SUMMARY
Gene regulatory networks (GRNs) comprising interactions between transcription factors (TFs) and regulatory loci control development and physiology. Numerous disease-associated mutations have been identified, the vast majority residing in non-coding regions of the genome. As current GRN mapping methods test one TF at a time and require the use of cells harboring the mutation(s) of interest, they are not suitable to identify TFs that bind to wild type and mutant loci. Here, we use gene-centered yeast one-hybrid (eY1H) assays to interrogate binding of 1,086 human TFs to 246 enhancers, as well as to 109 non-coding disease mutations. We detect both loss and gain of TF interactions with mutant loci that are concordant with target gene expression changes. This work establishes eY1H assays as a powerful addition to the toolkit of mapping human GRNs and for the high-throughput characterization of genomic variants that are rapidly being identified by genome-wide association studies.
INTRODUCTION
Gene regulatory networks (GRNs) comprising physical and functional interactions between transcription factors (TFs) and regulatory elements play a critical role in development and physiology (Davidson et al., 2002; Walhout, 2006). Consequently, inappropriate gene regulation underlies a variety of human diseases. A broad variety of disease-associated mutations have been uncovered, including mutations in TF-encoding genes as well as mutations in non-coding sequences such as enhancers and promoters. Importantly, ~90% of disease-associated variants identified by genome-wide association studies (GWAS) reside in the non-coding part of the genome (Hindorff et al., 2009; Maurano et al., 2012), and a main challenge is to determine the interactions with TFs that may be perturbed as a consequence of such mutations.
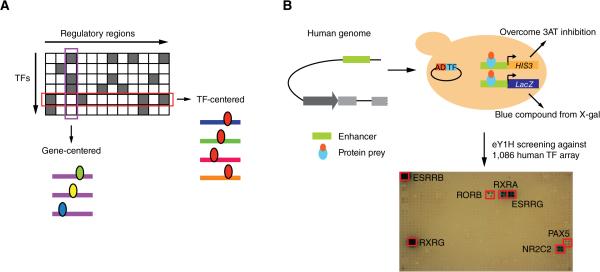
TF-DNA interactions can be mapped with either ‘TF-centered’ (protein-to-DNA) or ‘gene-centered’ (DNA-to-protein) methods (Figure 1A) (Arda and Walhout, 2009; Deplancke et al., 2006). Chromatin immunoprecipitation (ChIP) is the most widely used TF-centered method to identify the DNA regions with which a TF interacts in vivo. The last decade has seen an explosion of ChIP data. While progress has been impressive, several challenges remain. First, even for large consortia such as ENCODE, ChIP data have been generated for only ~150 of the ~1500 human TFs (Gerstein et al., 2012). This is because ChIP critically depends on suitable anti-TF antibodies, which are only available for a minority of human TFs. Second, each TF has been assayed only in a limited number of cell lines and conditions. Third, ChIP may work better for some TFs than for others. For instance, TFs with restricted expression patterns and/or expressed at low levels may be less amenable to ChIP compared to highly and broadly expressed TFs. Fourth, ChIP is not optimal for characterizing disease-relevant mutations in large-scale, high-throughput settings, because it requires disease cells or tissues that harbor the relevant mutation, which may be difficult to obtain. Finally, ChIP cannot be used to identify TFs with altered binding to mutant regulatory regions ab initio because the method is TF-centered and as a result one needs to first identify candidate TFs, and then test these one at a time.
Figure 1. Gene-Centered Yeast One-Hybrid Assays.
(A) Gene-centered versus TF-centered approaches for mapping protein-DNA interactions. Rectangles – regulatory regions; ellipses – TFs.
(B) Cartoon of eY1H assays. A DNA sequence of interest is cloned upstream of two reporter genes (HIS3 and LacZ) and integrated into the yeast genome (i.e., each DNA bait is tested in duplicate by activation of each reporter in the same yeast nucleus). The resulting yeast DNA bait strain is mated to a collection yeast strains harboring TFs fused to the Gal4 activation domain (AD). Positive interactions are determined by the ability of the diploid yeast to grow in the absence of histidine and overcome the addition of 3AT a competitive inhibitor of the HIS3 enzyme, and turn blue in the presence of X-gal. Each TF is tested in quadruplicate. Red boxes show positive interactions.
Enhanced yeast one-hybrid (eY1H) assays provide a gene-centered method for the detection and identification of TF-DNA interactions (Reece-Hoyes et al., 2011b); (Arda et al., 2010; Brady et al., 2011; Fuxman Bass et al., 2014; Martinez et al., 2008; Reece-Hoyes et al., 2013). Briefly, eY1H assays measure TF-DNA interactions in the milieu of the yeast nucleus. DNA regions to be assayed (DNA baits) are fused upstream of two reporter genes, LacZ and HIS3, and integrated into the yeast genome, enabling their incorporation into chromatin. TFs (preys) are introduced into the DNA bait strains by mating using a robotic platform, and are tested in quadruplicate, providing an inherent interaction retest (Figure 1B).
Here, we test our human eY1H platform (Reece-Hoyes et al., 2011a) to identify TFs interacting with human enhancers and to determine protein-DNA interaction changes caused by mutant TFs as well as non-coding disease-assoiated mutations. We find that eY1H assays more effectively retrieve TFs with limited expression patterns or levels when compared to ChIP. We provide examples of functional models of target sharing by TFs, including redundancy, which may provide robustness, and opposing function (activation vs repression), which can ascertain proper timing of enhancer activity during development. Finally, we demonstrate that eY1H assays can be effectively used to identify changes in TF binding conferred by disease-associated coding or non-coding mutations.
