Abstract
Objective:
To use in vivo neuroimaging and postmortem neuropathologic analysis in C9orf72 repeat expansion patients to investigate the hypothesis that C9orf72 promoter hypermethylation is neuroprotective and regionally selective.
Methods:
Twenty patients with a C9orf72 repeat expansion participating in a high-resolution MRI scan and a clinical examination and a subset of patients (n = 11) were followed longitudinally with these measures. Gray matter (GM) density was related to C9orf72 promoter hypermethylation using permutation-based testing. Regional neuronal loss was measured in an independent autopsy series (n = 35) of C9orf72 repeat expansion patients.
Results:
GM analysis revealed that hippocampus, frontal cortex, and thalamus are associated with hypermethylation and thus appear to be relatively protected from mutant C9orf72. Neuropathologic analysis demonstrated an association between reduced neuronal loss and hypermethylation in hippocampus and frontal cortex. Longitudinal neuroimaging revealed that hypermethylation is associated with reduced longitudinal decline in GM regions protected by hypermethylation and longitudinal neuropsychological assessment demonstrated that longitudinal decline in verbal recall is protected by hypermethylation.
Conclusions:
These cross-sectional and longitudinal neuroimaging studies, along with neuropathologic validation studies, provide converging evidence for neuroprotective properties of C9orf72 promoter hypermethylation. These findings converge with prior postmortem studies suggesting that C9orf72 promoter hypermethylation may be a neuroprotective target for drug discovery.
The C9orf72 hexanucleotide repeat expansion1,2 is associated with inclusions of TAR DNA binding protein of 43 kDa (TDP-43) as the primary pathologic substrate of neurodegeneration3 and accounts for the largest proportion of inherited forms of amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD).4 From a translational perspective, it is critical to improve our understanding of the C9orf72 expansion because expanded individuals have a known source of underlying pathology during life and may be good candidates for clinical trials of disease-modifying therapeutic agents.5
A candidate mechanism for therapeutic agents relates to recent molecular and neuropathologic studies suggesting that C9orf72 promoter hypermethylation can contribute to transcriptional silencing of mutant C9orf72. Hypermethylation is equally observed in ALS and FTD6,7 and at autopsy it has been associated with reduced pathologic inclusions.8 While postmortem studies suggest that C9orf72 promoter hypermethylation may be neuroprotective, in vivo longitudinal investigations of the potential neuroprotective properties of C9orf72 promoter methylation are lacking. Neuroimaging comparisons of C9orf72 expansion relative to C9orf72-negative patients provide preliminary evidence for regional selectivity of gray matter (GM) disease associated with C9orf72 expansion.9–14 In vivo demonstration of effects related to C9orf72 hypermethylation would provide an important method for monitoring response during a disease-modifying treatment trial.
In this report, we use cross-sectional and longitudinal neuroimaging to evaluate the hypothesis that hypermethylation of the C9orf72 promoter is neuroprotective and we provide neuropathologic validation of our findings in an independent autopsy cohort.
METHODS
Participants.
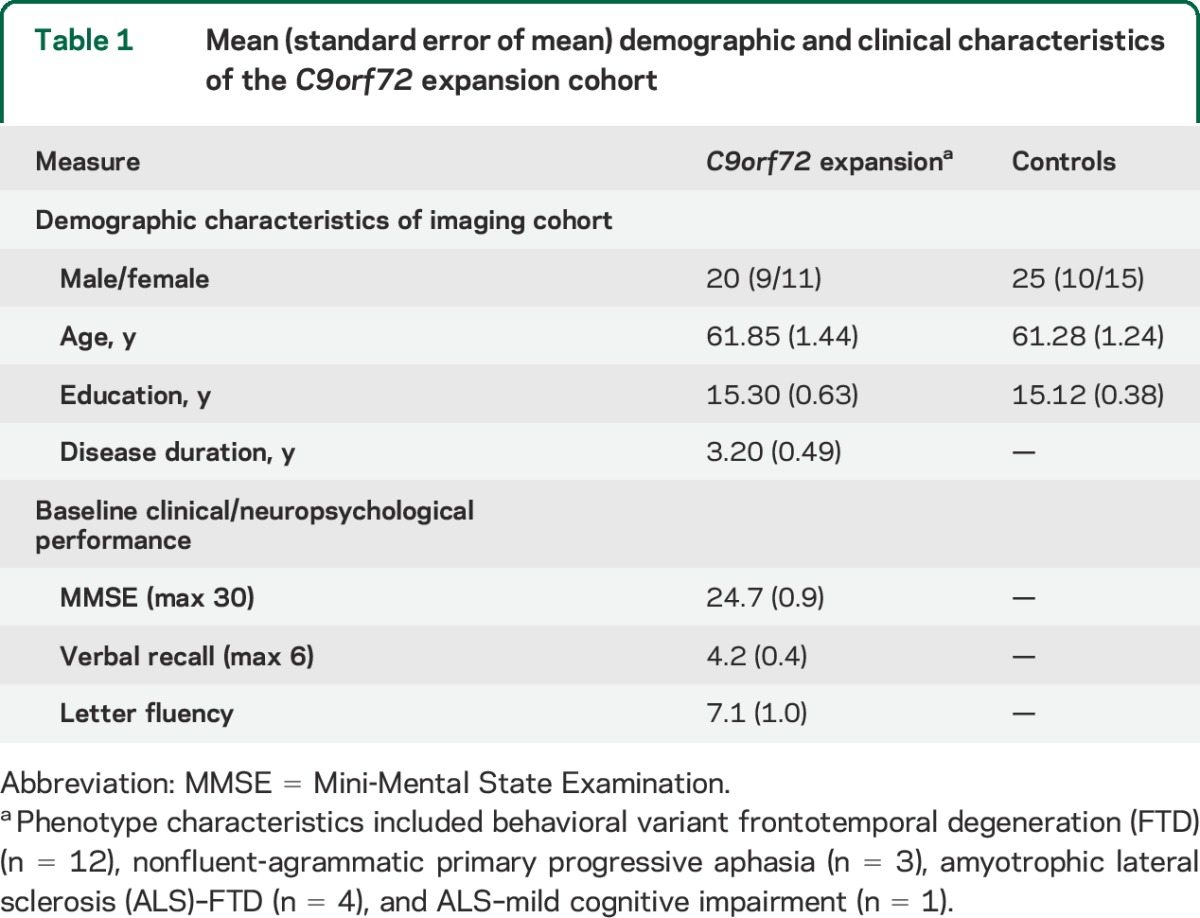
