Abstract
The communesins are a prominent class of indole alkaloids isolated from Penicillium species. Due to their daunting structural framework and potential as pharmaceuticals, communesins have inspired numerous total synthesis efforts. However, the genetic and biochemical basis of communesin biosynthesis has remained unexplored. Here, we report the identification and characterization of the communesin (cns) biosynthetic gene cluster from Penicillium expansum. We confirmed communesin is biosynthesized from the coupling of tryptamine and aurantioclavine, two building blocks derived from L-tryptophan. The post-modification steps were mapped by targeted-gene deletion experiments and structural elucidation of intermediates and new analogues. Our studies set the stage for biochemical characterization of the communesin biosynthesis towards understanding how nature generates remarkable structural complexity from simple precursors.
Keywords: Communesins, Biosynthesis, Alkaloids
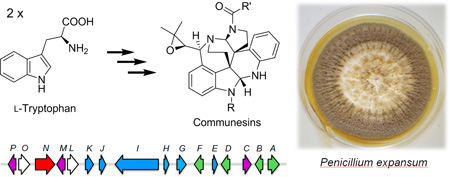
Communesins, a prominent class of complex indole alkaloids isolated from various Penicillium species, is shown to be biosynthesized from two different building blocks derived from l-tryptophan. The entire biosynthesis pathway is mapped by genetic inactivation studies, which led to the isolation of three new communesin analogues.
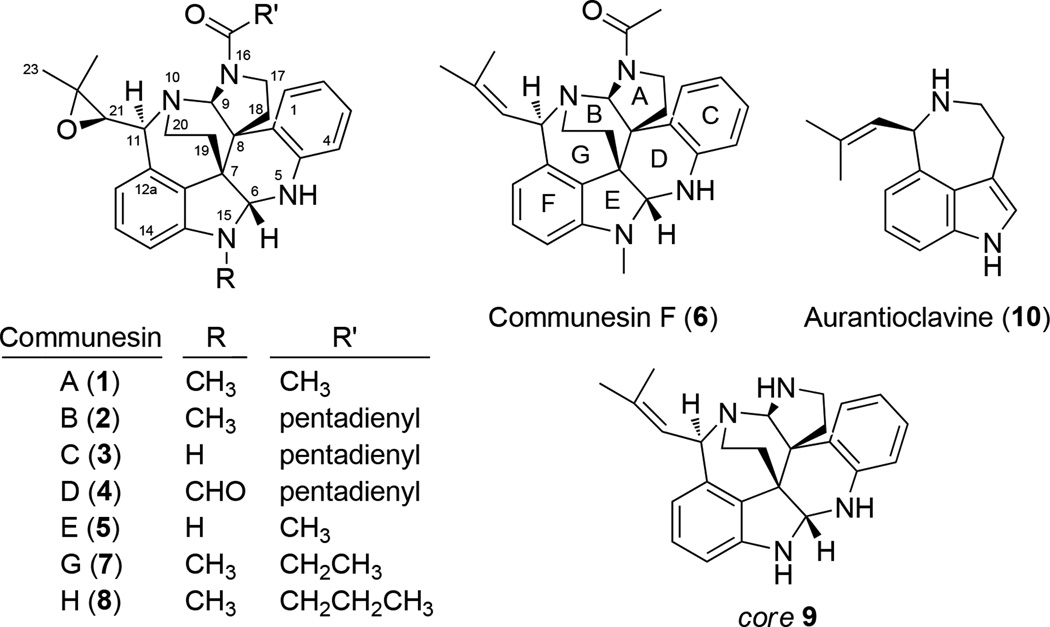
Many fungal indole alkaloids possess complex structures that are generated from concise biosynthetic pathways.[1] One of the most structurally complex family of indole alkaloids is the communesins (e.g., 1–8, Figure 1) isolated from marine and terrestrial Penicillium species.[2] The common core of the communesins, represented by structure 9, showcases the intricate molecular architectures of these polycyclic compounds. The natural products contain a total of 7 interconnected rings, two aminal linkages, and five (or six) stereocenters. The stereocenters at C7 and C8 are vicinal quaternary centers, which present a considerable synthetic challenge.