RESULTS
A Gene-Centered Human TF-Enhancer Interaction Network
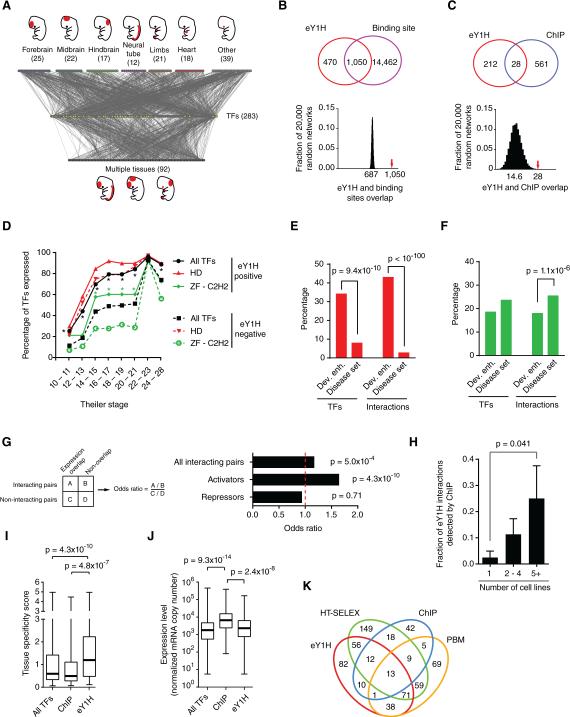
We first focused on a set of human developmental enhancers that were previously tested for embryonic activity in mouse transgenic assays at day E11.5 (Table S1) (Visel et al., 2007). We expanded the human eY1H platform to 1,086 full-length human TFs (76% of all 1,434), and examined interactions for 360 enhancers. Thus, in total we tested 390,960 putative TF-DNA interactions. To ensure the technical quality of the data, we only considered interactions in which both eY1H reporters and at least two of the four colonies of a TF quadrant tested positively. For the majority of cases, all four colonies score positively. Limitations and advantages of eY1H data are discussed extensively below. The resulting TF-enhancer interaction network contains 2,230 interactions between 246 enhancers and 283 TFs (Figures 2A and Table S2).
Figure 2. A Human Gene-Centered TF-Enhancer Interaction Network.
(A) The TF-enhancer interaction network comprises 2,230 interactions between 246 human developmental enhancers and 283 TFs. Enhancers that are active in a single tissue at day E11.5 (top nodes) or multiple tissues (bottom nodes) are connected to the TFs (middle yellow nodes) with which they interact.
(B, C) eY1H interactions significantly overlap with the occurrence of known TF binding sites (B) and ChIP peaks (C). The Venn diagrams on the left illustrate the number of overlapping interactions. The eY1H network was randomized 20,000 times by edge switching (Martinez et al., 2008) and the overlap in each randomized network was calculated (right panel). The numbers under the histogram peaks indicate the average overlap in the randomized networks. The red arrows indicate the observed overlap in the real network.
(D) Timing of expression during mouse development for homeodomain (HD) and ZF-C2H2 families. The fraction of TFs whose expression was detected at a particular Theiler Stage during development. *p < 0.01 by Fisher's exact test.
(E, F) Percentage of TFs or interactions involving homeodomains (E) or ZF-C2H2 TFs (F) for two datasets. Statistical significance determined by proportion comparison test.
(G) Overlap between enhancer activity and TF expression pattern. The fraction of TF-enhancer pairs that overlap in expression was compared between interacting and non-interacting pairs. The same analysis was performed for known activators and repressors. Statistical significance was determined using Fisher's exact test.
(H) The fraction of eY1H interactions that were also detected by ChIP were partitioned based on the number of cell lines in which a particular TF was tested by ChIP. p = 0.041 by Mann-Whitney's U test.
(I) Tissue specificity score for TFs detected by eY1H (n = 266), ChIP (n = 96) or all TFs present in the eY1H array (n = 896), based on their expression levels across 34 tissues (Ravasi et al., 2010). This score quantifies the departure of the observed TF expression pattern from the null distribution of uniform expression across all tissues, using relative entropy. Each box spans from the first to the third quartile, the horizontal lines inside the boxes indicate the median value and the whiskers indicate minimum and maximum values. Statistical significance determined by Mann-Whitney's U tests.
(J) The maximum expression level across 34 tissues were obtained from (Ravasi et al., 2010) for each TF detected eY1H (n = 266), ChIP (n = 96) or all TFs present in the eY1H array (n = 896) are plotted. Each box spans from the first to the third quartile, the horizontal lines inside the boxes indicate the median value and the whiskers indicate minimum and maximum values. Statistical significance determined by Mann-Whitney's U tests.
(K) Venn diagram depicting the overlap between TFs detected by eY1H and those detected by high-throughput SELEX (HT-SELEX), ChIP-seq and protein binding microarrays (PMBs).
We ascertained the quality of the network using several different metrics. First, we observed a statistically significant overlap between eY1H interactions and the presence of TF binding sites, which indicates that most interactions are likely direct (Figure 2B, p<0.0001). Second, we found a statistically significant overlap between eY1H and ENCODE ChIP interactions (Gerstein et al., 2012)(Figure 2C, p<0.0001). Third, TFs that interact with developmental enhancers are enriched for those expressed early in development, which is consistent with the timing of enhancer activity (Figure 2D). Finally, the network is enriched for homeodomain TFs, well-known regulators of developmental gene expression (Chi, 2005) (Figure S1). This enrichment is specific for the developmental enhancers because we did not observe it with the eY1H dataset related to disease mutations that is discussed below (Figure 2E and S1). The network is depleted for interactions involving ZF-C2H2 TFs (Figure 2F and S1), and, consistently, these TFs are overall expressed at later stages in development. Importantly, however, ZF-C2H2 TFs that do interact with the developmental enhancers are expressed at earlier stages than to those that do not (Figure 2D).
To further assess the quality of the network, we reasoned that if a TF truly binds an enhancer in vivo, the TF would be expressed at the same time and place where the enhancer is active. Indeed, we found a modest but significant overlap between enhancer activity and spatiotemporal TF expression (Figure 2G). We wondered if the enrichment was relatively modest because the TFs identified in eY1H assays are a collection of transcriptional activators and repressors. We observed a more striking enrichment for known transcriptional activators, while repressors are not enriched (Figure 2G, Table S3). Altogether, these findings provide general support for the quality of the human TF-enhancer interaction network.
eY1H Assays Provide a Powerful Addition to the GRN Mapping Toolkit
Several of our findings demonstrate that eY1H assays are complementary to other TF-DNA interaction mapping methods. For instance, we found that the fraction of eY1H interactions also detected by ChIP is larger for TFs that have been assayed by ChIP in multiple cell lines (Figure 2H). This underscores that ChIP in a given tissue/cell line only uncovers a subset of interactions in which the relevant TF engages, while eY1H assays interrogate the available repertoire of TFs for a given enhancer in a single experiment. Further, TFs that interact with developmental enhancers in eY1H assays exhibit more tissue-specific expression compared to all TFs tested or to TFs assayed by ChIP (Figure 2I). In addition, TFs assayed by ChIP are expressed at higher levels than those detected in eY1H assays (Figure 2J). These observations indicate that each method has particular strengths of detecting certain types of TFs. Indeed, we detected interactions for 82 TFs that had not been detected by any other high-throughput method (Figure 2K)(Badis et al., 2009; Jolma et al., 2013).