We report 20 patients recruited from the Cognitive Neurology or ALS Clinics at the University of Pennsylvania who screened positive for a C9orf72 expansion by repeat primed PCR as previously described,15 and were clinically diagnosed using a consensus procedure along with published criteria.16–18 All C9orf72 patients completed a neuroimaging study, venipuncture study, and a brief neuropsychological screening assessment during a routine clinical examination using a subset of materials from the Philadelphia Brief Assessment of Cognition.19,20 Baseline neuropsychological performance was assessed approximately 1 month (mean = 0.8 months, SEM = 1.0 months) from the baseline MRI acquisition, including (1) Mini-Mental State Examination (MMSE) (maximum score 30, prorated for motor weakness), (2) verbal recall following a delay after 3 trials of a 6-word list, and (3) category naming fluency for number of words beginning with “F” in 1 minute. One patient only had MMSE available for neuropsychological testing. We constrain our analyses to these neuropsychological tests in an effort to test regionally specific hypotheses generated from our neuroimaging and neuropathologic results (see below). Specifically, we hypothesized that hypermethylation in the hippocampus would be associated with verbal recall, hypermethylation in frontal cortex would be associated with category naming fluency, and if hypermethylation is regionally selective, it should not be associated with global impairments measured with the MMSE.
We additionally report 25 demographically comparable healthy controls who participated in a neuroimaging study (table 1), self-reported a negative history for neurologic or psychiatric disease, and completed an initial screening of MMSE >27.
Table 1.
Mean (standard error of mean) demographic and clinical characteristics of the C9orf72 expansion cohort
Standard protocol approvals, registrations, and patient consents.
All patients and controls participated in an informed consent procedure that was approved by an Institutional Review Board convened at the University of Pennsylvania.
Neuroimaging acquisition and preprocessing.
Each participant completed a volumetric T1-weighted magnetization-prepared rapid gradient echo scan acquired from a SIEMENS (Munich, Germany) 3.0 T Trio scanner with an 8-channel coil (repetition time = 1,620 ms; echo time = 3 ms; slice thickness = 1.0 mm; flip angle = 15°; matrix = 192 × 256; in-plane resolution = 0.9 × 0.9 mm). MRI volumes were preprocessed using Advanced Normalization Tools,21 as previously reported.22 A diffeomorphic deformation was used for registration to a standard local template space that is symmetric to minimize bias toward the reference space for computing the mappings, and topology-preserving to capture the large deformation necessary to aggregate images in a common space. GM probability images were calculated as a quantitative measure of GM density. Resulting images were downsampled to 2 mm3 resolution and smoothed (sigma = 2).
Neuroimaging analysis.
We performed 2 neuroimaging analyses using Randomise software implemented in FSL.23 Briefly, permutation testing is a statistical method for evaluating the relationship between a true assignment of covariates (signal) relative to many random assignments of covariates (noise). If the signal remains robust after 10,000 permutations of noise, then the null hypothesis can confidently be rejected. Our first analysis evaluated GM density within an explicit mask of GM voxels (>0.3) in C9orf72 expansion patients relative to controls to identify regional loci of disease (p < 0.0005 threshold-free cluster enhancement [TFCE]). Our second analysis evaluated the relationship between C9orf72 methylation and GM in diseased voxels, defined as suprathreshold regions from the prior analysis (p < 0.05 TFCE). This latter analysis was restricted to C9orf72 expansion patients and additionally included nuisance covariates for age and disease duration to control for other nonspecific measures that may correlate with disease.
Blood C9orf72 methylation.
