Abstract
There is currently no cure for neuron loss in the brain, which can occur due to traumatic injury or neurodegenerative disease. One method proposed to enhance neurogenesis in the brain is gene transfer to neural progenitor cells. In this work, a guanidine-based copolymer was synthesized and compared to an amine-based copolymer analog previously shown to effectively deliver genes in the murine brain. The guanidine-based copolymer was more efficient at gene transfer to immortalized, cultured cell lines; however, the amine-based copolymer was more effective at gene transfer in the brain. DNA condensation studies revealed that the nucleic acid complexes formed with the guanidine-based copolymer were more susceptible to unpackaging in the presence of heparin sulfate proteoglycans compared to complexes formed with the amine-based copolymer. Therefore, polyplexes formed from the amine-based copolymer may be more resistant to destabilization by the heparan sulfate proteoglycans present in the stem cell niches of the brain.
1. Introduction
Neural stem and progenitor cells (NSCs and NPCs) reside in two specialized niches in the adult mammalian brain: the subventricular zone (SVZ) of the lateral ventricles and the hippocampus.[1, 2] These cells contribute to neurogenesis and have been shown in rodent models to respond to cortical injury by proliferating.[3–5] Progenitors can be diverted to participate in scar tissue formation, however; replacement of diseased neuronal populations is very limited. Despite their capacity to regenerate cells, neurogenic regions cannot fully counteract neuron death in progressive neurodegenerative disease or trauma. Gene transfer has been proposed as a method to both direct and enhance neurogenesis in the brain.[6, 7] In order to realize the potential of this approach, effective nucleic acid transfer technologies to the central nervous system are needed. While non-viral vectors such as polymeric materials are preferred over viral vectors for their conceivable safety profiles and lower manufacturing cost, only a minority of clinical trials use this class of delivery modality due to their lower delivery efficiencies compared to viral vectors.[8, 9]
Most polymers used for gene transfer are polycations comprised of monomers containing primary, secondary, or tertiary amines.[10] Several groups, however, have demonstrated that guanidinylated polycations outperform their amine-based analogs in transfection to cultured cells.[11–14] Guanidiniums are protonated at physiological pH and have electrons that are delocalized, resulting in resonance-stabilized charge spread around the three nitrogen atoms.[15, 16] Polyarginines, which are cationic due to multiple guanidine groups, have been shown to bind with higher affinity to double-stranded DNA (dsDNA) than polylysines, which contain multiple primary amine groups.[17–19] The stronger DNA binding and the ability to interact with cell surface phosphates and sulfates to facilitate cell internalization are attributed to the efficient gene transfer capability of guanidine-containing polycations.[20]
Previously, our group designed a block-statistical copolymer comprised of a poly(ε-caprolactone) (PCL) block connected by a reducible disulfide to a statistical copolymer of tetraethylenepentamine (TEPA)-decorated poly(glycidyl methacrylate) (PGMA) monomers and oligo(ethylene glycol) monomethyl ether methacrylate (OEGMA) monomers.[21] This copolymer, PCL-SS-P[(GMA-TEPA)-st-OEGMA], complexes with nucleic acids to form polyplexes (cationic polymer/DNA particles) with diameters < 200 nm and was specifically designed to overcome many of the barriers to gene delivery. The hydrophilic OEGMA monomers and hydrophobic PCL monomers provide extracellular stability by reducing salt-induced aggregation and premature unpackaging, respectively. Once internalized into a cell, the protonatable amines in TEPA facilitate endosomal escape via the proton sponge effect and the internal disulfide bond can be reduced by glutathione in the cytosol to detach the PCL core. After the disulfide is reduced, the remaining statistical polycation is less stable and can therefore release its DNA cargo. We further demonstrated that this polymer transfects cultured HeLa cells more efficiently than branched polyethylenimine (PEI, 25k) and mediates effective gene transfer in the brain of mice.[21]
We hypothesized that the transfection efficiency of PCL-SS-P[(GMA-TEPA)-st-OEGMA] could be further increased by guanidinylating the primary amines in TEPA. In this study, guanidinylated PCL-SS-P[(GMA-TEPA)-st-OEGMA] copolymers were synthesized and tested both in vitro and in vivo for gene transfer ability. The results of this study highlight discordance between in vitro and in vivo efficiency and illustrate the importance of in vivo evaluation of new polymeric gene transfer materials.
2. Materials and Methods
2.1. Materials
Reagents for polymer synthesis were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. ε-Caprolactone (CL) was dried over CaH2 and distilled under reduced pressure prior to use. Glycidyl methacrylate was purified by vacuum distillation before use. Oligo(ethylene glycol) monomethyl ether methacrylate (OEGMA, Mn= 300 g/mol and pendent EO units DP~4.5) was purified by passing through a column filled with basic alumina to remove the inhibitor. Copper (I) chloride (CuCl) was washed with acetic acid and ethanol in turn to remove Cu2+. All cell culture reagents were purchased from Cellgro/Mediatech (Fisher Scientific, Pittsburgh PA). Endotoxin-free plasmid pCMV-Luc2 (Photinuspyralis luciferase under control of the cytomegalovirus (CMV) enhancer/promoter) was produced with the Qiagen Plasmid GigaPrep kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. The pmaxGFP® plasmid (green fluorescent protein from the copepod Pontellina p.) was purchased from Lonza. The bicinchoninic acid (BCA) protein quantification assay kit was purchased from Thermo Fischer Scientific (Waltham, MA) while the luciferase expression quantification kit was obtained from Promega (Madison, WI).
2.2. Cell lines
HeLa cells, human cervical carcinoma cells (ATCC® CCL-2™), were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics/antimyotics (AbAm) (100 IU of penicillin, 100 ug/mL of streptomycin, and 0.25 ug/mL of amphotericin B). Z310 cells were donated by Prof. Wei Zheng (Purdue) and cultured in Dulbecco’s minimum essential medium (DMEM) supplemented with 10% heat-inactivated FBS, 10% penicillin/streptomycin, 40 µg/mL gentamicin, and 10 ng/mL nerve growth factor (NGF). Primary neural progenitor cells (NPCs) were maintained in DMEM/F12 supplemented with 2 mM L-glutamine, 1% N-2 supplement, 5 µg/mL heparin, and 20 ng/mL of both endothelial growth factor and NGF.
2.3. Polymer synthesis
2.3.1. Synthesis of PCL-SS-P[(GMA-TEPA)-st-OEGMA
The reducible 2-hydroxyethyl-2’-(bromoisobutyryl)ethyl disulfide double-head initiator (HO-SS-iBuBr) was prepared according to literature.[21] PCL-SS-P[(GMA-TEPA)-st-OEGMA] (denoted as “Base copolymer”) was synthesized as reported previously.[21] Briefly, ring opening polymerization (ROP) of CL was performed using HO-SS-iBuBr as the initiator and Sn(Oct)2 as the catalyst. Then, a one-pot atom transfer radical polymerization (ATRP) of GMA and OEGMA was performed using PCL40-SS-iBuBr as the macroinitiator and CuCl/bpy as the catalyst. The copolymer composition was determined by 1H NMR. Integration of the NMR resonances assigned to the PCL block at 2.35 ppm (characteristic to the first carbon next to the carbonyl carbon), GMA block at 4.32 ppm (characteristic of the ethylene adjacent to the epoxy group), and OEGMA block at 3.40 ppm (characteristic of the methyl group) were compared. The GMA monomers in the polymer were then reacted with excess TEPA. Complete conversion was confirmed by monitoring the disappearance of the epoxy group peaks (3.26 ppm, 2.87 pp.m, 2.66 p.m.) and appearance of TEPA amine groups (2.5–3.0 pp.m.) in 1H-NMR.