The TF-Enhancer Network Reveals Functional Relationships between TFs
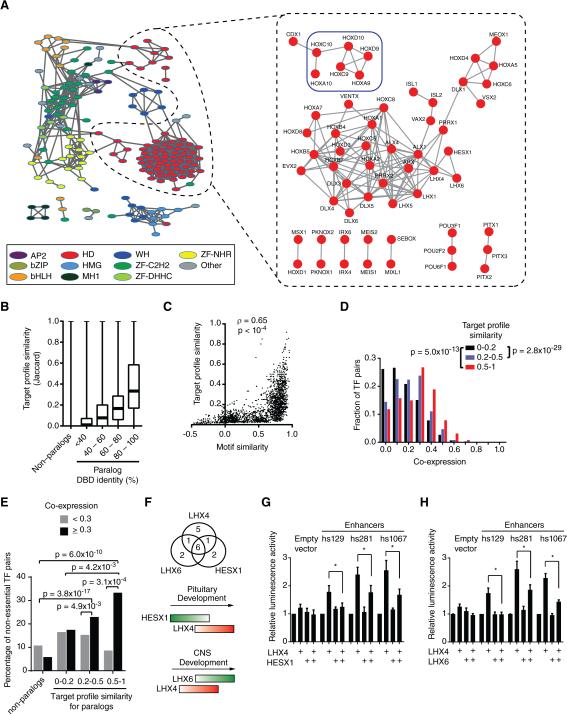
In eY1H assays, DNA baits often interact with multiple members of the same TF family (Reece-Hoyes et al., 2013). This is likely because such TFs have similar DNA binding domains and recognize similar DNA sequences (Badis et al., 2009; Grove et al., 2009; Weirauch et al., 2014). To visualize enhancer sharing by TFs, we calculated the target profile similarity for each pair of TFs: i.e., the number of overlapping enhancer targets relative to the number of targets that interact with either TF (Fuxman Bass et al., 2013). We delineated a TF association network in which TFs with target profile similarity ≥0.2 are connected (Figures 3A and S2). As expected, TFs generally cluster by family. Further, there is a significant correlation between DNA binding domain identity, DNA motif similarity and target profile similarity (Figures 3B, 3C and S3). However, similar to our observations in C. elegans, there are many examples of TF pairs with high DNA binding motif similarity but low target profile similarity (Figure 3C) (Reece-Hoyes et al., 2013).
Figure 3. TF Redundancy and Opposing Functions.
(A) TF association network. Each node represents a TF and edges connect TFs with a target profile similarity ≥0.2 (left, all TF families) or ≥0.45 (right, homeodomains). TFs with degree ≥ 3 in the eY1H network are shown. Node color indicates TF families. Colored squares highlight sets of TFs discussed in main text. AP2 – activating protein 2; bZIP – Basic Leucine Zipper Domain; bHLH – basic helix-loop-helix; HD – homeodomain; HMG – High-Mobility Group; MH1 – Mad homology 1; WH – Winged Helix; ZF-C2H2 – Zinc Finger C2H2; ZF-DHHC – Zinc Finger DHHC; ZF-NHR – Nuclear Hormone Receptor.
(B) Target profile similarity between TFs according to DNA binding domain identity. For each pair of TF paralogs with different DNA binding domain amino acid identity their target profile similarity was determined. Each box spans from the first to the third quartile, the horizontal lines inside the boxes indicate median value and the whiskers indicate minimum and maximum values. All pairwise comparisons between groups are significant (p < 0.01) by Dunn's multiple comparison test.
(C) Correlation between motif similarity and target profile similarity. For each TF pair target profile similarity was plotted against their DNA motif similarity determined as the Pearson correlation coefficient of the Z-scores obtained for all possible 8-mers in protein binding microarrays.
(D) Histogram of spatiotemporal co-expression for TF pairs according to their target profile similarity. Statistical significance determined by Mann-Whitney's U tests.
(E) Redundancy between TFs. Each pair of TF paralogs was binned according to their target profile similarity and according their spatiotemporal co-expression. The percentage of TF-pairs for which both TF knockouts are viable was determined. Statistical significance was determined using the proportion comparison test.
(F) Top: overlap between enhancers bound by LHX4, LHX6 and HESX1. Bottom: cartoon of developmental expression. Red – transcriptional activator; green – transcriptional repressor.
(G) HESX1 represses LHX4-induced enhancer activity. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. After 48 hrs, cells were harvested and luciferase assays were performed. Relative luminescence activity is plotted as fold change compared to cells co-transfected with control vector expressing GFP. Experiments were performed three times in 3-6 replicates. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
(H) LHX6 represses LHX4-induced enhancer activity. Experiments were performed three times in 3-6 replicates. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
See also Figures S2 and S3.
The sharing of enhancers by paralogous TFs begs the question of whether only one of these actually interacts with that DNA fragment in vivo, or if there could be a biological explanation for enhancer sharing between TFs. Conceptually, there are several possibilities. First, two TFs may share enhancers in the same tissue at the same time to provide redundancy that can lead to robustness of enhancer function when one TF is genetically or environmentally perturbed (MacNeil and Walhout, 2011). Second, TFs may bind the same enhancer, but in different tissues, or at different developmental times. Finally, TFs that share enhancers could have opposing regulatory effects where one activates and the other represses transcription, for instance at different developmental stages.
There are several examples of redundancy between TF paralogs. For instance, three ETS TFs share targets in human T cells and function redundantly (Hollenhorst et al., 2007). If redundancy is prevalent in human GRNs, one would expect that TFs that share targets would also tend to be co-expressed. Indeed, TFs that bind to highly overlapping sets of enhancers are generally more co-expressed than TFs that bind different enhancers (Figure 3D). An example is a group of six redundant Abdominal-B (Abd-B) HOX TFs (Maconochie et al., 1996) that bind a highly overlapping set of enhancers in eY1H assays (Figure 3A, blue box). These TFs are also highly co-expressed and neither of them is essential for viability, although overall 60% of TFs in the network confer lethality when knocked out in mouse. Importantly, TF pairs with both high target profile similarity and high co-expression similarity are overall enriched for pairs in which both TFs are non-essential (Figure 3E). Altogether, these results suggest a potentially widespread redundancy between TFs.