Blood was collected from all patients (n = 20) approximately 4 months (mean = 4.23; SEM = 2.33) from their MRI scan. For quantitative assessment of methylation levels, we used methylation-sensitive restriction enzyme DNA digestion coupled with quantitative PCR as described previously.7,8 Briefly, 100 ng of genomic blood DNA was digested for 6 hours with 2 units of HhaI (New England BioLabs, Ipswich, MA) and 2 units of HaeIII (New England BioLabs) or only with 2 units of HaeIII followed by heat inactivation. qPCR was done using 12.5 ng of digested DNA per reaction with 2× FastStart SYBR Green Master mix (Roche Applied Science, Indianapolis, IN) using primers amplifying the differentially methylated C9orf72 promoter region (5′-CAGTGTGAAAATCATGCTTGAGAGA-3′ and 5′-TTTGTGCTTGGTAGGCAGTG-3′). Prior evidence suggests that if the CpG cleavage site measured by our assay is methylated, then the entire promoter is densely hypermethylated.7 The difference in the number of cycles to threshold amplification (ΔCt) between double vs single digested DNA was used as measure of CpG methylation using the formula %methylation = 2^(−ΔCt). Additionally, we have previously demonstrated that blood DNA methylation and brain methylation are highly correlated.7 While we did not assess methylation in the control cases of the current study, a prior analysis in an independent control sample suggests that relatives without a C9orf72 repeat expansion have a mean C9orf72 promoter methylation rate of 1.559% (SD = 1.782%).7
Neuropathologic analysis.
We generated semiquantitative ratings of neuronal loss for an independent cohort of 35 individuals with a C9orf72 expansion and TDP-43 pathology. This includes patients with a clinical diagnosis consistent with FTD (n = 10), ALS-FTD (n = 7), ALS–mild cognitive impairment (MCI) (behavioral or executive n = 6), or ALS (n = 11). Neuron loss was graded absent (0), mild (1+), moderate (2+), or severe (3+) by a neuropathologist blinded to C9orf72 methylation status. Brain cerebellum methylation was available for all of these individuals and neuronal loss ratings were available for 35 hippocampus samples, 35 frontal cortex samples, and 32 cerebellum samples. Prior evidence suggests that brain cerebellum methylation is highly correlated with methylation in frontal cortex.7 A linear mixed effects model, which is a statistical method robust to missing data, was generated with a random factor for subject and fixed factors for regional neuronal loss (hippocampus, frontal cortex), cerebellum percent methylation, an interaction term for region by percent methylation, and a nuisance group covariate to control for the presence of motor neuron disease.
Longitudinal assessments.
Follow-up MRIs were available for a subset of C9orf72 expansion patients (n = 11; behavioral variant FTD [bvFTD] = 7, nonfluent-agrammatic primary progressive aphasia [naPPA] = 2, ALS-FTD = 2) approximately 1 year (mean = 12.99 months; SEM = 1.52) from baseline MRI. To evaluate regional longitudinal changes associated with percent methylation, we assessed GM density values in the widely reported Automated Anatomic Labels template24 for anatomic regions that overlapped with our prior cross-sectional analysis. This included the right hippocampus, right thalamus, and left middle frontal gyrus. By extending our analysis to include these more comprehensive regions, we were able to evaluate longitudinal change in voxels directly associated with methylation and in neighboring voxels within the same neuroanatomical loci. For each region and for each time point, we extracted the mean GM density values, as previously reported.25 A linear mixed effects regression model assessed fixed factors for months between MRI, percent methylation, disease duration, and an interaction term for months between MRI by percent methylation. We focus on the fixed effect interaction term significance in our results since our primary hypothesized outcome is for changes in GM density as a factor of increased duration between longitudinal MRIs. We included a random factor for each participant and for each region to account for slopes associated with individual decline and variation across neuroanatomical regions. For 8 regional measurements, we observed a >2% positive change whereby GM density appeared to increase and this is likely due to noise associated with scanner variation. Therefore these items were censored from the statistical analysis. An equal number of hypomethylated and hypermethylated measurements were censored and the interaction term remains significant (p = 0.034) if these items remained in the analysis.
A subset of 10 C9orf72 expansion patients (bvFTD = 7, naPPA = 1, ALS-FTD = 2) completed a follow-up brief longitudinal neuropsychological assessment within 6–24 months (mean = 16.03, SEM = 1.99 months) of baseline examination. We assessed longitudinal change in neuropsychological performance on available tests hypothesized to be associated with our regional neuroanatomic observations, as described above. For each neuropsychological test, we performed a linear mixed effects model that included subject as a random factor and included months between assessments, percent methylation, and an interaction term for months between assessments by percent methylation. To statistically control for presence of motor impairments associated with ALS and their potential impact on cognitive performance, we included a nuisance covariate in all neuropsychological statistical models that coded for presence/absence of motor symptoms. We focus on the interaction term significance since our primary hypothesized outcome is for changes in neuropsychological performance as a factor of increased disease duration.
RESULTS
Neuroimaging.
We identified widespread reductions in GM density for C9orf72 expansion patients (n = 20) compared to controls (n = 25) (figure 1). These regions of reduced GM density include bilateral medial and lateral frontotemporal cortices along with cerebellum and medial parietal cortex.
Figure 1. Neuroimaging results.