Figure 1.
Structures of the communesins and related compounds.
Given their remarkable structures, the communesins have inspired numerous total synthesis efforts in recent years,[3] including completed or formal total syntheses achieved by the groups of Qin,[3a, 3b] Ma,[3c, 3d] Weinreb,[3e, 3f] Funk,[3g, 3h] and Stoltz.[3i–k] In contrast, the biosynthetic pathway of communesins have remained unexplored since their isolation except preliminary feeding experiments that showed the communesin core is assembled from two indole precursors derived from tryptophan.[2e, 4] Stoltz and coworkers proposed several biosynthetic possibilities, all of which involved the coupling of tryptamine and aurantioclavine (10) as a key step towards building the core structure.[5] To understand how nature generates the structural complexity seen in the communesins, we set out to characterize the responsible alkaloid biosynthetic gene cluster. Here we report the genetic verification of the cns pathway from the known producer Penicillium expansum and isolation of new communesin analogues from blocked mutants.
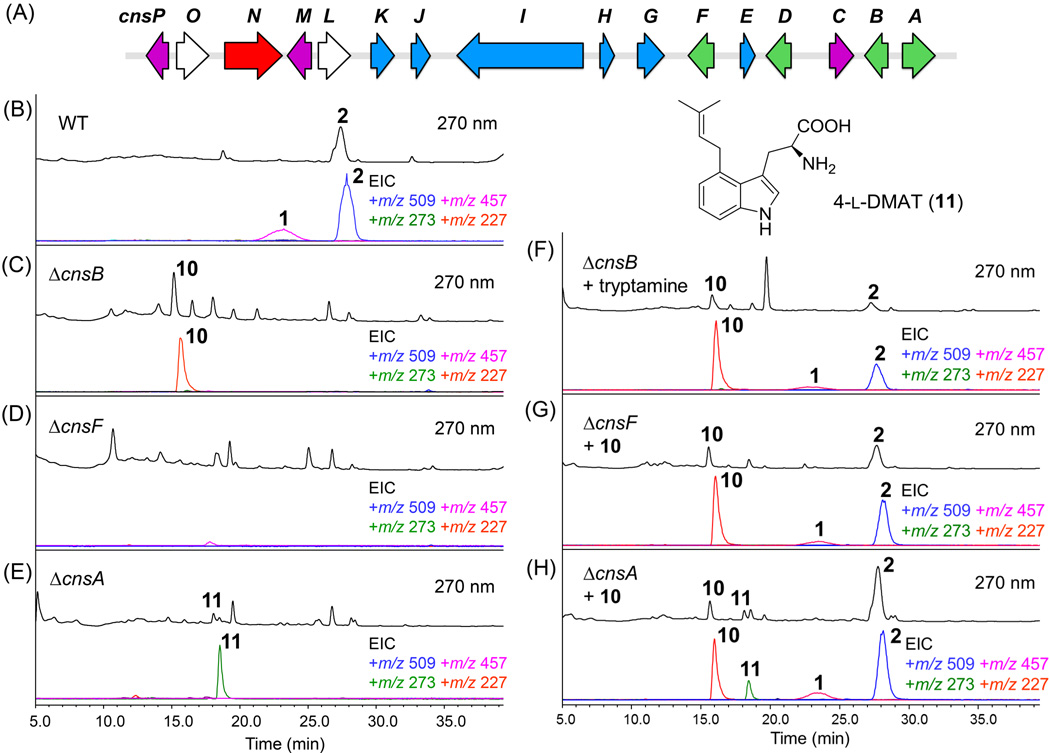
P. expansum NRRL 976 was confirmed to produce 1 and 2 (Figure 2B) by comparing the NMR spectra with reported data (Table S3–S4/Figures S11–S14).[2b, 2f, 6] Analysis of the sequenced genome using AntiSMASH2[7] revealed this strain is highly rich in secondary metabolism gene clusters, including 25 polyketide synthases (PKSs) and 17 nonribosomal peptide synthetases (NRPSs). We reasoned that the N-hexadienyl acyl chain in 2 should be derived from a highly reducing PKS, while the isoprenoid unit on the indolone should be installed by an enzyme homologous to 4-dimethylallyl tryptophan synthase (DMATS). Furthermore, precursor feeding studies showed that one equivalent of 6-fluoro-tryptamine can be incorporated into 1 and 2,[2e],[4] suggesting the likely presence of a tryptophan decarboxylase (TDC). We thus searched for a gene cluster that contained a PKS, DMATS and TDC in close proximity to each other in the P. expansum genome.