TFs that share a large proportion of targets could have opposing functions if one is an activator and the other is a repressor. For instance, the activator LHX4 and two repressors, LHX6 and HESX1, share a large proportion of enhancers (Figure 3F). HESX1 and LHX6 can both repress activation by LHX4 in transient transfection assays (Figures 3G, H). LHX4 is expressed after HESX1 in the developing pituitary and before LHX6 in the developing central nervous system (Figure 3F), suggesting that HESX1 may prevent precocious activation by LHX4 in the developing pituitary, while LHX6 repression prevents prolonged activation by LHX4 in the developing central nervous system. Thus, the network can identify TF pairs that bind to similar targets but that may have opposing functions in target gene regulation. This may be crucial to tightly control the expression of particular programs during development. Altogether, these data indicate that multiple TFs from the same family found to interact with overlapping sets of enhancers in eY1H assays may be relevant in vivo, and can provide different gene regulatory functionalities.
Human Disease and TF Network Connectivity
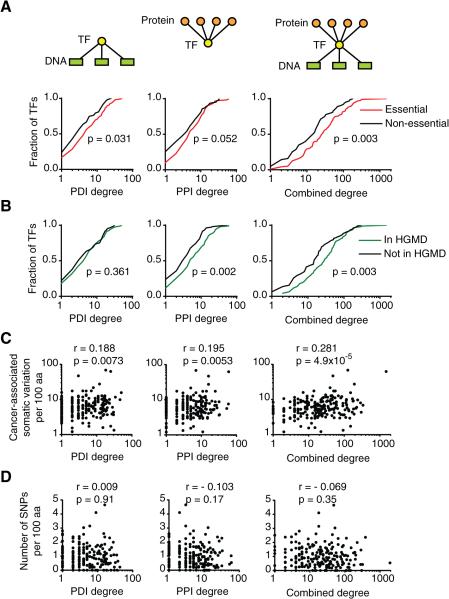
Mutations in TF coding sequences can cause a variety of diseases that could impact GRNs in different ways. Some mutations may abrogate DNA binding completely while others affect binding to only a subset of targets. We hypothesized that mutations in TFs that bind a large set of targets are more likely to affect an important biological function. It has been shown previously that TFs that bind to many promoters in C. elegans are more frequently essential for viability than TFs that only bind a few promoters (Deplancke et al., 2006). Similarly, protein-protein interaction hubs are more frequently essential (Goh et al., 2007). Interestingly, a combined protein-protein and protein-DNA interaction degree was more strongly associated with phenotypic output for human TFs than either degree alone. Essential and disease-associated TFs have a higher combined degree than non-essential TFs (Figure 4A, B). In addition, there is a significant correlation between combined TF degree and the density of somatic mutations in cancer (Figure 4C). This is specific to somatic mutations as no correlation was observed between TF degree and the density of protein altering variants in healthy individuals from the 1000 Genomes Project (Genomes Project et al., 2010)(Figure 4D). In sum, mutations in highly connected TFs more frequently affect phenotypic outcomes leading to disease.
Figure 4. Relationship Between TF Connectivity and Human Disease.
(A) Cumulative distribution of TF protein-DNA interaction (PDI), protein-protein interaction (PPI) and combined degrees for essential and non-essential TFs. Combined TF degree is defined as the product of PPI and PDI degrees and represents the number of paths connecting the protein interactors of a TF with its DNA targets. Statistical significance determined by Mann-Whitney's U tests.
(B) Cumulative distribution of TF degrees for TF reported as disease-associated genes in the Human Gene Mutation Databse (HGMD) and genes not reported in HGMD. Statistical significance determined by Mann-Whitney's U tests.
(C) Correlation between TF degree and the number of protein-altering SNPs and short indel variants per 100 amino acids in cancer samples obtained from the Catalogue of Somatic mutations in Cancer (COSMIC). Statistical significance was determined using Pearson correlation coefficient.
(D) Correlation between TF degree and the number of protein-altering SNPs and short indel variants per 100 amino acids in the 1000 genomes project. Statistical significance was determined using Pearson correlation coefficient.
Disease-Associated TF Coding Mutations
Both coding (in TFs) and non-coding (in regulatory DNA elements) mutations can cause human disease, likely by changing target gene expression in trans or cis, respectively. Such disease-associated mutations can potentially affect GRNs by: (1) complete loss of all TF-DNA interactions, (2) loss of a subset of interactions, (3) gain of interactions, or (4) a combination of interaction loss and gain. However, because no suitable methods were available to discriminate between these possibilities it remains unclear, in the vast majority of cases, which TF-DNA interactions are lost or gained as a result of specific mutations.
We hypothesized that eY1H assays would be highly suitable to interrogate differential TF binding caused by disease-associated mutations because: (1) mutations can readily be introduced in DNA baits or TF preys by molecular cloning, circumventing the need for patient samples harboring mutant TFs or mutant regulatory sequences; (2) eY1H assays enable direct, unbiased comparisons between wild type and mutant TFs, and (3) eY1H assays test all available TFs in parallel in one experiment, enabling the direct determination of differential TF binding to mutant regulatory DNA sequences.
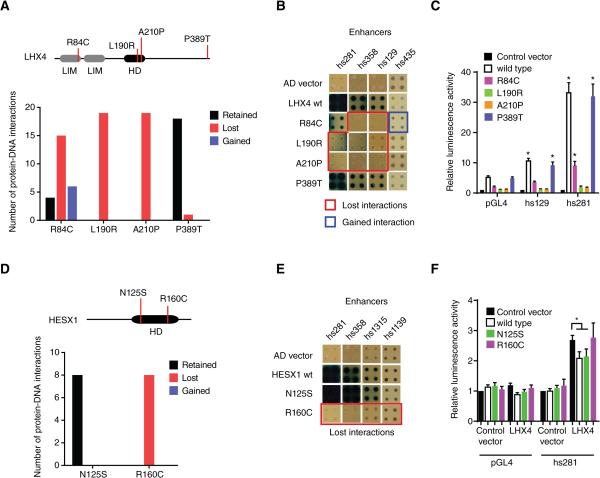
To determine the impact of TF coding mutations on enhancer binding, we first focused on four mutant LHX4 TFs that confer pituitary hormone deficiency (Pfaeffle et al., 2008; Tajima et al., 2007)(Figure 5A). The P389T mutation is located outside the DNA binding domain and this mutant retains most (18 of 19) protein-DNA interactions (Table S4). However, two mutations in the homeodomain (L190R and A210P) result in complete loss of interactions, which is consistent with previous in vitro experiments (Pfaeffle et al., 2008) (Figures 5A, 5B and Table S4). Interestingly, we detected partial loss and gain of weak interactions caused by the R84C mutation, which is located in the LIM domain and is known to modulate DNA binding (Pfaeffle et al., 2008). These results were further confirmed with luciferase assays in which we found changes in the transcriptional activation capacity that correlate with changes in DNA binding (Figure 5C).