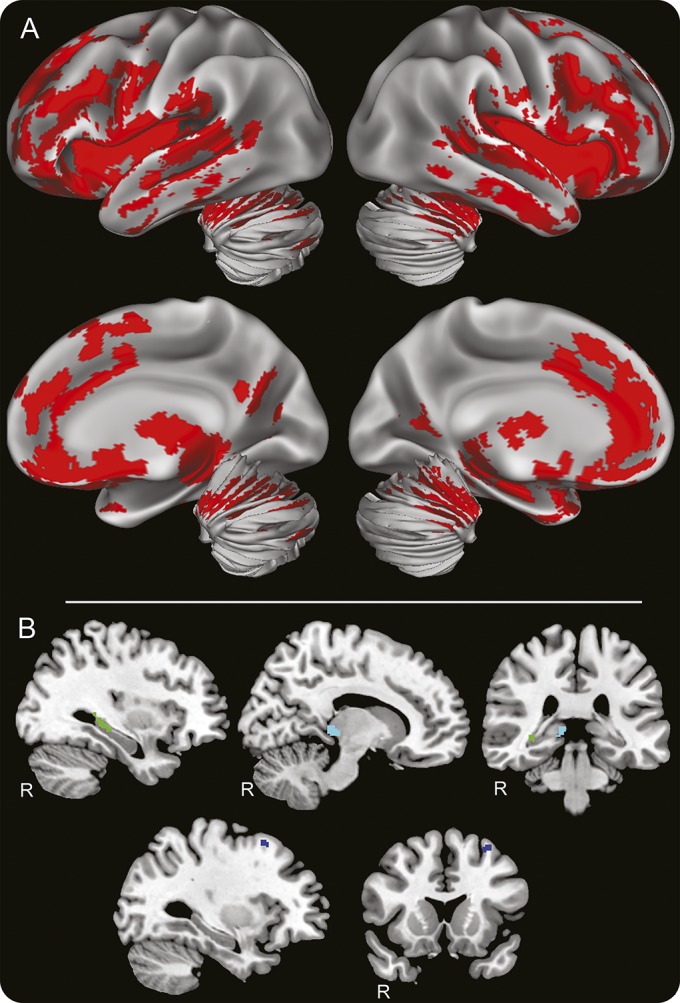
(A) Regions of significantly reduced gray matter density in C9orf72 repeat expansion patients (n = 20) relative to demographically comparable healthy controls (n = 25). (B) Regional loci protected from the C9orf72 expansion due to hypermethylation. Increased gray matter density was associated with increased C9orf72 methylation (n = 20) in the right hippocampus (green, 41 voxels, X: 30, Y: −34, Z: 0), right thalamus (cyan, 35 voxels, X: 12, Y: −31, Z: 1), and left premotor cortex region BA6 (blue, 19 voxels, X: −34, Y: 15, Z: 56). Coordinates are in Montreal Neurologic Institute stereotactic space. Right hemisphere marked with R, otherwise left hemisphere.
A nonparametric regression analysis correcting for age and disease duration at time of MRI acquisition identified 3 regional loci related to percent C9orf72 methylation (figure 1). Specifically, we observed less atrophic GM in the right hippocampus, right thalamus, and left premotor cortex associated with increased methylation. This suggested that C9orf72 hypermethylation is neuroprotective in these regions.
Neuropathologic validation.
To validate our neuroimaging observation that C9orf72 hypermethylation may be associated with neuroprotection, we evaluated whether C9orf72 hypermethylation in the cerebellum was associated with reduced neuron loss in hippocampus and frontal cortex of our independent postmortem autopsy sample of C9orf72 expansion cases (n = 35). Postmortem tissue for histologic analysis of the posterior thalamus was not available. A mixed effects model revealed significant interactions between C9orf72 cerebellum methylation and neuronal loss in the hippocampus (β = −0.058; p = 0.005) and frontal cortex (β = −0.053; p = 0.011) such that increased C9orf72 methylation was associated with reduced neuron loss in both regions (see table 2 for full statistical model and figure 2).
Table 2.
