Abstract
Biofilms are a predominant form of growth for bacteria in the environment and in the clinic. Critical for biofilm development are adherence, proliferation, and dispersion phases. Each of these stages includes reinforcement by, or modulation of, the extracellular matrix. Pseudomonas aeruginosa has been a model organism for the study of biofilm formation. Additionally, other Pseudomonas species utilize biofilm formation during plant colonization and environmental persistence. Pseudomonads produce several biofilm matrix molecules, including polysaccharides, nucleic acids, and proteins. Accessory matrix components shown to aid biofilm formation and adaptability under varying conditions are also produced by pseudomonads. Adaptation facilitated by biofilm formation allows for selection of genetic variants with unique and distinguishable colony morphology. Examples include rugose small-colony variants and wrinkly spreaders (WS), which over produce Psl/Pel or cellulose, respectively, and mucoid bacteria that over produce alginate. The well-documented emergence of these variants suggests that pseudomonads take advantage of matrix-building subpopulations conferring specific benefits for the entire population. This review will focus on various polysaccharides as well as additional Pseudomonas biofilm matrix components. Discussions will center on structure–function relationships, regulation, and the role of individual matrix molecules in niche biology.
Keywords: polysaccharide, eDNA, alginate, levan, Psl, Pel
Introduction
Cooperation among communities has significant biological advantages for individual members. Herds of mammals, flocks of birds, schools of fish, or colonies of insects are all prime examples of sociobiology existing to afford members of the population advantages they would not solely achieve. While in many cases the organisms that reside amidst a community can survive some period of time without their counterparts, life becomes simpler with the protection of the group. Prominent examples include protection from predators, adverse environmental conditions, and starvation. The latter is especially important for weaker members of the society who easily gain access to a consistent nutrient supply. In one of life's simplest forms, microorganisms are being increasingly studied in the context of community organization. Similar to the advantages that herds, schools, and flocks acquire, bacterial biofilms confer protection from external pressures while maintaining persistence from harsh environmental conditions (Klausen Costerton, Stewart & Costerton, 2001). Interestingly, cells within biofilm have diverse gene expression although the community is comprised of clonal members (Boles & Singh, 2008). Examples of polymicrobial biofilms are prevalent, adding an additional layer of complexity to the cooperative and competitive nature within biofilms. Sociobiology of community microorganisms is not a novel concept but proposed to explain heterogeneity in biofilm. While many advantages of sociobiology exist, members failing to provide a community asset, described as ‘selfish cheaters’ who take advantage of the community lifestyle regardless of input, also seem to be present (West et al., 2006). This review will discuss the components used by the predominant bacterial biofilm-forming genus Pseudomonas to build and maintain fundamental biofilm communities. Greater detail regarding the sociobiology perspective of biofilm cells can be gleamed from West et al., (2006) discussion. Specifically, production of virulence-relevant polysaccharides by Pseudomonas aeruginosa used for biofilm matrix molecules will be covered in greatest detail.
The pseudomonads are ubiquitous environmental organisms, occupying several niches. While some Pseudomonas species have the propensity to cause disease, others simply reside in their natural habitat. Frequently in either pathogenic or environmental conditions, the bacteria exist attached to a surface and encased in some form of polymeric substances, characteristic of a biofilm. Biofilm formation by P. aeruginosa is of particular interest because of its clinical relevance (Donlan & Costerton, 2002), but other Pseudomonas spp. have also been studied in conditions such as plant tissue (Osman et al., 1986; Fakhr, et al., 1999; Preston et al., 2001), soil (Schnider-Keel, et al., 2001; Dechesne et al., 2010) or fresh water streams (Costerton et al., 1987). Comparison of Pseudomonas spp. biofilm formation strategies will aid a more complete understanding of the individual species’ unique and conserved mechanisms.
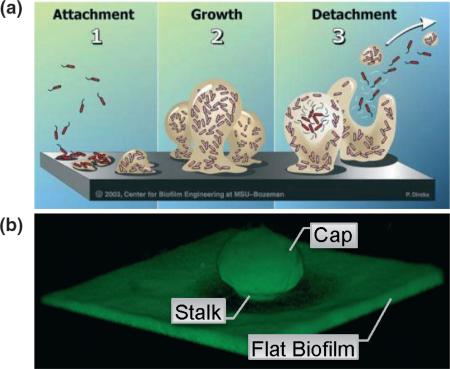
Biofilm-forming organisms rely on extracellular polymeric substance (EPS), also known as matrix, which is essential for colonization of surfaces and volumes (Sutherland, 2001a, b; Flemming et al., 2007). Biofilm development is highlighted in Box 1; where the P. aeruginosa biofilm paradigm describes individual bacteria cells initiating adherence to a substratum followed by clonal propagation, matrix building, and eventual biofilm maturation (Costerton et al., 1995; Costerton et al., 1999; Stoodley et al., 2002). Finally, biofilm populations release or disperse small aggregates or even individual cells for seeding of uncolonized sites and reinitiating the biofilm lifecycle (Costerton et al., 1995; Ma et al., 2009; Monds & O'Toole, 2009). Biofilm maturity is hallmarked by the presence of ‘capped’ mushroom-shaped structures (Box 1). The flat biofilm occurs during early surface colonization and propagation. Dense microcolonies initiate stalk formation that eventually forms the capped mushroom structures that are characteristic of P. aeruginosa biofilm (Box 1). Cell surface factors have been identified, which allow for initial interaction with surfaces and structure formation including pili (Klausen et al., 2003a, b), flagella (O'Toole & Kolter, 1998; Klausen et al., 2003a, b), proteins (Monds et al., 2007; Newell et al., 2009; Borlee et al., 2010), and extracellular polysaccha-rides (Nivens et al., 2001; Wozniak et al., 2003; Friedman & Kolter, 2004a, b; Jackson et al., 2004; Matsukawa & Greenberg, 2004; Ma et al., 2006; Ryder et al., 2007; Starkey et al., 2009; Byrd et al., 2010). These factors are commonly categorized as biofilm matrix components. Interestingly, in recent years, the predominance of nucleic acids among biofilm EPS has lead to the investigation of the importance of DNA in stabilizing the biofilm matrix (Whitchurch et al., 2002; Webb et al., 2003; Allesen-Holm et al., 2006; Yang, et al., 2007). In most cases, one or two of these components are the most abundant in the biofilm matrix at a given stage of biofilm development or in individual strains, although it is common to have accessory function from other components. Predominantly, P. aeruginosa requires polysaccha-ride in its biofilm matrix at several developmental stages, while taking advantage of nucleic acids during later maturation stages (Klausen et al., 2003a, b; Webb et al., 2003; Allesen-Holm et al., 2006; Yang et al., 2007; Ma et al., 2009). Two distinct classes of Pseudomonas polysaccharides have been shown to have a role in biofilm formation. Capsular polysaccharides maintain characteristics of protective dynamic polymers that decorate the exterior of one or more cells. Alternatively, aggregative polysaccharides offer structural integrity and interact with additional matrix components. Capsular polysaccharides provide a coat around a bacterium, while aggregative polysaccharides do not. Both capsular and aggregative polysaccharides will be the primary focus of this review as well as accessory biofilm matrix molecules influencing the function of Pseudomonas biofilms. The functions of each of these biofilm matrix components will also be discussed in the context of roles they play within the habitat that the microorganism resides (i.e. niche biology).
Capsular polysaccharides
Pseudomonas spp. has been isolated from diverse environmental conditions, and characterization of these isolates reveals that polysaccharide production is important for colonization of these niches (Osman et al., 1986; Keith & Bender, 1999; Gal et al., 2003; Laue et al., 2006; Chang et al., 2007; Li et al., 2010). Furthermore, the production of polysaccharides by biofilm-forming organisms supports colonization by facilitating aggregation, adherence, and surface tolerance (Osman et al., 1986; Penaloza-Vazquez et al., 1997; Keith et al., 2003; Laue et al., 2006). Most notably, P. aeruginosa produces alginate, and because of its correlation with adverse clinical outcomes, it has been closely monitored and investigated (Boucher et al., 1997; Hentzer et al., 2001; Nivens et al., 2001; Bjarnsholt, et al., 2009). Surveys of non-aeruginosa pseudomonads including phytopathogens have revealed that they produce alginate similar to what has been described for P. aeruginosa (Schnider-Keel et al., 2001; Laue et al., 2006). Additionally, various species produce levan, which also aids colonization and biofilm persistence (Laue et al., 2006). Levan has deviations in structure and composition that suggests it has unique functions distinct from those of alginate (Sutherland, 2001a, b).
Capsular polysaccharides: alginate
Molecular structure
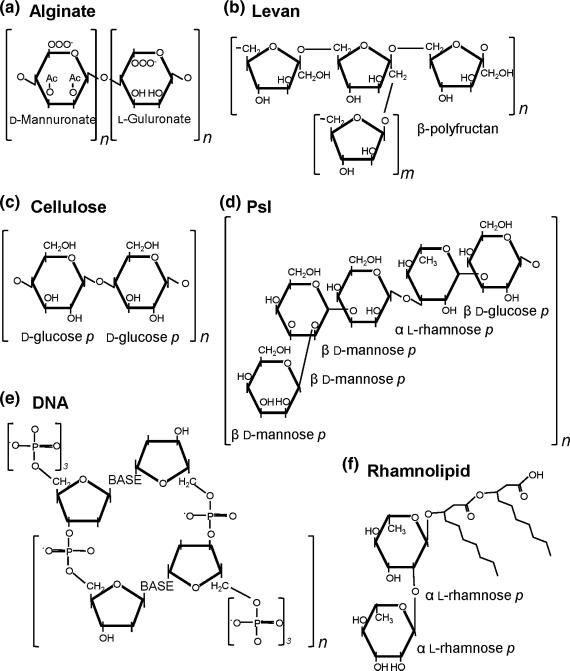
Alginate is an important matrix molecule for Pseudomonas biofilm formation by providing more than structural stability (Hentzer et al., 2001; Nivens et al., 2001). Specifically, alginate is a high molecular weight, acetylated polymer with nonrepetitive monomers of β-1,4 linked L-guluronic and D-mannuronic acids (Fig. 1a) (Evans & Linker, 1973). It is normally O-acetylated at the 2 and/or 3 position(s) on the D-mannuronate residues (Osman, et al., 1986). Both β-1,4 and β-1,3 linkages typically confer considerable rigidity (Fig. 1a) compared to β-1,2 and β-1,6 linkages found commonly in dextrans (Sutherland, 2001a, b). Alginate and the biosynthetic enzymes are well conserved across pseudomonads (Penaloza-Vazquez et al., 1997; Fakhr et al., 1999; Li et al., 2010).
Fig. 1.
Abundant biofilm matrix molecules. Adapted representative chemical structures of (a) alginate, (b) levan, (c) cellulose, (d) Psl, (e) DNA, and (f) rhamnolipid. Brackets depict repeating units of each molecule.
Regulatory factors
Concurrent with the identification of the alginate-related biofilm phenotype was the identification that specific genetic events lead to the overproduction of alginate in mucoid strains (Lam et al., 1980; Deretic et al., 1993; Martin et al., 1993; DeVries & Ohman, 1994; Mathee, et al., 1999; Hentzer et al., 2001; Qiu et al., 2007). Mucoid conversion occurs primarily through a selective process in patients with lungs cystic fibrosis (CF). CF is a complicated disease that has many manifestations, not the least of which is the buildup of dehydrated mucus in the airway allowing colonization of pathogens such as P. aeruginosa. It is also within the CF lung where spontaneous mutations arise allowing for successful competition amidst the rest of the population. The chronic inflamma-tory state of the CF lung likely facilitates mucoid conversion through reactive oxygen intermediates increasing DNA damage of P. aeruginosa (Deretic et al., 1993; Martin et al., 1993; Mathee et al., 1999). The overwhelming majority of mucoid strains from the CF airway possess mutations in mucA that results in a protein that is defective in interacting with the sigma factor (AlgT/U) controlling alginate expression (Martin et al., 1993). Under nonmucoid conditions, MucA sequesters with the sigma factor AlgT/U. Degradation or truncation of MucA results in a loss of the ability to interact with AlgT/U, allowing it to potentiate alginate production and ultimately a conversion to a mucoid phenotype (Martin et al., 1993; Mathee et al., 1997). While the prevalence of mucA mutants is noteworthy, especially mucA22 being the most abundant, stable non-mucA mutations have also been isolated that result in an identical mucoid phenotype (Martin et al., 1993; Boucher et al., 2000). Alginate regulation is complex, and perturbation of other regulatory factors, in addition to AlgT/U, may result in a similar phenotype.