2.3.2. Synthesis of guanidinylated copolymer
The guanidinylated copolymer, denoted as “Guan copolymer”, was synthesized by reacting Base copolymer with a 10-fold molar excess of o-methylisourea to primary amines in GMA-TEPA unit. Base copolymer was dissolved at 10 mg/mL in 1:1 (v/v) saturated NaCO3 and ddH2O and mixed with o-methylisourea dissolved in an equivalent amount of 1:1 (v/v) saturated NaCO3 and ddH2O in a round bottom flask with a stir bar and sealed with a septum. The solution was stirred at room temperature for 72 hours. After the reaction, the polymer was purified by dialysis (MW cutoff: 10,000 g/mol) against 4 L of distilled water, which was renewed 3 times per day over the course of 3 days, followed by lyophilization to yield a fluffy, white product. Polymer guanidinylation was confirmed spectroscopically by disappearance of primary amines (quantified by ninhydrin assay) and by appearance of guanidines assessed using a ligand-exchange assay developed by Weber.[22]. For the ninhydrin assay, the Base copolymer was used to make a standard curve. To quantify the amount of primary amines, the absorbance (420 nm) of polymer reacted with ninhydrinwas read on a Tecan Infinite M1000 PRO microplate reader. To confirm guanidinylation, 0.2g of potassium hexacyanoferrate and 0.2g of sodium nitroferricyanide(III) dihydrate were dissolved in 4 mL of 1M sodium hydroxide and then mixed with 3 mL of aqueous Guan copolymer at 3 mg/mL. After 1.5 minutes, the absorbance at 498 nm was measured. L-arginine, which contains a guanidine group, was used as the standard and Base copolymer, while O-methylisourea, and aqueous sodium carbonate were used as the negative controls.
2.4. Polyplex preparation and characterization
2.4.1. Polyplex formulation
Stock solutions of copolymers were prepared at 5 mg/mL in ddH2O and then acidified to pH = 6.4 using 1N HCl. The pCMV-Luc2 plasmid was diluted in ddH2O to a concentration of 0.1 mg/mL. Copolymer-DNA complexes (termed polyplexes) were formed by adding copolymer diluted in ddH2O to an equal volume of diluted pCMV-Luc2 plasmid at the desired charge ratio (N/P), followed by incubation at room temperature for 10 minutes.
2.4.2. Gel retardation testing
Copolymer complexation of plasmid was assessed by a gel retardation or electrophoretic mobility shift assay. Polyplexes (1 µg of pCMV-Luc2, 20 µL solution) were formed at various N/P ratios with 10% (v/v) of 10× BlueJuice™ Gel Loading Buffer and loaded into a 1% agarose gel made with TAE buffer (40 mMTris-acetate, 1 mMEDTA) and 5 mg/mL ethidium bromide. The gel was electrophoresed at 100 V for 50 minutes. The gel was then imaged on a Kodak (Rochester, NY) UV transilluminator (laser-excited fluorescence gel scanner).
2.4.3. Size and surface charge analysis
The average hydrodynamic diameter and surface charge of polyplexes were determined using dynamic light scattering (DLS, Brookhaven Instruments Corp ZetaPALS) and zeta potential analysis (Malvern Zetasizer). Polyplexes (N/P = 15, 1 µg of pCMV-Luc2, 20 µL solution) were diluted with 80 µL of ddH2O or 150 mM PBS (phosphate-buffered saline, pH 7.2). DLS was analyzed at room temperature using a wavelength of 659 nm and a detection angle of 90°. Measurements were taken over five 30-second intervals.
2.4.4. Polyplex unpackaging by heparin/heparan sulfate competition
The pCMV-Luc2 plasmid was mixed with the bis-intercalating dye YOYO-1 iodide (Invitrogen, Carlsbad, CA) at a dye to base pair ratio of 1:100 and incubated at room temperature for 1 hour. Polyplexes were prepared at an N/P = 15 by complexing YOYO-labeled plasmid with Base and Guan copolymers as previously mentioned. Polyplexes were treated with stated concentrations of heparin or heparan sulfate for 1 hour. The fluorescence (ex: 491 nm, ex: 509 nm) of each well was normalized to a DNA only control. To test polyplex unpackaging by gel retardation, heparin (10 µg/mL) was added to polyplexes for 10 minutes and the gel was electrophoresed as previously mentioned.
2.5. In vitro transfection and cytotoxicity analysis
2.5.1. Evaluation in HeLa and Z310 immortalized cells
HeLa cells and Z310 cells were seeded in complete cell culture medium at a density of 30,000 cells/well in a 24-well plate. Cells were allowed to attach for 16 hours at 37 °C, 5% CO2. Polyplexes were formed at different N/P ratios using 1 µg of pCMV-Luc2 in 20 µL total volume diluted in 180 µL of OptiMEM medium per well. The cells were washed once with PBS and then the polyplexes diluted in OptiMEM were added. The plates were incubated at 37 °C, 5% CO2 for 4 hours. After 4 hours, the polyplex solutions were aspirated, the cells were washed with PBS, and complete media was added to each well. After an additional 44 hours at 37 °C, 5% CO2, the wells were washed with PBS and 1× Reporter Lysis Buffer was added to each well. The plates were incubated at room temperature for 15 minutes, freeze-thawed, and then samples centrifuged at 15,000 g for 15 minutes at 4 °C to pellet the cell debris. The lysates were analyzed using a luciferase assay kit (Promega Corp.) according to the manufacturer’s instructions. Luminescence intensity was measured on a Tecan Infinite M1000 PRO microplate reader with integration for 1 second. The total protein content in each well was measured by a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) to assess cell viability and to normalize luciferase expression. Each copolymer at each N/P ratio was tested with a sample size of (n) = 4 for both cell lines.
2.5.2. Transfection of primary NPCs
NPCs were plated at a density of 26,300 cells/cm2 in growth media 24 hours prior to transfection. Polyplexes were prepared at an N/P ratio of 15 prior to dilution in NPC growth media with reduced heparin (1 µg/mL). After 4 hours of incubation with polyplexes at 37°C, NPCs were rinsed with DMEM/F12 before an exchange to fresh growth media. After an additional 44 hours, cells were incubated with reporter lysis buffer (Promega Corp.) for 15 minutes before storage at −80°C until analysis. Luciferase activity in cell lysates was measured as described for Z310 and HeLa cell lysates.
2.5.3 Flow cytometry analysis of GFP plasmid transfection
HeLa cells were transfected with polyplexes formed with Base or Guan copolymers and pmaxGFP™ (Lonza) as previously mentioned. For analysis, cells were washed with PBS, trypsinized, and pelleted at 500 g for 5 min at 4°C. The pellet was resuspended in 0.3 mL propidium iodide (PI) solution (1 µg/mL in PBS), kept on ice, and analyzed using flow cytometry, MACSQuant Analyzer (Miltenyi Biotec Inc., Auburn, CA). Intact cells were identified using the forward and side scatter data. The resulting cell population was gated into GFP+/PI+, GFP+/PI−, GFP−/PI+ and GFP−/PI− based on the green fluorescence and PI intensity from the control samples and with proper compensation.
2.6. In vivo polyplex delivery to murine brain
2.6.1. Intraventricular injections
All animal procedures were completed using protocols approved by the Institutional Animal Care and Use Committee at the University of Washington. Polyplexes were prepared as described above in 5% glucose using 2.5 µg of DNA in 10 µL total volume at an N/P of 15. Adult female C57BL/6J mice (Jackson Laboratories) were anesthetized by an intraperitoneal injection of Avertin. A 1 mm diameter craniotomy was made on the right side of the skull using a dental drill and 10 µL of polyplex solution or DNA only (n = 5 or 6 per group) was stereotaxically injected at 1 mm lateral, 0.5 mm caudal to bregma, and 1.9 mm depth from the dura using a 33 gauge 10 µL Hamilton syringe. The injection was made over 2.5 minutes and the syringe was kept in place for 2 minutes after injection to prevent backflow.