Figure 2.
Mapping the early stages of the communesin biosynthetic pathway. (A) The putative cns gene cluster from P. expansum NRRL 976. Genes are color-coded according to functions of encoded enzymes: precursor synthesis (green), post-core tailoring modifications (blue), other oxygenases (purple), fungal transcription factor (red) and those with unknown/unassigned function (white) (see Table S1). (B) Confirmation of production of 1 and 2 from the wild type strain. The UV (λ=270 nm) trace is shown above and the selected ion monitoring MS trace is shown below with the masses indicated in diffrent colors; (C)–(E) LC-MS analysis of metabolites extracted from ΔcnsA, ΔcnsB and ΔcnsF. (F)–(H) LC–MS analysis of metabolites produced from mutant strains shown in (C)–(E) after chemical complementation with proposed biosynthetic precursors. Tryptamine (F) or 10 (G and H) was supplemented at ~40 µg/mL.
A candidate gene cluster that encoded these three genes was found in Scaffold 19 (Figure 2A and Table S2). The putative cns gene cluster encodes, in additional to the PKS (CnsI), DMATS (CnsF) and TDC (CnsB), a methyltransferase (CnsE), an acyltransferase (CnsK), a serine hydrolase (CnsH) and a CoA-ligase (CnsG). In addition, an assortment of oxidative enzymes are found in the cluster, including a P450 (CnsC), a phytanoyl-CoA dioxygenase (CnsJ), and two non-heme iron-dependent oxygenases (CnsM and CnsP). Interestingly, the gene cluster contains a FAD-dependent monooxygenase (CnsA) and a putative catalase (CnsD) that share similarities to chanoclavine-I synthase (EasE) and its catalase partner (EasC), respectively (identity 51% and 59%, respectively).[8] EasC and EasE were reported to be involved in the cyclization of N-methyl-dimethylallyltryptophan (Me-DMAT) to form chanoclavine-I, a key intermediate in the biosynthesis of ergot alkaloids.[9] Hence CnsA and CnsD may be analogously involved in the synthesis of 10 through the cyclization of 4-DMAT 11.
We first inactivated cns genes that may be involved in precursor synthesis, such as cnsA, cnsB (TDC) and cnsF (DMATS), via double-crossover recombination using hygromycin-resistance gene hyg as a marker (see Supporting information, Figures S1–S8). In each knockout strain, the biosynthesis of both 1 and 2 were abolished, confirming the essential roles of these enzymes and the cns cluster in communesin biosynthesis (Figure 2C–E). The ΔcnsB mutant strain showed the accumulation of a new compound with λmax = 286 nm and m/z 227 [M+H]+, which was characterized to be aurantioclavine (10) by comparison to an authentic sample (Table S7 and Figures S19–S20). Complementing the ΔcnsB strain with tryptamine restored the biosynthesis of 1 and 2 (Figure 2F), confirming the role of CnsB in the generation of tryptamine.
