative stress (Romani et al. 2010), BBB damage (Bagashev and Sawaya 2013; Li et al. 2009; Strazza et al. 2011), up-regulation of genes such as monocyte chemoattractant protein-1 (MCP-1), TNF-α, and metalloprotease 9 (Li et al. 2009; Romani et al. 2010), its chemotaxic function (Li et al. 2009), inhibition of autophagy in macrophages (Van Grol et al. 2010), and activation of N-methyl-D-aspartate receptors (Campbell and Loret 2009; Li et al. 2009). Tat’s basic domain and cysteine-rich region could play an important role in these neurotoxic effects (Li et al. 2009), and patients infected with HIV-1 subtype C, containing a mutation in Tat’s cysteine region, tend to have less prevalence of HAND (Li et al. 2009). Extracellular Tat has also been associated with acquired immune deficiency syndrome (AIDS)-associated cancers such as Kaposi’s sarcoma, often developing more aggressively in HIV-infected patients (Johri et al. 2011; Nunnari et al. 2008; Romani et al. 2010). Tat’s basic domain, by competing with basic fibroblast growth factor for heparin sulfate proteoglycan, may induce spindle cell growth in Kaposi’s sarcoma (Campbell and Loret 2009). Compounds interacting with the TAR-binding domain of Tat [such as dCA, see Sect. 4.2 (Mousseau et al. 2012)] may have the ability to counteract the neurotoxic and cancer-promoting properties of Tat by either reducing its production at the transcription level and by directly inhibiting its pathogenic activity (Mediouni et al. 2015).
Here, we review a selection of new small-molecule inhibitors of the Tat/TAR and the Tat/cyclin T1/CDK9 complexes. For a more exhaustive collection of HIV transcription inhibitors, we suggest the following publications (Baba 2006; Massari et al. 2013; Mousseau and Valente 2012; Richter and Palu 2006; Stevens et al. 2006). When applicable, we will direct the reader to the most recent and appropriate review.
4 Tat/TAR Interaction
Tat recruits P-TEFb complex to the nascent TAR element of the viral RNA to promote transcriptional elongation, greatly enhancing HIV mRNA synthesis. As expected, disrupting Tat/TAR interaction dramatically reduces viral production and hence has been the focus for the development of numerous antiviral compounds (Massari et al. 2013; Mousseau and Valente 2012). However, none has yet reached the clinic, and only one has recently entered the clinical trial pipeline (NCT02219672). The discovery of a safe Tat-dependent transcriptional inhibitor would be of tremendous value, as it may reduce residual viremia from latently infected CD4+T cells and repress HIV reservoir replenishment, the major hurdle in the ongoing race for an HIV cure.
Here, we will focus on recent and specific small-molecule inhibitors targeting TAR or Tat. Other strategies including vaccine, ribozyme, aptamer, oligonucleotide-based antisense and non-small inhibitors such as aminoglycoside and peptide-based structure are reviewed elsewhere (Burnett and Rossi 2012; Campbell and Loret 2009; Eekels and Berkhout 2011; Massari et al. 2013; Mulhbacher et al. 2010; Richter and Palu 2006; Turner et al. 2006; Zeller and Kumar 2011).
4.1 TAR
TAR is a small non-coding hairpin RNA that folds into a well-defined three-dimensional structure. TAR is extremely well conserved among HIV clades and is a very attractive target for drug development. However, specifically targeting TAR with small molecules has proven challenging given the properties common to all RNAs such as negative charge, large flexibility, and reduced diversity of chemical composition. There is no crystal structure of the Tat/TAR complex, only structures of small-molecule ligands or Tat-derived peptides interacting with TAR exist to help guide the design of small-molecule inhibitors (Aboul-ela et al. 1995; Puglisi et al. 1992).
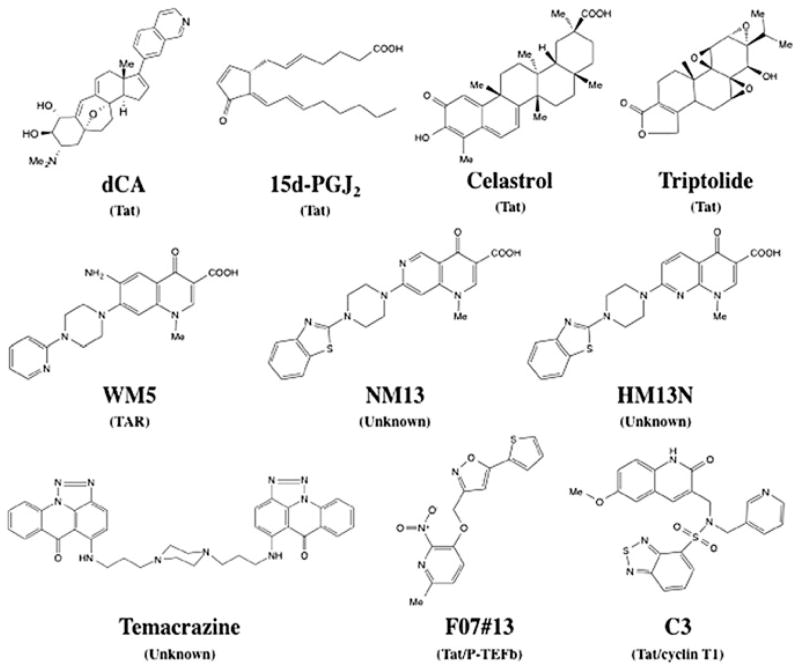
One of the most interesting TAR inhibitors reported to date is WM5 (Fig. 3), a 6-aminoquinolone, able to specifically block Tat/TAR interaction by binding to TAR at the micromolar range (Cecchetti et al. 2000; Parolin et al. 2003; Richter et al. 2004). This compound inhibits Tat-dependent LTR activity and inhibits acute viral replication in peripheral blood mononuclear cells (PBMCs), but with a low therapeutic index (TI) (Tabarrini et al. 2004). Successive structure-activity relationship studies resulted in several WM5 derivatives with anti-HIV properties (Tabarrini et al. 2010). HM13N and NM13 are the best quinolone-based inhibitors of Tat-dependent transcription described so far (Fig. 3) (Massari et al. 2010; Tabarrini et al. 2011). HM13N inhibits both HIV-1 and HIV-2 in MT-4 cells and displays a reasonably good TI in chronically and latently infected cell lines. NM13, is selective only to HIV-1 with an half-maximal inhibitory concentration (IC50) = 80 nM and a TI ≥ 3,707 in MT-4 cells, but has poor solubility (Tabarrini et al. 2011). Contrary to WM5, none of these two compounds bind specifically to the Tat/TAR complex, acting by an unknown mechanism. Two related 6-desfluoroquinolones, HM12 and HM13, inhibit in vivo TNF-α reactivation from latently infected OM-10.1 cells, a promyelocytic cell line, when engrafted in hu-SCID mice (Stevens et al. 2007; Tabarrini et al. 2008). These compounds are currently in development to improve their solubility and reduce cytotoxicity.
Fig. 3.
Structure of selected HIV transcription inhibitors. Brackets target of the compound
Recently, Stelzer et al. (2011) has developed a molecular dynamic software to screen novel TAR-binding small molecules. A library of 51,000 compounds were virtually screened onto a RNA dynamic ensemble of 20 conformers of TAR to help account for large degrees of RNA conformational adaptation during docking. A fluorescence-based binding assay confirmed several of the hits bound to TAR and inhibited binding to Tat. Netilmicin, an aminoglycoside antibiotic, bound TAR with the highest specificity and blocked viral replication in an HIV-1 indicator cell line with an IC50 of 23 μM. Netilmicin might, however, not be specific against HIV, just as previously observed for other aminoglycoside derivatives (Lapidot et al. 2008). Nevertheless, this study demonstrated the usefulness of high-throughput in silico screening to develop new TAR inhibitors.