2.6.2. Luciferase expression analysis
Brains were harvested from mice 48 hours post injection and separated into three sections: hindbrain, left hemisphere, and right hemisphere. Tissues were collected in 1× Reporter Lysis Buffer (Promega, Madison, WI) with 1× EDTA-free Roche’s Complete Protease Inhibitor Cocktail (Roche, Nutley, NJ) and three freeze-thaw cycles were performed in liquid nitrogen. Tissues were mechanically homogenized and cell debris were pelleted at 15,000 g for 15 minute at 4 °C. The lysates were collected and 20 µL of each lysate was analyzed using a luciferase assay kit (Promega Corp.) according to the manufacturer’s instructions. Luminescence intensity was measured on a Tecan Infinite M1000 PRO microplate reader with integration for 1 second. The total protein content in each well was measured by a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL) to normalize luciferase expression by protein content.
2.6.3. Immunohistochemistry and confocal microscopy
Injections were done as described above using polyplexes formulated with pmaxGFP™ (Lonza). Two days post injection, mice were euthanized with Avertin overdose and perfused intracardially with 0.9% saline followed by 4% paraformaldehyde in 0.1 M phosphate buffer. After perfusion and fixation, the brains were excised and equilibrated to 30% sucrose in phosphate buffer. Brains were embedded in OCT and sectioned into 40 um-thick coronal slices. For immunofluorescent labeling, slides were rinsed with PBS and blocked in PBS, 0.3% TritonX-100, 2% BSA for 1 hour. Primary antibodies (goat anti-Sox2, Santa Cruz Biotechnology; 1:250) were applied to the tissue sections in PBS, 0.3% TritonX-100, 2% BSA overnight at 4 °C. Sections were rinsed three times for 20 minutes in TBS, 0.1%Tween20 and species appropriate secondary antibodies conjugated with fluorophore were incubated in PBS, 0.1% Tween20, 2% donkey serum for 2 hours. Sections were, again, rinsed three times for 20 minutes in TBS-Tween, with the last rinse containing the nuclear marker, 4’,6-diamidino-2-phenylindole (DAPI; 1:1000). Sections were then mounted onto glass slides, sealed and coverslipped with gelvatol, and imaged using a confocal microscope.
2.6.4. Stereology
To quantify the population of GFP-transfected cells, cells populations surrounding the ventricles was calculated via fractionator stereology (an unbiased sampling method) by Stereo Investigator (Microbrightfield, Inc.). A grid size of 150 × 150 µm and counting frame of 50×50 µm was used to assure unbiased sampling of a randomized grid in a 1 in 6 series of tissue sections to generate averaged populations for each animal. Measurements were limited to the SVZ by creating electronic templates of a 100 µm margin around the ventricle-borders.
3. Results
3.1. Polymer synthesis and characterization
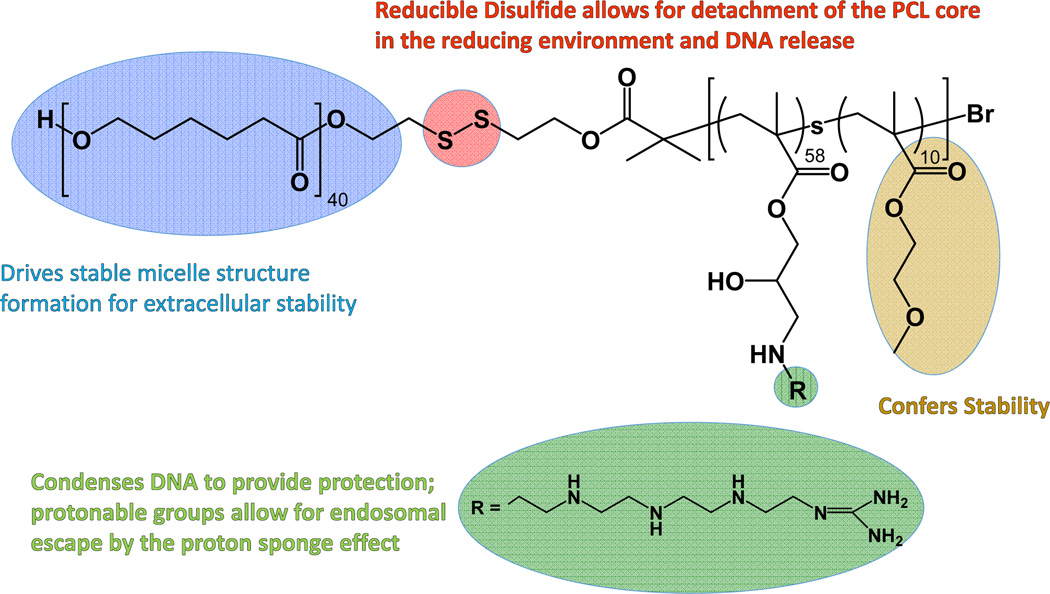
The PCL-SS-P[GMA-st-OEGMA] copolymer was synthesized using a combination of ring opening polymerization and atom transfer radical polymerization (ATRP) as we previously described.[21] Polymer composition was determined by 1H-NMR spectroscopy to be PCL40-SS-P[GMA60-st-OEGMA10]. The GMA monomers were then reacted with excess TEPA; 1H-NMR revealed complete disappearance of the GMA epoxy group resonances at 3.26 ppm, 2.87 ppm, and 2.66 ppm, and appearance of a broad peak characteristic of the amines in TEPA at 2.5–3.0 ppm, demonstrating complete reaction of GMA monomers. The molecular weight (MW) of the resulting polymer, PCL-SS-P[(GMA-TEPA)-st-OEGMA], called “Base copolymer”, was calculated to be 27.0 kDa. The Base copolymer was guanidinylated by reaction with 10-fold excess of o-methylisourea to TEPA as described previously,[11, 23] resulting in the polymer called “Guan copolymer” with MW ~ 29.0 kDa (Scheme 1). Guanidinylation of 90% of the TEPA primary amines was confirmed by ninhydrin assay and conversion of primary amines to guanidine was further confirmed by a ligand-exchange guanidine assay developed by Weber.[22]
Scheme 1.
Guanidinylated copolymer. The block copolymer contains a polycaprolactone block (blue), an internal disulfide bond (red), and a second hydrophilic block containing pendant guanidines (green) for DNA binding and oligoethylene glycols (yellow) for stability.
3.2. Polyplex formulation
Polyplexes were formed by adding polymer solutions to plasmid DNA at desired charge ratios. Gel electrophoresis showed that the Base copolymer and Guan copolymer complexed plasmid DNA at similar charge ratios (~ N/P = 3, Figure S1). The surface charge and average hydrodynamic diameter of polyplexes formed with both polymers at an N/P (defined as molar TEPA to molar phosphate) ratio of 15 were determined by zeta potential and dynamic light scattering measurements, respectively (Table 1). This charge ratio was selected because it was shown previously to be optimal for the Base copolymer.[21] The zeta potential of polyplexes formed with both Base and Guan copolymer were both positive (+23.5−41.6 mV) and their average sizes in water were comparable (160 nm). Particle size increased slightly in physiological salt concentrations (150 mM phosphate-buffered saline) to 226.4 ± 1.0 nm and 195.1 ± 4.4 nm for Base and Guan copolymer polyplexes, respectively.
Table 1.