In contrast, deletion of cnsF did not lead to the appearance of compounds that may be involved in the communesin pathway (Figure 2D). We hypothesized that this mutant cannot produce 4-DMAT 11 and subsequently 10. The absence of tryptamine in the extract may be due to fungal metabolic consumption. Chemical complementation of ΔcnsF with 10 restored the levels of 1 and 2, confirming that 10 is indeed a true intermediate en route to communesins as proposed by Stoltz et al.[3k] To assay the substrate specificity of CnsF towards either l-Trp or tryptamine, the recombinant protein was expressed and purified from E. coli BL21 (DE3) (Figure S9). CnsF displayed much higher turnover in converting l-Trp entirely into 11. In contrast, only partial conversion of tryptamine to 4-dimethylallyl tryptamine was observed (Figure S10). This is consistent with the assignment of CnsF as 4-DMATS based on its sequence similarly to FgaPT2.[10]
The ΔcnsA mutant strain accumulated a compound with λmax = 273 nm and m/z 273 [M+H]+, and was verified to be 11 (Table S8/Figure S21). Complementation of this strain with 10 restored the production of 1 and 2 (Figure 2H). Hence, CnsA was assigned as the aurantioclavine synthase that catalyses the conversion of 11 to 10. This conversion likely starts with flavin-dependent oxidation of the benzylic carbon in 11, followed by the intramolecular attack by the α-NH2. The subsequent decarboxylation then leads to the formation of 10 (Figure 4). This conversion may be similarly aided by the catalase CnsD as is the case during chanoclavine biosynthesis.[8b] Taken together, our knockout and chemical complementation experiments revealed the divergent synthesis of two different indole-containing building blocks, tryptamine and 10, from l-Trp.
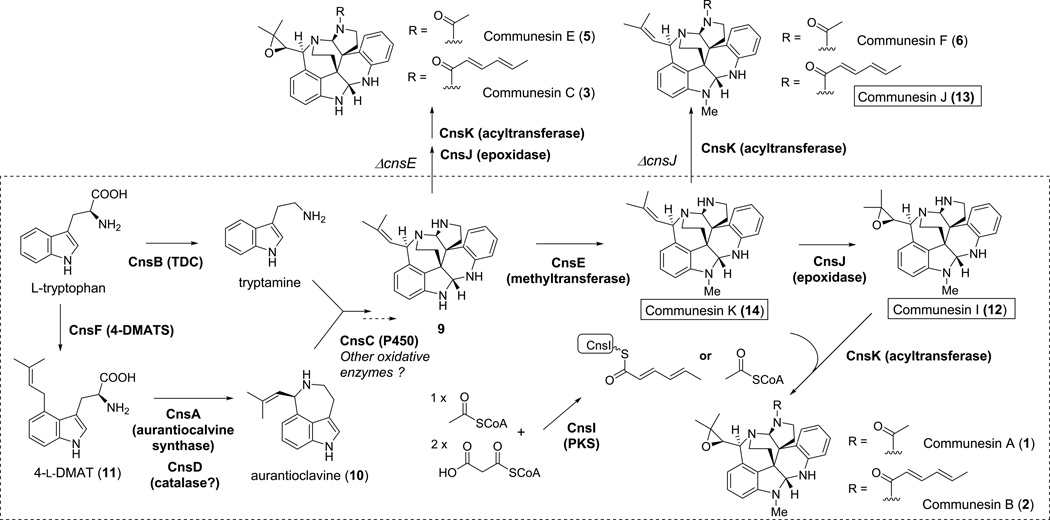
Figure 4.
The biosynthetic pathway of communesins mapped using knockout studies presenting in this work. The pathway in the dashed box represents the main pathway leading to 1 and 2 in P. expansum. The compounds outside of the box were isolated as shunt products using blocked mutants. Three new communesins, 12, 13 and 14 were produced.