The mutations resulting in constitutive alg operon expression and mucoid conversion can be recapitulated in vitro using specific strategies, including hydrogen peroxide-induced DNA damage (Mathee et al., 1999). Recent work has linked mucoid conversion mediated by oxidative stress with the SOS response and the error-prone DNA polymerase DinB (Sanders et al., 2006; Moyano et al., 2007). The frequency of which P. aeruginosa is able to adapt to its surroundings and persist is striking, and this also suggests that alginate provides an advantage during infections, especially in the CF lung (Hogardt & Heesemann, 2010). Alginate-aided protection of biofilm is important for colonization of surfaces by Pseudomonas environmental and phytopathogen species (Keith & Bender, 1999; Yu et al., 1999; Keith et al., 2003; Spiers et al., 2003). The virtually identical organization and conservation of the alginate biosynthesis loci in Pseudomonas putida and Pseudomonas syringae suggests that alginate overproduction in the non-aeruginosa pseudomonads is likely regulated similarly to P. aeruginosa (Penaloza-Vazquez et al., 1997; Fakhr et al., 1999; Li et al., 2010). One significant caveat is that the 5′ regulatory sequences of the P. syringae algD region, including the AlgT/U promoter regions, lack similarity to the P. aeruginosa algD promoter region (Fakhr et al., 1999). This could help explain some of the divergent environmental conditions affecting alginate production by P. syringae such as copper exposure (Kidambi et al., 1995; Penaloza-Vazquez et al., 1997). The algC gene, located distant from the alg operon and encoding a phosphomannomutase, is also essential for alginate biosynthesis (Zielinski, et al., 1990; Zielinski et al., 1991; Zielinski et al., 1992). Regardless of the high level of alginate gene conservation, it is unclear whether environmental or phytopathogenic strains undergo similar mucoid conversion processes as seen in P. aeruginosa. However, it was noted that plant tissue damage is associated with a hypersensitive response (HR), correlated with increased algD expression by P. syringae (Keith et al., 2003). The HR is generally characterized by brown spots with dead plant cells at the site of infection, often associated with some form of programmed cell death (Heath, 2000). HR is also characterized to have an abundance of reactive oxygen species, perhaps similar to the chronic inflammatory response, which facilitates mucoid conversion of P. aeruginosa at relatively high frequency (Mathee et al., 1999). Perhaps, a similar mechanism for alginate overproduction occurs during colonization of P. syringae on plant tissue.
Function in biofilm matrix
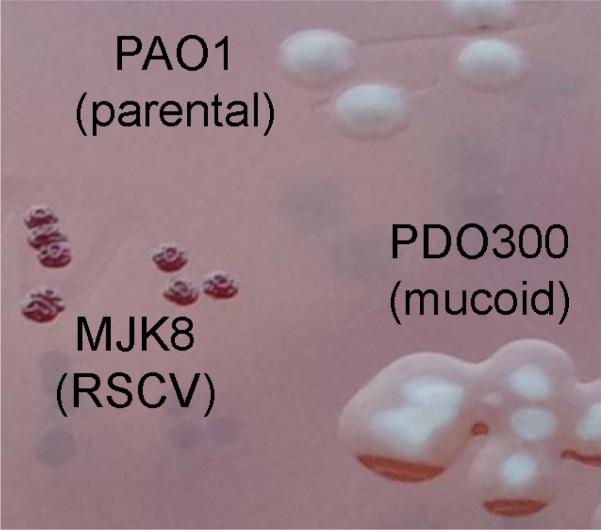
Early work investigating P. aeruginosa biofilms centered on mucoid strains capable of overproducing alginate (Ohman & Chakrabarty, 1982; Sá-Correia et al., 1987; Deretic et al., 1989; Pedersen et al., 1992; Deretic et al., 1993). Mucoid strains, commonly isolated from chronically infected patients with CF, generate enough alginate that it drips or runs off the culture dishes. The alginate polysaccharide makes colonies appear slimly compared to nonmucoid strains (Fig. 2); it is evident that they are encased in this viscous material (Hogardt & Heesemann, 2010). Alginate was once assumed to be the sole vital biofilm matrix component for P. aeruginosa, and this was likely due to its abundance and perceived protective capacity (Evans & Linker, 1973; Hoyle et al., 1993). More recent studies have challenged this paradigm.
Fig. 2.
Prominent Pseudomonas aeruginosa colony morphology variants. Cultures of PAO1, MJK8 (RSCV), and PDO300 (mucoid) were streaked on VBMM with Congo red. All strains grow at similar rates, yet MJK8 colonies are small and more aggregative and copious overproduction of alginate is obvious from PDO300. PAO1 has well-defined smooth colonies.
Structural stability is necessary for biofilm formation especially under high shear stress situations. Robust biofilm integrity relates to the ability to adhere to surfaces, either biotic or abiotic, and/or aggregation to neighboring biofilm cells (Hentzer et al., 2001; Webb et al., 2003; Yang et al., 2007; Ma et al., 2009). However, biofilm matrix components have functions in addition to simple structural capacity. Capsular polysaccharides like alginate and levan possess sufficient properties for water and nutrient retention, respectively. Alginate water retention has not been extensively studied, but it is predicted to be similar to polysaccharides like hyaluronic acid which can bind up to 1 kg of water g−1 polysaccharide (Sutherland, 2001a, b). Solubility of alginate is likely affected by the acetylation because acetyl groups inhibit interaction between polymer chains and cations, therefore enhancing gel formation (Sutherland, 2001a, b). These factors contribute to the highly viscous nature of alginate. Removal of O-acetyl residues alters the physical properties of alginate, resulting in increased binding of divalent cations and reduced polysaccharide solubility (Sutherland, 2001a, b). Alginate-containing biofilms have an obvious ability to survive harsh environments. Surviving desiccation stress is a limiting factor of the bacterial soil life cycle (Chang et al., 2007).
Role in niche biology
The contribution of alginate to persistence and immune evasion has been described (Ramphal & Vishwanath, 1987; Pier et al., 2001; Leid et al., 2005). Studies indicated that alginate production by mucoid strains conferred recalcitrance to antimicrobials and opsonophagocytosis (Schwarzmann & Boring, 1971; Simpson et al., 1988; Simpson et al., 1989; Simpson et al., 1993). Indeed, in support of this hypothesis, chronically infected patients with CF have circulating antibodies against alginate; however, it was noted that these antibodies fail to promote opsonic killing of P. aeruginosa in vitro (Pier et al., 2004). Alginate also has the ability to scavenge free radicals released by neutrophils and activated macrophages in vitro that are commonly able to kill bacteria (Simpson et al., 1989; Simpson et al., 1993; Govan & Deretic, 1996; Pier, 1998). Alternative studies found that mucoid strains and their revertant nonmucoid counterparts showed little evidence of variations in susceptibility to intrapulmonary killing by immune effectors or antibiotics (Blackwood & Pennington, 1981). In addition, a more recent survey comparing mucoid clinical isolates to nonmucoid clinical isolates confirms the finding that overproduction of alginate conferred no increased persistence in a murine lung infection model (Bragonzi et al., 2009). An explanation might be that clinical isolates revert to a nonmucoid morphology before or during infection in the mouse lungs used to assay for enhanced mucoid persistence. Alternatively, differences in lung infection models, microorganism and mouse strain backgrounds, or infectious doses used could account for these seemingly disparate findings. The nature of P. aeruginosa pathoadaptability makes determination of the direct contribution individual polysaccharides such as alginate have on persistence and immune evasion challenging. Clinical sputum samples from chronically infected patients with CF commonly include a mixture of both mucoid and nonmucoid strains. Along with polysaccharide production changes in response to AlgT/U activation, several additional loci are also regulated (Firoved et al., 2002). Perhaps, additional AlgT/ U-regulated factors contribute to the pathogenesis of mucoid strains, thereby clouding the individual role of alginate in some studies. In vitro model systems are helpful, but contain inherent flaws and are often limited to simple comparisons with individual strains. Regardless, the effect of alginate on individual aspects of the immune system needs to be investigated with the best models available. Alginate, in addition to other virulence factors present and participating in active infections, aids P. aeruginosa survival in the face of immune mediators and a chronic inflammatory state.
Pseudomonas aeruginosa-related CF infection is not the only situation where pseudomonads take advantage of alginate production. Evidence suggests in most situations that EPS production and biofilm formation generate a buffering zone for bacteria to maintain a controlled environment. An algU mutant strain of Pseudomonas fluorescens CHAO was more sensitive to osmotic stress and desiccation in vitro or in soil compared to the parental or mucA mutant strains (Schnider-Keel et al., 2001). Mucoid variants of P. fluorescens have enhanced adherence to plant roots, indicating a supplementary role for alginate in phytopathogenic biofilm formation (Bianciotto et al., 2001). Overall, it is clear that alginate produced under biofilm-relevant conditions aids environmental persistence and pathoadaptability, and further examination of precise mechanisms may aid what is known about P. aeruginosa biofilm-mediated capacities.
Capsular polysaccharides: levan
Molecular structure
In addition to alginate, the capsular polysaccharide levan is produced by a subset of pseudomonads, notably by the phytopathogen, P. syringae (Osman et al., 1986). Levan is a high molecular mass β-2,6 polyfructan with extensive branching through β-2,1 linkages (Fig. 1b) (Laue et al., 2006). Levan is produced exclusively from sucrose catalyzed by an extracellular levansucrase (Osman et al., 1986) (Li & Ullrich, 2001).
Regulatory factors
The function of levan has been predominantly studied among P. syringae strains; however, production of levan has been identified in other Pseudomonas spp. (Kang, et al., 1998; Gonzalez et al., 2003; Scarpellini et al., 2004; Pagès et al., 2007; Ivanova et al., 2009; Visnapuu et al., 2009). Pseudomonas syringae pv. Glycinea PG4180 possesses three genes lscA, lscB, and lscC, comprising the levansucrase biosynthetic operon, responsible for levan production (Osman et al., 1986). However, lscB and lscC appear to be primarily responsible for levansucrase functionality (Li & Ullrich, 2001). Compartmentalization of LscB in the extracellular and LscC in the periplasmic space was identified, while that of LscA was not identified in any protein fraction and thus not associated with levansucrase activities in P. syringae pv. Glycinea PG4180 (Li & Ullrich, 2001). Recent evidence suggests that levan production is regulated by LadS, which shares homology with the senor kinase-producing ladS locus of P. aeruginosa (Records & Gross, 2010). The specifics of levan production and levansucrase expression remain to be uncovered, but sensing external stimuli appears to be critical.
Biofilm matrix function
Based on spatiotemporal expression of levansucrase within biofilms, Laue et al. (2006) speculated that levan functions as a P. syringae storage molecule possibly protecting against starvation. They also showed that levan was not responsible for maintenance of biofilm structure, but that voids and blebs within the biofilm contained increased concentrations of levan, presumably as nutrient stores (Laue et al., 2006). This is an interesting finding that would provide an environmental organism a readily available nutrient store for occasions when other nutrients become depleted. The accumulation of levan in cell-free areas of biofilm had previously been suggested as a similar mechanism for nutrient retention in the oral cavity by Streptococcus mutans (Burne et al., 1996). Alternatively, observations of Pseudomonas brassicacearum adaptation to cadmium toxicity revealed that levan was downregulated by a subpopulation of cells, while a cadmium efflux pump and alginate were unregulated in response to cadmium exposure (Pagès et al., 2007). Together, these examples suggest that while alginate and levan share functional and regulatory properties, each possesses unique abilities that soil-borne pseudomonads take advantage of when necessary.
Aggregative polysaccharides
Continued biofilm studies in P. aeruginosa revealed that nonmucoid strains were adept at biofilm formation regardless of alginate production. In fact, several reports showed that alginate was not essential for biofilm formation in P. aeruginosa (Wozniak et al., 2003; Stapper, et al., 2004). Nonmucoid strains, harboring a native mucA gene while expressing low to absent levels of alginate, were able to produce mature robust biofilms (Wozniak et al., 2003). The lack of biofilm dependence on alginate led to a search for alternative polysaccharides that mediated biofilm formation in nonmucoid strains of P. aeruginosa. Matsukawa and Greenberg identified three alternative loci in P. aeruginosa PAO1, which they determined to have potential to generate polysaccharide matrix components (Matsukawa & Greenberg, 2004). Only one of the loci, termed polysaccharide synthesis locus (psl), was found to have relevance for aiding biofilm integrity (Matsukawa & Greenberg, 2004). Disruptions within the other two loci identified were similar to the parental strain in adherence and biofilm formation experiments (Matsukawa & Greenberg, 2004). Additionally, a different search identified a fourth locus later termed pel based on an obvious pellicle formation defect. Static and flow cell biofilms of PA14 Δpel also showed a defect in biofilm formation in P. aeruginosa PA14 (Friedman and Kolter, 2004a, b). Investigations of nonmucoid biofilms following these reports have been focused on the importance of psl and pel in biofilm formation.