Davidson et al. (2011) discovered novel non-charged drug-like small molecules targeting TAR using nuclear magnetic resonance (NMR) ligand-based screening. In this study, an arginine derivative was used as a probe to bind to the Tat binding region of TAR (similar to how arginine of the Tat basic domain would bind to TAR), to lock TAR into a specific conformation more favorable to the binding of drug-like compounds. Reorganization of the RNA structure led to the formation of a pocket in the major groove allowing the binding of six compounds. It will be important to validate these hits in HIV cellular models. This study was proof of concept for the use of ligand-based NMR screening to identify new small molecules to target TAR.
4.2 Tat
Given Tat’s crucial requirement for viral gene expression and its role in maintenance or reactivation of latency, the search for Tat inhibitors has been an intense area of research over the years. However, small molecules that inhibit Tat, either by direct interaction, by its degradation or by structurally modifying it, are rare (Kalantari et al. 2009; Mousseau et al. 2012; Narayan et al. 2011; Wan and Chen 2014). This lack of true Tat inhibitors in the literature might be a reflection of the employed screening strategies. For example, screening for Tat-dependent transcription inhibitors using the widely accepted cellular HIV LTR-reporter system may result in the identification of off-target inhibitors, affecting additional host cellular factors important to HIV transcription.
Nevertheless, in two separate studies, Prabhu and co-workers identified two small molecules inhibiting Tat-mediated activation of HIV-1 transcription using an HIV LTR-Luc reporter assay activated by Tat transfection (Kalantari et al. 2009; Narayan et al. 2011). The first compound, the cyclopentenone prostaglandin 15d-PGJ2 was previously reported to inhibit HIV replication (Fig. 3) (Rozera et al. 1996). This compound is an arachidonic acid-derived endogenous Michael acceptor electrophiles (MAEs) presenting an α,β-unsaturated carbonyl functionality (enone) and is capable of forming covalent Michael adducts by interacting with the cysteine sulfhydryls of certain proteins. At 3.1 μM, the compound 15d-PGJ2 reduces Tat-dependent transcription by 80 % and HIV replication by 75 % in acutely HIV-1 infected U937 cells. The second inhibitor discovered, celastrol, a tripernoid MAE, inhibits Tat-mediated transactivation by 80 % using a non-toxic concentration of 150 nM (Fig. 3). But, HIV replication inhibition measured by p24 enzyme-linked immunosorbent assay and quantitative reverse transcription polymerase chain reaction in acutely infected U937 was only 50 %. Celastrol is very toxic, since viability is reduced by 50 % in U937 cells at 250 nM. The relevance of these findings lies in the mechanism by which celastrol and 15d-PGJ2 interact with Tat, forming a covalent bond with the Cys thiols of the viral protein. In the case of celastrol, the secondary structure of Tat was altered even though it did not impair the binding to TAR, suggesting a probable deficient role of Tat in P-TEFb recruitment. Both of these two compounds need to be further optimized to increase their TI.
A bis-triazoloacridone compound, temacrazine, was found to inhibit HIV replication in acute, chronic, and latent cells in the nanomolar range (Fig. 3) (Turpin et al. 1998). The compound was first identified based on its antitranscriptional activity in cancer and appears to be a selective inhibitor of HIV transcription functioning via an unknown mechanism. It was suggested it affects a still unidentified highly specific viral target required for HIV-1 transcription. No additional follow-up on temacrazine has been reported since, suggesting that some undesirable properties prevented its development as an anti-HIV drug.
More recently, a group reported that triptolide, a diterpenoid epoxide isolated from Tripterygium wilfordii, was able to inhibit HIV-1 transcription by accelerating Tat degradation in a specific manner (Fig. 3) (Wan and Chen 2014). While not tested for direct interaction with Tat, the N-terminus of Tat (1–57) and the nuclear localization of Tat seemed to be required for Tat degradation. The reported IC50 in PBMCs averaged 1.3 nM, but the compound was quite toxic with a half-maximal cytotoxic concentration (CC50) around 13 nM limiting its use in its current form. Nonetheless, Triptolide has entered several clinical trials for different targets such as gastrointestinal diseases, rheumatoid arthritis, and HIV. The HIV clinical trial is currently in phase III (NCT02219672) and is performed in combination with a cocktail of ARVs to study its impact on the size of the HIV-1 reservoir, in naïve-ART Chinese patients currently in the acute phase of HIV-1 infection.
Finally, our laboratory has reported that didehydro-cortistatin A (dCA), an analogue of a natural steroidal alkaloid isolated from a marine sponge, inhibits Tat-mediated transactivation of the HIV-integrated provirus by binding specifically to the TAR-binding domain of Tat (Fig. 3) (Mousseau et al. 2012). We demonstrated that dCA inhibits transcription initiation and elongation from the viral promoter in chronically infected cells at subnanomolar concentrations without cell-associated toxicity (IC50 ranges from 1 pM to 2 nM; CC50 = 20 μM). Moreover, long-term treatment of chronically infected cells reduced viral mRNA to undetectable levels and dCA discontinuation does not result in viral rebound suggesting that dCA promotes rapid and prolonged silencing of the HIV promoter. Most importantly, we demonstrated that dCA could abrogate spontaneous as well as antigenic viral particle release from CD4+T cells explanted from virally suppressed subjects on ART.
Hence, by blocking Tat-dependent transcription, a Tat inhibitor such as dCA may reduce reactivation of latently infected cells and block low-ongoing replication, thereby controlling replenishment of the viral reservoir, which upon death of the long-lived latently infected memory CD4+T cells could result in a reduction of the reservoir pool over time.
5 Inhibition of P-TEFb, an Essential Cellular Complex for HIV-1-Activated Transcription
Several host cell factors have essential roles in HIV-1 transcriptional activation and may serve as potential targets for antiviral chemotherapy, as long as cell survival is not impaired [reviewed in (Coley et al. 2009; Massari et al. 2013; Mousseau and Valente 2012; Stevens et al. 2006)]. One advantage to this approach is that unlike current ARVs acting on viral proteins, targeting cellular factors would unlikely lead to viral resistance. In this section, we will focus on recent small-molecule inhibitors of the P-TEFb complex and its kinase activity.
5.1 CDK9
Finding a highly selective and non-cytotoxic CDK9 inhibitor is a difficult task due to its dual role in cellular and HIV transcription (Klebl and Choidas 2006; Nemeth et al. 2011; Wang and Fischer 2008). Nevertheless, major efforts have been made to find small-molecule inhibitors targeting specifically the CDK9 activity and the function of P-TEFb complex in HIV replication. The crystal structure of CDK9/cyclin T1/Tat complex has recently been resolved, revealing a main interaction between Tat and cyclin T1, but also with the T-loop of CDK9 (Gu et al. 2014; Tahirov et al. 2010). These studies suggested a conformational change in P-TEFb upon Tat binding, which opened the possibility to design inhibitors targeting specifically the interface of this viral protein/cellular host complex, without affecting Tat-free P-TEFb complexes, thereby avoiding toxicity (Narayanan et al. 2012; Ramakrishnan et al. 2012; Sedore et al. 2007).