Linear mixed-effects statistical models for regional neuronal loss associated with cerebellar methylation in an independent autopsy series (n = 35); individual longitudinal decline in regional gray matter density associated with methylation (n = 11); and individual neuropsychological decline associated with methylation (n = 10)
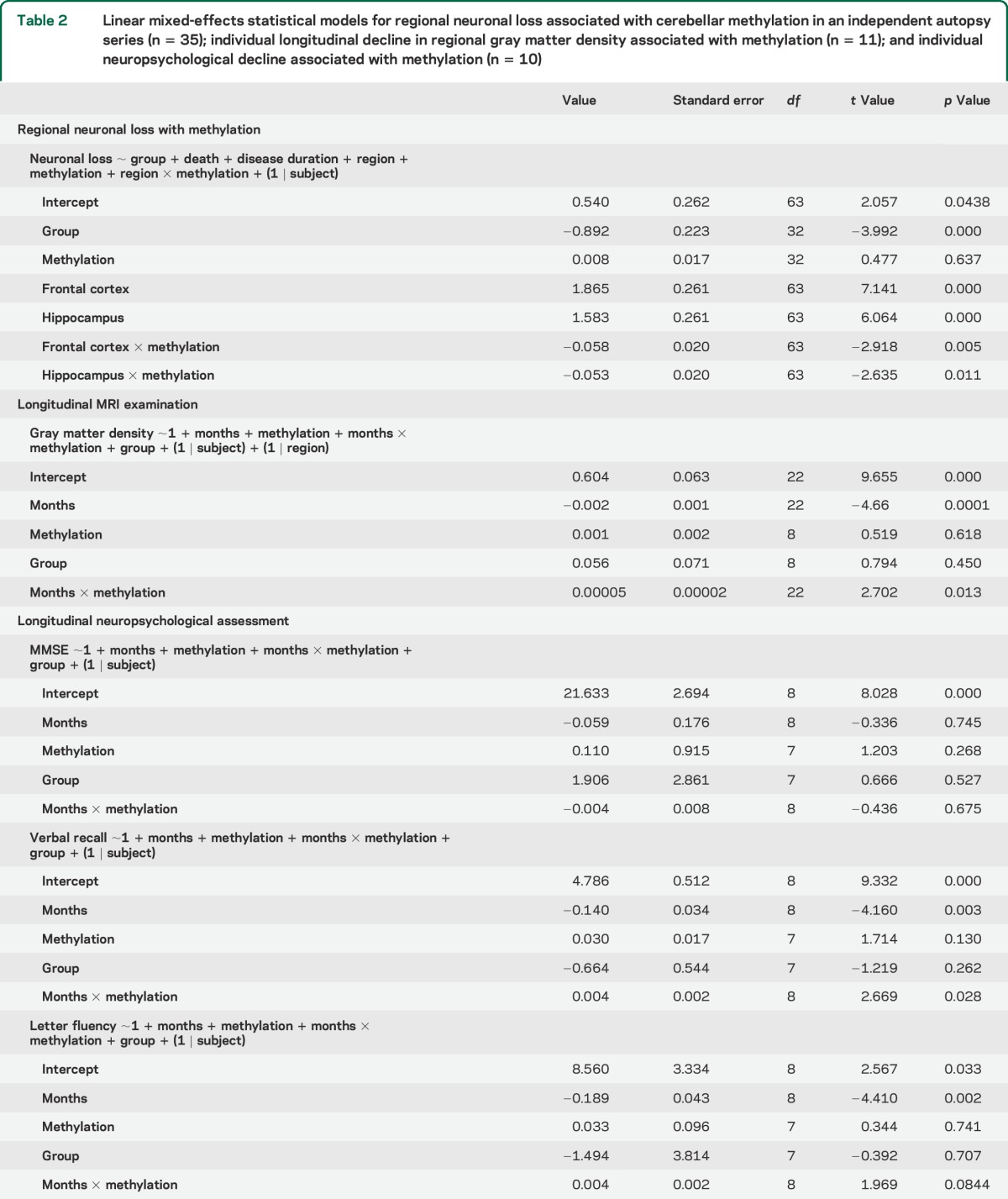
Figure 2. Neuron loss and C9orf72 methylation.
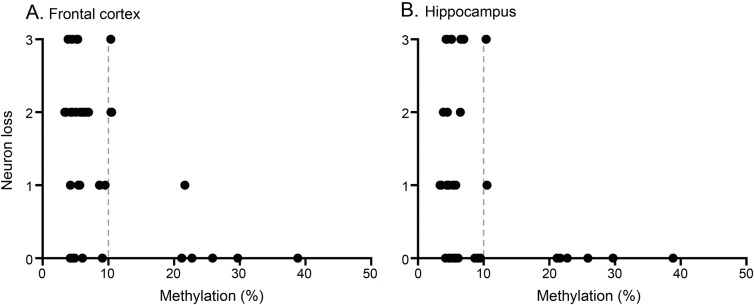
Semiquantitative neuropathology ratings of neuronal loss in the frontal cortex (A) and hippocampus (B) related to percent methylation (n = 35). Light gray bar denotes categorical boundary between hypomethylated and hypermethylated individuals.
Longitudinal results.
Together, the neuroimaging and neuropathologic results provide converging evidence suggesting that C9orf72 hypermethylation is neuroprotective of GM in the hippocampus and frontal cortex. However, this evidence is based on a single time point and could potentially be attributed to unintentional bias in the selection of time points at which individuals were assessed. We therefore performed 2 longitudinal analyses to evaluate the neuroprotective effects of hypermethylation in individuals over the disease course.
An evaluation of longitudinal GM change in right hippocampus, right thalamus, and left middle frontal cortex revealed a significant interaction demonstrating that GM atrophy progresses more rapidly over time with decreased C9orf72 methylation (β = 0.00005; p = 0.013; see table 2 for full statistical model and figure 3).