Aggregative polysaccharides: Psl
Molecular structure
Initial predictions were that Psl contained galactose, D-mannose, D-glucose, and L-rhamnose (Wozniak et al., 2003; Friedman & Kolter, 2004a, b; Matsukawa & Greenberg, 2004; Ma et al., 2007). More accurate biochemical determinations of the Psl polysaccharide have since been made. Psl is composed of a pentasaccharide repeating distinct from other known polysaccharides. Psl contains D-mannose, D-glucose, and L-rhamnose (Fig. 1d) (Byrd et al., 2009). Psl is made from the sugar nucleotide pool of precursors including GDP-D-mannose, UDP-D-glucose, and dTDP-L-rhamnose (Byrd et al., 2009). Psl is commonly found in at least two forms: a high molecular weight cell–associated component and a relatively smaller soluble form of Psl that can be isolated from cell-free culture supernatant (Byrd et al., 2009). Greater details regarding Psl biosynthesis can be found in the recent review from Franklin et al. (2011).
Regulatory factors
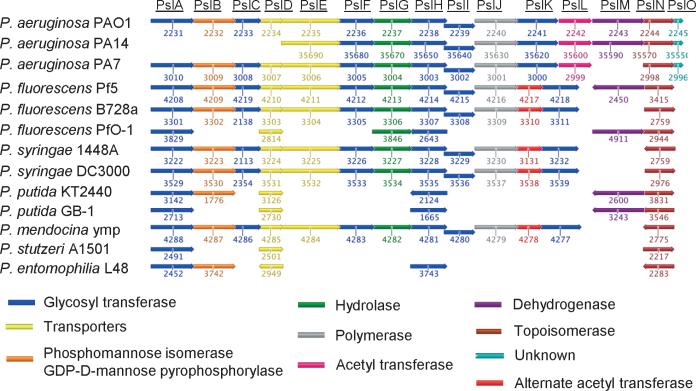
The psl locus in P. aeruginosa contains 15 co-transcribed genes with homology to known carbohydrate biosynthesis genes (Fig. 3) (Friedman & Kolter, 2004a, b; Jackson et al., 2004; Matsukawa & Greenberg, 2004). However, only 11 of the 15 psl genes are required to produce an adherent Psl-dependent biofilm (Byrd et al., 2009). One explainable exception is pslB, which shares redundant function with wbpW, a gene producing an enzyme with analogous function. The last three genes of the operon, pslMNO, are also not necessary for biofilm matrix functions (Byrd et al., 2009). The predicted functions of each of the psl products are depicted in Fig. 3 as well.
Fig. 3.
Polysaccharide synthesis locus (psl) in Pseudomonas spp. Relative gene organization and size are depicted for each pseudomonad and Psl component. The representative PAO1 Psl operon of approximately 18.4 kb is depicted with gene number designations for each comparison species given below individual components. Color coding represents predicted product function of each component. Information was gathered using the Pseudomonas database and prepared using the Geneious PRO software package (Stover et al., 2000; Nelson et al., 2002; Buell et al., 2003; Feil et al., 2005; Joardar et al., 2005; Paulsen et al., 2005; Lee et al., 2006; Winsor et al., 2009).
Induction of psl occurs in response to high intracellular levels of bis-(3′–5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) (Starkey et al., 2009; Borlee et al., 2010), an important intracellular signaling molecule in the bacterial world (Hengge, 2009). While the extent of c-di-GMP transcriptional effects remains to be elucidated, some transcriptional and posttranscriptional layers of psl regulation have been described involving RpoS and RsmA, respectively (Irie et al., 2010). RpoS, a stationary-phase σ-factor, induces psl transcription in response to changes in global physiology. Alternatively, the RNA-binding protein RsmA represses Psl translation through binding to the 5′ untranslated region of psl mRNA. RsmA repression of psl does not appear to be responsive to changes of growth phase (Irie et al., 2010).
Biofilm matrix function
While Psl is found in a larger cell-associated form and a smaller soluble form, the mechanism yielding the smaller form of Psl is not clear; possibly, it is a product of cleavage or breakdown of high molecular weight Psl. Alternatively, the processing of the larger and the smaller versions may be generated through a currently unappreciated mechanism. Breakdown of Psl could facilitate functions away from producing cell such as matrix building for biofilm cell-to-cell association, adherence to a surface, or even cell signaling. The cell-associated Psl can be visualized using scanning electron microscopy (SEM) where isolated blebs of Psl are seen on the surface of P. aeruginosa PAO1 (Byrd et al., 2010). Psl is extremely important for initial adherence of sessile cells to both biotic and abiotic substrates (Ma et al., 2006; Byrd et al., 2009; Byrd et al., 2010). Mature biofilms also take advantage of Psl for aiding structural stability and architecture (Ma et al., 2009). Psl also facilitates protection from innate immune effectors, complement and neutrophils (Mishra et al., 2012). Effects of Psl on immunity are likely to be intensified in biofilm where Psl is abundant. Ongoing investigations are focused on the role of both forms of Psl and its impact on single cells or biofilm communities. Regardless of the nature of Psl, it provides P. aeruginosa with an effective tool to establish a persistent biofilm infection.
Role in niche biology
The importance of Psl is particularly evident in rugose small-colony variants (RSCV) of P. aeruginosa. Converse from well-characterized mucoid P. aeruginosa strains producing massive quantities of alginate, overproduction of aggregative polysaccharides results in the rugose small-colony phenotype (Fig. 2). The RSCV phenotype was found to be prevalent in 33 of 86 P. aeruginosa-positive patients with CF over a 2-year period (Haussler et al., 1999). Stable variants that display a small, wrinkly colony phenotype on agar compared to wild-type PAO1 (Fig. 2) have been isolated from in vitro and in vivo biofilms (D'Argenio et al., 2002; Drenkard & Ausubel, 2002; Haussler et al., 2003; Kirisits et al., 2005; Starkey et al., 2009; Borlee et al., 2010). It is unclear why RSCV are so frequently isolated from in vivo and in vitro biofilms; perhaps, there is a selective role for aggregation during biofilm formation. Nonetheless, when biofilms of PAO1 and RSCV are mixed in various ratios, RSCV populations stabilize and are consistently found to represent c. 1/3 of the final biofilm population (Kirisits et al., 2005). This implies that the RSCV phenotype may have specific role or even a competitive advantage in the biofilm.
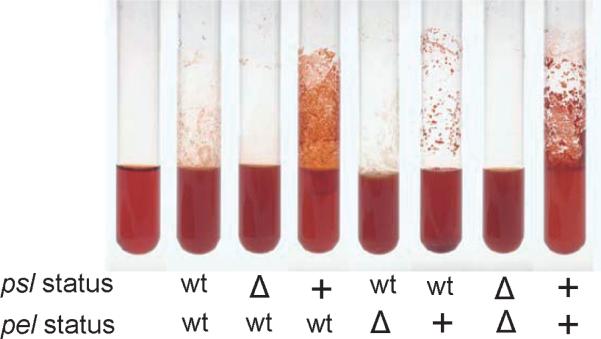
Diverse genotypic RSCV have been isolated, but all classes share the overabundance of accumulated intracellular c-di-GMP (Hickman et al., 2005; Starkey et al., 2009). MJK8 is a member of one specific class of RSCV, which has a mutation in the wsp operon resulting in a constitutively active diguanylate cyclase, WspR (Starkey et al., 2009). The increased c-di-GMP results in a decrease in motility and increased aggregation (Guvener & Harwood, 2007; Starkey et al., 2009). Additionally, Lee et al. (2007) described the requirement of c-di-GMP for the production of exopolysaccharides in P. aeruginosa biofilm. Exopolysaccharide biofilm matrix molecules Psl and Pel are induced in response to high intracellular c-di-GMP levels (Hickman et al., 2005; Guvener & Harwood, 2007; Starkey et al., 2009; Borlee et al., 2010). Investigation of the individual impact of enhanced Psl or Pel expression reveals that overexpression of either polysaccharide, but especially Psl, results in enhanced aggregation and biofilm formation. Wild-type, psl mutant, and psl overexpression strains were grown in a rolling culture tube overnight with Congo red supplemented media. Abundant aggregation and biofilm are noticeable in the tubes when Psl is over-expressed (Fig. 5). Conversely, wild type or strains lacking Psl resulted in moderate to absent adherence (Fig. 5). Artificially inducing psl with an arabinose-inducible promoter intensely mimics the RSCV aggregation pheno-type, which has a unique propensity to aggregate, thereby facilitating biofilm formation. Psl may also be important for mediating similar biofilm adherence during colonization of immunocompromised patients.
Fig. 5.
Psl and Pel influence aggregation, adherence, and colony morphology. Pseudomonas aeruginosa PAO1 strains with wild type, inactivated, or overexpressed status of Psl and Pel were grown in culture tubes containing Congo red to observe aggregation. Polysaccharide status is indicated below each tube.
While the majority of evidence describing the function of aggregative polysaccharides in biofilm formation has been made studying P. aeruginosa, the psl operon is also conserved throughout several other pseudomonads (Fig. 3) (Nelson et al., 2002; Buell et al., 2003; Vodovar et al., 2006; Winsor et al., 2009). For example, P. fluorescens B728a harbors 12 of the 15 genes in the psl operon, missing only L, M, and O genes (Fig. 3). Interestingly, the only allele among those that was shown to be necessary for Psl production was pslL (Byrd et al., 2009). However, an additional acyl transferase, Psyr3310, was found between pslJ and pslK in the genome that may be fulfilling the normal function of pslL in P. aeruginosa strains (Stover et al., 2000; Feil et al., 2005; Winsor, et al., 2009). Moreover, P. fluorescens Pf5, P. syringae strains 1448A, DC3000, and Pseudomonas mendocina ymp share the similar psl operon organization (Fig. 3) (Stover et al., 2000; Feil et al., 2005; Joardar et al., 2005; Paulsen et al., 2005; Winsor et al., 2009). Other psl genes that are well conserved in the strains examined include pslA, pslD, pslH, and pslN (Fig. 3). Conversely, the majority of the psl operon is missing in P. fluorescens Pf0-1 (Winsor et al., 2009), but unlike P. aeruginosa, which is limited in biofilm formation without psl components, it may have additional matrix components that play a compensatory role. Psl is not necessary for biofilms produced by P. aeruginosa PA14, which does not have pslABCD genes (Fig. 3) (Friedman & Kolter, 2004a, b; Lee et al., 2006).
Aggregative polysaccharides: Pel
Molecular structure
Another polysaccharide-producing locus, pel, is required for biofilm formation in P. aeruginosa PA14 (Friedman & Kolter, 2004a, b). Unlike the Psl polysaccharide, whose structure has been predicted (Fig. 1) (Byrd et al., 2009), the composition and nature of Pel polysaccharide are undefined. Some hints regarding the nature of Pel exist, including comparison of an air–liquid (A–L) biofilm matrix composition of PA14 and its pel mutant, which revealed that the mutant exhibited reduced glucose levels compared to the parental strain (Friedman & Kolter, 2004a, b). A recent survey of polysaccharides produced by P. aeruginosa PA14 did not provide the Pel structure (Coulon et al., 2010), but alluded to an alternate functional role for Pel.
Regulatory factors
The pel locus is comprised of a 7-gene operon producing proteins with predicted polysaccharide biosynthesis functions (Fig. 4) (Friedman & Kolter, 2004a, b). Similar to Psl, Pel is produced maximally when intracellular c-di-GMP levels are elevated (Lee et al., 2007). Surprisingly, unlike the psl operon, the pel operon is poorly conserved in P. syringae, P. putida, and P. mendocina (Fig. 4). The pel operon also appears to lack some genes necessary for complete biosynthesis of an exopolysaccharide, implying that the Pel machinery may be functioning with other polysaccharide enzymes (Franklin et al., 2011). Intriguingly, recent evidence suggests that there may be cross talk between factors affecting Pel production and Psl production. Ghafoor et al. (2011) show that greater Pel production occurs in the absence of Psl. Possible explanations may be that there is cross talk in the regulation of the polysaccharide loci or competition for precursors that account for limited production of individual polysaccha-rides. The latter has been proposed previously to affect production of polysaccharides like alginate (Pham et al., 2004; Rehm, 2010).
Fig. 4.

Pel polysaccharide synthesis locus among Pseudomonas spp. The conservation of individual components of the pel operon, which is responsible for Pel biosynthesis, is depicted for pel-containing Pseudomonas spp. The representative PAO1 Pel operon of approximately 12.2 kb and the corresponding gene number designation for each comparison species are given below representative loci. Color-coded boxes represent gene product functions as indicated on the top of this figure (Stover et al., 2000; Friedman & Kolter, 2004a, b; Paulsen et al., 2005; Winsor et al., 2009).