Physicochemical properties (diameter and surface charge in water and PBS) of polyplexes (N/P = 15) formed by the complexation of base copolymer or guan copolymer with luciferase plasmid.
| Polymer | Average Diameter (nm) in Water |
PDI in Water | Average Daimeter (nm) in PBS |
PDI in PBS | Zeta Potential (mV) |
|---|---|---|---|---|---|
| Base Copolymer | 161.1 ± 5.0 | 0.172 ± 0.015 | 226.4 ± 1.0 | 0.202 ± 0.014 | 41.6 ± 0.5 |
| Guan Copolymer | 158.3 ± 9.8 | 0.169 ± 0.022 | 195.1 ± 4.4 | 0.177 ± 0.012 | 23.5 ± 6.3 |
3.3 Transfection and cytotoxicity to cultured cells
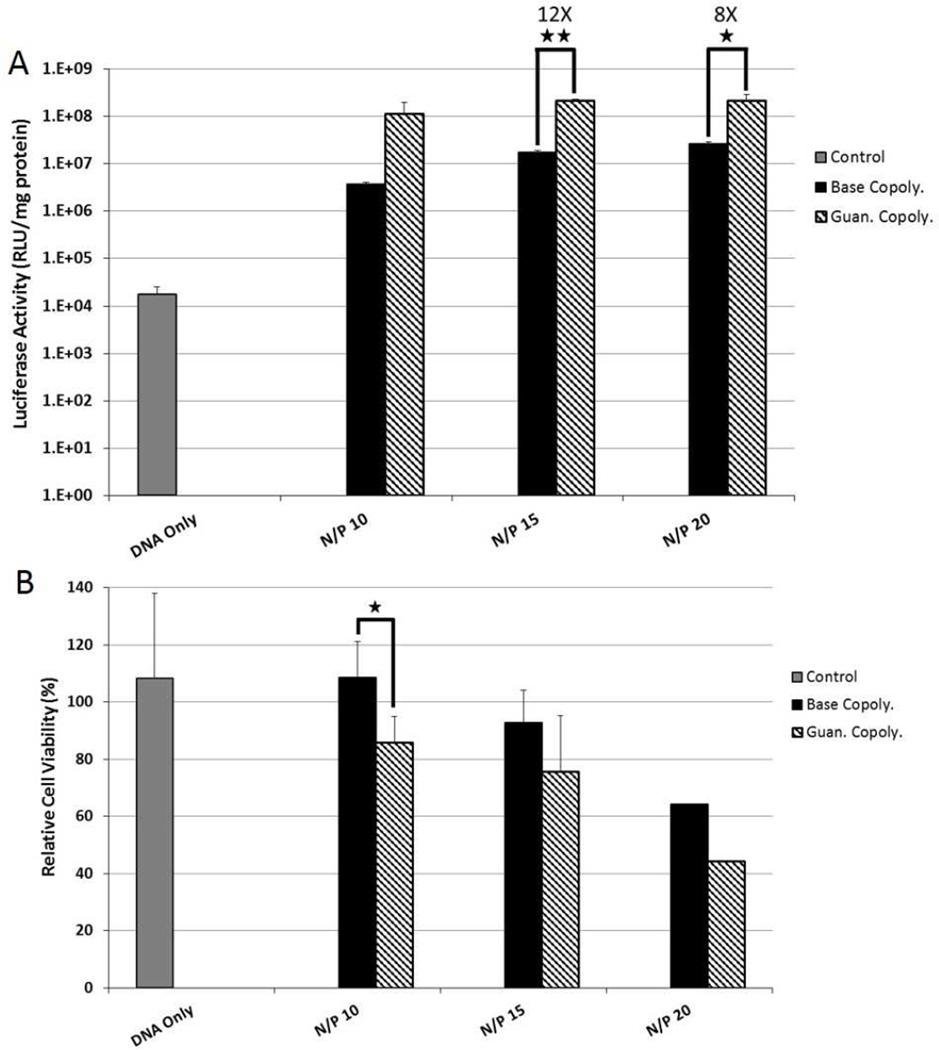
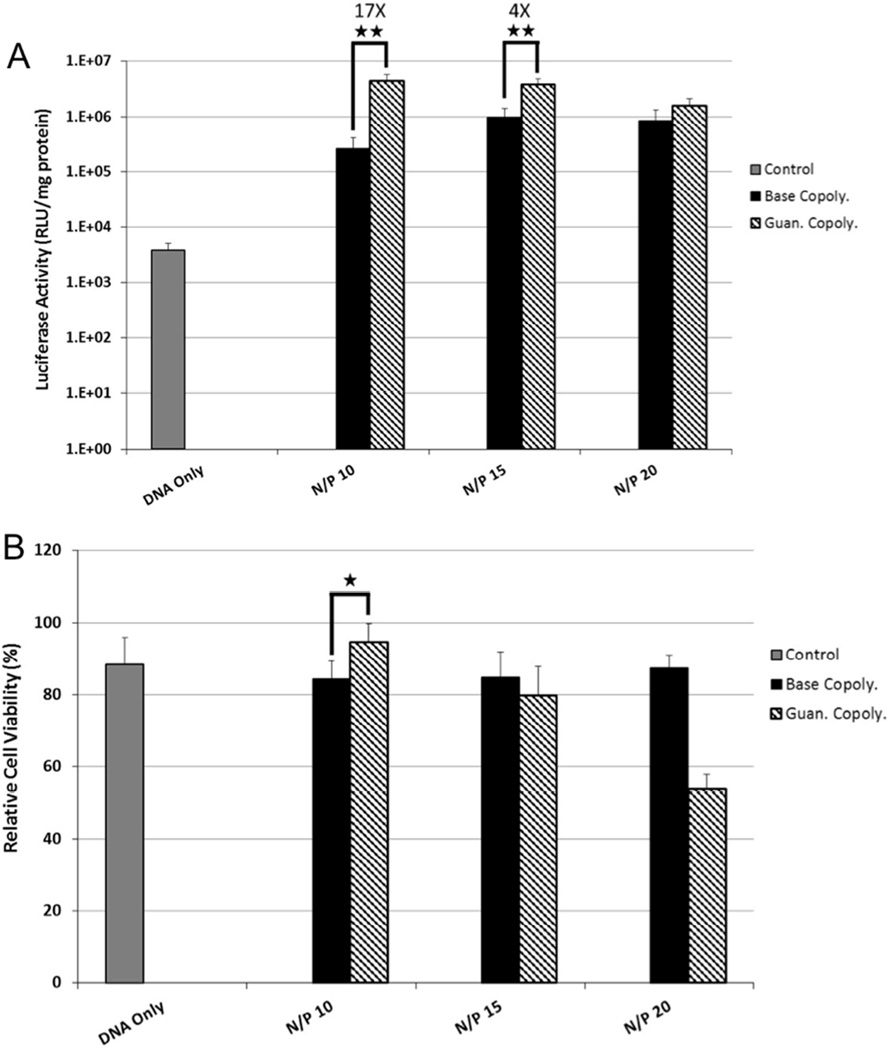
The luciferase plasmid was used as a reporter to track gene transfer efficiency with luciferase enzyme activity analyzed by light output. Polyplexes were prepared at N/P ratios of 10, 15, and 20 based on previous studies that showed N/P = 15 as an optimal formulation for the Base copolymer.[21] First, transfection efficiency and cytotoxicity to HeLa cervical carcinoma cells were evaluated as the immortalized cell line is a commonly-used standard for evaluation of transfection. The Guan copolymer transfected HeLa cells more efficiently (8 to 32-fold increased luciferase activity) than Base copolymer at all three charge ratios tested (Figure 1A). Transfection increased with increasing charge ratio at the cost of cell survival. Trends of reduced cell viability (decreased by ~5–10%) were observed for the guan copolymer compared to Base copolymer, but were not statistically significant (Figure 1B).
Figure 1.
(A) Transfection efficiency of Base and Guan copolymer polyplexes at various N/P ratios to cultured HeLa cells. Transfection efficiency was determined by delivering the luciferase plasmid and analyzing photon production from cell lysates. (B) Cytotoxicity of Base and Guan copolymer polyplexes at various N/P ratios to cultured HeLa cells as determined by protein content in cell lysate 48 hrs post transfection. Data shown as mean + SD (Student t-test, *p < 0.02, ** p < 0.002).