Among the first, CDK9 inhibitors reported to inhibit HIV were the nucleotide analogue DRB (Biglione et al. 2007; Marciniak and Sharp 1991), flavopiridol (Chao et al. 2000), and R-roscovitine (Wang et al. 2001). DRB and flavopiridol have a fair selectivity for CDK9 over other CDKs, but displayed a small TI. Flavopiridol potently inhibits both, CDK9 activity with an IC50 of 6 nM, and HIV replication at 10 nM, but chemical derivatives only improved selectivity and survival to a small degree (Ali et al. 2009). Based on the reported P-TEFb structure in complex with flavopiridol (Baumli et al. 2008), a recent computer-aided design study identified the small-molecule 2-phenylquinazolinone derivative #37, as an inhibitor of both CDK9 and CDK2 activity. This molecule inhibits phorbol myristate acetate—activated chronically infected OM-10.1 cells, with an IC50 of 4 μM and a CC50 of 345 μM (Sancineto et al. 2013).
A derivative of a Chinese antileukemia drug, indirubin-3′-monoxime (IM), was reported to be a specific CDK9 inhibitor, reducing Tat-induced viral expression in both PBMCs and macrophages, in the single-digit micromolar range with no associated toxicity (Heredia et al. 2005; Toossi et al. 2012). More recently, IM was shown to reduce viral replication of two multidrug-resistant HIV reverse transcriptase (RT) molecular clones in humanized mice (Heredia et al. 2014).
The third generation of R-roscovitine, an ATP analogue named CR8#13 (Carpio et al. 2010), inhibits Tat-activated transcription by targeting specifically CDK9 (Narayanan et al. 2012). CR8#13 displays an IC50 of 10 nM in TNF-α activated chronically infected OM-10.1 cells and inhibits RT activity by 90 % in PBMCs at 100 nM without displaying major toxicity.
Recently, a high-throughput docking of small molecules mimicking a Tat-peptide onto the binding pocket of CDK2 was used to find small molecules disrupting the cyclin/CDK interaction (Van Duyne et al. 2013). Upon optimization, the second-generation compound named F07#13 inhibited viral replication by 90 % in Tat-transfected HLM-1 cells (HLM-1 carries a provirus with a mutation in Tat) with an IC50 of 0.12 μM, without affecting basal transcription (Fig. 3). Given the close homology of the interface of cyclin/CDK interaction sites between CDK2 and CDK9, Van Duyne et al. tested this compound on the CDK9/cyclin T1/Tat interface pocket. F07#13 specifically inhibits HIV-1-activated transcription with preference toward targeting the complex when associated with Tat. Furthermore, HIV-1 infection in humanized mice was partially inhibited during a two-week treatment period without apparent toxicity (Van Duyne et al. 2013).
Of note, the phosphinic acid #93 was shown to be the most specific ATP-competitive inhibitor of CDK9/cyclin T1 kinase activity with an IC50 of 142 nM (Nemeth et al. 2014). This compound appeared to have some antiviral activity in one HIV-1 proliferation assay in MT4 cells even though further validation tests should be performed to ascertain efficacy against HIV in more relevant HIV-infected models.
The ability of compounds to distinguish between the different P-TEFb complexes (Narayanan et al. 2012; Ramakrishnan et al. 2012; Sedore et al. 2007) would favor specificity of HIV inhibition and limit off-target effects on cellular gene expression.
5.2 Cyclin T1
Cyclin T1 and cyclin T1/Tat interaction have also been sought after as potential targets to inhibit HIV transcription. Cyclin T1 interacts with the loop of TAR and with Tat through a critically conserved cysteine (Bieniasz et al. 1998; Garber et al. 1998).
Several non-small-molecule inhibitors have been explored such as cyclin T1 intrabodies (Bai et al. 2003), microRNA-198 (Sung and Rice 2009), cyclin T1-dominant negative mutants (Jadlowsky et al. 2008a, b), and the overexpression of HEXIM1/2 (Fraldi et al. 2005). Recently, an in silico screening targeting the Tat/TAR RNA recognition motif of cyclin T1 identified the compound C3, which inhibits association of cyclin T1/Tat, as well as Tat-mediated LTR transcription in a reporter assay (Fig. 3) (Hamasaki et al. 2013). C3 also inhibits HIV reactivation from TNF-α activated OM-10.1 and U1 cells at high nanomolar IC50 and with a CC50 > 10 μM.
The compounds described above such as dCA, triptolide, the WM5 derivatives, F07#13 or C3 targeting either Tat, TAR, P-TEFb represent great steps forward in the quest for efficacious HIV transcription inhibitors of integrated provirus in latently infected cells.
6 Inhibition of Extracellular Tat
Small molecules that inhibit extracellular Tat are scarce. Of note, the small-molecule inhibitor of Tat-dependent transcription 15d-PGJ2 (see Sect. 4.2 and Fig. 3), was shown to inhibit the inflammatory response by blocking Tat-induced MCP-1 production in the hippocampus (Kim et al. 2012). Several non-small molecules were reported to bind extracellular Tat and inhibit its related functions in HAND. Sulfated polymannuroguluronate (SPMG), a sulfated polysaccharide was shown to bind the Tat basic domain (Hui et al. 2006), a triad of basic amino-acids in Tat (K12, K41, R78) (Wu et al. 2011) as well as the CD4 receptor (Miao et al. 2004). This molecule was reported to have several positive effects on HAND. It inhibited T-lymphocyte apoptosis by combating oxidative damage of mitochondria (Miao et al. 2005) and blocked Tat-induced neuronal cell death implicated in HIV-associated dementia (Hui et al. 2008). SPMG also inhibits HIV-1 Tat-induced angiogenesis in Kaposi’s sarcoma (Lu et al. 2007).
It is known that heparin sulfate proteoglycan acts as a receptor of extracellular Tat to help internalization. Rusnati et al. (1998, 2001) have shown that polyanionic heparin-like compounds are able to block extracellular Tat uptake by interacting with its basic domain. For instance, polysulfonated distamycin A derivatives (PNU145156E and PNU153429) were found to interact and sequester extracellular Tat in the extracellular space as well as to inhibit intracellular Tat when these compounds were introduced by lipofection into the cells (Corallini et al. 1998). These compounds blocked Tat-induced neoangiogenesis in T53c14 cells (Corallini et al. 1998) and delay Tat-induced tumor growth and neovascularization in Kaposi’s sarcoma-like tumor model (Possati et al. 1999).
Recently, our laboratory has shown that the Tat inhibitor dCA was able to block extracellular Tat uptake in microglia-like and astrocyte cell line models by 40 and 45 %, respectively (Mediouni et al. 2015). In addition, in the astrocytic cell line U87MG, the Tat-mediated release of the key inflammatory signaling proteins IL-1β, TNF-α, and MCP-1 was reverted by dCA treatment. Finally, using a mouse model that specifically expresses Tat in astrocytes, we demonstrated that dCA reverses the potentiation by Tat of cocaine-mediated reward using conditioned place preference experiments (Mediouni et al. 2015).
Extracellular Tat can bind via its Arg78-Gly79-Asp80 (RGD) domain to αvβ3 integrin present in endothelial cells and induce cell proliferation, motility, and neovascularization, all processes required in cancer (Urbinati et al. 2005). SCH221153, an RGD-peptidomimetic compound, binds to Tat and prevents the interaction with αvβ3 integrin, thereby inhibiting angiogenic responses triggered by Tat in chick-embryo membrane (Urbinati et al. 2005). However, RGD domain is not present in all Tat variants limiting its wide spread use.