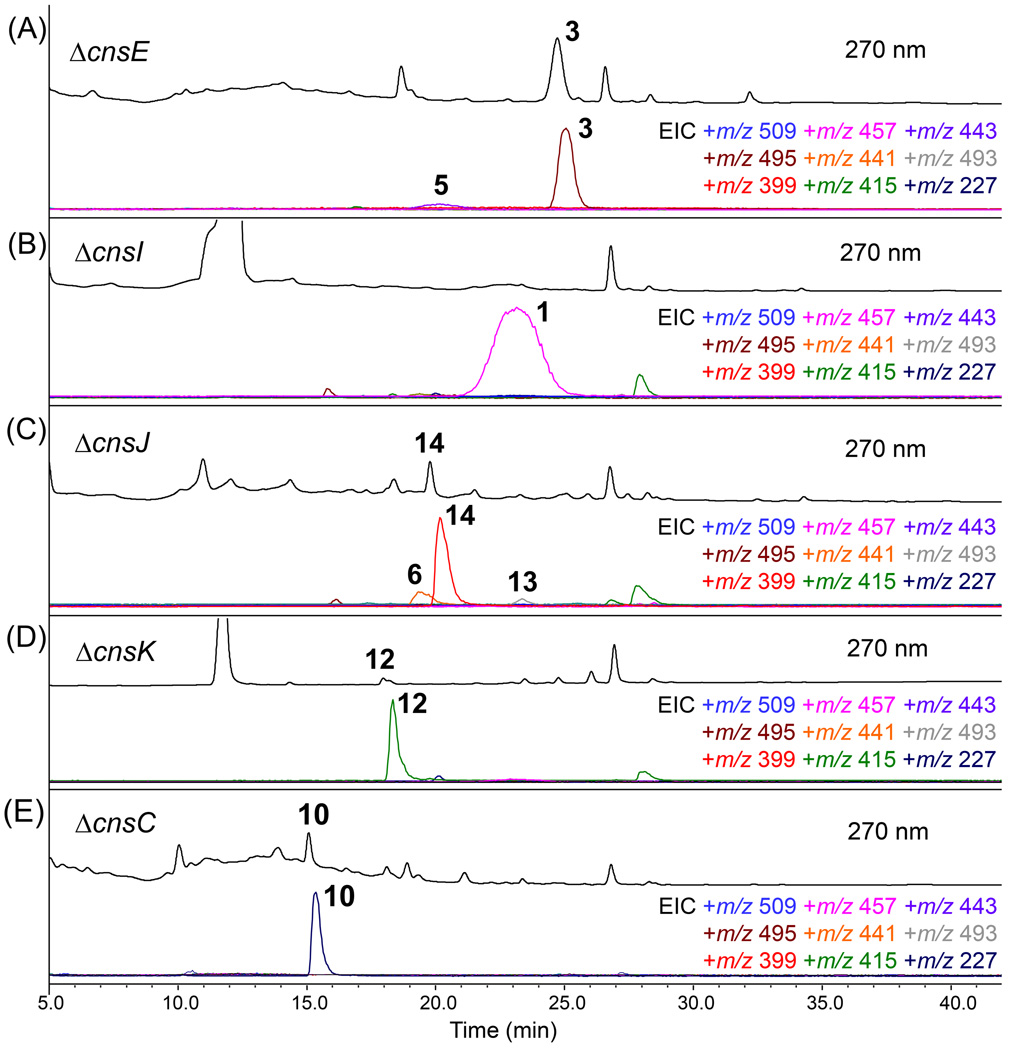
We then turned to the downstream enzymes that may modify the core 9 through methylation, acylation, and epoxidation. The N15-methylation and N16-acylation reactions take place on the secondary amines on opposite poles of the core, and may be important in stabilizing the otherwise labile aminal linkages. Deletion of cnsE, which encodes the sole methyltransferase in the cluster, led to the disappearance of 1 and 2, and the emergence of two compounds 3 (λmax = 268 nm, m/z 495 [M+H]+) and 5 (λmax = 269 nm, m/z 443 [M+H]+), which are the desmethyl versions of 2 and 1, respectively (Figure 3A). The structure of 3 was confirmed by NMR to be communesin C (Table S5/Figures S15–S16).