Biofilm matrix function
Interestingly, strain PA14 relies greatly on the Pel polysaccharide compared to other P. aeruginosa strains with fully intact psl loci (Fig. 3). The biofilm defect in a P. aeruginosa PA14 pel transposon insertion mutant was identified through a screen for poor A–L biofilm formation (Friedman & Kolter, 2004a, b). The original observation led to further investigating the importance of Pel during biofilm development. In addition to the formation of an A–L or pellicle biofilm, Pel promotes aggregative properties in broth culture. In PA14, or when overexpressed in PAO1 regardless of Psl expression, Pel aids adherence to culture tubes and aggregation in broth culture (Fig. 5). Generally, when both psl and pel are intact, it seems that Psl is predominant while Pel has only limited impact on biofilm phenotypes. However, in the instances where the psl operon is absent or disrupted, such as it is in PA14, or when c-di-GMP is maximally elevated, Pel has a clearer impact on biofilm formation. Yang et al. (2011) recently identified conditions in which PAO1 biofilms utilize Pel for greater structural stability in formation of microcolonies. In this instance, Pel and Psl together facilitate compactness of the biofilm and cell-to-cell association. Biofilm matrix dependence on Psl and/or Pel has been discussed in detail (Colvin et al., 2011b). Colvin et al. proposed four classes of P. aeruginosa strains based on their dependence on aggregative polysaccharides. Class I strains include those that rely predominantly on Pel for biofilm matrix stability. PA14 was the only identified member of this class, and although an exhaustive search was not carried out, the authors predict that other members of this class would also likely have psl mutations rendering them Psl negative. Class II strains rely predominantly on Psl for matrix stability even though they seem to be capable of producing the components necessary to generate the Pel polysaccharide, such as PAO1. Class III strains produce both Psl and Pel at low levels, and disruption of the ability to produce either polysaccharide independently did not dramatically reduce biofilm stability. Class IV matrix strains are Psl and Pel overproducers. Characteristically, this last class makes a rather dynamic matrix that does not rely on a single component, and these strains produce a robust biofilm (Colvin et al., 2011a, b). Future characterization of P. aeruginosa isolates will likely provide greater insights into these classes and the functional contributions of either Pel or Psl.
In addition to a structural role of Pel, recent evidence suggests Pel functions to mitigate antibiotic efficiency during biofilm formation (Colvin et al., 2011a, b). Coulon et al. (2010) examined a pel mutant with regard to modifications in lipopolysaccharide (LPS) O-antigen and found greater culture concentrations of soluble O-antigen (termed OPS) in the mutant compared to the wild-type strain. One might interrupt those data to indicate that Pel is a modifying element of O-antigen or LPS molecules rather than an exopolysaccharide. Nonetheless, the impact of Pel on biofilm stability and adherence is apparent, and continued studies will be required to understand the full nature and function of Pel during biofilm formation.
Role in niche biology
While adherence and overall structural stability of the biofilm have been hallmarks of polysaccharide contribution to biofilm function, more recently, polysaccharide-mediated tolerance to antibiotics and even impacts on cell signaling have been appreciated (Khan et al., 2010; Colvin et al., 2011a, b; Yang et al., 2011). A mutant strain lacking Pel production is more susceptible to tobramycin exposure compared with parental and complemented strains (Colvin et al., 2011a, b). While it is possible that Pel is directly binding tobramycin to inhibit the function of the aminoglycoside, it is also possible that a Pel-dependent biofilm forms in such a way that it further limits amino-glycoside penetration into the biofilm. Perhaps, the nature of the Pel-aided biofilm matrix is key because exposure of stationary-phase cultures to tobramycin failed to show any additional sensitivity of the pel mutant (Colvin et al., 2011a). Production of polysaccharides Pel and Psl, and therefore biofilm formation, also seems to affect quorum sensing and downstream extracellular DNA (eDNA) release (Yang et al., 2011). The lack of compactness among initial biofilm colonizers without aggregative polysaccharides could also affect quorum sensing expression and ultimately Pseudomonas quinolone signal (PQS)-mediated eDNA release as has been described previously (Klausen et al., 2003a, b; Yang et al., 2007; Barken et al., 2008; Yang et al., 2009). The connection between both polysaccharides Pel and Psl along with eDNA, type IV pili, quorum sensing, and biofilm organization is complex and requires extensive investigation to tease apart individual responsibilities and relationships. As a first step toward understanding these connections, recently, the TbpA (PA3885) protein, responding to quorum sensing and mitigating c-di-GMP levels in the cell, was shown to be a key component regulating the process of biofilm formation (Ueda & Wood, 2009). The levels of c-di-GMP ultimately impact aggregative polysaccharide production and downstream biofilm formation. Advances similar to this will benefit development of novel strategies to combat biofilm-related infections through targeting the regulation of biofilm formation.
Aggregative polysaccharides: cellulose
Similar to RSCV in P. aeruginosa, P. fluorescens SBW25 produces analogous variants referred to as a wrinkly spreader (WS) phenotype (Spiers et al., 2002). Characterization of the WS phenotype attributes a mutation in the chemosensory-like regulatory apparatus (wsp) for expression of the wss operon, whose products participate in the cellulose biosynthetic pathway (Spiers et al., 2002; Spiers et al., 2003). While cellulose-related biofilm function has not been described for P. putida and P. syringae pv. tomato str. DC3000, the genomes also contain homologs of the P. fluorescens SBW25 wss operon (Nelson et al., 2002; Buell et al., 2003). Cellulose production by P. fluorescens clearly impacts biofilm formation at the A–L interface (Gal et al., 2003; Spiers et al., 2003). Specifically, acetylation of cellulose is important for normal A–L biofilm formation, because the acetylation-defective mutant WS-18 produced a weak biofilm compared to the WS parental strain (Spiers et al., 2003). The dependence of the WS subpopulation that arises from the wild-type P. fluorescens smooth colony during biofilms formed at the A–L interface is strikingly similar to the phenotype of RSCV in P. aeruginosa. Both situations depend on the enhanced production of c-di-GMP, often from analogous pathways, and ultimately overexpression of exopolysaccharide (Spiers et al., 2002; Gal et al., 2003; Spiers et al., 2003; Kirisits et al., 2005; Spiers & Rainey, 2005; Starkey et al., 2009). It is intriguing, however, that the formation of natural biofilms by environmentally relevant pseudo-monads takes advantage of cellulose as opposed to Psl. The psl locus is widely conserved among pseudomonads that produce cellulose (Fig. 3), but the role of Psl in biofilms made by non-aeruginosa Pseudomonas species has not been described. Interestingly, some pseudomonads, like P. putida, lack a cognate psl locus and contain novel polysaccharide-producing loci important for biofilm stability (Nilsson et al., 2011). Two of the loci have also been described to be important for cell-to-cell interaction and pellicle formation. Pseudomonas putida exopolysaccharide A (pea) and bacterial cellulose (bcs) were each shown to play a role in biofilm-related phenotypes (Nielsen et al., 2011). The pea locus in P. putida KT4220 includes genes PP3132–3142, and only PP3142 shares an ortholog group with a psl gene, pslA (Stover et al., 2000; Nelson et al., 2002; Winsor et al., 2009). Like Psl, the pea-produced polysaccharide appears to contain mannose, rhamnose, and glucose and is cellulase sensitive. However, the pea-produced polysaccharide was not cross-reactive with antisera raised against P. aeruginosa Psl (Nielsen, et al., 2011). The absence of the Pel polysaccharide in several non-aeruginosa pseudomonads is also intriguing (Fig. 4). This suggests that while some matrix molecules share functions, others provide unique role in specific niches. Examples exist where polysaccharides have functional substitutes, such as where cellulose appears to substitute for P. aeruginosa's Pel-related functions in pellicle or A–L biofilm formation (Wozniak et al., 2003; Friedman & Kolter, 2004a, b; Ryder et al., 2007). Regardless of the matrix molecule produced by Pseudomonas spp., each species uses one or more of the following polysaccharides: alginate, levan, cellulose, Psl, or Pel, for proper biofilm and microcolony development.
Additional Pseudomonas biofilm matrix components
Nucleic acids
Another commonly abundant biofilm matrix building block is nucleic acid. Specifically, DNA is a critical component of the biofilm matrix (Whitchurch et al., 2002; Webb et al., 2003; Allesen-Holm et al., 2006; Rice et al., 2007; Yang et al., 2007; Ma et al., 2009). Initially, the importance of DNA in biofilm was shown by Whitchurch et al., (2002). DNA was mistakenly identified through a carbazole colorimetric assay, commonly used to quantitate uronic acid-containing polysaccharides (Whitchurch et al., 2002). Carbazole binds DNA (Tanious et al., 1997), similar to detection of alginate, highlighting the structural similarity of the two polysaccharide molecules (Fig. 1a and e). The deoxyribose backbone of DNA is a sugar in its most basic sense. Further examination indicated that DNA possesses a structural role in early P. aeruginosa biofilms (< 60 h) that were disrupted by exposure to DNase I, while older biofilms were more tolerant to DNase I treatment (Whitchurch et al., 2002). This insight suggests that DNA plays a role during biofilm development but that it is only a contributing component of a more diverse biofilm matrix during later biofilm development. Perhaps, the diverse nature of a mature biofilm matrix is better able to withstand the absence of DNA following DNase I treatment. Not surprisingly, extracellular DNA (eDNA) isolated from in vitro biofilms was identified to be derived from P. aeruginosa genomic DNA (Allesen-Holm et al., 2006). Allesen-Holm et al. (2006) further characterized the eDNA using confocal laser scanning microscopy and produced spectacular images showing organization of DNA in the biofilm matrix. After 48 h of growth, the biofilms appear to contain intense amounts of DNA in the matrix, especially in regions where microcolonies are forming. After 144 h, the biofilms contained proportionally less DNA in the matrix, visualized by nucleic acid-specific staining, but the location of DNA staining was striking (Allesen-Holm et al., 2006). Images captured the presence of DNA in the interior of the stalk after 5 days of growth and intensely concentrated in the interior of the mushroom-shaped cap after 6 days of growth (Allesen-Holm et al., 2006). Time points imaged after tower structures (Box 1) had formed indicated that DNA was located in specific areas of the biofilm, not throughout as it was at 2 days of growth. These data strengthen the conclusion that the biofilm matrix becomes less DNA dependent as the biofilm matures and also explains why DNase I treatment of older biofilms does not disrupt the biofilm adherence. The same authors also indicate that quorum sensing affects extracellular DNA accumulation in the P. aeruginosa biofilm (Allesen-Holm et al., 2006). Specifically, it is evident that PQS quorum sensing-controlled DNA release in the dense stalk microcolony is required for cap formation (Yang et al., 2009).
The role of DNA in the specific regions during development of the mushroom-like structures has been investigated further. A model where DNA- and pilin-dependent mushroom capping has been proposed states that twitching motility allows for motile cap-forming populations to move up the matrix of the stalk, along the DNA portion, to form the mushroom cap on the previously formed stalk (Box 1) (Klausen et al., 2003a, b; Barken et al., 2008). Ma et al. (2009) also investigated the role of polysaccharides and DNA during biofilm development. Although it appears that both DNA and polysaccharides are abundant in P. aeruginosa biofilms, there are divergent roles during tower formation and maturation process (Ma et al., 2009). Psl matrix-containing biofilm was found on the exterior of the cap of the mushroom-shaped structures, just outside of the nucleic acid-specific staining. Psl was markedly absent from the interior of the mushroom cap and was therefore termed a Psl cavity (Ma et al., 2009). Instead, genes controlling cell death and lysis seemed to be aiding Psl cavity formation and proposed to be contributing to DNA release. It is especially noteworthy that the biofilm matrix of pseudomonads is a dynamic process where coordinated changes in matrix structure correlate with biofilm development stages.
Regardless of the presence of an array of matrix molecules, mechanisms of matrix interaction remain under continued investigation. Hypotheses have been generated describing polysaccharides or nucleic acids interacting via surface charge of cells or simple nonspecific ‘trapping’ of biofilm aggregates. However, the spatial and temporal accumulation of DNA and production of polysaccharides during biofilm formation indicate that facilitated organization of cells exists in the biofilm. Identification of the mechanism whereby biofilm matrix molecules and cells interact will aid development of therapies for dispersion and clearance of biofilm infections.
Polysaccharide-containing matrix components
In addition to the predominant polysaccharides, accessory biofilm matrix components have been identified that aid biofilm matrix function. The impact of cyclic β glucans, LPS, and membrane vesicles (MV) has not been widely investigated in Pseudomonas spp., but evidence suggests that they have an accessory role with the biofilm matrix (Spiers & Rainey, 2005; Schooling & Beveridge, 2006; Nakamura et al., 2008; Coulon et al., 2010).
The primary responsibility of matrix components is to provide structural integrity to a biofilm, allowing for cell–cell interaction and formation of structures that convey accessibility to nutrients. However, another important role of the biofilm matrix is the potential to inhibit antimicrobials. Originally, it was speculated that the sticky, dense, and thick nature of biofilm cells and matrix inhibited penetration of antibiotics, resulting in the recalcitrance observed (Hoyle et al., 1992). Subsequently, it was determined that fluoroquinolone antibiotics penetrate biofilms readily, while aminoglycosides penetrate more poorly (Walters et al., 2003). Therefore, regardless of the accessibility of antibiotics to cells within the biofilm, sessile bacteria are physiologically distinct from their planktonic counterparts (Anderl et al., 2000; Whiteley et al., 2001). However, excreted factors like polysaccharides abundant in the biofilm matrix interact with aminoglyco-sides (Mah et al., 2003; Sadovskaya et al., 2010). Specifically, expression of a polysaccharide matrix molecule was identified that attributed antibiotic resistance of P. aeruginosa biofilm (Mah et al., 2003). The ndvB locus, expressed preferentially in biofilms, generates cyclic gly-cans (Mah et al., 2003). The resistance mechanism was proposed originally to rely on the ability of cyclic glycans to form a molecular complex with aminoglycoside antibiotics, abrogating the antibiotic's ability to reach its target (Athanassiou et al., 2003; Mah et al., 2003; Evrard et al., 2004; Sadovskaya et al., 2010). More recently, the Pel polysaccharide in both PAO1 and PA14 strains of P. aeruginosa has been shown to aid tolerance to amino-glycoside antibiotics, similar to the role of cyclic glucans (Khan et al., 2010; Colvin et al., 2011a, b). Independent of providing a penetration barrier, other matrix molecules have not been implicated in conferring recalcitrance directly for biofilm-contained cell populations, but continued studies will investigate the nature of biofilm-mediated resistance to antimicrobials.