Given the effects of Tat not only in transcription but most likely in AIDS associated pathologies, a Tat inhibitor would be of extreme benefit and hence more efforts should be put into advancing into the clinic such a long-awaited compound.
7 Conclusions
Despite two decades of research and the discovery of numerous compounds targeting Tat, TAR, P-TEFb, or their respective interactions, no small-molecule inhibitor has yet reached the clinic. The reason for this discrepancy may come from the methods employed to screen for Tat-dependent transcription inhibitors. Historically, the LTR-reporter assay with co-expression of Tat was widely used, but this type of assay leaves room for the identification of non-Tat specific inhibitors, eventually leading to cellular toxicity. So far, only two compounds have entered clinical trials: a benzodiazepine derivative, Ro 24-7429, developed by Roche in the 90s, and triptolide, a natural compound extracted from a vine used in traditional Chinese medicine. Ro 24-7429 was first evaluated, but side effects in the nervous system and absence of activity in patients cut these efforts short (Haubrich et al. 1995; Hsu et al. 1993). Currently, triptolide is in phase III and is being tested alongside ARVs to measure its impact in the HIV reservoir of naïve-ART patients in acute phase (NCT02219672).
Nevertheless, given the crucial role of Tat, TAR, and P-TEFb in viral transcription, these remain outstanding targets for the discovery of new small-molecule inhibitors of HIV replication and more so in the current context of treating latent HIV disease. A transcription inhibitor would be an exceptional addition to the current arsenal of ARVs as it would block viral reactivation from latently infected CD4+T cells, reduce low-ongoing viral replication from viral sanctuaries, and prevent reservoir replenishment.
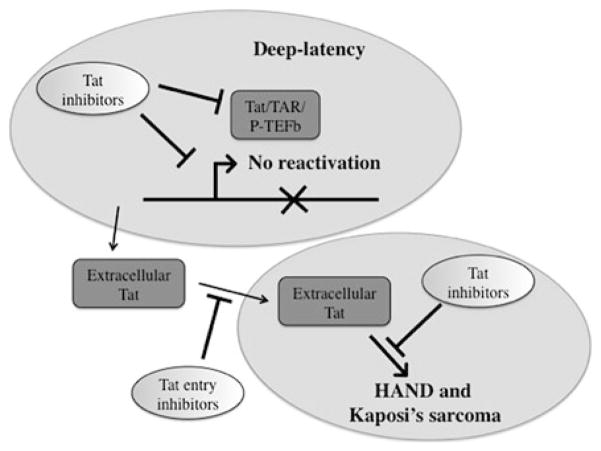
A transcription inhibitor could potentially control HIV reactivation from latency even in the absence of ART by establishing a state of deep-latency, which would be refractory to viral reactivation when ART is discontinued (Fig. 4). One could also speculate that such inhibitors would accelerate the rate of clearance of latently infected cells by reducing replication and replenishment of the latent reservoir. Thus, the latent pool of cells in an infected individual would be stabilized and death of the long-lived infected memory T cell would result in a continuous decay of this pool over time, possibly culminating in the long-awaited sterilizing cure.
Fig. 4.
Multiple targets of a Tat inhibitor. The primary role of a Tat inhibitor is to block HIV transcription to promote a state of deep-latency and inhibit HIV reactivation. A Tat inhibitor might also reduce uptake of extracellular Tat by blocking its interaction with cellular receptors. Finally, a Tat inhibitor might impact the effect of extracellular Tat in the induction of pathways resulting in HAND or Kaposi’s sarcoma
Among the recently identified HIV transcriptional inhibitors, dCA, an analogue of a natural compound isolated from a marine sponge, is one of the most promising. dCA binds directly to Tat and inhibits HIV transcription in a Tat-dependent manner in both acutely and chronically infected cells in the nanomolar range with no associated toxicity. Treatment of chronically infected cells with dCA resulted in a 2-log reduction in mRNA levels. More importantly, arrest of the treatment did not result in viral rebound upon drug removal for the duration of the experiment (one month) (Mousseau et al. 2012). In addition, dCA inhibits spontaneous viral release from latently infected CD4+T cells isolated from aviremic patients undergoing ART. As such, dCA might correspond to the type of the transcription inhibitor that would control reactivation and reservoir replenishment with the elusive goal of a cure.
Adding to the benefits, a Tat inhibitor targeting either the Tat basic domain or the cys-rich domain may directly impact the appearance of neurotoxicity and development of HAND as well as reduce Tat cancer-causing properties (Fig. 4). The addition of a transcription inhibitor to ART could be of immense value to control viral reactivation and decrease the viral reservoir, a key limitation in the current multi-therapy treatment.
Abbreviations
- ART
Antiretroviral therapy
- HIV
Human immunodeficiency virus
- ARVs
Antiretrovirals
- LTR
5′ long-terminal repeat
- RNAPII
RNA polymerase II
- TAR
Transactivation response element
- PCAF
p300/CBP-associated factor
- P-TEFb
Positive transcription elongation factor b
- CDK9
Cyclin-dependent kinase 9
- CTD
C-terminal domain
- HDAC
Histone deacetylase
- HAT
Histone acetyl transferase
- HAND
HIV-associated neurocognitive disorders
- BBB
Blood–brain barrier
- MCP-1
Chemoattractant protein-1
- PBMC
Peripheral blood mononuclear cell
- TI
Therapeutic index
- IC50
Half-maximal inhibitory concentration
- NMR
Nuclear magnetic resonance
- MAE
Michael acceptor electrophile
- CC50
Half-maximal cytotoxic concentration
- dCA
didehydro-cortistatin A
- RT
Reverse transcriptase
- SPMG
Sulfated polymannuroguluronate
- AIDS
Acquired immune deficiency syndrome
References
- Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253(2):313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- Albini A, Ferrini S, Benelli R, Sforzini S, Giunciuglio D, Aluigi MG, Proudfoot AE, Alouani S, Wells TN, Mariani G, Rabin RL, Farber JM, Noonan DM. HIV-1 Tat protein mimicry of chemokines. Proc Natl Acad Sci USA. 1998;95(22):13153–13158. doi: 10.1073/pnas.95.22.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, Soldi R, Giunciuglio D, Giraudo E, Benelli R, Primo L, Noonan D, Salio M, Camussi G, Rockl W, Bussolino F. The angiogenesis induced by HIV-1 tat protein is mediated by the Flk-1/KDR receptor on vascular endothelial cells. Nat Med. 1996;2(12):1371–1375. doi: 10.1038/nm1296-1371. [DOI] [PubMed] [Google Scholar]
- Aldovini A, Debouck C, Feinberg MB, Rosenberg M, Arya SK, Wong-Staal F. Synthesis of the complete trans-activation gene product of human T-lymphotropic virus type III in Escherichia coli: demonstration of immunogenicity in vivo and expression in vitro. Proc Natl Acad Sci USA. 1986;83(18):6672–6676. doi: 10.1073/pnas.83.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Ghosh A, Nathans RS, Sharova N, O’Brien S, Cao H, Stevenson M, Rana TM. Identification of flavopiridol analogues that selectively inhibit positive transcription elongation factor (P-TEFb) and block HIV-1 replication. Chembiochem. 2009;10(12):2072–2080. doi: 10.1002/cbic.200900303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M. Recent status of HIV-1 gene expression inhibitors. Antiviral Res. 2006;71(2–3):301–306. doi: 10.1016/j.antiviral.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Bachani M, Sacktor N, McArthur JC, Nath A, Rumbaugh J. Detection of anti-tat antibodies in CSF of individuals with HIV-associated neurocognitive disorders. J Neurovirol. 2013;19(1):82–88. doi: 10.1007/s13365-012-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagashev A, Sawaya BE. Roles and functions of HIV-1 Tat protein in the CNS: an overview. Virol J. 2013;10:358. doi: 10.1186/1743-422x-10-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Sui J, Zhu RY, Tallarico AS, Gennari F, Zhang D, Marasco WA. Inhibition of Tat-mediated transactivation and HIV-1 replication by human anti-hCyclinT1 intrabodies. J Biol Chem. 2003;278(3):1433–1442. doi: 10.1074/jbc.M208297200. [DOI] [PubMed] [Google Scholar]