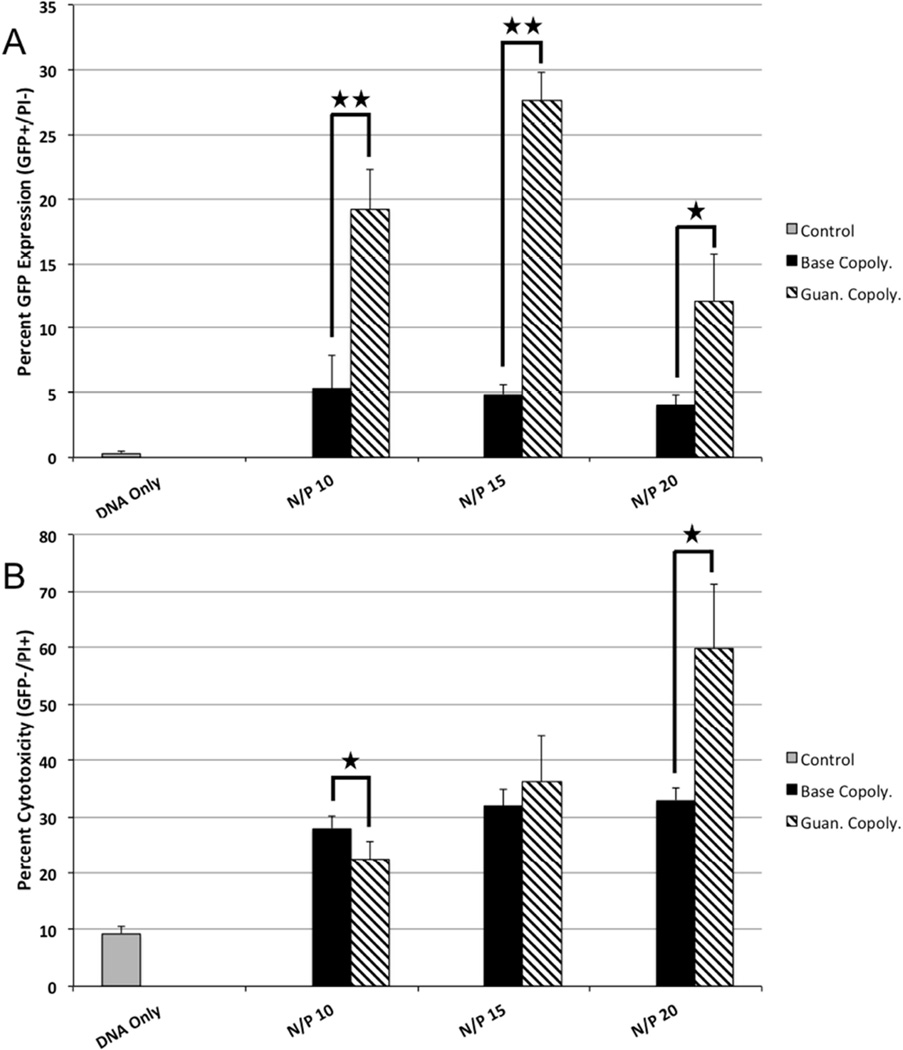
The luciferase reporter gene provides rapid information about total protein production from transgene delivery to a population of cells. To evaluate the percentage of transfected cells achieved using the Base versus Guan copolymer, the GFP plasmid was delivered using these polyplexes analysis by flow cytometry. Cells treated with polyplexes formed from the Guan copolymer had a higher percent GFP expression (GFP+/PI−) (3 to 5-fold increase) than cells treated with polyplexes from the Base copolymer at all charge ratios (Figure 2A). The Guan copolymer was more toxic to cells at high N/P ratios (Figure 2B).
Figure 2.
(A) Flow cytometry quantification of GFP plasmid transfection by Base and Guan copolymer polyplexes at various N/P ratios to cultured HeLa cells. (B) Cytotoxicity of Base and Guan copolymer polyplexes at various N/P ratios to cultured HeLa cells as determined by PI+/− staining. Data shown as mean + SD (Student t-test, *p < 0.04, ** p < 0.001).
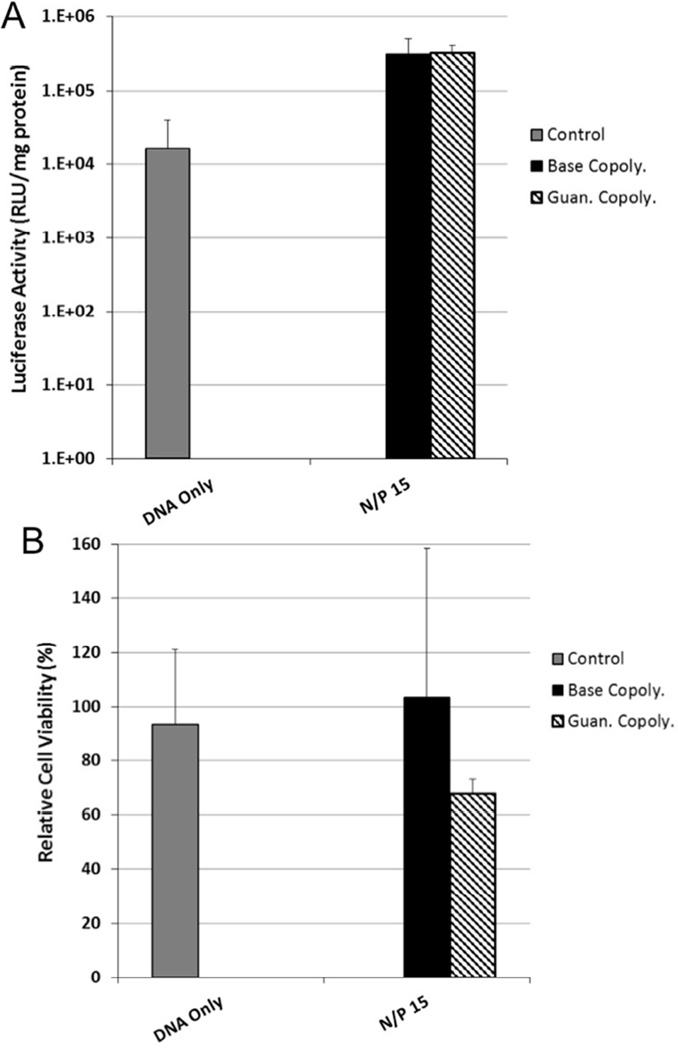
For intraventricular gene delivery to the brain, polyplexes first encounter the ependymal cells of the choroid plexus layer before reaching the NSCs and NPCs in the SVZ. Therefore, transfection efficiency of the polyplexes to both an immortalized rodent choroid plexus cell line, Z310, and to primary murine neural progenitor cells (NPCs) was next determined. As observed with HeLa cells, the Guan copolymer transfected Z310 cells more efficiently than the Base copolymer at both N/P = 10 and N/P = 15 (17-fold and 4-fold increase luciferase activity, respectively) (Figure 3A). No significant difference in luciferase activity was observed at N/P = 20, likely due to the increased toxicity from the Guan copolymer at this high charge ratio (Figure 3B). Based on this data, polyplexes at N/P = 15 were tested for transfection to primary NPCs in growth media at 37°C for 4 hours. In contrast to the results from the two epithelial cell lines, no increase in transfection efficiency was observed in NPCs by using Guan copolymer compared to Base copolymer (Figure 4). In addition, polyplexes formed using the Guan copolymer were more toxic to NPCs than polyplexes formed using Base copolymer.
Figure 3.
(A) Transfection efficiency of Base and Guan copolymer polyplexes at various N/P ratios to cultured Z310 immortalized choroid plexus cells. Transfection efficiency was determined by delivering the luciferase plasmid and analyzing photon production from cell lysates. (B) Cytotoxicity of Base and Guan copolymer polyplexes at various N/P ratios to cultured Z310 cells as determined by protein content in cell lysate 48 hrs post transfection. Data shown as mean + SD (Student t-test, *p < 0.05, ** p < 0.01).