The importance of LPS as a biofilm matrix molecule, or even its role in adhesion of Pseudomonas spp. to a surface, has not been defined completely. However, limited investigations have provided valuable clues into the nature of LPS-mediated biofilm stability. LPS was originally investigated because of its ability to affect surface charge or relative hydrophobicity of the cell (Rocchetta et al., 1999). Furthermore, LPS dictates alterations in attachment, transition to sessile growth, and colony morphology variations in several bacteria (Giwercman et al., 1992; Genevaux et al., 1999; Mireles et al., 2001; Ali et al., 2002; Landini & Zehnder, 2002; Rashid et al., 2003). Specifically, P. fluorescens WS variants rely on LPS interactions to aid overall biofilm integrity (Spiers & Rainey, 2005). Although the WS-5 stain possesses a mutation resulting in the loss of proper LPS expression (Gaspar, et al., 2000; Spiers & Rainey, 2005), WS-5 was able to form an A–L biofilm, albeit less robust than the wild-type smooth-colony strain. The phenotype was partially complemented by an LPS-expressing strain (Spiers & Rainey, 2005). Additional investigations of P. aeruginosa PAO1 and its LPS-mediated adherence and subsequent biofilm formation strongly suggest that modifications in the LPS core affect adherence. Changes in bacterial mechanical behavior during early biofilm formation as a result of ‘differential LPS core capping’ were correlated with mature biofilm matrix modifications (Lau et al., 2009). Although LPS appears not to be affected by quorum sensing after extended growth (De Kievit et al., 2001), but during early growth stages (< 4 h) (Nakamura et al., 2008), LPS responds to quorum sensing. Typically, LPS is anchored to the bacterial outer membrane through lipid A (Rocchetta et al., 1999), but can be released from the cells and accumulate in the culture media during normal growth (Cadieux et al., 1983; Al-Tahhan et al., 2000). It is likely that cell-free LPS associates with the complex biofilm EPS contributing to the architecture of the biofilm. These predictions are supported by the calculation that excreted polymers, products of cell lysis, and remaining intact cells account for 85–95% of total organic carbon in biofilms (Sutherland, 2001a, b). Continued investigation of the nature of LPS in the biofilm matrix is necessary for a complete understanding of the multifarious biofilm matrix, but it is intriguing to consider abundant LPS molecules as biofilm matrix components.
MV are multifunctional bilayered structures that bleb from the outer membranes of Gram-negative bacteria (Beveridge, 1999; Mashburn & Whiteley, 2005; Schooling & Beveridge, 2006; Mashburn-Warren et al., 2008). Importantly for the context of biofilm formation, MV interact with biofilm matrix components like eDNA (Schooling et al., 2009). MV have been proposed to provide a large portion of LPS in the biofilm matrix (Schooling & Beveridge, 2006) as they retain the intrinsic lipid asymmetry of the outer membrane with most of the LPS positioned within the outer leaflet of the membrane (Beveridge et al., 1997). Additionally, MV blebbing from the outer membrane results in a sampling of the periplasmic contents. Often, these include proteases, alkaline phosphatase, lipases, proelastase, autolysins, and toxins (Grenier & Mayrand, 1987; Kadurugamuwa & Beveridge, 1995; Fiocca et al., 1999; Kolling & Matthews, 1999; Keenan & Allardyce, 2000; Allan et al., 2003). The natural shedding of amphipathic LPS through MV formation could provide a usable framework for structural contribu tions of LPS that have been suggested (Wozniak et al., 2003; Spiers & Rainey, 2005). While it is apparent now that MV are abundant within a biofilm and confer obvious matrix-related function, additional investigations will more fully reveal the impact of MV for P. aeruginosa biofilm. Evidence suggests that formation of MV is controlled by the las quorum sensing system early in biofilm formation (Nakamura et al., 2008) and that PQS is required and sufficient for the formation of MV (Mashburn-Warren et al., 2008), suggesting that the las QS mainly regulates MV by PQS activation. Further investigation of quorum sensing and the generation of MV will provide greater insight as to how MV are used by biofilm-forming organisms.
Protein components of the biofilm matrix
In addition to polysaccharides and nucleic acids, the importance of biofilm matrix-related proteins has been increasingly appreciated. Proteins present in the biofilm matrix facilitate roles including surface adherence, interaction with other matrix molecules, and matrix stability. Proteins such as CdrA from P. aeruginosa possess carbohydrate-binding capacity, making it an interesting candidate for a mechanism promoting matrix molecule interaction (Borlee et al., 2010). The cdrAB locus comprises a two-partner secretion (TPS) system with a large adhesion and its transporter. The operon was named because of its response to cyclic diguanylate where in the presence of high c-di-GMP, robust expression is observed. The prototypical TPS system is the filamentous hemagglutinin (FHA) as well as its FhaC secretion partner from Bordetella pertussis. A key identifiable characteristic of CdrA making it a relevant biofilm matrix component is that it interacts with Psl from P. aeruginosa. Evidence suggests that CdrA-Psl binding could facilitate specific interactions among Psl molecules and with biofilm cells (Borlee et al., 2010). FHA from B. pertussis is an adhesion with multiple galactose-inhibitable binding targets including cilia (Tuomanen et al., 1988). FHA is required for B. pertussis to grow in characteristic aggregates (Menozzi et al., 1994). Like FHA, CdrA is predicted to be a rod-shaped protein with a β-helical tertiary structure and exposed integrin-binding motif (Borlee et al., 2010). Ongoing investigations of CdrA will elucidate the specific mechanism, whereby it stabilizes the biofilm matrix through its association with Psl.
Some Pseudomonas spp. transition from reversible to irreversible biofilm attachment through the function of LapA on the cell surface (Hinsa et al., 2003). Additionally, LapD provides a unique inside-out signaling mechanism for initiation of adherence, because it contains predicted GGDEF and EAL domains each functioning potentially to synthesize or degrade c-di-GMP, respectively. However, LapD lacks the ability to carry out either of these functions and was thus proposed to act as an effector protein linking intracellular c-di-GMP signaling to the extracellular protein LapA (Hinsa et al., 2003). LapA is a cell-to-cell interconnecting molecule as well as a surface adhesion (Gjermansen et al., 2010). Interestingly, a decrease in intracellular c-di-GMP levels in P. putida OUS82 led to dispersal of preformed biofilms that was LapG dependent. The authors of the study conclude that the LapG component acts on LapA in the biofilm matrix to cause dispersal (Gjermansen et al., 2010). The intricacies of the mechanism and the regulation of the lap locus are discussed in greater detail elsewhere, but it is critical to note that lap loci have been identified in only P. fluorescens and P. putida (Hinsa et al., 2003). The Lap components are absent in P. aeruginosa and P. syringae genomes, indicating that the nature of biofilm formation and irreversible attachment is distinct among various species of Pseudomonas (Hinsa et al., 2003). Interestingly, Pseudomonas species with the lap genes are in general lacking genes for synthesis of Pel polysaccharide.
Cytotoxic lectins produced by Pseudomonas spp. bind carbohydrates and aid biofilm stability and structure formation (Tielker et al., 2005; Diggle et al., 2006). In addition to virulence capabilities of lectins LecA and LecB in the lungs, individual mutants of either lecA or lecB exhibit defective biofilm phenotypes (Tielker et al., 2005; Diggle et al., 2006; Chemani et al., 2009). Further characterization identified that LecA has high specificity and affinity for hydrophobic galactosides (Diggle et al., 2006) while LecB binds L-fucose readily (Tielker et al., 2005). The impact of these lectins on P. aeruginosa cytotoxicity and biofilm formation led to the examination of LecA-and LecB-specific lectin-inhibiting carbohydrates as a therapeutic strategy (Chemani et al., 2009). Indeed, promising results were seen when co-administration of lectin-inhibiting carbohydrates was given in an animal model of infection. Markedly reduced lung injury and mortality were observed compared to infections without intervening carbohydrates (Chemani et al., 2009).
Cell appendages are also necessary for Pseudomonas biofilm formation although they are commonly not regarded as classical biofilm matrix molecules. Both flagella and type IV pili aid in biofilm formation (O'Toole & Kolter, 1998; Klausen et al., 2003a, b). During the biofilm maturation process, microcolonies are developed through clonal expansion of nonmotile cells, which develop the stalk of P. aeruginosa mushroom-like structures (Klausen et al., 2003a, b). Formation of the mushroom cap, however, requires the presence of type IV pili. In addition, flagellum-mediated motility along with the chemotaxis systems is required for cap formation (Barken et al., 2008). It is unclear whether flagella expression is mediating swimming or swarming motility, but speculation suggests swarming within the biofilm matrix, resulting in movement of cells into the mushroom cap (Barken et al., 2008). Swarming motility requires biosurfactant production (Kohler et al., 2000; Rashid & Kornberg, 2000), and the rhlA mutant displays reduced cap formation in mixed pilA/rhlA biofilms because of the lack of rhamnolipid biosurfactant (Pamp & Tolker-Nielsen, 2007). Investigating the interaction between multiple cell surface and matrix components is intriguing because Pseudomonas biofilms require multiple steps to produce intricate adherent structures containing sessile cells.
During a screen of a mutant library in a type IV pili-defective strain, Vallet et al., (2001) showed that an additional appendage made from an alternative fimbrial gene cluster encoding a chaperone usher pilus (cup) is important for adherence. Fimbrial adhesions other than type IV pili had not been described previous to this work, and the other members of the cup loci were also investigated for their potential contribution to biofilm adherence (Vallet et al., 2001). Using conditions permissible for cup fimbrial expression and assembly, it was shown to cooperate in cell–cell interactions and microcolony formation (Kulasekara et al., 2005; Ruer et al., 2007). Descriptions of the importance of Cup proteins for biofilm formation also suggest that perhaps the fimbriae are not redundant adherence proteins for the well-characterized type IV pili but that they provide a synergistic or conditional ability of Pseudomonas to form biofilms (Vallet et al., 2001; Giraud et al., 2011).
Additional relevant protein matrix molecules are being identified as required for biofilm growth. Recently, a functional amyloid-like protein from a P. fluorescens strain was identified. The locus responsible for amyloid production, fapA-F, when overexpressed significantly enhanced biofilm adherence (Dueholm et al., 2010). The major fibril subunit, produced from fapC, is well conserved even in P. aeruginosa. Amyloid production in P. fluorescens is similar to Escherichia coli csgAB-mediated curli formation (Dueholm et al., 2010). CsgAB is important for adherence and initiation of aggregation of E. coli. The fapC component is conserved in P. aeruginosa as well; this may represent another tool Pseudomonas possesses to efficiently form biofilms regardless of the environmental circumstance.