Figure 5. Disease-Associated Coding Mutations in TFs.
(A) Four missense mutations in LHX4 were tested for loss or gain of protein-DNA interactions in eY1H assays against 152 enhancers. The top panel depicts a cartoon of LHX4, including the location of the mutations and the homeodomain (HD) and LIM domains. The bottom panel shows the number of interactions retained (black bar), lost (red bar) or gained (blue blue) for each mutant compared to wild type interactions.
(B) Examples of interactions lost and gained for LHX4 missense mutations. Each TF-enhancer combination was tested in quadruplicate three times. One random quadruplicate test is shown corresponding to four enhancers. Red squares – interaction lost with TF mutant; blue square – retained interaction with TF mutant; AD vector – empty prey vector.
(C) Transcriptional activation mediated by wild type and mutant LHX4 alleles. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. Relative luminescence activity is plotted as fold change compared to cells co-transfected with empty expression vector. Experiments were performed four times with three replicates each. Average relative luminescence activity ± SEM is plotted. *p<0.05 vs empty expression vector by Student's t-test.
(D) Two missense mutations in HESX1 were tested for changes in protein-DNA interactions as in (A).
(E) Examples of interactions lost for HESX1 missense mutations.
(F) Repression of LHX4-induced enhancer activity by wild type and mutant HESX1 alleles. HEK293T cells were co-transfected with enhancer constructs cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. Relative luminescence activity is plotted as fold change compared to cells co-transfected with control vector expressing GFP. Experiments were performed six times with three replicates each. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
See also Table S4.
We also evaluated two missense mutations in the homeodomain of HESX1: the R160C mutation leads to septo-optic dysplasia (Dattani et al., 1998), whereas the N125S variant is a natural polymorphism in the Afro-Caribbean population (Brickman et al., 2001) that is not associated with disease. Interestingly, the R160C mutation completely abolishes all interactions, while the N125S variant has a wild type target profile (Figures 5D, 5E and Table S4). Wild type and the N125S variant of HESX1 repressed reporter gene expression in transient transfection assays while the R160C mutant did not, further confirming our findings (Figure 5F). Altogether, these data show that eY1H assays can be effectively used to determine the consequences of TF-coding mutations on DNA target binding.
Non-coding Mutations Associated with Human Disease
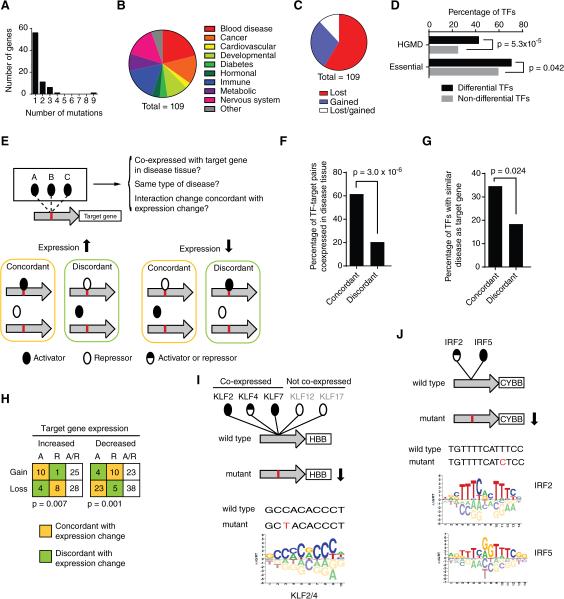
To determine the effect of non-coding mutations on TF binding, we selected 227 disease-associated mutations, affecting the expression of 137 genes. We identified interacting TFs for both wild type and mutant clones of each regulatory element with eY1H assays and detected differential TF binding for 109 mutations (75 genes) associated with a variety of diseases (Figure 6A, 6B and Table S5). Literature searches indicate that 66 of these mutations result in an increase while 39 confer a decrease in expression of the associated target gene (for four mutations the effect on gene expression is not known, Table S5). The majority of mutations resulted in interaction loss (64 of 109, or 59%). Remarkably, however, 32 mutations resulted in gain of interactions (29%) and 13 caused both interaction loss and gain (12%) (Figure 6C). Thus, gain of TF interactions may be a pervasive disease-causing mechanism. Overall, these mutations affect interactions with 111 TFs from all major families (Table S5). Strikingly, TFs involved in differential interactions are more frequently essential for viability and/or annotated as disease-associated in HGMD compared to TFs that are not involved in differential interactions (Figure 6D).
Figure 6. Disease-Associated Non-Coding Mutations.
(A) Number of mutations per gene for which differential TF interactions were detected by eY1H assays.
(B) Distribution of diseases associated with tested non-coding mutations.
(C) Distribution of mutations that result in loss of interactions, gain of interaction or both.
(D) Fraction of essential or disease-associated TFs (per HGMD) differentially interacting with non-coding mutations (differential TFs) and the remaining TFs in the eY1H human TF collection (non-differential TFs). Statistical significance determined by proportion comparison test.
(E) Cartoon depicting data integration used to obtain a supporting evidence score for differential eY1H interactions (see Extended Experimental Procedures).
(F, G) Percentage of differential TF-target gene pairs in which the TF is co-expressed with the target gene in the disease tissue (F) or is associated with a similar disease or mouse phenotype (G) for interaction changes concordant or discordant with target gene expression changes. Statistical significance determined by proportion comparison test.
(H) Number of interactions lost or gained involving activators (A), repressors (R) or bifunctional TFs (A/R, activators and repressors) for mutations that cause increased or decreased target gene expression. Only interactions in which the TF is co-expressed with the target gene in disease relevant tissue, or associated with a similar disease or phenotype are shown. Statistical significance was determined using Fisher's exact test.
(I, J) Examples of differential eY1H interactions with HBB promoter (I) and the promoter of the CYBB gene (J). Disease-associated mutations are indicated in red. Reported TF binding site logos are shown (Weirauch et al., 2014).
To validate the differential eY1H interactions, we first compared them to published differential interactions. Out of 227 mutations tested, 54 had reported differential interactions that were experimentally supported by reporter assays, in vitro binding assays and/or by ChIP. For 34 of these the reported TF was either absent from our collection (19 mutations) or was never detected in eY1H assays (15 mutations). We detected the reported differential interaction for four of the remaining 20 mutations and differential interaction with a close TF paralog for three additional mutations (Table S6). Importantly, the TFs detected by eY1H assays were not tested in the latter three studies, even though they have similar DNA binding specificity and are expressed in the relevant disease tissue (Table S5). Hence, it could be that either or both TF(s) contribute to the disease in vivo.