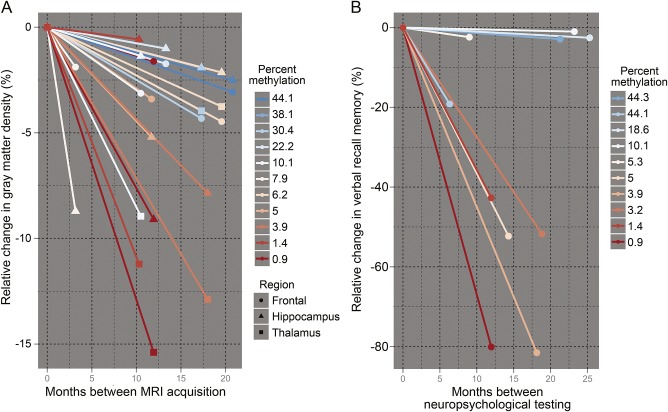
Figure 3. Relative longitudinal change in gray matter density and verbal recall.
Gray matter (GM) density (left) and verbal recall scores (right) for individual C9orf72 repeat expansion patients are shown as a function of time. For visualization purposes only, we plotted baseline GM density in each region at 0 and then plotted the relative percent change in GM density. In (A), each line represents relative GM change in one region for an individual (endpoints indicate each region being measured), and in (B), each line represents relative change in verbal recall. Line colors reflect percent C9orf72 methylation as shown in the graph legends with cold colors reflecting hypermethylated individuals (>10%) and warm colors indicating hypomethylated individuals (<10%). Cold color lines associated with hypermethylation have less of a negative slope and therefore illustrate neuroprotection.
In a second analysis, we assessed whether C9orf72 hypermethylation is neuroprotective against clinical decline by relating percent C9orf72 methylation to longitudinal neuropsychological assessments. We observed that verbal recall decline is significantly associated with hypomethylation (β = 0.004; p = 0.028; see table 2 for full statistical models), and category letter fluency decline was marginally associated with hypomethylation (β = 0.004; p = 0.084). However, global cognition measured with MMSE was not associated with methylation (β = −0.004; p = 0.675).
Exploratory subgroup results.
To evaluate if our neuroimaging and neuropsychological observations are potentially confounded by inclusion of heterogeneous clinical phenotypes, we performed several post hoc exploratory analyses. First, we evaluated whether the observed pattern of GM density was driven by a single phenotype. An analysis of variance (ANOVA) for each region revealed no significant differences in GM density across groups (naPPA, bvFTD, and FTD-ALS): thalamus (F3,16 = 1.79, p = 0.190), hippocampus (F3,16 = 1.213, p = 0.327), and frontal (F3,16 = 1.062, p = 0.393). Second, we performed an ANOVA to evaluate if C9orf72 methylation differed across phenotypes and this confirmed no difference across groups (F3,16 = 1.972, p = 0.156). While not significant, there is a trend toward higher C9orf72 methylation in naPPA (mean = 0.32) compared to bvFTD (mean = 0.18) and ALS-FTD (mean = 0.12). Therefore we repeated longitudinal analyses omitting naPPA and we still see a significant interaction between C9orf72 methylation for longitudinal decline in imaging (p = 0.045) and verbal recall (p = 0.050). Together, these exploratory findings suggest that our observations are not being driven by clinical heterogeneity in our study sample.
DISCUSSION
Molecular and neuropathologic studies have shown that C9orf72 promoter hypermethylation inhibits transcription of mutant C9orf72.6–8,26,27 We have shown that this transcriptional silencing of mutant C9orf72 is associated with reduced downstream neuropathologies, namely RNA foci and dipeptide repeat aggregates, raising the possibility that C9orf72 hypermethylation is neuroprotective.7,8 The current study provides in vivo neuroimaging and clinical evidence that hypermethylation is neuroprotective of hippocampus, frontal cortex, and thalamus, and ex vivo pathologic validation that hypermethylation is neuroprotective of hippocampus and frontal cortex.
Several prior neuroimaging studies have suggested that the regions observed in the current neuroimaging study are selectively reduced in C9orf72 expansion relative to sporadic forms of disease,9–14,28 including hippocampus,10 frontal cortex,12,13,28 and thalamus,13,14,29 among other cortical and subcortical regions. One study evaluated longitudinal neuroimaging in a small cohort (n = 6) of patients with C9orf72 expansion and observed that bilateral thalamus and left globus pallidus decline significantly more than controls, while hippocampus decline did not differ from controls.14 In the current study, we observed significant overall GM decline in hippocampus, thalamus, and frontal cortex, though we did not assess regions individually because each of these may have a different trajectory of decline. For example, studies of longitudinal GM decline associated with healthy aging suggest selective rates of decline across cortical and subcortical regions.30 Also, a neuropathologic staging study of TDP-43 in ALS suggests that there is early pathology in motor cortex (stage 1) that spreads to prefrontal cortex (stage 3) and does not reach hippocampus until later in the disease course (stage 4).31 One source of discrepancy of hippocampus decline between the prior imaging study and the current study is that group averages may obscure heterogeneous rates of decline across individuals who are hypomethylated or hypermethylated. Future studies with larger samples are warranted to better understand the association between longitudinal decline and C9orf72 methylation in each individual GM region.