Rhamnolipids
Rhamnolipids are surface-active glycolipid biosurfactants produced by various bacterial species, but they were originally identified from P. aeruginosa (Bergstrom et al., 1946a, b; Abdel-Mawgoud et al., 2010). Rhamnose and lipid moieties comprising rhamnolipids are linked by an O-glycoside (Abel et al., 1978). Originally, cultures of P. aeruginosa were found to produce an oily glycolipid named pyolipic acid following growth on glucose (Bergstrom et al., 1946a, b; Bergstrom et al., 1946a, b). Later, the structural subunits were identified as L-rhamnose and β-hydroxydecanoic acid (Jarvis, 1949; Hauser & Karnovsky, 1954). The specific biological role for the production of this dynamic biosurfactant has remained elusive. However, in vivo models have shown that an rhlA mutant of P. aeruginosa, which fails to product rhamnolipid, is more rapidly cleared from mice in models of intraperitoneal foreign body infection and pulmonary infection (Van Gennip et al., 2009). Sputum samples from P. aeruginosa-infected patients with CF had as much as 8 μg mL−1 of rhamnolipids, and the higher rhamnolipid concentration correlates with severity of clinical status (Kownatzki et al., 1987). Importantly, Read et al. suggest that these values may be an underestimate of the rhamnolipid concentration in explanted lung tissue secretions, which was 65 μg mL−1 (Read et al., 1992). It is unclear how rhamnolipid production leads to greater severity of disease, but in vitro biofilm studies suggest that rhamnolipid production affects biofilm formation (Davey et al., 2003; Boles et al., 2005; Lequette & Greenberg, 2005; Glick et al., 2010). Rhamnolipids are toxic to neutrophils, a characteristic also proposed to impact invasiveness and persistence (Alhede et al., 2009). Biosurfactants like rhamnolipid are important for motility-dependent capping of mushroom-shaped towers (Pamp & Tolker-Nielsen, 2007) and overall biofilm matrix remodeling (Davey et al., 2003). Alterations in swarming by rhlA mutants lead to defective biofilm formation (Harshey, 2003; Shrout et al., 2006; Overhage et al., 2007; Verstraeten et al., 2008; Yeung et al., 2009). Similar to remodeling that occurs for water channel clearance, it appears that rhamnolipids can facilitate dispersal mechanisms allowing for release of planktonic daughter populations (Boles et al., 2005). Final maturation of a biofilm is described as a transition back to a motile, dispersing state with the objective to seed new sites of infection and potential biofilm formation (Ma et al., 2009). During the colonization period, rhamnolipid potentially interacts with host cells exhibiting cytotoxicity (McClure & Schiller, 1992, 1996; Haussler et al., 1998). More aggressive released swimming populations may also activate the innate immune system unlike metabolically inert microorganisms in the biofilm state. In addition to releasing cells from the biofilm, rhamnolipids induce shedding of LPSs resulting in enhanced cell surface hydrophobicity, thereby favoring primary interaction with a surface (Zhang & Miller, 1994; Al-Tahhan et al., 2000). Modifying hydrophobicity and surface adherence through rhamnolipid secretion is especially important in environments where fewer nutrient sources are available, facilitating an adaptation to more rugged lifestyle (Neu, 1996; Deziel et al., 2003; Boles et al., 2005). Pseudomonads are particularly adept at acclimating to environmental stress through biofilm formation. Rhamnolipids aid adaptation through increased surface activity, wetting ability, detergency, and other amphipathic-related characteristics (Abdel-Mawgoud et al., 2010). For example, bacteria capable of degrading hydrophobic substances, such as linear alkanes (a nutrient source for P. aeruginosa), commonly secrete biosurfactants that facilitate uptake and assimilation of hydrocarbons (Hommel, 1994). Solubility of higher molecular weight n-alkanes is less than that of small molecular weight molecules, thereby interfering with bacterial uptake (Abdel-Mawgoud et al., 2010). Therefore, it remains undetermined how P. aeruginosa is able to use these high molecular weight hydrophobic compounds as an energy source; perhaps, rhamnolipid production is key in that role. Many other Pseudomonas spp. in addition to P. aeruginosa produce rhamnolipids (Gunther et al., 2005; Onbasli & Aslim, 2009a, b), but their specific functionality has not been well defined.
Conclusions and perspectives
Pseudomonas spp. can persist under extremely harsh conditions, including the environment and the human body, and this property has been linked to their ability to form biofilms. Pseudomonas aeruginosa pathoadaptation, in particularly, has driven a large degree of the early biofilm research relating to alginate and the production of ‘slime’ (Schwarzmann & Boring, 1971; Evans & Linker, 1973; Hussain et al., 1991; Ying et al., 1996; Licking, 1999). Alginate-overproducing mucoid strains confer a selective advantage in their niche, the CF lung. The conversion to, and selection for, mucoid strains within the CF lung continues to be investigated because of the negative clinical outcomes associated with consecutive mucoid-positive sputum cultures. Ultimately, the ability of mucoid strains to persist and evade clearance mechanisms results in patient mortality if aggressive therapies are not deployed. Prior to mucoid conversion, Psl- and Pel-expressing nonmucoid biofilm-producing strains are likely responsible for early intermittent colonization of the CF lung. Although mucoid variants are not found during this period, Pel- and Psl-overproducing variant strains are frequently isolated. CF sputum samples, and even in vitro biofilms, produce stable RSCV characterized as class IV (Pel and Psl high producers) matrix-producing strains (Haussler et al., 1999, 2003; Kirisits et al., 2005; Ma et al., 2006; Starkey et al., 2009; Colvin et al., 2011a, b). Both mucoid and RSCV morphotypes highlight the adaptive scenarios and the advantage of rapid evolution for P. aeruginosa. Additionally, it highlights the necessity for extensive biofilm formation and robust matrix production. While the importance of biofilm can be appreciated, combating the negative impact of biofilm requires greater elucidation.
Numerous questions remain unanswered surrounding Pseudomonas biofilm formation. The most pertinent questions include: What is the true nature of matrix components and how are these molecules interacting in the biofilm? How does biofilm formation occur and why do biofilm cells transition back to a free-swimming state? Finally, how do biofilm components confer recalcitrance to immune and environment antimicrobial factors?
Aggregative polysaccharides Pel and Psl were identified and appreciated more recently than the capsular polysaccharide alginate. Understandably, less is known about the basic nature of the aggregative polysaccharides. Byrd, et al., (2009) determined the polysaccharide repeating units and the general nature of how Psl is synthesized. However, the assembly of Psl on the cell surface and release of Psl to the extracellular environment remain under continued investigation. Similarly, predictions regarding Pel-containing sugars, such as glucose, have been proposed based on the comparison of pellicles formed by wild-type and pel mutants (Friedman & Kolter, 2004a, b). Increased evidence of cell association or release of these polysaccharides will also be insightful for full functional determination. Aggregative polysaccharides of emerging importance in environmental pseudomonads also need additional analysis for complete understanding of their capacities and functions.
What has been identified is the major cue signaling nonmucoid pseudomonads to form biofilms, high intra-cellular c-di-GMP levels (Guvener & Harwood, 2007; Lee et al., 2007; Monds et al., 2007; Jonas et al., 2009). The intracellular signaling molecule induces changes including aggregative exopolysaccharide overproduction (Kirisits, et al., 2005; Jenal & Malone, 2006; Ma et al., 2006; Lee et al., 2007; Jonas et al., 2009), downregulation of motility (Kirisits et al., 2005), and enhanced adherence protein production (Monds et al., 2007; Borlee et al., 2010).
Intermittent colonization of CF lungs prior to chronic infection likely occurs because of the exposure to environmental P. aeruginosa. Common class IV matrix-producing strains (Colvin et al., 2011a, b), such as RSCV, have an advantage during colonization. Enhanced persistence during initial infection is predicted to be due to polysaccharide-mediated aggregation of the c-di-GMP-overproducing strain MJK8 (Byrd et al., 2011). Continued studies are directed at evaluating the interaction of the P. aeruginosa RSCV and host innate immune system. Defining the process of initial and intermittent colonization with nonmucoid strains of P. aeruginosa in the CF lung prior to chronic infection will aid avoidance or treatment before chronic infections persist.
Interestingly, reports have described a matrix-free cavity within mature mushroom structures of P. aeruginosa biofilms, which also include swimming subpopulations poised for release from the biofilm (Purevdorj-Gage et al., 2005; Ma et al., 2009). Several reports suggest that reversal of c-di-GMP levels from high to low or inhibition of quorum sensing will achieve biofilm dispersal (Hickman et al., 2005; Barraud et al., 2009; Schleheck et al., 2009; Brackman et al., 2011). Understanding what conditions generate subpopulations of the biofilm that convert to a planktonic phenotype may aid in the understanding how to prevent or combat biofilms. Prematurely triggering the regulatory sensors responsible for initiation of the free-swimming subpopulation may result in release of the entire biofilm and be an attractive therapeutic strategy. Emerging evidence elucidating the dispersal agents (e.g. D-amino acids, cis-2-decenoic acid) offers promise for the future ability to treat devastating biofilm infections (Davies & Marques, 2009; Kolodkin-Gal et al., 2010).
Once chronic mucoid infections are established, treatment with antibiotics becomes extremely challenging. Greater understanding of chronic P. aeruginosa infections is essential, especially investigating how mucoid and nonmucoid biofilms confer recalcitrance to host immune factors. in vitro models can be used to address this need preliminarily, but mechanisms of evasion need to be tested in vivo. Currently, only limited chronic infection models have been established, but models in development hold promise (Welsh et al., 2009; Bragonzi, 2010). Additional novel approaches to studying chronic animal infection are nonetheless warranted; perhaps, models achieving bacterial persistence but not restricted to airway infection would be promising. Regardless of the specific modeling, interpretation of data from diverse models will ultimately aid in deciphering the complexity of biofilm infections in human disease.
What was once a simple ‘slime’ holding bacterial cells together has emerged to be a complex network including a wide diversity of molecules possessing functions in structure, signaling, and control of biofilm formation. For P. aeruginosa, biofilm matrix components are diverse and include polysaccharides like alginate, Psl, and Pel as well as nucleic acids and proteins. Less well-described biofilm components include LPS, cyclic β glucans, and MV with emerging importance for proper biofilm matrix function. Pseudomonas biofilm matrix components in addition to the ones described here will likely continue to be revealed and add to the abundant diverse nature of biofilm matrix. The theme of diversity in function and survivability is hardwired into Pseudomonas spp. and that carries through when discussing biofilm matrix components. Matrix components carry out functions that may be redundant, additive, or even synergistic with other co-produced matrix molecules, but individually, each likely contributes to the organisms’ ability to persist and adapt to environmental conditions.
Box 1. Descriptions of biofilm were popularized by the work of Costerton et al beginning more than a quarter century ago. Sessile bacterial populations were identified in several environmental niches (Geesey et al., 1977; Costerton et al., 1987). As the relevance of sessile bacteria in clinical pathologies garnered greater appreciation, a definition of biofilm was proposed as a community of adherent sessile bacteria encased in some form of polymeric matrix (Costerton et al., 1987; Costerton et al., 1994). Greater understanding of biofilm nature lead Pasek and Singh to set forth specific criteria for diagnosis of biofilm infections (Parsek & Singh, 2003). Briefly, (1) bacterial aggregates must be associated with a surface, (2) microscopically bacteria appear encased in a matrix (of host and/or bacteria origin), (3) the infection remains localized as opposed to systemic, (4) the infection is antibiotic recalcitrant, (5) culture negativity is often observed, and (6) chronic: symptoms of infection persist (Parsek & Singh, 2003).
Widely accepted models mimicking biofilm infections include those that can account for some of the above criteria and highlight the biofilm developmental process. Bacterial biofilm developmental stages have been well scrutinized and concluded to consist of key stages (Stoodley et al., 2002). Initially attachment of recently planktonic cells to a surface or location occurs. Then, production of an extra-polymeric substance corresponds with irreversible attachment and structured microcolonies. Finally, the hallmark of biofilm maturation is when a population of cells, often displaying motility characteristics (in P. aeruginosa), is released from the biofilm allowing for the cycle to continue (Stoodley et al., 2002; Ma et al., 2009). The structured microcolonies produced by P. aeruginosa have been compared to ‘mushroom’ shapes. During biofilm development initial clusters of cells forming microcolonies have been referred to popularly as ‘stalks’ of the mushroom (Klausen et al., 2003a, b). As the biofilm matures further motile sub-populations are important for the formation of ‘caps’ on the stalks thereby completing the mushroom shape (Klausen et al., 2003a, b). A sample image of a P. aeruginosa flow-cell biofilm is a classic example of the mushroom-shaped structure.
Psuedomonas biofilm development produces classic structure. (a) The stages of bioflm are represented: (1) Attachment as a transition from planktonic to sessile lifestyle. (2) Growth, during which the recognizable structures form. (3) Detachment and dispersal of cells for seeding new sites of attachment. Image was kindly provided by Montana State University Center for Biofilm Engineering: P. Dirckx. (b) A representative confocal micrograph of P. aeruginosa PAO1 biofilm grown in a flow-cell for 3 days.
Acknowledgements
This work was supported by Public Health Service grants AI061396 and HL058334 (D.J.W.) and T32 training grant 5T32HL007946-11 (E.M.). Matthew Byrd generated the image used in Fig. 5 with support from NRSA fellowship AI07870002. Special thanks to P. Dirckx and Montana State University Center for Biofilm Engineering for allowing us to use ‘Biofilm formation in 3 stages’ for Box 1.