Next, we devised a ‘supporting evidence score’ (see Extended Experimental Procedures) for each interaction involving a mutant regulatory element, in which we weighted the interactions according to: (1) co-expression of the differentially interacting TF and the target gene in disease-relevant tissues; (2) if the differentially bound TF is associated with a similar disease or mouse phenotype as the target gene mutation; and (3) if the target gene expression change caused by the mutation (increase or decrease) was concordant with gain/loss of a protein-DNA interaction with an activator/repressor (Figure 6E). It is of course important to note that the data used in this integration are not yet complete and have their own confidence issues. Out of the 294 differential interactions (with 109 non-coding mutants), 98 have a medium/high to high level of confidence (Table S5). Importantly, the differential interactions involving TFs expressed in disease-relevant tissues and/or associated with a similar disease are generally consistent with changes in target gene expression: an increase in expression is concordant with gain of interaction with an activator or loss of an interaction with a repressor, while a decrease in expression correlates with gain of interaction with a repressor or loss of interaction with an activator (Figure 6F-H). All differential interactions, gene expression changes, as well as expression and disease information are provided in Table S5.
Several mutations cause differential interactions with multiple TFs, often from the same family. Two examples illustrate how such interactions can be evaluated for in vivo relevance. The first example involves a C to T mutation in the beta globin gene promoter that results in reduced gene expression leading to thalassemia. This mutation results in loss of interactions with five paralogous TFs: KLF2, KLF4, KLF7, KLF12 and KLF17, that bind similar DNA sequences (Figure 6I). Two of these paralogs, KLF2 and KLF4, are more likely involved than the other three TFs, because they are expressed in erythroid cells and have been shown to activate beta globin gene expression (Alhashem et al., 2011; Gardiner et al., 2007). The second example involves a T to C mutation in the CYBB promoter that causes a reduction in expression leading to chronic granulomatous disease. eY1H assays identified loss of binding for IRF2 and IRF5, both of which are expressed in disease-relevant cells. Again the mutation occurs in the binding site of these TFs (Figure 6J). IRF2 has been shown to activate the CYBB promoter (Luo and Skalnik, 1996) and, therefore, it is likely that loss of this interaction is most relevant to the disease. However, IRF5 cannot be entirely excluded because these two TFs may share targets in vivo as discussed above for the developmental enhancers.
Dominant Mutations in the Sonic Hedgehog ZRS Enhancer
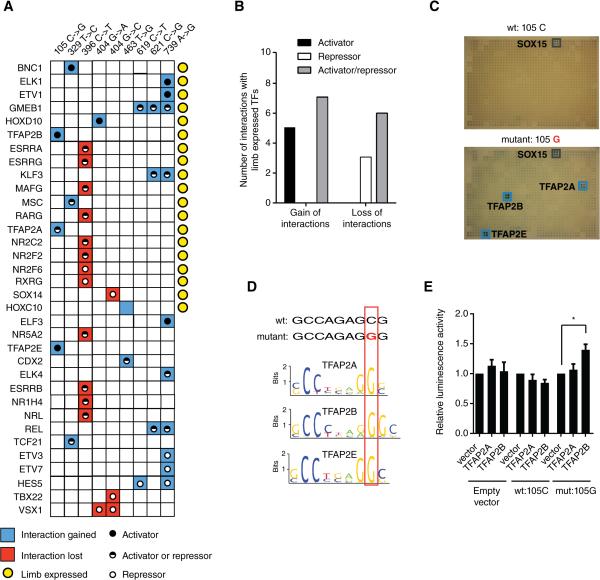
We identified differential interactions for nine dominant mutations in the ZRS enhancer of SHH that result in ectopic gene expression along the anterior margin of the limb bud, causing digit malformations and polydactyly (Sharpe et al., 1999)(Figure 7A, Table S5). Interestingly, we found both loss and gain of interactions with these mutations, involving many TFs that are expressed in the developing limb. Data integration showed that gain of interactions involving limb-expressed TFs mostly involves activators, while loss of interactions occurred more frequently with transcriptional repressors (p = 0.018, Figure 7B), both of which are concordant with the increased gene expression elicited by these dominant mutations. Thus, similar diseases can result from gain or loss of TF interactions caused by different mutations within an enhancer.
Figure 7. Mutations in the Limb SHH Enhancer.
(A) Summary of interactions lost (red) or gained (blue) for different mutations in the ZRS enhancer of sonic hedgehog. Yellow circles – TFs expressed in limb during development; black dots – activators; white dots – repressors; black/white dots – TF that can be both activators or repressors.
(B) Number of interaction changes occurring with limb-expressed activators, repressors or bifunctional TFs (activators/repressors) for interactions gained or lost in ZRS enhancer mutations. p = 0.018 by Fisher's exact test.
(C) Gain of interactions detected by eY1H assays in the 105C→G mutant in the ZRS enhancer of sonic hedgehog. Blue boxes indicate positive interactions.
(D) DNA binding motifs for TFAP2A, TFAP2B and TFAP2E discriminate wild type and mutant enhancer sequences.
(E) HEK293T cells were co-transfected with enhancer fragments containing wild type (105C) or mutant (105G) sequences cloned upstream of a Firefly luciferase reporter vector, and the indicated TF expression vectors. Relative luminescence activity is plotted as fold change compared to cells co-transfected with control vector expressing GFP. Experiments were performed four times in 3-6 replicates. Average relative luminescence activity ± SEM is plotted. *p<0.05 by Student's t-test.
See also Table S5.
We characterized the 105C→G mutation in more detail. This mutation results in gain of interactions with three AP2 TFs, two of which are expressed in the limb and could be responsible for the gain of SHH expression (Figures 7A and 7C). Indeed, this mutation creates a consensus AP2 binding site (Badis et al., 2009)(Figure 7D). TFAP2B is a transcriptional activator and activates the mutant, but not wild type enhancer in luciferase assays (Figure 7E). Together, these results show that TFAP2B can bind and activate 105C→G enhancer mutant, suggesting that aberrant binding of TFAP2B may result in the ectopic expression of SHH, thereby causing digit malformations.