- Barboric M, Yik JH, Czudnochowski N, Yang Z, Chen R, Contreras X, Geyer M, Peterlin BM, Zhou Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35(6):2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27(13):1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273(38):24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17 (23):7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19(10):1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19(1):60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, Loret EP. What does the structure-function relationship of the HIV-1 Tat protein teach us about developing an AIDS vaccine? Retrovirology. 2009;6:50. doi: 10.1186/1742-4690-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpio L, Klase Z, Coley W, Guendel I, Choi S, Van Duyne R, Narayanan A, Kehn-Hall K, Meijer L, Kashanchi F. microRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J RNAi Gene Silencing Int J RNA Gene Target Res. 2010;6(1):386–400. [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V, Parolin C, Moro S, Pecere T, Filipponi E, Calistri A, Tabarrini O, Gatto B, Palumbo M, Fravolini A, Palu G. 6-Aminoquinolones as new potential anti-HIV agents. J Med Chem. 2000;43(20):3799–3802. doi: 10.1021/jm9903390. [DOI] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275 (37):28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley W, Kehn-Hall K, Van Duyne R, Kashanchi F. Novel HIV-1 therapeutics through targeting altered host cell pathways. Expert Opin Biol Ther. 2009;9(11):1369–1382. doi: 10.1517/14712590903257781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corallini A, Betti M, Rusnati M, Campioni D, Ciomei M, Sola F, Calza N, Zauli G, Presta M, Barbanti-Brodano G, Caputo A. Characterization of the effects of two polysulfonated distamycin A derivatives, PNU145156E and PNU153429, on HIV type 1 Tat protein. AIDS Res Hum Retroviruses. 1998;14(17):1561–1571. doi: 10.1089/aid.1998.14.1561. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46(7):973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Regulation of HIV-1 gene expression. FASEB J. 1991;5(10):2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol. 2010;17(7):815–821. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A, Begley DW, Lau C, Varani G. A small-molecule probe induces a conformation in HIV TAR RNA capable of binding drug-like fragments. J Mol Biol. 2011;410(5):984–996. doi: 10.1016/j.jmb.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debaisieux S, Rayne F, Yezid H, Beaumelle B. The ins and outs of HIV-1 Tat. Traffic. 2012;13 (3):355–363. doi: 10.1111/j.1600-0854.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- Deeks SG. HIV: shock and kill. Nature. 2012;487(7408):439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun TW, Churchill M, Di Mascio M, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud MC, Peterlin M, Poli G, Routy JP, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Van Lint C, Verdin E, Woolfrey A, Zaia J, Barre-Sinoussi F. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol. 2012;12(8):607–614. doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Croul S, Morgello S, Amini S, Rappaport J, Khalili K. Detection of HIV-1 Tat and JCV capsid protein, VP1, in AIDS brain with progressive multifocal leukoencephalopathy. J Neurovirol. 2000;6(3):221–228. doi: 10.3109/13550280009015824. [DOI] [PubMed] [Google Scholar]
- Easley R, Carpio L, Dannenberg L, Choi S, Alani D, Van Duyne R, Guendel I, Klase Z, Agbottah E, Kehn-Hall K, Kashanchi F. Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription. Virology. 2010;405(2):322–333. doi: 10.1016/j.virol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eekels JJ, Berkhout B. Toward a durable treatment of HIV-1 infection using RNA interference. Prog Mol Biol Transl Sci. 2011;102:141–163. doi: 10.1016/B978-0-12-415795-8.00001-5. [DOI] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67(1):277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci USA. 1994;91(2):664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci USA. 2014;111 (6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraldi A, Varrone F, Napolitano G, Michels AA, Majello B, Bensaude O, Lania L. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology. 2005;2:42. doi: 10.1186/1742-4690-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon P, Khan MZ, Kolson DL. Current understanding of HIV-associated neurocognitive disorders pathogenesis. Curr Opin Neurol. 2011;24(3):275–283. doi: 10.1097/WCO.0b013e32834695fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol. 2000;20(18):6958–6969. doi: 10.1128/MCB.20.18.6958-6969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, Jones KA. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12(22):3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorantla S, Poluektova L, Gendelman HE. Rodent models for HIV-associated neurocognitive disorders. Trends Neurosci. 2012;35(3):197–208. doi: 10.1016/j.tins.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Babayeva ND, Suwa Y, Baranovskiy AG, Price DH, Tahirov TH. Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4. Cell Cycle. 2014;13(11):1788–1797. doi: 10.4161/cc.28756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki T, Okamoto M, Baba M. Identification of novel inhibitors of human immunodeficiency virus type 1 replication by in silico screening targeting cyclin T1/Tat interaction. Antimicrob Agents Chemother. 2013;57(3):1323–1331. doi: 10.1128/aac.01711-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubrich RH, Flexner C, Lederman MM, Hirsch M, Pettinelli CP, Ginsberg R, Lietman P, Hamzeh FM, Spector SA, Richman DD. A randomized trial of the activity and safety of Ro 24-7429 (Tat antagonist) versus nucleoside for human immunodeficiency virus infection. The AIDS clinical trials group 213 team. J Infect Dis. 1995;172(5):1246–1252. doi: 10.1093/infdis/172.5.1246. [DOI] [PubMed] [Google Scholar]
- Heredia A, Davis C, Bamba D, Le N, Gwarzo MY, Sadowska M, Gallo RC, Redfield RR. Indirubin-3′-monoxime, a derivative of a Chinese antileukemia medicine, inhibits P-TEFb function and HIV-1 replication. AIDS. 2005;19(18):2087–2095. doi: 10.1097/01.aids.0000194805.74293.11. [DOI] [PubMed] [Google Scholar]