Figure 4.
(A) Transfection efficiency of Base and Guan copolymer polyplexes at N/P=15 to primary murine neural progenitor cells. Transfection efficiency was determined by delivering the luciferase plasmid and analyzing photon production from cell lysates. (B) Cytotoxicity of Base and Guan copolymer polyplexes at N/P=15 to primary murine neural progenitor cells as determined by protein content in cell lysate 48 hrs post transfection. Data shown as mean + SD.
3.4 In vivo delivery via intraventricular injection to murine brain
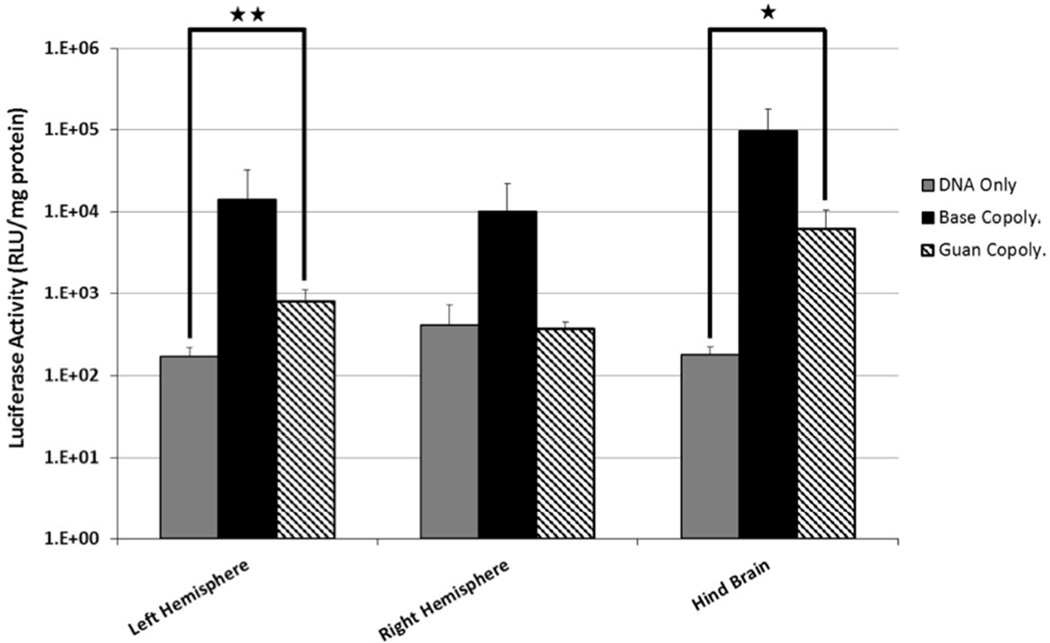
The N/P = 15 polyplexes were next tested for their ability to transfect cells in the murine SVZ. Polyplexes containing the luciferase plasmid were injected into the right lateral ventricle. Two days post injection, brain tissue was collected and separated into left hemisphere, right hemisphere, and hindbrain and then analyzed for luciferase expression. As observed previously, polyplexes formed with the Base copolymer transfected cells in vivo, resulting in luciferase activity ~104 relative light units (RLU)/mg protein in all three brain areas (Figure 5). Surprisingly, Guan copolymer polyplexes transfected cells less efficiently in vivo compared to Base copolymer polyplexes; luciferase activity was reduced by at least one order of magnitude in all sections of the brain. Statistically significant levels of reporter gene expression over DNA delivery alone were measured in the left hemisphere and hindbrains.
Figure 5.
Luciferase expression in murine brain sections 48 hrs post-administration of polyplexes (N/P=15) by intraventricular injection. (n = 6 per group; Student T-test, * p <0.02, ** p < 0.01).
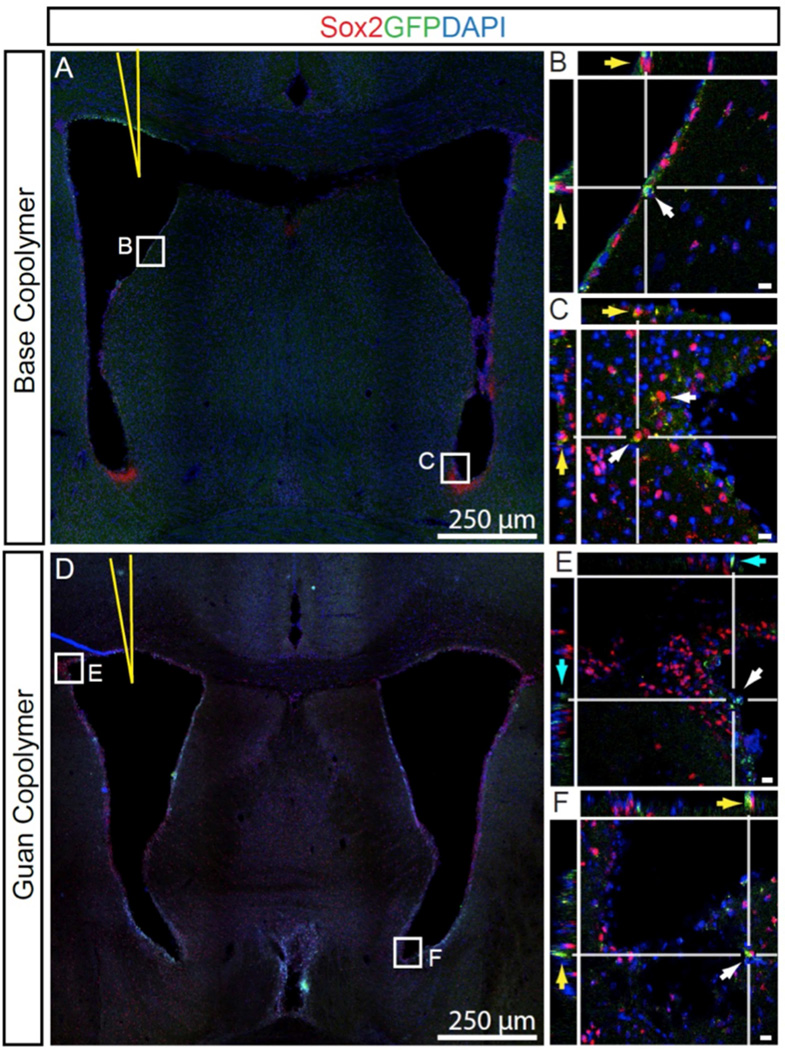
To further confirm these results, Base copolymer and Guan copolymer polyplexes (N/P = 15) containing the EGFP gene were prepared and administered by intraventricular injection as described above. The distribution of transfected cells in the brain two days after injection was determined by confocal imaging of coronal brain tissue sections stained with the neural stem cell marker, Sox2 and a nuclear stain, DAPI (Figure 6). As seen with luciferase expression, transfected, GFP+-expressing cells were reduced in number throughout the brain of Guan copolymer polyplex-treated mice. Brains injected with Base copolymer showed an abundance of the target cell population (GFP+ (green), Sox2+-cells (red)) within the injected ventricle (yellow arrows, Figure 6A, B) and within the contra lateral hemisphere (yellow arrows, Figure 6C). Conversely, few Sox2+-cells expressed GFP in brains injected with the Guan copolymer (Figure 6D–F). Importantly, Guan polymer treated brains had GFP expression primarily limited to the choroid and ependymal cells of the ventricle surface (Cyan arrow, Figure 6E).
Figure 6.
Confocal micrographs of GFP+ cells 48 hrs after delivery of polyplexes containing GFP plasmid. Base Copolymer complexes injected into the lateral ventricle (yellow needle) showed Sox2-cells transfected within the ipsilateral margin (B, yellow arrows) as well a numerous cells at the contralateral ventricle margin (C). Brains injected with Guan Copolymer showed markedly fewer Sox2+ GFP+-Cells at the ipsilateral (E, cyan arrow) and contralateral margin (F, yellow). Bar, 10 µm.