DISCUSSION
This study presents a gene-centered human TF-enhancer interaction network delineated by eY1H assays. The technical quality of this network is ensured by the inherent retest of interactions with two reporter genes, and the testing of TFs in quadruplicate, as well as due to the high demonstrated rate of reproducibility between independent experiments (~90%)(Reece-Hoyes et al., 2011b; Reece-Hoyes et al., 2013). The biological quality of this network is also high, as indicated by several metrics, including significant overlap with TF binding sites, ChIP interactions, TF expression and enhancer activity, enrichment for homeodomains and reporter assays. The relatively modest overlap with ChIP data reflects the notion that ChIP may retrieve indirect TF interactions, as well as a lack of sensitivity of ChIP data that were only obtained in one or two cell types. Like any other method, however, eY1H assays may also yield false positive and negative interactions with both enhancers and disease-causing mutant elements. False positive interactions may be retrieved when multiple members of the same TF family with highly similar consensus binding sites are found to bind to the same enhancer(s), and only a subset of these actually bind the enhancer in vivo. Importantly, however, we illustrate several mechanisms by which enhancer sharing can be biologically meaningful in attaining redundancy or in the precise timing of enhancer activity, for instance during development. The careful integration of eY1H interactions with high-resolution spatiotemporal expression and other types of data over time will provide protein-DNA interaction data of increasing validity and resolution.
The rate of false negatives in eY1H assays is likely to be considerable (Walhout, 2011). For instance, TFs that exclusively interact with DNA as heterodimers or after post-translational modification by another human protein will not be detected. In addition, eY1H assays cannot as of yet detect cooperative interactions with multiple TFs. Therefore, the retrieval of several known differential interactions with non-coding disease-causing mutations in a single experiment is highly encouraging.
A particularly powerful feature of the eY1H approach is that it uniquely enables the comparison of wild type and mutant TFs or regulatory elements, in a single experiment and in a high-throughput manner. Our findings show that both coding mutations in TF-encoding genes and non-coding mutations in regulatory sequences can result in rewiring of GRNs. While coding mutations cause mostly loss of protein-protein interactions (Sahni et al., under consideration), protein-DNA interaction changes caused by either coding or non-coding mutations involve both gain and loss of interactions, sometimes with the same mutation (Sahni et al., under consideration, this study). We provide a guide for interpreting eY1H data with non-coding disease-causing mutations. Specifically, we would prioritize differential interactions involving TFs that are co-expressed with the target gene, in the disease-relevant tissue. Further, we emphasize that concordant interactions, for instance increased gene expression and gain of interaction with an activator or loss with a repressor, are more likely relevant in vivo than other interactions. Obtaining additional, high-resolution gene expression and TF function data will be critical for the continued integration not only of eY1H data, but also of interaction data inferred by DNAse I hypersensitivity assays or predicted based on TF binding sites.
Most variants identified by GWAS reside in non-coding regions of the genome (Hindorff et al., 2009; Maurano et al., 2012). We propose that eY1H assays will provide a facile method with which differential TF interactions involving these variants can be analyzed. Overall this work provides an initial blueprint to study enhancer networks, as well as to determine how network connectivity is affected in disease.
EXPERIMENTAL PROCEDURES
eY1H Assays
Enhanced yeast one-hybrid (eY1H) assays were performed as described (Reece-Hoyes et al., 2011b). This method detects protein-DNA interactions and involves two components: a ‘DNA-bait’ (e.g. a gene promoter or enhancer) and a ‘TF-prey’. We generated DNA-bait strains for 360 human developmental enhancers selected from the Vista Enhancer Browser (http://enhancer.lbl.gov, Table S1). Enhancers (0.4-2.4 kb) were amplified by PCR (Table S7) from human genomic DNA (Clonetech) and were then Gateway-cloned (Reece-Hoyes et al., 2011b). Entry clones were sequenced using PacBio (Yale Center for Genomic Analysis, Table S8). The DNA-baits were cloned upstream of two Y1H reporter genes (LacZ and HIS3) and both DNA-bait::reporter constructs were integrated into the yeast genome to generate chromatinized ‘DNA-bait strains’. Yeast strains that express different TFs fused to the activation domain (AD) of yeast Gal4 were mated into the DNA bait strain. If a TF binds the regulatory region, the AD moiety activates reporter gene expression. LacZ activation was detected via the conversion of colorless X-gal into a blue compound, while His3 expression allows the yeast to grow on media lacking histidine and to overcome the addition of 3-amino-triazole (3AT), a competitive inhibitor of the His3 enzyme (Deplancke et al., 2004; Reece-Hoyes and Walhout, 2012). We updated the previously published arrayed collection of 988 human TFs (Reece-Hoyes et al., 2011a) by adding 146 TFs and removing 48 for which the clone turned out to be incorrect, was truncated or did not encode the DNA binding domain. The resulting collection contains one variant of 1,086 full-length TFs (76% of all 1,434 human TFs, Table S9).
eY1H assays were performed using a Singer robot that manipulates yeast strains in a 1536-colony format. Images of readout plates lacking histidine and containing 3AT and X-gal were processed using the Mybrid web-tool to automatically detect positive interactions (Reece-Hoyes et al., 2013). Each interaction was tested in quadruplicate and only those that were positive at least twice were considered genuine (Reece-Hoyes et al., 2011b). However, the vast majority of interactions detected (~90%) were supporter by all four colonies as previously published (Reece-Hoyes et al., 2011b). Interactions detected by Mybrid were then manually curated. False positives detected by Mybrid on plates with uneven background were removed. We included false negative interactions missed by Mybrid, for instance because they occur next to very strong positives or occur with baits that exhibit high background reporter gene expression. Positive colonies were sequenced to determine prey identity. Fourteen quads in the array were removed from the interaction list as they did not match the expected TF (see Extended Experimental Procedures). A total of 2,230 high-quality protein-DNA interactions between 246 enhancers and 283 TFs were included in the final dataset (Table S2).
Target Profile Similarity
Target profile similarity between TFs was calculated using the Jaccard index as the number of enhancer targets shared between two TFs A and B divided by the number of enhancers that interact with either A or B (Fuxman Bass et al., 2013). Target profile similarities range from 0 to 1, with 0 indicating no target overlap and 1 indicating complete target overlap.