Our longitudinal neuropsychological observation that C9orf72 promoter hypermethylation is neuroprotective converges with our neuroimaging findings. Hippocampus atrophy is the most widely reported source of verbal memory recall deficits, and we observed that C9orf72 promoter hypermethylation appears to be protective of decline in verbal memory recall. While reduced memory performance is not typically considered characteristic of FTD and ALS, a previous study observed that nearly all C9orf72 expansion patients in a cohort presented with an episodic memory deficit.29 Also, the hippocampus is commonly implicated as a site of pathology in C9orf72 expansion patients independent of an ALS or FTD phenotype3,32 and neuroimaging evidence suggests that the hippocampus is compromised in earliest stages (clinical dementia rating scale = 0.5) of bvFTD.33 We did not observe an association between global disease severity, measured with the MMSE, and C9orf72 promoter hypermethylation, suggesting that neuroprotection has some regional specificity. We did not assess clinical decline associated with thalamus because it is less clear what routine neuropsychological assessments are sensitive to thalamic disease. Future studies should evaluate clinical measures that capture thalamic contributions to FTD and ALS. While our neuropsychological analyses accounted for motor impairments in our patients, it will be important for larger scale studies to evaluate C9orf72 promoter hypermethylation in clinically homogeneous study cohorts.
Our neuropathologic analyses revealed regional selectivity of hippocampus and frontal cortex to hypermethylation. Specifically, we demonstrated that C9orf72 hypermethylation in the cerebellum is associated with relatively reduced neuronal loss in the hippocampus and frontal cortex, and this neuropathologic observation converges with our neuroimaging observations. Detailed neuropathologic studies have also implicated a special role for hippocampus in C9orf72 expansion. Specifically, the hippocampus is particularly vulnerable to the development of dipeptide repeat aggregates and TDP-43 aggregates in mutant C9orf72 cases.15,29,34,35 Together, the current study along with previous neuropathologic studies emphasizes the selective vulnerability of hippocampus in C9orf72 expansion cases, which corresponds to the hippocampus showing the strongest signal in terms of C9orf72 promoter hypermethylation-associated neuroprotection. Prospective studies that include collection of histologic data from the posterior thalamus, which was not evaluated in our series, are needed to validate the association between thalamic GM density and C9orf72 hypermethylation.
The current study only evaluated the neuroprotective modifying effects of C9orf72 hypermethylation, but it is possible that other genetic, epigenetic, or environmental factors also contribute to regional selectively of neuroanatomical regions. For example, recent work demonstrated several associations between neuroanatomical structure and single nucleotide polymorphisms in sporadic forms of FTD and ALS.36 Others have suggested that TMEM106B is a modifier of functional connectivity in FTD37 and that cognitive reserve mechanisms such as education may provide regionally protective modifiers of neuroanatomic structure.38
Other potential caveats to consider in the current study are related to our study population. We assessed C9orf72 promoter hypermethylation in a clinically heterogeneous cohort of individuals with some form of cognitive impairments that predominately included patients with bvFTD and additionally included patients with ALS-FTD, ALS-MCI, and naPPA. While our longitudinal neuroimaging and neuropsychological analyses revealed that the relationship between C9orf72 methylation and protection of decline was not associated with the presence or absence of motor impairment and our exploratory analyses suggest that our results were not confounded by clinical phenotype, it is possible that clinical phenotype is a contributing factor to our observed regional selectivity. While our sample size is the largest reported neuroimaging cohort of C9orf72 expansion patients to date, follow-up analyses and replication studies are required in larger and clinically homogeneous independent samples.
With these caveats in mind, we conclude that C9orf72 promoter hypermethylation is neuroprotective and regionally selective against mutant C9orf72. This observation is consistent with the growing body of evidence that demonstrates that C9orf72 hypermethylation is antagonistic to mutant C9orf72.8,26,27,39 We therefore suggest that therapeutic agents that aim to increase C9orf72 methylation or decrease C9orf72 transcription may have neuroprotective benefits in individuals with a C9orf72 expansion.
Supplementary Material
ACKNOWLEDGMENT
The authors thank John Q. Trojanowski and Virginia M.-Y. Lee for their resources and support.
GLOSSARY
- ALS
amyotrophic lateral sclerosis
- bvFTD
behavioral variant frontotemporal degeneration
- FTD
frontotemporal degeneration
- GM
gray matter
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
- naPPA
nonfluent-agrammatic primary progressive aphasia
- TDP-43
TAR DNA binding protein of 43 kDa
- TFCE
threshold-free cluster enhancement
Footnotes
Editorial, page 1616
AUTHOR CONTRIBUTIONS
Corey T. McMillan: study concept and design, acquisition of data, statistical analyses, interpretation of data, drafting of the manuscript, critical revision of the manuscript, obtained funding. Jenny Russ: acquisition of data, interpretation of data, drafting of the manuscript. Elisabeth M. Wood: acquisition of data, drafting of the manuscript. David J. Irwin: acquisition of data, interpretation of data, drafting of the manuscript, critical revision of the manuscript. Murray Grossman: acquisition of data, interpretation of data, drafting of the manuscript, critical revision of the manuscript, obtained funding. Leo McCluskey: acquisition of data, drafting of the manuscript, obtained funding. Lauren Elman: acquisition of data, drafting of the manuscript, obtained funding. Vivanna Van Deerlin: acquisition of data, interpretation of data, drafting of the manuscript, critical revision of the manuscript, obtained funding. Edward B. Lee: study concept and design, acquisition of data, statistical analyses, interpretation of data, drafting of the manuscript, critical revision of the manuscript, obtained funding.