Figure 3.
Mapping the late stages of communesin biosynthetic pathway. Confirming the roles of enzymes encoded by (A) cnsE (methyltransferase), (B) cnsI (PKS), (C) cnsJ (epoxidase) and (D) cnsK (acyltransferase). The role of the enzyme encoded by cnsC (P450) was also investigated and shown in (E). In each set, the UV (λ=270 nm) trace is shown above and the selected ion monitoring MS trace is shown below with the masses indicated in diffrent colors.
The co-isolation of 1 and 2 suggests that N16-acylation by acetyl-CoA and the hexadienyl unit should be in competition. Acylation to introduce the hexadienyl chain in 2 is reminiscent of the acylation of fumagillol by a pentaene polyketide in the biosynthesis of fumagillin.[11] In the cns pathway, cnsI encodes a highly reducing PKS that has ketoreductase and dehydratase domains, but no enoylreductase domain. As expected, knockout of cnsI led to the exclusive formation of 1 (Figure 3B). The gene cnsK encodes an enzyme belonging to the transferase family (PFAM02458), and may play a role in transferring the polyketide chain to N16 of the core. Deletion of cnsK abolished the production of both 1 and 2. Instead, a single product 12 (λmax = 267 nm, m/z 415 [M+H]+) was produced. The new compound was structurally characterized to be the non-acylated version of 1 and 2 (Table S9/Figures S22–S23). Production of 12 (communesin I) confirmed the role of CnsK as the N16 acyltransferase. It is possible that the hexadienyl product is first hydrolytically removed from the PKS by the serine hydrolase CnsH, converted to hexadienyl-CoA by the CoA-ligase CnsG, and then transferred to 12 by CnsK. Surprisingly, the simultaneous loss of 1 in this mutant strain suggests that CnsK may also be a promiscuous acyltransferase that can tolerate a range of acyl groups, including acetyl-, propionyl- and butyryl-CoA that leads to 1, 7 and 8, respectively. Exploration of different acyl groups could lead to production of additional unnatural communesin analogues.
The gene cnsJ encodes a putative oxidative enzyme that displays 40% sequence similarity to FtmF found in the fumitremorgin pathway.[12] CnsJ may therefore install the C21–C22-epoxide in the prenylated portion of 1 and 2. Indeed, deletion of cnsJ led to production of 6 (m/z 441 [M+H]+), 13 (m/z 493 [M+H]+) and 14 (m/z 399 [M+H]+), all of which were structurally characterized (Tables S6, S10, S11/Figures S24–S32). The predominant product 14 (communesin K) is a new compound that is not acylated at N16 and non-oxidized at C21–22. The two minor products 6 and 13, are communesin F and a new analogue (communesin J), respectively (Figure 4). The accumulation of non-acylated 14 as the major product suggests that the downstream acyltransfer reaction catalysed by CnsK may be sluggish in the absence of the C21–C22 epoxide, and most likely takes place after the actions of CnsJ (Figure 4). Communesin K (14) represents the simplest characterized communesin that contains the heptacyclic core. We also constructed a ΔcnsJ/ΔcnsE double mutant in an attempt to isolate 9. Although selective ion monitoring showed the presence of a new compound with UV and mass consistent to that of 9, isolation of was unsuccessful due to rapid degradation of the molecule. Although we cannot pinpoint the timing of the methylation at N15 with the knockout studies, the structure of 14 coupled with its significantly improved stability compared to 9, suggests that the methylation step should take place immediately after formation of 9 to yield 14.