3.5. Evaluation of polyplex stability in the presence of heparan sulfate
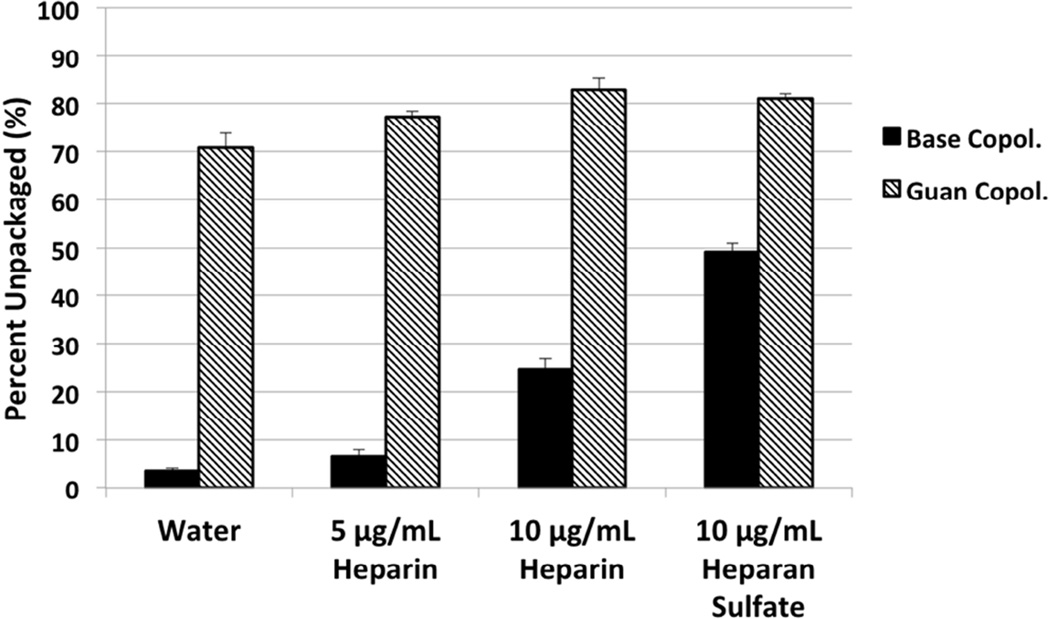
We hypothesized that the reduced transfection efficiency of Guan copolymer polyplexes in vivo might be due to differences in extracellular stability of the two formulations. Therefore, the stability of polyplexes in the presence of anionic glycosaminoglycans, heparin and heparan sulfate (HS), was determined using a YOYO-1 fluorescence quenching assay. The YOYO-1 dye fluoresces when intercalated in DNA; this fluorescence is quenched when the plasmid DNA is condensed in polyplex form. The compaction state of YOYO-1-labeled polyplexes incubated with HS for 1 hr was monitored by fluorescence emission measurements (Figure 7). While Base copolymer polyplexes treated with 5 µg/mL heparin remained condensed, with similar levels of YOYO-1 fluorescence as untreated polyplexes, Guan copolymer polyplexes showed nearly complete unpackaging, evidenced by the recovery of YOYO-1 fluorescence. Increasing heparin concentration to 10 µg/mL increased unpackaging in both copolymer polyplexes but Base copolymer polyplexes remained more condensed compared to Guan copolymer polyplexes. A similar result was obtained by competition with 10 µg/mL heparin in the gel retardation study of polyplexes (Figure S1). While the addition of heparin releases free plasmid from Guan polyplexes formed at N/P = 3, analogous polyplexes formed using Base copolymer remain stable in the presence of heparin. Thus, Base copolymer polyplexes are more resistant to competitive unpackaging by anionic glycosaminoglycans and proteoglycans compared to Guan copolymer polyplexes.
Figure 7.
Polyplex Unpackaging study. Polyplexes (N/P = 15) containing YOYO-1–labeled plasmids were incubated with heparin or heparan sulfate for 1 hr. The extent of DNA unpackaging was determined by measuring YOYO-1 fluorescence from polyplex solutions normalized to a control solution of YOYO-1 labeled plasmid at the same concentration.
4. Discussion
Peptides and polymers containing multiple guanidine groups, such as the TAT peptide from human immunodeficiency virus (HIV), have been shown to be effective materials for facilitating intracellular delivery to mammalian cells.[24, 25] In addition, an arginine-conjugated dendrimer has been used to deliver nucleic acids to brain by direct injection and by intranasal administration.[26, 27] In this work, we synthesized a guanidinylated analog of an efficient amine-based gene carrier that we previously developed.[21] Polyplexes formed from the two polymers were similar in physicochemical properties (Table 1), but consistent with several prior reports,[11–14] the guanidinylated materials transfected immortalized cultured cells with higher efficiency than the amine-based materials (Figures 1–3). Transfection efficiency to primary neural progenitor cells was comparable between the two polymers (Figure 4). However, when the polyplexes were injected intraventricularly to the murine brain, the amine-based polymers outperformed the guanidine-based polymers in gene transfer efficiency by over one order of magnitude (Figure 5).
The main type of cells transfected in the SVZ by intraventricular injection of polyplexes are NPCs due to their proliferative state and the strong preference of polyplexes to transfect dividing cells.[28] It was recently shown that the NPCs proliferate in niches directly in contact with “fractones”, extracellular matrix structures with branched fractal structures that are enriched in N-sulfate heparan sulfate (HS) proteoglycans, which bind and present the growth factor FGF-2 (fibroblast growth factor 2).[29, 30] We previously showed that HS proteoglycans in the liver prematurely unpackage polyplexes.[31] Juhasz and Biemann’s results with complexing oligoarginines with polyanionic biological molecules (such as oligonucleotides and proteoglycans) revealed that oligoarginines bind more strongly to sulfates than phosphates.[32] Therefore, we hypothesized that the guanidine-based polyplexes may be destabilized more by the SVZ fractones than lysine-based polyplexes. Indeed, several studies have compared the binding of the sulfates with either the guanidinium side chain of arginine or the amine side chain of lysine and found that greater affinity of sulfates to arginine.[33, 34] Fromm and coworkers, for example, demonstrated tighter interaction of arginine versus lysine for heparin, with Kd(lys)/Kd(arg) ~2.5, possibly due to stronger hydrogen bonding with guanidine, which can form parallel hydrogen bonds with sulfates, compared to lysine, which forms hydrogen bonds at 120°.
We tested this hypothesis by incubating polyplexes with both heparin and heparan sulfate, and monitoring unpackaging by a YOYO-1 dye-quenching assay (Figure 7). Guanidinylated polyplexes unpackaged readily in the presence of either heparin or heparan sulfate whereas amine-containing polyplexes resisted unpackaging under the experimental conditions. Similarly, Guan polyplexes were more destabilized by the presence of heparin than Base polyplexes as seen in the gel retardation electrophoresis (Figure S1). In addition, we found that neural progenitor cells could not be transfected with the guanidinylated polymer when heparin (used to bind FGF-2) is present in the media at 5 µg/mL (data not shown); instead, heparin had to be reduced to 1 µg/mL. Thus, the guanidine-based polyplexes are likely unpackaged to a greater extent in vivo compared to the amine-containing polyplexes due to stronger interactions with sulfated extracellular matrix components.
5. Conclusion
In this work, we compared the in vitro and in vivo gene transfer efficiency of an amine-and a guanidine-based polycation. While the guanidine-based polycation transfected cultured cells more efficiently, the amine-based polycation was more effective for in vivo gene transfer to the adult SVZ. Our data suggest that guanidinylated polyplexes may be more susceptible to premature, extracellular destabilization in vivo due to stronger affinity of the polymers for anionic proteoglycans encountered in the extracellular matrix. The molecular structure of the ventricular zone and the stem cell niche needs to be considered in the future design of materials for nucleic acid delivery to NPCs.