References
- Abdel-Mawgoud AM, Lepine F, Deziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol. 2010;86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel MH, Bass FG, Krane EJ, Thomas AL, Liggins GC. Pituitary stalk-section and some of its effects on endocrine function in the fetal lamb. Q J Exp Physiol Cogn Med Sci. 1978;63:211–219. doi: 10.1113/expphysiol.1978.sp002436. [DOI] [PubMed] [Google Scholar]
- Al-Tahhan RA, Sandrin TR, Bodour AA, Maier RM. Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl Environ Microbiol. 2000;66:3262–3268. doi: 10.1128/aem.66.8.3262-3268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhede M, Bjarnsholt T, Jensen PO, et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology. 2009;155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- Ali A, Rashid MH, Karaolis DKR. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl Environ Microbiol. 2002;68:5773–5778. doi: 10.1128/AEM.68.11.5773-5778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan ND, Kooi C, Sokol PA, Beveridge TJ. Putative virulence factors are released in association with membrane vesicles from Burkholderia cepacia. Can J Microbiol. 2003;49:613–624. doi: 10.1139/w03-078. [DOI] [PubMed] [Google Scholar]
- Allesen-Holm M, Barken KB, Yang L, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanassiou G, Michaleas S, Lada-Chitiroglou E, Tsitsa T, Antoniadou-Vyza E. Antimicrobial activity of beta-lactam antibiotics against clinical pathogens after molecular inclusion in several cyclodextrins. A novel approach to bacterial resistance. J Pharm Pharmacol. 2003;55:291–300. doi: 10.1211/002235702649. [DOI] [PubMed] [Google Scholar]
- Barken KB, Pamp SJ, Yang L, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom S, Theobell H, Davide H. Pyolipic acid. A metabolic product of Pseudomonas pyocyanea active against Mycobacterium tuberculosis. Arch Biochem Biophys. 1946a;10:165–166. [Google Scholar]
- Bergstrom S, Theobell H, Davide H. On a metabolic product of Ps. pyocyanea pyolipic acid, active against M. tuberculosis. Arkiv Chem Mineral Geol. 1946b;23A:1–12. [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ, Makin SA, Kadurugamuwa JL, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- Bianciotto V, Andreotti S, Balestrini R, Bonfante P, Perotto S. Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol Plant Microbe Interact. 2001;14:255–260. doi: 10.1094/MPMI.2001.14.2.255. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Jensen PO, Fiandaca MJ, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr Pulmonol. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- Blackwood LL, Pennington JE. Influence of mucoid coating on clearance of Pseudomonas aeruginosa from lungs. Infect Immun. 1981;32:443–448. doi: 10.1128/iai.32.2.443-448.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. P Natl Acad Sci USA. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- Borlee B, Goldman A, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JC, Schurr MJ, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- Boucher JC, Yu H, Mudd MH, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother. 2011;55:2655–2661. doi: 10.1128/AAC.00045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragonzi A. Murine models of acute and chronic lung infection with cystic fibrosis pathogens. Int J Med Microbiol. 2010;300:584–593. doi: 10.1016/j.ijmm.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Bragonzi A, Paroni M, Nonis A, et al. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med. 2009;180:138–145. doi: 10.1164/rccm.200812-1943OC. [DOI] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. P Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne RA, Chen YY, Wexler DL, Kuramitsu H, Bowen WH. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res. 1996;75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- Byrd MS, Pang B, Mishra M, Swords WE, Wozniak DJ. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappaB activation in A549 cells. MBio. 2010;1:e00140–10. doi: 10.1128/mBio.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Sadovskaya I, Vinogradov E, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd MS, Pang B, Hong W, et al. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect Immun. 2011;79:3087–3095. doi: 10.1128/IAI.00057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux JE, Kuzio J, Milazzo FH, Kropinski AM. Spontaneous release of lipopolysaccharide by Pseudomonas aeruginosa. J Bacteriol. 1983;155:817–825. doi: 10.1128/jb.155.2.817-825.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W-S, van de Mortel M, Nielsen L, Nino de Guzman G, Li X, Halverson LJ. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol. 2007;189:8290–8299. doi: 10.1128/JB.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemani C, Imberty A, de Bentzmann S, Pierre M, Wimmerova M, Guery BP, Faure K. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect Immun. 2009;77:2065–2075. doi: 10.1128/IAI.01204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GCL, Parsek MR. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011a;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, Howell PL, Wozniak DJ, Parsek MR. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2011b doi: 10.1111/j.1462-2920.2011.02657.x. DOI: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Coulon C, Vinogradov E, Filloux A, Sadovskaya I. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS ONE. 2010;5:e14220. doi: 10.1371/journal.pone.0014220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Marques CN. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechesne A, Owsianiak M, Bazire A, Grundmann GL, Binning PJ, Smets BF. Biodegradation in a partially saturated sand matrix: compounding effects of water content, bacterial spatial distribution, and motility. Environ Sci Technol. 2010;44:2386–2392. doi: 10.1021/es902760y. [DOI] [PubMed] [Google Scholar]
- Deretic V, Dikshit R, Konyecsni M, Chakrabarty AM, Misra TK. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989;171:1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Martin DW, Schurr MJ, Mudd MH, Hibler NS, Curcic R, Boucher JC. Conversion to mucoidy in Pseudomonas aeruginosa. Biotechnology. 1993;11:1133–1136. doi: 10.1038/nbt1093-1133. [DOI] [PubMed] [Google Scholar]
- DeVries CA, Ohman DE. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, Villemur R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology. 2003;149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2006;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416:740–743. doi: 10.1038/416740a. [DOI] [PubMed] [Google Scholar]
- Dueholm MS, Petersen SV, Sønderkær M, et al. Functional amyloid in Pseudomonas. Mol Microbiol. 2010;77:1009–1020. doi: 10.1111/j.1365-2958.2010.07269.x. [DOI] [PubMed] [Google Scholar]
- Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard B, Bertholet P, Gueders M, et al. Cyclodextrins as a potential carrier in drug nebulization. J Control Release. 2004;96:403–410. doi: 10.1016/j.jconrel.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Fakhr MK, Penaloza-Vazquez A, Chakrabarty AM, Bender CL. Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J Bacteriol. 1999;181:3478–3485. doi: 10.1128/jb.181.11.3478-3485.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil H, Feil WS, Chain P, et al. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. P Natl Acad Sci USA. 2005;102:11064–11069. doi: 10.1073/pnas.0504930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL, Solcia E. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Firoved AM, Boucher JC, Deretic V. Global genomic analysis of AlgU (σE)-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol. 2002;184:1057–1064. doi: 10.1128/jb.184.4.1057-1064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H-C, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aerguinosa biofilm matrix. J Bacteriol. 2004a;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004b;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol Ecol. 2003;12:3109–3121. doi: 10.1046/j.1365-294x.2003.01953.x. [DOI] [PubMed] [Google Scholar]
- Gaspar JA, Thomas JA, Marolda CL, Valvano MA. Surface expression of O-specific lipopolysaccharide in Escherichia coli requires the function of the TolA protein. Mol Microbiol. 2000;38:262–275. doi: 10.1046/j.1365-2958.2000.02094.x. [DOI] [PubMed] [Google Scholar]
- Geesey GG, Richardson WT, Yeomans HG, Irvin RT, Costerton JW. Microscopic examination of natural sessile bacterial populations from an alpine stream. Can J Microbiol. 1977;23:1733–1736. doi: 10.1139/m77-249. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Bauda P, DuBow MS, Oudega B. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch Microbiol. 1999;172:1–8. doi: 10.1007/s002030050732. [DOI] [PubMed] [Google Scholar]
- Ghafoor A, Hay ID, Rehm BH. The role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud C, Bernard CS, Calderon V, et al. The PprA–PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol. 2011;13:666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- Giwercman B, Meyer C, Lambert PA, Reinert C, Hoiby N. High-level beta-lactamase activity in sputum samples from cystic fibrosis patients during antipseudomonal treatment. Antimicrob Agents Chemother. 1992;36:71–76. doi: 10.1128/aac.36.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol. 2010;75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- Glick R, Gilmour C, Tremblay J, et al. Increase in rhamnolipid synthesis under iron-limiting conditions influences surface motility and biofilm formation in Pseudomonas aeruginosa. J Bacteriol. 2010;192:2973–2980. doi: 10.1128/JB.01601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez AJ, Rodicio MR, Mendoza MC. Identification of an emergent and atypical Pseudomonas viridiflava lineage causing bacteriosis in plants of agronomic importance in a Spanish region. Appl Environ Microbiol. 2003;69:2936–2941. doi: 10.1128/AEM.69.5.2936-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. 1987;55:111–117. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther NWT, Nunez A, Fett W, Solaiman DK. Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl Environ Microbiol. 2005;71:2288–2293. doi: 10.1128/AEM.71.5.2288-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- Hauser G, Karnovsky ML. Studies on the production of glycolipide by Pseudomonas aeruginosa. J Bacteriol. 1954;68:645–654. doi: 10.1128/jb.68.6.645-654.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler S, Nimtz M, Domke T, Wray V, Steinmetz I. Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect Immun. 1998;66:1588–1593. doi: 10.1128/iai.66.4.1588-1593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29:621–625. doi: 10.1086/598644. [DOI] [PubMed] [Google Scholar]
- Haussler S, Ziegler I, Lottel A, et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52:295–301. doi: 10.1099/jmm.0.05069-0. [DOI] [PubMed] [Google Scholar]
- Heath MC. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. P Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- Hogardt M, Heesemann J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol. 2010;300:557–562. doi: 10.1016/j.ijmm.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Hommel RK. In: Form and function of biosurfactants for degradation of water-insoluble substrates. Biochemistry of Microbial Degradation. Ratledge I, editor. Kluwer Academic Publishers; London: 1994. pp. 63–87. [Google Scholar]
- Hoyle BD, Alcantara J, Costerton JW. Pseudomonas aeruginosa biofilm as a diffusion barrier to piperacillin. Antimicrob Agents Chemother. 1992;36:2054–2056. doi: 10.1128/aac.36.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle BD, Williams LJ, Costerton JW. Production of mucoid exopolysaccharide during development of Pseudomonas aeruginosa biofilms. Infect Immun. 1993;61:777–780. doi: 10.1128/iai.61.2.777-780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Hastings JGM, White PJ. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa post-transcriptional regulator RsmA represses biofilm extracellular polysaccharide Psl synthesis. Mol Microbiol. 2010;78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EP, Christen R, Bizet C, et al. Pseudomonas brassicacearum subsp. neoaurantiaca subsp. nov., orange-pigmented bacteria isolated from soil and the rhizosphere of agricultural plants. Int J Syst Evol Microbiol. 2009;59:2476–2481. doi: 10.1099/ijs.0.009654-0. [DOI] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis FAJMJ. A glyco-lipide produced by Pseudomonas aeruginosa. J Am Chem Soc. 1949;71:4124–4126. [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Joardar V, Lindeberg M, Jackson RW, et al. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol. 2005;187:6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Melefors O, Romling U. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol. 2009;4:341–358. doi: 10.2217/fmb.09.7. [DOI] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SK, Lee SO, Lim YS, Jang KL, Lee TH. Purification and characterization of a novel levanoctaose-producing levanase from Pseudomonas strain K-52. Biotechnol Appl Biochem. 1998;27(Pt 2):159–166. [PubMed] [Google Scholar]
- Keenan JI, Allardyce RA. Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur J Gastroenterol Hepatol. 2000;12:1267–1273. doi: 10.1097/00042737-200012120-00002. [DOI] [PubMed] [Google Scholar]
- Keith LM, Bender CL. AlgT (σ22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J Bacteriol. 1999;181:7176–7184. doi: 10.1128/jb.181.23.7176-7184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith RC, Keith LM, Hernandez-Guzman G, Uppalapati SR, Bender CL. Alginate gene expression by Pseudomonas syringae pv. tomato DC3000 in host and non-host plants. Microbiology. 2003;149:1127–1138. doi: 10.1099/mic.0.26109-0. [DOI] [PubMed] [Google Scholar]
- Khan W, Bernier SP, Kuchma SL, Hammond JH, Hasan F, O'Toole GA. Aminoglycoside resistance of Pseudomonas aeruginosa biofilms modulated by extracellular polysaccharide. Int Microbiol. 2010;13:207–212. doi: 10.2436/20.1501.01.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidambi SP, Sundin GW, Palmer DA, Chakrabarty AM, Bender CL. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1995;61:2172–2179. doi: 10.1128/aem.61.6.2172-2179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003a;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003b;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Kohler T, Curty LK, Barja F, van Delden C, Pechere JC. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kownatzki R, Tummler B, Doring G. Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet. 1987;1:1026–1027. doi: 10.1016/s0140-6736(87)92286-0. [DOI] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Lam J, Chan R, Lam K, Costerton JRW. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol. 2002;184:1522–1529. doi: 10.1128/JB.184.6.1522-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PCY, Lindhout T, Beveridge TJ, Dutcher JR, Lam JS. Differential lipopolysaccharide core capping leads to quantitative and correlated modifications of mechanical and structural properties in Pseudomonas aeruginosa biofilms. J Bacteriol. 2009;191:6618–6631. doi: 10.1128/JB.00698-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue H, Schenk A, Li H, Lambertsen L, Neu TR, Molin S, Ullrich MS. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology. 2006;152:2909–2918. doi: 10.1099/mic.0.28875-0. [DOI] [PubMed] [Google Scholar]