STUDY FUNDING
Supported in part by the National Institutes of Health (AG043503, AG017586, AG039510, AG10124, AG032953) and the Wyncote Foundation. E.B.L. is supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award.
DISCLOSURE
The authors report no disclosures relevant to the manuscript. Go to Neurology.org for full disclosures.
REFERENCES
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011;72:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackenzie IRA, Frick P, Neumann M. The neuropathology associated with repeat expansions in the C9ORF72 gene. Acta Neuropathol 2014;127:347–357. [DOI] [PubMed] [Google Scholar]
- 4.Wood EM, Falcone D, Suh E, et al. Development and validation of pedigree classification criteria for frontotemporal lobar degeneration. JAMA Neurol 2013;70:1411–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boxer AL, Gold M, Huey E, et al. The advantages of frontotemporal degeneration drug development (part 2 of frontotemporal degeneration: the next therapeutic frontier). Alzheimers Dement 2013;9:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi Z, Rainero I, Rubino E, et al. Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet 2014;23:5630–5637. [DOI] [PubMed] [Google Scholar]
- 7.Russ J, Liu EY, Wu K, et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol 2015;129:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu EY, Russ J, Wu K, et al. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 2014;128:525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bede P, Bokde AL, Byrne S, et al. Multiparametric MRI study of ALS stratified for the C9orf72 genotype. Neurology 2013;81:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bede P, Elamin M, Byrne S, et al. Basal ganglia involvement in amyotrophic lateral sclerosis. Neurology 2013;81:2107–2115. [DOI] [PubMed] [Google Scholar]
- 11.Boxer AL, Mackenzie IR, Boeve BF, et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry 2011;82:196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol 2012;11:232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irwin DJ, McMillan CT, Brettschneider J, et al. Cognitive decline and reduced survival in C9orf72 expansion frontotemporal degeneration and amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2013;84:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahoney CJ, Downey LE, Ridgway GR, et al. Longitudinal neuroimaging and neuropsychological profiles of frontotemporal dementia with C9ORF72 expansions. Alzheimers Res Ther 2012;4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brettschneider J, Van Deerlin VM, Robinson JL, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol 2012;123:825–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain 2011;134:2456–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology 2011;76:1006–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strong MJ, Grace GM, Freedman M, et al. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2009;10:131–146. [DOI] [PubMed] [Google Scholar]
- 19.Libon DJ, Rascovsky K, Gross RG, et al. The Philadelphia Brief Assessment of Cognition (PBAC): a validated screening measure for dementia. Clin Neuropsychol 2011;25:1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avants BB, Libon DJ, Rascovsky K, et al. Sparse canonical correlation analysis relates network-level atrophy to multivariate cognitive measures in a neurodegenerative population. Neuroimage 2014;84:698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 2008;12:26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp 2002;15:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15:273–289. [DOI] [PubMed] [Google Scholar]
- 25.McMillan CT, Avants BB, Cook P, Ungar L, Trojanowski JQ, Grossman M. The power of neuroimaging biomarkers for screening frontotemporal dementia. Hum Brain Mapp 2014;35:4827–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L. Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res 2014;1584:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xi Z, Zinman L, Moreno D, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 2013;92:981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitwell JL, Weigand SD, Boeve BF, et al. Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 2012;135:794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahoney CJ, Beck J, Rohrer JD, et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain 2012;135:736–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raz N, Lindenberger U, Rodrigue KM, et al. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb Cortex 2005;15:1676–1689. [DOI] [PubMed] [Google Scholar]
- 31.Brettschneider J, Del Tredici K, Toledo JB, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol 2013;74:20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson YS, Barker H, Robinson AC, et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun 2014;2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitwell JL, Jack CR, Senjem ML, et al. MRI correlates of protein deposition and disease severity in postmortem frontotemporal lobar degeneration. Neurodegener Dis 2009;6:106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper-Knock J, Hewitt C, Highley JR, et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain 2012;135:751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackenzie IR, Arzberger T, Kremmer E, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol 2013;126:859–879. [DOI] [PubMed] [Google Scholar]
- 36.McMillan CT, Toledo JB, Avants BB, et al. Genetic and neuroanatomic associations in sporadic frontotemporal lobar degeneration. Neurobiol Aging 2014;35:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Premi E, Formenti A, Gazzina S, et al. Effect of TMEM106B polymorphism on functional network connectivity in asymptomatic GRN mutation carriers. JAMA Neurol 2014;71:216–221. [DOI] [PubMed] [Google Scholar]
- 38.Premi E, Gazzina S, Bozzali M, et al. Cognitive Reserve in granulin-related frontotemporal dementia: from preclinical to clinical stages. PLoS One 2013;8:e74762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belzil VV, Bauer PO, Prudencio M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 2013;126:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.