Supplementary Material
Acknowledgments
This work was supported by NIH 2R01NS064404; JKYT was supported by NSF GRFP (2011128558). We are grateful to Prof. Wei Zheng (Purdue University) for the generous donation of the Z310 cell line. We thank Prof. Patrick Stayton (University of Washington) for use of his ZetaPALS analyzer and Prof. Shaoyi Jiang (University of Washington) for use of his Malvern Zetasizer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 2.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 3.Chirumamilla S, Sun D, Bullock MR, Colello RJ. Traumatic brain injury induced cell proliferation in the adult mammalian central nervous system. J Neurotrauma. 2002;19:693–703. doi: 10.1089/08977150260139084. [DOI] [PubMed] [Google Scholar]
- 4.Dash PK, Mach SA, Moore AN. Enhanced neurogenesis in the rodent hippocampus following traumatic brain injury. Journal of Neuroscience Research. 2001;63:313–319. doi: 10.1002/1097-4547(20010215)63:4<313::AID-JNR1025>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 5.Ramaswamy S, Goings GE, Soderstrom KE, Szele FG, Kozlowski DA. Cellular proliferation and migration following a controlled cortical impact in the mouse. Brain Res. 2005;1053:38–53. doi: 10.1016/j.brainres.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 6.Hallbergson AF, Gnatenco C, Peterson DA. Neurogenesis and brain injury: managing a renewable resource for repair. The Journal of clinical investigation. 2003;112:1128–1133. doi: 10.1172/JCI20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimura S, Takagi Y, Harada J, Teramoto T, Thomas SS, Waeber C, et al. FGF-2 regulation of neurogenesis in adult hippocampus after brain injury. Proceedings of the National Academy of Sciences. 2001;98:5874–5879. doi: 10.1073/pnas.101034998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergen JM, Park I-K, Horner PJ, Pun SH. Nonviral approaches for neuronal delivery of nucleic acids. Pharmaceutical research. 2008;25:983–998. doi: 10.1007/s11095-007-9439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson BL, Breakefield XO. Viral vectors for gene delivery to the nervous system. Nature Reviews Neuroscience. 2003;4:353–364. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- 10.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 11.Carlson PM, Schellinger JG, Pahang JA, Johnson RN, Pun SH. Comparative study of guanidine-based and lysine-based brush copolymers for plasmid delivery. Biomaterials Science. 2013;1:736–744. doi: 10.1039/C3BM60079C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi JS, Nam K, Park J, Kim JB, Lee JK. Enhanced transfection efficiency of PAMAM dendrimer by surface modification with L-arginine. J Control Release. 2004;99:445–456. doi: 10.1016/j.jconrel.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 13.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Zheng N, Song Z, Yin L, Cheng J. The effect of side-chain functionality and hydrophobicity on the gene delivery capabilities of cationic helical polypeptides. Biomaterials. 2014;35:3443–3454. doi: 10.1016/j.biomaterials.2013.12.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gund P. Guanidine, trimethylenemethane, and" Y-delocalization." Can acyclic compounds have" aromatic" stability? Journal of Chemical Education. 1972;49:100. [Google Scholar]
- 16.Schug KA, Lindner W. Noncovalent binding between guanidinium and anionic groups: focus on biological-and synthetic-based arginine/guanidinium interactions with phosph [on] ate and sulf [on] ate residues. Chemical reviews. 2005;105:67–114. doi: 10.1021/cr040603j. [DOI] [PubMed] [Google Scholar]
- 17.Mascotti DP, Lohman TM. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry. 1997;36:7272–7279. doi: 10.1021/bi970272n. [DOI] [PubMed] [Google Scholar]
- 18.Standke KH, Brunnert H. Estimation of affinity constants for binding of model peptides to DNA by equilibrium dialysis. Nucleic Acids Res. 1975;2:1839–1849. doi: 10.1093/nar/2.10.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehling K, Arfmann HA, Standke KHC, Wagner KG. Specificity of DNA basic polypeptide interactions - influence of neutral residues incorporated into polylysine and polyarginine. Nucleic Acids Res. 1975;2:799–807. doi: 10.1093/nar/2.6.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Q, Huang Y, Zheng H, Wei T, Zheng S, Huo S, et al. The effect of guanidinylation of PEGylated poly (2-aminoethyl methacrylate) on the systemic delivery of siRNA. Biomaterials. 2013;34:3120–3131. doi: 10.1016/j.biomaterials.2013.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Wei H, Volpatti LR, Sellers DL, Maris DO, Andrews IW, Hemphill AS, et al. Dual Responsive, Stabilized Nanoparticles for Efficient In Vivo Plasmid Delivery. Angewandte Chemie International Edition. 2013;52:5377–5381. doi: 10.1002/anie.201301896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber CJ. The determination of the guanidine bases in urine. J Biol Chem. 1928;78:465–473. [Google Scholar]
- 23.Brancia FL, Oliver SG, Gaskell SJ. Improved matrix-assisted laser desorption/ionization mass spectrometric analysis of tryptic hydrolysates of proteins following guanidination of lysine-containing peptides. Rapid Commun Mass Spectrom. 2000;14:2070–2073. doi: 10.1002/1097-0231(20001115)14:21<2070::AID-RCM133>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 24.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Advanced drug delivery reviews. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Advanced drug delivery reviews. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim I-D, Lim C-M, Kim J-B, Nam HY, Nam K, Kim S-W, et al. Neuroprotection by biodegradable PAMAM ester (e-PAM-R)-mediated HMGB1 siRNA delivery in primary cortical cultures and in the postischemic brain. J Control Release. 2010;142:422–430. doi: 10.1016/j.jconrel.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Kim I-D, Shin J-H, Kim S-W, Choi S, Ahn J, Han P-L, et al. Intranasal delivery of HMGB1 siRNA confers target gene knockdown and robust neuroprotection in the postischemic brain. Mol Ther. 2012;20:829–839. doi: 10.1038/mt.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon EJ, Lasiene J, Jacobson BE, Park I-K, Horner PJ, Pun SH. Targeted nonviral delivery vehicles to neural progenitor cells in the mouse subventricular zone. Biomaterials. 2010;31:2417–2424. doi: 10.1016/j.biomaterials.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerever A, Schnack J, Vellinga D, Ichikawa N, Moon C, Arikawa-Hirasawa E, et al. Novel extracellular matrix structures in the neural stem cell niche capture the neurogenic factor fibroblast growth factor 2 from the extracellular milieu. Stem cells. 2007;25:2146–2157. doi: 10.1634/stemcells.2007-0082. [DOI] [PubMed] [Google Scholar]
- 30.Mercier F, Arikawa-Hirasawa E. Heparan sulfate niche for cell proliferation in the adult brain. Neuroscience letters. 2012;510:67–72. doi: 10.1016/j.neulet.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 31.Burke RS, Pun SH. Extracellular barriers to in vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chem. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 32.Juhasz P, Biemann K. Mass spectrometric molecular-weight determination of highly acidic compounds of biological significance via their complexes with basic polypeptides. Proceedings of the National Academy of Sciences. 1994;91:4333–4337. doi: 10.1073/pnas.91.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromm J, Hileman R, Caldwell E, Weiler J, Linhardt R. Differences in the interaction of heparin with arginine and lysine and the importance of these basic amino acids in the binding of heparin to acidic fibroblast growth factor. Archives of biochemistry and biophysics. 1995;323:279–287. doi: 10.1006/abbi.1995.9963. [DOI] [PubMed] [Google Scholar]
- 34.Wernersson E, Heyda J, Kubickova A, Krizek T, Coufal P, Jungwirth P. Effect of Association with Sulfate on the Electrophoretic Mobility of Polyarginine and Polylysine. J Phys Chem B. 2010;114:11934–11941. doi: 10.1021/jp1054342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.