- Lee D, Urbach J, Wu G, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-{gamma}-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ullrich MS. Characterization and mutational analysis of three allelic lsc genes encoding levansucrase in Pseudomonas syringae. J Bacteriol. 2001;183:3282–3292. doi: 10.1128/JB.183.11.3282-3292.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Nielsen L, Nolan C, Halverson LJ. Transient alginate gene expression by Pseudomonas putida biofilm residents under water-limiting conditions reflects adaptation to the local environment. Environ Microbiol. 2010;12:1578–1590. doi: 10.1111/j.1462-2920.2010.02186.x. [DOI] [PubMed] [Google Scholar]
- Licking E. Getting a grip on bacterial slime. Bus Week. 1999;13:98–100. [Google Scholar]
- Ma L, Jackson K, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak D. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–8356. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu H, Parsek MR, Bayles K, Wozniak DJ. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. P Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, McLean RJ, Whiteley M. Gram-negative outer membrane vesicles: beyond the cell surface. Geobiology. 2008;6:214–219. doi: 10.1111/j.1472-4669.2008.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, Ciofu O, Sternberg C, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure CD, Schiller NL. Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J Leukoc Biol. 1992;51:97–102. doi: 10.1002/jlb.51.2.97. [DOI] [PubMed] [Google Scholar]
- McClure CD, Schiller NL. Inhibition of macrophage phagocytosis by Pseudomonas aeruginosa rhamnolipids in vitro and in vivo. Curr Microbiol. 1996;33:109–117. doi: 10.1007/s002849900084. [DOI] [PubMed] [Google Scholar]
- Menozzi FD, Boucher PE, Riveau G, Gantiez C, Locht C. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun. 1994;62:4261–4269. doi: 10.1128/iai.62.10.4261-4269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireles II JR, Toguchi A, Harshey RM. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol. 2001;183:5848–5854. doi: 10.1128/JB.183.20.5848-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M, Byrd MS, Sergeant S, et al. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14:95–106. doi: 10.1111/j.1462-5822.2011.01704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- Moyano AJ, Lujan AM, Argarana CE, Smania AM. MutS deficiency and activity of the error-prone DNA polymerase IV are crucial for determining mucA as the main target for mucoid conversion in Pseudomonas aeruginosa. Mol Microbiol. 2007;64:547–559. doi: 10.1111/j.1365-2958.2007.05675.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Higashiyama Y, Izumikawa K, et al. The roles of the quorum-sensing system in the release of extracellular DNA, lipopolysaccharide, and membrane vesicles from Pseudomonas aeruginosa. Jpn J Infect Dis. 2008;61:375–378. [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, et al. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Neu TR. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev. 1996;60:151–166. doi: 10.1128/mr.60.1.151-166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PD, Monds RD, O'Toole GA. LapD is a bis-(3’,5’)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. P Natl Acad Sci USA. 2009;106:3461–3466. doi: 10.1073/pnas.0808933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L, Li X, Halverson LJ. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ Microbiol. 2011;13:1342–1356. doi: 10.1111/j.1462-2920.2011.02432.x. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Chiang W-C, Fazli M, Gjermansen M, Givskov M, Tolker-Nielsen T. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ Microbiol. 2011;13:1357–1369. doi: 10.1111/j.1462-2920.2011.02447.x. [DOI] [PubMed] [Google Scholar]
- Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Ohman DE, Chakrabarty AM. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect Immun. 1982;37:662–669. doi: 10.1128/iai.37.2.662-669.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onbasli D, Aslim B. Effects of some organic pollutants on the exopolysaccharides (EPSs) produced by some Pseudomonas spp. strains. J Hazard Mater. 2009a;168:64–67. doi: 10.1016/j.jhazmat.2009.01.131. [DOI] [PubMed] [Google Scholar]
- Onbasli D, Aslim B. Biosurfactant production in sugar beet molasses by some Pseudomonas spp. J Environ Biol. 2009b;30:161–163. [PubMed] [Google Scholar]
- Osman SF, Fett WF, Fishman ML. Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J Bacteriol. 1986;166:66–71. doi: 10.1128/jb.166.1.66-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J, Lewenza S, Marr AK, Hancock RE. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol. 2007;189:2164–2169. doi: 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès D, Sanchez L, Conrod S, et al. Exploration of intraclonal adaptation mechanisms of Pseudomonas brassicacearum facing cadmium toxicity. Environ Microbiol. 2007;9:2820–2835. doi: 10.1111/j.1462-2920.2007.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamp SJ, Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. J Bacteriol. 2007;189:2531–2539. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, et al. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen SS, Hoiby N, Espersen F, Koch C. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax. 1992;47:6–13. doi: 10.1136/thx.47.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penaloza-Vazquez A, Kidambi SP, Chakrabarty AM, Bender CL. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J Bacteriol. 1997;179:4464–4472. doi: 10.1128/jb.179.14.4464-4472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Webb JS, Rehm BH. The role of polyhydroxyalkanoate biosynthesis by Pseudomonas aeruginosa in rhamnolipid and alginate production as well as stress tolerance and biofilm formation. Microbiology. 2004;150:3405–3413. doi: 10.1099/mic.0.27357-0. [DOI] [PubMed] [Google Scholar]
- Pier GB. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News. 1998;6:339–347. [Google Scholar]
- Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pier GB, Boyer D, Preston M, et al. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J Immunol. 2004;173:5671–5678. doi: 10.4049/jimmunol.173.9.5671. [DOI] [PubMed] [Google Scholar]
- Preston LA, Bender CL, Schiller NL. Analysis and expression of algL, which encodes alginate lyase in Pseudomonas syringae pv. syringae. DNA Seq. 2001;12:455–461. doi: 10.3109/10425170109084474. [DOI] [PubMed] [Google Scholar]
- Purevdorj-Gage B, Costerton WJ, Stoodley P. Phenotypic differentiation and seeding dispersal in nonmucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology. 2005;151:1569–1576. doi: 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. P Natl Acad Sci USA. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramphal R, Vishwanath S. Why is Pseudomonas the colonizer and why does it persist? Infection. 1987;15:281–287. doi: 10.1007/BF01644139. [DOI] [PubMed] [Google Scholar]
- Rashid MH, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. P Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid MH, Rajanna C, Ali A, Karaolis DK. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol Lett. 2003;227:113–119. doi: 10.1016/S0378-1097(03)00657-8. [DOI] [PubMed] [Google Scholar]
- Read RC, Roberts P, Munro N, et al. Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. J Appl Phys. 1992;72:2271–2277. doi: 10.1152/jappl.1992.72.6.2271. [DOI] [PubMed] [Google Scholar]
- Records AR, Gross DC. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J Bacteriol. 2010;192:3584–3596. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol. 2010;8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. P Natl Acad Sci USA. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I, Darzins A, Wang S-K, Berry A, Chakrabarty AM. Alginate biosynthetic enzymes in mucoid and nonmucoid Pseudomonas aeruginosa: overproduction of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase by overexpression of the phosphomannose isomerase (pmi) gene. J Bacteriol. 1987;169:3224–3231. doi: 10.1128/jb.169.7.3224-3231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1->3)-glucans, which bind aminoglycosides. Glycobiology. 2010;20:895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- Sanders LH, Rockel A, Lu H, Wozniak DJ, Sutton MD. Role of Pseudomonas aeruginosa dinB-encoded DNA Polymerase IV in mutagenesis. J Bacteriol. 2006;188:8573–8585. doi: 10.1128/JB.01481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpellini M, Franzetti L, Galli A. Development of PCR assay to identify Pseudomonas fluorescens and its biotype. FEMS Microbiol Lett. 2004;236:257–260. doi: 10.1016/j.femsle.2004.05.043. [DOI] [PubMed] [Google Scholar]
- Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE. 2009;4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnider-Keel U, Lejbolle KB, Baehler E, Haas D, Keel C. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl Environ Microbiol. 2001;67:5683–5693. doi: 10.1128/AEM.67.12.5683-5693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooling SR, Hubley A, Beveridge TJ. Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol. 2009;191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzmann S, Boring JR. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect Immun. 1971;3:762–767. doi: 10.1128/iai.3.6.762-767.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- Simpson JA, Smith SE, Dean RT. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J Gen Microbiol. 1988;134:29–36. doi: 10.1099/00221287-134-1-29. [DOI] [PubMed] [Google Scholar]
- Simpson JA, Smith SE, Dean RT. Scavenging by alginate of free radicals released by macrophages. Free Radic Biol Med. 1989;6:347–353. doi: 10.1016/0891-5849(89)90078-6. [DOI] [PubMed] [Google Scholar]
- Simpson JA, Smith SE, Dean RT. Alginate may accumulate in cystic-fibrosis lung because the enzymatic and free-radical capacities of phagocytic-cells are inadequate for its degradation. Biochem Mol Biol Int. 1993;30:1021–1034. [PubMed] [Google Scholar]
- Spiers AJ, Rainey PB. The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology. 2005;151:2829–2839. doi: 10.1099/mic.0.27984-0. [DOI] [PubMed] [Google Scholar]
- Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. Biofilm formation at the air–liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol Microbiol. 2003;50:15–27. doi: 10.1046/j.1365-2958.2003.03670.x. [DOI] [PubMed] [Google Scholar]
- Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics. 2002;161:33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapper AP, Narasimhan G, Ohman DE, et al. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- Starkey M, Hickman JH, Ma L, et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001a;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. The biofilm matrix – an immobilized but dynamic microbial environment. Trends Microbiol. 2001b;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Tanious FA, Ding D, Patrick DA, Tidwell RR, Wilson WD. A new type of DNA minor-groove complex: carbazole dication–DNA interactions. Biochemistry. 1997;36:15315–15325. doi: 10.1021/bi971599r. [DOI] [PubMed] [Google Scholar]
- Tielker D, Hacker S, Loris R, et al. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology. 2005;151:1313–1323. doi: 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- Tuomanen E, Towbin H, Rosenfelder G, Braun D, Larson G, Hansson GC, Hill R. Receptor analogs and monoclonal antibodies that inhibit adherence of Bordetella pertussis to human ciliated respiratory epithelial cells. J Exp Med. 1988;168:267–277. doi: 10.1084/jem.168.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. P Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gennip M, Christensen LD, Alhede M, et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS. 2009;117:537–546. doi: 10.1111/j.1600-0463.2009.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Visnapuu T, Zamfir AD, Mosoarca C, Stanescu MD, Alamäe T. Fully automated chip-based negative mode nanoelectrospray mass spectrometry of fructoo-ligosaccharides produced by heterologously expressed levansucrase from Pseudomonas syringae pv. tomato DC3000. Rapid Commun Mass Spectrom. 2009;23:1337–1346. doi: 10.1002/rcm.4007. [DOI] [PubMed] [Google Scholar]
- Vodovar N, Vallenet D, Cruveiller S, et al. Complete genome sequence of the entomopathogenic and metabolically versatile soil bacterium Pseudomonas entomophila. Nat Biotechnol. 2006;24:673–679. doi: 10.1038/nbt1212. [DOI] [PubMed] [Google Scholar]
- Walters III MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh MJ, Rogers CS, Stoltz DA, Meyerholz DK, Prather RS. Development of a porcine model of cystic fibrosis. Trans Am Clin Climatol Assoc. 2009;120:149–162. [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Whiteley Y, Bangera MG, Bumgarnder RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Van Rossum T, Lo R, Khaira B, Whiteside MD, Hancock REW, Brinkman FSL. Pseudomonas genome database: facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 2009;37:D483–D488. doi: 10.1093/nar/gkn861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. P Natl Acad Sci USA. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol Microbiol. 2009;74:1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- Yang L, Hu Y, Liu Y, Zhang J, Ulstrup J, Molin S. Distinct roles of extracellular polymeric substances in Pseudomonas aeruginosa biofilm development. Environ Microbiol. 2011;13:1705–1717. doi: 10.1111/j.1462-2920.2011.02503.x. [DOI] [PubMed] [Google Scholar]
- Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE, Overhage J. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol. 2009;191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Kemme M, Simon SR. Alginate, the slime exopolysaccharide of Pseudomonas aeruginosa, binds human leukocyte elastase, retards inhibition by alpha 1-proteinase inhibitor, and accelerates inhibition by secretory leukoprotease inhibitor. Am J Respir Cell Mol Biol. 1996;15:283–291. doi: 10.1165/ajrcmb.15.2.8703486. [DOI] [PubMed] [Google Scholar]
- Yu J, Penaloza-Vazquez A, Chakrabarty AM, Bender CL. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Miller RM. Effect of a Pseudomonas rhamnolipid biosurfactant on cell hydrophobicity and biodegradation of octadecane. Appl Environ Microbiol. 1994;60:2101–2106. doi: 10.1128/aem.60.6.2101-2106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski NA, Chakrabarty AM, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]
- Zielinski NA, Maharaj R, Roychoudhury S, Danganan CE, Hendrickson W, Chakrabarty AM. Alginate synthesis in Pseudomonas aeruginosa: environmental regulation of the algC promoter. J Bacteriol. 1992;174:7680–7688. doi: 10.1128/jb.174.23.7680-7688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski NA, DeVault JD, Roychoudhury S, et al. Molecular genetics of alginate biosynthesis in Pseudomonas aeruginosa. In: Silver S, Chakrabarty AM, Iglewski B, Kaplan S, editors. Pseudomonas Biotransformations, Pathogenesis and Evolving Biotechnology. American Society for Microbiology; Washington, DC: 1990. pp. 15–27. [Google Scholar]