Mutated Regulatory Regions
Mutant DNA baits were generated by introducing mutations in the primers in the PCR step prior to generating entry clones (Table S10). Yeast DNA-bait strains were sequenced to verify the mutation and ensure the absence of additional mutations. eY1H screens were performed for two or three independent yeast strains per construct. Interactions that occurred with at least two out of three or two out of two of the strains were considered positive while interactions not occurring in any of the strains were considered negative (Table S5).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Walhout lab, R. McCord, N. Kaplan and J. Dekker for discussions and critical reading of the manuscript. We thank J. Gibcus for the pGL4 plasmid and expression vectors and advice on the luciferase assays. We thank N. Beittel and B. Parmentier for technical assistance. This work was supported by US National Institutes of Health grants GM082971 to A.J.M.W., HG004233 to M.V., and HG006066 to M.V. and D.E.H. J.I.F.B. was partially supported by a postdoctoral fellowship from the Pew Latin American Fellows Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.I.F.B. and A.J.M.W. conceived the project. J.I.F.B., S.S., N.B. and A.G-G performed eY1H experiments. J.I.F.B. performed the data analysis with assistance of A.M.; J.I.F.B. and S.S. performed the luciferase assays. J.I.F.B., N.S. and S.Y. performed the experiments and data analysis for Figure 5. N.S. and S.Y. were supervised by D.H. and M.V. J.I.F.B. and A.J.M.W. wrote the paper with contributions of the other authors.
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures and ten Supplemental tables.
REFERENCES
- Alhashem YN, Vinjamur DS, Basu M, Klingmuller U, Gaensler KM, Lloyd JA. Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding. The Journal of biological chemistry. 2011;286:24819–24827. doi: 10.1074/jbc.M111.247536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Taubert S, Conine C, Tsuda B, Van Gilst MR, Sequerra R, Doucette-Stam L, Yamamoto KR, Walhout AJM. Functional modularity of nuclear hormone receptors in a C. elegans gene regulatory network. Molecular Systems Biology. 2010;6:367. doi: 10.1038/msb.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arda HE, Walhout AJM. Gene-centered regulatory networks. Briefings in functional genomics and proteomics. 2009 doi: 10.1093/bfgp/elp049. doi:10.1093/elp049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, Liu W, Zeng A, Taylor-Teeples M, Kim D, et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol Syst Biol. 2011;7:459. doi: 10.1038/msb.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickman JM, Clements M, Tyrell R, McNay D, Woods K, Warner J, Stewart A, Beddington RS, Dattani M. Molecular effects of novel mutations in Hesx1/HESX1 associated with human pituitary disorders. Development. 2001;128:5189–5199. doi: 10.1242/dev.128.24.5189. [DOI] [PubMed] [Google Scholar]
- Chi YI. Homeodomain revisited: a lesson from disease-causing mutations. Hum Genet. 2005;116:433–444. doi: 10.1007/s00439-004-1252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattani MT, Martinez-Barbera JP, Thomas PQ, Brickman JM, Gupta R, Martensson IL, Toresson H, Fox M, Wales JK, Hindmarsh PC, et al. Mutations in the homeobox gene HESX1/Hesx1 associated with septo-optic dysplasia in human and mouse. Nature genetics. 1998;19:125–133. doi: 10.1038/477. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJM. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stam L, Reece-Hoyes JS, Hope IA, et al. A gene-centered C. elegansprotein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Fuxman Bass JI, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJ. Using networks to measure similarity between genes: association index selection. Nat Methods. 2013;10:1169–1176. doi: 10.1038/nmeth.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Tamburino AM, Mori A, Beittel N, Weirauch MT, Reece-Hoyes JS, Walhout AJ. Transcription factor binding to Caenorhabditis elegans first introns reveals lack of redundancy with gene promoters. Nucleic Acids Res. 2014;42:153–162. doi: 10.1093/nar/gkt858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner MR, Gongora MM, Grimmond SM, Perkins AC. A global role for zebrafish klf4 in embryonic erythropoiesis. Mech Dev. 2007;124:762–774. doi: 10.1016/j.mod.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, deMasi F, Barrasa MI, Newburger D, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Luo W, Skalnik DG. Interferon regulatory factor-2 directs transcription from the gp91phox promoter. The Journal of biological chemistry. 1996;271:23445–23451. doi: 10.1074/jbc.271.38.23445. [DOI] [PubMed] [Google Scholar]
- MacNeil LT, Walhout AJM. Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 2011;21:645–657. doi: 10.1101/gr.097378.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maconochie M, Nonchev S, Morrison A, Krumlauf R. Paralogous Hox genes: function and regulation. Annual Review of Genetics. 1996;30:529–556. doi: 10.1146/annurev.genet.30.1.529. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros V, Walhout AJM. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaeffle RW, Hunter CS, Savage JJ, Duran-Prado M, Mullen RD, Neeb ZP, Eiholzer U, Hesse V, Haddad NG, Stobbe HM, et al. Three novel missense mutations within the LHX4 gene are associated with variable pituitary hormone deficiencies. J Clin Endocrinol Metab. 2008;93:1062–1071. doi: 10.1210/jc.2007-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Barutcu AR, Patton McCord R, Jeong J, Jian L, MacWilliams A, Yang X, Salehi-Ashtiani K, Hill DE, Blackshaw S, et al. Yeast one-hybrid assays for high-throughput human gene regulatory network mapping. Nature Methods. 2011a;8:1050–1052. doi: 10.1038/nmeth.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Kent A, Shrestha S, Kadreppa S, Pesyna C, Lajoie B, Dekker J, Myers CL, Walhout AJM. Enhanced yeast one-hybrid (eY1H) assays for high-throughput gene-centered regulatory network mapping. Nature Methods. 2011b;8:1059–1064. doi: 10.1038/nmeth.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Pons C, Diallo A, Mori A, Shrestha S, Kadreppa S, Nelson J, DiPrima S, Dricot A, Lajoie BR, et al. Extensive rewiring and complex evolutionary dynamics in a C. elegans multiparameter transcription factor network. Mol Cell. 2013;51:116–127. doi: 10.1016/j.molcel.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Walhout AJ. Gene-centered yeast one-hybrid assays. Methods Mol Biol. 2012;812:189–208. doi: 10.1007/978-1-61779-455-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Lettice L, Hecksher-Sorensen J, Fox M, Hill R, Krumlauf R. Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr Biol. 1999;9:97–100. doi: 10.1016/s0960-9822(99)80022-0. [DOI] [PubMed] [Google Scholar]
- Tajima T, Hattori T, Nakajima T, Okuhara K, Tsubaki J, Fujieda K. A novel missense mutation (P366T) of the LHX4 gene causes severe combined pituitary hormone deficiency with pituitary hypoplasia, ectopic posterior lobe and a poorly developed sella turcica. Endocrine journal. 2007;54:637–641. doi: 10.1507/endocrj.k06-200. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic acids research. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM. Unraveling Transcription Regulatory Networks by Protein-DNA and Protein-Protein Interaction Mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Walhout AJM. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011;12:109. doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.