Oxidative coupling of 10 and tryptamine was proposed to be a possible route of forming the challenging vicinal quaternary centres of the communesins.[5]. We therefore investigated the role of CnsC, a cytochrome P450 encoded in the cluster. Protein sequence analysis suggests CnsC may be an atypical P450 with low sequence homology (<25%) to those in the P450 database.[13] Genetic inactivation of cnsC led to abolishment of 1 and 2, and the accumulation of 10 (Figure 3E). Since tryptamine is not detectable in the culture, we supplied tryptamine to ΔcnsC to ensure the block mutant is not defective in tryptamine biosynthesis. Supplementation of both tryptamine and 10 did not restore communesin biosynthesis. These results therefore hint that CnsC should act downstream of both 10 and tryptamine, and likely catalyses a key step in coupling these two indole derivatives together to form 9. The exact roles of this enzyme, along with other oxidative enzymes, such as CnsP, are currently under investigation using biochemical means.
In conclusion, the communesin biosynthesis pathway was mapped by genetic inactivation studies and led to the isolation of three new communesin analogues. The involvement of aurantioclavine as a building block was confirmed. Our studies provide a solution to the biosynthetic mystery surrounding this famous class of indole alkaloids, and set the stage for detailed biochemical characterization to understand how nature generates exceptional structural complexity from simple building blocks.
Supplementary Material
Acknowledgments
H-C. L. is supported by the National Science Council of Taiwan (102-2917-I-564-008) and G. C. was supported by a NSF Graduate Research Fellowship (DGE-0707424). This work was supported by NIH 1DP1GM106413 and 1R56AI101141. We thank Prof. Brian Stoltz for the gift of aurantioclavine. NMR instrumentation is supported by NSF (CHE-1048804) and the NCRR (S10RR025631).
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Hsiao-Ching Lin, Department of Chemical and Biomolecular Engineering Department of Chemistry and Biochemistry University of California, Los Angeles Los Angeles, CA 90095 (USA).
Grace Chiou, Department of Chemistry and Biochemistry University of California, Los Angeles.
Yit-Heng Chooi, Plant Sciences Division, Research School of Biology, The Australian National University, Australia.
Travis C. McMahon, Department of Chemistry and Biochemistry University of California, Los Angeles
Wei Xu, Department of Chemical and Biomolecular Engineering Department of Chemistry and Biochemistry University of California, Los Angeles Los Angeles, CA 90095 (USA).
Neil K. Garg, Email: neilgarg@chem.ucla.edu, Department of Chemistry and Biochemistry University of California, Los Angeles.
Yi Tang, Email: yitang@ucla.edu, Department of Chemical and Biomolecular Engineering Department of Chemistry and Biochemistry University of California, Los Angeles Los Angeles, CA 90095 (USA).
Reference
- 1.Xu W, Gavia DJ, Tang Y. Nat. Prod. Rep. 2014;31:1474–1487. doi: 10.1039/c4np00073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Jadulco R, Edrada RA, Ebel R, Berg A, Schaumann K, Wray V, Steube K, Proksch P. J. Nat. Prod. 2004;67:78–81. doi: 10.1021/np030271y. [DOI] [PubMed] [Google Scholar]; (b) Hayashi H, Matsumoto H, Akiyama K. Biosci. Biotechnol., Biochem. 2004;68:753–756. doi: 10.1271/bbb.68.753. [DOI] [PubMed] [Google Scholar]; (c) Andersen B, Smedsgaard J, Frisvad JC. J. Agric. Food Chem. 2004;52:2421–2428. doi: 10.1021/jf035406k. [DOI] [PubMed] [Google Scholar]; (d) Dalsgaard PW, Blunt JW, Munro MHG, Frisvad JC, Christophersen C. J. Nat. Prod. 2005;68:258–261. doi: 10.1021/np049646l. [DOI] [PubMed] [Google Scholar]; (e) Wigley LJ, Mantle PG, Perry DA. Phytochemistry. 2006;67:561–569. doi: 10.1016/j.phytochem.2005.10.011. [DOI] [PubMed] [Google Scholar]; (f) Numata A, Takahashi C, Ito Y, Takada T, Kawai K, Usami Y, Matsumura E, Imachi M, Ito T, Hasegawa T. Tetrahedron Lett. 1993;34:2355–2358. [Google Scholar]