- Heredia A, Natesan S, Le NM, Medina-Moreno S, Zapata JC, Reitz M, Bryant J, Redfield RR. Indirubin 3′-Monoxime, from a Chinese traditional herbal formula, suppresses viremia in humanized mice infected with multidrug-resistant HIV. AIDS Res Hum Retroviruses. 2014 doi: 10.1089/aid.2013.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman FM, Dohadwala MM, Wright AD, Hinton DR, Walker SM. Exogenous tat protein activates central nervous system-derived endothelial cells. J Neuroimmunol. 1994;54(1–2):19–28. doi: 10.1016/0165-5728(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Hsu MC, Dhingra U, Earley JV, Holly M, Keith D, Nalin CM, Richou AR, Schutt AD, Tam SY, Potash MJ, et al. Inhibition of type 1 human immunodeficiency virus replication by a tat antagonist to which the virus remains sensitive after prolonged exposure in vitro. Proc Natl Acad Sci USA. 1993;90(14):6395–6399. doi: 10.1073/pnas.90.14.6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L, Liu J, Nath A, Jones M, Raghavan R, Narayan O, Male D, Everall I. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol. 2000;6 (2):145–155. doi: 10.3109/13550280009013158. [DOI] [PubMed] [Google Scholar]
- Hui B, Li J, Geng MY. Sulfated polymannuroguluronate, a novel anti-acquired immune deficiency syndrome drug candidate, decreased vulnerability of PC12 cells to human immunodeficiency virus tat protein through attenuating calcium overload. J Neurosci Res. 2008;86 (5):1169–1177. doi: 10.1002/jnr.21566. [DOI] [PubMed] [Google Scholar]
- Hui B, Xia W, Li J, Wang L, Ai J, Geng M. Sulfated polymannuroguluronate, a novel anti-acquired immune deficiency syndrome drug candidate, blocks neuroinflammatory signalling by targeting the transactivator of transcription (Tat) protein. J Neurochem. 2006;97(2):334–344. doi: 10.1111/j.1471-4159.2006.03698.x. [DOI] [PubMed] [Google Scholar]
- Hunt PW. Th17, gut, and HIV: therapeutic implications. Curr Opin HIV AIDS. 2010;5 (2):189–193. doi: 10.1097/COH.0b013e32833647d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadlowsky JK, Nojima M, Okamoto T, Fujinaga K. Dominant negative mutant cyclin T1 proteins that inhibit HIV transcription by forming a kinase inactive complex with Tat. J Gen Virol. 2008a;89(Pt 11):2783–2787. doi: 10.1099/vir.0.2008/002857-0. [DOI] [PubMed] [Google Scholar]
- Jadlowsky JK, Nojima M, Schulte A, Geyer M, Okamoto T, Fujinaga K. Dominant negative mutant cyclin T1 proteins inhibit HIV transcription by specifically degrading Tat. Retrovirology. 2008b;5:63. doi: 10.1186/1742-4690-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeang KT, Chun R, Lin NH, Gatignol A, Glabe CG, Fan H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol. 1993;67 (10):6224–6233. doi: 10.1128/jvi.67.10.6224-6233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri MK, Mishra R, Chhatbar C, Unni SK, Singh SK. Tits and bits of HIV Tat protein. Expert Opin Biol Ther. 2011;11(3):269–283. doi: 10.1517/14712598.2011.546339. [DOI] [PubMed] [Google Scholar]
- Jones LE, Perelson AS. Transient viremia, plasma viral load, and reservoir replenishment in HIV-infected patients on antiretroviral therapy. J Acquir Immune Defic Syndr. 2007;45(5):483–493. doi: 10.1097/QAI.0b013e3180654836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantari P, Narayan V, Henderson AJ, Prabhu KS. 15-Deoxy-Delta12,14-prostaglandin J2 inhibits HIV-1 transactivating protein, Tat, through covalent modification. FASEB J. 2009;23 (8):2366–2373. doi: 10.1096/fj.08-124982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BO, Liu Y, Ruan Y, Xu ZC, Schantz L, He JJ. Neuropathologies in transgenic mice expressing human immunodeficiency virus type 1 Tat protein under the regulation of the astrocyte-specific glial fibrillary acidic protein promoter and doxycycline. Am J Pathol. 2003;162 (5):1693–1707. doi: 10.1016/s0002-9440(10)64304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SE, Lee EO, Yang JH, Kang JH, Suh YH, Chong YH. 15-deoxy-Delta(1)(2), (1)(4) -prostaglandin J(2) inhibits human immunodeficiency virus-1 tat-induced monocyte chemoattractant protein-1/CCL2 production by blocking the extracellular signal-regulated kinase-1/2 signaling pathway independently of peroxisome proliferator-activated receptor-gamma and heme oxygenase-1 in rat hippocampal slices. J Neurosci Res. 2012;90(9):1732–1742. doi: 10.1002/jnr.23051. [DOI] [PubMed] [Google Scholar]
- King JE, Eugenin EA, Buckner CM, Berman JW. HIV tat and neurotoxicity. Microbes Infect. 2006;8(5):1347–1357. doi: 10.1016/j.micinf.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Klebl BM, Choidas A. CDK9/cyclin T1: a host cell target for antiretroviral therapy. Future Virol. 2006;1(3):317–330. doi: 10.2217/17460794.1.3.317. [DOI] [Google Scholar]
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, Coulombe B, Price DH. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36(7):2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot A, Berchanski A, Borkow G. Insight into the mechanisms of aminoglycoside derivatives interaction with HIV-1 entry steps and viral gene transcription. FEBS J. 2008;275 (21):5236–5257. doi: 10.1111/j.1742-4658.2008.06657.x. [DOI] [PubMed] [Google Scholar]
- Li W, Li G, Steiner J, Nath A. Role of Tat protein in HIV neuropathogenesis. Neurotox Res. 2009;16(3):205–220. doi: 10.1007/s12640-009-9047-8. [DOI] [PubMed] [Google Scholar]
- Lu CX, Li J, Sun YX, Qi X, Wang QJ, Xin XL, Geng MY. Sulfated polymannuroguluronate, a novel anti-AIDS drug candidate, inhibits HIV-1 Tat-induced angiogenesis in Kaposi’s sarcoma cells. Biochem Pharmacol. 2007;74(9):1330–1339. doi: 10.1016/j.bcp.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Lusic M, Marcello A, Cereseto A, Giacca M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003;22(24):6550–6561. doi: 10.1093/emboj/cdg631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Nath A. Molecular determinants for cellular uptake of Tat protein of human immunodeficiency virus type 1 in brain cells. J Virol. 1997;71(3):2495–2499. doi: 10.1128/jvi.71.3.2495-2499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T. The BAF complex and HIV latency. Transcription. 2012;3(4):171–176. doi: 10.4161/trns.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10(13):4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM. Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep. 2010;7(1):37–43. doi: 10.1007/s11904-009-0033-9. [DOI] [PubMed] [Google Scholar]
- Massari S, Daelemans D, Barreca ML, Knezevich A, Sabatini S, Cecchetti V, Marcello A, Pannecouque C, Tabarrini O. A 1,8-naphthyridone derivative targets the HIV-1 Tat-mediated transcription and potently inhibits the HIV-1 replication. J Med Chem. 2010;53(2):641–648. doi: 10.1021/jm901211d. [DOI] [PubMed] [Google Scholar]
- Massari S, Sabatini S, Tabarrini O. Blocking HIV-1 replication by targeting the Tat-hijacked transcriptional machinery. Curr Pharm Des. 2013;19(10):1860–1879. doi: 10.2174/1381612811319100010. [DOI] [PubMed] [Google Scholar]
- Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr HIV Res. 2011;9(8):554–567. doi: 10.2174/157016211798998736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mediouni S, Jablonski J, Paris JJ, Clementz MA, Thenin-Houssier S, McLaughlin JP, Valente ST. Didehydro-Cortistatin a Inhibits HIV-1 Tat mediated neuroinflammation and prevents potentiation of cocaine reward in Tat transgenic mice. Curr HIV Res. 2015 doi: 10.2174/1570162x13666150121111548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Geng M, Li J, Li F, Chen H, Guan H, Ding J. Sulfated polymannuroguluronate, a novel anti-acquired immune deficiency syndrome (AIDS) drug candidate, targeting CD4 in lymphocytes. Biochem Pharmacol. 2004;68(4):641–649. doi: 10.1016/j.bcp.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Miao B, Li J, Fu X, Gan L, Xin X, Geng M. Sulfated polymannuroguluronate, a novel anti-AIDS drug candidate, inhibits T cell apoptosis by combating oxidative damage of mitochondria. Mol Pharmacol. 2005;68(6):1716–1727. doi: 10.1124/mol.105.015412. [DOI] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology. 2007;4:36. doi: 10.1186/1742-4690-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau G, Clementz MA, Bakeman WN, Nagarsheth N, Cameron M, Shi J, Baran P, Fromentin R, Chomont N, Valente ST. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe. 2012;12(1):97–108. doi: 10.1016/j.chom.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau G, Valente ST. Strategies to block HIV transcription: focus on small molecule Tat inhibitors. Biology. 2012;1(3):668–697. doi: 10.3390/biology1030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol. 2010;10(5):551–556. doi: 10.1016/j.coph.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Narayan V, Ravindra KC, Chiaro C, Cary D, Aggarwal BB, Henderson AJ, Prabhu KS. Celastrol inhibits Tat-mediated human immunodeficiency virus (HIV) transcription and replication. J Mol Biol. 2011;410(5):972–983. doi: 10.1016/j.jmb.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Sampey G, Van Duyne R, Guendel I, Kehn-Hall K, Roman J, Currer R, Galons H, Oumata N, Joseph B, Meijer L, Caputi M, Nekhai S, Kashanchi F. Use of ATP analogs to inhibit HIV-1 transcription. Virology. 2012;432(1):219–231. doi: 10.1016/j.virol.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth G, Varga Z, Greff Z, Bencze G, Sipos A, Szantai-Kis C, Baska F, Gyuris A, Kelemenics K, Szathmary Z, Minarovits J, Keri G, Orfi L. Novel, selective CDK9 inhibitors for the treatment of HIV infection. Curr Med Chem. 2011;18(3):342–358. doi: 10.2174/092986711794839188. [DOI] [PubMed] [Google Scholar]
- Nemeth G, Greff Z, Sipos A, Varga Z, Szekely R, Sebestyen M, Jaszay Z, Beni S, Nemes Z, Pirat JL, Volle JN, Virieux D, Gyuris A, Kelemenics K, Ay E, Minarovits J, Szathmary S, Keri G, Orfi L. Synthesis and evaluation of phosphorus containing, specific CDK9/CycT1 inhibitors. J Med Chem. 2014;57(10):3939–3965. doi: 10.1021/jm401742r. [DOI] [PubMed] [Google Scholar]
- Nunnari G, Smith JA, Daniel R. HIV-1 Tat and AIDS-associated cancer: targeting the cellular anti-cancer barrier? J Exp Clin Cancer Res CR. 2008;27:3. doi: 10.1186/1756-9966-27-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10(5):426–435. doi: 10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci USA. 2008;105(10):3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolin C, Gatto B, Del Vecchio C, Pecere T, Tramontano E, Cecchetti V, Fravolini A, Masiero S, Palumbo M, Palu G. New anti-human immunodeficiency virus type 1 6-aminoquinol-ones: mechanism of action. Antimicrob Agents Chemother. 2003;47(3):889–896. doi: 10.1128/AAC.47.3.889-896.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessler F, Cron RQ. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 2004;5(3):158–167. doi: 10.1038/sj.gene.6364047. [DOI] [PubMed] [Google Scholar]
- Possati L, Campioni D, Sola F, Leone L, Ferrante L, Trabanelli C, Ciomei M, Montesi M, Rocchetti R, Talevi S, Bompadre S, Caputo A, Barbanti-Brodano G, Corallini A. Antiangiogenic, antitumoural and antimetastatic effects of two distamycin A derivatives with anti-HIV-1 Tat activity in a Kaposi’s sarcoma-like murine model. Clin Exp Metastasis. 1999;17 (7):575–582. doi: 10.1023/a:1006737029616. [DOI] [PubMed] [Google Scholar]
- Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R, Liu H, Donahue H, Malovannaya A, Qin J, Rice AP. Identification of novel CDK9 and Cyclin T1-associated protein complexes (CCAPs) whose siRNA depletion enhances HIV-1 Tat function. Retrovirology. 2012;9:90. doi: 10.1186/1742-4690-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Parolin C, Gatto B, Del Vecchio C, Brocca-Cofano E, Fravolini A, Palu G, Palumbo M. Inhibition of human immunodeficiency virus type 1 tat-trans-activation-responsive region interaction by an antiviral quinolone derivative. Antimicrob Agents Chemother. 2004;48 (5):1895–1899. doi: 10.1128/AAC.48.5.1895-1899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter SN, Palu G. Inhibitors of HIV-1 Tat-mediated transactivation. Curr Med Chem. 2006;13 (11):1305–1315. doi: 10.2174/092986706776872989. [DOI] [PubMed] [Google Scholar]
- Romani B, Engelbrecht S, Glashoff RH. Functions of Tat: the versatile protein of human immunodeficiency virus type 1. J Gen Virol. 2010;91(Pt 1):1–12. doi: 10.1099/vir.0.016303-0. [DOI] [PubMed] [Google Scholar]
- Rozera C, Carattoli A, De Marco A, Amici C, Giorgi C, Santoro MG. Inhibition of HIV-1 replication by cyclopentenone prostaglandins in acutely infected human cells. Evidence for a transcriptional block. J Clin Invest. 1996;97(8):1795–1803. doi: 10.1172/JCI118609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnati M, Presta M. HIV-1 Tat protein and endothelium: from protein/cell interaction to AIDS-associated pathologies. Angiogenesis. 2002;5(3):141–151. doi: 10.1023/A:1023892223074. [DOI] [PubMed] [Google Scholar]
- Rusnati M, Tulipano G, Urbinati C, Tanghetti E, Giuliani R, Giacca M, Ciomei M, Corallini A, Presta M. The basic domain in HIV-1 Tat protein as a target for polysulfonated heparin-mimicking extracellular Tat antagonists. J Biol Chem. 1998;273(26):16027–16037. doi: 10.1074/jbc.273.26.16027. [DOI] [PubMed] [Google Scholar]
- Rusnati M, Urbinati C, Caputo A, Possati L, Lortat-Jacob H, Giacca M, Ribatti D, Presta M. Pentosan polysulfate as an inhibitor of extracellular HIV-1 Tat. J Biol Chem. 2001;276(25):22420–22425. doi: 10.1074/jbc.M010779200. [DOI] [PubMed] [Google Scholar]
- Sancineto L, Iraci N, Massari S, Attanasio V, Corazza G, Barreca ML, Sabatini S, Manfroni G, Avanzi NR, Cecchetti V, Pannecouque C, Marcello A, Tabarrini O. Computer-aided design, synthesis and validation of 2-phenylquinazolinone fragments as CDK9 inhibitors with anti-HIV-1 Tat-mediated transcription activity. ChemMedChem. 2013;8(12):1941–1953. doi: 10.1002/cmdc.201300287. [DOI] [PubMed] [Google Scholar]
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35(13):4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Sklar PA, Ward DJ, Baker RK, Wood KC, Gafoor Z, Alzola CF, Moorman AC, Holmberg SD. Prevalence and clinical correlates of HIV viremia (‘blips’) in patients with previous suppression below the limits of quantification. AIDS. 2002;16(15):2035–2041. doi: 10.1097/00002030-200210180-00008. [DOI] [PubMed] [Google Scholar]
- Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol. 2011;7(8):553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, De Clercq E, Balzarini J. The regulation of HIV-1 transcription: molecular targets for chemotherapeutic intervention. Med Res Rev. 2006;26(5):595–625. doi: 10.1002/med.20081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, Pollicita M, Pannecouque C, Verbeken E, Tabarrini O, Cecchetti V, Aquaro S, Perno CF, Fravolini A, De Clercq E, Schols D, Balzarini J. Novel in vivo model for the study of human immunodeficiency virus type 1 transcription inhibitors: evaluation of new 6-desfluoroquinolone derivatives. Antimicrob Agents Chemother. 2007;51(4):1407–1413. doi: 10.1128/AAC.01251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazza M, Pirrone V, Wigdahl B, Nonnemacher MR. Breaking down the barrier: the effects of HIV-1 on the blood-brain barrier. Brain Res. 2011;1399:96–115. doi: 10.1016/j.brainres.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung TL, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 2009;5(1):e1000263. doi: 10.1371/journal.ppat.1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabarrini O, Massari S, Cecchetti V. 6-desfluoroquinolones as HIV-1 Tat-mediated transcription inhibitors. Future Med Chem. 2010;2(7):1161–1180. doi: 10.4155/fmc.10.208. [DOI] [PubMed] [Google Scholar]
- Tabarrini O, Massari S, Daelemans D, Stevens M, Manfroni G, Sabatini S, Balzarini J, Cecchetti V, Pannecouque C, Fravolini A. Structure-activity relationship study on anti-HIV 6-desfluoroquinolones. J Med Chem. 2008;51(17):5454–5458. doi: 10.1021/jm701585h. [DOI] [PubMed] [Google Scholar]
- Tabarrini O, Massari S, Sancineto L, Daelemans D, Sabatini S, Manfroni G, Cecchetti V, Pannecouque C. Structural investigation of the naphthyridone scaffold: identification of a 1,6-naphthyridone derivative with potent and selective anti-HIV activity. ChemMedChem. 2011;6 (7):1249–1257. doi: 10.1002/cmdc.201100073. [DOI] [PubMed] [Google Scholar]
- Tabarrini O, Stevens M, Cecchetti V, Sabatini S, Dell’Uomo M, Manfroni G, Palumbo M, Pannecouque C, De Clercq E, Fravolini A. Structure modifications of 6-aminoquinolones with potent anti-HIV activity. J Med Chem. 2004;47(22):5567–5578. doi: 10.1021/jm049721p. [DOI] [PubMed] [Google Scholar]
- Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465(7299):747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey MG, Jones KA. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev. 1989;3 (3):265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- Toossi Z, Wu M, Hirsch CS, Mayanja-Kizza H, Baseke J, Aung H, Canaday DH, Fujinaga K. Activation of P-TEFb at sites of dual HIV/TB infection, and inhibition of MTB-induced HIV transcriptional activation by the inhibitor of CDK9, Indirubin-3′-monoxime. AIDS Res Hum Retroviruses. 2012;28(2):182–187. doi: 10.1089/AID.2010.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JJ, Fabani M, Arzumanov AA, Ivanova G, Gait MJ. Targeting the HIV-1 RNA leader sequence with synthetic oligonucleotides and siRNA: chemistry and cell delivery. Biochim Biophys Acta. 2006;1758(3):290–300. doi: 10.1016/j.bbamem.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Turpin JA, Buckheit RW, Jr, Derse D, Hollingshead M, Williamson K, Palamone C, Osterling MC, Hill SA, Graham L, Schaeffer CA, Bu M, Huang M, Cholody WM, Michejda CJ, Rice WG. Inhibition of acute-, latent-, and chronic-phase human immunodeficiency virus type 1 (HIV-1) replication by a bistriazoloacridone analog that selectively inhibits HIV-1 transcription. Antimicrob Agents Chemother. 1998;42(3):487–494. doi: 10.1128/aac.42.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbinati C, Mitola S, Tanghetti E, Kumar C, Waltenberger J, Ribatti D, Presta M, Rusnati M. Integrin alphavbeta3 as a target for blocking HIV-1 Tat-induced endothelial cell activation in vitro and angiogenesis in vivo. Arterioscler Thromb Vasc Biol. 2005;25(11):2315–2320. doi: 10.1161/01.ATV.0000186182.14908.7b. [DOI] [PubMed] [Google Scholar]
- Van Duyne R, Guendel I, Jaworski E, Sampey G, Klase Z, Chen H, Zeng C, Kovalskyy D, El Kouni MH, Lepene B, Patanarut A, Nekhai S, Price DH, Kashanchi F. Effect of mimetic CDK9 inhibitors on HIV-1-activated transcription. J Mol Biol. 2013;425(4):812–829. doi: 10.1016/j.jmb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Grol J, Subauste C, Andrade RM, Fujinaga K, Nelson J, Subauste CS. HIV-1 inhibits autophagy in bystander macrophage/monocytic cells through Src-Akt and STAT3. PLoS One. 2010;5 (7):e11733. doi: 10.1371/journal.pone.0011733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Z, Chen X. Triptolide inhibits human immunodeficiency virus type 1 replication by promoting proteasomal degradation of Tat protein. Retrovirology. 2014;11(1):88. doi: 10.1186/preaccept-1002681664135027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, de la Fuente C, Deng L, Wang L, Zilberman I, Eadie C, Healey M, Stein D, Denny T, Harrison LE, Meijer L, Kashanchi F. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J Virol. 2001;75(16):7266–7279. doi: 10.1128/jvi.75.16.7266-7279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Fischer PM. Cyclin-dependent kinase 9: a key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharmacol Sci. 2008;29(6):302–313. doi: 10.1016/j.tips.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Weeks BS, Lieberman DM, Johnson B, Roque E, Green M, Loewenstein P, Oldfield EH, Kleinman HK. Neurotoxicity of the human immunodeficiency virus type 1 tat transactivator to PC12 cells requires the Tat amino acid 49–58 basic domain. J Neurosci Res. 1995;42 (1):34–40. doi: 10.1002/jnr.490420105. [DOI] [PubMed] [Google Scholar]
- Wesselingh SL, Power C, Glass JD, Tyor WR, McArthur JC, Farber JM, Griffin JW, Griffin DE. Intracerebral cytokine messenger RNA expression in acquired immunodeficiency syndrome dementia. Ann Neurol. 1993;33(6):576–582. doi: 10.1002/ana.410330604. [DOI] [PubMed] [Google Scholar]
- Wiley CA, Baldwin M, Achim CL. Expression of HIV regulatory and structural mRNA in the central nervous system. AIDS. 1996;10(8):843–847. doi: 10.1097/00002030-199607000-00007. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Wu YL, Ai J, Zhao JM, Xiong B, Xin XJ, Geng MY, Xin XL, Jiang HD. Sulfated polymannuroguluronate inhibits Tat-induced SLK cell adhesion via a novel binding site, a KKR spatial triad. Acta pharmacologica Sinica. 2011;32(5):647–654. doi: 10.1038/aps.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, Duan L, Choi AL, Girling V, Ho T, Li P, Fujimoto K, Lampiris H, Hare CB, Pandori M, Haase AT, Gunthard HF, Fischer M, Shergill AK, McQuaid K, Havlir DV, Wong JK. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202(10):1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller SJ, Kumar P. RNA-based gene therapy for the treatment and prevention of HIV: from bench to bedside. Yale J Biol Med. 2011;84(3):301–309. [PMC free article] [PubMed] [Google Scholar]
- Zucchini S, Pittaluga A, Brocca-Cofano E, Summa M, Fabris M, De Michele R, Bonaccorsi A, Busatto G, Barbanti-Brodano G, Altavilla G, Verlengia G, Cifelli P, Corallini A, Caputo A, Simonato M. Increased excitability in tat-transgenic mice: role of tat in HIV-related neurological disorders. Neurobiol Dis. 2013;55:110–119. doi: 10.1016/j.nbd.2013.02.004. [DOI] [PubMed] [Google Scholar]