- 3.(a) Wu HX, Xue F, Xiao X, Qin Y. J. Am. Chem. Soc. 2010;132:14052–14054. doi: 10.1021/ja1070043. [DOI] [PubMed] [Google Scholar]; (b) Yang J, Wu HX, Shen LQ, Qin Y. J. Am. Chem. Soc. 2007;129:13794-+. doi: 10.1021/ja075705g. [DOI] [PubMed] [Google Scholar]; (c) Zuo Z, Xie W, Ma D. J. Am. Chem. Soc. 2010;132:13226–13228. doi: 10.1021/ja106739g. [DOI] [PubMed] [Google Scholar]; (d) Zuo Z, Ma D. Angew. Chemie. Intl. Ed. 2011;50:12008–12011. doi: 10.1002/anie.201106205. [DOI] [PubMed] [Google Scholar]; (e) Liu P, Seo JH, Weinreb SM. Angew. Chemie. Intl. Ed. 2010;49:2000–2003. doi: 10.1002/anie.200906818. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Seo JH, Liu P, Weinreb SM. J. Org. Chem. 2010;75:2667–2680. doi: 10.1021/jo100339k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Belmar J, Funk RL. J. Am. Chem. Soc. 2012;134:16941–16943. doi: 10.1021/ja307277w. [DOI] [PubMed] [Google Scholar]; (h) Crawley SL, Funk RL. Org. Lett. 2003;5:3169–3171. doi: 10.1021/ol034407v. [DOI] [PubMed] [Google Scholar]; (i) Han SJ, Vogt F, May JA, Krishnan S, Gatti M, Virgil SC, Stoltz BM. J. Org. Chem. 2014 doi: 10.1021/jo502534g. DOI: 10.1021/jo502534g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Han SJ, Vogt F, Krishnan S, May JA, Gatti M, Virgil SC, Stoltz BM. Org. Lett. 2014;16:3316–3319. doi: 10.1021/ol5013263. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) May JA, Zeidan RK, Stoltz BM. Tetrahedron Lett. 2003;44:1203–1205. [Google Scholar]; (l) Siengalewicz P, Gaich T, Mulzer J. Angew. Chemie. Intl. Ed. 2008;47:8170–8176. doi: 10.1002/anie.200801735. [DOI] [PubMed] [Google Scholar]
- 4.Wigley LJ, Perry DA, Mantle PG. Mycol. Res. 2008;112:131–137. doi: 10.1016/j.mycres.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 5.May JA, Stoltz B. Tetrahedron. 2006;62:5262–5271. [Google Scholar]
- 6.Kerzaon I, Pouchus YF, Monteau F, Le Bizec B, Nourrisson M-R, Biard J-F, Grovel O. Rapid Commun. Mass Spectrom. 2009;23:3928–3938. doi: 10.1002/rcm.4330. [DOI] [PubMed] [Google Scholar]
- 7.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Lorenz N, Olsovska J, Sulc M, Tudzynski P. Appl. Environ. Microbiol. 2010;76:1822–1830. doi: 10.1128/AEM.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goetz KE, Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. Curr. Genet. 2011;57:201–211. doi: 10.1007/s00294-011-0336-4. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen CAF, Folly C, Hatsch A, Molt A, Schroder H, O'Connor SE, Naesby M. Microb. Cell Fact. 2014;13 doi: 10.1186/s12934-014-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unsold IA, Li SM. Microbiology-Sgm. 2005;151:1499–1505. doi: 10.1099/mic.0.27759-0. [DOI] [PubMed] [Google Scholar]
- 11.Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y. J. Am. Chem. Soc. 2013;135:4616–4619. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffan N, Grundmann A, Afiyatullov S, Ruan H, Li S-M. Org. Biomol. Chem. 2009;7:4082–4087. doi: 10.1039/b908392h. [DOI] [PubMed] [Google Scholar]
- 13.Nelson DR. Hum. Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.